Abstract
Human cellular apoptosis susceptibility (chromosomal segregation 1-like, CSE1L) gene plays a role in nuclear-to-cytoplasm transport and chromosome segregation during mitosis, cellular proliferation, and apoptosis. CSE1L is involved in colon carcinogenesis. CSE1L gene expression was assessed with three data sets using Affymetrix U133 + gene chips on normal human colonic mucosa (NR), adenomas (ADs), and colorectal carcinoma (CRC). CSE1L protein expression in CRC, AD, and NR from the same patients was measured by immunohistochemistry using a tissue microarray. We evaluated CSE1L expression in CRC cells (HCT116, SW480, and HT29) and its biological functions. CSE1L mRNA was significantly increased in all AD and CRC compared with NR (P < 0.001 and P = 0.02, respectivly). We observed a change in CSE1L staining intensity and cellular localization by immunohistochemistry. CSE1L was significantly increased during the transition from AD to CRC when compared with NR in a CRC tissue microarray (P = 0.01 and P < 0.001). HCT116, SW480, and HT29 cells also expressed CSE1L protein. CSE1L knockdown by shRNA inhibited protein, resulting in decreased cell proliferation, reduced colony formation in soft agar, and induction of apoptosis. CSE1L protein is expressed early and across all stages of CRC development. shRNA knockdown of CSE1L was associated with inhibition of tumorigenesis in CRC cells. CSE1L may represent a potential target for treatment of CRC.
Colorectal adenocarcinoma accounts for approximately 10% of all cancer-related deaths in the United States.1 Colorectal oncogenesis follows the classic dysplasia-adenoma-carcinoma sequence as genetic mutations accumulate in the colonic epithelium,2 and, as malignant tumors progress, they associate with nodal and distant metastatic disease. Most deaths occur in patients with late-stage tumors, which are usually incurable.3, 4 Therefore, the discovery of biomarkers for early detection and of potential targets for colorectal prevention is an important research priority.
Human chromosomal segregation 1-like (CSE1L) gene, alias human cellular apoptosis susceptibility and exportin-2 gene, is located on chromosome 20q13. CSE1L is implicated in the regulation of multiple cellular mechanisms, including the mitotic spindle checkpoint, as well as proliferation and apoptosis, two opposing processes that are tightly linked in mammalian cells.5 The CSE1L gene consists of 25 exons and encodes a protein of 100-kDa molecular weight. This protein is usually found in the cytoplasm and nuclei of cells. It functions as a key component in cytoplasmic-to-nuclear transport and is in charge of the translocation of importin-α from the nucleus to the cytoplasm to be recycled and used again in the nuclear protein import pathway.6 Defects in CSE1L lead to abnormal chromosome segregation, as suggested by the observation that the locus where this gene resides is a region frequently harboring amplifications, aneuploidies, or aberrant chromosomes that are frequently observed in cancer cells.7 In addition, CSE1L has been reported to be highly expressed in a variety of cancer types and to positively correlate with tumor grade and worse outcome.5, 8
CSE1L has also been shown to have a role in apoptosis, with studies showing that increased CSE1L protein expression facilitates apoptosis induced by cypermethrin9 and chemotherapeutic agents.10 It has been observed that, although CSE1L protein enhances the apoptotic effect of doxorubicin, 5-fluorouracil, cisplatin, and 4OH-tamoxifen, the same protein decreases apoptosis induced by paclitaxel in colon cancer tumor cells.10
CSE1L mRNA expression in colorectal cancer shows a significant correlation with 20q DNA copy number status. CSE1L protein expression is not associated with 20q gain.11 The expression of CSE1L mRNA and protein in human colorectal cancer resection specimens and its role in the progression of colorectal cancer are currently being investigated. Zhu et al5 examined the expression levels of CSE1L mRNA using semiquantitative RT-PCR. CSE1L expression in the human colon cancer cell line RKO was knocked down using a lentivirus-mediated siRNA. The inhibition of expression in RKO cells caused cell cycle arrest and decreased cell colony formation and apoptosis.
The effect of CSE1L on the viability, adhesion, and migration of colorectal cancer cell lines HCT116 and SW480 has been previously reported. It was shown that CSE1L depletion via siRNA knockdown significantly reduced the adhesive capacity of HCT116 and SW480, decreasing their proliferation and inducing apoptosis.12
In this study, we assessed the expression of CSE1L at the mRNA and protein level in a subset of human colorectal cancers, as compared to adenomas and normal mucosa. The CSE1L mRNA expression levels were evaluated against three publicly available data sets (accession numbers GSE4183, GSE8671, and GSE14333) retrieved from the Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo). We show evidence that CSE1L protein expression increases significantly in the early stages of colorectal cancer progression. We also observed a shift in CSE1L cellular localization during the transition from adenomas to carcinomas. Moreover, we evaluated the functional role of this protein in select colorectal cancer cell lines (HCT115, HT29, and SW480) through the knockdown of its expression by shRNA.
Materials and Methods
This study was approved by the Institutional Review Board at the H. Lee Moffitt Cancer Center and Research Institute (Tampa, FL).
