Abstract
We report the case of a pediatric patient with a Salmonella enterica serotype Infantis infection. Detailed microbiological investigation revealed that this isolate carries four β-lactamase genes (blaTEM-1b variant, blaSHV-5, blaCTX-M-15, and blaCMY-2) conferring resistance to all β-lactams but imipenem. This is the first report of a Salmonella isolate with CTX-M and AmpC enzymes on the American continent, the first report of blaCMY-2 in Salmonella serotype Infantis, and the first report of blaCTX-M-15 in the genus Salmonella.
CASE REPORT
In August 2002, an 18-month-old infant male with a clinical syndrome indicating an enteritis-like infection was admitted to the Santa Teresa Hospital in Comaguaya (Honduras). The patient, whose symptoms began 3 days before admission, presented with a fever up to 40°C, mucous bloody diarrhea, nausea, and vomiting. His urine was cloudy, with a pH of 5, and laboratory analysis showed elevated numbers of red blood cells (4/field), white blood cells (6/field), and granular casts (6/field). Laboratory analysis of fecal samples revealed the presence of erythrocytes (4/field) and neutrophils (15/field). Because of limited resources available at the hospital, no microbiological techniques were performed on these samples. An empirical treatment without a confirmatory microbiological diagnosis was immediately initiated with intravenous amikacin (50 mg every 12 h) and intravenous ceftriaxone (350 mg every 12 h) and continued for 9 days. Oral anti-inflammatory drugs (acetaminophen) to control pain and fever were also administered, and the patient was rehydrated intravenously as required. The condition of the patient improved (fever decreased to normal temperature values, and vomiting and diarrhea ceased), and he was discharged from the hospital. Two weeks later, the patient returned with a relapse of the symptoms and severe malnutrition. A second empirical treatment consisting of intravenous amikacin (40 mg every 12 h) and cefoxitin (250 mg every 6 h) was administered for 9 days. At the time of the second admission, urine and stool samples were collected and sent to the Laboratorio Central de Microbiologia in Tegucigalpa, Honduras, for microbiological analysis. Both samples were confirmed, by following standard methods, as positive for the isolation of Salmonella sp. (preenrichment of the sample in tetrathionate broth, followed by plating on Taylor's xylose lysine deoxycholate agar). The laboratory values for the hemogram were 3.18 million red blood cells/mm3, 8,800 white blood cells/mm3, 2,200 lymphocytes/mm3, 165,000 platelets/mm3, a hematocrit of 21.9%, a hemoglobin concentration of 7.6 g/dl, a mean corpuscular hemoglobin level of 23.9 pg, a mean corpuscular hemoglobin concentration of 34.7 g/dl, and a mean corpuscular volume of 69 fl. All of these values were consistent with a loss of blood and reflected the presence of anemia in the patient. Screening for human immunodeficiency virus by enzyme-linked immunosorbent assay was negative. The isolates were tested for antimicrobial resistance in the Laboratorio Central de Microbiologia in Tegucigalpa, where an antibiogram was performed. The antibiogram determined that these isolates were resistant to amikacin, gentamicin, chloramphenicol, sulfamethoxazole-trimethoprim, ceftriaxone, and cefoxitin and were sensitive to ciprofloxacin. Unfortunately, these results were not available before the patient was discharged for the second time. After the second empirical treatment, the patient showed an improvement in the clinical condition and was discharged. However, microbiological eradication of the Salmonella infection was not confirmed before discharge, and attempts to locate the patient after his second stay in the hospital have failed.
Microbiology and genetic characterization.
