Abstract
Circulating cytokines and growth factors are regulators of inflammation and have been implicated in autoimmune and metabolic diseases. In this genome-wide association study (GWAS) of up to 8,293 Finns we identified 27 genome-widely significant loci (p < 1.2 × 10−9) for one or more cytokines. Fifteen of the associated variants had expression quantitative trait loci in whole blood. We provide genetic instruments to clarify the causal roles of cytokine signaling and upstream inflammation in immune-related and other chronic diseases. We further link inflammatory markers with variants previously associated with autoimmune diseases such as Crohn disease, multiple sclerosis, and ulcerative colitis and hereby elucidate the molecular mechanisms underpinning these diseases and suggest potential drug targets.
Introduction
Genome-wide association studies (GWASs) have identified multiple loci for inflammatory diseases.1, 2 However, the biochemical pathways underlying the link from locus to complex disease have often remained elusive. Cytokines and growth factors (hereafter cytokines) are inflammatory regulators, and therefore important intermediate phenotypes for inflammatory diseases.3 These intermediate phenotypes can be exploited in GWASs to elucidate the biochemical pathways underlying the link from locus to disease susceptibility. Cytokines have been implicated in the progression of inflammatory bowel diseases (MIM: 266600), multiple sclerosis (MIM: 126200), atherosclerosis (MIM: 108725), and various forms of cancer. Here, we studied the genetic basis for blood levels of multiple cytokines to gain insights to the molecular intermediates and causal pathways related to inflammatory diseases.
Human genetics can guide the prioritization of pharmaceutical targets and inform drug development. Individuals harboring loss of functional alleles are in some ways analogous to participants in a randomized trial receiving an inhibitor because both presumably result in reduced protein function. Pharmaceutical targets with human genetic support have twice the probability for eventual regulatory approval as compared to non-supported targets.4 Cytokines are attractive targets since they exert their effects via cell surface receptors that are readily druggable. Furthermore, since inflammatory diseases often share common pathology, the indications for an approved drug can be expanded. For example, adalimumab, a tumor necrosis factor alpha (TNF-α) antibody, originally gained FDA approval for rheumatoid arthritis (MIM: 180300) but was later approved for juvenile idiopathic arthritis (MIM: 604302), ankylosing spondylitis (MIM: 106300), psoriatic arthritis (MIM: 607507), and Crohn disease (MIM: 266600).5, 6, 7
To gain insights into the molecular intermediates and causal pathways related to autoimmune and metabolic diseases, we studied the genetic basis for circulating levels of 41 cytokines. Our results provide information on the genetic regulation of normal physiological variation of cytokines among healthy individuals and potentially inform drug development.
Material and Methods
Study Populations
The Cardiovascular Risk in Young Finns Study
The Cardiovascular Risk in Young Finns Study (YFS) is a multicenter follow-up study with randomly chosen subjects from the Finnish cities of Helsinki, Kuopio, Oulu, Tampere, and Turku and their rural surroundings. The study began in 1980 when 3,596 children and young adults participated in the first cross-sectional survey. The follow-up visits have been conducted in 1983, 1986, 1989, 2001, 2007, and 2011. The present cross-sectional study includes 2,019 unrelated individuals who participated in the 2007 follow-up and who had both cytokine measurements and genotype data available. In addition, gene expression data from 1,664 participants of the 2011 follow-up were analyzed for the present study. All participants gave written informed consent and the study was approved by local ethics committees.8
FINRISK
FINRISK surveys are population-based cross-sectional studies conducted every 5 years to monitor the levels of chronic disease risk factors in Finland. Each survey includes 25- to 74-year-old randomly chosen subjects from five geographical areas of Finland. The present study analyzes cytokine data from participants of the 1997 and 2002 surveys. The study visit includes a clinical examination and semi-fasting blood sampling. For eQTL analyses, a peripheral blood sample was drawn to quantify mRNA expression profiles from a subset of 513 FINRISK2007 participants living in the Helsinki area.9 All participants gave written informed consent.
Cytokine Quantification
From YFS and FINRISK2002, a total of 48 cytokines were measured by using Bio-Rad’s premixed Bio-Plex Pro Human Cytokine 27-plex Assay and 21-plex Assay, and Bio-Plex 200 reader with Bio-Plex 6.0 software.9 The assays were performed according to manufacturer’s instructions, except that the amount of beads, detection antibodies, and streptavidin-phycoerythrin conjugate were used with 50% lower concentrations than recommended. Only measures within the cytokine-specific detection range were included in the analyses. Cytokines with >90% of values missing were excluded (7 out of 48).
In FINRISK1997, a total of 17 cytokines overlapped with those measured in FINRISK2002 and YFS and were thus included in the GWAS. Genome-wide and cytokine data were available from up to 4,608 FINRISK1997 participants and up to 1,705 FINRISK2002 participants. The cytokine quantification was performed from EDTA plasma in FINRISK1997, from heparin plasma in FINRISK2002, and from serum in YFS.
Statistical Analyses
Subjects whose cytokine concentration were below or above laboratory analysis detection limits were omitted from the analyses. Cytokine distributions were first normalized with inverse transformation. The transformed phenotypes were then adjusted for age, sex, body mass index, and the first ten genetic principal components by calculating residuals of linear regression model. Subsequently, another inverse transformation was performed for model residuals to ensure normally distributed phenotypes. All effect sizes are hereby in SD-scaled units.
Genome-wide association testing was performed using Snptest2 software v.2.5beta.10, 11 Imputation inaccuracy was assessed with missing data likelihood score test. Allele effects were estimated from additive model (-frequentist 1). To prevent centering and scaling of phenotypes, use_raw_phenotypes option was enabled.
Meta-analyses were performed using METAL software (v.2011-03-25).12 Prior to meta-analysis, all markers with imputation info < 0.7, model fit info < 0.7, and minor allele count < 10 were excluded. After filtering, 10.7 million markers were included in the meta-analysis. Each association test was weighted by sample size. Genomic control correction was used to account for population stratification and cryptic relatedness. Heterogeneity statistics based on Cochrane’s Q-test were calculated for all markers included in meta-analysis to estimate heterogeneity of effect sizes across the three cohorts. Statistical significance was set at p < 1.2 × 10−9, i.e., dividing the established genome-wide statistical significance (p < 5 × 10−8) by 41 (number of cytokines analyzed). Random effect meta-analyses were performed with R software. To assess whether cytokine-associated loci harbor multiple variants independently associated with cytokine concentration, the models were further adjusted with locus-specific lead variant and the model was fitted against the concentrations of all included cytokines. All independent variants were used to calculate the total variance explained by the identified loci. The proportion of total variance explained by independent variants was calculated by the following formula:
where β is the regression coefficient and MAF is minor allele frequency.13
Instrumental variable analysis of the VEGF cascade effects were performed with two-stage least-squares regression using ivreg from the R-package AER.14 A weighted gene score based on the sum of cytokine-increasing alleles of the two independent variants associated with VEGF (rs6921438 and rs12214617) was used as the instrument. The estimates from the three individual cohorts were combined with inverse variance weighted meta-analyses. The observational associations were calculated as linear regression between the cytokines after adjusting for age, sex, body-mass index, and the first ten genetic principal components.
eQTL Analyses
Gene expression data from peripheral blood leukocytes were available from 1,664 participants from the YFS 30-year follow-up survey in 2011. After preprocessing, the expression levels were analyzed with an Illumina HumanHT-12 v.4 Expression BeadChip. Raw Illumina probe data was exported from Beadstudio and analyzed in R using the Bioconductor packages. The HT-12 v4 BeadChip contains 47,231 expression probes. Probes expressed by <5% of the participants were excluded, resulting in 19,637 probes.15
Gene expression data from peripheral blood leukocytes were analyzed for 518 individuals from FINRISK2007 with Illumina HumanHT-12 v.3 Expression BeadChips. Probes that were not autosomal, were complementary to cDNA from erythrocyte globin components, and were mapped to more than one genomic position were excluded. The filtering resulted in total of 35,419 probes included in the analyses.
