Abstract
The small heterodimer partner (SHP) nuclear receptor is an important regulator of nonalcoholic fatty liver disease. However, little is known about the role of SHP in alcoholic fatty liver. In this study, we used a modified chronic ethanol–binge model to examine cyclic alterations of lipid metabolism in wild-type (WT) and Shp−/− mice over a 24-hour period after binge. The serum and hepatic lipid profiles, as well as the expression of lipid synthesis genes and markers of endoplasmic reticulum stress, exhibited distinct variations in WT and Shp−/− mice in response to ethanol diet plus ethanol binge (ED+E) and control diet plus maltose binge. ED+E induced steatosis in WT mice, which correlated with a marked up-regulation of activating transcription factor 4 protein (ATF4) but down-regulation of C/EBP homologous protein (CHOP) and sterol regulatory element-binding transcription factor 1c protein (SREBP-1c). On the contrary, the control diet plus maltose binge caused lipid accumulation in Shp−/− mice, which was accompanied by a sharp elevation of CHOP, SREBP-1c, and REV-ERBα proteins but a diminished ATF4. REV-ERBα activated CHOP promoter activity and gene transcription, which were inhibited by SHP. Knockdown Rev-Erbα in Shp−/− mice prevented steatosis induced by ED+E. Our study revealed a critical role of SHP and REV-ERBα in controlling rhythmic CHOP expression in alcoholic fatty liver.
Liver is a major organ to detoxify alcohol and thus is prone to be damaged by alcohol abuse. Research has been well established that excessive consumption of alcohol results in the development and progression of chronic liver disease, including liver steatosis, steatohepatitis, cirrhosis, and hepatocellular carcinoma.1, 2 Although mechanisms that lead to these pathologies have been studied extensively, the development of an effective therapy for alcoholic liver disease (ALD) remains challenging.
The endoplasmic reticulum (ER) stress response is a fundamental reaction seen even in healthy liver and has been noted to increase after alcohol intake.3 The ER stress response, at steady state, plays a protective role against such toxic factors by removing misfolded and/or unfolded proteins from the ER. However, when the stress becomes persistent and strong, the response switches to the induction of liver steatosis, insulin resistance, and apoptosis.4 Various factors have been identified as causes for alcohol-induced ER stress response, such as oxidative stress, acetoaldehyde, free fatty acid, insulin resistance, disturbance of calcium homeostasis, and disruption of one carbon metabolism, all of which are known to impair ER functions.3, 5
Small heterodimer partner (SHP; NR0B2) is an atypical orphan nuclear receptor mainly expressed in the liver.6, 7 Unlike many other nuclear receptors, SHP lacks DNA-binding domain and inhibits transcriptional activity of other transcription factors.8 Our previous studies revealed a central role of SHP in liver metabolism, including bile acid, lipid, and one carbon metabolism.5, 9, 10, 11 Specifically, SHP inhibited very-low-density lipoprotein secretion by repressing microsomal triglyceride transfer protein11 expression and played a key role to control diurnal regulation of plasma triglyceride by the circadian locomotor output cycles kaput gene.12 Consequently, Shp−/− mice were resistant to high-fat diet induced fatty liver.13 We recently demonstrated that SHP was an essential component of the liver circadian clock machinery through interaction with RAR-related orphan receptor (RORγ) and nuclear receptor subfamily 1 group D1 (REV-ERBα) to regulate neuronal PAS domain-containing protein 2–mediated hepatic lipid metabolism.14 Neuronal PAS domain-containing protein 2 was induced in Shp−/− mice and knockdown neuronal PAS domain-containing protein 2 attenuated very-low-density lipoprotein production and sensitized Shp−/− mice to develop steatosis.14 Despite these previous studies that elucidated a critical role of SHP in nonalcoholic fatty liver disease (NAFLD), the regulatory function of SHP in alcoholic fatty liver (AFL) remained elusive.
Although the role of the ER stress response has been established in alcoholic fatty liver diseases, the importance of circadian clock in the ER stress response in ALD, and the role of diet although intriguing, remains largely unexplored. The objective is to understand the function of SHP in integrating the liver circadian clock with alcohol-induced ER stress response and steatosis. The hypothesis is that genes in ER stress and lipid metabolism are under the control of liver clock that is modulated by alcohol and SHP. In the present study, we elucidated a critical cross talk between SHP and REV-ERBα in the regulation of alcohol-induced steatosis involving C/EBP homologous protein (CHOP). Our study provides novel insight into SHP and REV-ERBα control of ER stress signaling associated with the development of AFL.
Materials and Methods
Cell Culture and Animals
Mouse hepatoma cell line Hepa 1-6 (ATCC, Manassas, VA; CRL-1830) was maintained in Dulbecco's modified Eagle's medium (11965-118; Invitrogen, Carlsbad, CA) with 100 U of penicillin G–streptomycin sulfate/mL and 10% heat-inactivated fetal bovine serum (10437-028; Gibco, Waltham, MA). C57BL/6J mice were purchased from Jackson Laboratory (Bar Harbor, ME). Shp−/− on C57BL/6J background was described previously.10, 14 Mice were fed a standard rodent chow with free access to water and maintained in a 12-hour light/dark cycle (light on 6 am to 6 pm), temperature-controlled (23°C), and virus-free facility. Experiments were performed on male mice at the age of 8 to 12 weeks unless stated otherwise (n = 5 per group). For ethanol feeding, the Gao-binge model was used with slight modification.1, 5, 15 In brief, the mice were acclimated to a control liquid diet (F1259SP; Bio-Serv, Flemington, NJ) for 5 days and followed by control liquid diet or 5% Lieber-DeCarli ethanol liquid diet (F1258SP; Bio-Serv) for 10 days. On day 11 at 9 am the mice were challenged by oral gavage of a single dose of maltose dextrin (9 g/kg of body weight) or ethanol (5 g/kg of body weight) solution. Nine hours after the binge, blood samples and liver tissues were collected every 6 hours over a 24-hour light/dark cycle at Zeitgeber time (ZT) 12, ZT18, ZT0, and ZT6. All experiments were performed in accordance with relevant guidelines and regulations approved by the Institutional Animal Care and Use Committee at the University of Connecticut (Storrs).
Plasmids and Viruses
The mouse Chop (Gene ID 13198) promoter luciferase reporter (pGL3-mChop-pro) and its mutation construct (pGL3-mChop-pro-mut) were engineered in our laboratory. In brief, the mouse Chop promoter was amplified by Takara LA Taq Polymerase (RR002A; Takara Bio, Shiga, Japan) using wild-type (WT) mice genomic DNA as a template and cloned into pGL3-basic (E1751; Promega, Madison, WI). pGL3-mChop-pro-mut was generated by a QuikChange Site-Directed Mutagenesis Kit (200521; Stratagene, San Diego, CA). The integrity of the recombinant plasmids was verified by sequence analysis. Two shRNAs for knockdown mouse Rev-Erbα (TRCN0000027094 and TRCN0000027070) and one shRNA control plasmid in pLKO.1 vector (SHC002) were purchased from Sigma (St. Louis, MO). The sequences of the shRNAs were provided in Table 1. For adeno-associated virus-8 (AAV8)–mediated shRNA knockdown of Rev-Erbα, U6-Rev-Erbα-shRNA-2 cassette was cloned into an AAV8 vector and virus particles were produced by Vector BioLabs (Malvern, PA). Mice were injected via tail vein with purified AAV8 virus at 1 × 1011 virus particles per mouse once a week for 2 weeks and subjected to Gao-binge model. Serum, liver, and brain tissues were collected at 21 hours (ZT0) after oral gavage. The in vivo adenoviral transduction of SHP (adSHP) and its control green fluorescent protein (adGFP) into Shp−/− mice has been described previously.14, 16 The Shp−/−/adGFP and Shp−/−/adSHP mice were fed with standard chow diet and the liver tissues were collected every 4 hours over a 24-hour light/dark cycle at ZT2, ZT6, ZT10, ZT14, ZT18, and ZT22.
Table 1.
