Abstract
Pathways of liver fibrosis are controlled by connective tissue growth factor (CCN2). In this study, CCN2 was identified as a target of miR-199a-5p, which was principally expressed in quiescent mouse hepatic stellate cells (HSCs) and directly suppressed production of CCN2. Up-regulated CCN2 expression in fibrotic mouse livers or in activated primary mouse HSCs was associated with miR-199a-5p down-regulation. MiR-199a-5p in quiescent mouse HSCs inhibited the activity of a wild-type CCN2 3′ untranslated region (3′-UTR) but not of a mutant CCN2 3′-UTR lacking the miR-199a-5p-binding site. In activated mouse HSCs, CCN2, α-smooth muscle actin, and collagen 1(α1) were suppressed by a miR-199a-5p mimic, whereas in quiescent mouse HSCs, the inhibited CCN2 3′-UTR activity was blocked by a miR-199a-5p antagomir. CCN2 3′-UTR activity in human HSCs was reduced by a miR-199a-5p mimic. MiR-199a-5p was present at higher levels in exosomes from quiescent versus activated HSCs. MiR-199a-5p–containing exosomes were shuttled from quiescent mouse HSCs to activated mouse HSCs in which CCN2 3′-UTR activity was then suppressed. Exosomes from quiescent HSCs caused miR-199a-5p–dependent inhibition of CCN2, α-smooth muscle actin, or collagen 1(α1) in activated HSCs in vitro and bound to activated HSCs in vivo. Thus, CCN2 suppression by miR-199a-5p accounts, in part, for low-level fibrogenic gene expression in quiescent HSCs and causes dampened gene expression in activated HSCs after horizontal transfer of miR-199a-5p in exosomes from quiescent HSCs.
Chronic liver diseases, such as hepatitis, alcoholic liver disease, and nonalcoholic steatosis, are the cause of considerable morbidity worldwide. These diseases are frequently characterized by the presence of fibrosis, a debilitating condition that involves excessive deposition of insoluble collagen that predisposes patients toward more severe conditions, such as cirrhosis, cancer, and end-stage disease.1 Liver fibrosis is a major unsolved medical challenge because there are currently no approved drugs to directly treat it and, furthermore, liver fibrosis still persists in many individuals even after successful treatment of the primary disease (eg, antiviral therapy for hepatitis). It is now well recognized that hepatic stellate cells (HSCs) are the principal fibrogenic cell type in the liver, despite constituting only approximately 5% of the total liver cell population.1, 2 During liver injury, HSCs transition from quiescent vitamin A–storing interstitial cells into highly activated collagen-producing myofibroblastic cells that produce a provisional matrix for hepatocyte repopulation. Whereas this function is curtailed after acute injury, the activated phenotype persists during chronic injury and results in the excessive collagen deposition that is a hallmark of fibrosis. This phenomenon has spawned a major scientific effort to understand the role of HSCs in fibrosis and the value of targeting their profibrotic pathways to identify rational therapeutic targets.3, 4, 5, 6 Furthermore, mechanisms that function to naturally suppress fibrogenic gene expression in quiescent HSCs or that promote HSC senescence or apoptosis during regression of fibrosis may provide additional leads for developing new therapeutic strategies.6, 7, 8 There is optimism that these various approaches will result in the development of effective antifibrotic therapies even when the underlying primary disease persists.5
Many studies have found that alterations in HSC activation are orchestrated by dynamic changes in the expression and/or action of a host of soluble or cell-associated signaling molecules,6, 9 but an additional mechanism of HSC regulation has recently come to light with our finding that fibrogenic pathways in HSCs are modulated by exosomes.10 Exosomes, produced by most if not all cells of the body, are nanosized membranous vesicles that are capable of delivering complex molecular payloads to recipient cells, which may become functionally reprogrammed according to the molecular cues received.11, 12, 13 HepG2 hepatoblastoma cells were the first liver cell type for which exosome production was documented,14 and recent reports have described exosomal pathways for a variety of hepatic cancer cells that result in delivery of RNAs or proteins and subsequent regulation of cancer cell proliferation, chemosensitivity or migration,15, 16, 17, 18 intercellular transmission of the hepatitis C virus,19 or acquisition of a proangiogenic phenotype by endothelial cells.20, 21 However, a more expansive and highly complex role for exosomes in the liver is now emerging, with the recognition that a broad variety of hepatic cells, including hepatocytes, macrophages, cholangiocytes, and HSCs, produce exosomes that can influence a broad spectrum of cellular processes involved in liver repair after hepatectomy or pathogenesis associated with fibrosis, alcoholic liver disease, nonalcoholic steatohepatitis, or nonalcoholic fatty liver disease.10, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31 A question of particular interest is whether quiescent HSCs produce exosomes that contain a cargo that has the ability to attenuate the function of activated HSCs. Such a mechanism might contribute to the transient role of activated HSCs during normal wound healing and would have therapeutic potential in chronic fibrosing liver injury.
In this report, we found that miR-199a-5p is produced at high levels by quiescent HSCs and is delivered exosomally from quiescent HSCs to activated HSCs in which fibrogenic gene expression is then attenuated. This effect is, at least in part, due to the direct targeting by miR-199a-5p of the 3′ untranslated region (3′-UTR) of the profibrotic molecule, connective tissue growth factor (CCN2; also known as CTGF),32 resulting in reduced expression of CCN2 and of its downstream targets α-smooth muscle actin (α-SMA) and collagen 1(α1). These studies identify exosomal miR-199a-5p as a natural suppressor of the activated HSC phenotype.
Materials and Methods
Animal Procedures and Tissue Specimens
Animal protocols were approved by the Institutional Animal Care and Use Committee of Nationwide Children's Hospital (Columbus, OH). Normal male Swiss Webster mice (6 to 8 weeks) (n = 10) received i.p. carbon tetrachloride (CCl4; 175 μL in 1325 μL/kg of corn oil; Sigma-Aldrich, St Louis, MO) three times per week for 5 weeks. Control mice received i.p. corn oil (1500 μL/kg of corn oil) alone. On death, individual liver lobes were harvested and either snap-frozen in liquid nitrogen for subsequent RNA extraction or perfused using phosphate-buffered saline (PBS) followed by 4% paraformaldehyde (Sigma-Aldrich) for histologic analysis of fixed tissue. In some experiments, mice that had received CCl4 three times a week for 2 weeks were sacrificed 4 hours after tail vein injection of 40 μg of PKH26-labeled exosomes that had been purified from day 2 (D2) primary mouse HSCs, which had been maintained for 2 days in medium that contained 1% serum from which serum exosomes had been removed by prior ultracentifugation (see below). Livers lobes were either perfused for subsequent isolation of HSCs or hepatocytes or immediately harvested for fluorescence imaging using a Xenogen IVIS 200 instrument (PerkinElmer, Waltham, MA). In an alternative model of liver injury, male FVB mice (6 to 8 weeks) (n = 10) received i.p. thioacetic acid (100 mg/kg; Sigma-Aldrich) in saline three times per week for 5 weeks. Control mice received i.p. saline alone. Mice were sacrificed 72 hours after the last injection, livers were harvested, and RNA was isolated and processed for quantitative RT-PCR (RT-qPCR). Some livers were fixed for in situ analysis.
Culture of Primary Mouse HSCs or Hepatocytes or of Human LX-2 HSCs
Primary HSCs were isolated, essentially as we have previously described,33 by buoyant-density centrifugation from normal male Swiss Webster mice (6 to 8 weeks), and spent medium from the cultured cells was replaced with fresh Dulbecco’s modified Eagle’s medium, F12, and 10% fetal bovine serum (FBS) medium on D1 and every other day as needed. HSCs were split 1:3 every 5 days and used at passages 0 to 6 (P0 to P6). Our previous studies found that, when isolated from normal (nonfibrotic) animals, cells studied within 24 hours of brief culture do not exhibit characteristics typical of activated cells (eg, α-SMA, CCN2) and thereafter gradually transition to a highly activated phenotype during the ensuing 7 to 20 days of culture.10, 22, 23, 33, 34 In this study, CCN2 and miR-199a-5p expression were measured in primary HSCs until D20 of culture. In some experiments, triplicate wells of cells in serum-free medium were incubated for up to 48 hours in the presence of 0 to 3 ng/mL of transforming growth factor (TGF)-β1. Cells were then evaluated for CCN2, miR-199a-5p, α-SMA, or collagen 1(α1) expression by RT-qPCR.