Selection of Human Tissues
Using human colorectal cancer tissue microarrays (TMAs) (prepared at the Moffitt Cancer Center Tissue Core Facility), 150 tissue samples were analyzed for CSE1L expression by immunohistochemistry. The tissue samples included 88 (59%) colorectal cancers (CRCs), 40 (27%) histologically normal colonic mucosa (NR), and 22 (15%) adenomas (ADs). The diagnosis of the tissues included on the TMA was confirmed by a senior pathologist (D.C.) with experience in gastrointestinal pathology. The tumors were staged using the Astler-Coller modification of the Duke's classification.
Tumors occurring in patients with genetic cancer syndromes as well as arising in the background of inflammatory bowel disease were excluded from the study. The NR samples were taken near the resected colorectal margin, away from the tumor site, from the CRC colon resection specimens included in this study.
TMA Design
The colon 1 TMA was generated at the Moffitt Cancer Center Tissue Core and for this study it contained core samples from 40 histologically normal colon, 22 colonic adenomas, 88 colon adenocarcinoma, and 26 human colon cancer cell lines. The hematoxylin and eosin slides corresponding to the samples were first examined by two pathologists (K.G.N. and D.C.) to confirm the type of tissue (normal colon, adenoma, or adenocarcinoma) and to outline with a permanent marker the tissue area to be sampled for the TMA. For the TMA construction, 1 mm in diameter cores were sampled from the original formalin-fixed, paraffin-embedded tissue block, with cores taken at each sampling from the area previously outlined by the pathologists. These cores were then implanted into the recipient TMA using a Tissue Arrayer (Beecher Instruments, Inc., Sun Prairie, MI) to produce the colon TMA block. Tissue sections from this TMA were then subjected to the CSE1L immunostain. The colon 1 TMA data, including TMA maps and raw scores, are provided in Supplemental Table S1 (details of tissues included), Supplemental Table S2 (map), and Supplemental Table S3 (raw immunohistochemistry scores).
Immunohistochemistry
The tissues were stained for CSE1L using a rabbit polyclonal antibody (4°C overnight; dilution, 1:300; Spring Bioscience, Pleasanton, CA). The CSE1L-stained TMAs were examined by two independent observers (D.C. and E.H.-J.) experienced in immunohistochemical methods, and a consensus score was reached for each sample. CSE1L was scored into four grades, according to the intensity: 0, 1+, 2+, and 3+. The percentage of CSE1L-positive cells was also scored into four categories: 0 (0%), 1 (1% to 33%), 2 (34% to 66%), and 3 (67% to 100%). The product of the intensity by percentage scores was used as the final score and was classified as follows: 0 indicates negative; 1 to 3, weak; 4 to 6, moderate; and 7 to 9, strong.
Statistical Analysis
Descriptive statistics for the scores were generated and reported for each tissue group. The initial method used to compare CSE1L expression in CRC and NR was the one-way analysis of variance. Tukey's honestly significant difference method was used to adjust P value for pair-wise comparison. Spearman correlation was used to test any correlation between CSE1L score and tumor grade and stage. A Cox proportional hazard model was used to test whether CSE1L was associated with overall survival and disease-free survival. All statistical analyses were two-sided and considered significant at P < 0.05.
mRNA Microarray Analysis
To evaluate whether the variation in CSE1L protein expression between NR and CRC reflected a corresponding modulation of CSE1L mRNA, gene expression was tested in three publicly available data sets arrayed on the Affymetrix HG-U133 + 2.0 GeneChip. These data sets are described below and include GEO data sets GSE8671, GSE4183, and GSE14333. The probe set 201112_s_at was selected as the best representative on the basis of transcript-level sequence matching to the National Center for Biotechnology Information Reference Sequence13 NM_001316 using the IGB tool.14 The R statistical software was used for expression analysis (freely available open source statistical package: www.r-project.org, last accessed September 2010). GSE418315, 16 involves a comparison of NR, AD, and CRC and tissue from patients with irritable bowel syndrome from biopsies. These data were MAS5.0 normalized and scaled to 500, and the CSE1L probe set (201112_s_at) was tested for differences in expression (NR versus AD; and NR versus CRC). The Mann-Whitney test was used. Expression plots are displayed after log2 transformation as mean and SEM. GSE8671 is a comparison of matched normal colon mucosa and adenomas.17 The expression data were MAS5.0 normalized, scaled to 500, and log2 transformed. Statistically significant differences in CSE1L expression were tested using a Wilcoxon signed-rank test for paired samples. The fold-changes between adenoma and normal colon mucosa were represented in bar graphs. GSE1433318 is a study of 290 primary colorectal cancer tumors of different stages. The Moffitt subset of these cases (n = 205) was used together with 10 normal colon mucosa samples from patients treated at the Moffitt Cancer Center under a University of South Florida Institutional Review Board–approved protocol. The expression data were MAS5.0 normalized, scaled to 500, and log2 transformed. Differences were tested between normal and tumor (all stages). Data were visualized using mean and SEM.