Linear regression with probe as a dependent variable was used to test associations between cytokine-associated variants and transcripts. Age and sex were used as covariates. Genotype dosage was calculated for each included variant with Qctool software. Probes with p < 0.05 in both YFS and FINRISK07 were included in the eQTL meta-analyses. The eQTL results were combined by inverse variance weighted meta-analysis.
Prioritizing Variants from the Identified Loci
An individual locus might harbor hundreds of cytokine-associated variants. To highlight the potentially causal variants, we explored the identified loci using FINEMAP software.16 FINEMAP utilizes shotgun stochastic search algorithm to identify the most likely causal variants within a cytokine-associated locus. Variants within ±500 kb of the lead variant were included in the analyses. In addition to meta-analysis association statistics, FINEMAP software requires a variant correlation matrix as an input which was here calculated by using YFS genotype data.
Database Searches
To assess whether cytokine concentration could contribute to disease pathogenesis, we searched the GWAS catalog for complex trait-associated variants within 1Mb window from the cytokine lead variants.17 The catalog was downloaded on July 6, 2015 and catalog variants with p < 5 × 10−8 were selected for further analyses. The statistical significance for variant-cytokine association was inferred at p < 1.2 × 10−9.
Results
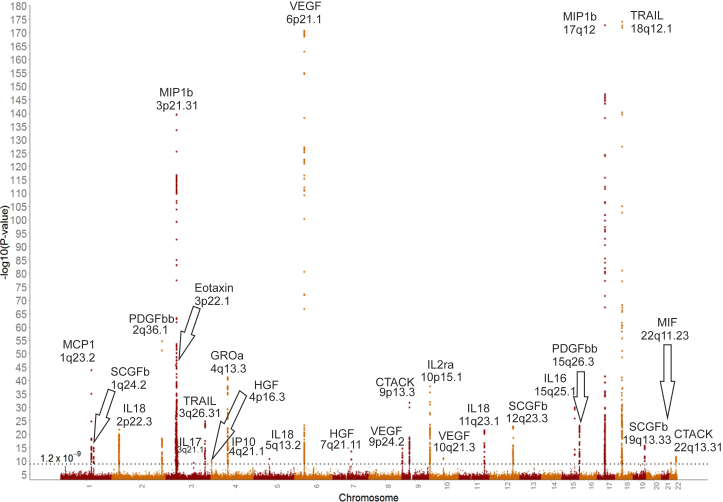
We performed a GWAS for the circulating concentrations of 41 cytokines (listed in Table S1). The GWAS meta-analysis included up to 8,293 Finnish individuals from three independent population cohorts: the Cardiovascular Risk in Young Finns Study (YFS), FINRISK1997, and FINRISK2002. Study cohort characteristics are reported in Table S1. On average, the YFS participants are younger than the FINRISK1997 and FINRISK2002 participants (37 versus 60 years). Correlations between the cytokine levels are shown in Figure S1. Four cytokines (VEGF, IL-12p70, IL-13, and IL-10) form a tightly correlated cluster. Most of the cytokines included in the study are positively correlated with each other.
GWAS Identifies 27 Loci Associated with Circulating Cytokines
Using an additive genetic model adjusted for the first ten genetic principal components, age, sex, and body mass index, we tested for univariate associations between 10.7 million genetic polymorphisms and 41 cytokine concentrations. Genotype imputation was performed based on reference haplotypes provided by the 1000 Genomes Project September 2013 release.18 The meta-analyses identified 27 genome-wide significant loci (p < 1.2 × 10−9 accounting for 41 measures), 17 of which have not been associated with cytokines, C-reactive protein, or white blood cell count in previous GWASs (Table 1 and Figure 1). With the aforementioned threshold for genome-wide significance and sample size, we had 84% power to detect an association with effect size of 0.11 standard deviations per one additional copy of effect allele assuming minor allele frequency (MAF) of 50%. The potential candidate genes of the 27 genetic variants are also reported in Table 1. Results from the random effect meta-analysis for heterogenic variants (HetPval < 0.1) are reported in Table S2. Effect estimates from the random effect models were coherent with effect estimates from the fixed effect models; however, for ten of the heterogenic loci, the p value did not reach the genome-wide significance (1.2 × 10−9) with the random effect model. The strongest association signals without heterogeneity are reported in Table S3. Complete summary statistics from meta-analyses are available online. Cytokine concentration means per allele in absolute concentration units for lead variants are reported in Table S4 and genomic lambda values in Table S5. The lambda values ranged from 0.98 to 1.02, indicating that there was no overall inflation in the test statistics.
Table 1.
All Loci Associated with the Concentration of at least One of the Studied Cytokines
| Cytokine | Rs-id | Gene | Locus | Position | A1 | A2 | MAF | Info | Beta (SD) | SE (SD) | p Value | HetPVal |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MCP1 | rs12075 | ACKR1 | 1q23.2 | 159175354 | G | A | 0.47 | 1.00 | 0.219 | 0.015 | 1.44 × 10−44 | 8.76 × 10−78 |
| SCGFb | rs4656185 | F5 | 1q24.2 | 169476326 | G | A | 0.29 | 1.00 | 0.205 | 0.026 | 1.16 × 10−15 | 7.37 × 10−6 |
| IL-18 | rs385076 | NLRC4 | 2p22.3 | 32489851 | T | C | 0.34 | 0.97 | 0.243 | 0.025 | 1.66 × 10−22 | 0.07 |
| PDGFbb | rs13412535 | SERPINE2 | 2q36.1 | 224874874 | G | A | 0.19 | 0.87 | 0.335 | 0.021 | 2.46 × 10−55 | 1.86 × 10−10 |
| Eotaxin | rs 2228467 | ACKR2 | 3p22.1 | 42906116 | T | C | 0.07 | 1.00 | 0.416 | 0.029 | 2.27 × 10−46 | 0.02 |
| MIP1b | rs113010081 | CCRL2 | 3p21.31 | 46457412 | T | C | 0.13 | 0.89 | 0.595 | 0.024 | 3.85 × 10−140 | 0.33 |
| IL-17 | rs1530455 | PDIA5 | 3q21.1 | 122854899 | T | C | 0.36 | 0.95 | −0.108 | 0.017 | 4.71 × 10−10 | 0.81 |
| TRAIL | esv2656942 | TNFSF10 | 3q26.31 | 172274209 | A | – | 0.04 | 0.98 | −0.431 | 0.041 | 1.74 × 10−25 | 0.74 |
| HGF | rs3748034 | HGFAC | 4p16.3 | 3446091 | G | T | 0.12 | 0.97 | 0.150 | 0.023 | 1.79 × 10−10 | 0.50 |
| GROa | rs508977 | CXCL1 | 4q13.3 | 74762383 | T | G | 0.24 | 1.00 | 0.380 | 0.028 | 7.56 × 10−42 | 0.03 |
