Abstract
Neddylation is a crucial post-translational modification that depends on the E3 cullin ring ligase (CRL). The E2-adapter component of the CRL, sensitive to apoptosis gene (SAG), is critical for the function of CRL-mediated ubiquitination; thus, the deletion of SAG regulates neddylation. We examined the role of SAG-dependent neddylation in T-cell–mediated immunity using multiple approaches: a novel T-cell–specific, SAG genetic knockout (KO) and chemical inhibition with small-molecule MLN4924. The KO animals were viable and showed phenotypically normal mature T-cell development. However, in vitro stimulation of KO T cells revealed significantly decreased activation, proliferation, and T-effector cytokine release, compared with WT. Using in vivo clinically relevant models of allogeneic bone marrow transplantation also demonstrated reduced proliferation and effector cytokine secretion associated with markedly reduced graft-versus-host disease. Similar in vitro and in vivo results were observed with the small-molecule inhibitor of neddylation, MLN4924. Mechanistic studies demonstrated that SAG-mediated effects in T cells were concomitant with an increase in suppressor of cytokine signaling, but not NF-κB translocation. Our studies suggest that SAG is a novel molecular target that regulates T-cell responses and that inhibiting neddylation with the clinically available small-molecule MLN4924 may represent a novel strategy to mitigate T-cell–mediated immunopathologies, such as graft-versus-host disease.
T cells are key mediators of immune pathologies associated with autoimmunity and alloimmunity. As such, T cells are central to the efficacy of allogeneic bone marrow transplantation (allo-BMT), a critical and curative interventional therapy for patients with aggressive hematological malignancies.1, 2, 3, 4 However, 40% to 50% of allo-BMT recipients experience severe transplant-related complications, known as graft-versus-host disease (GVHD), leading to high transplant-related mortality.1, 5, 6
Post-translational modifications (PTMs) modulate cellular processes1, 7; however, little is known about the role of PTMs in T-cell responses. Neddylation, a type of PTM, modulates ubiquitination of some proteins8, 9, 10, 11 through activation of the cullin ring ligase (CRL) degradation scaffolding (an E3 ligase). This is achieved via attachment of the ubiquitin-like molecule—neural precursor cell expressed developmentally down-regulated protein 8 (NEDD8)—to the cullin protein backbone of the CRL complex.1, 12 The NEDD8 molecule is activated by an ATP-dependent and specific E1 enzyme known as NEDD8 activating enzyme, before its transfer to the E2 molecule called NEDD8-conjugating enzyme (Ubc12) that then transfers NEDD8 to the cullin E3 ligase backbone. The attachment of NEDD8 to cullin serves as a critical activating step in the recruitment of the multisubunit components that compose the CRL complex.1, 12 A key component of the CRL complex is the adaptor molecule, sensitive to apoptosis gene (SAG)/really interesting new gene box protein 2 (Rbx2).13, 14, 15, 16 Previous studies have shown that silencing SAG/Rbx2 via siRNA inhibits the CRL complex, thereby regulating the neddylation pathway.1, 14, 15, 16, 17 Another method for regulating the neddylation pathway is achieved through the use of a pharmacological small-molecule inhibitor MLN4924. MLN4924 is a specific and potent inhibitor of NEDD8 activating enzyme.1, 6 Studies using MLN4924 have demonstrated its ability to potently suppress cancer in experimental models.8, 18 Recent data have suggested that targeting neddylation with MLN4924 regulated the inflammatory functions of dendritic cells and macrophages, whereas inhibition of neddylation by knocking down Ubc12 mitigated CD4 T-cell responses.1, 19, 20 Collectively, these studies point to an emerging role for neddylation in the regulation of immune cell subsets. However, the exclusive utilization of either the small molecule or the shRNA knockdown approaches does not rule out the possibility of potential off-target effects.21, 22, 23, 24, 25 Furthermore, the role of neddylation in T-cell–mediated alloimmunity and disease processes is not known.
To determine the role of neddylation, and to selectively target only a subset of CRL complexes (CRL 2 and 5), we generated novel T-cell–specific SAG/Rbx2 deficient mice and determined the impact of SAG-dependent neddylation in T-cell responses and the alloimmune disease process, GVHD. We found that SAG-deficient mature naïve T cells showed reduced activation, proliferation, and cytokine secretion in vitro and in vivo and furthermore caused less severe GVHD. Similar results were observed with small-molecule inhibitor MLN4924. Furthermore, mechanistic studies suggested that in contrast to neddylation inhibition–mediated effects on dendritic cells (DCs) and macrophages, the observed decrease in T-cell functions was negligibly because of inhibition of NF-κB1, 3; rather, these were achieved through an accumulation of the suppressor of cytokine signaling (SOCS) proteins SOCS 1 and 3. We thus report a novel molecular mechanism for neddylation in T cells. In addition, the results of our studies using the small-molecule inhibitor of neddylation (MLN4924), in light of its clinical availability, suggest that this strategy could be tested in carefully designed human clinical trials to attenuate GVHD.
Materials and Methods
Mice
Female C57BL/6 (H-2b; CD45.2+), BALB/c (H-2d) mice were purchased from the National Cancer Institute (Frederick, MD), and C3H.sw (H-2b) mice were purchased from The Jackson Laboratory (Bar Harbor, ME). The Sagfl/fl were generated as previously described.26, 27 The LCK-Cre mice were purchased from Taconic (Hudson, NY). The age of mice used for experiments ranged between 7 and 12 weeks. All animals were cared for under regulations reviewed and approved by the University Committee on Use and Care of Animals of the University of Michigan, based on University Laboratory Animal Medicine guidelines.
Cell Isolation and Cultures
Briefly, DCs were generated from bone marrow cells flushed from the femurs of 8- to 12-week-old female C57BL/6 mice and cultured in 150 × 15-mm CytoOne petri dishes (USA Scientific, Ocala, FL) at 1 × 107 in 20 mL of RPMI 1640 supplemented with 10% fetal calf serum, 4 mmol/L l-glutamine, 10 U/mL penicillin, 100 μg/mL streptomycin, 0.5 mmol/L 2-mercaptoethanol, and 20 ng/mL granulocyte-macrophage colony-stimulating factor. On day 4, 20 mL of fresh complete RPMI 1640 containing 20 ng/mL granulocyte-macrophage colony-stimulating factor was added to each culture. After 7 days of culture, the loosely adherent cells were harvested and purified using anti-CD11c magnetic microbeads (Miltenyi Biotec Ltd, Auburn, CA) and the autoMACS (Miltenyi Biotec Ltd.).
T cells were isolated from whole splenoctye homogenates, incubated with CD90.2+ microbeads (Miltenyi Biotec Ltd.), and separated using the autoMACS, (Miltenyi Biotec Ltd.) following the manufacturer's instructions.
BMT
BMTs were performed as previously described.28 Briefly, syngeneic (BALB/c → BALC/c or C57BL/6 → C57BL/6) and allogeneic (C57BL/6 → BALB/c or C3H.sw → C57BL/6) recipients received lethal irradiation. On day -1, BALB/c recipients received a total of 800 cGy of irradiation (split dose separated by 3 hours) and B6 animals received a single dose of 1000 cGy. Donor splenic CD90.2+ T cells were magnetically separated using an autoMACs (Miltenyi Biotec, Bergisch Gladbach, Germany) and 0.5 × 106 to 1 × 106 T cells were transferred to BALB/c recipients and 2 × 106 T cells were transferred to C57BL/6 recipients. A total of 5 × 106 donor whole bone marrow or T-cell–depleted bone marrow was transferred to recipients as indicated. Survival was monitored daily, and the recipient body weight and GVHD clinical scores were determined weekly, as described previously.28 Animals received vehicle (phosphate-buffered saline) or 20 mg/kg MLN4924 by sub-q injection daily for indicated time.
Cytokine Detection
Isolated CD90.2+ T cells were seeded onto 60-mm culture dishes at 3 × 106 cells per well and a 24-well plate at 2 × 105 cells per well, respectively. Pretreated cells were cultured with dimethyl sulfoxide (Sigma-Aldrich, St. Louis, MO) and MLN4924 (Active Biochem, Maplewood, NJ) at the indicated dosages for 2 hours. Cells were then stimulated by either irradiated allosplenocytes or α-CD3 and α-CD28 at the indicated concentrations for indicated time period. Supernatants were subsequently collected and stored at −20°C until analysis. Tumor necrosis factor-α and IL-6 enzyme-linked immunosorbent assay kits were purchased from R&D Systems (Minneapolis, MN) and performed as per the manufacturers' instructions and read at 450 nm by a SpectraMax microplate reader (Molecular Devices, Sunnyvale, CA).