Primary mouse hepatocytes were isolated from normal male FVB mice (approximately 5 weeks) using a modified collagenase perfusion procedure.35 Briefly, mouse livers were perfused through the portal vein with perfusion buffer (10 mmol/L HEPES, 3 mmol/L KCl, 130 mmol/L NaCl, 1 mmol/L NaH2PO4, 10 mmol/L glucose, and 0.5 mmol/L EGTA) followed by a second perfusion with digestion buffer (10 mmol/L HEPES, 3 mmol/L KCl, 130 mmol/L NaCl, 1 mmol/L NaH2PO4, 10 mmol/L glucose, 0.0279% CaCl2 and 0.6 mg/mL collagenase type I; Sigma-Aldrich). The livers were then removed, cut into pieces, and homogenized in 10 mL of complete William E medium (Gibco, Billings, MT) containing 10% FBS and 10 mmol/L HEPES buffer for 2 to 3 minutes. The liver homegenate was first passed through a 120-μm-pore mesh filter and then through a 70-μm-pore mesh filter. The resulting cell suspension was centrifuged at 50 × g for 4 minutes and then resuspended in 90% Percoll in PBS and centrifuged at 200 × g for 10 minutes. The pellet was collected seeded at 1 × 106 cells/mL with complete William E medium. Human LX-2 cells (a kind gift from Dr. Scott Friedman, Icahn School of Medicine at Mount Sinai, New York, NY) were maintained in culture as described.33
Fluorescence-Activated Cell Sorting
Cells obtained after perfusion collagenase digestion of whole liver were subjected to a Percoll density gradient centrifugation (see above). Cells collected at the 25%/50% interface were then stained with antibodies to CD45 (Allophycocyanin Fire750, 30-F11; Biolegend, San Diego, CA), CD31 (Allophycocyanin, 390; Biolegend), CD146 (phycoerythrin, ME-9F1; Biolegend), F4/80 (Allophycocyanin, BM8; Biolegend), and CD11b (PE-Cy7, M1/70; BD Pharmingen, San Diego, CA) to identify luminal sinusoidal endothelial cells (LSECs) and Kupffer cell subsets. Cells were incubated in staining buffer (PBS and 2% bovine serum albumin) in the presence of antibodies for 20 minutes at 4°C, washed twice, and filtered through a 70-μm cell strainer. LSECs and Kupffer cell subsets were sorted on an Influx cell sorter (BD Biosciences, San Jose, CA), and subset purity was verified to be >95%.
RNA Extraction and RT-qPCR
Total RNA from liver tissues or individual liver cell types was extracted using a microRNeasy Plus kit (Qiagen, Valencia, CA) and reverse transcribed using a miScript II RT kit (Qiagen) according to the manufacturers' protocols. Quantification of RNA levels for CCN2, miR-199a-5p, α-SMA, or collagen 1(α1) was achieved by RT-qPCR using an Eppendorf Mastercycler System and SYBR Green Master Mix (Eppendorf, Hauppauge, NY). Primers are listed in Table 1. Each reaction was run in triplicate, and all samples were normalized to glyceraldehyde-3-phosphate dehydrogenase. Negative controls were a nonreverse transcriptase reaction and a nonsample reaction.
Table 1.
Primers Used for RT-qPCR
| Gene (mouse) | GenBank accession number∗ | Sense primer | Antisense primer | Size (bp) |
|---|---|---|---|---|
| CCN2 | NM_010217 | 5′-CACTCTGCCAGTGGAGTTCA-3′ | 5′-AAGATGTCATTGTCCCCAGG-3′ | 111 |
| 199a-5p | MIMAT0000229 | 5′-CCCAGUGUUCAGACUACCUGUUC-3′ | Universal antisense | 23 |
| Collagen 1(α1) | NM_007742 | 5′-GCCCGAACCCCAAGGAAAAGAAGC-3′ | 5′-CTGGGAGGCCTCGGTGGACATTAG-3′ | 148 |
| α-SMA | NM_007392 | 5′-GGCTCTGGGCTCTGTAAGG-3′ | 5′-CTCTTGCTCTGGGCTTCATC-3′ | 148 |
| GAPDH | NM_008084 | 5′-TGCACCACCAACTGCTTAGC-3′ | 5′-GGCATGGACTGTGGTCATGAG-3′ | 87 |
CCN2, connective tissue growth factor; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; α-SMA, α-smooth muscle actin.
Accession numbers are for the NCBI Nucleotide database, available at http://www.ncbi.nlm.nih.gov/nuccore.
In Situ Hybridization
Fixed liver sections were hybridized with a miR-199a-5p antisense probe (5′-DIGGAACAGGTAGTCTGAACACTGGG-3′ DIG; Exiqon Inc., Woburn, MA) or a scramble probe for 60 minutes at 55°C (Exiqon), followed by washes with varying concentrations of standard saline citrate buffer. Probes were detected with a monoclonal anti–digoxigenin-alkaline phosphatase antibody (1:800) (Roche, Indianapolis, IN) for 60 minutes followed by nitro-blue tetrazolium and 5-bromo-4-chloro-3′-indolyphosphate substrate (Roche) at 30°C for 2 hours. The slides were counterstained with Nuclear Fast Red, mounted with Eukitt Medium (VWR, Radnor, PA), and examined by light microscopy.
Immunohistochemistry
Fixed liver sections were incubated with NH1 anti-CCN2 IgY (5 μg/mL33), anti–α-SMA (1:100; Dako Cytomatio, Denmark), anti–collagen 1(α1) (1:250; Abcam, Cambridge, MA), or anti–platelet-derived growth factor receptor-β (1:500 Cell Signaling, Beverly MA) followed by Alexa Fluor 568 goat–anti-chicken IgY, Alexa Fluor 488 goat–anti-mouse IgG, or Alexa Fluor 647 goat–anti-rabbit IgG (all at 1:1000; Life Technologies) for 1 hour at room temperature. The slides were mounted with Vectashield Mounting Medium containing DAPI nuclear stain (Vector Laboratories, Burlingame, CA) and examined by confocal microscopy.
Western Blot Analysis
Equivalent amounts of proteins in cell lysates were run on 12% or 15% SDS-PAGE gels and transferred to nitrocellulose membranes, which were then probed with NH1 anti-CCN2 IgY (5 μg/mL), anti–α-SMA (1:100; Dako Cytomatio), or anti–collagen 1(α1) (1:250; Abcam) and developed using an enhanced chemiluminescence detection system (GE Health Care, Piscataway, NJ).
Transfection of Primary Mouse HSCs with miR-199a-5p Mimic
P6 mouse HSCs were transfected with 100 nmol/L miR-199a-5p mimic or a scrambled mimic (Qiagen) by electroporation (Nucleofector Kit, Lonza, Koln, Germany). Transfected cells were placed in 6-well plates, incubated in Dulbecco’s modified Eagle’s medium, F12, and 10% FBS medium for 24 hours, and then processed for RNA isolation.
Transfection of Primary Mouse HSCs with miR-Selection Fire-Ctx Lentivector-CCN2 3′-UTR and a miR-199a-5p Antagomir
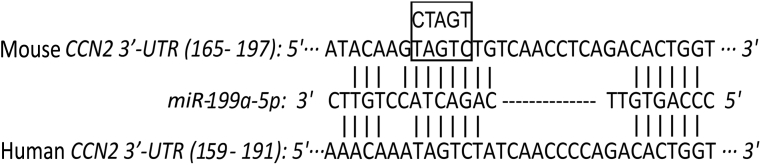
A predicted binding site for miR-199a-5p in mouse CCN2 3′-UTR was identified using BLAST (Figure 1). The full length of CCN2 3′-UTR (http://www.ncbi.nlm.nih.gov/genbank; accession number BC006783.1) was amplified by PCR from primary mouse HSC genomic DNA using forward primer 5′-GAGGGATCCGTCACACTCTCAACAAATAAACTGCCC-3′ and reverse primer 5′-GAGGAATTCAGCCAGGAAGTAAGGGAACCGAACTCA-3′. The PCR fragment was digested with BamHI and EcoRI, subcloned into Fire-Ctx sensor lentivector (SBI, Mountain View, CA), downstream of the Firefly luciferase reporter and cytotoxin (CTX) drug sensor genes, and verified by DNA sequencing. A mutant CCN2 3′-UTR that contained a 5–base point mutation in the binding region for miR-199a-5p (TAGTC → CTAGT) (Figure 1) was amplified from the wild-type mouse CCN2 3′-UTR using forward primer 5′-ACAAGCTAGTTGTCAACCTCAGACACTGG-3′ (underlined section represents an altered sequence that allows a mutation to be inserted in the final product) and reverse primer 5′-CCAGTGTCTGAGGTTGACAGACTACTTGT-3′ and verified by DNA sequencing.
Figure 1.
miR-199a-5p binding site in connective tissue growth factor (CCN2) 3′ untranslated region (3′-UTR). Sequence alignment of miR-199a-5p with its binding site in the 3′-UTR of human (http://www.ncbi.nlm.nih.gov/genbank; accession number BC087839.1) or mouse (http://www.ncbi.nlm.nih.gov/genbank; accession number BC006783.1) CCN2. The region in the 3′-UTR selected for substitution mutagenesis (nt 172–176) is shown in the boxed area.
Primary D4 HSCs were co-transfected with a miR-199a-5p antagomir (Qiagen) and Fire-Ctx sensor lentivectors that contained CCN2 wild-type or mutant 3′-UTR or parental vector alone. To control for transfection efficiency, cells were also transfected with pRL-CMV vector (Promega, Madison WI) containing Renilla luciferase reporter gene. After 24 hours, luciferase activity was measured in triplicate using an E1910 Dual Luciferase Reporter Assay System (Promega). Renilla luciferase activity was used for normalization, and Firefly luciferase activity in miR-199a-5p antagomir-transfected cells was compared with that in mock-transfected cells.
