Abstract
Giant cell arteritis (GCA) is the most common form of vasculitis in individuals older than 50 years in Western countries. To shed light onto the genetic background influencing susceptibility for GCA, we performed a genome-wide association screening in a well-powered study cohort. After imputation, 1,844,133 genetic variants were analyzed in 2,134 case subjects and 9,125 unaffected individuals from ten independent populations of European ancestry. Our data confirmed HLA class II as the strongest associated region (independent signals: rs9268905, p = 1.94 × 10−54, per-allele OR = 1.79; and rs9275592, p = 1.14 × 10−40, OR = 2.08). Additionally, PLG and P4HA2 were identified as GCA risk genes at the genome-wide level of significance (rs4252134, p = 1.23 × 10−10, OR = 1.28; and rs128738, p = 4.60 × 10−9, OR = 1.32, respectively). Interestingly, we observed that the association peaks overlapped with different regulatory elements related to cell types and tissues involved in the pathophysiology of GCA. PLG and P4HA2 are involved in vascular remodelling and angiogenesis, suggesting a high relevance of these processes for the pathogenic mechanisms underlying this type of vasculitis.
Introduction
During the last decade, genome-wide association studies (GWASs), in which common genetic variation across the whole genome is interrogated in a hypothesis-free fashion, were a breakthrough in biomedical research methodology and have led to the identification of thousands of robust genetic associations within a wide spectrum of complex human diseases.1 However, some diseases of low prevalence have received less attention due to the difficulty in recruiting well-powered study cohorts, even though rare diseases result in a significant disease burden. An example is giant cell arteritis (GCA [MIM: 187360]), the most common form of vasculitis in Western countries in people over 50 years old. GCA is characterized by chronic inflammation of large arteries, such as the aorta and the carotid arteries and its extracranial branches, which may lead to severe clinical sequelae if not treated promptly, including vision loss, scalp and tongue necrosis, aortic dissection/rupture, and cerebral infarction.2, 3, 4 Although the genetic component of GCA has previously been investigated following a candidate gene approach, most of the described genetic associations were based on underpowered analyses and usually failed to be replicated in independent populations.5
Recently, an international collaborative effort involving European and North American research consortia has made possible more powerful studies, including an Immunochip study, that have identified firm risk signals for GCA predisposition, such as HLA molecules and key genes of the immune response like protein tyrosine phosphatase, non-receptor type 22 (PTPN22 [MIM: 600716]), and interleukin 17A (IL17A [MIM: 603149]).6, 7, 8 Taking advantage of the large sample collection that this collaboration has enabled, comprising ten independent populations of European ancestry that cover the whole gradient for prevalence of the disease, we performed an agnostic genetic study in GCA at the genome-wide level.
Subjects and Methods
Study Population
This study included a total of 2,134 GCA-affected case subjects and 9,125 unaffected control subjects from ten independent populations of European ancestry: Spain (805 case and 1,323 control subjects), UK (352 case and 2,965 control subjects), Italy (271 case and 960 control subjects), North America (176 case and 1,181 control subjects from USA and Canada), Germany (160 case and 667 control subjects), France (114 case and 488 control subjects), Norway (104 case and 121 control subjects), the Netherlands (69 case and 638 control subjects), Switzerland (46 case and 500 control subjects), and Ireland (37 case and 282 control subjects). The diagnosis of GCA was established according to the 1990 American College of Rheumatology classification criteria for this disease.9 In addition, the diagnosis was subsequently confirmed by either a biopsy of the temporal artery (89.83%) or arterial imaging (10.17%) consistent with GCA. A detailed description of the main clinical characteristics of the different case cohorts is provided in Table S1. All participants signed an informed consent form before being enrolled in the study. The procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) of all participant centers.
Genotyping and Quality Controls
Genomic DNA from peripheral blood samples of all GCA-affected case subjects as well as the Spanish and Irish control subjects were screened using the GWAS platform “Infinium HumanCore Beadchip” in an iScan System and the Genotyping Module (v.1.9) of the GenomeStudio software (Illumina). The genotyping was conducted in the Genomics and Genotyping Unit of the Pfizer-University of Granada-Junta de Andalucía Centre for Genomics and Oncological Research (GENYO, Granada, Spain), according to the manufacturer’s protocol. SNPs with a cluster separation < 0.4 were removed after the calling.
Genotyping data from the remaining control population were obtained from nine cohorts of geographically matched European ancestries included in previous studies10, 11, 12, 13, 14, 15, 16, 17 (Table S2).
All studies were subjected to stringent quality-control measures separately based on the same analytical protocol using PLINK v.1.07.18 Specifically, we filtered out SNPs with call rates < 0.98 and minor allele frequencies (MAF) < 0.01 and those that deviated from Hardy-Weinberg equilibrium (HWE; p < 0.001 in both case and control subjects). Similarly, samples with less than 95% of successfully called SNPs, and one subject per pair of first-degree relatives (identity by descent > 0.4) were removed. Sex chromosomes were also excluded from the analysis.
Finally, to check the consistency of the results, we re-genotyped the Spanish samples for the associated signals using predesigned TaqMan 5′ SNP genotyping assays (assay IDs: C__16222465_10, C__25614474_20, and C___2397211_10) in a 7900HT Fast Real-Time PCR System (Applied Biosystems) and compared the TaqMan types with the corresponding imputed data.
Imputation Methods
After applying the QC filters, whole-genome SNP genotype imputation was carried out with IMPUTE v.219 using the combined 1000 Genome Project Phase III (1KGPh3) data as reference panel, which includes 2,504 individuals.20 In brief, for each individual dataset, the strand orientation, chromosome position, and SNP identification were updated in PLINK to match build 37 (hg19) of the 1KGPh3. Next, PLINK-formatted files were converted to the appropriate format with GTOOL and the genotyping data were split into chunks of 50,000 Mb covering whole-genome regions. Imputation was done separately for each study. To ensure quality of imputed SNPs, the most likely call was used for merging genotypes, but only if the most likely call exceeded a probability threshold of 0.9 (the genotype was set to missing otherwise). As the individual imputation of each case/control set could lead to stratification, imputed data were also subjected to rigorous quality filters in PLINK, including removal of SNPs with call rates < 0.98, MAFs < 0.01, and those that were not in HWE (p < 0.001). Moreover, after merging case/control sets, singleton SNPs and those showing strong evidence of discordance in the genotype distribution between cases and controls due to possible miscalling were removed using an in-house Perl script. Finally, principal component (PC) analyses were conducted to control for possible population stratification using PLINK and the GCTA64 and R-base software under GNU Public license v.2. For that, we calculated and plotted the ten first PCs of each individual and those samples located at >4 standard deviations from the cluster centroids of each cohort were considered outliers and, consequently, excluded from the analyses.
