Abstract
Objectives. To investigate the potential association of tumor necrosis factor-α T-857C polymorphism with susceptibility to the five common malignant tumors. Materials and Methods. A comprehensive search of PubMed/Medline, Embase, and Web of Science databases was performed up to November 2015. Pooled odds ratios (ORs) and 95% confidence intervals (95% CIs) were calculated to assess the strength of the association. Subgroup analysis, heterogeneity analyses, and publication bias were also texted in the meta-analysis. Results. A total of twenty-two publications involving 5215 cases and 6755 controls were recruited. Overall, the meta-analysis revealed an increased risk between the TNF-α T-857C polymorphism and gastric cancer susceptibility in T versus C model, heterozygote genetic model, and dominant genetic model. An increased risk between the TNF-α T-857C polymorphism and hepatocellular cancer susceptibility in homozygote genetic model and recessive genetic model was also found. No significant association was found between the TNF-α T-857C polymorphism and colorectal cancer, cervical cancer, and prostate cancer. Conclusions. Our meta-analyses suggest that TNF-α T-857C polymorphism may be associated with increased risk of gastric cancer and hepatocellular cancer development. Therefore, the TNF-α T-857C polymorphism could be considered as one possible risk factor of gastric cancer and hepatocellular cancer according to our study.
1. Introduction
Cancer has been a disease which endangers human physical and psychosocial wellbeing, causing a significant public health and economic burden all over the world. Cancer is a multifactorial disease, and the etiology of these cancers is extremely complex. In order to understand its pathology, numerous susceptibility genes and external environmental factors appear to be considered.
Chronic inflammation has long been associated with the development of cancer. Recent evidences have reignited the interest of cancer researchers in the exciting concept of an association between chronic inflammation and cancer. Rather than protecting against cancer, growing evidence indicates that TNF-α can promote the development of cancer [1]. Previously our study also found that targeting TNF-a suppressed breast cancer growth and TNF-α monoclonal antibody exerted effectively antitumor activity [2], which further supported this assertion.
The length of TNF gene is 12 kilobases (kb) and it is located on the short arm of chromosome 6 (p21.1–p21.3) [3]. As transcription of TNF-α is regulated under genetic control, recent studies have shown that its promoter polymorphisms at TNF-α G-238A (rs361525), TNF-α G-308A (rs1800629), TNF-α T-857C (rs1799724), and TNF-α T-1031C (rs1799964) positions could regulate TNF-a production, thus affecting the risk of cancers [4–7]. Therefore, genetic polymorphisms of TNF-α gene have been supposed as candidate risk factors of cancer.
There are a large number of studies on the association between TNF-α G-308A (rs1800629), TNF-α G-238A (rs361525), and cancers [8, 9]; TNF-α G-308A (rs1800629) and TNF-α G-238A (rs361525) have been successfully identified as risk factors of cancer. Molecular epidemiological research suggests that TNF-α T-857C (rs1799724) polymorphisms may be associated with an increased risk of cancers [10–13], but results remain controversial. TNF-α T-857C is a C to T transition in the promoter at position −857, and previous data have shown that TNF-α T-857C allele T increases the transcription of TNF-α [14–16]. Therefore, TNF-α T-857C polymorphism may be associated with cancer risk and represents candidate risk marker of cancers. To explore a more precise estimation of the relationship between TNF-α T-857C polymorphism and cancers, we performed a meta-analysis.
2. Materials and Methods
2.1. Study Selection
To identify eligible studies published before November 2015, we applied a systematic literature search strategy to the following electronic databases: PubMed/Medline, Embase, and Web of Science. We used the following keywords and subject headings in combination to identifying relevant articles in electronic databases: (tumor necrosis factor alpha OR TNF-α OR 857 C/T OR rs1799724) AND (polymorphism OR variant OR genotype OR mutation) AND (cancer OR carcinoma OR neoplasm). Duplicate articles were manually filtered using the “find duplicate function” of EndNote X7. Two of the authors reviewed results of each of the database searches to make sure that published papers were not missed.
2.2. Inclusion and Exclusion Criteria
The following inclusion criteria were used for the literature selection: (1) genotype distributions of both cases and controls were available; (2) the studies might be cohort or case-control studies; (3) articles about TNF-α T-857C polymorphism and cancer risk; (4) sufficient published data for estimating an odds ratio (OR) with 95% confidence interval (CI); (5) when several publications were available for the same study group, we retained the most recent one for analysis. We excluded publications as follows: (1) based on pedigree data that were excluded; (2) retrospective or cross sectional studies; (3) nonoriginal research (reviews, editorials, or commentaries), abstracts, unpublished studies, and duplicated studies; and (4) studies on animals.
2.3. Data Extraction
Two authors (Ping Wang and June Wang) independently extracted characteristics of studies and resolved any uncertainty through discussion. If these two authors could not reach a consensus, a third author was consulted to resolve the dispute and a final majority decision was made. After excluding the overlap studies and including the additional ones, this meta-analysis covered a total of 22 articles on TNF-α T-857C polymorphism. From each identified article, we extracted the first author's name, publication year, country, ethnicity, type of disease, sample size, source of control, genotyping method, and Hardy-Weinberg equilibrium (HWE) for controls.
2.4. Statistical Analysis
The Hardy-Weinberg equilibrium (HWE) in the controls was tested by the Chi-square test for goodness of fit. The following contrasts for the associations between TNF-α T-857C polymorphism and the cancers mentioned above were evaluated: T allele versus C allele, homozygote comparison (TT versus CC), heterozygote comparison (TC versus CC), and recessive (TT versus TC + CC) and dominant (TT + TC versus CC) genetic model, respectively. The strength of association between TNF-α T-857C polymorphism and cancer risk was assessed using the pooled odds ratio (OR) with 95% confidence intervals (CIs). If the number of included studies was applicable, subgroup analysis was performed based on HWE status of controls, ethnicity, source of control, and genotyping method. The heterogeneity of the data was quantified using Chi-square statistics. Heterogeneity among studies was considered significant when P < 0.1 or I2 > 50%. If there was significant heterogeneity among studies, the random effects model (DerSimonian and Laird) was used; otherwise, the fixed-effects model (Mantel and Haenszel) was acceptable. We plotted Begg's funnel plot to examine the underlying publication bias. We conducted sensitivity analysis by deleting each included study in turn to evaluate the overall robustness of the study's results. All analyses were conducted using Review manager 5.3 and Stata 12.0. All the P values were two-sided.