Transfection of Human LX-2 HSCs with miR-Selection Fire-Ctx Lentivector-CCN2 3′-UTR and a miR-199a-5p Mimic
Human LX-2 cells were cultured as previously described.33 LX-2 cells were co-transfected with miR-199a-5p mimic and either the parental lentivector or the vector containing the wild-type CCN2 3′-UTR (see above). Cells were also transfected with pRL-CMV vector to control for transfection efficiency. After 24 hours, luciferase activity was measured as described for above for mouse HSCs.
Characterization of Exosomes in Conditioned Medium from Mouse HSCs
Primary mouse D2 or P6 HSCs were incubated in T-75 culture flasks for 48 hours in Dulbecco’s modified Eagle’s medium and F-12 medium containing, respectively, low (1%) serum or no serum. Before use, the serum was depleted of its instrinsic exosome components by serial stepwise centrifugation and ultracentrifugation steps as previously described.36 HSC conditioned medium underwent serial centrifugation steps37 to isolate exosomes, which were then resuspended in 50 to 100 μL of PBS. Purified exosomes were evaluated for size and frequency by NanoSight nanoparticle tracking analysis (Malvern Instruments, Westborough, MA). Videography was used to visualize and record the features of the particle analysis. Accuracy of measurement was ensured by prior dilution of the sample to approximately 106 particles/mL in 0.1× PBS. Western blots were performed on 10 μg of purified exosomes as previously described,10, 23 except the primary antibody was raised against CD81 (1:400; ProSci, Poway, CA). Exosomal RNAs were isolated, and the presence of miR-199a-5p was determined by RT-qPCR and normalized to let-7a, which we have previously validated.23 These exosomes were also used to treat P6 recipient HSCs with or without transfection of a miR-199a-5p antagomir. Triplicate wells of cells were incubated for up to 48 hours in the presence or absence of 8 μg/mL of exosomes, and RNA levels for CCN2, α-SMA, or collagen 1(α1) were measured by RT-qPCR as described above.
Analysis of Exosome-Mediated miR-199a-5p Communication in HSC Co-Cultures
With the use of our previously described co-culture system,10, 23 exosomal delivery of miR-199a-5p was determined by assessment of dual luciferase activity in P6 mouse primary HSCs transfected with parental miR-Selection Fire-Ctx lentivector or the same vector that contained either wild-type or mutant CCN2 3′-UTR, when co-cultured for 24 hours with D1 HSCs that had been treated for 24 hours with or without 10 μmol/L of GW4869, an exosome inhibitor that blocks neutral sphingomyelinase 2, which is required for the biosynthesis of ceramide on which exosome production is dependent.38, 39
Statistical Analysis
All experiments were performed at least three times with triplicate measurements, with data expressed as means ± SEM. The data from RT-qPCR or luciferase activity assays were analyzed by the t-test using Sigma plot software version 12.5 (SPSS Inc., Chicago, IL), and P < 0.05 was considered statistically significant.
Results
MiR-199a-5p Expression in Experimental Liver Fibrosis
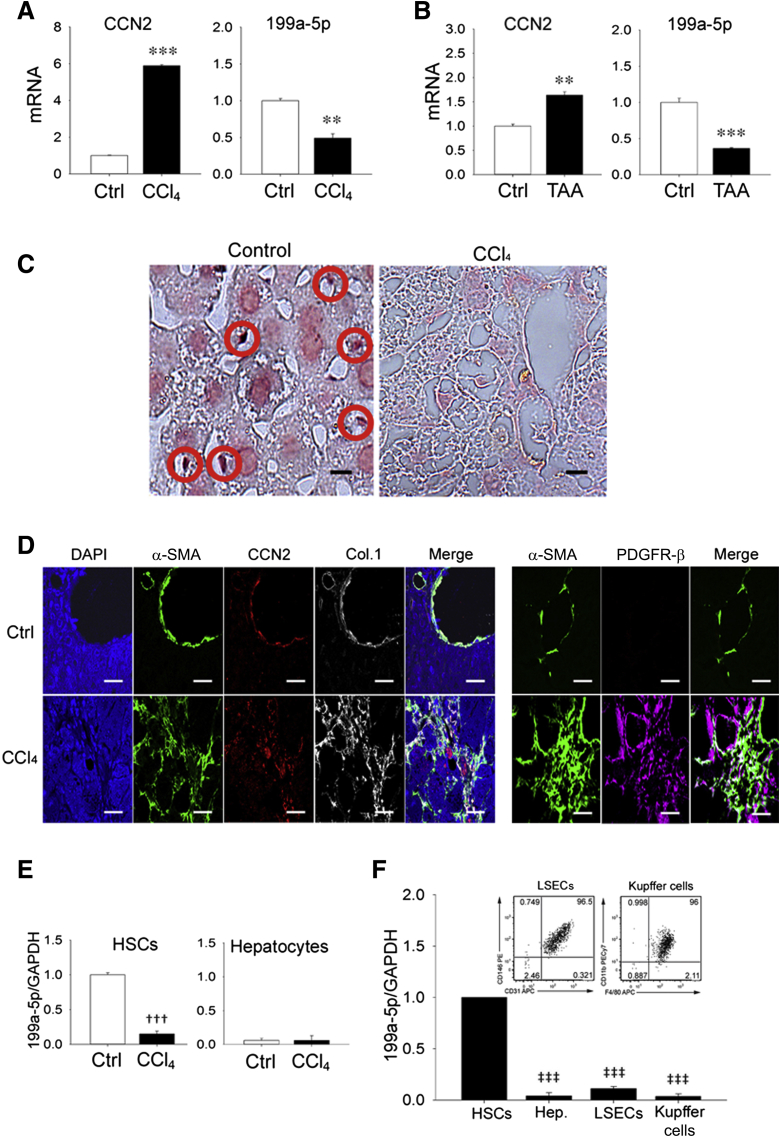
Analysis of total hepatic RNA revealed that CCN2 was highly expressed in mice with CCl4-induced liver fibrosis but not in oil-treated controls (Figure 2A), consistent with many previous observations.32 Because we determined that the 3′UTR of CCN2 contains a potential binding site for miR-199a-5p that is conserved in mouse and human (Figure 1), we analyzed the same tissues for miR-199a-5p expression and found that its levels were reduced in fibrotic liver compared with healthy controls and thus inversely proportional to those of CCN2 (Figure 2A). To ensure that these findings were consistent in a different experimental model of hepatic fibrosis, RNA was analyzed in livers from mice with thioacetic acid–induced fibrosis with the result that CCN2 or miR-199a-5p were, respectively, up- or down-regulated compared with control animals (Figure 2B). In situ hybridization suggested that miR-199a-5p in normal tissue was predominantly localized to small nonparenchymal cells in which expression was lost during CCl4 injury (Figure 2C) and that were identified as presumptive HSCs based on their expression of CCN2, α-SMA, collagen 1(α1), or platelet-derived growth factor-β in experimental fibrosis in vivo (Figure 2D). To verify the cellular distribution of miR-199a-5p, freshly isolated HSCs or hepatocytes from control or fibrotic livers were individually analyzed for their respective miR-199a-5p levels. As shown in Figure 2E, HSCs from normal livers contained levels of miR-199a-5p that were much higher than those in hepatocytes; furthermore, miR-199a-5p levels were substantially reduced in HSCs from fibrotic livers, unlike those in hepatocytes, which remained unchanged from their low levels in control livers. The principal miR-199a-5p–producing cells in normal liver were substantiated using density gradient centrifugation and fluorescence-activated cell sorting, which revealed that >95% of miR-199a-5p expression was attributable to HSCs, with only minimal contributions from hepatocytes, Kupffer cells, or LSECs (Figure 2F). These results indicate that miR-199a-5p expression is a principal characteristic of quiescent HSCs but that its expression is highly diminished during HSC activation in vivo and is reciprocal to the presence of CCN2, α-SMA, and collagen 1(α1) in HSCs in vivo (Figure 2D) or to their respective RNA levels in HSCs isolated from control versus fibrotic livers.10, 23
Figure 2.
Suppression of miR-199a-5p during liver fibrosis. Hepatic expression of connective tissue growth factor (CCN2) mRNA or miR-199a-5p assessed by RT-qPCR and normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) after 5-week administration of CCl4 in Swiss Webster mice (A) or thioacetic acid (TAA) in FVB mice (B) compared with the respective carrier controls. C:In situ hybridization for miR-199a-5p with positive nonparenchymal cells [presumptive quiescent hepatic stellate cells (HSCs)] indicated with red circles [staining with scrambled probe was negative (not shown)]. D: Immunohistochemical detection of α-smooth muscle actin (α-SMA), CCN2, collagen 1(α1) (Col.1), or platelet-derived growth factor receptor (PDGFR)-β in livers from CCl4- or oil-treated mice. E: RT-qPCR analysis of miR-199a-5p expression in primary HSCs or primary hepatocytes cultured for 24 hours in medium that contained 10% fetal bovine serum after being isolated from control mice or mice that were treated for 5 weeks with CCl4 to induce liver fibrosis. F: Relative miR-199a-5p expression in HSCs, hepatocytes, luminal sinusoidal endothelial cells (LSECs), or Kupffer cells isolated from healthy mouse liver. The insets show fluorescence-activated cell sorting for LSECs or Kupffer cells. Data are typical of 3 independent experiments measured in triplicate (C and D). n = 3 independent experiments performed in triplicate (A, B, E and F). Scale bars: 20 μm (C); 50 μm (D). ∗∗P < 0.01, ∗∗∗P < 0.001 versus carrier control; †††P < 0.001 versus oil-treated (control) HSCs; ‡‡‡P < 0.001 versus HSCs. Ctrl, control; Hep., heptocytes.