Cellular Studies
Cell Culture and shRNA Treatment
Human colon cancer cells HCT116, SW480, and HT29 were acquired from ATCC (Manassas, VA) and grown in Dulbecco's modified Eagle's medium and McCoy 5A media with 10% fetal bovine serum, 50 IU/mL penicillin, and 50 mg/mL streptomycin. The cells were cultured at 37°C in a humidified atmosphere of 5% CO2. HCT116, HT29, and SW480 cells in OPTI-MEM medium (Life Technologies, Carlsbad, CA) were seeded in 6-well plates and cultured at 37°C in a humidified atmosphere of 5% CO2 to get 70% confluence. Then, cells were treated with scrambled shRNA (SC-108080), CSE1L shRNA (SC-29908-v; Santa Cruz Biotechnology, Dallas, TX) using Polybrene reagent (SC-134220), and incubated at 37°C in a humidified atmosphere of 5% CO2 for 48 hours. After 48 hours, the cells were harvested and used for proliferation, colony formation, and apoptosis assays.
Proliferation Assay
Cells transfected with CSE1L shRNA, scrambled shRNA, and reagent were seeded in 96-well plates at a density of 3000 cells per well in Dulbecco's modified Eagle's medium and McCoy 5A media and allowed to grow for 72 hours at 37°C in a humidified atmosphere of 5% CO2. After 72 hours, media were aspirated and replaced with 20 μL of 1 mg/mL MTT and incubated for 2 to 4 hours at 37°C in a humidified atmosphere of 5% CO2. Media were aspirated, 200 μL of dimethyl sulfoxide was added to each well, plates were incubated for 5 minutes with shaking, and absorbance was read at 540 nm.
Soft Agar Colony Formation Assay
A standard soft agar colony formation assay was performed in CSE1L shRNA, scrambled shRNA, and reagent-treated cells. The cells were seeded at a density of 5000 per well in a 12-well plate in 0.3% agar over a 0.6% bottom agar layer. Colonies were fed with growth media and the drugs (5 × 10−5 mol/L), and growth of colony formation was observed for 10 to 14 days. Colonies were photographed after overnight incubation with 1 mg/mL MTT in the wells. The colonies were counted under stereomicroscope. The experiments were performed at least twice, each in triplicate.
Apoptosis Assay
The cytoplasmic DNA fragments and histones were detected in the cells treated with CSE1L shRNA, scrambled shRNA, and reagent using Cell Death Detection enzyme-linked immunosorbent assay kit (Roche Diagnostic, Indianapolis, IN), as per the manufacturer's instructions. Standard soft agar colony formation assays were performed in HCT116, SW480, and HT29 cells, and the cells were treated with scrambled or CSE1L shRNA. The cells were seeded at a density of 5000 per well in a 12-well plate in 0.3% agar over a 0.6% bottom agar layer. Colonies were fed with 10% enriched media and were observed for 14 days. Colonies were photographed after overnight incubation with 1 mg/mL MTT in the wells. The colonies were counted under stereomicroscope and compared with control. The experiments were performed at least twice, each in triplicate.
Western Blot Analysis
Proteins were extracted from cells using radioimmunoprecipitation assay buffer with protease inhibitor. Total protein (40 μg) was resolved on 12.5% SDS-PAGE running gel and a 5% stacking gel. Proteins were then electrotransferred onto nitrocellulose membranes. After blocking in 5% nonfat powdered milk for 1 hour, the membranes were washed and then treated with antibodies to CSE1L (CAS) and β-actin (1:1000 and 1:5000, respectively) overnight at 4°C (Santa Cruz Biotechnology, Santa Cruz, CA). After washing, blots were incubated with horseradish peroxidase–conjugated secondary antibody IgG (1:5000) for 1 hour at room temperature. The washed blot was then treated with SuperSignal West Pico chemiluminescent substrate (Pierce Biotechnology, Rockford, IL) for positive antibody reaction. Membranes were exposed to X-ray film (Eastman Kodak Company, Rochester, NY) for visualization of protein bands.
Results
Clinical Pathologic Findings
Patients with colorectal cancer (n = 88) were an average age of 64 years (range, 24 to 92 years). Forty-eight patients (54%) were male. Tumors ranged from 1.4 to 14.5 cm. Most tumors had a polypoid and ulcerated appearance. To correlate CSE1L expression with tumor grade, we used a three-tier grading system. Six tumors (7%) were well differentiated, 67 (76%) moderately differentiated, and 15 (17%) poorly differentiated (Table 1). Three (3%) colorectal cancers were Duke's stage A, 34 (39%) Duke's stage B, 35 (40%) Duke's stage C, and 16 (18%) Duke's stage D. Only two patients, both with rectal cancer, received preoperative radiation to reduce the size of their tumors.
Table 1.