Primer Sequences
| Quantitative PCR primers | Primer type | Sequences (5′ to 3′) |
|---|---|---|
| Srebp1c | Sense | 5′-TTGCTGGCTTGGTGATGCTATG-3′ |
| Antisense | 5′-CTGGTGGAGGGCTGGAAGG-3′ | |
| Fasn | Sense | 5′-TCGGGTGTGGTGGGTTTGG-3′ |
| Antisense | 5′-GCGTGAGATGTGTTGCTGAGG-3′ | |
| Chop | Sense | 5′-GCACCTATATCTCATCCCCAG-3′ |
| Antisense | 5′-TGCGTGTGACCTCTGTTG-3′ | |
| Rev-Erbα | Sense | 5′-GACCTTTCTCAGCACGACC-3′ |
| Antisense | 5′-CATCACTGTCTGGTCCTTCAC-3′ | |
| Actin | Sense | 5′-GCTCCGGCATGTGCAA-3′ |
| Antisense |
5′-AGGATCTTCATGAGGTAGT-3′ |
|
| shRNAs |
Region |
Sequences (5′ to 3′) |
| Rev-Erbα-shRNA-1 | CDS | 5′-CCGGCCATCGAGAAATCTTCACCTACTCGAGTAGGTGAAGATTTCTCGATGGTTTTT-3′ |
| Rev-Erbα-shRNA-2 |
CDS |
5′-CCGGGCGCTTTGCATCGTTGTTCAACTCGAGTTGAACAACGATGCAAAGCGCTTTTT-3′ |
| Cloning primers |
Sequences (5′ to 3′) |
|
| mChop promoter | Forward | 5′-GCTCACGCGTAGGGCGTAAGAGCATCACAC-3′ |
| mChop promoter |
Reverse |
5′-CGTCCTCGAGTCGACGTGTTAGGCTCAAGA-3′ |
| Mutagenesis primers |
Sequences (5′ to 3′) |
|
| mChop promoter-Mut | Forward | 5′-CCGCGAAGCCGCGTGATTTAAAGCCACTTCCGGGT-3′ |
| mChop promoter-Mut | Reverse | 5′-ACCCGGAAGTGGCTTTAAATCACGCGGCTTCGCGG-3′ |
CDS, coding sequence; Chop, C/EBP homologous protein; Fasn, fatty acid synthase; mChop, mouse Chop; Mut, mutation; Srebp1c, sterol regulatory element-binding transcription factor 1c.
In Vivo Plasmid Transfection
The expression plasmids pcDNA3 and pcDNA3-Flag-Rev-Erbα were described previously.14 shRNAs for mouse Atf4 (TRCN0000301646) were purchased from Sigma. In vivo plasmid transfection was performed using TurboFect in vivo Transfection Reagent (R0541; Thermo Scientific, Waltham, MA). Plasmids (50 μg) were diluted in 400 μL 5% glucose solution, mixed with 6 μL of in vivo transfection reagent, and incubated for 20 minutes before injected to WT mice through tail vein. Serum and liver tissues were collected at day 4 after injection. For shAtf4 knockdown, pLKO-shAtf4 or control vector pLKO plasmids were injected at day 1 and day 5 during the ethanol feeding period. After ethanol binge, serum and liver tissues were collected at ZT12.
Measurements of TG and ALT/AST Levels
Serum and liver triglyceride (TG) levels were determined as described previously.14 Briefly, lipid was extracted from 200 mg liver tissue into 600 μL chloroform-methanol mix (2:1) and redissolved in 500 μL phosphate-buffered saline (pH 7.4) with 5% Triton X-100. Serum TG or dissolved liver TG was measured using Pointe Scientific Triglycerides Liquid Reagents (Thermo Fisher Scientific; 23-666-410), according to the manufacturer's instructions. Each sample was run in duplicate. Serum levels of aspartate aminotransferase (AST) and alanine aminotransferase (ALT) were determined by Infinity AST (glutamate oxalacetate transaminase) and ALT (glutamate pyruvate transaminase) reagents (Thermo Fisher Scientific; TR70121 and TR71121).
Histological Analysis of Liver Sections
Liver tissue fixing with formalin was described previously.10 Paraffin sections were cut at thickness of 4 μm and subjected to hematoxylin and eosin staining and periodic acid-Schiff staining. The images were captured using Olympus BX41 microscope (Olympus, Lake Success, NY). The random fields from each liver section were captured and analyzed.
RNA Isolation and Quantitative PCR
Total RNA was isolated using TRIzol Reagent (15596-018; Invitrogen), and cDNA synthesis was performed with High-Capacity cDNA Reverse Transcription Kit (4368813; Applied Biosystems, Foster City, CA). The quantitative PCR was performed using the iTaq Universal SYBR Green Supermix (1725124; Bio-Rad, Hercules, CA) with primers shown in Table 1. The relative ratio of specific genes to internal control β-actin for each sample was then calculated.
Western Blots
Hepa1 cells or liver tissues were lysed as described previously.8 The concentration of protein was measured using Pierce BCA Protein Assay Kit (Thermo Fisher; 23227). Cell lysates (30 μg) were resolved by SDS-PAGE and transferred to nitrocellulose membranes by standard procedures. Antibody binding was visualized with SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific; 34080), according to the manufacturer's protocol. Equal loading of protein was verified with β-actin, glyceraldehyde-3-phosphate dehydrogenase, or tubulin. Quantitative analysis of the band intensity was performed by ImageJ software version 1.50 (NIH, Bethesda, MD; http://imagej.nih.gov/ij).
Western blots of CHOP and activating transcription factor 4 (ATF4) proteins were performed in 97H cells treated with tunicamycin (ER stress inducer). MHC97H cells were fasted for 24 hours before treatment with tunicamycin (1 ng/mL) for 24 hours. As MHC97H cells have low basal levels of ATF4 and CHOP, they were chosen to determine their induction by tunicamycin.
The following antibodies were used for primary antibody incubation: β-actin (sc-47778; Santa Cruz Biotechnology, Dallas, TX), sterol regulatory element-binding transcription factor 1c (SREBP-1c; MA5-11685; Thermo Fisher Scientific), acetyl-CoA carboxylase (sc-30212; Santa Cruz Biotechnology), fatty acid synthase (sc-20140; Santa Cruz Biotechnology), PRKR-like endoplasmic reticulum kinase (3192; Cell Signaling Technology, Danvers, MA), inositol-requiring enzyme α (3294; Cell Signaling Technology), ATF6 (ab122897; Abcam, Cambridge, UK), total eukaryotic translation initiation factor 2 α (5324p; Cell Signaling Technology), phosphorylated eukaryotic translation initiation factor 2α (3398p; Cell Signaling Technology), ATF4 (11815s; Cell Signaling Technology), CHOP (2895p; Cell Signaling Technology), p-AKT (9271; Cell Signaling Technology), REV-ERBα (SAB2101632; Sigma), and FLAG (F3165-1MG; Sigma).
Knockdown Experiment
REV-ERBα
After acclimatization for 5 days, mice were given control or ethanol-containing Lieber-DeCarli liquid diets for 10 days, followed by oral gavage of maltose (control) or ethanol solutions at 9 am on day 10. WT mice were injected with AAV8-driven control or shRNA for Rev-Erbα through the tail vain. Two weeks later, the mice were injected again and subject to the Gao-binge protocol after 3 days. Livers and sera were collected at ZT0.
ATF4
WT mice were injected with pLKO or pLKO-sh-Atf4 plasmids (50 μg per mice) on days 1 and 5 of ethanol feeding using the TurboFect in vivo Transfection Reagent following the manufacturer's instructions. On day 11, mice were orally gavaged with maltose or ethanol. Livers and sera were collected at ZT12.
Luciferase Assay
Hepa1-6 cells in 24-well plates were cotransfected with mouse Chop promoter (pGL3-mChop-pro), mouse Chop promoter mutant (pGL3-mChop-pro-mut), pcDNA3-Flag-Shp, pcDNA3-Flag-Rev-Erbα, or pcDNA3-RORα, as indicated in figure legends using transfection reagent X-tremeGENE HP (06366236001; Roche, Basel, Switzerland).17 In the ethanol group, 48 hours after transfection, cells were cultured in fetal bovine serum–free Dulbecco's modified Eagle's medium for 3 hours and followed by 400 mmol/L ethanol treatment for an additional 6 hours. Luciferase and renilla activities were determined by Dual-Luciferase Reporter Assay Systems (Promega; E1960). Luciferase activity was normalized by renilla activity expressed from cotransfected pSV-renilla. Each data point was the average of triplicate assays and repeated three times.
Statistical Analysis
All of the experiments were performed in triplicate and repeated at least three times. The data are presented as the means ± SEM. Statistical analysis was performed using the Student's t-test for unpaired data to compare the values between the two groups. P < 0.05 was considered statistically significant.
Results
Shp−/− Mice Developed Different Types of Steatosis in Response to ED+E and CD+M
The recently developed Gao-binge model15 represents an acute model of alcohol-induced liver injury. It is easy to perform, thus has been widely used by other research groups.18, 19 One observation was that the changes in gene expression and serum enzymes were transient after binge.15 To have a better understanding of the phenotypic changes, we collected samples over a 24-hour light/dark cycle (Supplemental Figure S1).15 To establish the physiological relevance of SHP in AFL, we first examined Shp expression in WT mice after ethanol diet plus ethanol binge (ED+E). Interestingly, the expression of Shp in the control diet plus maltose binge (CD+M) group appeared rhythmic, which was low during the night and high during the day (Figure 1A). Shp mRNA was markedly reduced 9 hours after ethanol binge compared with maltose binge (ZT12) (Figure 1A), which was gradually recovered to the same level as in the CD+M group after 24 hours. We did not detect Shp mRNA in Shp−/− mice (data not shown). The quantitative PCR results were in agreement with the RNA-sequencing data (Supplemental Figure S2A).