The final numbers of individuals and SNPs that remained in the filtered datasets are shown in Table S2.
Statistical Analyses
Table S3 shows the estimation of the overall statistical power of this study accordingly with the CaTS Power Calculator for Genetic Studies software, which implements the methods described in Skol et al. assuming an additive genetic model.21
PLINK was used to conduct all the case/control analyses. First, the genotype frequencies of all markers were compared between case and control subjects of every individual dataset by logistic regression on the best-guess genotypes (>0.9 probability) assuming an additive model and using the ten first study-specific PCs and the gender as covariates. Next, a combined analysis of all studies was performed using the inverse variance weighted meta-analysis under a fixed effects model. In order to maximize the total number of SNPs analyzed without compromising the consistency of the results, we tested all SNPs present in the largest GCA sample set (Spain) and in one or more additional studies (i.e., ranging from 2 to 10 studies but including always the Spanish set). A total of 1,844,133 were evaluated in the meta-analysis. To identify independent effects across associated regions, dependency analyses at the cohort level genome-wide scans were conducted in PLINK by stepwise logistic regression with adjustment for the most associated signals in the common set of SNPs, followed by inverse variance weighted meta-analysis under a fixed effects model. The heterogeneity of the odds ratios (OR) across the different studies was estimated using both I2 and Cochran’s Q tests. The Manhattan plots were obtained with an in-house R script, and the zooms of the associated regions were obtained with LocusZoom v.1.1.22
Functional Annotation of Associated Variants
We evaluated the putative functional implications of the GCA risk loci by implementing our data with publicly available functional annotation data.
Despite the high efficiency of the imputation process, it was not possible to obtain imputed types for every known polymorphism. Therefore, in a first step, we identified all the SNP taggers (r2 > 0.8) of the associated signals of our GWAS using the 1KGPh3 data for the European populations and PLINK. All taggers were considered equally as candidates for prioritizing casualty or hypothesizing possible molecular causes of the observed associations in the subsequent bioinformatic approaches. Then, we explored whether the taggers of each GWAS signal had possible functional implications. The tools PolyPhen-223 and Combined Annotation Dependent Depletion (CADD)24 were used to evaluate possible damaging effects on the protein sequence of coding non-synonymous SNPs. Regarding the intronic and intergenic variants, we explored whether they lay within known or predicted regulatory DNA elements (including regions of DNAase hypersensitivity, binding sites of transcription factors, promoter regions, chromatin marks, etc.) and whether they had predictive effects on clinical phenotypes using the online tools for exploring annotations of the noncoding genome RegulomeDB25 and HaploReg v.4.1.26 Sources of these databases include public datasets from the projects Gene Expression Omnibus (GEO), the Roadmap Epigenomics, the Encyclopedia of DNA Elements (ENCODE), and 1KGPh3, as well as published literature.
In addition, to provide an illustrative picture of the current knowledge on the GCA genetics, we conducted a molecular pathway enrichment analysis, considering both previously suggested GCA genes and those showing an association with disease susceptibility in this study, using the tool for that purpose of the Gene Ontology (GO) reference genome project,27, 28 powered by the Protein Analysis Through Evolutionary Relationships (PANTHER) Classification System.29 Biological pathways showing p values lower than 0.05 after Bonferroni correction were considered associated with the disease. Finally, predictive protein-protein relationships among these same genes were also tested using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database.30 Only candidate genes that showed statistically significant signals after correction for multiple testing (which was performed with a previously validated method to control the genome-wide type 1 error rate at 0.0531) were included in the above analyses.
Results
Testing for Association with Disease Susceptibility
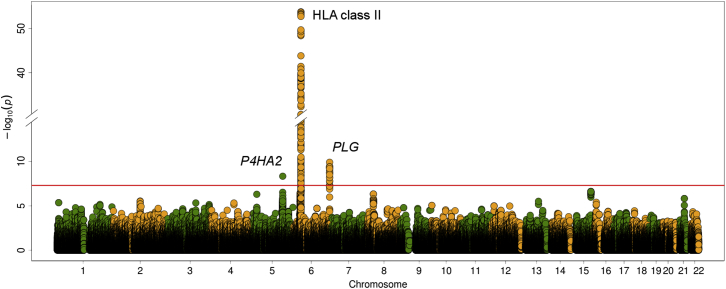
Three genomic regions contained association signals at the genome-wide level of significance in the overall meta-analysis: human leukocyte antigen (HLA) class II, plasminogen (PLG [MIM: 173350]), and prolyl 4-hydroxylase subunit alpha 2 (P4HA2 [MIM: 600608]) (Tables 1 and S4; Figure 1).
Table 1.
Independent Association Signals with Giant Cell Arteritis Susceptibility at the Genome-wide Level of Significance
| SNP | Location (GRCh37) | Locus | Change |
Meta-analysis |
Minor Allele Frequency (GCA/CTRL)a |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p Value | OR (CI 95%) | P(Q) | I2 | Spain | UK | Italy | North America | Germany | France | Norway | Netherlands | Switzerland | Ireland | ||||
| rs9268905 | 6:32432077 | HLA-DRA/HLA-DRB1 | C<G | 1.94 × 10−54 | 1.79 (1.67–1.93) | 0.75 | 0 | 0.46/0.32 | 0.52/0.37 | 0.33/0.23 | 0.48/0.32 | 0.39/0.28 | 0.48/0.29 | 0.47/0.33 | 0.37/0.28 | 0.42/0.34 | 0.45/0.34 |
| rs9275592 | 6:32680620 | HLA-DQA1/HLA-DQA2 | T<G | 1.14 × 10−40 | 2.08 (1.87–2.32) | 0.98 | 0 | 0.15/0.08 | 0.18/0.11 | 0.09/0.05 | 0.20/0.09 | 0.18/0.10 | 0.18/0.10 | 0.28/0.16 | 0.18/0.10 | 0.19/0.11 | 0.15/0.10 |
| rs4252134 | 6:161153527 | PLG | C<T | 1.23 × 10−10 | 1.28 (1.19–1.39) | 0.04 | 49 | 0.33/0.29 | 0.36/0.28 | 0.36/0.33 | 0.32/0.30 | 0.32/0.28 | 0.34/0.33 | 0.38/0.23 | 0.38/0.28 | 0.47/0.28 | 0.41/0.30 |
| rs128738 | 5:131540875 | P4HA2 | T<G | 4.60 × 10−9 | 1.32 (1.20–1.45) | 0.49 | 0 | 0.19/0.14 | 0.22/0.17 | 0.16/0.14 | 0.17/0.16 | 0.20/0.17 | 0.18/0.16 | 0.22/0.18 | 0.23/0.15 | 0.18/0.16 | 0.16/0.18 |
Abbreviations are as follows: GCA, giant cell arteritis; CTRL, controls; GRCh37, genome reference consortium human genome build 37; OR, per-allele odds ratio for the minor allele; CI, confidence interval; Q, Cochran’s Q test.