3. Results
3.1. Study Selection and Characteristic
There were 723 papers relevant to the search words. The flowchart of selection of studies and reasons for exclusion is presented in Figure 1. Finally, a total of 22 studies were included in this meta-analysis. All those 22 studies were reported in English. The details of included studies were shown in Table 1. In the initial search, 723 articles were retrieved, and 553 studies were excluded after reading titles and abstracts. 5215 cases and 6755 healthy controls were included in this study. There are 9 studies for gastric cancer [17–25], 4 studies for colorectal cancer [26–29], 4 studies for hepatocellular cancer [30–33], 3 studies for cervical cancer [34–36], and 2 studies for prostate cancer [37, 38]. Among those 22 studies, 10 studies were from Caucasian populations, and 10 studies were from Asian populations, while 2 studies were from African populations. The sample size of cases ranged from 84 to 1139, while the sample size ranged from 55 to 1378 in the controls. Cases were histological diagnosed in almost all studies. Most studies used healthy subjects as controls. All studies indicated that the distribution of genotypes in the controls was consistent with Hardy-Weinberg equilibrium, except 3 studies [19, 25, 33]. Several genotyping methods were used, including TaqMan, PCR-RFLP, and PCR.
Figure 1.

Flow diagram of the study selection process.
Table 1.
Characteristics of published studies included in this meta-analysis.
| First author | Country | Ethnicity | Cancer type | Sample size | Control source | Casea | Controla | Methods | HWE |
|---|---|---|---|---|---|---|---|---|---|
| de Oliveira 2015 | Brazilian | Caucasian | Gastric cancer | 262/240 | HB | 157/95/10 | 157/64/19 | PCR-RFLP | <0.01 |
| Yang 2009 | Korean | Asian | Gastric cancer | 84/325 | PB | 49/33/2 | 227/92/6 | PCR | 0.34 |
| Hou 2007 | Poland | Caucasian | Gastric cancer | 304/416 | PB | 226/74/4 | 312/99/5 | TaqMan | 0.36 |
| Sugimoto 2007 | Japanese | Asian | Gastric cancer | 105/172 | HB | 66/27/12 | 125/40/7 | PCR-RFLP | 0.11 |
| Shirai 2006 | Japanese | Asian | Gastric cancer | 168/479 | PB | 102/62/4 | 316/146/17 | TaqMan | 0.98 |
| Zambon 2005 | Italian | Caucasian | Gastric cancer | 129/644 | HB | 83/44/2 | 404/227/13 | PCR-RFLP | <0.01 |
| Wu 2004 | Chinese | Asian | Gastric cancer | 204/210 | PB | 148/51/5 | 152/56/2 | PCR | 0.20 |
| Ohyama 2004 | Japanese | Asian | Gastric cancer | 300/472 | HB | 193/98/9 | 312/144/16 | TaqMan | 0.90 |
| Lee 2004 | Korean | Asian | Gastric cancer | 341/261 | PB | 229/97/15 | 185/69/7 | PCR | 0.85 |
| Hamadien 2016 | Arabian | African | Colorectal cancer | 100/100 | HB | 85/15/0 | 85/15/0 | TaqMan | 0.42 |
| Kapitanović 2014 | Croatian | African | Colorectal cancer | 200/200 | PB | 130/64/6 | 126/67/7 | TaqMan | 0.60 |
| Garrity-Park 2008 | American | Caucasian | Colorectal cancer | 114/114 | HB | 98/16/0 | 92/22/0 | PCR | 0.25 |
| Landi 2006 | Spanish | Caucasian | Colorectal cancer | 281/268 | HB | 219/58/4 | 220/45/3 | TaqMan | 0.68 |
| Yang 2015 | Chinese | Asian | HCC | 298/889 | HB | 203/79/16 | 622/228/39 | PCR-RFLP | <0.01 |
| Shin 2015 | Korean | Asian | HCC | 157/201 | PB | 116/35/6 | 142/53/6 | PCR–RFLP | 0.70 |
| Jung 2009 | Korean | Asian | HCC | 227/ 365 | HB | 137/75/15 | 258/100/7 | PCR | 0.45 |
| Wang 2003 | Japanese | Asian | HCC | 125/55 | PB | 80/44/1 | 35/19/1 | PCR | 0.38 |
| Kohaar 2014 | Indian | Caucasian | Cervical cancer | 150/200 | HB | 99/44/7 | 102/85/13 | PCR-RFLP | 0.40 |
| Nieves-Ramirez 2011 | Mexican | Caucasian | Cervical cancer | 191/205 | PB | 93/82/16 | 114/76/15 | PCR | 0.64 |
| Deshpande 2005 | Hispanic | Caucasian | Cervical cancer | 139/115 | HB | 116/22/1 | 84/26/5 | PCR | 0.12 |
| Kesarwani 2009 | Indian | Caucasian | Prostate cancer | 197/256 | PB | 136/57/4 | 196/56/4 | PCR | 1.00 |
| Danforth 2008 | Hispanic | Caucasian | Prostate cancer | 1139/1378 | HB | 923/203/13 | 1110/254/14 | TaqMan | 0.90 |
a(A/B/C): A, B, and C represented the number of genotypes CC, TC, and TT, respectively; HCC, hepatocellular cancer; HWE, P value for Hardy-Weinberg equilibrium for TNF-α T-857C polymorphism among controls; PCR, polymerase chain reaction; RFLP, restriction fragment length polymorphism; TaqMan, a fluorogenic exonuclease assay.
3.2. Quantitative Synthesis
The summary results of meta-analysis of the association between the TNF-α T-857C polymorphism and cancer risk are displayed in Table 2.