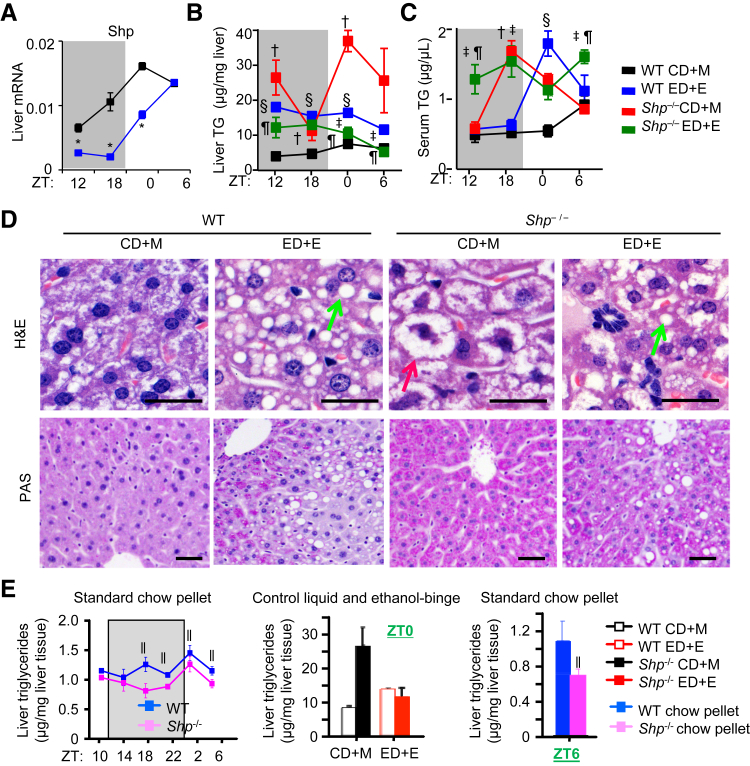
Figure 1.
Shp deficiency altered maltose- and ethanol-mediated alteration of lipid metabolism. A: Quantitative PCR of mRNA expression of Shp in WT mice fed control diet + maltose binge (CD+M; black) or ethanol diet + ethanol binge (ED+E; blue). Each lane represents triplicate assays from equal amounts of mRNA pooled from five individual mice. The Gao-binge model was used. The samples were collected over a 12-hour/12-hour light/dark cycle 9 hours after the binge at Zeitgeber time (ZT) 12, 18, 0, and 6. Gray shading indicates dark cycle; white indicates light cycle. B and C: Liver (B) and serum (C) triglyceride (TG) contents in WT (black and blue) and Shp−/− (red and green) mice fed CD+M (black and red) or ED+E (blue and green). D: Representative images of hematoxylin and eosin (H&E; top row) and periodic acid-Schiff (PAS; bottom row) staining of liver sections from WT and Shp−/− mice fed CD+M or ED+E at ZT0. We use different magnification to convey the results more clearly. Green arrows indicate macrosteatosis; red arrow, microsteatosis. E: Left panel: Liver TG contents in WT (blue) and Shp−/− (magenta) mice fed a standard chow pellet diet. The livers were collected over a 12-hour/12-hour light/dark cycle at ZT10, 14, 18, 22, 2, and 6. Middle panel: Liver TG contents in WT (blank) and Shp−/− (filled) mice fed CD+M (black) or ED+E (red) at ZT0. Right panel: Liver TG contents in WT (blue) and Shp−/− (magenta) mice fed a standard chow pellet diet at ZT6. Data are expressed as means ± SD (A–C and E). n = 5 per time point per group. ∗P < 0.05 WT CD+M versus WT ED+E; †P < 0.05 WT CD+M versus Shp−/− CD+M; ‡P < 0.05 WT ED+E versus Shp−/− ED+E; §P < 0.05 WT CD+M versus WT ED+E; ¶P < 0.05 Shp−/− CD+M versus Shp−/− ED+E; ‖P < 0.05 WT chow versus Shp−/− chow (E). Scale bar = 20 μm (D). Original magnifications: ×40 (D, top row); ×20 (D, bottom row).
Liver and serum TG levels exhibited distinct variations in both WT and Shp−/− mice after the maltose or ethanol binge over a 24-hour light/dark cycle. The liver TG content in WT mice was increased 9 hours after ethanol binge (ZT12), and was decreased to the normal level after 24 hours (ZT6), as compared to the CD+M (Figure 1B). Remarkably, the liver TG content in Shp−/− mice showed a striking induction and oscillation in CD+M, which was decreased in ED+E. Serum TG showed dissimilar rhythmic changes (Figure 1C). ED+E-induced serum TG reached the highest level at ZT0 in WT mice, which remained in a constant high level from ZT12 to ZT6 in Shp−/− mice. The decreased liver TG at several time points in WT and Shp−/− mice correlated with the increased serum TG, suggesting a dynamic oscillation in hepatic lipid secretion during the 24-hour light/dark cycle. The induction in serum TG in Shp−/−-CD+M mice also correlated with high serum ALT and AST levels, as shown in our recent publication.5
Hematoxylin and eosin staining revealed steatosis in WT-ED+E and Shp−/−-ED+E groups (Figure 1D). Interestingly, we observed a variant form of hepatic fat accumulation in Shp−/−-CD+M, which was characterized by small intracytoplasmic fat vacuoles accumulated in the cell, similar to microvesicular steatosis. Furthermore, periodic acid-Schiff staining revealed increased glycogen storage in Shp−/−-CD+M, and to a lesser extent in Shp−/−-ED+E (Figure 1D).
Previous studies have shown that Shp deficiency prevented the development of steatosis under normal chow and high-fat diet.11, 13, 14, 20 Because liver lipid metabolism is under the control of circadian clock, we detected liver TG levels in WT and Shp−/− mice fed a standard chow over a 24-hour light/dark cycle. Overall, the liver TG showed a diurnal rhythm and was diminished in Shp−/− versus WT mice (Figure 1E). In contrast, we observed an induction of liver TG in Shp−/− fed with CD+M. Therefore, Shp−/− mice had reduction of hepatic TG contents under regular chow diet but induction of liver TG in CD+M (Figure 1E). Taken together, the results suggest that ED+E transiently induces hepatic steatosis in WT mice and Shp−/− mice.
Ethanol Diet Plus Ethanol Binge Inactivated SREBP-1c Pathway in WT Mice, whereas Control Diet Plus Maltose Binge Activated SREBP-1c Pathway in Shp−/− Mice
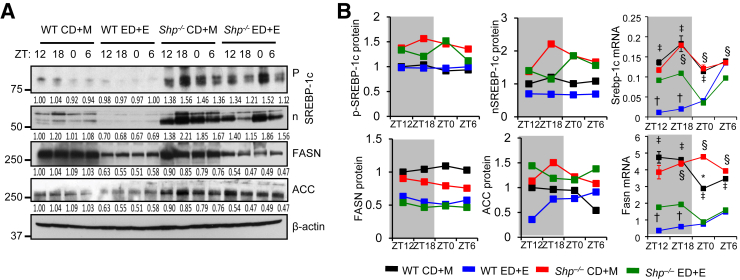
SREBP-1c is a key regulator of lipid metabolism21 by activating fatty acid synthase and acetyl-CoA carboxylase to induce lipid accumulation.22 Surprisingly, both the precursor (p) and processed nuclear (n) forms of SREBP-1c proteins were decreased in WT-ED+E versus WT-CD+M; on the other hand, they were highly increased in Shp−/−-CD+M versus WT-CD+M, and decreased in Shp−/−-ED+E versus Shp−/−-CD+M (Figure 2A). Fatty acid synthase and acetyl-CoA carboxylase proteins showed similar changes that correlated with SREBP-1c protein. The changes at SREBP-1c and fatty acid synthase proteins also occurred at the mRNA level, as determined by quantitative PCR (Figure 2B) and RNA-sequencing (Supplemental Figure S2B). The activation of SREBP-1c pathway by maltose binge may contribute to the accumulation of liver TG in Shp−/−-CD+M, whereas the inactivation of this pathway by ethanol binge does not explain the increased liver TG in WT-ED+E. The results suggest that alternative pathways may contribute to the observed phenotype.
Figure 2.
Shp deficiency altered hepatic lipogenic markers in response to maltose and ethanol feeding. A: Western blot of hepatic lipogenic proteins in WT and Shp−/− mice fed control diet + maltose binge (CD+M) or ethanol diet + ethanol binge (ED+E). Quantification of the intensity of each band was provided under each line. Each lane represents equal amounts of protein pooled from five individual mice. Results are quantified in panel B. B: Quantitative PCR of mRNA expression of hepatic lipogenic genes in WT (black and blue) and Shp−/− (red and green) mice fed CD+M (black and red) or ED+E (blue and green). Each lane represents triplicate assays from equal amounts of mRNA pooled from five individual mice. Gray shading indicates dark cycle; white indicates light cycle. Data are expressed as means (left and middle column) or means ± SD (right column). ∗P < 0.05 WT CD+M versus Shp−/− CD+M; †P < 0.05 WT ED+E versus Shp−/− ED+E; ‡P < 0.05 WT CD+M versus WT ED+E; §P < 0.05 Shp−/− CD+M versus Shp−/− ED+E. ACC, acetyl-CoA carboxylase; FASN, fatty acid synthase; SREBP-1c, sterol regulatory element-binding transcription factor 1c.