N (GCA/CTRL): Spain = 805/1,323, UK = 352/2,965, Italy = 271/960, North America = 176/1,181, Germany = 160/667, France = 114/488, Norway = 104/121, Netherlands = 69/638, Switzerland = 46/500, Ireland = 37/282, combined = 2,134/9,125.
Figure 1.
Manhattan Plot Representation of the GWAS Results
The −log10 of the inverse variance-weighted meta-analysis p values are plotted against their physical chromosomal position. The red line represents the genome-wide level of significance (p < 5 × 10−8). The most relevant associations are highlighted.
Within the HLA region, the top hit was rs9268905 (p = 1.94 × 10−54, OR = 1.79, 95% CI 1.67–1.93), located between HLA-DRA (MIM: 142860) and HLA-DRB1 (MIM: 142857). Dependence analyses suggested a possible independent effect on disease susceptibility within the HLA-DQA1 (MIM: 146880)/HLA-DQA2 (MIM: 613503) genomic region after controlling for the rs9268905 signal (lead SNP = rs9275592, adjusted p = 1.78 × 10−10, OR = 1.48, 95% CI 1.31–1.66). No association at the genome-wide level of significance remained within the HLA after conditioning on both rs9268905 and rs9275592 (Table S5 and Figure S1).
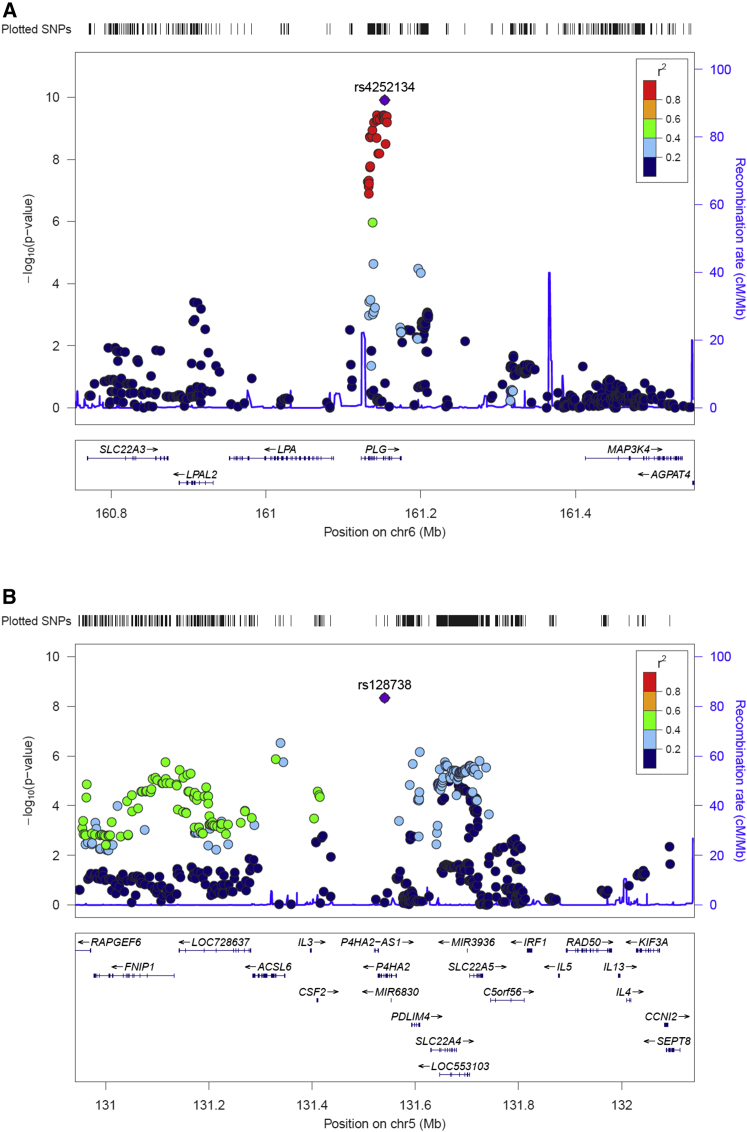
Outside the HLA region, rs4252134, located in an intron of PLG at chromosome 6, represented the most strongly associated variant (p = 1.23 × 10−10, OR = 1.28, 95% CI 1.19–1.39, Figure 2). Although some heterogeneity was observed between studies (I2 = 48.8), consistent OR directions of the minor allele (toward risk) were observed in all sets (Tables 1 and S4; Figure S2). Other SNPs within the gene (both intronic and exonic) also showed significant p values, but their statistical significance was lost when conditioned on rs4252134 (Table S6 and Figure S3). Finally, another intronic SNP of P4HA2 at chromosome 5, rs128738, surpassed the statistical threshold (p = 4.60 × 10−9, OR = 1.32, 95% CI 1.20–1.45). This genomic region also showed additional suggestive signals, but none of them was significant at the genome-wide level (Figure 2).
Figure 2.
Regional Plots of the Associated Loci with GCA outside the HLA Region in the Overall Meta-analysis
(A) Plasminogen (PLG) region.
(B) Prolyl 4-hydroxylase subunit alpha 2 (P4HA2) region.
Lead variants are highlighted in violet.
To confirm the above results, we obtained direct genotypes of the whole Spanish cohort using TaqMan probes for rs128738 and two perfect proxies (r2 = 1) of rs4252134 and rs9268905 (rs4252125 and rs2395185, respectively). The overall concordance reached after comparing TaqMan types with the corresponding imputed data was 99.94% for rs128738, 99.81% for rs4252134/rs4252125, and 99.94% for rs9268905/rs2395185.
Functional Annotations of Proxies of the Non-HLA Hits
To prioritize variants that could drive the observed non-HLA associations, we identified all the SNPs in high linkage disequilibrium (LD, r2 > 0.8) with the lead signals in the European populations of 1KGPh3 (Tables S7 and S8) and used public online annotation tools to evaluate their possible functional implications. One PLG polymorphism (rs4252125) was annotated as missense in the dbSNP database. However, this variant was predicted to be benign according to PolyPhen-2 and showed no evidence of being potentially pathogenic after scoring the deleteriousness with CADD (raw score = −0.63, PHRED-like scaled C-score = 0.104). Then, using RegulomeDB, we identified those SNPs overlapping with known and predicted DNA elements with a higher probability of regulatory effects (score ≤ 3) (Tables S7 and S8) and performed a detailed functional annotation of these tagger variants and the lead SNPs using HaploReg (Tables 2 and S9–S11). Most of them overlapped with DNase hypersensitivity sites and histone marks enriched at promoters and enhancers (Table 2). Interestingly, some of these annotations were related to cell types and tissues involved in GCA pathophysiology. For example, PLG rs4252135 co-localized with DNase peaks in different immune cell lines and had a predicted enhancer chromatin state in lymphoblastoid cells (Tables S9 and S10). A higher enrichment of promoter and enhancer epigenetic marks in these tissues was observed for the prioritized P4HA2 variants (Table S9). Specifically, the lead signal rs128738 overlapped with enhancer histone marks in immune cells and with the imputed Transcription 3′ Enhancer mark in the aorta (Table S10). Additionally, P4HA2 rs156023 showed evidence of influencing enhancer activity in hematopoietic stem cells and neutrophils as well as promoter activity in mononuclear cells and monocytes (Tables S9 and S10).