Table 2.
Results of overall and subgroups analyses.
| No | T versus C | TT versus CC | TC versus CC | TT versus TC + CC | TT + TC versus CC | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR [95% CI] | P(Z) | P a | OR [95% CI] | P(Z) | P a | OR [95% CI] | P(Z) | P a | OR [95% CI] | P(Z) | P a | OR [95% CI] | P(Z) | P a | ||
| Gastric cancer | ||||||||||||||||
| Total | 9 | 1.12 [1.01, 1.25] | 0.04 | 0.46 | 1.12 [0.79, 1.58] | 0.52 | 0.20 | 1.16 [1.02, 1.33] | 0.02 | 0.58 | 1.05 [0.75, 1.48] | 0.78 | 0.15 | 1.16 [1.02, 1.32] | 0.02 | 0.64 |
| HWE, yes | 7 | 1.01 [0.70, 1.45] | 0.96 | <0.01 | 1.42 [0.95, 2.12] | 0.08 | 0.42 | 1.16 [1.00, 1.34] | 0.06 | 0.68 | 1.36 [0.89, 2.07] | 0.16 | 0.40 | 1.18 [1.02, 1.36] | 0.02 | 0.60 |
| HWE, no | 2 | 0.99 [0.79, 1.25] | 0.96 | 0.64 | 0.57 [0.28, 1.16] | 0.12 | 0.69 | 1.19 [0.76, 1.85] | 0.45 | 0.11 | 0.51 [0.26, 1.03] | 0.06 | 0.56 | 1.10 [0.81, 1.48] | 0.54 | 0.27 |
| Caucasian | 3 | 1.01 [0.84, 1.21] | 0.94 | 0.88 | 0.66 [0.35, 1.22] | 0.18 | 0.63 | 1.13 [0.91, 1.40] | 0.28 | 0.23 | 0.60 [0.33, 1.12] | 0.11 | 0.52 | 1.07 [0.87, 1.32] | 0.50 | 0.52 |
| Asian | 6 | 1.19 [1.04, 1.37] | 0.01 | 0.38 | 1.46 [0.96, 2.22] | 0.08 | 0.32 | 1.18 [1.00, 1.40] | 0.04 | 0.63 | 1.38 [0.91, 2.09] | 0.13 | 0.29 | 1.21 [1.03, 1.42] | 0.02 | 0.56 |
| HB | 4 | 1.09 [0.93, 1.28] | 0.28 | 0.15 | 1.04 [0.46, 2.34] | 0.93 | 0.04 | 1.17 [0.96, 1.42] | 0.12 | 0.42 | 0.98 [0.42, 2.27] | 0.96 | 0.03 | 1.14 [0.95, 1.38] | 0.16 | 0.41 |
| PB | 5 | 1.15 [0.99, 1.34] | 0.06 | 0.71 | 1.34 [0.77, 2.32] | 0.30 | 0.71 | 1.16 [0.97, 1.38] | 0.10 | 0.44 | 1.26 [0.73, 2.18] | 0.40 | 0.65 | 1.17 [0.99, 1.39] | 0.07 | 0.54 |
| PCR | 3 | 1.22 [0.99, 1.50] | 0.06 | 0.50 | 1.82 [0.89, 3.75] | 0.10 | 0.90 | 1.17 [0.91, 1.49] | 0.22 | 0.24 | 1.73 [0.84, 3.54] | 0.13 | 0.83 | 1.21 [0.96, 1.54] | 0.11 | 0.31 |
| PCR-RFLP | 3 | 1.16 [0.83, 1.61] | 0.38 | 0.08 | 1.10 [0.32, 3.77] | 0.88 | 0.02 | 1.21 [0.94, 1.55] | 0.13 | 0.27 | 1.03 [0.29, 3.67] | 0.97 | 0.01 | 1.18 [0.93, 1.50] | 0.16 | 0.26 |
| TaqMan | 3 | 1.07 [0.90, 1.26] | 0.44 | 0.89 | 0.89 [0.49, 1.61] | 0.70 | 0.89 | 1.13 [0.93, 1.38] | 0.22 | 0.63 | 0.85 [0.47, 1.53] | 0.59 | 0.84 | 1.11 [0.92, 1.35] | 0.28 | 0.73 |
| HCC | ||||||||||||||||
| Total | 4 | 1.16 [0.89, 1.51] | 0.27 | 0.08 | 1.65 [1.06, 2.57] | 0.03 | 0.14 | 1.10 [0.90, 1.34] | 0.35 | 0.33 | 1.61 [1.04, 2.49] | 0.03 | 0.19 | 1.16 [0.96, 1.40] | 0.13 | 0.16 |
| HWE, yes | 3 | 1.16 [0.77, 1.77] | 0.48 | 0.05 | 2.29 [1.16, 4.51] | 0.02 | 0.14 | 1.13 [0.87, 1.48] | 0.37 | 0.19 | 2.21 [1.12, 4.34] | 0.02 | 0.20 | 1.22 [0.94, 1.57] | 0.13 | 0.09 |
| HB | 2 | 1.32 [0.90, 1.94] | 0.15 | 0.04 | 2.13 [0.68, 6.67] | 0.19 | 0.04 | 1.19 [0.95, 1.50] | 0.14 | 0.24 | 2.00 [0.70, 5.69] | 0.20 | 0.05 | 1.30 [0.90, 1.87] | 0.16 | 0.10 |
| PB | 2 | 0.93 [0.66, 1.29] | 0.65 | 0.90 | 1.06 [0.36, 3.09] | 0.92 | 0.51 | 0.87 [0.59, 1.30] | 0.51 | 0, 59 | 2.00 [0.70, 5.69] | 0.86 | 0.48 | 0.89 [0.61, 1.31] | 0.56 | 0.72 |
