Abstract
Identification of over 500 epigenetic regulators in humans raises an interesting question regarding how chromatin dysregulation contributes to different diseases. Bromodomain and PHD finger-containing protein 1 (BRPF1) is a multivalent chromatin regulator possessing three histone-binding domains, one non-specific DNA-binding module, and several motifs for interacting with and activating three lysine acetyltransferases. Genetic analyses of fish brpf1 and mouse Brpf1 have uncovered an important role in skeletal, hematopoietic, and brain development, but it remains unclear how BRPF1 is linked to human development and disease. Here, we describe an intellectual disability disorder in ten individuals with inherited or de novo monoallelic BRPF1 mutations. Symptoms include infantile hypotonia, global developmental delay, intellectual disability, expressive language impairment, and facial dysmorphisms. Central nervous system and spinal abnormalities are also seen in some individuals. These clinical features overlap with but are not identical to those reported for persons with KAT6A or KAT6B mutations, suggesting that BRPF1 targets these two acetyltransferases and additional partners in humans. Functional assays showed that the resulting BRPF1 variants are pathogenic and impair acetylation of histone H3 at lysine 23, an abundant but poorly characterized epigenetic mark. We also found a similar deficiency in different lines of Brpf1-knockout mice. These data indicate that aberrations in the chromatin regulator gene BRPF1 cause histone H3 acetylation deficiency and a previously unrecognized intellectual disability syndrome.
Keywords: developmental disorder, intellectual disability, epigenetic regulator, bromodomain, BRPF2, PHD finger, PWWP domain, PZP domain, histone acetylation
Introduction
Epigenetic regulation is essential for human development and becomes aberrant in different diseases. The human genome encodes over 500 epigenetic regulators.1, 2, 3 To date, ∼30 chromatin modifiers have been associated with human diseases, including the ATP-dependent helicases CHD4, CHD7, CHD8, SMARCA2, SMARCA4, and ATRX;4, 5, 6 the DNA methyltransferases DNMT1 and DNMT3b;4 the histone methyltransferases EZH2 and KMT3B;4 the lysine acetyltransferases CREBBP, EP300, KAT6A, KAT6B, and ESCO2;4, 7, 8, 9, 10, 11, 12, 13, 14 and the histone deacetylases HDAC4 and HDAC8.15, 16 By contrast, disease association remains much less clear for hundreds of chromatin readers that utilize structural modules to sense DNA and histone modification states for chromatin-based signaling.17, 18 Thus, it is important to identify disease links for such chromatin readers. Bromodomain and PHD finger-containing protein 1 (BRPF1) is a multivalent chromatin reader composed of multiple nucleosome-binding modules, including double PHD fingers (linked with a C2HC zinc knuckle), a bromodomain, and a PWWP domain.19, 20, 21 The PHD fingers and zinc knuckle form a bivalent nucleosome-binding unit, known as the PZP (PHD finger–zinc knuckle–PHD finger) domain.22, 23 In addition, BRPF1 possesses two enhancer of polycomb (EPC)-like motifs flanking the PZP domain.19, 20, 21 Through one of these EPC-like motifs and its N-terminal region, BRPF1 interacts with and activates three histone acetyltransferases, KAT6A, KAT6B, and KAT7.19, 20, 21 Thus, BRPF1 is a unique chromatin reader that also possesses sequence motifs mediating interaction with histone acetyltransferases.
Zebrafish and mouse genetic studies have shown that Brpf1 is essential for embryo survival, hematopoiesis, head patterning, and brain development.24, 25, 26, 27, 28, 29 Moreover, mutations in KAT6A (MIM: 601408) and KAT6B (MIM: 605880) have recently been discovered in multiple individuals with several developmental disorders characterized by intellectual disability and other abnormalities.7, 8, 9, 10, 11, 12, 13 Thus, an interesting question is whether mutations of human BRPF1 (MIM: 602410) cause similar developmental anomalies. Here, we report that heterozygous missense, nonsense, and reading frameshift mutations in this gene cause a neurodevelopmental disorder characterized by congenital hypotonia, global developmental delay, intellectual disability, and facial dysmorphisms. We show that these mutations cause deficiency of histone H3K23 acetylation and provide evidence that BRPF1 acts through KAT6A and KAT6B to govern this chromatin modification in vitro and in vivo. Together, this study and the recent reports on individuals with KAT6A and KAT6B mutations7, 8, 9, 10, 11, 12, 13 indicate an emerging group of developmental disorders caused by aberrant histone H3 acetylation.