Table 2.
Prioritized Variants of the Giant Cell Arteritis-Associated Non-HLA Regions Accordingly to the Functional Annotations of the Lead Signals and Their Proxies using ENCODE Data
| SNP | Location (GRCh37) | Locus | Distance from Hit (bp) | r2 | RDB Score |
Regulatory Chromatin Marks |
Protein Binding and Regulatory Motifs |
GRASP QTL and eQTL Annotations |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Promoter Histone Marks | Enhancer Histone Marks | DNase Hypersensitivity | Proteins Bound (ChIP-Seq) | Regulatory Motifs Altered (PWM) | GRASP QTL Traits | p Value | Ref (PMID) | eQTL Hits | ||||||
| rs4252134 | 6:161153527 | PLG (intronic) | 0 | NA | 6 | – | – | – | – | – | gene expression of DDR1 in peripheral blood monocytes | 3.31 × 10−6 | 20502693 | – |
| rs4252126 | 6:161152294 | PLG (intronic) | 1,233 | 0.995 | 1f | yes | – | yes | CTCF | Pax-6 | – | – | – | – |
| rs4252135 | 6:161154232 | PLG (intronic) | 705 | 1.000 | 1f | yes | yes∗ | yes∗ | CTCF, RAD21, SMC3, ZNF143, NFKB, FOXA1, FOXA2, ZNF263 | COMP1 | gene expression of DDR1 in peripheral blood monocytes | 2.92 × 10−6 | 20502693 | – |
| rs34126283 | 6:161130002 | PLG (intronic) | 23,525 | 0.920 | 3a | yes | yes | yes | GATA2 | Cdc5, E2F, Evi-1, HDAC2, PLZF, Pou2f2, TATA | – | – | – | – |
| rs4252125 | 6:161152240 | PLG (missense) | 1,287 | 0.995 | 3a | yes | – | yes | CTCF, RAD21 | AP-3 | gene expression of UNQ9391 in liver | 4.09 × 10−14 | 22006096 | – |
| rs128738 | 5:131540875 | P4HA2 (intronic) | 0 | NA | ND | yes | yes∗ | – | – | BDP1 | – | – | – | 24 |
| rs101194 | 5:131515413 | P4HA2 (intergenic) | 25,462 | 0.901 | 3a | yes | yes∗ | yes | GATA2, P300 | CEBPB | gene expression of P4HA2 in lymphoblastoid cell lines | 2.00 × 10−7 | 17873874 | 30 |
| gene expression of SLC22A5 in peripheral blood monocytes | 1.16 × 10−8 | 20502693 | – | |||||||||||
| rs152054 | 5:131519540 | P4HA2 (intergenic) | 21,335 | 0.967 | 3a | yes∗ | yes∗ | yes∗ | CFOS | BAF155, BCL, Egr-1, Ets, FEV, GATA, NERF1a, Nrf-2, PU.1, Pax-5, SETDB1, SIX5, Tel2, Znf143 | – | – | – | 25 |
| rs152051 | 5:131539025 | P4HA2 (intronic) | 1,850 | 1.000 | 3a | yes | yes∗ | yes∗ | YY1, NFKB | AIRE, Foxa, GR, VDR | gene expression of P4HA2 in CEU-CHB-JPT-YRI lymphoblastoid cell lines | 4.66 × 10−9 | 17873874 | 28 |
| rs156023 | 5:131545168 | P4HA2 (intronic) | 4,293 | 0.967 | 3a | yes∗ | yes∗ | – | – | AIRE, LBP-1, LBP-9 | – | – | – | 22 |
Chromatin marks in blood tissues are indicated with an asterisk. Abbreviations are as follows: SNP, single nucleotide polymorphism; GRCh37, genome reference consortium human genome build 37; bp, base pair; RDB, Regulome database; PWM, position weight matrix ID; GRASP, genome-wide repository of associations between SNPs and phenotypes; PMID, PubMed identifier; eQTL, expression quantitative trait loci. RDB scores: 1f, eQTL + TF binding / DNase peak; 3a, TF binding + any motif + DNase peak; 6, other; ND, no data; NA, not applicable.
In addition, key regulatory proteins (some of them related to the immune response) bound by ChIP-seq experiments as well as relevant motif disruptions were reported in most cases (Table 2).
Consequently, most prioritized SNPs also correlated with eQTL effects in peripheral blood monocytes and lymphoblastoid cells (Tables 2 and S11). Interestingly, rs101194 and rs152054 were reported to affect P4HA2 expression in arterial tissues in the Genotype-Tissue Expression (GTEx) study,32 with rs101194 specifically acting as cis-eQTL in the aorta (Table S11).