| PCR | 2 | 1.08 [0.86, 1.35] | 0.51 | 0.66 | 1.20 [0.66, 2.17] | 0.55 | 0.47 | 1.05 [0.80, 1.39] | 0.71 | 0.90 | 1.18 [0.66, 2.12] | 0.58 | 0.47 | 1.07 [0.83, 1.39] | 0.60 | 0.78 |
| PCR-RFLP | 2 | 1.24 [0.70, 2.18] | 0.46 | 0.02 | 2.35 [0.73, 7.51] | 0.15 | 0.11 | 1.10 [0.64, 1.89] | 0.74 | 0.07 | 2.45 [1.22, 4.93] | 0.01 | 0.17 | 1.18 [0.65, 2.18] | 0.58 | 0.04 |
| Colorectal cancer | ||||||||||||||||
| Total | 4 | 1.01 [0.80, 1.27] | 0.94 | 0.45 | 0.99 [0.40, 2.41] | 0.97 | 0.62 | 1.01 [0.78, 1.31] | 0.93 | 0.45 | 1.01 [0.41, 2.45] | 0.99 | 0.69 | 1.01 [0.78, 1.30] | 0.93 | 0.43 |
| African | 2 | 0.94 [0.68, 1.29] | 0.69 | 0.85 | 0.83 [0.27, 2.54] | 0.75 | NA | 0.94 [0.65, 1.36] | 0.75 | 0.86 | 0.88 [0.29, 2.67] | 0.82 | NA | 0.93 [0.65, 1.34] | 0.71 | 0.85 |
| Caucasian | 2 | 1.01 [0.58, 1.76] | 0.97 | 0.14 | 1.34 [0.30, 6.05] | 0.70 | NA | 1.00 [0.54, 1.85] | 1.00 | 0.13 | 1.29 [0.29, 5.84] | 0.74 | NA | 1.00 [0.54, 1.86] | 0.99 | 0.12 |
| HB | 3 | 1.06 [0.76, 1.48] | 0.72 | 0.33 | 1.34 [0.30, 6.05] | 0.70 | NA | 1.07 [0.77, 1.49] | 0.69 | 0.31 | 1.29 [0.29, 5.84] | 0.74 | NA | 1.08 [0.78, 1.49] | 0.65 | 0.30 |
| PB | 1 | 0.92 [0.65, 1.31] | 0.66 | NA | 0.83 [0.27, 2.54] | 0.75 | NA | 0.93 [0.61, 1.41] | 0.72 | NA | 0.88 [0.29, 2.67] | 0.82 | NA | 0.92 [0.61, 1.38] | 0.68 | NA |
| PCR | 1 | 0.71 [0.36, 1.38] | 0.31 | NA | NA | NA | NA | 0.68 [0.34, 1.38] | 0.29 | NA | NA | NA | NA | 0.68 [0.34, 1.38] | 0.29 | NA |
| TaqMan | 3 | 1.06 [0.83, 1.35] | 0.65 | 0.49 | 0.99 [0.40, 2.41] | 0.97 | 0.62 | 1.08 [0.81, 1.43] | 0.60 | 0.54 | 1.01 [0.41, 2.45] | 0.99 | 0.69 | 1.07 [0.82, 1.41] | 0.61 | 0.50 |
| Cervical cancer | ||||||||||||||||
| Total | 3 | 0.75 [0.44, 1.29] | 0.30 | 0.003 | 0.59 [0.23, 1.54] | 0.28 | 0.11 | 0.77 [0.42, 1.42] | 0.40 | 0.01 | 0.81 [0.47, 1.39] | 0.44 | 0.20 | 0.75 [0.40, 1.40] | 0.36 | 0.005 |
| Prostate cancer | ||||||||||||||||
| Total | 2 | 1.05 [0.89, 1.24] | 0.57 | 0.11 | 1.20 [0.62, 2.35] | 0.59 | 0.74 | 1.04 [0.86, 1.25] | 0.68 | 0.08 | 1.16 [0.60, 2.27] | 0.66 | 0.85 | 1.05 [0.87, 1.25] | 0.62 | 0.08 |
P a was derived from Chi-square statistics.
3.2.1. Association between the TNF-α T-857C Polymorphism and Gastric Cancer Risk
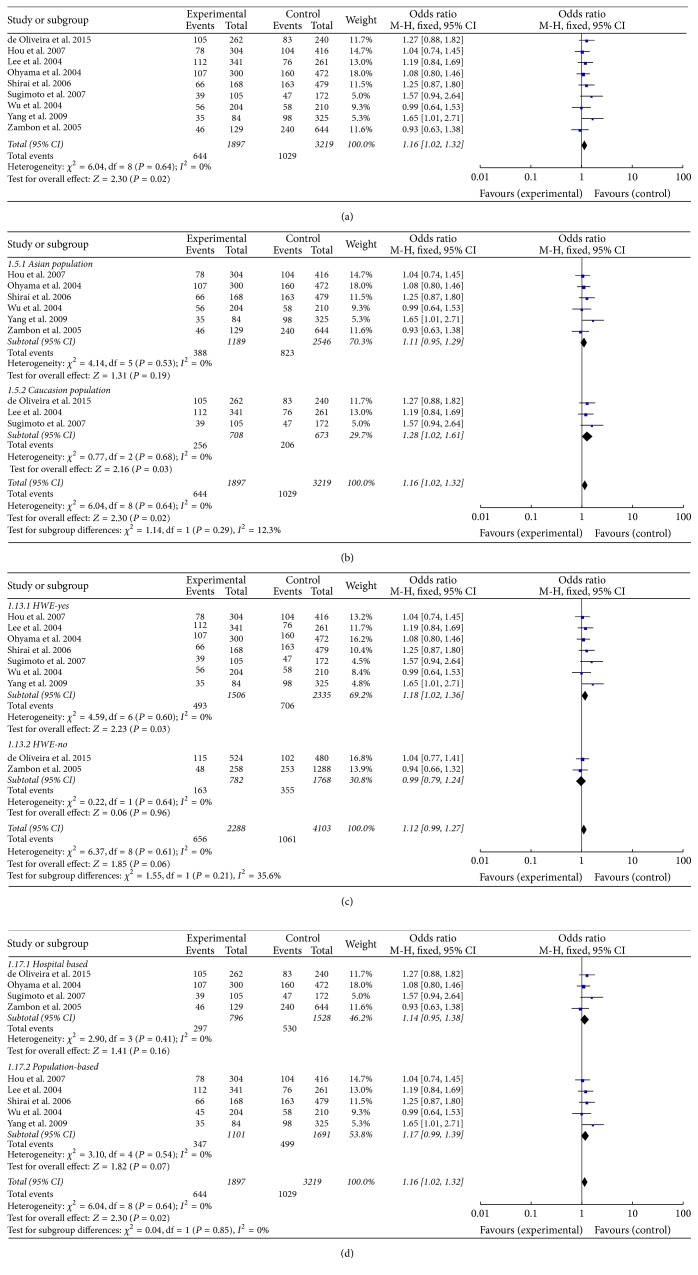
A total of 9 relevant studies, consisting of 1897 patients and 3219 controls, were examined for the association between the TNF-α T-857C polymorphism and gastric cancer risk. In all subjects, meta-analysis showed an increased risk between the TNF-α T-857C polymorphism and gastric cancer susceptibility in three genetic models (T versus C: OR = 1.12, 95% CI = 1.01–1.25, P = 0.04, I2 = 0%, fixed-effects model; TC versus CC: OR = 1.16, 95% CI = 1.02–1.33, P = 0.02, I2 = 0%, fixed-effects model; TT + TC versus CC: OR = 1.16, 95% CI = 1.02–1.32, P = 0.02, I2 = 0%, fixed-effects model, Figure 2). Further stratified analyses based on ethnic subgroups revealed similar results in Asian populations for the T versus C model, TC versus CC model, and the dominant model. The sensitivity analysis showed the results without substantive change (Figure 3 for TT + TC versus CC model).