Functional Association Between miR-199a-5p and CCN2 in HSCs
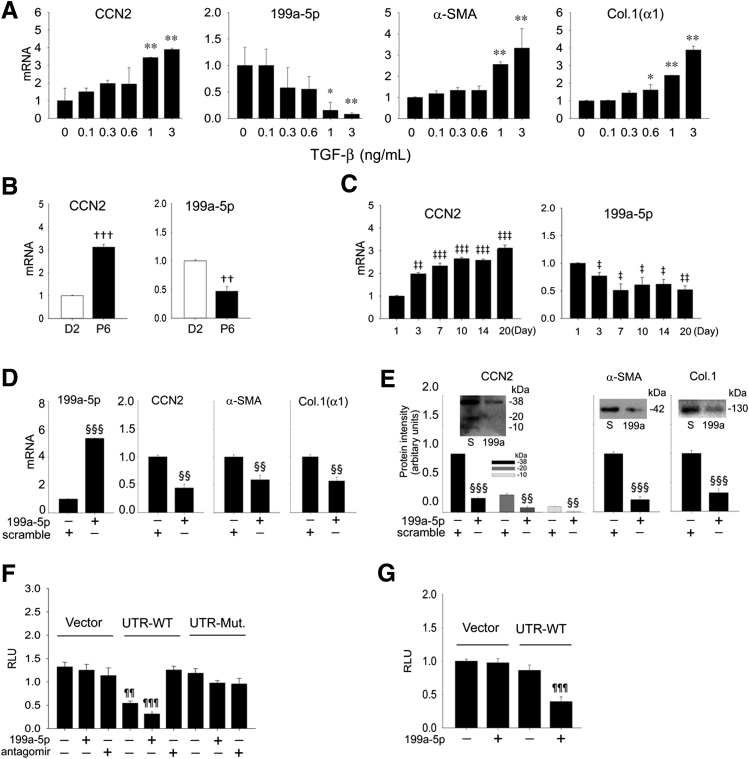
Primary HSCs from healthy mice exhibited a dose-dependent stimulation of CCN2, α-SMA, or collagen 1(α1) expression and a corresponding dose-dependent inhibition of miR-199a-5p expression after treating D2 HSCs for 48 hours with 0 to 3 ng/mL of TGF-β1 (Figure 3A). The reciprocal association between miR-199a-5p and CCN2 was also apparent on comparison of their respective concentrations in primary mouse HSCs on D2 of culture (high expression of miR-199a-5p and low expression of CCN2) to those in highly activated mouse P6 HSCs (low expression of miR-199a-5p and high expression of CCN2) (Figure 3B). Similarly, whereas CCN2 expression was progressively increased in the first 20 days of primary HSCs culture (during which time the cells undergo autonomous activation), the expression of miR-199a-5p progressively decreased during the same period (Figure 3C). To determine the functional properties of miR-199a-5p in HSCs, fully activated P6 HSCs were transfected with a miR-199a-5p mimic; the resulting sixfold increase in miR-199a-5p expression was associated with a significant attenuation of CCN2, α-SMA, or collagen 1(α1) mRNA (Figure 3D) or protein (Figure 3E) from their usual high levels. Because the CCN2 3′-UTR contains a potential miR-199a-5p binding site (Figure 1), we next determined whether this accounted for the observed reciprocity of mir-199a-5p and CCN2. This was accomplished using early-culture (D4) mouse HSCs to analyze the luciferase reporter activities of a parental lentivector versus the same vector that harbored either a wild-type CCN2 3′-UTR or a mutant form of the 3′-UTR that contained a substitution mutation in the presumptive miR-199a-5p binding site (Figure 1). Compared with cells transfected with the parental vector, luciferase activity of the vector that contained the full-length wild-type CCN2 3′-UTR was diminished by approximately 40% (Figure 3F). This was attributable to the presence of endogenous mir-199a-5p in the D4 cells because suppression of CCN2 3′-UTR activity did not occur when the cells were i) transfected with a mutant CCN2 3′-UTR lacking the miR-199a-5p site (Figure 3F), ii) co-transfected with wild type CCN2 3′-UTR and a miR-199a-5p antagomir (the latter of which had no effect on the parental vector or the mutant UTR), or iii) transfected with a miR-199a-5p mimic, which caused robust inhibition of wild-type, but not mutant, CCN2 3′-UTR (Figure 3F). These results indicate that CCN2 transcriptional activity in HSCs was suppressed by the direct binding of miR-199a-5p to its predicted binding site in the CCN2 3′-UTR. As miR-199a-5p is conserved between mouse and human and the human CCN2 3′-UTR also contains a predicted miR-199a-5p binding site (Figure 1), we performed similar studies in the human LX-2 HSC line. Because these cells are fully activated and express high CCN2 levels, they were co-transfected with a miR-199a-5p mimic with the result that luciferase activity of the CCN2 3′-UTR vector was suppressed, whereas the parental vector was unaffected (Figure 3G). Thus, CCN2 3′-UTR activity is miR-199a-5p-dependent in human HSCs and mouse HSCs.
Figure 3.
Reciprocal expression of miR-199a-5p and connective tissue growth factor (CCN2) mRNA during hepatic stellate cell (HSC) activation in vitro. Expression of miR-199a-5p and fibrogenic genes [CCN2, α-smooth muscle actin (α-SMA), collagen 1(α1) (Col.[α1])] in day 2 (D2) primary mouse HSCs treated for 48 hours with 0 to 3 ng/mL transforming growth factor (TGF)-β1 in serum-free medium (A), D2 or passage 6 (P6) mouse HSCs cultured in medium that contained 1% fetal bovine serum (FBS) for 24 hours (B), or primary HSCs cultured for up to 20 days in culture after isolation, with RNA isolation performed after a 24-hour incubation in medium that contained 1% FBS (C). D: Effect on miR-199a-5p levels or mRNA expression of fibrogenic gene expression after treating P6 HSCs (maintained in medium containing 10% FBS) for 24 hours with 100 nmol/L miR-199a-5p mimic or a scramble sequence (S). E: Protein production in the cells treated in (D) assessed by Western blot and densitometric scanning of immunoreactive bands. The various isoforms of CCN2 represent the full-length CCN2 protein (38 kDa) and several low-mass (10 to 20 kDa) stable proteolytic products that retain bioactivity and have been extensively documented in many systems, including HSCs.32, 40F: D4 mouse HSCs (maintained in medium that contained 10% FBS) were transfected with parental Fire-Ctx (vector) or Fire-Ctx that contained either the wild-type (WT) mouse CCN2 3′ untranslated region (UTR) (UTR-WT) or a mutant form of mouse CCN2 3′-UTR lacking the predicted miR-199a-5p binding site (UTR-Mut.) (Figure 1). Some cells were co-transfected with a miR-199a-5p mimic or a miR-199a-5p antagomir. Activities of the dual luciferase reporters were measured 36 hours after transfection. Firefly luciferase activity in cell lysates was normalized to that of Renilla luciferase and is expressed in relative luminescence units (RLU). G: LX-2 cells were maintained in medium that contained 10% FBS and transfected for 24 hours with parental or CCN2 3′-UTR-WT lentivectors, with or without co-transfection with miR-199a-5p mimic; data are expressed in RLU. ∗P < 0.05, ∗∗P < 0.01 versus 0 ng/mL of TGF-β1; ††P < 0.01, †††P < 0.001 versus D2; ‡P < 0.05, ‡‡P < 0.01, and ‡‡‡P < 0.001 versus D1; §§P < 0.01, §§§P < 0.001 versus scramble; and ¶¶P < 0.01, ¶¶¶P < 0.001 versus control.