Ethanol Diet Plus Ethanol Binge Induced ATF4 Protein in WT Mice, whereas Control Diet Plus Maltose Binge Activated CHOP and REV-ERBα Proteins in Shp−/− Mice
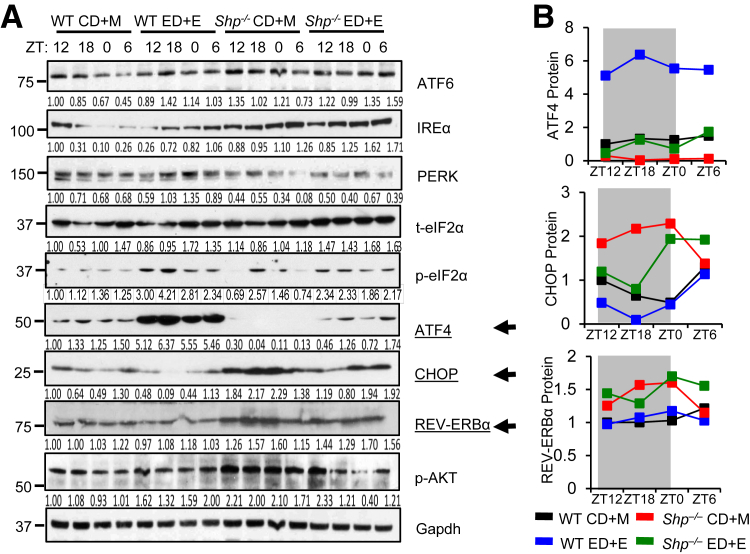
The ER stress signaling pathways consist of three arms, inositol-requiring enzyme 1 α, PRKR-like endoplasmic reticulum kinase, and ATF6 (Supplemental Figure S3), which were often dysregulated in AFL.3, 23 To identify proteins that were regulated by SHP, we analyzed major ER stress markers and observed interesting expression profiles in WT and Shp−/− mice fed with CD+M and ED+E. Overall, most proteins showed modest to strong cyclic variations over a 24-hour dark (ZT12 to ZT18)/light (ZT0 to ZT6) cycle in WT-CD+M, which were disrupted to various extents in WT-ED+E, Shp−/−-CD+M, and Shp−/−-ED+E (Figure 3A). Specifically, ATF6 and inositol-requiring enzyme 1 α proteins were moderately increased, whereas the PRKR-like endoplasmic reticulum kinase protein was modestly down-regulated in Shp−/− versus WT. Furthermore, total eukaryotic translation initiation factor 2 α was induced in Shp−/−-ED+E versus WT-ED+E, whereas phosphorylated eukaryotic translation initiation factor 2α was noticeably induced in WT-ED+E relative to other groups. The ATF4 protein showed the most striking induction in WT-ED+E versus WT-CD+M, whereas in Shp−/− mice ATF4 protein was largely impeded. Notably, despite that ATF4 protein is known as an activator of CHOP transcription,4 we observed opposite changes in the expression of CHOP protein. The CHOP protein was highly induced in Shp−/−-CD+M versus WT-CD+M and was diminished in both WT and Shp−/− by ethanol (Shp−/−-ED+E versus Shp−/−-CD+M and WT-ED+E versus WT-CD+M) (Figure 3B). Although the inactivation of AKT by ER stress was reported to induce CHOP expression,24 we observed an induction in p-AKT in Shp−/−-CD+M versus WT-CD+M. Most interestingly, REV-ERBα protein showed cyclic changes similar to the CHOP protein, which was highest in Shp−/−-CD+M but decreased in Shp−/−-ED+E. Overall, there are two major observations. First, the distinct expression pattern of various ER stress markers suggests differential and unique responses of WT and Shp−/− mice to the maltose and ethanol binge. Second, the results suggest a potential connection between CHOP and REV-ERBα.
Figure 3.
Shp deficiency disrupted maltose- and ethanol-mediated endoplasmic reticulum (ER) stress response. A: Western blot of hepatic proteins in the ER stress signaling pathways in WT and Shp−/− mice fed control diet + maltose binge (CD+M) or ethanol diet + ethanol binge (ED+E). Each lane represents equal amounts of protein pooled from five individual mice. Gray shading indicates dark cycle; white indicates light cycle. Arrows indicate most obviously changed proteins. Quantification of the intensity of each band was provided under each line and in panel B. ATF, activating transcription factor; CHOP, C/EBP homologous protein; Gapdh, glyceraldehyde-3-phosphate dehydrogenase; IRE, inositol-requiring enzyme; p-eIF, phosphorylated eukaryotic translation initiation factor; PERK, PRKR-like endoplasmic reticulum kinase; t-eIF, total eukaryotic translation initiation factor; ZT, Zeitgeber time.
The Expression of Hepatic CHOP and ATF4 Exhibited Different Alteration by Diet and Feeding Conditions in Mice
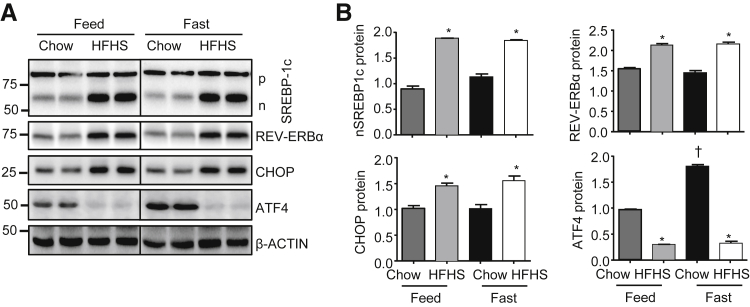
The unexpected opposite expression pattern between CHOP and ATF4 protein in WT and Shp−/− mice observed above suggests a critical influence of maltose and ethanol feeding in combination with Shp deficiency. To further determine the relationship between CHOP and ATF4, we examined their expression levels using a different model (ie, in high-fat and high-sucrose fed WT mice). Hepatic cleaved nSREBP-1c, REV-ERBα, and CHOP proteins were similarly up-regulated by a high-fat and high-sucrose diet in both fed and fasted mice. In contrast, ATF4 protein was down-regulated by the high-fat and high-sucrose diet in both conditions (Figure 4). To confirm the previous report that ATF4 can activate Chop transcription directly,25 we overexpressed ATF4 and peroxisome proliferator-activated receptor γ, the known activators of Chop, in Hepa1 cells and observed the induction of Chop mRNA by both genes (Supplemental Figure S4A). In addition, tunicamycin (ER stress activator) induced both ATF4 and CHOP protein in MHC97H cells (Supplemental Figure S4B). These results suggest that, although ATF4 can function as a transcriptional activator of Chop, Chop expression in vivo is also tightly controlled by other transcription factors and nutrient status. As a result, Shp deficiency plus maltose binge appear to play a dominant role in increasing CHOP protein in Shp−/−-CD+M, which completely masked the effect of ATF4, as seen in Figure 3.
Figure 4.
Hepatic C/EBP homologous protein (CHOP) and activating transcription factor (ATF4) expression was altered differentially by diets in mice. A: Western blot of hepatic sterol regulatory element-binding transcription factor 1c (SREBP-1c), REV-ERBα, CHOP, and ATF4 protein in WT mice fed control chow or a high-fat and high-sucrose (HFHS) diet under feeding and fasting conditions. B: The quantification of protein levels of SREBP-1c, REV-ERBα, CHOP, and ATF4. Samples were pooled and run in duplicate. Data are expressed as means ± SD (B). N = 5 individual mice per group (B). ∗P < 0.05, chow versus HFHS; †P < 0.05, feed chow versus fast chow. n, cleaved; p, precursor.