Figure 2.
Calculated OR and 95% CIs for the associations TNF-α T-857C polymorphism and gastric cancer risk in the TT + TC versus CC model ((a) for overall populations; (b) for ethnicity subgroup; (c) based on HWE for controls; (d) for control sources subgroup).
Figure 3.

The plot of sensitivity analysis for evaluating the association between TNF-α T-857C polymorphism and gastric cancer risk in the TT + TC versus CC model.
3.2.2. Association between the TNF-α T-857C Polymorphism and Hepatocellular Cancer Risk
Four studies consisting of 807 cases and 1510 controls were included in this analysis. All these four studies are based on Asian population. A significant increase in hepatocellular cancer risk was observed in the Asian population in two gene models (TT versus CC: OR = 1.65, 95% CI = 1.06–2.57, P = 0.03, I2 = 46%, fixed-effects model; TT versus TC + CC: OR = 1.61, 95% CI = 1.04–2.49, P = 0.03, I2 = 37%, fixed-effects model, Figure 4). Furthermore, analysis by excluding studies with control inconsistent with HWE showed elevated cancer risk in homozygote comparison and recessive genetic model (TT versus CC: OR = 2.29, 95% CI = 1.16–4.51, P = 0.02, I2 = 46%, fixed-effects model; TT versus TC + CC: OR = 2.21, 95% CI = 1.12–4.93, P = 0.02, I2 = 37%, fixed-effects model). Sensitivity analyses were also conducted, and no conspicuous change of the pooled ORs was detected.
Figure 4.
Calculated OR and 95% CIs for the associations TNF-α T-857C polymorphism and hepatocellular cancer risk in the TT versus CC model.
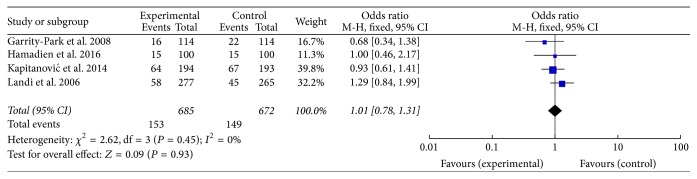
3.2.3. Association between the TNF-α T-857C Polymorphism and Colorectal Cancer Risk
We included four studies to describe the association between the TNF-α T-857C polymorphism and colorectal cancer risk (Figure 5). However, our analysis showed no association between the polymorphism and colorectal cancer risk. The result of stratified analyses based on ethnic subgroups also revealed negative association. The sensitivity analysis showed the results without substantive change.
Figure 5.
Calculated OR and 95% CIs for the associations TNF-α T-857C polymorphism and colorectal cancer risk in the TC versus CC model.
3.2.4. Association between the TNF-α T-857C Polymorphism and Cervical Cancer and Prostate Cancer Risk
Of all the included studies, only three case-control studies involving 480 cases and 520 controls focused on cervical cancer and two studies involving 1336 cases and 2517 controls on prostate cancer. Meta-analysis of our result revealed negative association between the polymorphism and these two cancers. Table 2 showed the result of our analysis.
3.3. Publication Bias
Due to limitations of the quantity of included studies, we just test the publication bias between target gene polymorphism and gastric cancer. Funnel plots were conducted to assess the publication bias, and no evidence of asymmetry was observed (Figure 6 for TT + TC versus CC model). This result was further supported by the analysis using Egger's test (T versus C: P = 0.06; TT versus CC: P = 0.14; TC versus CC: P = 1.00; TT versus TC + CC: P = 0.14; TT + TC versus CC: P = 0.83).
Figure 6.
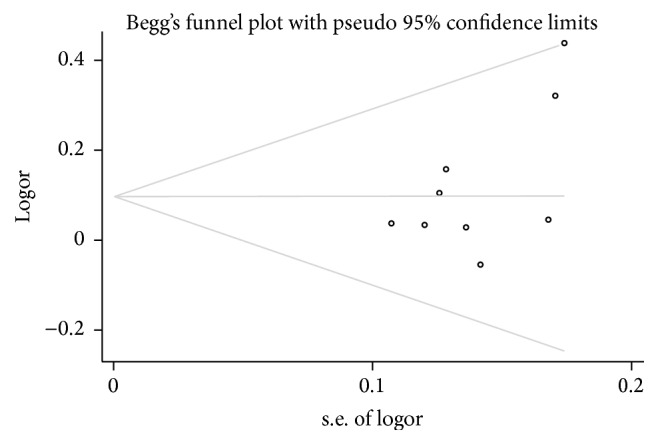
Funnel plot analysis to detect publication bias for TT + TC versus CC.