Quantitative PCR
Using 1 μg of each RNA template, cDNA was synthesized using SuperScript VILO (Invitrogen, Carlsbad, CA). Quantitative PCR primers were as follows: murine glyceraldehyde-3-phosphate dehydrogenase (forward, 5′-CCACAGTCCATGCCATCACTGC-3′; reverse, 5′-GCCCAAGATGCCCTTCAGTGGG-3′), Rbx2 (forward, 5′-GGACGTTGAGTGCGATACCT-3′; reverse, 5′-TGCCGATTCTTTGGACTACC-3′), SOCS1 (forward, 5′-CAGCCGACAATGCGATCTC-3′; reverse, 5′-AAGGTGCGGAAGTGAGTGT-3′), and SOCS3 (forward, 5′-GTCACCCACAGCAAGTTTCC-3′; reverse, 5′-GCTGTCGCGGATAAGAAAGG-3′).
Flow Cytometry
To analyze immunophenotype surface markers, lymphocytes contained in the spleen were harvested, stained, and gated on CD4-conjugated PerCP/Cy5.5 (clone: GK1.5) or CD8-conjugated APC (clone: 53-6.7) and configurations of the following per mouse in duplicate: CD69-PE (clone: H1.2F3), CD62L-PE (clone: MEL-14), CD44-PerCP/Cy5.5 (clone: IM7), and CD44-APC (clone: IM7). Intracellular cytokines were elicited by stimulating splenocytes with cell stimulation cocktail with protein transport inhibitors (eBiosciences, San Diego, CA) for 6 hours. Annexin V was purchased from BD Biosciences (San Jose, CA). All flow cytometry antibodies were purchased from eBiosciences. Stained cells were then analyzed with an Accuri C6 Flow Cytometer (BD Biosciences).
Western Blot and Subcellular Fractionation
T cells were seeded in 60-mm dishes with 3 × 106 cells per dish and were cultured with dimethyle sulfoxide (Sigma-Aldrich) or MLN4924 (Active Biochem) at the indicated dosages. Where stimulation is indicated, T cells were incubated for 2 hours with MLN4924, followed by concurrent stimulation with α-CD3 and α-CD-28 for 24 hours. After treatment/stimulation, whole cell lysates were obtained and protein concentrations determined with Pierce BCA Protein Assay (Thermo Scientific). Equal amounts of protein were separated by SDS-PAGE gel (120 V, 1.5 hours) and subsequently transferred to nitrocellulose membrane (20 V, 1 hour). The following antibodies were used to analyze the membranes: α-tubulin (11H10), NF-κB p65 (D14E12), Lamin A/C (4C11), pIκBα (5A5), and IκBα (L35A5), and were purchased from Cell Signaling (Danvers, MA). Cullin1 (EPR3103Y), SOCS1, SOCS3, and LDH (EP1566Y) were purchased from abcam (Cambridge, MA). Monoclonal SAG antibody was raised against the really interesting new gene domain (AA44-113), as previously described.16 Secondary antibodies conjugated to horseradish peroxidase (Jackson ImmunoResearch, West Grove, PA) or Alexa Fluor 488 (Life Technologies, Carlsbad, CA) were used to detect primary antibodies. Densitometric analysis was performed using ImageJ software version 1.5i (NIH, Bethesda, MD). Subcellular fractionation was performed on 3 × 106 T cells seeded in 60-mm dishes. Cells were pretreated in the presence or absence of vehicle or MLN4924 for 2 hours, followed by concurrent α-CD3 and α-CD28 stimulation for indicated time. The cytoplasmic and nuclear fractions were isolated using the Nuclear Extract Kit (Active Motif, Carlsbad, CA) per the manufacturer's instructions. The extracts were then analyzed via Western blot.
Cell Proliferation Assay and Mixed Lymphoctye Reaction
Splenic T cells (2 × 105/well) were magnetically separated from WT-B6 or WT-BALB/c mice by autoMACS using CD90.2 microbeads and subsequently cultured with irradiated (30 Gy) WT-B6 DC at 40:1 (5 × 103/well) and 100:1 (2 × 103/well) for 72 and 96 hours. Before plating, T cells were stained with 2 μmol/L carboxyfluorescein diacetate succinimyl ester (Life Technologies) where indicated. Incorporation of 3H-thymidine (1 μCi/well) by proliferating T cells during the final 6 hours of culture was measured by a TopCount (PerkinElmer, Boston, MA).
Confocal Microscopy
T cells were seeded onto Corning glass coverslips (1 × 105 cells/slip) (Fisher Scientific, Waltham, MA) using cytospin technique. Pretreated cells were cultured with dimethyl sulfoxide (Sigma-Aldrich) or MLN4924 (Active Biochem) at the indicated dosages for 2 hours, followed by concurrent stimulation with α-CD3 and α-CD28 for an additional 24 hours. Coverslips were then washed, fixed with 4% paraformaldehyde for 20 minutes, and subsequently permeabilized with 0.3% Triton X. Cells were stained with NF-κB p65 (D14E12) primary (1:500) and Alexa Fluor 488 (Molecular Probes) secondary (1:1000), DAPI (Invitrogen), and Alexa Fluor 555 phalloidin (1:500). Coverslips were then mounted using ProLong Gold Antifade Reagent (Molecular Probes) and Z-stack images were acquired at room temperature using a Nikon A-1 confocal microscope (Nikon, Mellville, NY) using an oil immersion 60× objective with a numerical aperture of 1.4 and imported into NIS-Elements Software version 4.11.0 (Nikon). Excitation lasers 405, 488, and 561 nm were used. Microscope laser exposure and settings were obtained using appropriate isotype controls and were retained for each experimental group.
Affymetrix Microarrays and Analysis
The tcRNA was extracted from purified WT B6 T cells and miR-142 knockout (KO) T cells with RNeasy Mini Kit (Qiagen, Hilden, Germany). After the quality of the total RNA was verified by an Agilent 2100 Bioanalyzer, the samples were processed using the WT-Ovation Pico System (Affymetrix, Santa Clara, CA), and a single round of amplification for samples with even stricter concentration restraints. This system incorporates oligo(dT) and random primers for amplification at the 3′ end and throughout the whole transcriptome. Affymetrix mouse genome 430 2.0 Arrays (Affymetrix), which contain 45,000 transcripts for annotated genes and expressed sequence tags, were used. The stained arrays were scanned on an Agilent Gene Array Scanner (Affymetrix) with a 560-nm filter. The data were analyzed using the R statistical environment version 3.0.0 (https://cran.r-project.org/bin/windows/base, last accessed June 3, 2016) provided by Bioconductor and were examined for the quality control by showing the same distribution of the PM probes for each chip and no degradation. The expression values were then calculated for each gene using a robust multiarray average.29, 30 This modeling strategy converts the PM probe values into an expression value (log2-transformed) for each gene. A total of 41,346 Affy IDs differentially expressed were converted to Human Entrez gene ID (ConceptGen only uses each gene once) yielding 22,066 genes and analyzed for gene function concept based on multiple database, including GO and MeSH database. The probe sets with a fold-change of two or greater were selected. The gene set that is involved in specific function concept was analyzed for enrichment by Gene Set Enrichment Analysis developed at the Broad Institute of Massachusetts Institute of Technology and Harvard (http://www.broadinstitute.org/gsea/index.jsp, last accessed April 17, 2014), which determines whether a set of genes are statistically significant between two biological states.31, 32 The microarray data were deposited in Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo; accession number GSE83593).
Statistical Analysis
Bars and error bars represent the means and SEM, respectively. Nonsurvival analysis was performed using unpaired t-test between two groups. Analysis of variance was used for comparisons with more than two groups. Survival data analysis was performed using a Mantel-Cox log-rank test.