Characterization of Exosomal miR-199a-5p
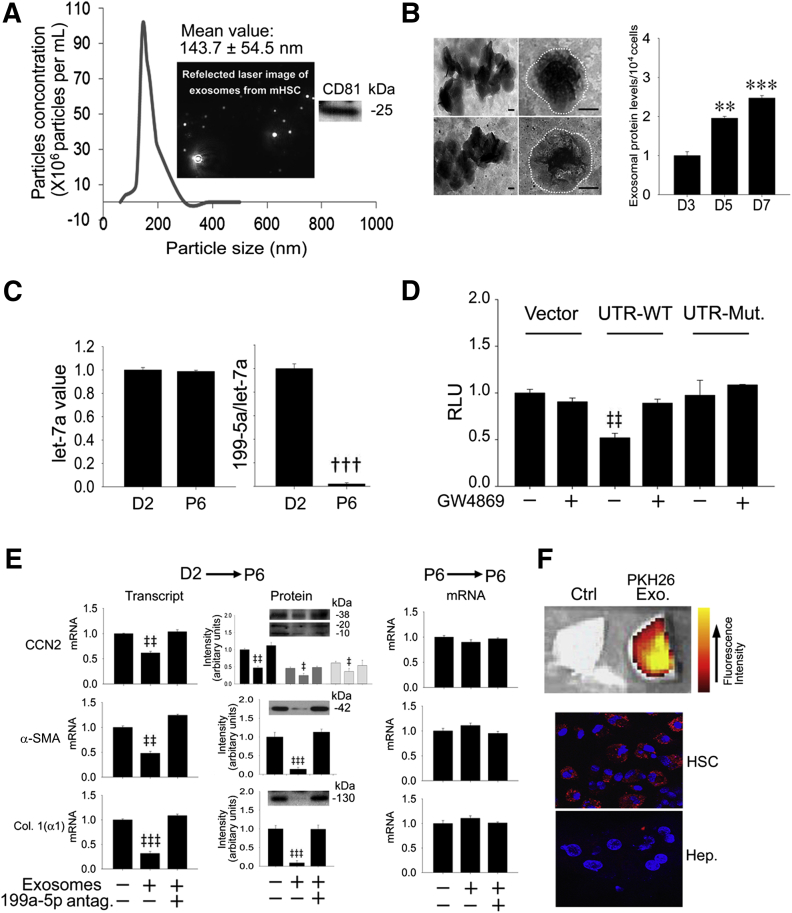
Because we have reported previously that exosomes from activated or quiescent HSCs are involved in intercellular shuttling of, respectively, CCN234 or miR-214 and Twist1,10, 23 it was of interest to determine whether miR-199a-5p is also an exosomal constituent. We have previously reported that HSC-derived exosomes are bimembrane vesicles (assessed by transmission electron microscopy) that have a charge of −26 mV and a mean diameter of approximately 140 nm (assessed by zeta potential analysis and dynamic light scattering, respectively) and are positive for the exosome markers CD9 and flotillin (assessed by Western blot).10 To reinforce these findings, we undertook additional characterization of HSC-derived exosomes and found that they had a mean diameter of approximately 143 nm assessed by nanoparticle tracking analysis and were positive by Western blot analysis for CD81 (Figure 4A), which is a more specific marker for exosomes than CD9.41 Transmission electron microscopy revealed comparable size and morphologic features of exosomes from D2 versus P6 HSCs and an increased rate of exosome production was documented during progressive HSC activation (Figure 4B). As assessed by RT-qPCR—for which exosomal let-7a was used as a housekeeping gene because its levels were unchanged during HSC activation (Figure 4C)—miR-199a-5p levels were substantially reduced in exosomes purified from activated P6 HSCs compared with those purified from quiescent D2 cells (Figure 4C), thus reflecting their differential expression at the cellular level (Figure 3B). To establish whether miR-199a-5p–containing exosomes from quiescent HSCs regulate the CCN2 gene in activated HSCs, a co-culture approach10, 23 was used, which allowed this question to be addressed for exosomes at their endogenous levels of production by donor cells and subsequent action in recipient cells. Exosomal regulation of CCN2 was revealed by assessment of luciferase activity in P6 HSCs transfected with wild-type or mutant CCN2 3′-UTR luciferase vectors (Figure 1) or vector alone when co-cultured with D1 HSCs. Luciferase activity in the activated recipient HSCs was significantly suppressed for the wild-type CCN2 3′-UTR compared with the vector control when they were co-cultured with D1 HSCs for 24 hours (Figure 4D). This suppressed activity was not apparent when D0 HSCs were treated with GW4869 or when activated recipient HSCs were transfected with the mutated CCN2 3′-UTR (Figure 4D). However, treatment of D0 HSCs with GW4869 had no effect in P6 HSCs on the activity of either the parental vector or the mutant CCN2 3′-UTR. These data thus indicate that exosomes mediate the intercellular transfer of miR-199a-5p from quiescent HSCs to activated HSCs in which CCN2 3′-UTR activity is then down-regulated. To verify that this interaction resulted in alterations in gene expression, RT-qPCR analysis was performed on recipient P6 HSCs after the addition for 24 hours of purified exosomes from serum-starved D2 cells. Expression in P6 HSCs of CCN2, α-SMA, or collagen 1(α1) mRNA (assessed by RT-qPCR) or production of their respective proteins (assessed by Western blot) was suppressed by exosomes from D2 HSCs, but this was reversed by transfection of the recipient cells with a miR-199a-5p antagomir (Figure 4E). In contrast, gene expression in recipient P6 HSCs was not affected by addition of exosomes purified from donor P6 HSCs in the presence or absence of the miR-199a-5p antagomir (Figure 4E). Thus exosomes from quiescent HSCs regulate CCN2-dependent fibrogenic signaling in activated HSCs through a mechanism that involves the intercellular transfer and action of exosomal miR-199a-5p, including its targeting of the CCN2 3′-UTR and subsequent regulation of CCN2 expression and its downstream mediators α-SMA and collagen 1(α1). Finally, to verify that exosomes from quiescent HSCs bind to activated HSCs in vivo, exosomes purified from D2 HSCs in vitro and labeled with PKH26 dye were administered i.v. to mice that had been treated for 2 weeks with CCl4. After 4 hours, exosomes had a predominant hepatic localization, with no detectable presence in other organ systems (Figure 4F) (data not shown). Examination of isolated liver cells revealed that the exosomes had bound to the HSC population, with essentially no binding to hepatocytes (Figure 4F). These data thus confirmed that activated HSCs are a physiologic target of exosomes from quiescent HSCs.
Figure 4.
Exosomal miR-199a-5p dampens fibrogenic signaling in activated hepatic stellate cells (HSCs). A: Nanoparticle tracking analysis of exosomes isolated from the conditioned medium of passage 6 (P6) mouse HSCs that were cultured for 48 hours in serum-free medium. The insets show a typical video still obtained during data collection (left inset) and a Western blot for CD81 of the purified exosome preparation (right inset). B: Transmission electron microscopy of exosomes from quiescent [day 2 (D2)] or activated (P6) HSCs collected from the cells after 48-hour culture in, respectively, medium that contained 1% fetal bovine serum (FBS) (exosome-depleted) or serum-free medium (left column). The outer margins of the exosomes are shown by the white dotted line. Production of exosomes during HSC activation in vitro; cells were maintained for 48 hours in 1% FBS (exosome-depleted) before harvesting the exosomes (right column). C: RT-PCR analysis of miR-199a-5p levels in exosomes purified from D2 HSCs [cultured for 48 hours in medium that contained 1% FBS (exosome depleted)] or P6 HSCs (cultured for 48 hours in serum-free medium), with data normalized to let-7a expression as previously validated.23D: Using the co-culture system previously described,10 recipient P6 mouse HSCs (maintained in medium that contained 10% FBS) were transfected with Fire-Ctx plasmids containing either wild-type or mutant connective tissue growth factor (CCN2) 3′ untranslated region (3′-UTR) (Figure 1) and co-cultured for 24 hours with D1 mouse HSCs, some of which had been pretreated with 10 μmol/L GW4869. The activity of firefly luciferase in lysates of recipient cells, normalized to that of Renilla luciferase, is shown 24 hours after the central divider had been removed from the microwells, as described.10E: Fibrogenic gene expression and protein production in P6 HSCs, with or without transfection of the cells with a miR-199a-5p antagomir for 24 hours in medium that contained 10% FBS, followed by 24-hour serum-starvation and then treatment under serum-free conditions with exosomes (8 μg/mL) purified from D2 HSCs [cultured for 48 hours in medium that contained 1% FBS (exosome depleted)] or P6 HSCs (cultured for 48 hours in serum-free medium). CCN2 isoforms are discussed in Figure 3E. F: PKH26 Exo is hepatic uptake of PKH26-stained exosomes [collected from D2 HSCs that were maintained for 2 days in medium that contained exosome-depleted serum (1%)] 4 hours after i.v. injection in CCl4-treated mice (175 μL in 1325 μL of corn oil/kg, 3 times per week for 2 weeks) (top image). Control (Ctrl) is liver from a CCl4-treated mouse that was not injected with exosomes. Imaging of DAPI-stained isolated liver cells (blue) revealing uptake in vivo of PKH26-stained exosomes (red) by activated HSCs but not hepatocytes (middle and bottom image). Data are typical of 3 independent experiments performed in triplicate. ∗∗P < 0.01, ∗∗∗P < 0.001 versus D3; †††P < 0.001 versus D2; ‡P < 0.05, ‡‡P 0.01, and ‡‡‡P < 0.001 versus Ctrl. Scale bars = 100 nm (B). Col. 1(α1), collagen 1(α1); Hep., hepatocytes; Mut, mutant; RLU, relative luminescence units; WT, wild-type.