4. Discussion
Genetic factors have been shown to influence the susceptibility of patients to various diseases and have attracted increasing attention. Chronic inflammation and cytokines are thought to play the most important role in tumor promotion and progression by driving angiogenesis, cell metastasis, and immune-suppression. TNF is an important infectious agent in inflammation progression as well as cancer development [39]. In our meta-analysis, we aggregated data from published studies to estimate genetic associations between the TNF-α T-857C polymorphism and the susceptibility of five common cancers. Our analysis provided some evidence to support an elevated risk between the TNF-α T-857C polymorphism and gastric cancer and hepatocellular cancer susceptibility. But no associations were found in the remained three cancers (colorectal cancer, cervical cancer, and prostate cancer). In the stratified analysis by ethnicity for the TNF-α T-857C polymorphism and gastric cancer susceptibility, we found a significant risk in Asian populations rather than Caucasian populations, suggesting that the increased gastric risk may be ethnospecific. In the stratified analysis based on HWE, we found an elevated risk between the TNF-α T-857C polymorphism and hepatocellular cancer susceptibility. The results of HWE indicated that studies out of HWE might bias the result. Hence, more high quality primary studies are needed. We did not found any meaningful associations in the stratified analysis in all included cancer types by source of control and genotyping method.
To date, numerous related studies have been conducted to investigate the association between the TNF-α T-857C polymorphism and disease risk; however, the exact role of TNF-α T-857C as a carcinogen is still controversial. The previous meta-analysis by Wei et al. in 2011 also investigated the relationship between the TNF-α T-857C polymorphism and hepatocellular cancer susceptibility [40]. However, no association was observed between TNF-α T-857C polymorphism and hepatocellular cancer susceptibility in their study. Compared with their study, our meta-analysis had a lager sample size than them, which increased the statistic power. We included 4 studies, consisting of 807 patients and 1510 health controls, and revealed that TNF-α T-857C polymorphism is associated with a significant increased risk of HCC in homozygote model and recessive genetic model. With regard to the TNF-α T-857C polymorphism and gastric cancer susceptibility, the study of Cen and Wu [41] was published in 2013. Compared with the previous meta-analysis, we retained the most recent study which increased new cases for analysis when two publications [25, 42] were available. Moreover, in our meta-analysis, we conducted four subgroups analysis based on ethnicity, source of control, genotyping method, and HWE for controls and explained these roundly, while they just conducted subgroups analysis based on race. Thus, our results are more reliable and dependable. Hence, this is the most comprehensive meta-analysis that investigated the relationship between the TNF-α T-857C polymorphism and gastric cancer susceptibility.
It is important to note the limitations of our meta-analysis. First, heterogeneity is one of the important issues in genetic association meta-analysis. In our study, some genetic models showed clear homogeneity while others had various heterogeneities, either in total populations or in subgroup analysis. Heterogeneity may be caused by different environment or lifestyle; however, we could not study these factors due to lack of individual data. Second, it should be noted that publication bias is a potential threat to the validity of our meta-analysis because of limitations of the quantity of included studies. In addition, only articles published in English were selected and this may result in language bias leading to an overestimation of effect sizes. Therefore, the statistic power of this meta-analysis might be affected and false positive or false negative rate might occur. Third, in some cancer types, since the number of relevant original documents was limited, there was not enough power to identify the relationship between TNF-α T-857C variant and cancer risk. Thus, further identification based on well-designed studies with large sample sizes is needed. Fourth, as we know, cancer is a multifactorial disease and genetic mutations, environmental changes, lifestyle, diet, age, and gender may be factors in the development of cancer. Like all meta-analyses, it is a secondary retrospective study; therefore, we could not explain the fundamental underlying mechanisms clearly due to unadjusted data.
5. Conclusions
This meta-analysis with published data suggested that the TNF-α T-857C polymorphism is a risk factor for gastric cancer, especially in Asian populations. Our result also indicated that the TNF-α T-857C polymorphism also plays an important role in hepatocellular cancer development. There is lack of association between the TNF-α T-857C polymorphism and colorectal cancer, cervical cancer, prostate cancer, and breast cancer. However, considering the limited objectives of this meta-analysis, further studies providing adjusted data, large sample size, and gene-environment detailed information are needed to assess the findings.
Acknowledgments
This study was supported by National Natural Science Funds (nos. 81573459, 81472033, and 30901308), the National Science Foundation of Hubei Province (nos. 2013CFB233 and 2013CFB235), the project of Wuhan Science and Technology Bureau (nos. 2014070404010223 and 2014060101010045), and Hubei Province Health and Family Planning Scientific Research Project (WJ2015Q021).
Competing Interests
The authors have no financial conflict of interests.