Results
Inhibition of Neddylation with Small-Molecule MLN4924 Regulates T-Cell Responses
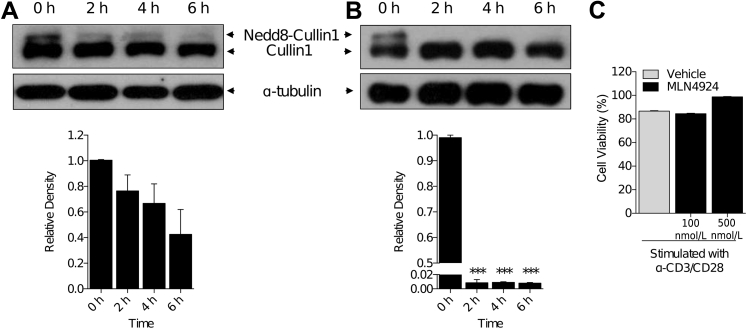
We determined whether the small-molecule MLN4924 that is known to inhibit neddylation in a variety of cell types1, 6 affects neddylation of Cul-1 in T cells. We incubated CD90.2+ T cells in the presence of MLN4924 at 100 nmol/L. We observed a 25% decrease in neddylation, as indicated by the presence of the higher-molecular-weight band of Cul-1 at the 100 nmol/L concentration (Figure 1A), over a time period of 6 hours. However, when T cells were treated with higher MLN4924 concentrations (500 nmol/L), we observed that the higher-molecular-weight band was absent within 2 hours of treatment (Figure 1B), suggesting that MLN4924 inhibited neddylation in T cells. Although MLN4924 did not affect the viability of DCs,1 but because it is cytotoxic to cancer cells,8, 10, 11 we next determined whether it was cytotoxic to the T cells at the doses at which it deneddylated Cul-1. We cultured T cells in the presence or absence of the indicated concentrations of MLN4924 and examined those cells for annexin V positivity. There was no difference in the viability of T cells at either 100 or 500 nmol/L when compared to diluent-treated controls (Figure 1C).
Figure 1.
Neddylation inhibition in T cells. Analysis by Western blot of Cullin-1 protein in CD90.2+ splenic T cells. Loss of higher-molecular-weight band of Cullin-1 by treatment with 100 nmol/L (A) and 500 nmol/L (B) MLN4924 indicates level of inhibition of nedd8 attachment to cullin protein. α-Tubulin protein levels were analyzed as an equal loading control. C: CD90.2+ T-cell viability by annexin V staining after incubation with MLN4924 at indicated concentrations for 72 hours. One representative experiment each of three is shown. ∗∗∗P < 0.001 versus 0 h.
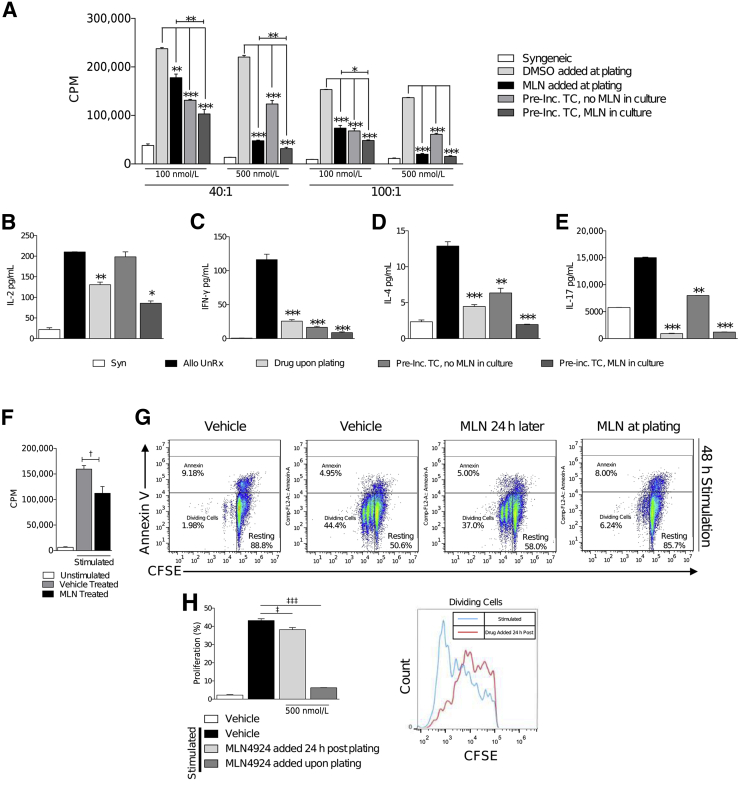
We have previously shown that culturing DCs with MLN4924 decreases allo-T-cell responses; furthermore, we have also demonstrated that MLN4924 has a DC-intrinsic effect by decreasing their allostimulatory capacity.1 However, whether inhibition of neddylation in T cells diminishes their intrinsic proliferative capacity is unknown. To test this, we used a mixed lymphocyte reaction in which [3]H is taken up by, and incorporated into, proliferating cells. We cultured irradiated C57BL/6 (H-2b) bone marrow–derived dendritic cells with BALB/c (H-2d) CD90.2+ T cells in the following experimental culture groups: i) the vehicle-treated group; ii) the group in which MLN4924 was added to the culture at the time of plating; iii) the group in which T cells had been pretreated with MLN4924 for 6 hours and were co-cultured with DCs in the absence of the drug; and iv) the group in which MLN4924-pretreated T cells were cultured with MLN4924 present throughout the duration of the co-culture. We observed a significant decrease in all groups cultured with MLN4924, as compared to vehicle-treated controls cultured for 72 hours (Figure 2A) and 96 hours (Supplemental Figure S1). Interestingly, cultures that contained pretreated T cells, but that did not have MLN4924 present during co-culture, still exhibited a significant decrease in T-cell proliferation, suggesting that inhibition of neddylation has a T-cell–intrinsic effect (Figure 2A). Furthermore, the culture of MLN4924-pretreated T cells with the drug present throughout co-culture resulted in an even greater decrease in T-cell proliferation, suggesting that MLN4924 also affects DCs (Figure 2A), confirming the findings observed from our previous data.1
Figure 2.
T-cell functions are attenuated by neddylation blockade. A: BALB/c T cells were cultured with irradiated (30 Gy) C57BL/6 bone marrow–derived dendritic cells (BMDCs) at 40:1 and 100:1 ratios (T cell/BMDC) in the presence or absence of MLN4924 for 72 hours. Where indicated, T cells were preincubated with MLN4924 at indicated dosage for 6 hours before plating. Enzyme-linked immunosorbent assay quantification of IL-2 (B), interferon (IFN)-γ (C), IL-4 (D), and IL-17 (E) release from T cells, cultured in the presence or absence of MLN4924 with allogeneic BMDCs as indicated in A for 72 hours. One representative experiment of three is shown in cells treated in triplicate. Analysis of T-cell stimulation with 5 μg/mL α-CD3 and 2.5 μg/mL α-CD28 in the presence or absence of vehicle or 500 nmol/L MLN4924 by 3[H] thymidine incorporation for the last 6 hours of 72-hour culture (F) or carboxyfluorescein diacetate succinimyl ester (CFSE) dilution for 48 hours with annexin V viability staining (G). G and H: Inhibition of neddylation by MLN4924 significantly reduces the proliferation of T cells when treated at time of plating or by treatment 24 hours after stimulation. H: Quantification of representative T cells after 48 hours total stimulation between T cells treated with vehicle or MLN4924 24 hours after stimulation. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 versus syngeneic control; †P < 0.05 versus unstimulated; ‡P < 0.05, ‡‡‡P < 0.001 versus vehicle control. Allo UnRx, Allo-BMT untreated; CPM, count per minute; DMSO, dimethyl sulfoxide; Pre-Inc. TC, preincubated T-cells; Syn, syngeneic.
T cells release lineage-specific cytokines that have many paracrine effects on other immune cell subsets.13 Thus, we then examined IL-2 (an activation cytokine) and interferon-γ, IL-4, and IL-17, which are type 1 helper T cell (Th1)–, Th2-, and Th17-associated cytokines, respectively. All groups treated with MLN4924 exhibited decreased cytokine production (Figure 2, B–E). To definitively determine that the effects on T-cell proliferation was only because of the deneddylation of Cul-1 in T cells by MLN4924, we stimulated purified T cells with TCR/coreceptor stimulation (via α-CD3 and α-CD28). We observed a similar decrease in T-cell proliferation by treatment with MLN4924 when stimulated with anti-CD3/28 as with allostimulation (Figure 2F).