Baseline Patient Characteristics (n = 88)
| Characteristic | Value |
|---|---|
| Age (years) | |
| Range | 24-92 |
| Mean | 64 |
| Sex | |
| Male | 48 |
| Female | 40 |
| Tumor size (cm) | |
| Range | 1.4-14.5 |
| Mean | 5.2 |
| Tumor grade | |
| LG | 73 |
| HG | 15 |
| Pathological stage | |
| I | 3 |
| II | 32 |
| III | 37 |
| IV | 16 |
HG, high-grade dysplasia; LG, low-grade dysplasia.
mRNA Expression of CSE1L in Colorectal Oncogenesis
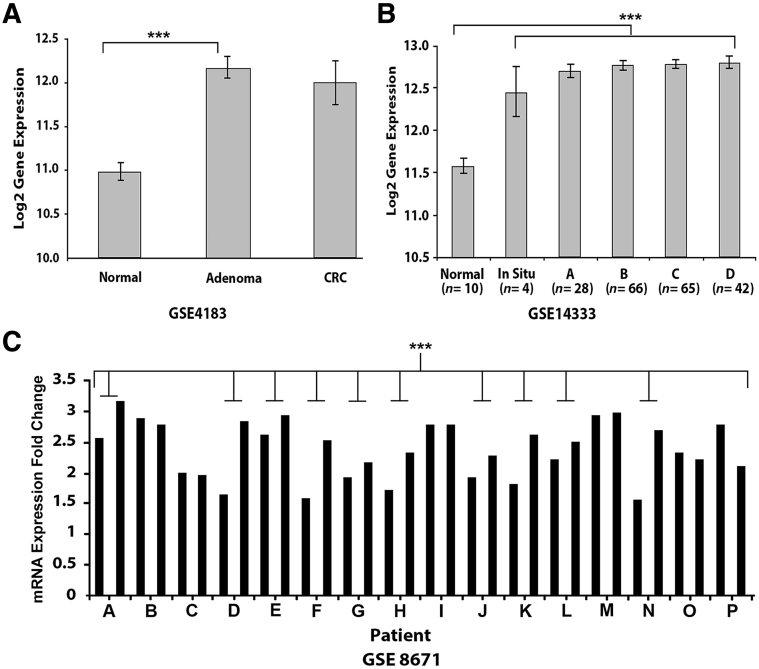
To determine whether CSE1L changes in protein level during human colorectal cancer development reflect changes in gene expression, we compared CSE1L mRNA levels by gene expression microarray in three publicly available data sets (GEO DataSets GSE4183, GSE14333, and GSE8671) taken from GEO (http://www.ncbi.nlm.nih.gov/gds). In GSE4183, the expression of CSE1L mRNA increased significantly between NR and AD (P < 0.001) (Figure 1A) and NR and CRC (P = 0.02) (Figure 1A). The same increase in CSE1L mRNA from NR compared to all stages of CRC was seen in GSE14333 (P < 0.001) (Figure 1B). Using paired samples from GSE8671 (Figure 1C), it was shown that adenomas consistently overexpress CSE1L (P < 0.001).
Figure 1.
mRNA expression analysis of CSE1L in colorectal carcinoma (CRC) tissues. A: CSE1L mRNA expression increases approximately twofold between normal human colonic mucosa (NR) and adenomas (ADs) or NR and CRC (P = 0.02). B: CSE1L mRNA expression increases from NR and all stages of CRC (Astler-Coller stages A, B, C, and D) in a large cohort of CRC patients. C: CSE1L mRNA expression is significantly up-regulated in NR/AD pairs in different patients. ∗∗∗P < 0.001.
CSE1L Protein Expression in Colorectal Cancer Progression and Patient Survival
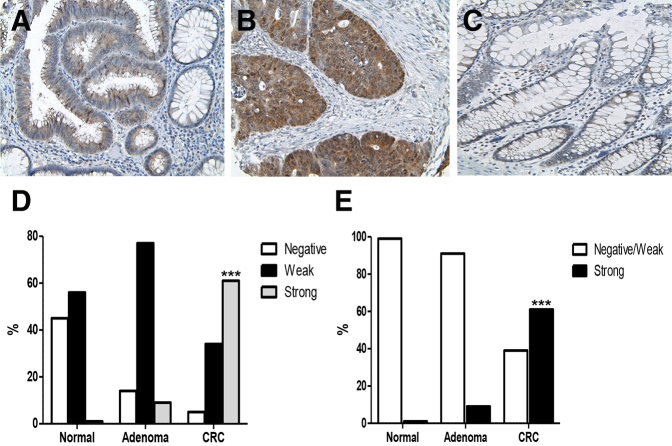
The CSE1L staining was localized to the cytoplasm in ADs, whereas CRCs showed both cytoplasmic and nuclear staining (Figure 2, A–C). In CRC specimens, approximately 34% (30/88) had weak CSE1L staining, 61% (54/88) exhibited moderate to strong CSE1L staining, and 5% (4/88) were CSE1L negative. In contrast, in NR samples, 45% (18/40) were CSE1L negative and 55% (22/40) had weak CSE1L staining. In the AD samples, 77% (17/22) showed weak CSE1L staining, 9% (2/22) were CSE1L moderate to strongly positive, and 14% (3/22) were CSE1L negative. There was a statistically significant difference found in CSE1L staining score between NR and CRC (P < 0.001), between NR and AD (P = 0.01), and between AD and CRC (P < 0.001) (Figure 2, D and E). There was no statistically significant trend between CSE1L score and tumor grade (P = 0.77) or stage (P = 0.25). No statistically significant association was found between CSE1L score and the patients' overall survival (P = 0.99) or disease-free survival (P = 0.66) using the Cox proportional hazards model.
Figure 2.