Discussion
MiR-199a-3p and miR-199a-5p are processed from, respectively, the 5′ and 3′ arms of the same precursor, miR-199a-2, which is encoded together with miR-214 by the antisense strand of the DNM3 gene, termed DNM3 opposite strand (DNM3os).42, 43 MiR-199a-2 has high sequence identity among zebrafish, puffer fish, mice, and humans, suggesting that its functions have been evolutionarily conserved.44 DNM3os is required for normal growth, development, and survival as indicated by the finding that DNM3os-null mice die within a month of birth because of skeletal abnormalities that include osteopenia and defects in the skull and spine.45 MiR-199a exhibits highly variable patterns of expression during development, differentiation, and in various diseases, and its actions are also very variable even with respect to the same gene target because such genes have cell- and context-specific functions.42
Our data indicate that miR-199a-5p suppresses CCN2 in human or mouse HSCs via its direct targeting of the CCN2 3′-UTR and that diminished expression of miR-199a-5p in activated HSCs allows CCN2 and its downstream targets to be expressed. Our data are supported by the decreased expression of miR-199a-5p in keloid tissue compared with normal human skin46 and the very recent finding that miR-199a-5p functions as a tumor suppressor in follicular cancer by suppressing CCN2.47 Furthermore, the concomitant inhibition of expression of α-SMA and/or collagen 1(α1) is consistent with their CCN2 dependency in HSCs33 but may also result from the regulation of other miR-199a-5p targets. Although our mechanistic data are consistent with decreased miR-199a-5p accounting for increased CCN2 expression during HSC activation, other studies have found miR-199a-5p expression to be increased in fibrotic livers or in activated HSCs.48, 49 However, variable expression during liver fibrosis has been reported for miR-214, the other miR encoded by DNM3os, which independently targets the 3′-UTR of CCN2 and inhibits expression of CCN2 and other fibrosis- or inflammation-related molecules in HSCs.10, 23, 50 Although further experiments are needed to fully clarify the sites of expression and action of the miR-199a-2/214 cluster during progression of chronic liver disease, a principal mechanism regulating its expression in many biological systems involves the basic helix-loop-helix transcription factor, Twist1.45, 51, 52, 53, 54 We recently reported that, through its binding of the E-box response element in the upstream region, Twist1 suppresses CCN2 expression in quiescent HSCs by driving, at least in part, miR-214 promoter activity and expression.23 Because Twist1 is decreasingly expressed and decreasingly interacts with DNM3os during HSC activation,23 this causes activation-associated decreases in expression of miR-21423 or miR-199a-5p (these results), thereby allowing expression of their common CCN2 target and its downstream fibrogenic mediators (α-SMA, collagen). Future studies will investigate other control mechanisms of the miR-199a-5p/miR-214-CCN2 axis in activated HSCs, including transcriptional regulation of miR-199a-2 by Egr-1 and STAT3 transcription factors55, 56 and methylation or histone modifications of miR-199a-2 or miR-199a-1,57, 58, 59 the latter of which is a separate precursor of miR-199a-5p that is not regulated by Twist1 or Egr-1.42
Exosomes are membranous nanovesicles that arise by inward budding of multivesicular bodies, which then fuse with the plasma membrane and cause exosomes to be released from the cell. Exosomes contain a complex mixture of miRs, mRNAs, and proteins, and their ability to traverse the intercellular space and to be taken up by neighboring cells is becoming increasingly recognized as an important mechanism for delivering genetic, epigenetic, or proteomic information from donor cells and effecting functional modifications in recipient cells.11, 19, 60, 61 Whereas most previous studies have focused on these mechanisms as they relate to exosomal pathways in cancer and immune cells, including HCCs,15, 17, 18 emerging evidence from this and other laboratories has revealed that fibrogenic cells are also regulated by exosomal networks.10, 22, 23, 31, 34 In addition to revealing that miR-199a-5p targets the CCN2 3′-UTR in the same cells as which it is produced, the studies reported here indicate that miR-199a-5p is released from HSCs in exosomes and that exosomal miR-199a-5p concentrations reflect those of their producer cells, being expressed at high levels in exosomes from quiescent HSCs and at low levels in exosomes from activated HSCs. We found that delivery of endogenous exosomal miR-199a-5p from quiescent HSCs to activated HSCs resulted in down-regulation of CCN2 3′-UTR activity and that exosomes from quiescent HSCs are preferentially targeted to the activated HSC population in vivo in CCl4-injured liver and suppress fibrogenic gene expression in activated HSCs in vitro in a miR-199a-5p–dependent manner. These data extend our previous findings in mouse and human systems revealing that quiescent HSCs shuttle exosomes to activated HSCs in which CCN2 3′-UTR activity and gene expression are attenuated by miR-214, which was delivered exosomally or transcriptionally activated in the recipient cells after exosomal delivery of Twist1.10, 23 The actions of cellular or exosomal Twist1 and the miR-199a/214 cluster in regulating CCN2 is summarized in Figure 5. We propose that exosomal dampening mechanisms help to maintain HSC quiescence and that exosomes from populations of quiescent HSCs contribute to suppression of HSC activation during localized or acute liver injury, during diminished periods of fibrogenic activity in progressive liver diseases, such as nonalcoholic steatohepatitis, and/or during therapy-induced fibrosis regression. Because exosomes from activated HSCs can deliver CCN2 mRNA to quiescent HSCs, thus increasing their fibrogenic potential,34 it is likely that differentially activated HSCs engage in a functional exosome exchange, which allows their phenotypic status to be locally communicated and for the recipient cells to respond accordingly. Because it is now becoming recognized that HSC function can be stimulated by exosomes from other cell types, such as injured hepatocytes,31 complex exosome communication networks likely exist in the liver that serve to choreograph and fine-tune the response to injury and maintain homeostasis under normal healthy conditions. In the future, analytical tools and animal models need to be developed so that the precise interplay of these exosome pathways can be elucidated in vivo and to determine whether components of the exosomal payload from quiescent HSCs (eg, Twist1, miR-199a-2, miR-214) can be used as therapeutic agents to dampen fibrogenic signaling in HSCs.
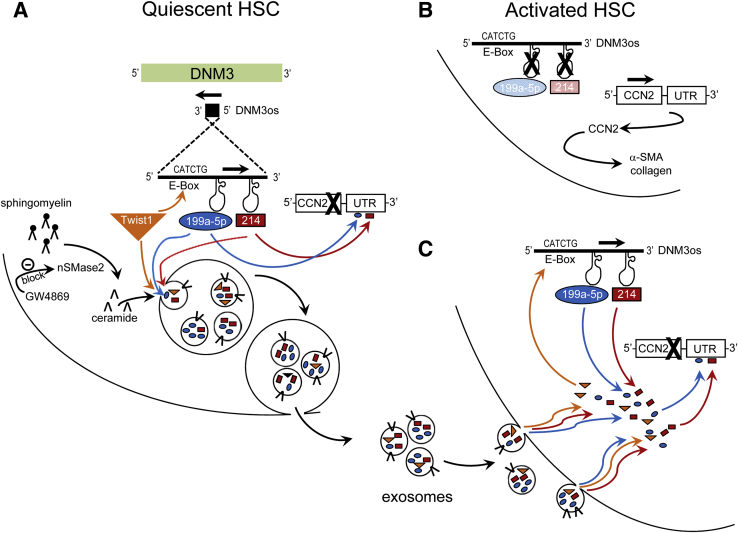
Figure 5.
Regulation of connective tissue growth factor (CCN2) 3′ untranslated region (3′-UTR) in hepatic stellate cells (HSCs) by endogenous or exosomal components of the Twist1-miR-199a-5p/miR-214 axis. On the basis of the data presented here and in our previous publications,10, 23 the following mechanism has been identified. A: In quiescent HSCs, CCN2 expression is suppressed, at least in part, by direct binding of miR-199a-5p or miR-214 to their respective binding sites in the 3′-UTR region of CCN2. MiR-199a-5p and miR-214 are encoded by DNM3 opposite strand (DNM3os) and are maintained at high levels in quiescent HSCs by the action of Twist1 transcription factor. In addition, Twist1, miR-199a-5p, and/or miR-214 become incorporated into internal vesicles, which are released from the cells as exosomes. Quiescent HSCs are thus donors of exosomes that are enriched in transcription factors and microRNAs that inhibit fibrogenic gene expression. The early stages of vesicle biogenesis in the cells are dependent on neutral sphingomyelinase 2 (nSMase2), which generates ceramide from sphingomyelin and can be blocked by GW4869. B: Activated HSCs contain low levels of Twist1, miR-199a-5p, or miR-214, resulting in high CCN2 3′-UTR activity and high levels of CCN2 expression. Through their induction downstream of CCN2 (and/or induction by other miR-199a-2/214–sensitive targets), α-smooth muscle actin (α-SMA), and collagen 1(α1) levels are also increased. C: The activated HSC phenotype is reversed on binding to and uptake by the cells of exosomes from quiescent HSCs. CCN2 expression becomes suppressed in recipient activated HSCs because of the action of exosomal miR-199a-5p and/or miR-214, which directly bind to the CCN2 3′-UTR, and exosomal Twist1, which transcriptionally drives production of endogenous miR-199a-5p and/or miR-214 from DNM3os. Exosomes also play a role in delivery of profibrogenic molecules to quiescent or partially activated HSCs from injured hepatocytes31 or fully activated HSCs22 (not illustrated).