References
- 1.Balkwill F. TNF-α in promotion and progression of cancer. Cancer and Metastasis Reviews. 2006;25(3):409–416. doi: 10.1007/s10555-006-9005-3. [DOI] [PubMed] [Google Scholar]
- 2.Yu M., Zhou X., Niu L., et al. Targeting transmembrane TNF-α suppresses breast cancer growth. Cancer Research. 2013;73(13):4061–4074. doi: 10.1158/0008-5472.can-12-3946. [DOI] [PubMed] [Google Scholar]
- 3.Lindholm E., Bakhtadze E., Cilio C., Agardh E., Groop L., Agardh C.-D. Association between LTA, TNF and AGER polymorphisms and late diabetic complications. PLoS ONE. 2008;3(6) doi: 10.1371/journal.pone.0002546.e2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huizinga T. W. J., Westendorp R. G. J., Bollen E. L. E. M., et al. TNF-α promoter polymorphisms, production and susceptibility to multiple sclerosis in different groups of patients. Journal of Neuroimmunology. 1997;72(2):149–153. doi: 10.1016/S0165-5728(96)00182-8. [DOI] [PubMed] [Google Scholar]
- 5.Lippitz B. E. Cytokine patterns in patients with cancer: a systematic review. The Lancet Oncology. 2013;14(6):e218–e228. doi: 10.1016/s1470-2045(12)70582-x. [DOI] [PubMed] [Google Scholar]
- 6.Persson C., Canedo P., MacHado J. C., El-Omar E. M., Forman D. Polymorphisms in inflammatory response genes and their association with gastric cancer: a HuGE systematic review and meta-analyses. American Journal of Epidemiology. 2011;173(3):259–270. doi: 10.1093/aje/kwq370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watts T. H. TNF/TNFR family members in costimulation of T cell responses. Annual Review of Immunology. 2005;23:23–68. doi: 10.1146/annurev.immunol.23.021704.115839. [DOI] [PubMed] [Google Scholar]
- 8.Lv K., Chen R., Cai Q., Fang M., Sun S. Effects of a single nucleotide polymorphism on the expression of human tumor necrosis factor-alpha. Scandinavian Journal of Immunology. 2006;64(2):164–169. doi: 10.1111/j.1365-3083.2006.01786.x. [DOI] [PubMed] [Google Scholar]
- 9.Serretti A., Liappas I., Mandelli L., et al. Interleukin-1 alpha and beta, TNF-alpha and HTTLPR gene variants study on alcohol toxicity and detoxification outcome. Neuroscience Letters. 2006;406(1-2):107–112. doi: 10.1016/j.neulet.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Hernández-Díaz Y., Tovilla-Zárate C., Juárez-Rojop I., López-Narváez M., Álvarez-Cámara J., González-Castro T. Association between CRP and TNF-α genes variants and cardiovascular heart disease in a Mexican population: protocol for a case-control study. International Journal of Environmental Research and Public Health. 2016;13(1):p. 103. doi: 10.3390/ijerph13010103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hou R., Ren X., Wang J., Guan X. TNF-α and MTHFR polymorphisms associated with cerebral palsy in Chinese infants. Molecular Neurobiology. 2015:1–6. doi: 10.1007/s12035-015-9566-7. [DOI] [PubMed] [Google Scholar]
- 12.Yu G.-I., Ha E., Park S.-H., et al. Association of tumor necrosis factor-α (TNF-α) promoter polymorphisms with overweight/obesity in a Korean population. Inflammation Research. 2011;60(12):1099–1105. doi: 10.1007/s00011-011-0372-z. [DOI] [PubMed] [Google Scholar]
- 13.Yoshida T., Kato K., Yokoi K., et al. Association of candidate gene polymorphisms with chronic kidney disease in Japanese individuals with hypertension. Hypertension Research. 2009;32(5):411–418. doi: 10.1038/hr.2009.22. [DOI] [PubMed] [Google Scholar]
- 14.Puthothu B., Bierbaum S., Kopp M. V., et al. Association of TNF-α with severe respiratory syncytial virus infection and bronchial asthma. Pediatric Allergy and Immunology. 2009;20(2):157–163. doi: 10.1111/j.1399-3038.2008.00751.x. [DOI] [PubMed] [Google Scholar]
- 15.Giardina E., Hüffmeier U., Ravindran J., et al. Tumor necrosis factor promoter polymorphism TNF∗-857 is a risk allele for psoriatic arthritis independent of the PSORS1 locus. Arthritis and Rheumatism. 2011;63(12):3801–3806. doi: 10.1002/art.30591. [DOI] [PubMed] [Google Scholar]
- 16.Shi K.-Q., Cai X.-H., Xiao D.-D., et al. Tumour necrosis factor-α-857T allele reduces the risk of hepatitis B virus infection in an Asian population. Journal of Viral Hepatitis. 2012;19(2):e66–e72. doi: 10.1111/j.1365-2893.2011.01540.x. [DOI] [PubMed] [Google Scholar]
- 17.Lee S.-G., Kim B., Yook J.-H., Oh S.-T., Lee I., Song K. TNF/LTA polymorphisms and risk for gastric cancer/duodenal ulcer in the Korean population. Cytokine. 2004;28(2):75–82. doi: 10.1016/j.cyto.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 18.Ohyama I., Ohmiya N., Niwa Y., et al. The association between tumour necrosis factor-α gene polymorphism and the susceptibility to rugal hyperplastic gastritis and gastric carcinoma. European Journal of Gastroenterology and Hepatology. 2004;16(7):693–700. doi: 10.1097/01.meg.0000108315.52416.bf. [DOI] [PubMed] [Google Scholar]
- 19.Wu M.-S., Chen L.-T., Shun C.-T., et al. Promoter polymorphisms of tumor necrosis factor-α are associated with risk of gastric mucosa-associated lymphoid tissue lymphoma. International Journal of Cancer. 2004;110(5):695–700. doi: 10.1002/ijc.20199. [DOI] [PubMed] [Google Scholar]
- 20.Zambon C.-F., Basso D., Navaglia F., et al. Pro- and anti-inflammatory cytokines gene polymorphisms and Helicobacter pylori infection: interactions influence outcome. Cytokine. 2005;29(4):141–152. doi: 10.1016/j.cyto.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 21.Shirai K., Ohmiya N., Taguchi A., et al. Interleukin-8 gene polymorphism associated with susceptibility to non-cardia gastric carcinoma with microsatellite instability. Journal of Gastroenterology and Hepatology. 2006;21(7):1129–1135. doi: 10.1111/j.1440-1746.2006.04443.x. [DOI] [PubMed] [Google Scholar]