CSE1L expression in normal, adenoma, and colorectal carcinoma (CRC). A–C: CSE1L expression is induced in adenomas (A) and colorectal carcinoma (B), but not in normal colon epithelial cells (C). D: Quantification of CSE1L expression using the Allred scoring system shows a significantly increased staining score between normal and CRC and between normal and adenoma (P = 0.01). E: Quantification of CSE1L expression is also significantly increased between adenoma and CRC. ∗∗∗P < 0.001. Original magnification: ×100 (A); ×200 (B and C).
CSE1L Knockdown Inhibits Proliferation and Colony Formation and Induces Apoptosis
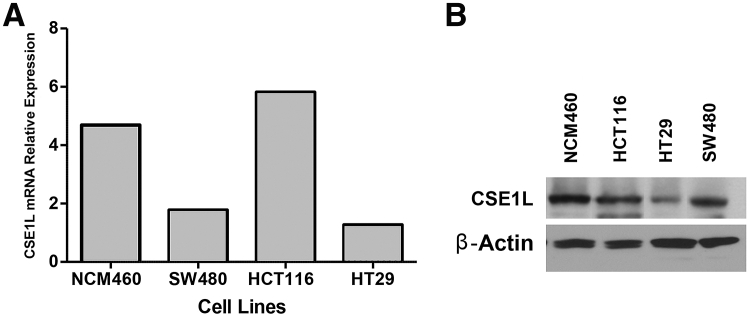
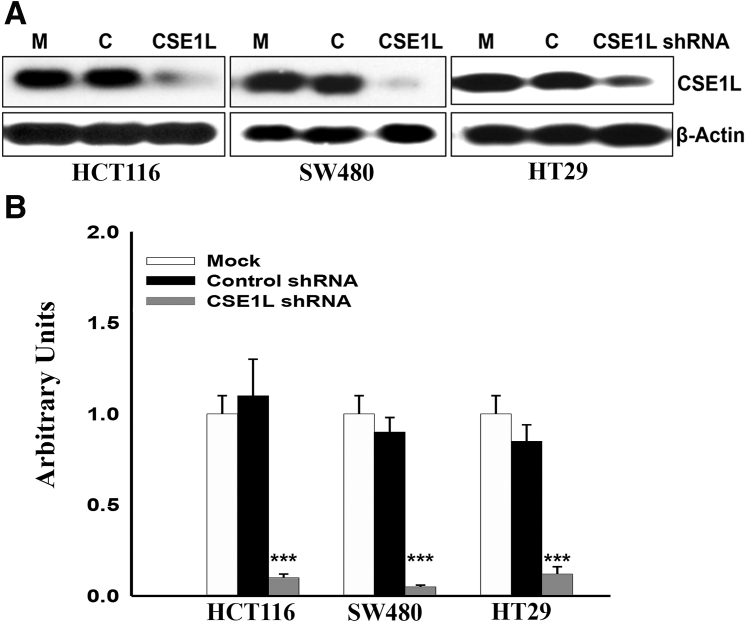
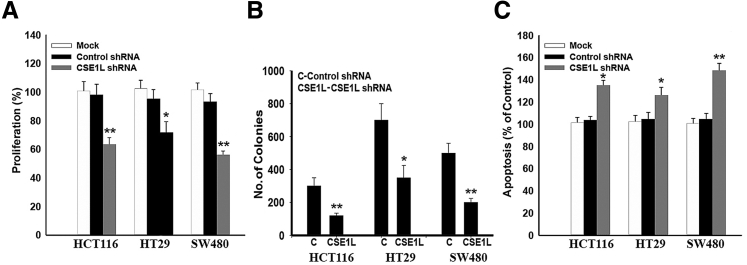
Both CSE1L mRNA and protein were expressed at baseline in multiple colorectal cancer cell lines (HCT116, SW480, and HT29) (Figure 3, A and B). Using CSE1L shRNA, we were able to significantly inhibit the protein expression in these cell lines and evaluated the functional implication of this inhibition. The percentage of CSE1L protein inhibition in HCT116 was 90% knockdown; SW480, 94% knockdown; and HT-29, 86% knockdown (Figure 4A). Densitometry analysis of protein bands in arbitrary units was significant in all three cell lines after CSE1L shRNA knockdown (P < 0.001) (Figure 4B). Proliferation was significantly inhibited in HCT116, HT29, and SW480 cells (Figure 5A). Down-regulation of CSE1L was also associated with inhibition of colony formation in all three cell lines (P < 0.01, P < 0.05, and P < 0.01) (Figure 5B). Apoptosis, after inhibition of CSE1L, was induced, as measured by cell death enzyme-linked immunosorbent assay, in HCT116, HT29, and SW480 cell lines (Figure 5C) compared with scramble shRNA (P < 0.05, P < 0.05, and P < 0.01, respectively).
Figure 3.
CSE1L baseline expression in NCM460, SW480, HCT116, and HT29 colorectal cancer cell lines. A: CSE1L mRNA is expressed at baseline in all four cell lines. The relative mRNA expression for NCM460 and HCT116 is higher (greater than four) than HT29 and SW480 (less than two) cell lines. B: Western blot analysis of CSE1L baseline protein expression shows strong expression in NCM460, HCT116, and SW480 and lower expression in the HT29 cell line. β-Actin was used as a loading control.