Next, we used carboxyfluorescein diacetate succinimyl ester (CFSE) and annexin V staining to measure the proliferation and viability dynamics of T cells treated in the presence or absence of vehicle or MLN4924 when stimulated with α-CD3 and α-CD28. We saw that the proliferation of T cells treated with MLN4924 at the time of plating was significantly reduced when compared to the vehicle control but did not exhibit an increase in cell apoptosis (Figure 2G). Furthermore, when T cells were stimulated and allowed to proliferate for 24 hours before the addition of MLN4924, we still observed a significant decrease in T-cell proliferation (Figure 2G). Indeed, a comparison of the T cells that proliferated unimpeded for 24 hours with the T cells treated with the vehicle alone showed a dramatic decrease in proliferation on inhibition of neddylation with MLN4924 (Figure 2H). These data demonstrate that after T-cell stimulation, MLN4924 inhibits proliferation without significantly altering apoptosis.
We next determined whether T cells with inhibited neddylation displayed an activated phenotype on stimulation. After the treatment of T cells with MLN4924 and stimulation with α-CD3 and α-CD28 for 72 hours, we observed decreased activation markers (CD44hi, CD69+) and increased naïve T cells (CD44−CD62L+) (Supplemental Figure S2). Similar results were seen at 48 hours after activation (data not shown). Likewise, cultures in which T cells were allowed to proliferate for 24 hours before the addition of MLN4924 contained fewer CD69+ and CD44hi T cells (Supplemental Figure S2). These data suggest that inhibition of neddylation in T cells mitigates their proliferation and activation after stimulation.
Genetic Deficiency of SAG Does Not Affect T-Cell Development
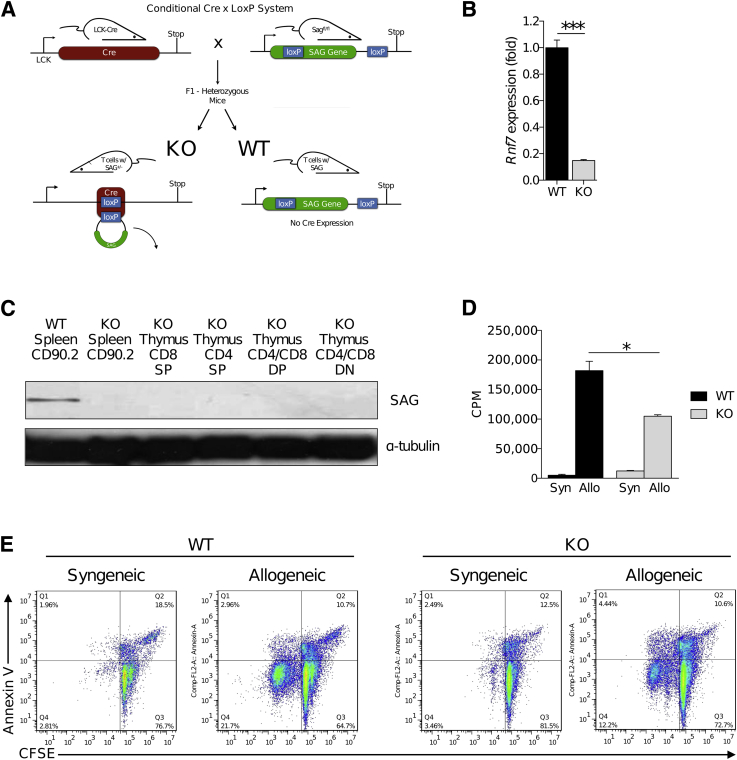
The loss of SAG/Rbx2 protein regulates the Cul-1 neddylation pathway.1, 14, 15, 16 The functional effects of neddylation inhibition with MLN4294 from above could potentially be because of the off-target effects of the small-molecule approach. To better and more clearly determine the effect of Cul-1 neddylation in T-cell responses, we next generated KO mice that were deficient in SAG protein only in the T cells. To this end, we crossed LCK-Cre mice with Sagfl/fl (Figure 3A). We then crossed the F1 heterozygous generations to obtain LCK-Cre−/Sagfl/fl (WT) and LCK-Cre+/Sagfl/fl (KO) mice (Figure 3A) and confirmed that they had Sag deletion only in the T cells (Figure 3B). We next ensured that SAG was absent from peripheral T cells in the spleen as well as all stages of T-cell development in the thymus by examining CD8 single positive cells, CD4 single positive cells, CD4/CD8 double positive cells, and CD4/CD8 double negative cells (Figure 3C). These results indicated that no detectable SAG was present in T cells of the KO animals.
Figure 3.
Conditional knockout of SAG protein in T cells reduces allogeneic (Allo) T-cell proliferation. A: Breeding scheme of C57BL/6 LCK-Cre mice with C57BL/6 SAGfl/fl mice. F1 heterozygous generations were subsequently bred producing animals with [knockout (KO)] and without [wild type (WT)] the T-cell–specific deficiency of SAG. Mice were continually bred and used on the basis of confirmed genotype of Rnf7 deficiency by quantitative PCR (B) or SAG protein deficiency shown by immunoblot detection in peripheral splenic CD90.2+ T cells and in developing T-cell populations in the thymus in WT or KO animals, as indicated (C); CD8 single positive (SP), CD4 SP, CD4/CD8 double positive (DP), and CD4/CD8 double negative (DN). WT or KO T cells co-cultured with irradiated (30 Gy) syngeneic (Syn; C57BL/6 H-2b) or allogeneic (BALB/c H-2d) splenocytes for 96 hours measured by 3[H] thymidine incorporation during the last 6 hours of culture (D) or carboxyfluorescein diacetate succinimyl ester (CFSE) dilution with annexin V viability staining (E). ∗P < 0.05, ∗∗∗P < 0.001. CPM, count per minute.
On confirming that Sag was deleted in the T cells (Figure 3, B and C), we first examined whether the loss of SAG protein affected the development of T cells. We did not observe any significant differences in the total number of thymocytes, nor in the total number of double-positive T cells (CD4+CD8+) (Supplemental Figure S3, A and B), suggesting that absence of SAG did not substantially alter thymopoesis. We next performed detailed phenotypical analysis of the T-cellular contents of the secondary lymphoid organs—the spleen and peripheral lymph nodes. There was no significant difference in the total number of CD3+ T cells (Supplemental Figure S3C), nor was there a difference in the total number of naïve T cells (CD62L+CD44−) (Supplemental Figure S3, D and E), or CD4+Foxp3+ cells (data not shown), suggesting that normal T-cell development occurs in the conditional KO mouse.
SAG Deficiency Attenuates T-Cell Responses
We next examined the proliferative capacity of SAG-deficient T cells after allogeneic stimulation. We cultured WT or KO T cells with irradiated BALB/c splenocytes for 96 hours. Consistent with the pharmacological inhibition of neddylation, we observed that significantly fewer SAG-deficient T cells had proliferated when compared with WT-SAG competent controls (Figure 3D). To examine the viability dynamics of the proliferating cells, we stained KO T cells with carboxyfluorescein diacetate succinimyl ester (CFSE) and subjected them to allogeneic stimulation by irradiated splenocytes. We observed a decrease in proliferation but without an increase in annexin V (Figure 3E), suggesting that although the cells were live, there was a defect in their ability to proliferate. Similar results were obtained after α-CD3 and α-CD28 stimulation (data not shown).