Subjects and Methods
Human Subjects
Families gave consent for studies approved by the CHU Sainte-Justine Institutional Review Board or by the institutional review boards of the DDD (Decipher Developmental Disorder) Study, the CAUSES (Clinical Assessment of the Utility of Sequencing and Evaluation as a Service) Study, or the Leiden University Medical Center and Maastricht University Medical Center. Written informed consent was obtained for all ten individuals involved in this study.
Exome and Sanger Sequencing
Individual P1’s exome sequencing was performed as a part of the CAUSES study. Clinical exome sequencing of individual P2 was carried out at Baylor Genetics. For individual P3 (sibling of individual P2), only Sanger sequencing of a candidate mutation was done at Baylor Genetics. Clinical exome sequencing for individuals P4 and P5 was performed at GeneDx. Exome sequencing of individuals P6 and P10 was carried out at Leiden University Medical Center and Maastricht University Medical Center, respectively. Individuals P7, P8, and P9 were subject to research exome sequencing as a part of the DDD study. After exome sequencing, PCR fragments were amplified from genomic DNA or reversely transcribed cDNA to confirm BRPF1 mutations.
Lymphoblastoid Cell Line Preparation
Lymphoblastoid cell lines were established and cultured as described previously.30
Animal Study Approval
Mice used were in the C57BL/6J genetic background. The Brpf1f allele contains two loxP sites flanking exons 4–6 of Brpf1.25, 27, 28 EIIa-Cre-mediated total knockouts, as well as forebrain-specific and hematopoietic-specific knockouts, have been described elsewhere.26, 27, 28, 29 To generate epiblast-specific knockouts, Brpf1f/f mice were mated with the Meox2-Cre strain (Jackson Laboratory, 003755; Bar Harbor). Knockout efficiency was verified by genomic PCR and/or quantitative reverse-transcription PCR (qRT-PCR) as previously described.26, 27, 28, 29, 31 Mouse-related procedures were performed according to the Animal Use Protocol 5786, which was reviewed and approved by the Facility Animal Care Committee of McGill University.
Immunoprecipitation and Histone Acetylation Assays
An expression plasmid for FLAG-tagged KAT6A was transfected into HEK293 cells, along with expression vectors for HA-tagged BRPF1, ING5, and MEAF6, as specified. About 48 hr after transfection, cells were washed twice with PBS, and soluble protein extracts were prepared for affinity purification on anti-FLAG M2 agarose (Sigma) as previously described.20, 31 The FLAG peptide was used to elute bound proteins for immunoblotting with anti-FLAG and -HA antibodies or for acetyltransferase activity determination. Acetylation of HeLa oligonucleosomes was performed as previously reported.20, 31 After acetylation reactions, immunoblotting was carried out to detect histone H3 or its site-specific acetylation and was performed with anti-histone H3 (Abcam, ab1791), anti-H3K9ac (Abcam, ab10812), anti-H3K14ac (EMD Millipore, 07-353), anti-H3K18ac (EMD Millipore, 07-354), and anti-H3K23ac (EMD Millipore, 07-355) antibodies.