Figure 4.
Knockdown of protein expression in HCT116, SW480, and HT29 colorectal cell lines. A: Western blot assessment of CSE1L shRNA (SC-29908-v) knockdown of protein expression in HCT116, SW480, and HT29 cell lines compared to mock (M) and scramble shRNA (C). β-Actin was used as a loading control. There is inhibition of CSE1L protein expression when cells are treated with shRNA compared to the mock and scramble shRNA. B: The percentage of inhibition: HCT116 (90% knockdown), SW480 (94% knockdown), and HT29 (86% knockdown), respectively. Densitometry analysis of protein bands (arbitrary units) is significant in all three cell lines after CSE1L shRNA knockdown. ∗∗∗P < 0.001.
Figure 5.
CSE1L shRNA interference in HCT116, HT29, and SW480 cell line proliferation and survival. A: CSE1L shRNA interference in human colorectal cancer cells leads to significant inhibition of proliferation. B: Cell colonies treated with CSE1L shRNA show significant reduction in anchorage-independent growth versus colonies treated with control shRNA. C: shRNA causes significant induction of apoptosis, as measured by death cell enzyme-linked immunosorbent assay in HCT116, HT29, and SW480 cell lines compared to the mock and scramble shRNA groups. ∗P < 0.05, ∗∗P < 0.01.
Discussion
CSE1L expression is necessary for accurate alignment of chromosomes, which assures genomic stability during cell division. Therefore, aberrant CSE1L expression may affect genomic stability, a feature that is lost in cancer. Previous studies of CSE1L protein expression in colorectal cancer have also been investigated in selected cell lines.19 In this study, we show mRNA and protein expression of CSE1L in human colorectal tissues using multiple data sets and multiple cell lines. Interestingly, when comparing the progression from normal mucosa to adenomas to colorectal cancers, we noticed a significant increase in CSE1L expression at both the mRNA and protein level during the transition from normal mucosae to adenoma. High expression of CSE1L protein was also noted in selected human colon cancer cell lines (HCT116, HT29, and SW480). These results are in agreement with prior studies12, 19 and are consistent with the notion that CSE1L protein expression may play a role in the early stages of colorectal tumorigenesis. Interestingly, we observed a change in CSE1L staining intensity and cellular localization during the transition from adenoma to colorectal cancer.
Our observation of concomitant nuclear CSE1L stain in colorectal cancers may reflect the CSE1L function as a transport factor. It is known that the N-terminal domain of CSE1L is homologous to the Ran-binding domain in β-importin.20, 21 Furthermore, Kutay et al22 have shown that CSE1L is necessary for recycling of importin-α from the nucleus to the cytosol. Importin recycling is required for nuclear transport of a variety of proteins, including those necessary for mitosis or apoptosis.23 Seiden-Long et al24 studied a human colorectal cancer TMA containing samples from normal colonic mucosa, primary, and metastatic colorectal tumors; adenomas were not included. Similar to our study, the investigators reported nuclear and cytoplasmic distribution of the stain in the tumors, but the authors did not specify whether such localization was seen in the normal and adenomatous samples as well.
Tai et al25 previously reported that high cytoplasmic staining of CSE1L in a CRC tumor was associated with advanced cancer stage and depth of tumor penetration. There was no relationship between nuclear CSE1L staining and clinical manifestations, including tumor stage.
In our study, we found no correlation between CSE1L expression with tumor grade and stage. This lack of correlation may be because of the fact that our TMA contained approximately 67% of moderately differentiated and only 7% of well-differentiated tumor samples. Similarly, the TMA had only 16 Duke's stage D primary CRC and no metastatic CRC samples. This did not allow appropriate correlation with tumor grade and stage.
CSE1L expression has been previously studied in metastatic tumors. A significant reduction of CSE1L levels has been observed when comparing primary to metastatic tumors.12 In primary CRC, it has been shown that depletion of CSE1L expression significantly reduced the adhesive capacity, while increasing migration and invasion of HCT116 and SW480 CRC tumor cell lines compared with negative controls.12 It has been shown that the COOH-terminal domain of CSE1L interacts with metalloproteinases-2–containing vesicles.26 Interestingly, these authors demonstrated an increased level of CSE1L in the sera of patients with metastatic colorectal cancer.26 E-cadherin mRNA levels and intercellular adhesion molecule-1 were significantly reduced after siRNA knockdown, potentially increasing tumor invasion and metastasis. In contrast, there was a sevenfold to eightfold increase in matrix metalloproteinase-9 levels in HCT116 and SW480, respectively.12 These findings are significant in light of previous studies showing that CRC tumors expressing high cytoplasmic CSE1L were associated with an overall poor patient survival rate.25
Our study was designed to test the changes in CSE1L mRNA and protein expression during the progression from normal colonic mucosa to carcinoma using human CRC gene expression profiling and human colorectal tissue samples.