| IP10 | rs141053179 | CXCL10 | 4q21.1 | 77589911 | C | G | 0.01 | 0.99 | 1.103 | 0.140 | 2.81 × 10−15 | 0.97 |
| IL-18 | rs17229943 | OCLN | 5q13.2 | 68682536 | A | C | 0.08 | 0.84 | 0.312 | 0.046 | 1.62 × 10−11 | 0.98 |
| VEGF | rs6921438 | VEGFA | 6p21.1 | 43925607 | G | A | 0.47 | 0.98 | −0.490 | 0.018 | 2.09 × 10−171 | 2.19 × 10−47 |
| HGF | rs5745687 | HGF | 7q21.11 | 81359051 | C | T | 0.04 | 1.00 | −0.307 | 0.041 | 2.71 × 10−14 | 0.29 |
| VEGF | rs7030781 | VLDLR | 9p24.2 | 2686273 | A | T | 0.42 | 0.98 | −0.137 | 0.017 | 2.57 × 10−15 | 0.01 |
| CTACK | rs2070074 | IL11RA | 9p13.3 | 34649442 | A | G | 0.11 | 1.00 | −0.447 | 0.037 | 1.79 × 10−32 | 5.55 × 10−5 |
| IL-2ra | rs12722497 | IL2RA | 10p15.1 | 6095928 | C | A | 0.06 | 0.99 | 0.628 | 0.049 | 1.57 × 10−38 | 0.18 |
| VEGF | rs10761731 | JMJD1C | 10q21.3 | 65027610 | A | T | 0.36 | 0.99 | 0.119 | 0.017 | 1.01 × 10−11 | 0.54 |
| IL-18 | rs71478720 | IL18 | 11q23.1 | 112009605 | C | T | 0.24 | 0.99 | −0.267 | 0.028 | 3.71 × 10−22 | 0.59 |
| SCGFb | rs187503377 | STAB2 | 12q23.3 | 104261835 | C | T | 0.02 | 0.93 | 0.965 | 0.097 | 1.34 × 10−23 | 0.07 |
| IL-16 | rs4778636 | IL16 | 15q25.1 | 81591639 | G | A | 0.04 | 1.00 | −0.727 | 0.063 | 1.11 × 10−30 | 0.22 |
| PDGFbb | rs4965869 | PCSK6 | 15q26.3 | 101990320 | C | T | 0.26 | 1.00 | 0.184 | 0.018 | 5.66 × 10−24 | 7.2 × 10−3 |
| MIP1b | rs113877493 | CCL4L1 | 17q12 | 34812273 | C | T | 0.17 | 0.92 | −0.612 | 0.022 | 1.62 × 10−173 | 3.58 × 10−7 |
| TRAIL | rs192989810 | MEP1B | 18q12.1 | 29783353 | C | T | 0.01 | 0.96 | 2.221 | 0.078 | 8.40 × 10−175 | 5.92 × 10−3 |
| SCGFb | rs116924815 | CLEC11A | 19q13.33 | 51230733 | C | T | 0.03 | 0.97 | 0.608 | 0.074 | 1.74 × 10−16 | 0.03 |
| MIF | rs2330634 | MIF | 22q11.23 | 24250795 | C | G | 0.37 | 0.99 | −0.156 | 0.025 | 4.53 × 10−10 | 0.11 |
| CTACK | rs201003839 | PPARA | 22q13.31 | 46534944 | A | ACGGGC | 0.27 | 0.99 | 0.190 | 0.027 | 2.42 × 10−12 | 1.36 × 10−3 |
The same locus is occasionally associated with more than one cytokine but only the strongest trait association is reported in the table. Allele 2 is the effect allele. Beta and SE are in SD units. All positions refer to human genome build 37. Info column reports the imputation quality metric calculated by the Impute2 software. HetPVal reports the effect size heterogeneity statistics between the cohorts.
Figure 1.
Combined Manhattan Plot of Meta-analysis Results from the GWAS of 41 Circulating Cytokines
Loci not previously associated with cytokine concentration in the GWAS are bolded. The horizontal dashed line indicates the genome-wide significance threshold (p < 1.2 × 10−9) accounting for the number of cytokines tested in the study.
Loci with large effects included rs192989810 from 18q12.1 for TNF-related apoptosis inducing ligand (TRAIL) with β = 2.2 SD at MAF of 1%. This variant is located within MEP1B (MIM: 600389), and leukocytes from Mep1b−/− mice have been shown to have impaired migration in response to monocyte chemotactic protein-1 (MCP1) and macrophage inflammatory protein-1α (MIP1a).19 Additional low-frequency variants with large effects included rs141053179 (β = 1.1 SD) for interferon gamma-induced protein 10 (IP10), rs116924815 (β = 0.61 SD) for stem cell growth factor beta (SCGFb), and rs4778636 (β = −0.73 SD) for interleukin-16 (IL-16). Common variants (MAF > 5%) with large effects (β up to −0.49) included rs2070074 and rs13412535 for cutaneous T cell attracting (CTACK) and platelet derived growth factor BB (PDGFbb).
Our results confirmed associations for ten genome-wide significant loci previously identified. These include two loci with interleukin-18 (IL-18) concentration (2p22.3 and 11q23.1).20, 21 We also replicated two loci previously associated with VEGF (6p21.1 and 9p24.2)22 and one additional locus previously associated with monocyte chemotactic protein-1 (MCP1; 1q23.2).23 Manhattan plots, QQ plots, and detailed plots from heterogeneity for each cytokine are provided in the supplement (Figures S2–S124).
At 6p21.1, the signal associated with VEGF was narrow and suggested that a few causal variants might explain the signal (Figure S55). After conditioning on rs6921438, the analysis revealed another independent variant in the locus (rs12214617) that is located 46 kb from rs6921438. According to Ensembl, rs12214617 is located in the promoter flanking region, suggesting a potential role in gene regulation. A previous GWAS of VEGF levels identified the same lead variant (rs6921438) in 6p21 associated with VEGF concentration.22 Our results confirm this association but the secondary associations signal was different from ours, potentially due to different imputation reference.
For some of the genetic loci associated with cytokines, the association signal is much wider, making the identification of the potential functional variant more challenging. For instance, the signal at 2p22.3 associated with IL-18 levels spans approximately 600 kb.
The association peak for PDGFbb in 2q36.1 appeared to be formed by two variants (rs13412535 and rs68066031) in strong linkage disequilibrium. Conditional analysis demonstrated that the signal could be fully explained by rs13412535. According to Ensembl, rs13412535 is located in a transcription factor binding cite and DNase peak near SERPINE2 (MIM: 177010) and is potentially affecting gene expression.