Inhibition of Neddylation in T Cells Minimally Affects NF-κB
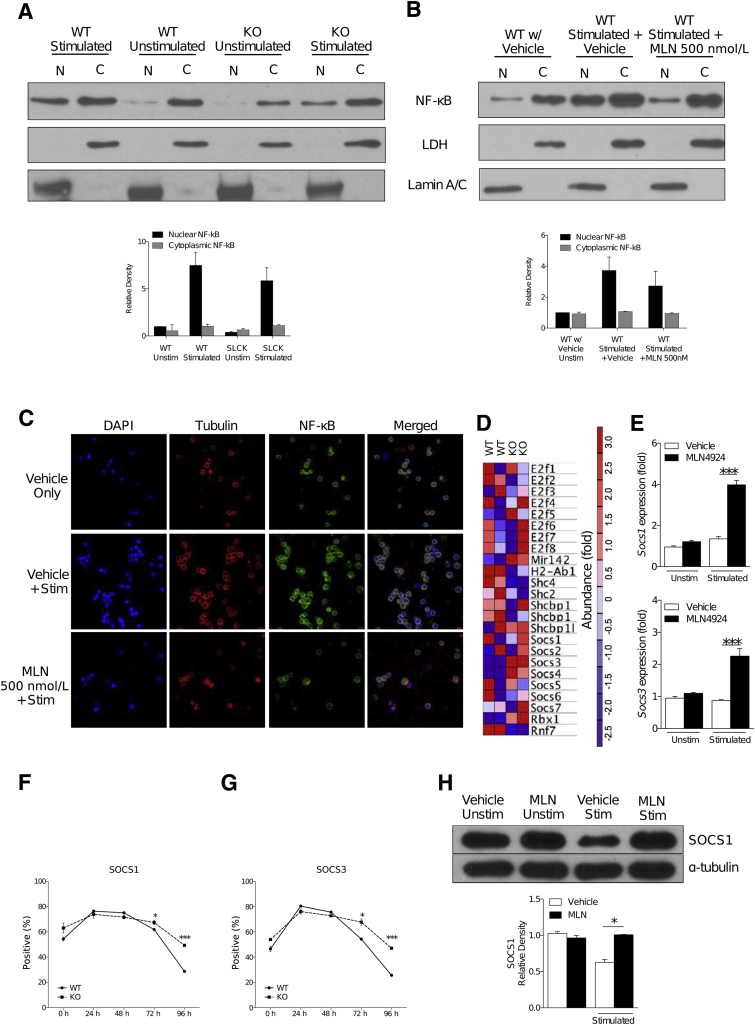
Inhibition of neddylation—by either MLN4924 treatment or molecular knockdown of SAG—is known to regulate the degradation of inhibitor-κB (IκB), thereby preventing the function of the transcription factor NF-κB.1, 18 TCR signaling is known to trigger the NF-κB pathway33; therefore, we determined whether the inhibition of NF-κB translocation was responsible for the decrease in SAG-deficient T-cell functions. To test this, we assessed p65 NF-κB translocation to the nucleus in WT and KO T cells after TCR stimulation. In contrast to the aforementioned results from other immune cell subsets, namely macrophages18 and DCs,1 we saw that NF-κB translocation to the nucleus was not significantly affected by the deletion of SAG (Figure 4A) after α-CD3 and α-CD28 stimulation. Likewise, T cells treated with MLN4924 exhibited only a partial inhibition of NF-κB translocation after similar stimulation (Figure 4B). These data suggest that inhibition of neddylation by either SAG deficiency or MLN4924 does not significantly affect NF-κB in T cells.
Figure 4.
SOCS proteins are degraded less in T cells with dysfunctional neddylation pathway. Protein analysis by Western blot of p65 isoform of NF-κB protein using nuclear (N) and cytosolic (C) fractions from wild-type (WT) and knockout (KO) T cells (A) or WT T cells cultured in the presence or absence of vehicle or 500 nmol/L MLN4924 (B), subjected to or withheld from concurrent stimulation with 5 μg/mL α-CD3 and 2.5 μg/mL α-CD28. Plots below Western blot images show densitometric and statistical analysis of p65 NF-κB presence in the nuclear fraction and cytosolic fraction. Lamin A/C and lactate dehydrogenase proteins were analyzed to demonstrate the presence of nuclear and cytosolic fractions, respectively. One representative experiment of three is shown. C: Immunocytochemistry analysis of p65 NF-κB (column 3) localization in bone marrow–derived dendritic cells cultured in the presence or absence of MLN4924 and stimulated with 5 μg/mL α-CD3 and 2.5 μg/mL α-CD28. Cell nuclei were stained using DAPI (column 1). Cytoplasmic actin was stained with phalloidin (column 2). Merged images of staining (column 4). One representative experiment of three is shown. D: Gene expression in WT and KO T cells identified in Affymetrix microarray analysis. The data set and top 24 enriched genes shown were obtained from two biological replicates. E: SOCS1 and SOCS3 gene expression in CD90.2+ T cells, stimulated with 5 μg/mL α-CD3 and 2.5 μg/mL α-CD28 where indicated, and treated with 500 nmol/L MLN4924 or vehicle. Flow cytometric analysis of SOCS1 protein (F) and SOCS3 protein levels (G) in WT and KO C57BL/6 T cells stimulated with irradiated (30 Gy) allogeneic BALB/c splenocytes for indicated time. One representative experiment of two is shown in groups treated in triplicate. H: Western blot analysis of SOCS1 protein levels in WT T cells treated with MLN4924 500 nmol/L and stimulated with 5 μg/mL α-CD3 and 2.5 μg/mL α-CD28 for 72 hours. One representative experiment of three is shown. ∗P < 0.05, ∗∗∗P < 0.001. Original magninfication, ×60 (C). SLCK, SAGfl/fl × Lck-Crel; Stim, stimulated; Unstim, unstimulated.
Next, we visualized the dynamics of p65 translocation using a complementary method featuring immunocytochemistry and confocal microscopy analysis. We treated T cells with vehicle or MLN4924 and stimulated these cells with α-CD3 and α-CD28. Both the vehicle- and MLN4924-treated cells exhibited similar NF-κB localization patterns (Figure 4C). Taken together, these data suggest that inhibition of neddylation in T cells uses a disparate mechanism in T-cell function regulation when compared to macrophages and DCs.
T-Cell Deficiency of SAG Increases SOCS Signaling
Because neddylation is a pathway that regulates the degradation of proteins, and given that proteins are known to exhibit positive and negative feedback on their own transcription and translation (or those of other proteins),21, 23 we then examined the mRNA transcripts present in WT and KO T cells using microarray analysis. After microarray analysis, we found lower levels of RNF7/SAG in the KO T cells, confirming that SAG was deficient in these cells (Figure 4D). More important, we saw the increased expression of several SOCS mRNA (SOCS 1 to 7) in KO T cells (Figure 4D). To validate the microarray results, we next examined the gene expression of SOCS1 and SOCS3. Indeed, we saw increased expression of both SOCS1 and SOCS3 in stimulated T cells that were treated with MLN4924, compared to vehicle-treated controls (Figure 4E). We next examined SOCS4 protein in vehicle- or MLN4924-treated T cells. Although SOCS4 expression was significantly higher in unstimulated MLN4924-treated T cells (similar to the microarray results), no differences in SOCS4 levels were observed between vehicle-treated and MLN4924-treated T cells after stimulation (Supplemental Figure S3F).
SOCS proteins are involved in the negative regulation of intracellular signaling after TCR and cytokine receptor ligation.34 Furthermore, specifically SOCS 1 and 3 proteins are known to recruit the Cul5/SAG complex (ie, known targets of neddylation), thereby regulating the ubiquitination of substrate proteins.34 SOCS 1 and 3 increase after T-cell stimulation and are subsequently degraded by ubiquitination. Therefore, we next examined the protein expression of SOCS 1 and 3 in T cells from KO animals and WT animals after stimulation with irradiated (30 Gy) allogeneic BALB/c whole splenocytes over the course of 4 days. Although the initial level of both SOCS1 (Figure 4F) and SOCS3 (Figure 4G) was greater in KO T cells, early after activation (up to 48 hours), the levels of both these proteins increased and was comparable between the KO and WT T cells. However, the level of SOCS 1 and 3 remained higher in the KO T cells at later time points (72 and 96 hours), indicating reduced degradation in the KO T cells when compared to WT T cells (Figure 4, F and G). The increased presence in SOCS1 (Figure 4H) was also confirmed in WT T cells treated with MLN4294. These data collectively suggest that the absence of SAG leads to sustained negative regulation of T cells through reduced degradation of negative signaling molecules SOCS 1 and 3.