- 22.Hou L., El-Omar E. M., Chen J., et al. Polymorphisms in Th1-type cell-mediated response genes and risk of gastric cancer. Carcinogenesis. 2007;28(1):118–123. doi: 10.1093/carcin/bgl130. [DOI] [PubMed] [Google Scholar]
- 23.Sugimoto M., Furuta T., Shirai N., et al. Different effects of polymorphisms of tumor necrosis factor-alpha and interleukin-1 beta on development of peptic ulcer and gastric cancer. Journal of Gastroenterology and Hepatology. 2007;22(1):51–59. doi: 10.1111/j.1440-1746.2006.04442.x. [DOI] [PubMed] [Google Scholar]
- 24.Yang J. J., Ko K.-P., Cho L. Y., et al. The role of TNF genetic variants and the interaction with cigarette smoking for gastric cancer risk: a nested case-control study. BMC Cancer. 2009;9, article 238 doi: 10.1186/1471-2407-9-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Oliveira J. G., Rossi A. F. T., Nizato D. M., et al. Influence of functional polymorphisms in TNF-α, IL-8, and IL-10 cytokine genes on mRNA expression levels and risk of gastric cancer. Tumor Biology. 2015;36(12):9159–9170. doi: 10.1007/s13277-015-3593-x. [DOI] [PubMed] [Google Scholar]
- 26.Landi S., Gemignani F., Bottari F., et al. Polymorphisms within inflammatory genes and colorectal cancer. Journal of Negative Results in BioMedicine. 2006;5, article 15 doi: 10.1186/1477-5751-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garrity-Park M. M., Loftus E. V., Jr., Bryant S. C., Sandborn W. J., Smyrk T. C. Tumor necrosis factor-alpha polymorphisms in ulcerative colitis-associated colorectal cancer. American Journal of Gastroenterology. 2008;103(2):407–415. doi: 10.1111/j.1572-0241.2007.01572.x. [DOI] [PubMed] [Google Scholar]
- 28.Kapitanović S., Čačev T., Catela Ivković T., Lončar B., Aralica G. TNFα gene/protein in tumorigenesis of sporadic colon adenocarcinoma. Experimental and Molecular Pathology. 2014;97(2):285–291. doi: 10.1016/j.yexmp.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 29.Hamadien M.-A., Khan Z., Vaali-Mohammed M.-A., et al. Polymorphisms of tumor necrosis factor alpha in Middle Eastern population with colorectal cancer. Tumor Biology. 2016;37(4):5529–5537. doi: 10.1007/s13277-015-4421-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y., Kato N., Hoshida Y., et al. Interleukin-1β gene polymorphisms associated with hepatocellular carcinoma in hepatitis C virus infection. Hepatology. 2003;37(1):65–71. doi: 10.1053/jhep.2003.50017. [DOI] [PubMed] [Google Scholar]
- 31.Jung K. W., Ha E., Yu G. I., et al. TNFα promoter polymorphism is a risk factor for susceptibility in hepatocellular carcinoma in Korean population. Clinica Chimica Acta. 2009;407(1-2):16–19. doi: 10.1016/j.cca.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 32.Shin S. P., Kim N. K., Kim J. H., et al. Association between hepatocellular carcinoma and tumor necrosis factor alpha polymorphisms in South Korea. World Journal of Gastroenterology. 2015;21(46):13064–13072. doi: 10.3748/wjg.v21.i46.13064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang M.-D., Hsu C.-M., Chang W.-S., et al. Tumor necrosis factor-α genotypes are associated with hepatocellular carcinoma risk in Taiwanese males, smokers and alcohol drinkers. Anticancer Research. 2015;35(10):5417–5423. [PubMed] [Google Scholar]
- 34.Deshpande A., Nolan J. P., White P. S., et al. TNF-α promoter polymorphisms and susceptibility to human papiflomavirus 16-associated cervical cancer. Journal of Infectious Diseases. 2005;191(6):969–976. doi: 10.1086/427826. [DOI] [PubMed] [Google Scholar]
- 35.Nieves-Ramirez M. E., Partida-Rodriguez O., Alegre-Crespo P. E., Tapia-Lugo M. D. C., Perez-Rodriguez M. E. Characterization of single-nucleotide polymorphisms in the tumor necrosis factor α promoter region and in lymphotoxin α in squamous intraepithelial lesions, precursors of cervical cancer. Translational Oncology. 2011;4(6):336–344. doi: 10.1593/tlo.11226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kohaar I., Hussain S., Kumar A., Singhal P., Roy S., Choudhury B. C. Impact of Haplotype TNF-LTA Locus with Susceptibility to Cervical Cancer in Indian Population. 2014. [Google Scholar]
- 37.Danforth K. N., Rodriguez C., Hayes R. B., et al. TNF polymorphisms and prostate cancer risk. Prostate. 2008;68(4):400–407. doi: 10.1002/pros.20694. [DOI] [PubMed] [Google Scholar]
- 38.Kesarwani P., Mandhani A., Mittal R. D. Polymorphisms in tumor necrosis factor-a gene and prostate cancer risk in North Indian cohort. Journal of Urology. 2009;182(6):2938–2943. doi: 10.1016/j.juro.2009.08.016. [DOI] [PubMed] [Google Scholar]
- 39.Anderson G. M., Nakada M. T., DeWitte M. Tumor necrosis factor-α in the pathogenesis and treatment of cancer. Current Opinion in Pharmacology. 2004;4(4):314–320. doi: 10.1016/j.coph.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 40.Wei Y., Liu F., Li B., et al. Polymorphisms of tumor necrosis factor-alpha and hepatocellular carcinoma risk: a HuGE systematic review and meta-analysis. Digestive Diseases and Sciences. 2011;56(8):2227–2236. doi: 10.1007/s10620-011-1617-y. [DOI] [PubMed] [Google Scholar]
- 41.Cen G., Wu W. Association between tumor necrosis factor-alpha 857C/T polymorphism and gastric cancer: a meta-analysis. Tumor Biology. 2013;34(6):3383–3388. doi: 10.1007/s13277-013-0910-0. [DOI] [PubMed] [Google Scholar]
- 42.de Oliveira J. G., Rossi A. F. T., Nizato D. M., Miyasaki K., Silva A. E. Profiles of gene polymorphisms in cytokines and toll-like receptors with higher risk for gastric cancer. Digestive Diseases and Sciences. 2013;58(4):978–988. doi: 10.1007/s10620-012-2460-5. [DOI] [PubMed] [Google Scholar]