The clinical isolate was obtained by the Veterinary Laboratories Agency in the United Kingdom as part of a project focusing on the genetic characterization of β-lactam resistance. An international collaborative effort with groups in the Universidad de La Rioja (La Rioja, Spain) and Creighton University (Omaha, Nebr.) was adopted to complete this study. The serotype of the isolate (UCM267) was identified as Infantis by following standard methods. It was screened for susceptibility to a panel of 12 β-lactams plus 12 other antibiotics by using a disk diffusion method (10) and was found to be resistant to ampicillin, amoxicillin plus clavulanic acid, ceftiofur, cefuroxime, ceftazidime, cefotaxime, ceftriaxone, cefoperazone, cefoxitin, cefpodoxime, aztreonam, amikacin, chloramphenicol, colistin sulfate, gentamicin, sulfamethoxazole-trimethoprim, and a triple-sulfonamide solution and sensitive to imipenem, nalidixic acid, neomycin, tetracycline, furazolidone, and streptomycin. MICs of cefoxitin were 128 mg/liter and determined as previously described (6). The resistance phenotype suggested the presence of an extended-spectrum β-lactamase (ESBL) enzyme and/or the presence of an AmpC β-lactamase.
The presence of β-lactamases was assessed by isoelectric focusing (6). Four bands with approximate pI values of 5.2, 8.2, 8.6, and 9 were detected, indicating the production of four enzymes.
The isolate was analyzed by an AmpC multiplex PCR (6) and produced a product consistent with the CIT group (comprises LAT-1 to LAT-4, CMY-2 to CMY-7, and BIL-1). Subsequently, the full-length blaCMY PCR amplicon was sequenced as described before (6) and identified as blaCMY-2. Identification of TEM-, SHV-, and CTX-M-type β-lactamases was carried out by amplification and sequencing of the respective genes by using methods previously described (1, 11). The amplicons were sequenced on both strands in an Applied Biosystems ABI 310 sequencer, and analysis revealed that the isolate carried a blaTEM-1B variant gene with a silent mutation at base 739 (C to T), a blaSHV-5 gene, and a blaCTX-M-15 gene. In addition, the isolate was positive for class 1 integrons by PCR with primers L2 and L3 located at the conserved region (5). Furthermore, a single amplicon of approximately 1,300 bp was generated by using primers L2 and R1 (7), revealing that the integron contained a single gene cassette encoding the blaSHV-5 determinant.
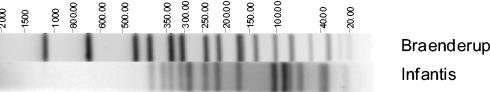
Preparation of DNA for pulsed-field gel electrophoresis (PFGE) was as described by the U.S. Centers for Disease Control and Prevention (2). Figure 1 shows the XbaI-PFGE fingerprint for the isolate. Plasmid extraction was performed as described previously (6); two plasmids with approximate sizes of 86 and 59.6 MDa were identified.
FIG. 1.
Image generated by the Bionumerics software showing the XbaI-PFGE restriction profile for Salmonella serotype Infantis UCM267. The sizes (in kilobases) of the fragments generated were calculated by comparing them to a Salmonella serotype Braenderup PulseNet universal control strain (H9812).
Assessment of the transferability of resistance.
Conjugations were performed with the Salmonella serotype Infantis isolate and a rifampin-resistant recipient Escherichia coli K-12 20R764, by use of in-broth and filter-mating methods (6). Conjugation mixtures were plated on CHROMagar ECC (M-Tech Diagnostics) containing rifampin (100 mg/liter) and cefotaxime (1 mg/liter) or CHROMagar ECC containing rifampin (100 mg/liter) and cefoxitin (32 mg/liter) and then incubated for 24 and 48 h at 37°C. Transconjugants were obtained only on the cefotaxime plates. Attempts to transfer resistance to cefoxitin from the Salmonella serotype Infantis isolate by conjugation consistently failed. Transformation experiments were conducted as follows: plasmid DNA was prepared with a QIAGEN high-speed MIDI kit. Electrocompetent E. coli cells (ElectroMAX DH10B; Invitrogen) were transformed by electroporation with a Bio-Rad GenePulser II electroporator, under standard conditions (2 kW, 200 Ω, and 25 μF). Transformants were selected on nutrient agar containing 32 mg of cefoxitin/liter after 16 h of incubation at 37°C. Plasmid analysis demonstrated the acquisition of plasmids of 59.6 and 86 MDa by the transconjugants and the transformants, respectively. Antimicrobial susceptibility testing of transconjugants and transformants showed that both plasmids in isolation were able to confer resistance to the same β-lactams. However, the 86-MDa plasmid also conferred resistance to cefoxitin and aztreonam.