Candidate Genes and Pathway Analysis
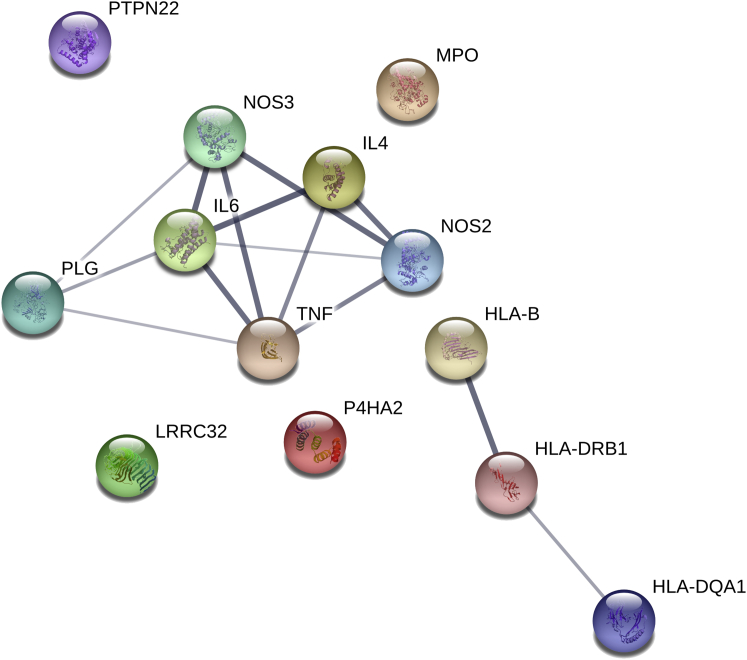
We also checked the statistical significance in our GWAS of previously described GCA-associated genomic regions (±100 kbp 3′ and 5′ of the reported gene) through candidate gene and Immunochip studies.5, 8 Significant associations after controlling for multiple testing were observed across most of the analyzed regions, with the myeloperoxidase (MPO [MIM: 606989]), tumor necrosis factor (TNF [MIM: 191160]), interleukin 6 (IL6 [MIM: 147620]), nitric oxide synthase 2 (NOS2 [MIM: 163730]), and PTPN22 regions harboring the strongest non-HLA hits (MPO rs10853005, p = 7.19 × 10−5, OR = 0.84; TNF rs4959077, p = 2.08 × 10−4, OR = 1.42; IL6 rs77741999, p = 2.17 × 10−4, OR = 1.20; NOS2 rs4255826, p = 5.75 × 10−4, OR = 0.87; and PTPN22 rs2476601, p = 7.88 × 10−4, OR = 1.24) (Tables S12 and S13). Subsequently, we accomplished a protein-protein interaction (PPI) and biological pathway enrichment analysis with those candidate hits showing statistically significant signals after correction for multiple testing in our GWAS (Table S13). The molecular network of the selected proteins had significantly more interactions than expected (number of nodes, 13; number of edges, 14; average node degree, 2.15; clustering coefficient, 0.851; expected number of edges, 3; PPI enrichment, p = 1.13 × 10−6; Figure 3). Interestingly, PLG represented a relevant node showing evidence of interaction with different proteins (e.g., NOS3, IL-6, and TNF) (Figure 3). Regarding the functional enrichments of the network, the most significantly associated GO processes were those related to regulation of both cell-cell adhesion and the immune/inflammatory response (Table S14).
Figure 3.
Interaction Network Formed for GCA Risk Loci
GWAS genes of this study and those previously identified through Immunochip and consistent candidate gene studies were included in the analysis. STRING database was used to look for both direct and indirect interactions among selected genes. The width of the blue lines indicates the reliability of each interaction.
Discussion
This study represents an unbiased screening of genetic variation in GCA at the genome-wide level. GCA was one of the few types of vasculitis in which GWAS data were not available. Therefore, the results presented here may help to better understand the pathogenic mechanisms underlying this condition and its genetic similarities with other vasculitides. In this sense, our data reinforce the idea of GCA as an archetypal HLA class II disease mediated by an antigen-driven immune response,33 which is in contrast not only to Takayasu’s arteritis (TAK [MIM: 207600]), another large-vessel vasculitis, but also to other forms of vasculitis associated with class I molecules like Behçet disease (MIM: 109650).34
Two independent association signals with GCA predisposition were observed within the HLA region, one located between HLA-DRA and HLA-DRB1 and another one between HLA-DQA1 and HLA-DQA2. This is consistent with the amino acid model that we proposed using imputed Immunochip data to explain the HLA class II association with GCA, which comprised the positions 13 and 56 of the DRβ1 and DQα1 molecules, respectively.8 Indeed, the strongest hit in the Immunochip study (which had partial overlap of the sample collections with this one) was a SNP that tagged the model (rs477515) in high LD with the two independent HLA lead SNPs that we observed in our GWAS (rs9268923: r2 = 0.84, D’ = 0.95; rs3957146: r2 = 0.23, D’ = 0.99).
Regarding the non-HLA associations, several variants within PLG were firmly associated with risk to develop GCA at the genome-wide level of significance in this study, although they all were in high LD and represented a single signal according to the dependence analysis. This gene encodes a secreted blood zymogen that can be converted through a complex conformational modification into two different active proteins, plasmin and angiostatin.35 The plasminogen system has an important role in a wide spectrum of physiological processes, including wound healing, fibrinolysis, angiogenesis, and lymphocyte recruitment and inflammation via production of cytokines and reactive oxygen species,36, 37 all of them relevant processes in GCA.38 Considering the opposite roles of plasmin and angiostatin in the induction of pro-angiogenic processes,36, 37 we hypothesize that the PLG risk alleles could unbalance the metabolism of its encoded protein leading to the characteristic pro-inflammatory phenotypes of GCA, although there is a lack of experimental support for this assumption. Interestingly, anti-plasminogen antibodies have been correlated with systemic disease activity in ANCA-associated vasculitis (MIM: 608710),39 a type of vasculitis involving small- to medium-sized blood vessels that is also strongly associated with HLA class II molecules.40 In addition, PLG has been shown to be a shared risk gene for coronary artery disease (MIM: 608320) and periodontitis (MIM: 260950), characterized by chronic inflammation,41 and multiple sclerosis (MIM: 126200), which is also an immune-mediated condition.42 Future studies aimed at improving the understanding of the zymogen activation may shed light into the PLG association with GCA.
P4HA2 represents the second non-HLA hit in our GWAS. This gene encodes an isoform of the alpha subunit of the collagen prolyl 4-hydroxylase, which catalyzes the formation of 4-hydroxyproline from proline residues that is essential for collagen biosynthesis, as it is required for the proper three-dimensional folding of newly synthesized procollagen chains.43 P4HA2 is considered an important hypoxia response gene and its expression is regulated by hypoxia-inducible factor-1 (HIF-1).44 Other relevant HIF-1-induced genes include serpin family E member 1 (SERPINE1 [MIM: 173360], which is the principal inhibitor of the plasmin activation) and genes that have been previously associated with GCA risk through candidate gene studies, such as vascular endothelial growth factor A (VEGFA [MIM: 192240], a potent endothelial growth factor), matrix metalloproteinase 9 (MMP9 [MIM: 120361], involved in the breakdown of extracellular matrix), and IL6 (a pro-inflammatory cytokine).5, 45, 46 Indeed, the hypoxic induction of all these HIF-1 target genes (and, indirectly, the inhibition of plasmin formation) is also related to the typical processes involved in the lesions of GCA-affected individuals, i.e., fibrosis, inflammation, destruction of the internal elastic lamina, and vascular remodeling, with proliferation and migration of medial myofibroblasts and neoangiogenesis.38, 45 The prioritized P4HA2 SNPs proposed here correlated with cis-eQTLs in immune cells and arterial tissues. In particular, expression data indicate that rs101194 may influence P4HA2 expression in whole blood, lymphoblastoid cells, and the aorta, which is one of the most severely affected vessels in GCA.32, 38, 47, 48, 49
Overall, our results are consistent with the currently accepted understanding of the pathophysiology of GCA, in which vascular remodeling and angiogenesis, either under hypoxic conditions or by dysregulation of hypoxia-sensitive genes, are critical to the development of the clinical presentations.50 Future analysis of the genetic overlap between GCA and other forms of vasculitis using GWAS data may help to elucidate whether these pathogenic processes are a common feature in vasculitides and to identify other relevant pathways for the development of GCA.