Inhibition of Neddylation Mitigates T-Cell Responses in Vivo and Reduces GVHD
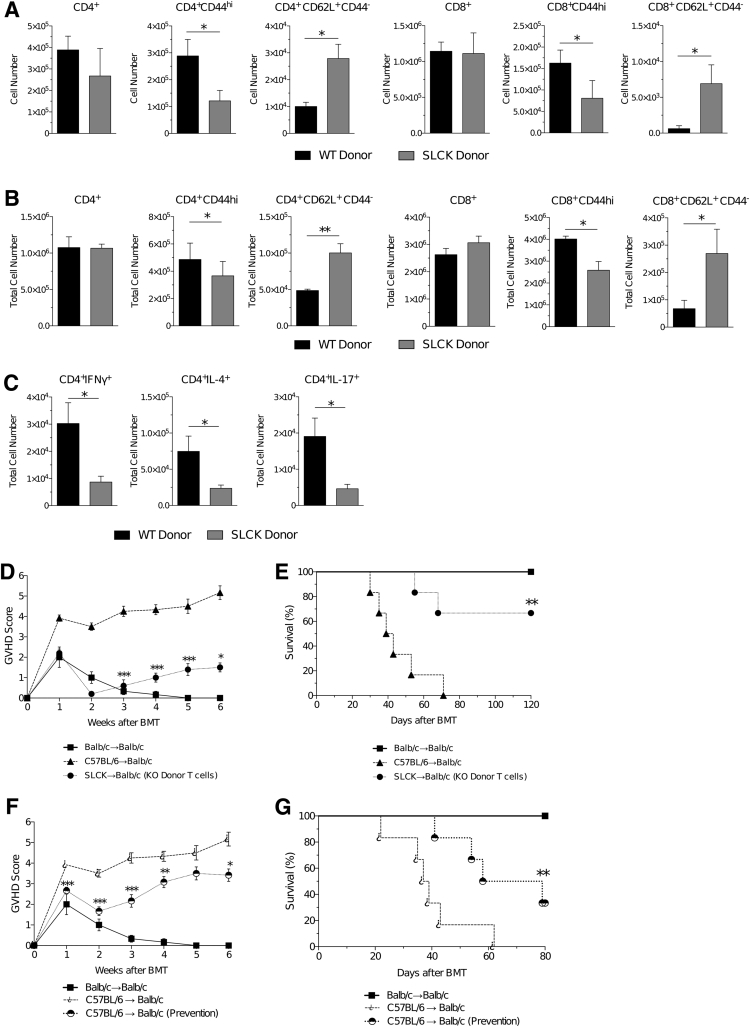
We next determined whether the functional defects in SAG-deficient T cells were also relevant after in vivo allogeneic stimulation. Specifically, to analyze whether SAG-deficient T cells proliferated to a lesser degree in vivo, we performed a clinically relevant major histocompatibility complex (MHC) mismatch allo-BMT in which C57BL/6 (H-2b) cells were transferred to BALB/c (H-2d) mice after lethal irradiation with 800 cGy. Syngeneic BALB/c to BALB/c transplant served as the controls. The recipients received 5 × 106 T-cell–depleted WT bone marrow and 0.5 × 106 CD90.2+ T cells from either conditional KO SAG-deficient mice or WT SAG-competent mice on day +0. We then analyzed the donor-cell immunophenotype in the recipients 7 and 14 days after transplant. On day +7 after transplant, we saw that there were significantly fewer activated donor T cells (CD4+CD44hi and CD8+CD44hi) but significantly greater unactivated naïve donor T cells (CD4+CD62L+CD44− and CD8+CD62L+CD44−) (Figure 5A). A similar immunophenotype was seen on day +14 after transplant (Figure 5B). We also examined the production of cytokines by donor T cells. We observed that similar to in vitro observations, the levels of Th1 (interferon-γ+ CD4+), Th2 (IL-4+ CD4+), and Th17 (IL-17+ CD4+) cytokine production were significantly decreased in the donor KO T cells (Figure 5C).
Figure 5.
SAG deficiency mitigates T-cell functions in vivo and reduces graft-versus-host disease (GVHD). Flow cytometric analysis of splenic donor (H-2b) wild-type (WT) and knockout (KO) T cells shown on day 7 (A) and day 14 (B) after C57BL/6 → BALB/c (H-2d) allogeneic transplant. Similar levels of CD4+ and CD8+ were observed at both time points with a significant decrease in activated (CD44hi) and significant increase in naïve (CD62L+ CD44−) T cells. C: Significantly fewer SAG-deficient (KO) CD4+ T cells produced interferon (IFN)-γ, IL-4, and IL-17 on day 14 after allogeneic bone marrow transplantation (BMT); determined by flow cytometry gating and the total number of splenocytes isolated on analysis. Recipients of allogeneic transplant that received KO donor T cells exhibit significantly less severe GVHD (D) and improved survival (E), compared to recipients of WT donor allogeneic (allo)-T cells. Treatment of allo-BMT recipients that received WT donor T cells with 20 mg/kg MLN4924 sub-q resulted in less severe GVHD (F) and significantly improved survival (G). Animals treated with MLN4924 were treated for 5 days starting day −1 through day +3, relative to transplant on day 0. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 versus control. SLCK, SAGfl/fl × Lck-Cre.
Next, we determined whether the changes in donor T-cell expansion led to biologically significant differences between the allogeneic BALB/c recipients of the B6 KO and WT T cells. The allogeneic recipients that received WT donor T cells exhibited signs of severe GVHD (Figure 5D) and greater mortality (Figure 5E) when compared with recipients of syngeneic transplant. By contrast, allo-BMT recipients that received donor KO T cells exhibited decreased signs of GVHD and significantly improved survival (Figure 5, D and E) when compared with recipients of WT donor T cells.
To test for potential clinical and therapeutic implications of these data, we next determined whether in vivo administration of MLN4924 may decrease T-cell proliferative responses and affect GVHD severity after allo-BMT. Therefore, we performed a similar MHC mismatch allo-BMT as above and transferred WT C57BL/6 T cells (0.5 × 106 CD90.2+) and whole bone marrow (5 × 106) to BALB/c mice after lethal irradiation of the recipients. We began injecting recipient mice with either vehicle or 20 mg/kg MLN4924 sub-q starting on day –1, and continued daily through day +3 after BMT. All recipients of syngeneic BMT survived, indicating that administration of MLN4924, at the doses noted above, was not toxic. More important, allo-BMT recipients treated with MLN4924 exhibited decreased GVHD clinical scores (Figure 5F) and increased survival (Figure 5G) when compared with allo-BMT recipients of the vehicle control. We found similar improved survival rates using a second clinical model of BMT, which used a partial MHC mismatched parent in an F1 model, in which C57BL/6 (H-2b) cells were transferred to B6D2F1 (H-2b/d) mice, thus demonstrating strain-independent results (Supplemental Figure S4). Taken together, these data suggest that the inhibition of neddylation in T cells may hold possible therapeutic relevance and could be used as a novel agent through which to mitigate GVHD in clinical settings.
Discussion
Alteration of existing proteins by PTMs is a key regulatory process. Neddylation has emerged as one of the key PTMs that regulates several cellular processes. Its role in regulation of DC and macrophage immune responses has been recently appreciated.1, 3 These data were in the context of global inhibition of neddylation by the small-molecule MLN49246. Herein, we demonstrate that SAG/Rbx2-dependent Cul-1 neddylation is a critical regulator of T-cell proliferative responses in vitro and in vivo. SAG deficiency causes a sustained increase in SOCS 1 and 3, key negative regulators of T-cell responses after their activation. Furthermore, targeting SAG mitigated severity of T-cell–mediated immune pathology, GVHD. All of these effects were also recapitulated by the more global inhibition of neddylation by MLN4924, suggesting that a more targeted SAG-dependent regulation of neddylation could potentially be a better approach.
Recently, Jin et al20 have shown that the global inhibition of all CRL complexes through knocking down of Ubc12 inhibits neddylation and regulates T-cell functions. Our data—which used the pharmacological small-molecule inhibitor of neddylation, MLN4924, and a more targeted genetic approach of SAG−/− T cells—confirm and extend these observations. Specifically, our use of MLN4924 revealed an intrinsic effect of neddylation in T cells, decreased the release of T-cell–specific cytokines, and exhibited a defect in T-cell proliferation. Furthermore, when T cells were stimulated unimpeded for 24 hours before the addition of MLN4924, proliferation was still dramatically halted by the addition of neddylation inhibitor. These data suggest that T-cell responses can be modulated via regulation of the neddylation pathway, even after onset of TCR stimulation.
However, the impacts that specific CRL complexes have on T-cell functions are, thus far, unknown. In contrast to global CRL inhibition, our results provide a novel perspective on the role that certain CRL complexes, which need SAG—CRL2 and CRL5—play in T-cell proliferative responses. Herein, we generated a novel KO mouse featuring conditional SAG protein deletion in T cells, thus enabling the selective inhibition of specific E3 ligases involved in the neddylation pathway. Allogeneic stimulation of SAG-deficient T cells also resulted in decreased proliferation, as well as in the diminished release of the T-cell lineage and the activation cytokines.
Previous studies have shown that the regulation conferred by neddylation in innate immune cell subsets, DCs and macrophages, is achieved through the prevention of IκB degradation, thus inhibiting NF-κB translocation to the nucleus.1, 18 However, on examination of NF-κB dynamics in T cells (using either MLN4924 or SAG-deficient T cells), we found that the NF-κB pathway was only minimally affected. This suggests that in contrast to innate immune cells, DCs and macrophages, a disparate pathway may be responsible for the diminished T-cell functions. We profiled the total mRNA expression and found that transcripts for SOCS proteins were increased in KO T cells when compared with WT SAG competent controls. SOCS play a critical role in negative regulation of T-cell responses after their activation.