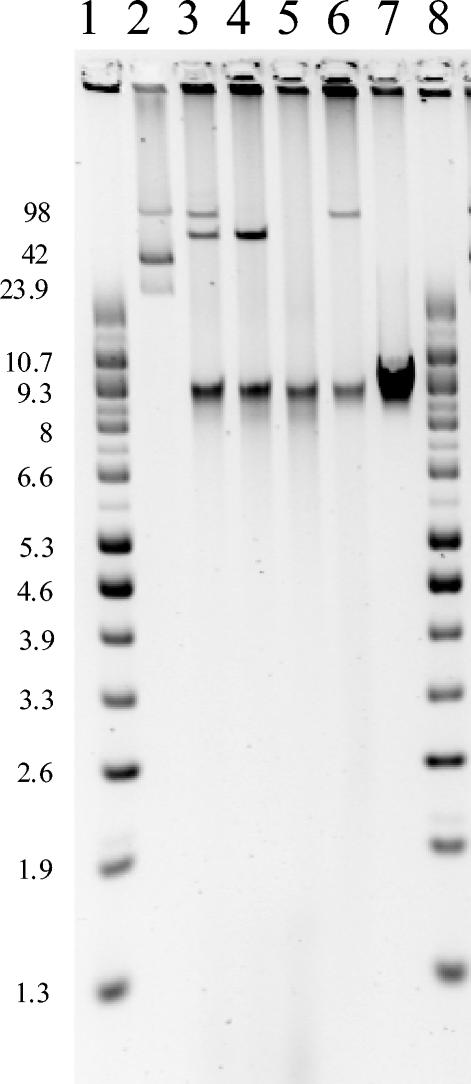
PCR analysis of the transformants carrying the 86-MDa plasmid indicated that the genetic determinants for blaCMY-2 and the class 1 integron with the blaSHV-5 cassette were colocalized on the same plasmid. Also, PCR analysis of the transconjugants carrying the 59.6-MDa plasmid indicated that the genetic determinants for blaTEM-1b and blaCTX-M-15 were colocalized on the same plasmid. Figure 2 illustrates the transfer (by transformation or conjugation) of plasmids of approximately 86 and 59.6 MDa from the Salmonella serotype Infantis isolate to E. coli recipients.
FIG. 2.
Plasmid profiles of isolates resulting from conjugation and transformation experiments. Lanes 1 and 8, supercoiled DNA ladder (Gibco BRL); lane 2, E. coli 39R861; lane 3, Salmonella serotype Infantis UCM267; lane 4, E. coli transconjugant; lane 5, E. coli recipient K-12 20R764; lane 6, E. coli transformant; lane 7, E. coli recipient DH10B. Sizes are expressed in megadaltons.
Conclusion.
Salmonella serotype Infantis has previously been reported to occur in children's infections in hospitals. The first outbreak due to multiresistant Salmonella serotype Infantis (including resistance to several ESBL) affecting hospitalized children was reported in 1996 in Brazil (9). These isolates had a single conjugative plasmid and were sensitive to cefoxitin. There is a second report, from Brazil in 1999, of a nosocomial outbreak in a neonatal unit caused by ESBL-producing Salmonella serotype Infantis (12). Salmonella serotype Infantis has been the second-most-common serotype in Argentina in recent years, being isolated mostly from hospitalized pediatric patients (8). Data presented in our study document the first description of a Salmonella isolate from Central America producing four β-lactamases. This strain originated from a pediatric patient that received two empirical treatments with cephalosporins. It is impossible to know if the patient was infected with a resistant strain carrying those genes or if the emergence of resistance was due to clinical treatment or in vivo transfer of the genetic determinants from resident flora to the Salmonella serotype Infantis isolates. All the genetic determinants responsible for the β-lactam resistance were located on potentially transferable plasmids. Treatment failure of a patient infected with a Salmonella serotype Anatum isolate resistant to ceftriaxone has been observed due to the in vivo acquisition of a plasmid containing the blaCTX-M-3 gene (15).