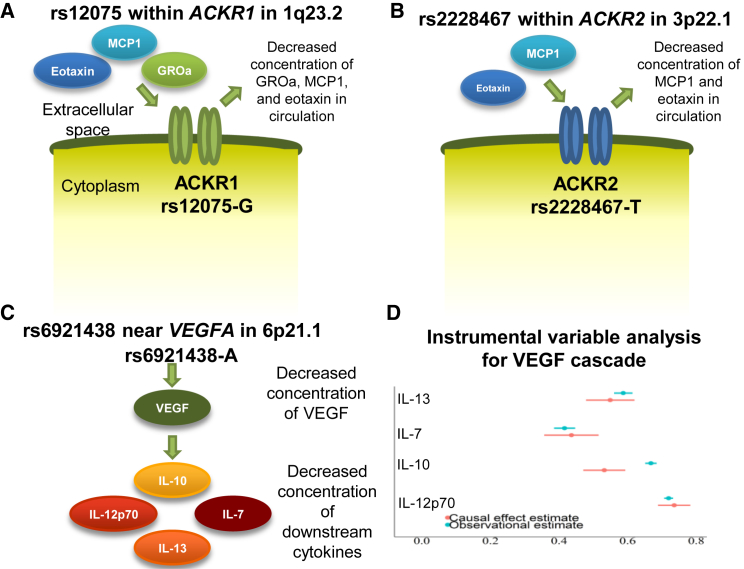
Four of the identified loci were associated with the concentration of more than one cytokine (1q23.2, 6p21.1, 3p22.1, and 10q21.3). The multiple associations observed at 1q23.2 and 3p22.1 appeared to be mediated through altering a cytokine receptor. rs12075, located in 1q23.2, is significantly associated with three cytokines: eotaxin, MCP1, and growth-regulated oncogene-α (GROa). rs12075 is located within ACKR1 (MIM: 613665), encoding a receptor for multiple cytokines and is also a human erythrocyte blood group antigen.24, 25 This suggests that the effect of rs12075 on the three cytokines is a result of altered binding of these cytokines to the ACKR1 receptor (Figure 2A). Similarly, the effect of rs2228467 at 3p22.1 on eotaxin and MCP1 appears to be mediated via chemokine-binding proteinatypical chemokine receptor 2 (ACKR2 [MIM: 602648]) (Figure 2B). rs6921438, located at 6p21.1, is associated with concentration of five cytokines (VEGF, IL-12p70, IL-7, IL-10, and IL-13). This locus contains VEGFA (MIM: 192240) encoding VEGF and suggests that VEGF might regulate the concentration of IL-12p70, IL-7, IL-10, and IL-13. To test this directional mediation, we performed Mendelian randomization analysis using a genetic score from rs6921438 and rs12214617 (variants at 6p21.1 independently associated with VEGF) as the instrumental variable.14 The observational and causal effect estimates matched, supporting the role of VEGF as an upstream regulator of IL-12p70, IL-7, IL-10, and IL-13 (Figures 2C and 2D).
Figure 2.
Variants Genome-widely Associated with More than One Cytokine
Variant rs12075-A is associated with increased concentrations of eotaxin, monocyte chemotactic protein-1 (MCP1), and growth regulated oncogene-α (GROa). In addition, rs12075-A is associated with increased ACKR1 mRNA. Since ACKR1 is a cell surface receptor, the cluster effect on the three cytokines is unlikely to be mediated (in a cascade) through any of the three cytokines associated with rs12075 (A). Similarly, effect of rs2228467 is mediated through a cell surface receptor (ACKR2) (B). Variant rs6921438 in 6p21.1 is located near VEGFA. This suggests that VEGF is causally regulating the concentrations of IL-7, IL-10, IL-12p70, and IL-13 (C). Instrumental variable analyses indicate that the inter-correlations between the cytokines match the causal effect estimates; this suggests a causal role of VEGF in mediating a cascade effect on IL-7, IL-10, IL-12p70, and IL-13 (D).
Variants in 10q21.3 were associated with VEGF and IL-12p70. The lead variant within this locus is rs10761731, which has the smallest association test p value for VEGF. The ratio of the IL-12p70 effect size divided by the effect size of VEGF remains approximately constant in both loci (6p21.1 and 10q21.3), suggesting tight regulatory effect of VEGF on IL-12p70 concentration.
To assess whether the genetic loci identified harbor additional independent variants, we performed conditional meta-analyses for each cytokine-lead variant pairs. The 17q12 locus harbored five variants independently associated with macrophage inflammatory protein-1β (MIP1b) concentration (Table S6). These five statistically independent variants together explained 34% of the variation in circulating MIP1b. The variance explained by the independent variants for each cytokine are listed in Table S7. The variants independently associated with VEGF explain 15% of the variation in circulating VEGF. The MEP1B locus had a large influence on circulating TRAIL and the variants identified here explain 14% of observed variance in TRAIL concentration. In addition, GROa was shown to be under a strong genetic control as the identified variants explained 10% of the circulating GROa variance.
Expression Analyses Link Three Variants, Cytokine mRNA, and Circulating Cytokine Concentrations
To further elucidate the molecular mechanisms underlying the genome-wide association peaks, we performed peripheral blood expression quantitative trait loci (eQTL) analyses for lead variants. Transcriptomic data were meta-analyzed for 2,177 participants from YFS and FINRISK2007.
These analyses identified 26 significant eQTLs for 15 variants. The eQTLs for variants in Table 1 are presented in Table 2. Significant eQTLs for variants not listed in Table 1, are reported in Table S8, including variants with eQTLs to more than one gene. For example, rs2070074 was associated with the expression of two genes (GALT [MIM: 606999] and IL11RA [MIM: 600939]).
Table 2.
Results from eQTL Analyses for Locus-Specific Lead Variants
| Variant Locus | Variant | Probe | Probe Locus | Gene | Other Allele | Effect Allele | Beta (SD) | SE (SD) | p Value | HetPval |
|---|---|---|---|---|---|---|---|---|---|---|
| 1q23.2 | rs12075 | ILMN_1723684 | 1q23.2 | ACKR1 | G | A | 0.111 | 0.017 | 6.95 × 10−11 | 1.00 |
| 2q36.1 | rs13412535 | ILMN_1655595 | 2q36.1 | SERPINE2 | G | A | −0.309 | 0.018 | 9.65 × 10−63 | 1.00 |
| 3p21.31 | rs113010081 | ILMN_1763322 | 3p21.31 | CCR3 | T | C | 0.372 | 0.031 | 4.80 × 10−33 | 0.87 |
| 4q13.3 | rs508977 | ILMN_1745522 | 4q13.3 | PF4V1 | T | G | 0.257 | 0.031 | 7.54 × 10−17 | 0.66 |
| 5q13.2 | rs17229943 | ILMN_1679620 | 5q13.2 | LOC728519 | A | C | 0.342 | 0.036 | 6.72 × 10−22 | 0.35 |
| 5q13.2 | rs17229943 | ILMN_1760189 | 5q13.2 | NAIP | A | C | 0.256 | 0.031 | 3.05 × 10−16 | 0.36 |
| 5q13.2 | rs17229943 | ILMN_2260082 | 5q13.2 | NAIP | A | C | 0.228 | 0.032 | 8.09 × 10−13 | 0.79 |
| 5q13.2 | rs17229943 | ILMN_1767377 | 5q13.2 | LOC153561 | A | C | 0.226 | 0.037 | 6.88 × 10−10 | 0.25 |
| 9p13.3 | rs2070074 | ILMN_1664912 | 9p13.3 | IL11RAa | A | G | 0.192 | 0.018 | 2.07 × 10−26 | 0.36 |
| 9p13.3 | rs2070074 | ILMN_1720024 | 9p13.3 | IL11RAa | A | G | 0.081 | 0.013 | 8.30 × 10−10 | 1.00 |
| 9p13.3 | rs2070074 | ILMN_1657475 | 9p13.3 | GALT | A | G | −0.155 | 0.016 | 2.70 × 10−21 | 0.90 |
| 15q25.1 | rs4778636 | ILMN_2290628 | 15q25.1 | IL16a | G | A | 0.636 | 0.023 | 2.70 × 10−161 | 1.00 |
| 17q12 | rs113877493 | ILMN_2100209 | 17q12 | CCL4L1 | C | T | −0.389 | 0.020 | 2.14 × 10−87 | 1.00 |
| 17q12 | rs113877493 | ILMN_2105573 | 17q12 | CCL3L3 | C | T | −0.391 | 0.039 | 4.51 × 10−23 | 0.47 |
| 17q12 | rs113877493 | ILMN_2218856 | 17q12 | CCL3L1 | C | T | −0.180 | 0.021 | 1.72 × 10−18 | 0.70 |
| 17q12 | rs113877493 | ILMN_1716276 | 17q12 | CCL4L2 | C | T | −0.164 | 0.021 | 1.27 × 10−14 | 0.15 |
| 19q13.33 | rs116924815 | ILMN_1807359 | 19q13.33 | CLEC11Aa | C | T | 0.450 | 0.023 | 1.99 × 10−85 | 1.00 |
| 22q11.23 | rs2330634 | ILMN_1690982 | 22q11.23 | DDT | C | G | 0.196 | 0.006 | 4.75 × 10−211 | 0.77 |
Probe IDs are from Illumina HumanHT-12 v.4 Expression BeadChip.