In summary, miR-199a-5p binds to its target in the 3′-UTR of CCN2, resulting in reduced expression of CCN2. This suppressive pathway is constitutively present in quiescent HSCs but is also elaborated in activated HSCs after horizontal transfer of miR-199a-5p in exosomes from quiescent HSCs.
Acknowledgments
We thank Scott Friedman for kindly providing LX-2 cells.
D.R.B. is the guarantor of this work and, as such, had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
L.C. conducted study design, data acquisition, data analysis and manuscript preparation; R.C. conducted data acquisition and data analysis; V.M.V. conducted study design, data acquisition and data analysis; D.R.B. conducted funding acquisition, study design, data analysis, and manuscript preparation.
Footnotes
Supported by NIH grants R01AA021276 and R21AA023626 (D.R.B.).
L.C. and R.C. contributed equally to this work.
Disclosures: D.R.B. is a stockholder of FibroGen Inc.
References
- 1.Friedman S.L. Hepatic fibrosis – overview. Toxicology. 2008;254:120–129. doi: 10.1016/j.tox.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 2.Friedman S.L. Mechanisms of hepatic fibrogenesis. Gastroenterology. 2008;134:1655–1669. doi: 10.1053/j.gastro.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoon Y.J., Friedman S.L., Lee Y.A. Antifibrotic therapies: where are we now? Semin Liver Dis. 2016;36:87–98. doi: 10.1055/s-0036-1571295. [DOI] [PubMed] [Google Scholar]
- 4.Wang P., Koyama Y., Liu X., Xu J., Ma H.Y., Liang S., Kim I.H., Brenner D.A., Kisseleva T. Promising therapy candidates for liver fibrosis. Front Physiol. 2016;7:47. doi: 10.3389/fphys.2016.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedman S.L. Hepatic fibrosis: emerging therapies. Dig Dis. 2015;33:504–507. doi: 10.1159/000374098. [DOI] [PubMed] [Google Scholar]
- 6.Lee Y.A., Wallace M.C., Friedman S.L. Pathobiology of liver fibrosis: a translational success story. Gut. 2015;64:830–841. doi: 10.1136/gutjnl-2014-306842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu X.J., Hong Q., Wang Z., Yu Y.Y., Zou X., Xu L.H. MicroRNA21 promotes interstitial fibrosis via targeting DDAH1: a potential role in renal fibrosis. Mol Cell Biochem. 2016;411:181–189. doi: 10.1007/s11010-015-2580-2. [DOI] [PubMed] [Google Scholar]
- 8.Kisseleva T., Brenner D.A. Inactivation of myofibroblasts during regression of liver fibrosis. Cell Cycle. 2013;12:381–382. doi: 10.4161/cc.23549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puche J.E., Saiman Y., Friedman S.L. Hepatic stellate cells and liver fibrosis. Compr Physiol. 2013;3:1473–1492. doi: 10.1002/cphy.c120035. [DOI] [PubMed] [Google Scholar]
- 10.Chen L., Charrier A., Zhou Y., Chen R., Yu B., Agarwal K., Tsukamoto H., Lee L.J., Paulaitis M.E., Brigstock D.R. Epigenetic regulation of connective tissue growth factor by microRNA-214 delivery in exosomes from mouse or human hepatic stellate cells. Hepatology. 2014;59:1118–1129. doi: 10.1002/hep.26768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thery C. Exosomes: secreted vesicles and intercellular communications. F1000 Biol Rep. 2011;3:15. doi: 10.3410/B3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thery C., Ostrowski M., Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 13.Thery C., Zitvogel L., Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 14.Calzolari A., Raggi C., Deaglio S., Sposi N.M., Stafsnes M., Fecchi K., Parolini I., Malavasi F., Peschle C., Sargiacomo M., Testa U. TfR2 localizes in lipid raft domains and is released in exosomes to activate signal transduction along the MAPK pathway. J Cell Sci. 2006;119:4486–4498. doi: 10.1242/jcs.03228. [DOI] [PubMed] [Google Scholar]
- 15.Kogure T., Lin W.L., Yan I.K., Braconi C., Patel T. Intercellular nanovesicle-mediated microRNA transfer: a mechanism of environmental modulation of hepatocellular cancer cell growth. Hepatology. 2011;54:1237–1248. doi: 10.1002/hep.24504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Basu S., Bhattacharyya S.N. Insulin-like growth factor-1 prevents miR-122 production in neighbouring cells to curtail its intercellular transfer to ensure proliferation of human hepatoma cells. Nucleic Acids Res. 2014;42:7170–7185. doi: 10.1093/nar/gku346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi K., Yan I.K., Kogure T., Haga H., Patel T. Extracellular vesicle-mediated transfer of long non-coding RNA ROR modulates chemosensitivity in human hepatocellular cancer. FEBS Open Bio. 2014;4:458–467. doi: 10.1016/j.fob.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He M., Qin H., Poon T.C., Sze S.C., Ding X., Co N.N., Ngai S.M., Chan T.F., Wong N. Hepatocellular carcinoma-derived exosomes promote motility of immortalized hepatocyte through transfer of oncogenic proteins and RNAs. Carcinogenesis. 2015;36:1008–1018. doi: 10.1093/carcin/bgv081. [DOI] [PubMed] [Google Scholar]
- 19.Ramachandran S., Palanisamy V. Horizontal transfer of RNAs: exosomes as mediators of intercellular communication. Wiley Interdiscip Rev RNA. 2012;3:286–293. doi: 10.1002/wrna.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang A., Dong J., Li S., Wang C., Ding H., Li H., Su X., Ge X., Sun L., Bai C., Shen X., Fang T., Li J., Shao N. Exosomal transfer of vasorin expressed in hepatocellular carcinoma cells promotes migration of human umbilical vein endothelial cells. Int J Biol Sci. 2015;11:961–969. doi: 10.7150/ijbs.11943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conigliaro A., Costa V., Lo Dico A., Saieva L., Buccheri S., Dieli F., Manno M., Raccosta S., Mancone C., Tripodi M., De Leo G., Alessandro R. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol Cancer. 2015;14:155. doi: 10.1186/s12943-015-0426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Charrier A., Chen R., Chen L., Kemper S., Hattori T., Takigawa M., Brigstock D.R. Exosomes mediate intercellular transfer of pro-fibrogenic connective tissue growth factor (CCN2) between hepatic stellate cells, the principal fibrotic cells in the liver. Surgery. 2014;156:548–555. doi: 10.1016/j.surg.2014.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen L., Chen R., Kemper S., Charrier A., Brigstock D.R. Suppression of fibrogenic signaling in hepatic stellate cells by Twist1-dependent microRNA-214 expression: role of exosomes in horizontal transfer of Twist1. Am J Physiol Gastrointest Liver Physiol. 2015;309:G491–G499. doi: 10.1152/ajpgi.00140.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Csak T., Bala S., Lippai D., Satishchandran A., Catalano D., Kodys K., Szabo G. microRNA-122 regulates hypoxia-inducible factor-1 and vimentin in hepatocytes and correlates with fibrosis in diet-induced steatohepatitis. Liver Int. 2015;35:532–541. doi: 10.1111/liv.12633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dolganiuc A., Petrasek J., Kodys K., Catalano D., Mandrekar P., Velayudham A., Szabo G. MicroRNA expression profile in Lieber-DeCarli diet-induced alcoholic and methionine choline deficient diet-induced nonalcoholic steatohepatitis models in mice. Alcohol Clin Exp Res. 2009;33:1704–1710. doi: 10.1111/j.1530-0277.2009.01007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gyorgy B., Szabo T.G., Pasztoi M., Pal Z., Misjak P., Aradi B., Laszlo V., Pallinger E., Pap E., Kittel A., Nagy G., Falus A., Buzas E.I. Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell Mol Life Sci. 2011;68:2667–2688. doi: 10.1007/s00018-011-0689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Momen-Heravi F., Saha B., Kodys K., Catalano D., Satishchandran A., Szabo G. Increased number of circulating exosomes and their microRNA cargos are potential novel biomarkers in alcoholic hepatitis. J Transl Med. 2015;13:261. doi: 10.1186/s12967-015-0623-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirsova P., Ibrahim S.H., Krishnan A., Verma V.K., Bronk S.F., Werneburg N.W., Charlton M.R., Shah V.H., Malhi H., Gores G.J. Lipid-induced signaling causes release of inflammatory extracellular vesicles from hepatocytes. Gastroenterology. 2016;150:956–967. doi: 10.1053/j.gastro.2015.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masyuk A.I., Huang B.Q., Ward C.J., Gradilone S.A., Banales J.M., Masyuk T.V., Radtke B., Splinter P.L., LaRusso N.F. Biliary exosomes influence cholangiocyte regulatory mechanisms and proliferation through interaction with primary cilia. Am J Physiol Gastrointest Liver Physiol. 2010;299:G990–G999. doi: 10.1152/ajpgi.00093.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masyuk A.I., Masyuk T.V., Larusso N.F. Exosomes in the pathogenesis, diagnostics and therapeutics of liver diseases. J Hepatol. 2013;59:621–625. doi: 10.1016/j.jhep.2013.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Povero D., Panera N., Eguchi A., Johnson C.D., Papouchado B.G., de Araujo Horcel L., Pinatel E.M., Alisi A., Nobili V., Feldstein A.E. Lipid-induced hepatocyte-derived extracellular vesicles regulate hepatic stellate cell via microRNAs targeting PPAR-gamma. Cell Mol Gastroenterol Hepatol. 2015;1:646–663e4. doi: 10.1016/j.jcmgh.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang G., Brigstock D.R. Regulation of hepatic stellate cells by connective tissue growth factor. Front Biosci. 2012;17:2495–2507. doi: 10.2741/4067. [DOI] [PubMed] [Google Scholar]