Fluorescence Microscopy
For analysis of subcellular localization, expression plasmids for EGFP-BRPF1 and its mutants were transiently transfected into HEK293 cells with or without those for KAT6A, ING5, and MEAF6, as specified in Figure S4. About 16 hr after transfection, live green fluorescence signals were examined under a fluorescence microscope, and fluorescence images were captured for further processing as previously described.31
qRT-PCR and Regular PCR
For analysis of nonsense-mediated mRNA decay (NMD), control and mutant lymphoblastoid cells and fibroblasts were prepared for RNA extraction. RNA was reversely transcribed with the QuantiTect Reverse Transcription Kit (QIAGEN). qRT-PCR was performed on Realplex2 (Eppendorf) with the Green-2-Go qPCR Mastermix (BioBasic). Primers were designed on the web-based IDT real-time PCR primer designer and synthesized by IDT Biotechnology. BR27 (5′-TTT CCC GAA TTC AAG CTG CCA GAG GTG GTC TA-3′) and BR20 (5′-GGG CCC AAG CTT AGT CCT CCT CGT CCA TGT C-3′) were for amplification of the region corresponding to the N-terminal part of BRPF1, whereas BR11 (5′-G GCA CGG AAG AAT TCC TGG CAG AGA A-3′) and BR10 (5′-TTT CCC AAG CTT AGCT GTG CAG CCT CTG CAT-3′) were for a region encoding the C-terminal part of BRPF1.
To determine whether NMD definitely degradates the mutated BRPF1 transcript, cDNA prepared from fibroblasts cultured from a skin biopsy of individual P5 was used for PCR amplification with primers BR419 (5′-TTG AGC ACA TCC CAC CAG CT-3′) and BR421 (5′-TGT TCT TTA AGG GCC CAG TT-3′). The amplified fragment was gel purified for Sanger sequencing. To verify her heterozygous mutation, genomic DNA from this individual and her parents was used for PCR amplification with primers BR419 and BR420 (5′-GTG GTG GGA TGC AGG GCA CT-3′). DNA fragments were gel purified for sequencing.
Statistical Analysis
Statistical analysis was performed with an unpaired two-tailed Student’s t test. p < 0.05 was considered to be statistically significant. Graphs were generated with GraphPad Prism 6 (Graphpad Software).
Results
Identification and Clinical Features of Ten Subjects with BRPF1 Mutations
Mutations in KAT6A and KAT6B have recently been discovered in dozens of individuals with intellectual disability and congenital abnormalities.7, 8, 9, 10, 11, 12, 13 Our recent mouse genetic studies have shown that Brpf1 is essential for embryo survival, hematopoiesis, and brain development.25, 26, 27, 28, 29 Together, these new findings raise the interesting question of whether BRPF1 mutations cause developmental anomalies. To address this question, we sought to leverage exome sequencing that had been carried out on a clinical or research basis at different institutions around the world. In addition to the use of prior collaborations, we employed new web-based matching tools such as Genematcher32 and Decipher33 to identify individuals with mutations in BRPF1. In the process, we identified ten subjects carrying nine heterozygous mutations in the gene (Table 1). These subjects were from nine different families. Seven mutations were de novo (individuals P1 and P5–P10; Table 1 and Figure S1). As for the remaining two mutations, individual P4 inherited the c.1108C>T mutation (GenBank: NM_001003694.1) from his mother, who was mosaic for the mutation (mosaicism, 7% in blood DNA; Tables 1 and S1). Individuals P2 and P3 are adopted siblings whose biological mother is affected with intellectual disability. The siblings share the same variant (c.942_955del; Tables 1 and S1), which is presumed to be maternal in origin, although their biological mother is unavailable for testing.
Table 1.