We observed a statistically significant increase in CSE1L expression during the progression of colorectal cancer. The results of our in vitro experiments are in agreement with previous studies.5, 12, 27 The knockdown of CSE1L using shRNA on multiple colorectal cancer cell lines was associated with inhibition of cell proliferation and cell growth in soft agar and in induction of apoptosis.
Our findings provide additional support for CSE1L as a potential target for treatment of CRC.
Acknowledgments
We thank the Histology Section of the Tissue Core at the Moffitt Cancer Center and Research Institute for the support in performing the immunohistochemical stains, and Rasa Hamilton (Moffitt Cancer Center) for editorial assistance.
Footnotes
Supported in part by NIH grant 1R01 CA-129227-01A1 and Bankhead Coley grant 08BR-02 (M.P.M.). The Tissue Core Facility at the H. Lee Moffitt Cancer Center and Research Institute and National Cancer Institute designated Comprehensive Cancer Center is supported under NIH grant P30-CA76292.
J.M.P. and K.G.N. contributed equally to this work.
Disclosures: None declared.
Current address of D.S., Department of Surgery, University of Tennessee Health Sciences Center, Memphis, TN.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.ajpath.2016.06.016.
Supplemental Data
References
- 1.Siegel R., Naishadham D., Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Morson B. President's address: the polyp-cancer sequence in the large bowel. Proc R Soc Med. 1974;67:451–457. doi: 10.1177/00359157740676P115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kountouras J., Boura P., Lygidakis N.J. New concepts of molecular biology for colon carcinogenesis. Hepatogastroenterology. 2000;47:1291–1297. [PubMed] [Google Scholar]
- 4.Temple L.K., Hsieh L., Wong W.D., Saltz L., Schrag D. Use of surgery among elderly patients with stage IV colorectal cancer. J Clin Oncol. 2004;22:3475–3484. doi: 10.1200/JCO.2004.10.218. [DOI] [PubMed] [Google Scholar]
- 5.Zhu J.H., Hong D.F., Song Y.M., Sun L.F., Wang Z.F., Wang J.W. Suppression of cellular apoptosis susceptibility (CSE1L) inhibits proliferation and induces apoptosis in colorectal cancer cells. Asian Pac J Cancer Prev. 2013;14:1017–1021. doi: 10.7314/apjcp.2013.14.2.1017. [DOI] [PubMed] [Google Scholar]
- 6.Kohler M., Speck C., Christiansen M., Bischoff F.R., Prehn S., Haller H., Gorlich D., Hartmann E. Evidence for distinct substrate specificities of importin alpha family members in nuclear protein import. Mol Cell Biol. 1999;19:7782–7791. doi: 10.1128/mcb.19.11.7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brinkmann U., Brinkmann E., Gallo M., Pastan I. Cloning and characterization of a cellular apoptosis susceptibility gene, the human homologue to the yeast chromosome segregation gene CSE1. Proc Natl Acad Sci U S A. 1995;92:10427–10431. doi: 10.1073/pnas.92.22.10427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aust D.E., Muders M., Kohler A., Schmidt M., Diebold J., Muller C., Lohrs U., Waldman F.M., Baretton G.B. Prognostic relevance of 20q13 gains in sporadic colorectal cancers: a FISH analysis. Scand J Gastroenterol. 2004;39:766–772. doi: 10.1080/00365520410003191. [DOI] [PubMed] [Google Scholar]
- 9.Izaguirre M.F., Vergara M.N., Casco V.H. CAS role in the brain apoptosis of Bufo arenarum induced by cypermethrin. Biocell. 2006;30:309–320. [PubMed] [Google Scholar]
- 10.Liao C.F., Luo S.F., Shen T.Y., Lin C.H., Chien J.T., Du S.Y., Jiang M.C. CSE1L/CAS, a microtubule-associated protein, inhibits taxol (paclitaxel)-induced apoptosis but enhances cancer cell apoptosis induced by various chemotherapeutic drugs. BMB Rep. 2008;41:210–216. doi: 10.5483/bmbrep.2008.41.3.210. [DOI] [PubMed] [Google Scholar]
- 11.Sillars-Hardebol A.H., Carvalho B., Belien J.A., de Wit M., Delis-van Diemen P.M., Tijssen M., van de Wiel M.A., Ponten F., Meijer G.A., Fijneman R.J. CSE1L, DIDO1 and RBM39 in colorectal adenoma to carcinoma progression. Cell Oncol (Dordr) 2012;35:293–300. doi: 10.1007/s13402-012-0088-2. [DOI] [PubMed] [Google Scholar]
- 12.Alnabulsi A., Agouni A., Mitra S., Garcia-Murillas I., Carpenter B., Bird S., Murray G.I. Cellular apoptosis susceptibility (chromosome segregation 1-like, CSE1L) gene is a key regulator of apoptosis, migration and invasion in colorectal cancer. J Pathol. 2012;228:471–481. doi: 10.1002/path.4031. [DOI] [PubMed] [Google Scholar]