- 33.Chen L., Charrier A.L., Leask A., French S.W., Brigstock D.R. Ethanol-stimulated differentiated functions of human or mouse hepatic stellate cells are mediated by connective tissue growth factor. J Hepatol. 2011;55:399–406. doi: 10.1016/j.jhep.2010.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Charrier A., Chen R., Chen L., Kemper S., Hattori T., Takigawa M., Brigstock D.R. Connective tissue growth factor (CCN2) and microRNA-21 are components of a positive feedback loop in pancreatic stellate cells (PSC) during chronic pancreatitis and are exported in PSC-derived exosomes. J Cell Commun Signal. 2014;8:147–156. doi: 10.1007/s12079-014-0220-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghosh A., Sil P.C. A 43-kDa protein from the leaves of the herb Cajanus indicus L. modulates chloroform induced hepatotoxicity in vitro. Drug Chem Toxicol. 2006;29:397–413. doi: 10.1080/01480540600837944. [DOI] [PubMed] [Google Scholar]
- 36.Shelke G.V., Lässer C., Gho Y.S., Lötvall J. Importance of exosome depletion protocols to eliminate functional and RNA-containing extracellular vesicles from fetal bovine serum. J Extracell Vesicles. 2014;3 doi: 10.3402/jev.v3.24783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thery C., Amigorena S., Raposo G., Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;Chapter 3:Unit 3.22. doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 38.Chairoungdua A., Smith D.L., Pochard P., Hull M., Caplan M.J. Exosome release of beta-catenin: a novel mechanism that antagonizes Wnt signaling. J Cell Biol. 2010;190:1079–1091. doi: 10.1083/jcb.201002049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kosaka N., Iguchi H., Yoshioka Y., Takeshita F., Matsuki Y., Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285:17442–17452. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams E.J., Gaca M.D., Brigstock D.R., Arthur M.J., Benyon R.C. Increased expression of connective tissue growth factor in fibrotic human liver and in activated hepatic stellate cells. J Hepatol. 2000;32:754–761. doi: 10.1016/s0168-8278(00)80244-5. [DOI] [PubMed] [Google Scholar]
- 41.Kowal J., Arras G., Colombo M., Jouve M., Morath J.P., Primdal-Bengtson B., Dingli F., Loew D., Tkach M., Thery C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci U S A. 2016;113:E968–E977. doi: 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gu S., Chan W.Y. Flexible and versatile as a chameleon-sophisticated functions of microRNA-199a. Int J Mol Sci. 2012;13:8449–8466. doi: 10.3390/ijms13078449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Landgraf P., Rusu M., Sheridan R., Sewer A., Iovino N., Aravin A. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lim L.P., Glasner M.E., Yekta S., Burge C.B., Bartel D.P. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 45.Watanabe T., Sato T., Amano T., Kawamura Y., Kawamura N., Kawaguchi H., Yamashita N., Kurihara H., Nakaoka T. Dnm3os, a non-coding RNA, is required for normal growth and skeletal development in mice. Dev Dyn. 2008;237:3738–3748. doi: 10.1002/dvdy.21787. [DOI] [PubMed] [Google Scholar]
- 46.Wu Z.Y., Lu L., Liang J., Guo X.R., Zhang P.H., Luo S.J. Keloid microRNA expression analysis and the influence of miR-199a-5p on the proliferation of keloid fibroblasts. Genet Mol Res. 2014;13:2727–2738. doi: 10.4238/2014.April.14.2. [DOI] [PubMed] [Google Scholar]
- 47.Sun D., Han S., Liu C., Zhou R., Sun W., Zhang Z., Qu J. Microrna-199a-5p functions as a tumor suppressor via suppressing connective tissue growth factor (CTGF) in follicular thyroid carcinoma. Med Sci Monit. 2016;22:1210–2117. doi: 10.12659/MSM.895788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murakami Y., Toyoda H., Tanaka M., Kuroda M., Harada Y., Matsuda F., Tajima A., Kosaka N., Ochiya T., Shimotohno K. The progression of liver fibrosis is related with overexpression of the miR-199 and 200 families. PLoS One. 2011;6:e16081. doi: 10.1371/journal.pone.0016081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ogawa T., Enomoto M., Fujii H., Sekiya Y., Yoshizato K., Ikeda K., Kawada N. MicroRNA-221/222 upregulation indicates the activation of stellate cells and the progression of liver fibrosis. Gut. 2012;61:1600–1609. doi: 10.1136/gutjnl-2011-300717. [DOI] [PubMed] [Google Scholar]
- 50.Iizuka M., Ogawa T., Enomoto M., Motoyama H., Yoshizato K., Ikeda K., Kawada N. Induction of microRNA-214-5p in human and rodent liver fibrosis. Fibrogenesis Tissue Repair. 2012;5:12. doi: 10.1186/1755-1536-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee Y.B., Bantounas I., Lee D.Y., Phylactou L., Caldwell M.A., Uney J.B. Twist-1 regulates the miR-199a/214 cluster during development. Nucleic Acids Res. 2009;37:123–128. doi: 10.1093/nar/gkn920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Loebel D.A., O'Rourke M.P., Steiner K.A., Banyer J., Tam P.P. Isolation of differentially expressed genes from wild-type and Twist mutant mouse limb buds. Genesis. 2002;33:103–113. doi: 10.1002/gene.10091. [DOI] [PubMed] [Google Scholar]
- 53.Loebel D.A., Tsoi B., Wong N., Tam P.P. A conserved noncoding intronic transcript at the mouse Dnm3 locus. Genomics. 2005;85:782–789. doi: 10.1016/j.ygeno.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 54.Yin G., Chen R., Alvero A.B., Fu H.H., Holmberg J., Glackin C., Rutherford T., Mor G. TWISTing stemness, inflammation and proliferation of epithelial ovarian cancer cells through MIR199A2/214. Oncogene. 2010;29:3545–3553. doi: 10.1038/onc.2010.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sakurai K., Furukawa C., Haraguchi T., Inada K., Shiogama K., Tagawa T., Fujita S., Ueno Y., Ogata A., Ito M., Tsutsumi Y., Iba H. MicroRNAs miR-199a-5p and -3p target the Brm subunit of SWI/SNF to generate a double-negative feedback loop in a variety of human cancers. Cancer Res. 2011;71:1680–1689. doi: 10.1158/0008-5472.CAN-10-2345. [DOI] [PubMed] [Google Scholar]
- 56.Haghikia A., Missol-Kolka E., Tsikas D., Venturini L., Brundiers S., Castoldi M., Muckenthaler M.U., Eder M., Stapel B., Thum T., Petrasch-Parwez E., Drexler H., Hilfiker-Kleiner D., Scherr M. Signal transducer and activator of transcription 3-mediated regulation of miR-199a-5p links cardiomyocyte and endothelial cell function in the heart: a key role for ubiquitin-conjugating enzymes. Eur Heart J. 2011;32:1287–1297. doi: 10.1093/eurheartj/ehq369. [DOI] [PubMed] [Google Scholar]
- 57.Kim S., Lee U.J., Kim M.N., Lee E.J., Kim J.Y., Lee M.Y., Choung S., Kim Y.J., Choi Y.C. MicroRNA miR-199a* regulates the MET proto-oncogene and the downstream extracellular signal-regulated kinase 2 (ERK2) J Biol Chem. 2008;283:18158–18166. doi: 10.1074/jbc.M800186200. [DOI] [PubMed] [Google Scholar]
- 58.Cheung H.H., Lee T.L., Davis A.J., Taft D.H., Rennert O.M., Chan W.Y. Genome-wide DNA methylation profiling reveals novel epigenetically regulated genes and non-coding RNAs in human testicular cancer. Br J Cancer. 2010;102:419–427. doi: 10.1038/sj.bjc.6605505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hou J., Lin L., Zhou W., Wang Z., Ding G., Dong Q., Qin L., Wu X., Zheng Y., Yang Y., Tian W., Zhang Q., Wang C., Zhuang S.M., Zheng L., Liang A., Tao W., Cao X. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell. 2011;19:232–243. doi: 10.1016/j.ccr.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 60.Johnstone R.M. Exosomes biological significance: a concise review. Blood Cells Mol Dis. 2006;36:315–321. doi: 10.1016/j.bcmd.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 61.Vlassov A.V., Magdaleno S., Setterquist R., Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. 2012;1820:940–948. doi: 10.1016/j.bbagen.2012.03.017. [DOI] [PubMed] [Google Scholar]