To summarize, through the analysis of common variation across the whole genome, we have identified PLG and P4HA2 as the main non-HLA genetic factors underlying GCA predisposition. Their crucial role in neoangiogenesis highlights the high relevance of this process in the pathogenic mechanisms leading to this form of vasculitis.
Consortia
The members of the Spanish GCA Group are José Luis Callejas, Luis Caminal-Montero, Marc Corbera-Bellalta, Eugenio de Miguel, J. Bernardino Díaz López, María Jesús García-Villanueva, Carmen Gómez-Vaquero, Mercedes Guijarro-Rojas, Ana Hidalgo-Conde, Begoña Marí-Alfonso, Agustín Martínez Berriochoa, Aleida Martínez Zapico, Víctor Manuel Martínez-Taboada, José A. Miranda-Filloy, Jordi Monfort, Norberto Ortego-Centeno, Mercedes Pérez-Conesa, Sergio Prieto-González, Enrique Raya, Raquel Ríos Fernández, Julio Sánchez-Martín, Bernardo Sopeña, Laura Tío, and Ainhoa Unzurrunzaga.
The members of the UKGCA Consortium include Andrew Gough, John D. Isaacs, Michael Green, Neil McHugh, Lesley Hordon, Sanjeet Kamath, Mohammed Nisar, Yusuf Patel, Cee-Seng Yee, Robert Stevens, Pradip Nandi, Anupama Nandagudi, Stephen Jarrett, Charles Li, Sarah Levy, Susan Mollan, Abdel Salih, Oliver Wordsworth, Emma Sanders, Esme Roads, Anne Gill, Lisa Carr, Christine Routledge, Karen Culfear, Asanka Nugaliyadde, Lynne James, Jenny Spimpolo, Andy Kempa, Felicity Mackenzie, Rosanna Fong, Genessa Peters, Bridie Rowbotham, Zahira Masqood, Jane Hollywood, Prisca Gondo, Rose Wood, Steve Martin, Lubna Haroon Rashid, James I. Robinson, Mike Morgan, Louise Sorensen, and John Taylor.
The members of the Vasculitis Clinical Research Consortium are Simon Carette, Sharon Chung, David Cuthbertson, Lindsy J. Forbess, Ora Gewurz-Singer, Gary S. Hoffman, Curry L. Koening, Kathleen M. Maksimowicz-McKinnon, Carol A. McAlear, Larry W. Moreland, Christian Pagnoux, Philip Seo, Ulrich Specks, Robert F. Spiera, Antoine Sreih, Kenneth J. Warrington, and Michael Weisman.
Acknowledgments
The authors thank Sofía Vargas, Sonia García, Gema Robledo, Lubna Haroon Rashid, and Steve Martin for technical support and the participants for their collaboration. Dr. Gregory Pugnet is thanked for participating actively in the recruitment of French samples. The Norwegian Systemic Vasculitis and Connective Tissue Disease Registry (NOSVAR) at Oslo University Hospital is acknowledged for providing data on the Norwegian case subjects.
F.D.C. was supported by Instituto de Salud Carlos III (ISCIII), Spain, through the RETICS Program RD12/0009/0004 (RIER), and by the Ramón y Cajal program of the Spanish Ministry of Economy and Competitiveness through the grant RYC-2014-16458. J.M. was founded by Junta de Andalucía through PAI group CTS180. The Vasculitis Clinical Research Consortium (VCRC) received support from the United States National Institute of Arthritis and Musculoskeletal and Skin Diseases (U54AR057319), the National Center for Research Resources (U54 RR019497), the Office of Rare Diseases Research, and the National Center for Advancing Translational Science. The VCRC is part of the Rare Diseases Clinical Research Network (RDCRN).
The UKGCA Consortium has been funded by Research into Ageing and The Wellcome Trust and is currently supported by the National Institute for Health Research (Clinician Scientist Fellowship for S.L.M., Leeds Musculoskeletal Biomedical Research Unit [A.W.M. and J.H.B.], and Diagnostic Evidence Co-operative [A.W.M.]), the Medical Research Council, and the Ann Wilks Memorial Fund. This article presents independent research funded in part by the National Institute for Health Research (NIHR). The views expressed are those of the authors and not necessarily those of the UK National Health Service, the NIHR, or the Department of Health.
Published: December 29, 2016
Footnotes
Supplemental Data include 3 figures and 14 tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.11.013.