AmpC enzymes confer resistance to a wide spectrum of β-lactams. In Salmonella serotypes, the majority of AmpC-like enzymes have been reported to be CMY-2. The first report (Salmonella serotype Senftenberg) came from Algeria in 1997 (3), and since then, CMY-2 has been found in the following S. enterica serotypes: Heidelberg, Newport, Typhimurium, Bredeney, Mikawasima, and Montevideo. Also, S. enterica serotype Wien has been found to carry blaCMY-4. This case report represents the first incident of a blaCMY-2 gene in serotype Infantis. The fact that plasmid-mediated AmpC enzymes have been reported by many countries suggests that a global problem has developed.
CTX-M β-lactamases efficiently hydrolyze many newer broad-spectrum oximino-β-lactams. CTX-M-producing enterobacteria are endemic in Latin America and in some areas of northeastern Europe. CTX-M enzymes have increasingly been found in Salmonella organisms over recent years. Reports exist for Salmonella serotype Wien with CTX-M-3; Salmonella serotype Virchow with CTX-M-9; Salmonella serotype Infantis with CTX-M-2; Salmonella serotype Typhimurium with CTX-M-4, -5, and -6; Salmonella serotype Anatum with CTX-M-3; Salmonella serotype Oranienburg with CTX-M-3; and Salmonella serotype Enteritidis with CTX-M-3. To the best of our knowledge, we are presenting the first report of CTX-M-15 within the genus Salmonella. Unlike the majority of CTX-M enzymes, CTX-M-15 confers resistance to ceftazidime (13). This enzyme has recently been found in Enterobacteriaceae in India, Poland, Russia, and Turkey, but it had never before been detected on the American continent.
The ESBL SHV-5 was first described to occur in Salmonella serotype Senftenberg in India (14) and has recently been described to occur in Salmonella serotype Typhimurium in Romania. However, it has never been reported to occur in Salmonella serotypes on the American continent.
To date, a total of seven blaTEM-1 molecular variants have been reported due to the pattern of silent mutations in the blaTEM-1 structural gene and in its promoter (from blaTEM-1A to blaTEM-1G) (4). The molecular variant blaTEM-1B has frequently been found in the family Enterobacteriaceae, including in the genus Salmonella (1). The new molecular variant found in our study, with a silent mutation with respect to blaTEM-1b at nucleotide position 739, could be named blaTEM-1H.
In conclusion, this report provides a good example of the emergence of β-lactamase genes among bacterial species and especially Salmonella serotypes. In addition, this emergence of β-lactam resistance is not limited to any one country or continent; it is worldwide. This situation is of particular concern to public health, as these mechanisms of resistance now threaten the value of cephalosporin treatment against pathogenic enterobacteria.
Acknowledgments
We thank E. John Threfall (Health Protection Agency UK) for careful reading of the manuscript and helpful comments.
REFERENCES
- 1.Briñas, L., M. Zarazaga, Y. Sáenz, F. Ruiz-Larrea, and C. Torres. 2002. β-Lactamases in ampicillin-resistant Escherichia coli isolates from foods, humans, and healthy animals. Antimicrob. Agents Chemother. 46:3156-3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2002. Standardized molecular subtyping of foodborne bacterial pathogens by pulsed-field gel electrophoresis. Centers for Diseases Control and Prevention, Atlanta, Ga.