SHP Alters the Rhythmic Expression of CHOP via a REV-ERBα–Dependent Mechanism
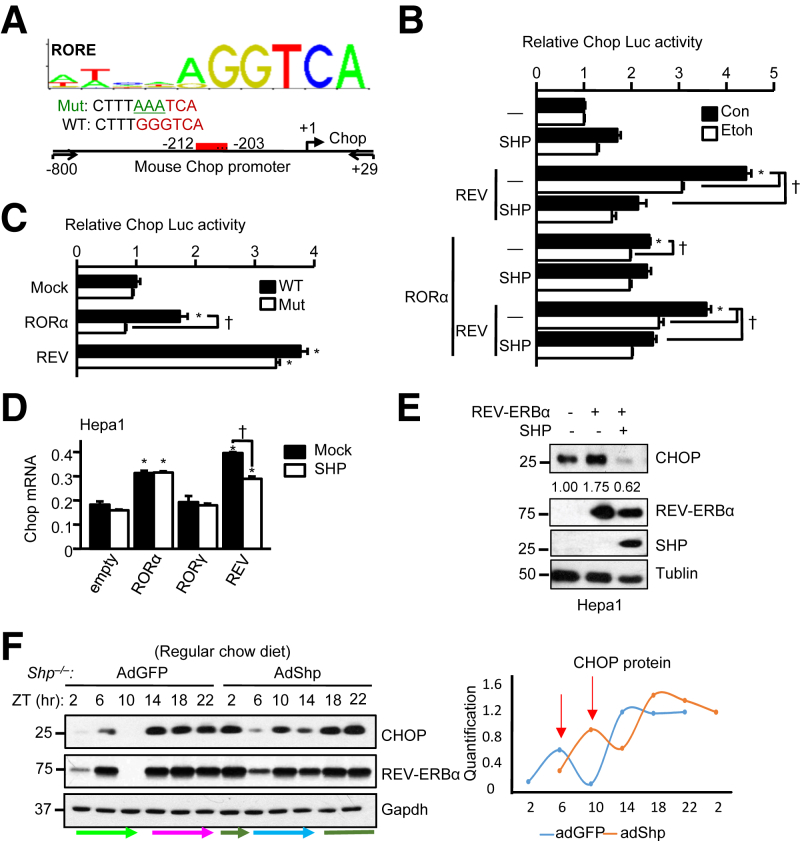
REV-ERBα was reported being both a transcriptional activator26 and repressor.27 The similar expression pattern between CHOP and REV-ERBα (Figure 3) raised an interesting question whether REV-ERBα acts as a transcription activator of CHOP. Because REV-ERBα can compete with ROR binding to the ROR element,28 we searched ROR element and identified it in mouse Chop promoter (Figure 5A). We cloned the native Chop promoter from −800 to 29 into a pGL3 vector for luciferase reporter assay. Overexpression of REV-ERBα significantly induced Chop promoter activity, which was inhibited by SHP coexpression and ethanol treatment (Figure 5B). Interestingly, RORα transactivated Chop promoter with less potency, but antagonized the activation of REV-ERBα. The antagonism of RORα to REV-ERBα activation was likely because of their competition for binding. Consistent with our previous observation,14 SHP did not affect RORα activity. Mutation of ROR element in the Chop promoter greatly diminished RORα transactivation, and to a lesser extent, the REV-ERBα activity (Figure 5C). The results indicate that ROR element in the Chop promoter may have a stronger binding for REV-ERBα than for RORα, consistent with the higher potency of REV-ERBα to activate Chop promoter.
Figure 5.
C/EBP homologous protein (Chop) gene transcription was activated by REV-ERBα and inhibited by SHP. A: Sequence logo of RAR-related orphan receptor (ROR) element (RORE; MA0071.1; JASPAR CORE Vertebrata database, http://jaspar.genereg.net) and RORE (red) in the mouse Chop promoter. Arrows indicate the location of primers for the construction of Chop promoter luciferase reporter. B: Luciferase reporter assay of Chop promoter activity in Hepa1 cells. The cells were transfected with mock (–), RORα, REV-ERBα (Rev), and SHP expression vectors, and treated with 0 (Con, gray) or 400 mmol/L (light blue) ethanol (Etoh) for 6 hours before the luciferase assay. C: Luciferase reporter assay of wild-type and mutant Chop promoter activity in Hepa1 cells. D: Quantitative PCR of Chop mRNA in RORα-, RORγ-, or REV-ERBα–overexpressing Hepa1 cells transfected with mock (black) or SHP (magenta) expression vectors. E: Western blot of proteins in Hepa1 cells transfected with expression plasmids of REV-ERBα, SHP, or shRNA-REV-ERBα. F: Western blot of hepatic CHOP and REV-ERBα in Shp−/− mice reexpressed with adenoviral transduction of GFP (AdGFP) and SHP (AdSHP) on a regular chow diet (left panel) and the quantification of CHOP protein level (right panel). Livers were collected over a 12-hour/12-hour light/dark cycle. Colored arrows show the shift of the circadian pattern. Neon green arrow: peak on ZT6 in Shp−/−/AdGFP; blue arrow: peak on ZT10 in Shp−/−/AdShp; magenta arrow: remain high expression from ZT14 to ZT22 in Shp−/−/AdGFP; green arrow and green line: remain high expression from ZT18 to ZT2 in Shp−/−/AdShp. Data are expressed as means ± SD (B–D), or means (F). ∗P < 0.05 versus empty vector; †P < 0.05. Con, control; luc, lucicerase; Mut, mutation; ZT, Zeitgeber time.
Chop mRNA was remarkably induced by both RORα and REV-ERBα (Figure 5D). On the other hand, Shp repressed the effect of REV-ERBα but not RORα. In addition, CHOP protein was induced by REV-ERBα and its induction was abolished by SHP (Figure 5E). Thus, our results identified REV-ERBα as a new activator of CHOP.
Thus far, we observed that the maltose liquid diet, which is widely used as a control of ethanol diet in the alcohol research field, had a significant differential effect on TG metabolism in Shp−/− mice as compared to the regulator chow diet (Figure 1). To further determine the impact of SHP on the expression correlation between CHOP and REV-ERBα, we overexpressed SHP using adenovirus in Shp−/− mice and collected samples over a 24-hour light/dark cycle under the standard chow-fed condition. CHOP protein showed the same rhythmicity as REV-ERBα protein in control AdGFP mice (Figure 5F), which displayed a diurnal rhythm during the day (ZT: 2-6-10, low-high-low) with peak levels occurring constant during the night. Ectopic overexpression of SHP (AdSHP) disrupted the daily rhythm by causing a 4-hour shift in CHOP and REV-ERBα protein rhythmicity (ZT: 6-10-14: low-high-low) without altering the nocturnal rhythm. The quantification of CHOP protein level was shown on the right. Taken together, our results demonstrate that SHP alters the rhythmic expression of CHOP via a REV-ERBα–dependent regulation.
Knockdown REV-ERBα Prevented Ethanol-Induced Steatosis in Shp−/− Mice
Next, we asked whether REV-ERBα played a physiological role in AFL in the presence (WT) or absence of Shp (Shp−/− mice) by knockdown REV-ERBα in vivo (Supplemental Figure S5). We generated two shRNAs against REV-ERBα (shRev). The second shRev showed a strong effect in reducing SREBP-1c protein in Hepa1 cells (Supplemental Figure S6A), which was used in subsequent in vivo studies. ShRev down-regulated REV-ERBα protein and mRNA in liver, but not in WT mouse brain (Supplemental Figure S6, B–D), suggesting a liver-specific effect. Consistent with the results presented in Figure 3, ATF4 protein was highly induced by ethanol binge in WT/null mice (null: shRev control), whereas it was not affected by shRev (Supplemental Figure S6E). Rev-Erbα mRNA was also down-regulated by shRev in Shp−/− mice (Supplemental Figure S6F).
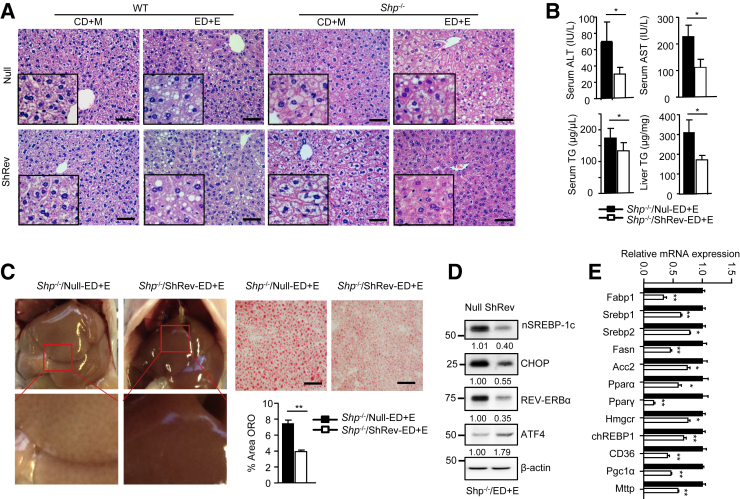
Histology analysis by hematoxylin and eosin staining revealed several interesting observations. In WT mice, shRev-ED+E moderately increased steatosis compared with null-ED+E (Figure 6A). However, in Shp−/− mice, the steatosis (more in the form of microsteatosis) in shRev-CD+M was somewhat increased compared with null-CD+M. However, these differences did not reach statistical significance, as determined by liver TG content (Supplemental Figure S7). The steatosis induced by ethanol binge in Shp−/−/null-ED+E liver was largely prevented in Shp−/−/shRev-ED+E liver, which was accompanied by the decrease in serum ALT, serum AST, serum TG, and liver TG in Shp−/−/shRev-ED+E versus Shp−/−/null-ED+E (Figure 6B). We also observed alleviated pale liver color and markedly reduced neutral lipid staining in Shp−/−/shRev-ED+E versus Shp−/−/null-ED+E (Figure 6C). The nSREBP-1c and CHOP protein (Figure 6D) and many genes involved in lipid metabolism (Figure 6E) were down-regulated by shRev. Therefore, Shp and Rev-Erbα double deficiency prevented ethanol-induced alcoholic fatty liver.
Figure 6.