Contributor Information
F. David Carmona, Email: dcarmona@ipb.csic.es.
Spanish CGA Group:
José Luis Callejas, Luis Caminal-Montero, Marc Corbera-Bellalta, Eugenio de Miguel, J. Bernardino Díaz López, María Jesús García-Villanueva, Carmen Gómez-Vaquero, Mercedes Guijarro-Rojas, Ana Hidalgo-Conde, Begoña Marí-Alfonso, Agustín Martínez Berriochoa, Aleida Martínez Zapico, Víctor Manuel Martínez-Taboada, José A. Miranda-Filloy, Jordi Monfort, Norberto Ortego-Centeno, Mercedes Pérez-Conesa, Sergio Prieto-González, Enrique Raya, Raquel Ríos Fernández, Julio Sánchez-Martín, Bernardo Sopeña, Laura Tío, and Ainhoa Unzurrunzaga
UKGCA Consortium:
Andrew Gough, John D. Isaacs, Michael Green, Neil McHugh, Lesley Hordon, Sanjeet Kamath, Mohammed Nisar, Yusuf Patel, Cee-Seng Yee, Robert Stevens, Pradip Nandi, Anupama Nandagudi, Stephen Jarrett, Charles Li, Sarah Levy, Susan Mollan, Abdel Salih, Oliver Wordsworth, Emma Sanders, Esme Roads, Anne Gill, Lisa Carr, Christine Routledge, Karen Culfear, Asanka Nugaliyadde, Lynne James, Jenny Spimpolo, Andy Kempa, Felicity Mackenzie, Rosanna Fong, Genessa Peters, Bridie Rowbotham, Zahira Masqood, Jane Hollywood, Prisca Gondo, Rose Wood, Steve Martin, Lubna Haroon Rashid, James I. Robinson, Mike Morgan, Louise Sorensen, and John Taylor
Vasculitis Clinical Research Consortium:
Simon Carette, Sharon Chung, David Cuthbertson, Lindsy J. Forbess, Ora Gewurz-Singer, Gary S. Hoffman, Curry L. Koening, Kathleen M. Maksimowicz-McKinnon, Carol A. McAlear, Larry W. Moreland, Christian Pagnoux, Philip Seo, Ulrich Specks, Robert F. Spiera, Antoine Sreih, Kenneth J. Warrington, and Michael Weisman
Web Resources
1000 Genomes, http://www.internationalgenome.org/
BEAGLE, http://faculty.washington.edu/browning/beagle/beagle.html
CaTS – Power Calculator, http://csg.sph.umich.edu/abecasis/CaTS/
ENCODE, https://www.encodeproject.org/
GEC: Genetic Type I Error Calculator, http://statgenpro.psychiatry.hku.hk/gec/
Gene Ontology Consortium, http://geneontology.org/
HaploReg, http://www.broadinstitute.org/mammals/haploreg/haploreg.php
IMPUTE2, http://mathgen.stats.ox.ac.uk/impute/impute_v2.html
LocusZoom, http://locuszoom.sph.umich.edu/locuszoom/
OMIM, http://www.omim.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
RegulomeDB, http://RegulomeDB.org/
Roadmap, http://www.roadmapepigenomics.org/
STRING, http://www.string-db.org/
Supplemental Data
References
- 1.Manolio T.A. Bringing genome-wide association findings into clinical use. Nat. Rev. Genet. 2013;14:549–558. doi: 10.1038/nrg3523. [DOI] [PubMed] [Google Scholar]
- 2.Jennette J.C., Falk R.J., Bacon P.A., Basu N., Cid M.C., Ferrario F., Flores-Suarez L.F., Gross W.L., Guillevin L., Hagen E.C. 2012 revised International Chapel Hill Consensus Conference Nomenclature of Vasculitides. Arthritis Rheum. 2013;65:1–11. doi: 10.1002/art.37715. [DOI] [PubMed] [Google Scholar]
- 3.Gonzalez-Gay M.A., Barros S., Lopez-Diaz M.J., Garcia-Porrua C., Sanchez-Andrade A., Llorca J. Giant cell arteritis: disease patterns of clinical presentation in a series of 240 patients. Medicine (Baltimore) 2005;84:269–276. doi: 10.1097/01.md.0000180042.42156.d1. [DOI] [PubMed] [Google Scholar]
- 4.Gonzalez-Gay M.A., Vazquez-Rodriguez T.R., Lopez-Diaz M.J., Miranda-Filloy J.A., Gonzalez-Juanatey C., Martin J., Llorca J. Epidemiology of giant cell arteritis and polymyalgia rheumatica. Arthritis Rheum. 2009;61:1454–1461. doi: 10.1002/art.24459. [DOI] [PubMed] [Google Scholar]
- 5.Carmona F.D., González-Gay M.A., Martín J. Genetic component of giant cell arteritis. Rheumatology (Oxford) 2014;53:6–18. doi: 10.1093/rheumatology/ket231. [DOI] [PubMed] [Google Scholar]
- 6.Serrano A., Márquez A., Mackie S.L., Carmona F.D., Solans R., Miranda-Filloy J.A., Hernández-Rodríguez J., Cid M.C., Castañeda S., Morado I.C., UK GCA Consortium Spanish GCA Consortium Identification of the PTPN22 functional variant R620W as susceptibility genetic factor for giant cell arteritis. Ann. Rheum. Dis. 2013;72:1882–1886. doi: 10.1136/annrheumdis-2013-203641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Márquez A., Hernández-Rodríguez J., Cid M.C., Solans R., Castañeda S., Fernández-Contreras M.E., Ramentol M., Morado I.C., Narváez J., Gómez-Vaquero C., Spanish GCA Consortium Influence of the IL17A locus in giant cell arteritis susceptibility. Ann. Rheum. Dis. 2014;73:1742–1745. doi: 10.1136/annrheumdis-2014-205261. [DOI] [PubMed] [Google Scholar]
- 8.Carmona F.D., Mackie S.L., Martín J.E., Taylor J.C., Vaglio A., Eyre S., Bossini-Castillo L., Castañeda S., Cid M.C., Hernández-Rodríguez J., Spanish GCA Group A large-scale genetic analysis reveals a strong contribution of the HLA class II region to giant cell arteritis susceptibility. Am. J. Hum. Genet. 2015;96:565–580. doi: 10.1016/j.ajhg.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hunder G.G., Bloch D.A., Michel B.A., Stevens M.B., Arend W.P., Calabrese L.H., Edworthy S.M., Fauci A.S., Leavitt R.Y., Lie J.T. The American College of Rheumatology 1990 criteria for the classification of giant cell arteritis. Arthritis Rheum. 1990;33:1122–1128. doi: 10.1002/art.1780330810. [DOI] [PubMed] [Google Scholar]
- 10.Barrett J.H., Iles M.M., Harland M., Taylor J.C., Aitken J.F., Andresen P.A., Akslen L.A., Armstrong B.K., Avril M.F., Azizi E., GenoMEL Consortium Genome-wide association study identifies three new melanoma susceptibility loci. Nat. Genet. 2011;43:1108–1113. doi: 10.1038/ng.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bentham J., Morris D.L., Cunninghame Graham D.S., Pinder C.L., Tombleson P., Behrens T.W., Martín J., Fairfax B.P., Knight J.C., Chen L. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat. Genet. 2015;47:1457–1464. doi: 10.1038/ng.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Consortium W.T.C.C., Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubois P.C., Trynka G., Franke L., Hunt K.A., Romanos J., Curtotti A., Zhernakova A., Heap G.A., Adány R., Aromaa A. Multiple common variants for celiac disease influencing immune gene expression. Nat. Genet. 2010;42:295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gregersen P.K., Kosoy R., Lee A.T., Lamb J., Sussman J., McKee D., Simpfendorfer K.R., Pirskanen-Matell R., Piehl F., Pan-Hammarstrom Q. Risk for myasthenia gravis maps to a (151) Pro→Ala change in TNIP1 and to human leukocyte antigen-B∗08. Ann. Neurol. 2012;72:927–935. doi: 10.1002/ana.23691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Radstake T.R., Gorlova O., Rueda B., Martin J.E., Alizadeh B.Z., Palomino-Morales R., Coenen M.J., Vonk M.C., Voskuyl A.E., Schuerwegh A.J., Spanish Scleroderma Group Genome-wide association study of systemic sclerosis identifies CD247 as a new susceptibility locus. Nat. Genet. 