- 3.Koeck, J. L., G. Arlet, A. Philippon, S. Basmaciogullari, H. V. Thien, Y. Buisson, and J. D. Cavallo. 1997. A plasmid-mediated CMY-2 beta-lactamase from an Algerian clinical isolate of Salmonella Senftenberg. FEMS Microbiol. Lett. 152:255-260. [DOI] [PubMed] [Google Scholar]
- 4.Leflon-Guibout, V., B. Heym, and M.-H. Nicolas-Chanoine. 2000. Updated sequence information and proposed nomenclature for blaTEM genes and their promoters. Antimicrob. Agents Chemother. 44:3232-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liebana, E., C. Clouting, C. A. Cassar, L. P. Randall, R. A. Walker, E. J. Threlfall, F. A. Clifton-Hadley, A. M. Ridley, and R. H. Davies. 2002. Comparison of gyrA mutations, cyclohexane resistance, and the presence of class I integrons in Salmonella enterica from farm animals in England and Wales. J. Clin. Microbiol. 40:1481-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liebana, E., M. Gibbs, C. Clouting, L. Barker, F. A. Clifton-Hadley, E. Pleydell, B. Abdalhamid, N. D. Hanson, L. Martin, C. Poppe, and R. H. Davies. 2004. Characterization of beta-lactamases responsible for resistance to extended-spectrum cephalosporins in Escherichia coli and Salmonella enterica strains from food-producing animals in the United Kingdom. Microb. Drug Resist. 10:1-9. [DOI] [PubMed] [Google Scholar]
- 7.Maguire, A. J., D. F. J. Brown, J. J. Gray, and U. Desselberger. 2001. Rapid screening technique for class 1 integrons in Enterobacteriaceae and nonfermenting gram-negative bacteria and its use in molecular epidemiology. Antimicrob. Agents Chemother. 45:1022-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Merino, L. A., M. C. Ronconi, M. M. Navia, J. Ruiz, J. M. Sierra, N. B. Cech, N. S. Lodeiro, and J. Vila. 2003. Analysis of the clonal relationship among clinical isolates of Salmonella enterica serovar Infantis by different typing methods. Rev. Inst. Med. Trop. Sao Paulo 45:119-123. [DOI] [PubMed] [Google Scholar]
- 9.Moraes, B. A., C. A. Cravo, M. M. Loureiro, C. A. Solari, and M. D. Asensi. 2000. Epidemiological analysis of bacterial strains involved in hospital infection in a university hospital from Brazil. Rev. Inst. Med. Trop. Sao Paulo 42:201-207. [DOI] [PubMed] [Google Scholar]
- 10.NCCLS. 1999. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. Approved standard. NCCLS document M31-A. NCCLS, Wayne, Pa.
- 11.Pagani, L., E. Dell'Amico, R. Migliavacca, M. M. D'Andrea, E. Giacobone, G. Amicosante, E. Romero, and G. M. Rossolini. 2003. Multiple CTX-M-type extended-spectrum β-lactamases in nosocomial isolates of Enterobacteriaceae from a hospital in northern Italy. J. Clin. Microbiol. 41:4264-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pessoa-Silva, C. L., C. M. Toscano, B. M. Moreira, A. L. Santos, A. C. Frota, C. A. Solari, E. L. Amorim, G. Carvalho Mda, L. M. Teixeira, and W. R. Jarvis. 2002. Infection due to extended-spectrum beta-lactamase-producing Salmonella enterica subsp. enterica serotype Infantis in a neonatal unit. J. Pediatr. 141:381-387. [DOI] [PubMed] [Google Scholar]
- 13.Poirel, L., M. Gniadkowski, and P. Nordmann. 2002. Biochemical analysis of the ceftazidime-hydrolysing extended-spectrum beta-lactamase CTX-M-15 and of its structurally related beta-lactamase CTX-M-3. J. Antimicrob. Chemother. 50:1031-1034. [DOI] [PubMed] [Google Scholar]
- 14.Revathi, G., K. P. Shannon, P. D. Stapleton, B. K. Jain, and G. L. French. 1998. An outbreak of extended-spectrum, beta-lactamase-producing Salmonella Senftenberg in a burns ward. J. Hosp. Infect. 40:295-302. [DOI] [PubMed] [Google Scholar]
- 15.Su, L.-H., C.-H. Chiu, C. Chu, M.-H. Wang, J.-H. Chia, and T.-L. Wu. 2003. In vivo acquisition of ceftriaxone resistance in Salmonella enterica serotype Anatum. Antimicrob. Agents Chemother. 47:563-567. [DOI] [PMC free article] [PubMed] [Google Scholar]