SOCS 1 and 3 are known components of the CRL2 and CRL5 complexes12, 34 and are known to require neddylation to be degraded by the ubiquitination pathway. When T cells were stimulated, we found a greater and more sustained increase in the SOCS 1 and 3 proteins in neddylation-inhibited cells—by either MLN4924 or SAG deficiency. These results suggest that the increased presence of the SOCS 1 and 3 may be responsible for decreased T-cell functions after the inhibition of neddylation. SOCS4 levels were significantly different in unstimulated T cells, but not stimulated T cells, suggesting that SOCS4 and SOCS 1 and 3 proteins may have differential responses after T-cell receptor ligation, although this cannot be definitively ascertained from our data. Future studies will explore the specific nature and role of neddylation sites in SOCS 1 and 3 proteins and their role in regulation of T-cell responses. Nonetheless, these data suggest that the key molecular mechanisms through which neddylation regulates T-cell responses is not through NF-κB, unlike in DCs and macrophages.
T cells play a central role in many immune-mediated diseases, including GVHD.2, 5 On transfer of SAG-deficient T cells into MHC-mismatched recipients, we observed significantly less severe GVHD clinical scores and increased survival. These results confirmed our in vitro observations that SAG-deficient T cells have reduced responses after allostimulation. Using the same MHC-mismatched model, but transferring WT T cells instead, we treated recipient mice with MLN4924. We recapitulated the results of decreased GVHD scores and further improved survival after the transfer of KO T cells.
In conclusion, our novel finding that the in vivo regulation of specific neddylation E3 ligases—CRL2 and CRL5—plays a specific role in T-cell regulation provides novel insights into the understanding of a heretofore unknown mechanism through which neddylation confers regulatory effects on T-cell responses. Furthermore, our findings may have important clinical implications, and suggest that targeting neddylation with MLN4924 (currently in phase 1/2 clinical trials) may mitigate T-cell–mediated diseases, such as GVHD.
Acknowledgment
We thank the Microscopy & Image-Analysis Laboratory of the University of Michigan's Biomedical Research Core Facilities for preparation of the samples and images.
Footnotes
Supported by NIH National Heart, Lung, and Blood Institute grants HL-090775 and HL-128046 (P.R.) and National Cancer Institute grants CA-173878 and CA-171277 (P.R.). Support for the Microscopy and Image-Analysis Laboratory core is provided by the University of Michigan Cancer Center (NIH grant CA46592) and The University of Michigan Gut Peptide Research Center (NIH grant DK34933).
Disclosures: None declared.
Current address of N.D.M., Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.ajpath.2016.06.014.
Supplemental Data
Inhibition of neddylation decreases T-cell proliferative responses. BALB/c T cells were cultured with irradiated (30 Gy) C57BL/6 bone marrow–derived dendritic cells at 40:1 and 100:1 ratios in the presence or absence of MLN4924 for 96 hours. Where indicated, T cells were preincubated with MLN4924 at indicated dosage for 6 hours. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. CPM, count per minute; DMSO, dimethyl sulfoxide; Pre-Inc. TC, preincubated T-cells.
Inhibition of neddylation results in less activated T cells after stimulation. CD90.2+ T cells treated with vehicle (dimethyl sulfoxide) or 500 nM MLN4924 and stimulated with 5 μg/mL α-CD3 and 2.5 μg/mL α-CD28 for 72 hours, as indicated. Phenotypic analysis by flow cytometry of activated CD44hi T cells (column 1), naïve and effector T cells (column 2), and activated CD69+ T cells (column 3). MLN4924 was added either at the time of plating or 24 hours after T-cell stimulation commencement.
Naïve phenotype of T cells in SAG-deficient animals is not different. Total number of thymocytes (A), CD4+CD8+ T cells in the thymus (B), CD3+ T cells in the spleen and peripheral lymph node (C), and naïve (CD62L+ CD44−) T cells in the spleen (D) and peripheral lymph nodes (E) of wild-type (WT) and knockout (KO) animals. Data analyzed by flow cytometry. F: Representative immunoblot of SOCS4 protein in T cells unstimulated or stimulated for 96 hours and cultured with or without 500 nmol/L MLN4924. Densitometric analysis of immunoblots of unstimulated (Unstim) T cells or stimulated (Stim) for 48, 72, or 96 hours, treated with vehicle or MLN4924 as indicated. ∗P < 0.05.
Inhibition of neddylation with MLN4924 improves survival of allogeneic bone marrow transplantation (BMT). Recipients of allogeneic BMT were treated with vehicle or 20 mg/kg MLN4924 sub-q daily, for 5 days starting day −1 through day +3, relative to transplant on day 0. Significantly improved survival is observed in recipient animals of C3H.SW → C57BL/6 minor-antigen mismatch allogeneic BMT that were treated with MLN4924. ∗∗P < 0.01.
References
- 1.Mathewson N., Toubai T., Kapeles S., Sun Y., Oravecz-Wilson K., Tamaki H., Wang Y., Guoqing H., Sun Y., Reddy P. Neddylation plays an important role in the regulation of murine and human dendritic cell function. Blood. 2013;122:2062–2073. doi: 10.1182/blood-2013-02-486373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choi S., Reddy P. Graft-versus-host disease. Panminerva Med. 2010;52:111–124. [PubMed] [Google Scholar]
- 3.Chang F.M., Reyna S.M., Granados J.C., Wei S.J., Innis-Whitehouse W., Maffi S.K., Rodriguez E., Slaga T.J., Short J.D. Inhibition of neddylation represses lipopolysaccharide-induced proinflammatory cytokine production in macrophage cells. J Biol Chem. 2012;287:35756–35767. doi: 10.1074/jbc.M112.397703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferrara J., Antin J.H. Wiley; Malden, MA: 2009. The pathophysiology of graft-versus-host disease. Thomas' Hematopoietic Cell. [Google Scholar]
- 5.Choi S.W., Reddy P. Current and emerging strategies for the prevention of graft-versus-host disease. Nat Rev Clin Oncol. 2014;11:536–547. doi: 10.1038/nrclinonc.2014.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soucy T.A., Smith P.G., Milhollen M.A., Berger A.J., Gavin J.M., Adhikari S., Brownell J.E., Burke K.E., Cardin D.P., Critchley S., Cullis C.A., Doucette A., Garnsey J.J., Gaulin J.L., Gershman R.E., Lublinsky A.R., McDonald A., Mizutani H., Narayanan U., Olhava E.J., Peluso S., Rezaei M., Sintchak M.D., Talreja T., Thomas M.P., Traore T., Vyskocil S., Weatherhead G.S., Yu J., Zhang J., Dick L.R., Claiborne C.F., Rolfe M., Bolen J.B., Langston S.P. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y.C., Peterson S.E., Loring J.F. Protein post-translational modifications and regulation of pluripotency in human stem cells. Cell Res. 2014;24:143–160. doi: 10.1038/cr.2013.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei D., Li H., Yu J., Sebolt J.T., Zhao L., Lawrence T.S., Smith P.G., Morgan M.A., Sun Y. Radiosensitization of human pancreatic cancer cells by MLN4924, an investigational NEDD8-activating enzyme inhibitor. Cancer Res. 2012;72:282–293. doi: 10.1158/0008-5472.CAN-11-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emanuele M.J., Elia A.E.H., Xu Q., Thoma C.R., Izhar L., Leng Y., Guo A., Chen Y.N., Rush J., Hsu P.W.C., Yen H.C.S., Elledge S.J. Global identification of modular cullin-RING ligase substrates. Cell. 2011;147:459–474. doi: 10.1016/j.cell.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang D., Tan M., Wang G., Sun Y. The p21-dependent radiosensitization of human breast cancer cells by MLN4924, an investigational inhibitor of NEDD8 activating enzyme. PLoS One. 2012;7:e34079. doi: 10.1371/journal.pone.0034079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo Z., Yu G., Lee H.W., Li L., Wang L., Yang D., Pan Y., Ding C., Qian J., Wu L., Chu Y., Yi J., Wang X., Sun Y., Jeong L.S., Liu J., Jia L. The Nedd8-activating enzyme inhibitor MLN4924 induces autophagy and apoptosis to suppress liver cancer cell growth. Cancer Res. 2012;72:3360–3371. doi: 10.1158/0008-5472.CAN-12-0388. [DOI] [PubMed] [Google Scholar]