Identification of BRPF1 Mutations in Ten Individuals
| Individual | Mutation (GenBank:NM_001003694.1) | Substitution in BRPF1 | Origin of Mutation |
|---|---|---|---|
| P1 | c.362_363delAG | p.Glu121Glyfs∗2 | de novo |
| P2 | c.942_955del | p.Trp315Leufs∗26 | parental samples not available; biological mother affected by intellectual disability and father by cerebral palsy |
| P3 | c.942_955del | p.Trp315Leufs∗26 | |
| P4 | c.1108C>T | p.Pro370Ser | mosaic in unaffected mother; mosaicism, 7% in blood DNA, based on exome sequencing reads (21/293) |
| P5 | c.1363C>T | p.Arg455∗ | de novo |
| P6 | c.1688_1689del | p.His563Profs∗8 | de novo |
| P7 | c.1883_1886dup | p.Gln629Hisfs∗34 | de novo |
| P8 | c.2497C>T | p.Arg833∗ | de novo |
| P9 | c.2915dupC | p.Met973Asnfs∗24 | de novo |
| P10 | c.3298C>T | p.Arg1100∗ | de novo |
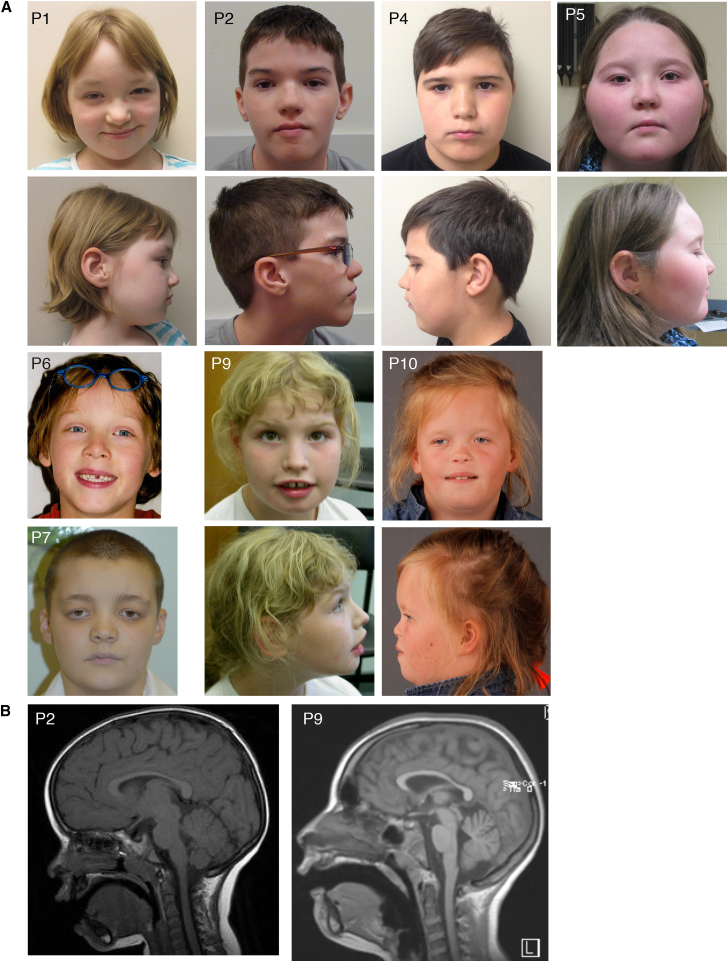
Global developmental delay, expressive language impairment, and intellectual disability were observed in all individuals (Tables 2 and S1). As shown in Figure 1A, facial dysmorphisms include a flat facial profile, a round face, a broad root of the nose, widely spaced eyes (with down-slanting palpebral fissures), ptosis, and blepharophimosis. A majority of the ten individuals have hypotonia and gross and fine motor delay. Five of them had neonatal feeding issues. Four have epilepsy. In addition, one individual had episodes of staring spells consistent with absence seizures, but electroencephalogram (EEG) only showed high voltage hypnogogic hypersynchrony, without epileptiform activity (Tables 1 and S1). Another individual displayed EEG abnormalities when treated with oxcarbazepine, but did not have clinical seizures (Tables 1 and S1). Some individuals had MRI anomalies; two had white matter hyper-intensity and two had a paucity of white matter, including thinning of the corpus callosum in one of them (Figure 1B and Table 2). These results suggest abnormal myelination in patients with BRPF1 mutations. Interestingly, myelination and corpus callosum defects are present in forebrain-specific Brpf1-knockout mice.28 Three individuals also displayed C2-C3 spinal fusions (Table S1), which is consistent with deregulated Hox expression upon Brpf1 deletion in mice28 and with skeletal abnormalities in Brpf1-null zebrafish.24 Additional dysmorphisms, malformations, and clinical issues in the ten individuals are detailed in Table S1.