The variant is associated with cytokine concentration and with mRNA coding the cytokine. CTACK is also known as IL-11RA and stem cell growth factor beta (SCGFb) as CLEC11A.
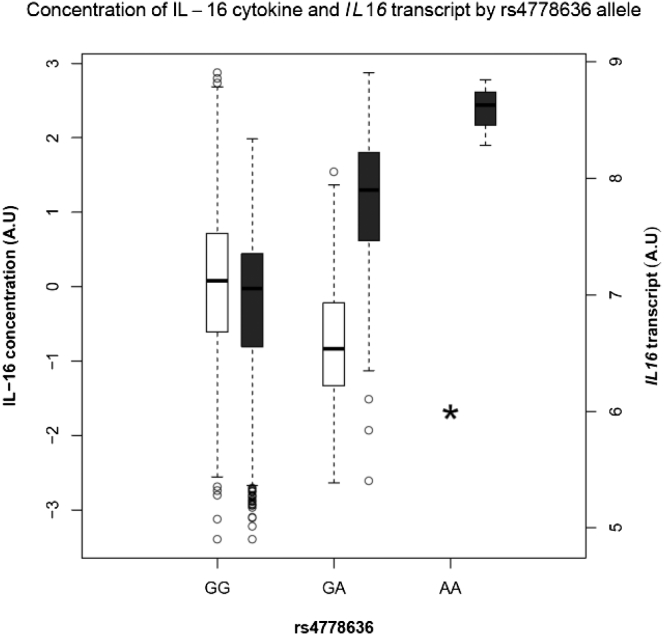
Three variants were associated with both the mRNA encoding the cytokine and the pertinent circulating cytokine concentration. Allele rs4778636-A in IL16 (MIM: 603035) was associated with decreased concentration of circulating IL-16 (Table 1) but increased concentration of IL16 mRNA (Figure 3). The Ensembl’s Variant Effect Predictor suggests that the A-allele creates a splice site variant leading to nonsense-mediated decay of IL16 mRNA.26 Increased IL16 mRNA concentration may therefore be a compensatory mechanism to account for missing circulating IL-16. The genome-wide association of rs4778636 with circulating IL-16 confirms results from a small candidate gene study.27 The MIP1b-associated variant (rs113877493) is located approximately 200 kb away from a cluster of cytokine-coding genes in 17q21. According to our results, the MIP1b-associatied variant is an eQTL for four of the genes (CCL4L1 [MIM: 603782], CCL4L2 [MIM: 610757], CCL3L1 [MIM: 601395], CCL3L3 [MIM: 609468]) belonging to this cluster. The variant that was associated with concentration of PDGFbb and resided at a transcription factor binding site (rs13412535) was associated with expression of SERPINE2. The variant rs13412535 is located within the first intron of SERPINE2, a typical place for transcriptional regulation to occur. According to ENCODE data, the variant is located with the DNase hypersensitivity peak, which suggests that the site is accessible for transcription factors.
Figure 3.
Boxplots of Interleukin-16 Cytokine and IL16 mRNA Concentrations
The mRNA and circulating cytokine concentration are depicted by the dark gray and white boxes, respectively. Variant rs4778636 is located within IL16. The asterisk indicates undetectable concentration of circulating IL-16. The A allele of rs4778636 leads to nonsense-mediated decay of IL16 mRNA and thus circulating concentration of IL-16 is undetectable in rs4778636-AA homozygotes. The box indicates the 25th and 75th percentile. The black horizontal lines within the boxes represent median. The whiskers indicate the least and the maximum value excluding outliers, indicated by dots.
Fine Mapping Suggests Causal Mutations Affecting Cytokine Concentrations
To pinpoint individual variants and genes within the identified loci, we used FINEMAP software which utilizes Bayesian inference to estimate most likely causal mutation responsible from the association signal. For 8 of the 27 identified loci, the association test lead variant was among the most likely causal mutations according to FINEMAP. For IL-17, the fine-mapping analysis identified a potentially causal variant placed in a potential transcription factor binding site (rs3804750), which was different from the association test lead variant (rs1530455) residing in an intron of the same gene without any potential functional annotation. The new variant identified by FINEMAP is also predicted to create a splice site according to Ensembl variant effect predictor leading to nonsense-mediated decay of PDIA5 (MIM: 616942) mRNA, thus offering a potential functional explanation for the association signal (Table S9).
Connecting Cytokines to Complex Diseases
To bridge the connection between the cytokine traits and disease, we searched the GWAS catalog for variant-trait and variant-disease associations within a 1 Mb window from cytokine lead variants.17 The search window approach was selected since many GWASs for the complex diseases have been conducted with HapMap2 and therefore missed variants present in the 1000 Genomes imputation panel.28 The results for cytokine-linked variants previously associated with disease status are listed in Table 3; the corresponding associations for cytokine-linked variants previously associated with quantitative traits are listed in Table S10. The circulating interleukin-2 receptor alpha subunit (IL-2ra) increasing variant (rs12722489-C) has previously been shown to increase the risk for Crohn disease and multiple sclerosis.1, 29 The variant rs943072-G here associated with increased concentrations of VEGF and IL-12p70 has previously been linked to increased risk for ulcerative colitis (MIM: 266600).30 In addition, both cytokines have been shown to be increased in persons with an inflammatory bowel disease.31, 32 These results link the VEGF cascade depicted in Figure 2C to inflammatory bowel disease. Variant rs7616215-T here associated with increased concentration of MIP1b has previously been shown to decrease the risk for Behcet disease (MIM: 109650).33 In addition to disease associations, the cytokine loci identified here were linked to 15 different molecular traits. For example, rs2251746 located at 1q23.2 within FCER1A (MIM: 147140) and previously linked to IgE concentration was here associated with circulating concentration of GROa and MCP1.
Table 3.
Variants Previously Associated with Disease and Here Found to Be Associated with Cytokine Concentrations
| Locus | Rs-id | Disease | Cytokine | Other Allele | Effect Allele/Disease Risk Increasing Allele | EAF | Beta (SD) | SE (SD) | p Value |
|---|---|---|---|---|---|---|---|---|---|
| 3p21.31 | rs7616215 | Behcet disease | MIP1b | T | C | 0.40 | −0.142 | 0.016 | 4.8 × 10−19 |
| 3p21.31 | rs13096142 | celiac disease | MIP1b | C | T | 0.29 | −0.163 | 0.017 | 4.8 × 10−21 |
| 6p21.1 | rs943072 | ulcerative colitis | IL-12p70 | T | G | 0.11 | 0.155 | 0.025 | 3.3 × 10−10 |
| 6p21.1 | rs943072 | ulcerative colitis | VEGF | T | G | 0.11 | 0.176 | 0.027 | 2.0 × 10−11 |
| 10p15.1 | rs12722489 | Crohn disease and multiple sclerosis | IL-2ra | T | C | 0.87 | 0.325 | 0.034 | 4.1 × 10−21 |
| 10p15.1 | rs12722515 | inflammatory bowel disease | IL-2ra | A | C | 0.87 | 0.328 | 0.034 | 1.6 × 10−21 |
Each variant reported in the table has been previously associated with corresponding disease or trait. p values denote the variant-cytokine association. Abbreviation: EAF, effect allele frequency.