- 13.Pruitt K.D., Tatusova T., Maglott D.R. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. Epub 2006 Nov 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nicol J.W., Helt G.A., Blanchard S.G., Jr., Raja A., Loraine A.E. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics. 2009;25:2730–2731. doi: 10.1093/bioinformatics/btp472. Epub 2009 Aug 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galamb O., Sipos F., Solymosi N., Spisak S., Krenacs T., Toth K., Tulassay Z., Molnar B. Diagnostic mRNA expression patterns of inflamed, benign, and malignant colorectal biopsy specimen and their correlation with peripheral blood results. Cancer Epidemiol Biomarkers Prev. 2008;17:2835–2845. doi: 10.1158/1055-9965.EPI-08-0231. [DOI] [PubMed] [Google Scholar]
- 16.Galamb O., Spisak S., Sipos F., Toth K., Solymosi N., Wichmann B., Krenacs T., Valcz G., Tulassay Z., Molnar B. Reversal of gene expression changes in the colorectal normal-adenoma pathway by NS398 selective COX2 inhibitor. Br J Cancer. 2010;102:765–773. doi: 10.1038/sj.bjc.6605515. Epub 2010 Jan 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sabates-Bellver J., Van der Flier L.G., de Palo M., Cattaneo E., Maake C., Rehrauer H., Laczko E., Kurowski M.A., Bujnicki J.M., Menigatti M., Luz J., Ranalli T.V., Gomes V., Pastorelli A., Faggiani R., Anti M., Jiricny J., Clevers H., Marra G. Transcriptome profile of human colorectal adenomas. Mol Cancer Res. 2007;5:1263–1275. doi: 10.1158/1541-7786.MCR-07-0267. [DOI] [PubMed] [Google Scholar]
- 18.Jorissen R.N., Gibbs P., Christie M., Prakash S., Lipton L., Desai J., Kerr D., Aaltonen L.A., Arango D., Kruhoffer M., Orntoft T.F., Andersen C.L., Gruidl M., Kamath V.P., Eschrich S., Yeatman T.J., Sieber O.M. Metastasis-associated gene expression changes predict poor outcomes in patients with Dukes stage B and C colorectal cancer. Clin Cancer Res. 2009;15:7642–7651. doi: 10.1158/1078-0432.CCR-09-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsao T.Y., Tsai C.S., Tung J.N., Chen S.L., Yue C.H., Liao C.F., Wang C.C., Jiang M.C. Function of CSE1L/CAS in the secretion of HT-29 human colorectal cells and its expression in human colon. Mol Cell Biochem. 2009;327:163–170. doi: 10.1007/s11010-009-0054-0. Epub 2009 Feb 18. [DOI] [PubMed] [Google Scholar]
- 20.Chi N.C., Adam E.J., Adam S.A. Different binding domains for Ran-GTP and Ran-GDP/RanBP1 on nuclear import factor p97. J Biol Chem. 1997;272:6818–6822. doi: 10.1074/jbc.272.10.6818. [DOI] [PubMed] [Google Scholar]
- 21.Gorlich D., Dabrowski M., Bischoff F.R., Kutay U., Bork P., Hartmann E., Prehn S., Izaurralde E. A novel class of RanGTP binding proteins. J Cell Biol. 1997;138:65–80. doi: 10.1083/jcb.138.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kutay U., Bischoff F.R., Kostka S., Kraft R., Gorlich D. Export of importin alpha from the nucleus is mediated by a specific nuclear transport factor. Cell. 1997;90:1061–1071. doi: 10.1016/s0092-8674(00)80372-4. [DOI] [PubMed] [Google Scholar]
- 23.Moroianu J., Blobel G., Radu A. Nuclear protein import: Ran-GTP dissociates the karyopherin alphabeta heterodimer by displacing alpha from an overlapping binding site on beta. Proc Natl Acad Sci U S A. 1996;93:7059–7062. doi: 10.1073/pnas.93.14.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seiden-Long I.M., Brown K.R., Shih W., Wigle D.A., Radulovich N., Jurisica I., Tsao M.S. Transcriptional targets of hepatocyte growth factor signaling and Ki-ras oncogene activation in colorectal cancer. Oncogene. 2006;25:91–102. doi: 10.1038/sj.onc.1209005. [DOI] [PubMed] [Google Scholar]
- 25.Tai C.J., Su T.C., Jiang M.C., Chen H.C., Shen S.C., Lee W.R., Liao C.F., Chen Y.C., Lin S.H., Li L.T., Shen K.H., Yeh C.M., Yeh K.T., Lee C.H., Shih H.Y., Chang C.C. Correlations between cytoplasmic CSE1L in neoplastic colorectal glands and depth of tumor penetration and cancer stage. J Transl Med. 2013;11:29. doi: 10.1186/1479-5876-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tung M.C., Tsai C.S., Tung J.N., Tsao T.Y., Chen H.C., Yeh K.T., Liao C.F., Jiang M.C. Higher prevalence of secretory CSE1L/CAS in sera of patients with metastatic cancer. Cancer Epidemiol Biomarkers Prev. 2009;18:1570–1577. doi: 10.1158/1055-9965.EPI-08-0948. Epub 2009 Apr 21. [DOI] [PubMed] [Google Scholar]
- 27.Behrens P., Brinkmann U., Wellmann A. CSE1L/CAS: its role in proliferation and apoptosis. Apoptosis. 2003;8:39–44. doi: 10.1023/a:1021644918117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.