2010;42:426–429. doi: 10.1038/ng.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rotger M., Dang K.K., Fellay J., Heinzen E.L., Feng S., Descombes P., Shianna K.V., Ge D., Günthard H.F., Goldstein D.B., Telenti A., Swiss HIV Cohort Study. Center for HIV/AIDS Vaccine Immunology Genome-wide mRNA expression correlates of viral control in CD4+ T-cells from HIV-1-infected individuals. PLoS Pathog. 2010;6:e1000781. doi: 10.1371/journal.ppat.1000781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stahl E.A., Raychaudhuri S., Remmers E.F., Xie G., Eyre S., Thomson B.P., Li Y., Kurreeman F.A., Zhernakova A., Hinks A., BIRAC Consortium. YEAR Consortium Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat. Genet. 2010;42:508–514. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howie B.N., Donnelly P., Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skol A.D., Scott L.J., Abecasis G.R., Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat. Genet. 2006;38:209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 22.Pruim R.J., Welch R.P., Sanna S., Teslovich T.M., Chines P.S., Gliedt T.P., Boehnke M., Abecasis G.R., Willer C.J. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boyle A.P., Hong E.L., Hariharan M., Cheng Y., Schaub M.A., Kasowski M., Karczewski K.J., Park J., Hitz B.C., Weng S. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ward L.D., Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44(D1):D877–D881. doi: 10.1093/nar/gkv1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gene Ontology Consortium Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., The Gene Ontology Consortium Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas P.D., Campbell M.J., Kejariwal A., Mi H., Karlak B., Daverman R., Diemer K., Muruganujan A., Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li M.X., Yeung J.M., Cherny S.S., Sham P.C. Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum. Genet. 2012;131:747–756. doi: 10.1007/s00439-011-1118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.GTEx Consortium Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carmona F.D., Martín J., González-Gay M.A. New insights into the pathogenesis of giant cell arteritis and hopes for the clinic. Expert Rev. Clin. Immunol. 2016;12:57–66. doi: 10.1586/1744666X.2016.1089173. [DOI] [PubMed] [Google Scholar]
- 34.Carmona F.D., Martín J., González-Gay M.A. Genetics of vasculitis. Curr. Opin. Rheumatol. 2015;27:10–17. doi: 10.1097/BOR.0000000000000124. [DOI] [PubMed] [Google Scholar]
- 35.Law R.H., Abu-Ssaydeh D., Whisstock J.C. New insights into the structure and function of the plasminogen/plasmin system. Curr. Opin. Struct. Biol. 2013;23:836–841. doi: 10.1016/j.sbi.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 36.Miles L.A., Parmer R.J. Plasminogen receptors: the first quarter century. Semin. Thromb. Hemost. 2013;39:329–337. doi: 10.1055/s-0033-1334483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moser T.L., Stack M.S., Asplin I., Enghild J.J., Højrup P., Everitt L., Hubchak S., Schnaper H.W., Pizzo S.V. Angiostatin binds ATP synthase on the surface of human endothelial cells. Proc. Natl. Acad. Sci. USA. 1999;96:2811–2816. doi: 10.1073/pnas.96.6.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ly K.H., Régent A., Tamby M.C., Mouthon L. Pathogenesis of giant cell arteritis: More than just an inflammatory condition? Autoimmun. Rev. 2010;9:635–645. doi: 10.1016/j.autrev.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 39.Hao J., Wang C., Gou S.J., Zhao M.H., Chen M. The association between anti-plasminogen antibodies and disease activity in ANCA-associated vasculitis. Rheumatology (Oxford) 2014;53:300–306. doi: 10.1093/rheumatology/ket345. [DOI] [PubMed] [Google Scholar]
- 40.Lyons P.A., Rayner T.F., Trivedi S., Holle J.U., Watts R.A., Jayne D.R., Baslund B., Brenchley P., Bruchfeld A., Chaudhry A.N. Genetically distinct subsets within ANCA-associated vasculitis. N. Engl. J. Med. 2012;367:214–223. doi: 10.1056/NEJMoa1108735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schaefer A.S., Bochenek G., Jochens A., Ellinghaus D., Dommisch H., Güzeldemir-Akçakanat E., Graetz C., Harks I., Jockel-Schneider Y., Weinspach K. Genetic evidence for PLASMINOGEN as a shared genetic risk factor of coronary artery disease and periodontitis. Circ Cardiovasc Genet. 2015;8:159–167. doi: 10.1161/CIRCGENETICS.114.000554. [DOI] [PubMed] [Google Scholar]
- 42.Sadovnick A.D., Traboulsee A.L., Bernales C.Q., Ross J.P., Forwell A.L., Yee I.M., Guillot-Noel L., Fontaine B., Cournu-Rebeix I., Alcina A. Analysis of plasminogen genetic variants in multiple sclerosis patients. G3 (Bethesda) 2016;6:2073–2079. doi: 10.1534/g3.116.030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Myllyharju J. Prolyl 4-hydroxylases, the key enzymes of collagen biosynthesis. Matrix Biol. 2003;22:15–24. doi: 10.1016/s0945-053x(03)00006-4. [DOI] [PubMed] [Google Scholar]
- 44.Gilkes D.M., Bajpai S., Chaturvedi P., Wirtz D., Semenza G.L. Hypoxia-inducible factor 1 (HIF-1) promotes extracellular matrix remodeling under hypoxic conditions by inducing P4HA1, P4HA2, and PLOD2 expression in fibroblasts. J. Biol. Chem. 2013;288:10819–10829. doi: 10.1074/jbc.M112.442939. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45.Deng W., Feng X., Li X., Wang D., Sun L. Hypoxia-inducible factor 1 in autoimmune diseases. Cell. Immunol. 2016;303:7–15. doi: 10.1016/j.cellimm.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 46.Dimova E.Y., Samoylenko A., Kietzmann T. Oxidative stress and hypoxia: implications for plasminogen activator inhibitor-1 expression. Antioxid. Redox Signal. 2004;6:777–791. doi: 10.1089/1523086041361596. [DOI] [PubMed] [Google Scholar]
- 47.Stranger B.E., Nica A.C., Forrest M.S., Dimas A., Bird C.P., Beazley C., Ingle C.E., Dunning M., Flicek P., Koller D. Population genomics of human gene expression. Nat. Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Westra H.J., Peters M.J., Esko T., Yaghootkar H., Schurmann C., Kettunen J., Christiansen M.W., Fairfax B.P., Schramm K., Powell J.E. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 2013;45:1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lappalainen T., Sammeth M., Friedländer M.R., ’t Hoen P.A., Monlong J., Rivas M.A., Gonzàlez-Porta M., Kurbatova N., Griebel T., Ferreira P.G., Geuvadis Consortium Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaiser M., Younge B., Björnsson J., Goronzy J.J., Weyand C.M. Formation of new vasa vasorum in vasculitis. Production of angiogenic cytokines by multinucleated giant cells. Am. J. Pathol. 1999;155:765–774. doi: 10.1016/S0002-9440(10)65175-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.