Table 2.
Summary of Major Clinical Features in Ten Individuals with BRPF1 Mutations
| P1 | P2 | P3 | P4 | P5 | P6 | P7 | P8 | P9 | P10 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Neonatal feeding difficulties | N | Y | NA | Y | N | N | Y | N | N | Y |
| Hypotonia | Y | Y | NA | Y | Y | Y | NA | N | Y | Y |
| Gross motor delay | Y | Y | Y | Y | Y | Y | Y | N | Y | Y |
| Fine motor delay | Y | Y | Y | N | Y | N | U | Y | Y | U |
| Language delay | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| GDD or ID | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Epilepsy | N | Y | Y | EEG anomaly | probable absence seizures | N | Y | N | Y | N |
| Brain MRI | WM hyper-intensity | decreased WM and thin CC | nl | nl | nl | NA | NA | NA | decreased WM | nl |
| Flat facial profile | Y | N | Y | Y | Y | N | U | Y | Y | Y |
| Round face | Y | Y | Y | N | Y | N | Y | Y | N | Y |
| Broad root of the nose | Y | Y | Y | Y | Y | N | Y | Y | Y | Y |
| Widely spaced eyes | Y | Y | Y | Y | Y | N | Y | Y | Y | Y |
| Down-slanting palpebral fissures | N | Y | N | N | N | N | Y | N | Y | Y |
| Ptosis | Y | N | Y | N | Y | Y | Y | N | N | Y |
| Blepharophimosis | U | N | Y | Y | Y | N | NA | N | N | Y |
| Spinal anomalies | Y | Y | Y | N | Y | NA | NA | N | N | NA |
| Joint hypermobility | Y | N | N | N | Y | Y | Y | Y | Y | N |
See Table S1 for more details. Abbreviations are as follows: CC, corpus callosum; GDD, global developmental delay; ID, intellectual disability; N, no; NA, data not available, nl, normal; U, unknown; WM, white brain matter; Y, yes.
Figure 1.
Facial and Brain Characteristics of Individuals with BRPF1 Mutations
(A) Photos for eight individuals. The photos are not in the same scale. Ages of the individuals when the photos were taken are as follows: P1, 6 years and 5 months; P2, 13 years and 3 months; P4, 10 years and 6 months; P5, 8 years and 3 months; P6, 4 years; P7, 12 years and 6 months; P9, 8 years; P10, 12 years.
(B) Brain magnetic resonance imaging (MRI) images of individuals P2 and P9.
BRPF1 Mutations Are Predicted to Generate Different Types of Variants
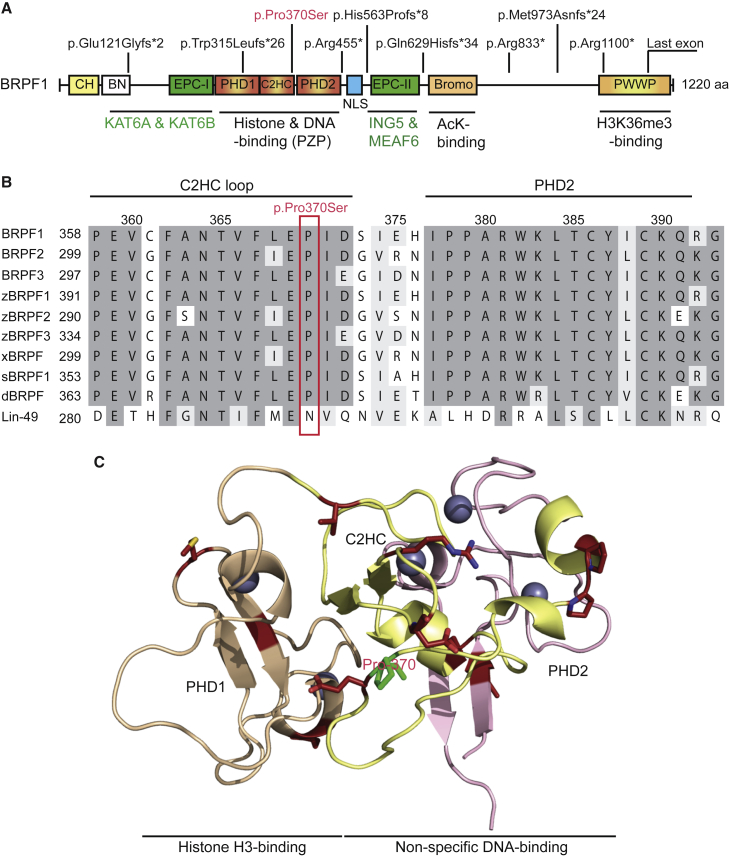
Among the BRPF1 mutations, one is missense and the remaining eight are nonsense mutations or reading frameshifts that lead to C-terminal truncations of the protein (Table 1 and Figure 2A). The missense mutation in individual P4 (p.Pro370Ser [c.1108C>T]; Table 1 and Figure 2A) alters the codon for Pro370, which is invariant from Drosophila to humans (Figure 2B) and is located within a loop of the PZP domain important for non-specific binding to the DNA backbone of nucleosomes (Figure 2C).23 Moreover, it is almost invariant in PZP domains from