Discussion
This study identified 27 genetic loci contributing to circulating concentration of one or more cytokines in the general population. In three cases, the analyses linked the same gene, gene expression, and protein, thus likely pinpointing the gene mediating the association. The results indicated a potential drug target for treatment of Behcet disease and celiac disease, since variants associated with decreased concentration of MIP1b in this study have previously been linked with increased risk of these diseases.33, 34 Furthermore, our results point to an alternative indication for IL-2ra-antagonist, since the IL-2-ra variant identified in our study has previously been associated with increased risk of Crohn disease. The IL-2ra-targeting antibody daclizumab reduces annualized relapse rate in individuals with multiple sclerosis by 50% compared to placebo.35 Our results suggest that daclizumab might be beneficial for persons with Crohn disease as well.
The association of one locus to multiple circulating cytokines might help researchers understand clustering of autoimmune diseases. For example, a person with rheumatoid arthritis has more than 1.6-fold risk for autoimmune thyroiditis (MIM: 608173) compared to the general population.36 A genetic variant in VEGFA was here associated with increased circulating VEGF and shown to have a causal cascade effect on IL-7, IL-10, IL-13, and IL-12p70 (Figures 2C and 2D). Axitinib, a VEGF-receptor inhibitor, has been shown to reduce IL-12p70 production in cultured monocyte-derived dendritic cells,37 which support our findings. Furthermore, VEGFA transcription follows the same temporal pattern with IL7 (MIM: 146660) transcription during wound healing, and it has been suggested that the secretion of these two cytokines might be connected by the same pathway.38 These results from prior functional studies support our genetic evidence in humans that VEGF causally regulates secretion of IL-7, IL-10, IL-13, and IL-12p70 under normal physiological conditions in healthy individuals. VEGF contributes to pathogenesis of asthma (MIM: 600807) by stimulating angiogenesis, edema, inflammation, and airway remodeling. These effects are partly mediated via VEGF-mediated upregulation of IL-13.39 Most importantly, by linking the VEGF cascade with the risk for ulcerative colitis, our results suggest that drugs targeting VEGF might have potential in treatment of ulcerative colitis.30 This is further substantiated by GWASs of inflammatory bowel diseases, where the identified loci were enriched in pathways related to VEGF cascade cytokines (IL-12 and IL-10).2 Furthermore, variants located 530 kb from rs10761731 in 10q21.3 (i.e., also associated with VEGF as the case of 6p21.1) were associated with ulcerative colitis.2 The 6p21.1 region had suggestive association with age at diagnosis of inflammatory bowel disease in another immunochip-based study.40
Another result with direct implications for drug development was found for the TRAIL-associated MEP1B locus. Drugs targeting TRAIL signaling include Conatumumab, which is currently in phase II trials for treatment of a variety of cancers (ClinicalTrials.gov: NCT01327612). The low-frequency variant here identified to have a major influence on circulating TRAIL levels could help to clarify the causality of TRAIL signaling in the development of cancer and assess potential side-effects from TRAIL-lowering similarly as done for IL-1α/β and other targets.41
Plasma IL-16 levels have previously been associated to HIV (MIM: 609423) progression,42 but the locus has not been linked to HIV-related traits in a GWAS.43 This might be due to the small allele frequency in Europeans or possible recessive effects. However, our results offer a possibility to examine HIV progression in human knock-outs, since the frequency of rs4778636-AA homozygote is up to 25% in the Yoruba population of Nigeria.44 Circulating IL-16 has been suggested to serve as a biomarker for impaired kidney transplant function, emphysema, and the efficacy of interferon-β treatment in multiple sclerosis.45, 46, 47 If IL-16 is used as a biomarker, the relatively high frequency of rs4778636-AA homozygotes in African and Asian ethnicities must be accounted for.
Although TNF-α antibodies have been successfully used to treat various autoimmune diseases,5, 6 no associations with cytokine concentrations were detected within the TNF (MIM: 191160) locus (encoding TNF-α) in the MHC class III region at 6p21.33. The MHC region displays a unique pattern of linkage disequilibrium: a subset of MHC haplotypes have moderate correlation with each other whereas variants within same haplotype block form a stronger correlation structure.48 This distinct linkage disequilibrium pattern may create complex epistasis to MHC locus, which might impede the association studies in this region. Furthermore, the cytokines were measured in conditions without stimulation. Measuring the cytokine concentrations after antigen stimulus might reveal additional loci contributing to circulating cytokine concentrations.
Variable degree of heterogeneity was found for of the 15 of the 27 genetic loci identified. For most of the loci displaying heterogeneity for the lead variant, another variant with genome-wide association but no heterogeneity could be found (see Figures S111–S124). The reason for occasional heterogeneity is probably differences in stability and accuracy of the cytokine experimental assay. In the case of the association between rs12075 and MCP1, differences in blood sample processing may also potentially contribute: heparin treatment has been shown to release significant amounts of MCP1 from ACKR1 and thus affect the association signal caused by altered receptor binding properties.49 In two loci, MIF (MIM: 153620) and HGF (MIM: 142409), either the lead variant or a variant detected by using Bayesian fine mapping analysis was identified as potential causal missense variant for the respective proteins MIF and HGF. It remains unclear whether the missense variants are actually causal for these loci and whether they alter the respective protein levels in the circulation. An alternative mechanism for these associations cannot be ruled out where the change in amino acid sequence could interfere with the probe binding and thereby lead to a false positive association. The validation of these associations with circulating protein levels requires further studies and caution should be taken if these variants are used for causal estimation without the functional role being further established.
In conclusion, we identified a total of 27 loci contributing to the genetic regulation of circulating concentrations of cytokines. Improved understanding of the genetic basis of these inflammatory markers will help to clarify the causal roles of cytokine signaling and upstream inflammation in immune-related and other chronic diseases. By linking rs4778636 to undetectable concentration of IL-16, these results enable studies of HIV and other inflammatory diseases in IL-16 knock-out humans. In addition, we identified a potential drug target for treatment of Behcet disease and celiac disease as well as indicated a possibility to expand indication of daclizumab from multiple sclerosis to Crohn disease. These results provide the basis for further studies on the molecular regulation of the immune system in health and disease.
Acknowledgments
The Young Finns Study has been financially supported by the Academy of Finland: grants 286284 (T.L.), 134309, 126925, 121584, 124282, 129378, 117787, and 41071; the Social Insurance Institution of Finland; Kuopio, Tampere, and Turku University Hospital Medical Funds (grant X51001 for T.L.); Juho Vainio Foundation; Paavo Nurmi Foundation; Finnish Foundation of Cardiovascular Research; Finnish Cultural Foundation; Tampere Tuberculosis Foundation (T.L.); Emil Aaltonen Foundation; and Yrjö Jahnsson Foundation. P.W. was supported by Finnish Diabetes Research Foundation, Novo Nordisk Foundation, and Yrjö Jahnsson Foundation. V.S. was supported by the Academy of Finland, grant number 139635, and the Finnish Foundation for Cardiovascular Research. We gratefully acknowledge Ville Aalto and Irina Lisinen for expert technical assistance in the data management.
Published: December 15, 2016
Footnotes
Supplemental Data include 124 figures and 10 tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.11.007.