- 12.Enchev R.I., Schulman B.A., Peter M. Protein neddylation: beyond cullin-RING ligases. Nat Rev Mol Cell Biol. 2015;16:30–44. doi: 10.1038/nrm3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bevan M.J. Helping the CD8+ T-cell response. Nat Rev Immunol. 2004;4:595–602. doi: 10.1038/nri1413. [DOI] [PubMed] [Google Scholar]
- 14.Duan H., Wang Y., Aviram M., Swaroop M., Loo J.A., Bian J., Tian Y., Mueller T., Bisgaier C.L., Sun Y. SAG, a novel zinc RING finger protein that protects cells from apoptosis induced by redox agents. Mol Cell Biol. 1999;19:3145–3155. doi: 10.1128/mcb.19.4.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swaroop M., Bian J., Aviram M., Duan H., Bisgaier C.L., Loo J.A., Sun Y. Expression, purification, and biochemical characterization of SAG, a ring finger redox-sensitive protein. Free Radic Biol Med. 1999;27:193–202. doi: 10.1016/s0891-5849(99)00078-7. [DOI] [PubMed] [Google Scholar]
- 16.Jia L., Yang J., Hao X., Zheng M., He H., Xiong X., Xu L., Sun Y. Validation of SAG/RBX2/ROC2 E3 ubiquitin ligase as an anticancer and radiosensitizing target. Clin Cancer Res. 2010;16:814–824. doi: 10.1158/1078-0432.CCR-09-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan M., Davis S.W., Saunders T.L., Zhu Y., Sun Y. RBX1/ROC1 disruption results in early embryonic lethality due to proliferation failure, partially rescued by simultaneous loss of p27. Proc Natl Acad Sci U S A. 2009;106:6203–6208. doi: 10.1073/pnas.0812425106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan M., Zhu Y., Kovacev J., Zhao Y., Pan Z.Q. Disruption of Sag/Rbx2/Roc2 induces radiosensitization by increasing ROS levels and blocking NF-κB activation in mouse embryonic stem cells. Free Radic Biol Med. 2010;49:976–983. doi: 10.1016/j.freeradbiomed.2010.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li L., Liu B., Dong T., Lee H.W., Yu J., Zheng Y., Gao H., Zhang Y., Chu Y., Liu G., Niu W., Zheng S., Jeong L.S., Jia L. Neddylation pathway regulates the proliferation and survival of macrophages. Biochem Biophys Res Commun. 2013;432:494–498. doi: 10.1016/j.bbrc.2013.02.028. [DOI] [PubMed] [Google Scholar]
- 20.Jin H.S., Liao L., Park Y., Liu Y.C. Neddylation pathway regulates T-cell function by targeting an adaptor protein Shc and a protein kinase Erk signaling. Proc Natl Acad Sci U S A. 2013;110:624–629. doi: 10.1073/pnas.1213819110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho J.H., Dimri M., Dimri G.P. A positive feedback loop regulates the expression of polycomb group protein BMI1 via WNT signaling pathway. J Biol Chem. 2013;288:3406–3418. doi: 10.1074/jbc.M112.422931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baek S.T., Kerjan G., Bielas S.L., Lee J.E., Fenstermaker A.G., Novarino G., Gleeson J.G. Off-target effect of doublecortin family shRNA on neuronal migration associated with endogenous microRNA dysregulation. Neuron. 2014;82:1255–1262. doi: 10.1016/j.neuron.2014.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodrigues G.A., Falasca M., Zhang Z., Ong S.H., Schlessinger J. A novel positive feedback loop mediated by the docking protein Gab1 and phosphatidylinositol 3-kinase in epidermal growth factor receptor signaling. Mol Cell Biol. 2000;20:1448–1459. doi: 10.1128/mcb.20.4.1448-1459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gu S., Jin L., Zhang Y., Huang Y., Zhang F., Valdmanis P.N., Kay M.A. The loop position of shRNAs and pre-miRNAs is critical for the accuracy of dicer processing in vivo. Cell. 2012;151:900–911. doi: 10.1016/j.cell.2012.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh S., Narang A.S., Mahato R.I. Subcellular fate and off-target effects of siRNA, shRNA, and miRNA. Pharm Res. 2011;28:2996–3015. doi: 10.1007/s11095-011-0608-1. [DOI] [PubMed] [Google Scholar]
- 26.Li H., Tan M., Jia L., Wei D., Zhao Y., Chen G., Xu J., Zhao L., Thomas D., Beer D.G., Sun Y. Inactivation of SAG/RBX2 E3 ubiquitin ligase suppresses KrasG12D-driven lung tumorigenesis. J Clin Invest. 2014;124:835–846. doi: 10.1172/JCI70297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan M., Li H., Sun Y. Endothelial deletion of Sag/Rbx2/Roc2 E3 ubiquitin ligase causes embryonic lethality and blocks tumor angiogenesis. Oncogene. 2014;33:5211–5220. doi: 10.1038/onc.2013.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reddy P., Maeda Y., Liu C., Krijanovski O.I., Korngold R., Ferrara J.L.M. A crucial role for antigen-presenting cells and alloantigen expression in graft-versus-leukemia responses. Nat Med. 2005;11:1244–1249. doi: 10.1038/nm1309. [DOI] [PubMed] [Google Scholar]
- 29.Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 30.Mathew J.P., Taylor B.S., Bader G.D., Pyarajan S., Antoniotti M., Chinnaiyan A.M., Sander C., Burakoff S.J., Mishra B. From bytes to bedside: data integration and computational biology for translational cancer research. PLoS Comput Biol. 2007;3:e12. doi: 10.1371/journal.pcbi.0030012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., Houstis N., Daly M.J., Patterson N., Mesirov J.P., Golub T.R., Tamayo P., Spiegelman B., Lander E.S., Hirschhorn J.N., Altshuler D., Groop L.C. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 33.Paul S., Schaefer B.C. A new look at T cell receptor signaling to nuclear factor-κB. Trends Immunol. 2013;34:269–281. doi: 10.1016/j.it.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshimura A., Naka T., Kubo M. SOCS proteins, cytokine signalling and immune regulation. Nat Rev Immunol. 2007;7:454–465. doi: 10.1038/nri2093. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Inhibition of neddylation decreases T-cell proliferative responses. BALB/c T cells were cultured with irradiated (30 Gy) C57BL/6 bone marrow–derived dendritic cells at 40:1 and 100:1 ratios in the presence or absence of MLN4924 for 96 hours. Where indicated, T cells were preincubated with MLN4924 at indicated dosage for 6 hours. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. CPM, count per minute; DMSO, dimethyl sulfoxide; Pre-Inc. TC, preincubated T-cells.
Inhibition of neddylation results in less activated T cells after stimulation. CD90.2+ T cells treated with vehicle (dimethyl sulfoxide) or 500 nM MLN4924 and stimulated with 5 μg/mL α-CD3 and 2.5 μg/mL α-CD28 for 72 hours, as indicated. Phenotypic analysis by flow cytometry of activated CD44hi T cells (column 1), naïve and effector T cells (column 2), and activated CD69+ T cells (column 3). MLN4924 was added either at the time of plating or 24 hours after T-cell stimulation commencement.
Naïve phenotype of T cells in SAG-deficient animals is not different. Total number of thymocytes (A), CD4+CD8+ T cells in the thymus (B), CD3+ T cells in the spleen and peripheral lymph node (C), and naïve (CD62L+ CD44−) T cells in the spleen (D) and peripheral lymph nodes (E) of wild-type (WT) and knockout (KO) animals. Data analyzed by flow cytometry. F: Representative immunoblot of SOCS4 protein in T cells unstimulated or stimulated for 96 hours and cultured with or without 500 nmol/L MLN4924. Densitometric analysis of immunoblots of unstimulated (Unstim) T cells or stimulated (Stim) for 48, 72, or 96 hours, treated with vehicle or MLN4924 as indicated. ∗P < 0.05.
Inhibition of neddylation with MLN4924 improves survival of allogeneic bone marrow transplantation (BMT). Recipients of allogeneic BMT were treated with vehicle or 20 mg/kg MLN4924 sub-q daily, for 5 days starting day −1 through day +3, relative to transplant on day 0. Significantly improved survival is observed in recipient animals of C3H.SW → C57BL/6 minor-antigen mismatch allogeneic BMT that were treated with MLN4924. ∗∗P < 0.01.