Knockdown Rev-Erbα impeded ethanol-induced steatosis in Shp−/− mice. A: Hematoxylin and eosin staining of liver sections in WT and Shp−/− mice with Rev-Erbα knockdown and then fed control diet + maltose binge (CD+M) or ethanol diet + ethanol binge (ED+E). Sampling at Zeitgeber time (ZT) 12. Null: control vector for Sh-Rev-Erbα. B: Serum alanine transaminase (ALT), aspartate transaminase (AST), triglyceride (TG), and liver TG in Shp−/−/Null and Shp−/−/Sh-Rev mice fed with ED+E. C: Gross morphology (left panels) and oil red O staining of liver sections (right panels) in Shp−/−/Null and Shp−/−/Sh-Rev mice fed with ED+E. D: Western blot of liver protein in Shp−/−/Null and Shp−/−/Sh-Rev mice fed ED+E. E: Quantitative PCR of hepatic lipogenic gene mRNA in Shp−/−/Null and Shp−/−/Sh-Rev mice fed ED+E. Data are expressed as means ± SD (B, C, and E). ∗P < 0.05, ∗∗P < 0.01 versus Shp−/−/Null-ED+E. Scale bar = 20 μm (A and C). Original magnifications: ×20 (A, main images); ×40 (A, insets). Acc, acetyl-CoA carboxylase; ATF, activating transcription factor; CD36, cluster of differentiation 36; CHOP, C/EBP homologous protein; chREBP, carbohydrate-responsive element-binding protein; Fabp, fatty acid-binding protein; Fasn, fatty acid synthase; Hmgcr, 3-hydroxy-3-methylglutaryl-CoA reductase; Mttp, microsomal triglyceride transfer protein; Pgc1α, peroxisome proliferator-activated receptor γ coactivator 1-α; Ppar, peroxisome proliferator-activated receptor; Srebp, sterol regulatory element-binding transcription factor.
Discussion
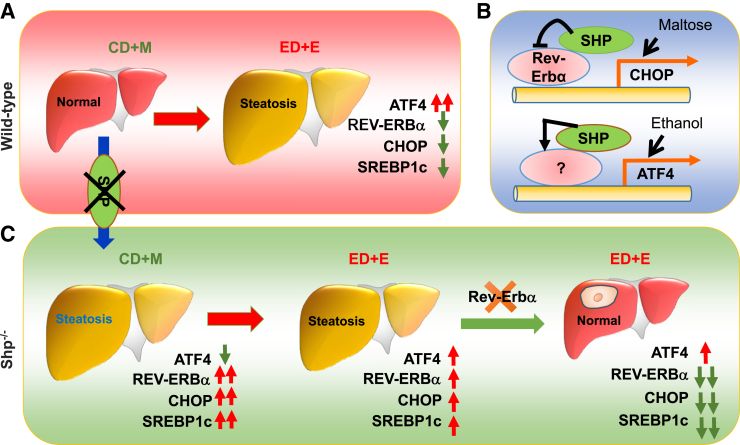
Fatty liver (steatosis) is the collection inside hepatocytes of excessive amounts of fat, which consists mainly of TG. An important cause of fatty liver is alcohol abuse, referred to as AFL. Macrovesicular alcoholic steatosis is a common form of AFL and is featured by the presence of large sharp fat droplets in hepatocytes, and the nuclei are in a peripheral location.29 Although alcohol impairs circadian rhythm in liver,30 the influence of nuclear receptor circadian regulators on alcoholic fatty liver remains unclear. Our studies identified two new players (ie, nuclear receptors SHP and REV-ERBα) that are important in mediating the development of AFL (Figure 7).
Figure 7.
Schematic summarizing major findings in this study. A: WT mice: ethanol diet + ethanol binge (ED+E) induces steatosis, which correlates with a marked up-regulation of activating transcription factor 4 (ATF4) protein but down-regulation of C/EBP homologous protein (CHOP) and sterol regulatory element-binding transcription factor 1c (SREBP-1c) protein. However, knockdown ATF4 does not prevent steatosis induced by ED+E. B: Top image: SHP inhibits Chop transcriptional activation by REV-ERBα. Chop induction by loss of SHP repression is augmented by control diet + maltose binge (CD+M). Bottom image: SHP activates ATF4 via an unknown mechanism. ED+E induces ATF4 while transiently down-regulating SHP expression. C:Shp−/− mice: CD+M causes microvesicular steatosis, which correlates with a sharp elevation of CHOP, SREBP-1c, and REV-ERBα protein but a diminished ATF4 protein. On the contrary, ED+E-induced steatosis correlates with moderate up-regulation of ATF4, CHOP, SREBP-1c, and REV-ERBα protein relative to WT-ED+E. Knockdown REV-ERBα prevents ED+E-induced steatosis.
The most widely used model for alcoholic liver injury is feeding with the Lieber-DeCarli diet containing ethanol for 4 to 6 weeks31; however, this model only induces mild steatosis and slight elevation of serum ALT with low or no liver inflammation.15 The Gao-binge model is a relatively new model, which represents an acute model of alcohol-induced liver injury and has been widely used by other research groups.18, 19 One limitation done by other groups is that samples are collected at one time point 9 hours after binge and the liver phenotype (inflammation and lipid accumulation) may represent transient responses to acute injury. A novel aspect of our study is to examine lipid metabolic changes by collecting samples over a 24-hour light/dark cycle. Another novelty of our study is to determine the impact of nuclear receptors SHP and REV-ERBα in AFL, both of which are important regulators of the circadian clock machinery in the liver.
The function of SHP has been well characterized by us and others in cholesterol and lipid metabolism associated with NAFLD.11, 14, 32 However, SHP's role in ALD remains largely unknown. Therefore, the studies were conducted using both WT and Shp−/− mice. Phenotypically, several unexpected findings were observed. The serum and hepatic lipid profile, as well as the expression of lipid synthetic genes and ER stress markers, exhibit strong but distinct cyclic variations in WT and Shp−/− mice. ED+E induces steatosis in WT mice, which correlates with a marked up-regulation of ATF4 protein; it leads to a down-regulation of CHOP and SREBP-1c protein. On the contrary, CD+M seems to cause a variant of steatosis (namely, microvesicular steatosis) in Shp−/− mice, which is accompanied by a sharp elevation of CHOP, SREBP-1c, and REV-ERBα protein but a diminished ATF4 protein. The differential activation of ATF4 and CHOP, which corresponds to steatosis in WT mice by ethanol and microvesicular steatosis in Shp−/− mice by maltose, respectively, suggest a divergent function of both proteins to potentially regulate two different forms of steatosis. Unfortunately, knockdown ATF4 (Supplemental Figure S8) did not diminish ED+E-induced steatosis in WT mice (data not shown), suggesting its activation by ethanol is likely the consequence but not the cause of steatosis. One possibility is that ED+E may cause mitochondria damage and decrease mitochondria β oxidation.1 The microvesicular fat deposition in the short timelines of these studies may reflect an intermediate stage of lipid formation. Nonetheless, because liver transplant donors with macrosteatosis (common) and microsteatosis (less common) respond differently to intermittent hepatic inflow occlusion during tumor hepatectomy,33 our results may contribute to a better understanding of the molecular basis of microsteatosis.
REV-ERBα is an integrator of circadian rhythms and metabolism in the liver.34 Dual depletion of Rev-Erbα and Rev-Erbβ disrupts lipid homeostasis gene networks,35 and single deletion of Rev-Erbα leads to modest hepatosteatosis.36 However, these early studies were performed in NAFLD and no detailed studies have been done to elucidate the function of REV-ERBα in AFL. Interestingly, REV-ERBα interacts with SHP and exerts as a transcriptional repressor of NPAS2 expression in NAFLD.14 In this study, we demonstrate that REV-ERBα acts as a transcriptional activator of CHOP expression and contributes to the development of AFL under Shp-deficient conditions. Therefore, REV-ERBα appears to have bidirectional transcriptional effects as a repressor or an activator, depending on the gene promoter as well as the context of lipid metabolic dysregulation (NAFLD versus AFL). Of critical importance, REV-ERBα knockdown prevents ethanol diet plus ethanol binge–induced steatosis in Shp−/− mice. Interestingly, knockdown of REV-ERBα shows minimal effect in altering lipid profile in WT mice fed with maltose or ethanol, signifying the importance of REV-ERBα and SHP cross talk in the control of ALD.37
In conclusion, our study elucidates a crucial cross talk between two circadian clock modulators, SHP and REV-ERBα, to regulate AFL by the integration of ER stress marker CHOP. SHP inhibits CHOP activation by REV-ERBα; thus, the permanent loss of SHP repression in Shp−/− mice enhances CHOP induction by maltose, which may contribute to the development of steatosis. In addition, double deficiency of Shp and Rev-Erbα prevented mice from developing ethanol-induced steatosis. Because most researchers in the alcohol field study ALD using samples collected from a single time point, our study represents an important shift in alcohol research by encompassing the liver clock component. The knowledge gained from this study is of clinical importance when developing therapeutic agents to target SHP and REV-ERBα in NAFLD versus AFL.
Acknowledgments
Z.Y. and H.T. performed experiments and prepared the manuscript; Y.Z., S.L., C.H., Y.H., and G.M.V. performed experiments; L.W. supervised the work and wrote the manuscript.
Footnotes
Supported by NIHR01DK104656, R01DK080440, R01ES025909, R21AA022482, R21AA024935, VA Merit Award 1I01BX002634, and P30 DK34989 (Yale Liver Center) (L.W.); and National Natural Scientific Foundation of China grant 81572443 (Y.H.).