Web Resources
ClinicalTrials.gov, http://clinicaltrials.gov
OMIM, http://www.omim.org/
Summary statistics from the meta-analyses, http://computationalmedicine.fi/data#Cytokine_GWAS
Variant Effect Predictor, http://useast.ensembl.org/Homo_sapiens/Tools/VEP
Supplemental Data
References
- 1.Patsopoulos N.A., Esposito F., Reischl J., Lehr S., Bauer D., Heubach J., Sandbrink R., Pohl C., Edan G., Kappos L., Bayer Pharma MS Genetics Working Group. Steering Committees of Studies Evaluating IFNβ-1b and a CCR1-Antagonist. ANZgene Consortium. GeneMSA. International Multiple Sclerosis Genetics Consortium Genome-wide meta-analysis identifies novel multiple sclerosis susceptibility loci. Ann. Neurol. 2011;70:897–912. doi: 10.1002/ana.22609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jostins L., Ripke S., Weersma R.K., Duerr R.H., McGovern D.P., Hui K.Y., Lee J.C., Schumm L.P., Sharma Y., Anderson C.A., International IBD Genetics Consortium (IIBDGC) Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akdis M., Burgler S., Crameri R., Eiwegger T., Fujita H., Gomez E., Klunker S., Meyer N., O’Mahony L., Palomares O. Interleukins, from 1 to 37, and interferon-γ: receptors, functions, and roles in diseases. J. Allergy Clin. Immunol. 2011;127 doi: 10.1016/j.jaci.2010.11.050. 701–21.e1, 70. [DOI] [PubMed] [Google Scholar]
- 4.Nelson M.R., Tipney H., Painter J.L., Shen J., Nicoletti P., Shen Y., Floratos A., Sham P.C., Li M.J., Wang J. The support of human genetic evidence for approved drug indications. Nat. Genet. 2015;47:856–860. doi: 10.1038/ng.3314. [DOI] [PubMed] [Google Scholar]
- 5.Miller A.V., Ranatunga S.K.M. Immunotherapies in rheumatologic disorders. Med. Clin. North Am. 2012;96:475–496. doi: 10.1016/j.mcna.2012.04.003. ix–x. [DOI] [PubMed] [Google Scholar]
- 6.Ahluwalia J.P. Immunotherapy in inflammatory bowel disease. Med. Clin. North Am. 2012;96:525–544. doi: 10.1016/j.mcna.2012.04.009. x. [DOI] [PubMed] [Google Scholar]
- 7.Weinblatt M.E., Keystone E.C., Furst D.E., Moreland L.W., Weisman M.H., Birbara C.A., Teoh L.A., Fischkoff S.A., Chartash E.K. Adalimumab, a fully human anti-tumor necrosis factor alpha monoclonal antibody, for the treatment of rheumatoid arthritis in patients taking concomitant methotrexate: the ARMADA trial. Arthritis Rheum. 2003;48:35–45. doi: 10.1002/art.10697. [DOI] [PubMed] [Google Scholar]
- 8.Raitakari O.T., Juonala M., Rönnemaa T., Keltikangas-Järvinen L., Räsänen L., Pietikäinen M., Hutri-Kähönen N., Taittonen L., Jokinen E., Marniemi J. Cohort profile: the cardiovascular risk in Young Finns Study. Int. J. Epidemiol. 2008;37:1220–1226. doi: 10.1093/ije/dym225. [DOI] [PubMed] [Google Scholar]
- 9.Ritchie S.C., Würtz P., Nath A.P., Abraham G., Havulinna A.S., Fearnley L.G., Sarin A.-P., Kangas A.J., Soininen P., Aalto K. The biomarker GlycA is associated with chronic inflammation and predicts long-term risk of severe infection. Cell Syst. 2015;1:293–301. doi: 10.1016/j.cels.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 10.Marchini J., Howie B., Myers S., McVean G., Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 11.Marchini J., Howie B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 12.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park J.-H., Wacholder S., Gail M.H., Peters U., Jacobs K.B., Chanock S.J., Chatterjee N. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat. Genet. 2010;42:570–575. doi: 10.1038/ng.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davey Smith G., Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum. Mol. Genet. 2014;23(R1):R89–R98. doi: 10.1093/hmg/ddu328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turpeinen H., Seppälä I., Lyytikäinen L.-P., Raitoharju E., Hutri-Kähönen N., Levula M., Oksala N., Waldenberger M., Klopp N., Illig T. A genome-wide expression quantitative trait loci analysis of proprotein convertase subtilisin/kexin enzymes identifies a novel regulatory gene variant for FURIN expression and blood pressure. Hum. Genet. 2015;134:627–636. doi: 10.1007/s00439-015-1546-5. [DOI] [PubMed] [Google Scholar]
- 16.Benner C., Spencer C.C.A., Havulinna A.S., Salomaa V., Ripatti S., Pirinen M. FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics. 2016;32:1493–1501. doi: 10.1093/bioinformatics/btw018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L., Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abecasis G.R., Altshuler D., Auton A., Brooks L.D., Durbin R.M., Gibbs R.A., Hurles M.E., McVean G.A., 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crisman J.M., Zhang B., Norman L.P., Bond J.S. Deletion of the mouse meprin beta metalloprotease gene diminishes the ability of leukocytes to disseminate through extracellular matrix. J. Immunol. 2004;172:4510–4519. doi: 10.4049/jimmunol.172.7.4510. [DOI] [PubMed] [Google Scholar]
- 20.Matteini A.M., Li J., Lange E.M., Tanaka T., Lange L.A., Tracy R.P., Wang Y., Biggs M.L., Arking D.E., Fallin M.D. Novel gene variants predict serum levels of the cytokines IL-18 and IL-1ra in older adults. Cytokine. 2014;65:10–16. doi: 10.1016/j.cyto.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He M., Cornelis M.C., Kraft P., van Dam R.M., Sun Q., Laurie C.C., Mirel D.B., Chasman D.I., Ridker P.M., Hunter D.J. Genome-wide association study identifies variants at the IL18-BCO2 locus associated with interleukin-18 levels. Arterioscler. Thromb. Vasc. Biol. 2010;30:885–890. doi: 10.1161/ATVBAHA.109.199422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Debette S., Visvikis-Siest S., Chen M.-H., Ndiaye N.-C., Song C., Destefano A., Safa R., Azimi Nezhad M., Sawyer D., Marteau J.-B. Identification of cis- and trans-acting genetic variants explaining up to half the variation in circulating vascular endothelial growth factor levels. Circ. Res. 2011;109:554–563. doi: 10.1161/CIRCRESAHA.111.243790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Voruganti V.S., Laston S., Haack K., Mehta N.R., Smith C.W., Cole S.A., Butte N.F., Comuzzie A.G. Genome-wide association replicates the association of Duffy antigen receptor for chemokines (DARC) polymorphisms with serum monocyte chemoattractant protein-1 (MCP-1) levels in Hispanic children. Cytokine. 2012;60:634–638. doi: 10.1016/j.cyto.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller L.H., Mason S.J., Dvorak J.A., McGinniss M.H., Rothman I.K. Erythrocyte receptors for (Plasmodium knowlesi) malaria: Duffy blood group determinants. Science. 1975;189:561–563. doi: 10.1126/science.1145213. [DOI] [PubMed] [Google Scholar]
- 25.Horuk R., Chitnis C.E., Darbonne W.C., Colby T.J., Rybicki A., Hadley T.J., Miller L.H. A receptor for the malarial parasite Plasmodium vivax: the erythrocyte chemokine receptor. Science. 1993;261:1182–1184. doi: 10.1126/science.7689250. [DOI] [PubMed] [Google Scholar]