Disclosures: None declared.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.ajpath.2016.07.014.
Supplemental Data
Mouse model of Gao-binge.15 After acclimatization for 5 days, the mice were given control or ethanol (EtOH)-containing Lieber-DeCarli liquid diets for 10 days, followed by oral gavage of maltose (control) or EtOH solutions at 9 am on day 10. Nine hours after the binge [Zeitgeber time (ZT) 12; 6 pm], liver tissues and blood samples were collected every 6 hours for 24 hours. Please note that both control and EtOH diets were provided throughout the sample collection period after the binge.
RNA-sequencing analysis of SHP mRNA (A) and sterol regulatory element-binding transcription factor 1c (SREBP-1c) and fatty acid synthase (Fasn) mRNA (B) in wild-type and Shp−/− mice fed with control diet plus maltose binge (CD+M) and ethanol diet plus ethanol binge (ED+E). Samples were collected at Zeitgeber time (ZT) 12, ZT18, ZT0, and ZT6. The purple arrow in A indicates the deleted exon 1 in the Shp locus in Shp−/− mice.
Endoplasmic reticulum (ER) stress signaling pathways. ATF4/6, activating transcription factor 4/6; CHOP (alias DDIT3), C/EBP homologous protein; eIF2a, eukaryotic translation initiation factor 2A; IRE1, inositol-requiring enzyme-1; JNK, c-Jun N-terminal kinase; PERK, PRKR-like endoplasmic reticulum kinase; XBP1, X-box binding protein 1.
A: Quantitative PCR of C/EBP homologous protein (CHOP) mRNA in Hepa1 cells transfected with mock (control; blue), sterol regulatory element-binding transcription factor 1c (SREBP1c; red), activating transcription factor (ATF) 4 (green), and peroxisome proliferator-activated receptor (PPAR) γ (purple) expression vectors (1 μg per well). B: Western blot of CHOP and ATF4 proteins in 97H cells treated with tunicamycin (TM; endoplasmic reticulum stress inducer). Data are expressed as the means ± SD (A). ∗P < 0.05 versus control.
Diagram showing the timeline for REV-ERBα knockdown experiment. The feeding procedure is the same as in Supplemental Figure S1. AAV, adeno-associated virus; Etoh, ethanol.
A: Western blot of sterol regulatory element-binding transcription factor 1c (SREBP-1c) protein in Hepa1 cells transfected with shRev. B: Western blot of REV-ERBα protein in WT mouse liver transduced with shRev. C and D: Quantitative PCR of Rev-Erbα mRNA in liver (C) and brain (D) of WT mice injected with adeno-associated virus (AAV) 8-Null (Null) or AAV8-shRev (shRev), and subjected to the Gao-binge protocol. E: Western blot of activating transcription factor (ATF) 4 protein in WT mice liver injected with AAV8-Null (Null) or AAV8-shRev (shRev), and subjected to the Gao-binge protocol. For Western blot, equal amounts of protein were pooled from individual mice and loaded in duplicate. F: Quantitative PCR of Rev-Erbα mRNA in Shp−/− mice injected with AAV8-Null (Null) or AAV8-shRev (shRev), and subjected to the Gao-binge protocol. Data are expressed as means ± SD (C, D, and F). N = 5 per group (E). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. CD+M, diet plus maltose binge; ED+E, ethanol diet plus ethanol binge.
A–D: Serum alanine transaminase (ALT; A), aspartate transaminase (AST; B), triglyceride (TG; C), and liver TG (D) in WT/Null and WT/shRev, Shp−/−/Null and Shp−/−/shRev mice fed with control diet plus maltose binge (CD+M) and ethanol diet plus ethanol binge (ED+E). The experimental procedures are the same as in Figure 6. ∗P < 0.05 versus Shp−/−/Null-ED+E.
Diagram showing the timeline for activating transcription factor 4 knockdown experiment. The feeding procedure is the same as in Supplemental Figure S1. Etoh, ethanol; ZT, Zeitgeber time.
References
- 1.Williams J.A., Manley S., Ding W.X. New advances in molecular mechanisms and emerging therapeutic targets in alcoholic liver diseases. World J Gastroenterol. 2014;20:12908–12933. doi: 10.3748/wjg.v20.i36.12908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.You M., Jogasuria A., Lee K., Wu J., Zhang Y., Lee Y.K., Taylor C., Sadana P. Signal transduction mechanisms of alcoholic fatty liver disease: emerging role of lipin-1. Curr Mol Pharmacol. 2015 doi: 10.2174/1874467208666150817112109. [Epub ahead of print] doi: 10.2174/1874467208666150817112109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ji C. Mechanisms of alcohol-induced endoplasmic reticulum stress and organ injuries. Biochem Res Int. 2012;2012:216450. doi: 10.1155/2012/216450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malhi H., Kaufman R.J. Endoplasmic reticulum stress in liver disease. J Hepatol. 2011;54:795–809. doi: 10.1016/j.jhep.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsuchiya H., da Costa K.A., Lee S., Renga B., Jaeschke H., Yang Z., Orena S.J., Goedken M.J., Zhang Y., Kong B., Lebofsky M., Rudraiah S., Smalling R., Guo G., Fiorucci S., Zeisel S.H., Wang L. Interactions between nuclear receptor SHP and FOXA1 maintain oscillatory homocysteine homeostasis in mice. Gastroenterology. 2015;148:1012–1023.e14. doi: 10.1053/j.gastro.2015.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rudraiah S., Zhang X., Wang L. Nuclear receptors as therapeutic targets in liver disease: are we there yet? Annu Rev Pharmacol Toxicol. 2016;56:605–626. doi: 10.1146/annurev-pharmtox-010715-103209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y., Hagedorn C.H., Wang L. Role of nuclear receptor SHP in metabolism and cancer. Biochim Biophys Acta. 2011;1812:893–908. doi: 10.1016/j.bbadis.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou T., Zhang Y., Macchiarulo A., Yang Z., Cellanetti M., Coto E., Xu P., Pellicciari R., Wang L. Novel polymorphisms of nuclear receptor SHP associated with functional and structural changes. J Biol Chem. 2010;285:24871–24881. doi: 10.1074/jbc.M110.133280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang L., Lee Y.K., Bundman D., Han Y., Thevananther S., Kim C.S., Chua S.S., Wei P., Heyman R.A., Karin M., Moore D.D. Redundant pathways for negative feedback regulation of bile acid production. Dev Cell. 2002;2:721–731. doi: 10.1016/s1534-5807(02)00187-9. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y., Xu N., Xu J., Kong B., Copple B., Guo G.L., Wang L. E2F1 is a novel fibrogenic gene that regulates cholestatic liver fibrosis through the Egr-1/SHP/EID1 network. Hepatology. 2014;60:919–930. doi: 10.1002/hep.27121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang J., Iqbal J., Saha P.K., Liu J., Chan L., Hussain M.M., Moore D.D., Wang L. Molecular characterization of the role of orphan receptor small heterodimer partner in development of fatty liver. Hepatology. 2007;46:147–157. doi: 10.1002/hep.21632. [DOI] [PubMed] [Google Scholar]
- 12.Pan X., Zhang Y., Wang L., Hussain M.M. Diurnal regulation of MTP and plasma triglyceride by CLOCK is mediated by SHP. Cell Metab. 2010;12:174–186. doi: 10.1016/j.cmet.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L., Liu J., Saha P., Huang J., Chan L., Spiegelman B., Moore D.D. The orphan nuclear receptor SHP regulates PGC-1alpha expression and energy production in brown adipocytes. Cell Metab. 2005;2:227–238. doi: 10.1016/j.cmet.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 14.Lee S.M., Zhang Y., Tsuchiya H., Smalling R., Jetten A.M., Wang L. Small heterodimer partner/neuronal PAS domain protein 2 axis regulates the oscillation of liver lipid metabolism. Hepatology. 2015;61:497–505. doi: 10.1002/hep.27437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bertola A., Mathews S., Ki S.H., Wang H., Gao B. Mouse model of chronic and binge ethanol feeding (the NIAAA model) Nat Protoc. 2013;8:627–637. doi: 10.1038/nprot.2013.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y., Liu C., Barbier O., Smalling R., Tsuchiya H., Lee S., Delker D., Zou A., Hagedorn C.H., Wang L. Bcl2 is a critical regulator of bile acid homeostasis by dictating Shp and lncRNA H19 function. Sci Rep. 2016;6:20559. doi: 10.1038/srep20559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Z., Zhang Y., Wang L. A feedback inhibition between miRNA-127 and TGFbeta/c-Jun cascade in HCC cell migration via MMP13. PLoS One. 2013;8:e65256. doi: 10.1371/journal.pone.0065256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen P., Miyamoto Y., Mazagova M., Lee K.C., Eckmann L., Schnabl B. Microbiota protects mice against acute alcohol-induced liver injury, alcoholism. Clin Exp Res. 2015;39:2313–2323. doi: 10.1111/acer.12900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roh Y.S., Zhang B., Loomba R., Seki E. TLR2 and TLR9 contribute to alcohol-mediated liver injury through induction of CXCL1 and neutrophil infiltration. Am J Physiol Gastrointest Liver Physiol. 2015;309:G30–G41. doi: 10.1152/ajpgi.00031.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watanabe M., Houten S.M., Wang L., Moschetta A., Mangelsdorf D.J., Heyman R.A., Moore D.D., Auwerx J. Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J Clin Invest. 2004;113:1408–1418. doi: 10.1172/JCI21025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferre P., Foufelle F. Hepatic steatosis: a role for de novo lipogenesis and the transcription factor SREBP-1c, diabetes. Obesity Metab. 2010;12(Suppl 2):83–92. doi: 10.1111/j.1463-1326.2010.01275.x. [DOI] [PubMed] [Google Scholar]