- 26.McLaren W., Pritchard B., Rios D., Chen Y., Flicek P., Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim S., Swaminathan S., Inlow M., Risacher S.L., Nho K., Shen L., Foroud T.M., Petersen R.C., Aisen P.S., Soares H., Alzheimer’s Disease Neuroimaging Initiative (ADNI) Influence of genetic variation on plasma protein levels in older adults using a multi-analyte panel. PLoS ONE. 2013;8:e70269. doi: 10.1371/journal.pone.0070269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang J., Ellinghaus D., Franke A., Howie B., Li Y. 1000 Genomes-based imputation identifies novel and refined associations for the Wellcome Trust Case Control Consortium phase 1 Data. Eur. J. Hum. Genet. 2012;20:801–805. doi: 10.1038/ejhg.2012.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Franke A., McGovern D.P.B., Barrett J.C., Wang K., Radford-Smith G.L., Ahmad T., Lees C.W., Balschun T., Lee J., Roberts R. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat. Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson C.A., Boucher G., Lees C.W., Franke A., D’Amato M., Taylor K.D., Lee J.C., Goyette P., Imielinski M., Latiano A. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat. Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fuss I.J., Becker C., Yang Z., Groden C., Hornung R.L., Heller F., Neurath M.F., Strober W., Mannon P.J. Both IL-12p70 and IL-23 are synthesized during active Crohn’s disease and are down-regulated by treatment with anti-IL-12 p40 monoclonal antibody. Inflamm. Bowel Dis. 2006;12:9–15. doi: 10.1097/01.mib.0000194183.92671.b6. [DOI] [PubMed] [Google Scholar]
- 32.Kanazawa S., Tsunoda T., Onuma E., Majima T., Kagiyama M., Kikuchi K. VEGF, basic-FGF, and TGF-beta in Crohn’s disease and ulcerative colitis: a novel mechanism of chronic intestinal inflammation. Am. J. Gastroenterol. 2001;96:822–828. doi: 10.1111/j.1572-0241.2001.03527.x. [DOI] [PubMed] [Google Scholar]
- 33.Kirino Y., Bertsias G., Ishigatsubo Y., Mizuki N., Tugal-Tutkun I., Seyahi E., Ozyazgan Y., Sacli F.S., Erer B., Inoko H. Genome-wide association analysis identifies new susceptibility loci for Behçet’s disease and epistasis between HLA-B∗51 and ERAP1. Nat. Genet. 2013;45:202–207. doi: 10.1038/ng.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garner C., Ahn R., Ding Y.C., Steele L., Stoven S., Green P.H., Fasano A., Murray J.A., Neuhausen S.L. Genome-wide association study of celiac disease in North America confirms FRMD4B as new celiac locus. PLoS ONE. 2014;9:e101428. doi: 10.1371/journal.pone.0101428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gold R., Giovannoni G., Selmaj K., Havrdova E., Montalban X., Radue E.-W., Stefoski D., Robinson R., Riester K., Rana J., SELECT study investigators Daclizumab high-yield process in relapsing-remitting multiple sclerosis (SELECT): a randomised, double-blind, placebo-controlled trial. Lancet. 2013;381:2167–2175. doi: 10.1016/S0140-6736(12)62190-4. [DOI] [PubMed] [Google Scholar]
- 36.Cooper G.S., Bynum M.L.K., Somers E.C. Recent insights in the epidemiology of autoimmune diseases: improved prevalence estimates and understanding of clustering of diseases. J. Autoimmun. 2009;33:197–207. doi: 10.1016/j.jaut.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heine A., Held S.A.E., Daecke S.N., Riethausen K., Kotthoff P., Flores C., Kurts C., Brossart P. The VEGF-receptor inhibitor axitinib impairs dendritic cell phenotype and function. PLoS ONE. 2015;10:e0128897. doi: 10.1371/journal.pone.0128897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palagummi S., Harbison S., Fleming R. A time-course analysis of mRNA expression during injury healing in human dermal injuries. Int. J. Legal Med. 2014;128:403–414. doi: 10.1007/s00414-013-0941-5. [DOI] [PubMed] [Google Scholar]
- 39.Lee C.G., Link H., Baluk P., Homer R.J., Chapoval S., Bhandari V., Kang M.J., Cohn L., Kim Y.K., McDonald D.M., Elias J.A. Vascular endothelial growth factor (VEGF) induces remodeling and enhances TH2-mediated sensitization and inflammation in the lung. Nat. Med. 2004;10:1095–1103. doi: 10.1038/nm1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cleynen I., Boucher G., Jostins L., Schumm L.P., Zeissig S., Ahmad T., Andersen V., Andrews J.M., Annese V., Brand S., International Inflammatory Bowel Disease Genetics Consortium Inherited determinants of Crohn’s disease and ulcerative colitis phenotypes: a genetic association study. Lancet. 2016;387:156–167. doi: 10.1016/S0140-6736(15)00465-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Interleukin 1 Genetics Consortium Cardiometabolic effects of genetic upregulation of the interleukin 1 receptor antagonist: a Mendelian randomisation analysis. Lancet Diabetes Endocrinol. 2015;3:243–253. doi: 10.1016/S2213-8587(15)00034-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baier M., Werner A., Bannert N., Metzner K., Kurth R. HIV suppression by interleukin-16. Nature. 1995;378:563. doi: 10.1038/378563a0. [DOI] [PubMed] [Google Scholar]
- 43.Johnson E.O., Hancock D.B., Gaddis N.C., Levy J.L., Page G., Novak S.P., Glasheen C., Saccone N.L., Rice J.P., Moreau M.P. Novel genetic locus implicated for HIV-1 acquisition with putative regulatory links to HIV replication and infectivity: a genome-wide association study. PLoS ONE. 2015;10:e0118149. doi: 10.1371/journal.pone.0118149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alachkar N., Ugarte R., Huang E., Womer K.L., Montgomery R., Kraus E., Rabb H. Stem cell factor, interleukin-16, and interleukin-2 receptor alpha are predictive biomarkers for delayed and slow graft function. Transplant. Proc. 2010;42:3399–3405. doi: 10.1016/j.transproceed.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 46.Bowler R.P., Bahr T.M., Hughes G., Lutz S., Kim Y.-I., Coldren C.D., Reisdorph N., Kechris K.J. Integrative omics approach identifies interleukin-16 as a biomarker of emphysema. OMICS. 2013;17:619–626. doi: 10.1089/omi.2013.0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nischwitz S., Faber H., Sämann P.G., Domingues H.S., Krishnamoorthy G., Knop M., Müller-Sarnowski F., Yassouridis A., Weber F. Interferon β-1a reduces increased interleukin-16 levels in multiple sclerosis patients. Acta Neurol. Scand. 2014;130:46–52. doi: 10.1111/ane.12215. [DOI] [PubMed] [Google Scholar]
- 48.Blomhoff A., Olsson M., Johansson S., Akselsen H.E., Pociot F., Nerup J., Kockum I., Cambon-Thomsen A., Thorsby E., Undlien D.E., Lie B.A. Linkage disequilibrium and haplotype blocks in the MHC vary in an HLA haplotype specific manner assessed mainly by DRB1∗03 and DRB1∗04 haplotypes. Genes Immun. 2006;7:130–140. doi: 10.1038/sj.gene.6364272. [DOI] [PubMed] [Google Scholar]
- 49.Schnabel R.B., Baumert J., Barbalic M., Dupuis J., Ellinor P.T., Durda P., Dehghan A., Bis J.C., Illig T., Morrison A.C. Duffy antigen receptor for chemokines (Darc) polymorphism regulates circulating concentrations of monocyte chemoattractant protein-1 and other inflammatory mediators. Blood. 2010;115:5289–5299. doi: 10.1182/blood-2009-05-221382. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.