- 22.Purohit V., Gao B., Song B.J. Molecular mechanisms of alcoholic fatty liver, alcoholism. Clin Exp Res. 2009;33:191–205. doi: 10.1111/j.1530-0277.2008.00827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ji C. New insights into the pathogenesis of alcohol-induced ER stress and liver diseases. Int J Hepatol. 2014;2014:513787. doi: 10.1155/2014/513787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hyoda K., Hosoi T., Horie N., Okuma Y., Ozawa K., Nomura Y. PI3K-Akt inactivation induced CHOP expression in endoplasmic reticulum-stressed cells. Biochem Biophys Res Commun. 2006;340:286–290. doi: 10.1016/j.bbrc.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 25.Averous J., Bruhat A., Jousse C., Carraro V., Thiel G., Fafournoux P. Induction of CHOP expression by amino acid limitation requires both ATF4 expression and ATF2 phosphorylation. J Biol Chem. 2004;279:5288–5297. doi: 10.1074/jbc.M311862200. [DOI] [PubMed] [Google Scholar]
- 26.Harding H.P., Lazar M.A. The orphan receptor Rev-ErbA alpha activates transcription via a novel response element. Mol Cell Biol. 1993;13:3113–3121. doi: 10.1128/mcb.13.5.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J., Yin L., Lazar M.A. The orphan nuclear receptor Rev-erb alpha regulates circadian expression of plasminogen activator inhibitor type 1. J Biol Chem. 2006;281:33842–33848. doi: 10.1074/jbc.M607873200. [DOI] [PubMed] [Google Scholar]
- 28.Guillaumond F., Dardente H., Giguere V., Cermakian N. Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J Biol Rhythms. 2005;20:391–403. doi: 10.1177/0748730405277232. [DOI] [PubMed] [Google Scholar]
- 29.Yerian L. Histopathological evaluation of fatty and alcoholic liver diseases. J Dig Dis. 2011;12:17–24. doi: 10.1111/j.1751-2980.2010.00472.x. [DOI] [PubMed] [Google Scholar]
- 30.Udoh U.S., Valcin J.A., Gamble K.L., Bailey S.M. The molecular circadian clock and alcohol-induced liver injury. Biomolecules. 2015;5:2504–2537. doi: 10.3390/biom5042504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Everitt H., Hu M., Ajmo J.M., Rogers C.Q., Liang X., Zhang R., Yin H., Choi A., Bennett E.S., You M. Ethanol administration exacerbates the abnormalities in hepatic lipid oxidation in genetically obese mice. Am J Physiol Gastrointest Liver Physiol. 2013;304:G38–G47. doi: 10.1152/ajpgi.00309.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Datta S., Wang L., Moore D.D., Osborne T.F. Regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase promoter by nuclear receptors liver receptor homologue-1 and small heterodimer partner: a mechanism for differential regulation of cholesterol synthesis and uptake. J Biol Chem. 2006;281:807–812. doi: 10.1074/jbc.M511050200. [DOI] [PubMed] [Google Scholar]
- 33.Han S., Kim G., Lee S.K., Kwon C.H., Gwak M., Lee S., Ha S., Park C.K., Ko J.S., Joh J. Comparison of the tolerance of hepatic ischemia/reperfusion injury in living donors: macrosteatosis versus microsteatosis. Liver Transpl. 2014;20:775–783. doi: 10.1002/lt.23878. [DOI] [PubMed] [Google Scholar]
- 34.Duez H., Staels B. Rev-erb-alpha: an integrator of circadian rhythms and metabolism. J Appl Physiol. 2009;107:1972–1980. doi: 10.1152/japplphysiol.00570.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cho H., Zhao X., Hatori M., Yu R.T., Barish G.D., Lam M.T., Chong L.W., DiTacchio L., Atkins A.R., Glass C.K., Liddle C., Auwerx J., Downes M., Panda S., Evans R.M. Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature. 2012;485:123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bugge A., Feng D., Everett L.J., Briggs E.R., Mullican S.E., Wang F., Jager J., Lazar M.A. Rev-erbalpha and Rev-erbbeta coordinately protect the circadian clock and normal metabolic function. Genes Dev. 2012;26:657–667. doi: 10.1101/gad.186858.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duez H., van der Veen J.N., Duhem C., Pourcet B., Touvier T., Fontaine C., Derudas B., Bauge E., Havinga R., Bloks V.W., Wolters H., van der Sluijs F.H., Vennstrom B., Kuipers F., Staels B. Regulation of bile acid synthesis by the nuclear receptor Rev-erbalpha. Gastroenterology. 2008;135:689–698. doi: 10.1053/j.gastro.2008.05.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mouse model of Gao-binge.15 After acclimatization for 5 days, the mice were given control or ethanol (EtOH)-containing Lieber-DeCarli liquid diets for 10 days, followed by oral gavage of maltose (control) or EtOH solutions at 9 am on day 10. Nine hours after the binge [Zeitgeber time (ZT) 12; 6 pm], liver tissues and blood samples were collected every 6 hours for 24 hours. Please note that both control and EtOH diets were provided throughout the sample collection period after the binge.
RNA-sequencing analysis of SHP mRNA (A) and sterol regulatory element-binding transcription factor 1c (SREBP-1c) and fatty acid synthase (Fasn) mRNA (B) in wild-type and Shp−/− mice fed with control diet plus maltose binge (CD+M) and ethanol diet plus ethanol binge (ED+E). Samples were collected at Zeitgeber time (ZT) 12, ZT18, ZT0, and ZT6. The purple arrow in A indicates the deleted exon 1 in the Shp locus in Shp−/− mice.
Endoplasmic reticulum (ER) stress signaling pathways. ATF4/6, activating transcription factor 4/6; CHOP (alias DDIT3), C/EBP homologous protein; eIF2a, eukaryotic translation initiation factor 2A; IRE1, inositol-requiring enzyme-1; JNK, c-Jun N-terminal kinase; PERK, PRKR-like endoplasmic reticulum kinase; XBP1, X-box binding protein 1.
A: Quantitative PCR of C/EBP homologous protein (CHOP) mRNA in Hepa1 cells transfected with mock (control; blue), sterol regulatory element-binding transcription factor 1c (SREBP1c; red), activating transcription factor (ATF) 4 (green), and peroxisome proliferator-activated receptor (PPAR) γ (purple) expression vectors (1 μg per well). B: Western blot of CHOP and ATF4 proteins in 97H cells treated with tunicamycin (TM; endoplasmic reticulum stress inducer). Data are expressed as the means ± SD (A). ∗P < 0.05 versus control.
Diagram showing the timeline for REV-ERBα knockdown experiment. The feeding procedure is the same as in Supplemental Figure S1. AAV, adeno-associated virus; Etoh, ethanol.
A: Western blot of sterol regulatory element-binding transcription factor 1c (SREBP-1c) protein in Hepa1 cells transfected with shRev. B: Western blot of REV-ERBα protein in WT mouse liver transduced with shRev. C and D: Quantitative PCR of Rev-Erbα mRNA in liver (C) and brain (D) of WT mice injected with adeno-associated virus (AAV) 8-Null (Null) or AAV8-shRev (shRev), and subjected to the Gao-binge protocol. E: Western blot of activating transcription factor (ATF) 4 protein in WT mice liver injected with AAV8-Null (Null) or AAV8-shRev (shRev), and subjected to the Gao-binge protocol. For Western blot, equal amounts of protein were pooled from individual mice and loaded in duplicate. F: Quantitative PCR of Rev-Erbα mRNA in Shp−/− mice injected with AAV8-Null (Null) or AAV8-shRev (shRev), and subjected to the Gao-binge protocol. Data are expressed as means ± SD (C, D, and F). N = 5 per group (E). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. CD+M, diet plus maltose binge; ED+E, ethanol diet plus ethanol binge.
A–D: Serum alanine transaminase (ALT; A), aspartate transaminase (AST; B), triglyceride (TG; C), and liver TG (D) in WT/Null and WT/shRev, Shp−/−/Null and Shp−/−/shRev mice fed with control diet plus maltose binge (CD+M) and ethanol diet plus ethanol binge (ED+E). The experimental procedures are the same as in Figure 6. ∗P < 0.05 versus Shp−/−/Null-ED+E.
Diagram showing the timeline for activating transcription factor 4 knockdown experiment. The feeding procedure is the same as in Supplemental Figure S1. Etoh, ethanol; ZT, Zeitgeber time.