nine other human proteins, including three JADE proteins, three histone demethylases, and three zinc finger proteins (AF-10, AF-17, and PHF14) (Figure S2). All eight truncating mutations encode variants lacking important structural domains of BRPF1 (Figure 2A). The variants p.Glu121Glyfs∗2 (c.362_363delAG) (individual P1; Table 1), p.Trp315Leufs∗26 (c.942_955del) (individuals P2 and P3), p.Arg455∗ (c.1363C>T) (individual P5), and p.His563Profs∗8 (c.1688-1689del) (individual P6) are devoid of the ING5- and MEAF6-interacting domain (Figure 2A). On the basis of published data,20 these variants are predicted to be unable to form tetrameric complexes with KAT6A (or KAT6B), ING5, and MEAF6. By contrast, the ING5- and MEAF6-interacting domain is intact in the remaining four truncation variants, p.Gln629Hisfs∗34 (c.1883_1886dup), p.Arg833∗ (c.2497C>T), p.Met973Asnfs∗24 (c.2915dupC), and p.Arg1100∗ (c.3298C>T) (individuals P7–P10, respectively; Table 1 and Figure 2A). These four mutants are expected to form tetrameric complexes with KAT6A (or KAT6B), ING5, and MEAF6. Therefore, the nine mutations appear to generate three distinct groups of variants, suggesting that the mutations might deregulate BRPF1 functions through different mechanisms.
Figure 2.
Domain Organization of BRPF1 and Its Variants from Individuals with Developmental Anomalies
(A) Schematic representation of BRPF1 and nine variants as identified in ten individuals. See Table 1 for DNA sequence changes in the individuals. BRPF1 possesses multiple modules, including the PZP domain, bromodomain, and PWWP domain, for chromatin association. The PZP domain comprises two PHD fingers linked with a C2HC zinc finger. The first PHD finger recognizes the N terminus of histone H3.21, 23, 34 The C2HC zinc knuckle and the second PHD finger form a non-specific DNA binding domain.22, 23 The bromodomain has acetyllysine-binding ability,35 and the PWWP domain targets trimethylated histone H3.36, 37 The EPC-like motif C-terminal to the PZP domain is essential for formation of a stable trimeric complex with ING5 and MEAF6.19, 20, 21 Through the EPC-like motif N-terminal to the PZP domain and a conserved region further N-terminal to this motif, BRPF1 interacts with and activates KAT6A, KAT6B, and KAT7.19, 20, 21 Unlike p.Pro370Ser, the other eight variants contain C-terminal truncations due to nonsense or reading-frameshift mutations. These mutations are not located within the last coding exon and might trigger NMD in vivo. Abbreviations are as follows: CH, C2H2 zinc finger; BN, conserved BRPF N-terminal domain; EPC, enhancer of polycomb-like motif; NLS, nuclear localization signal.
(B) Sequence alignment of human BRPF1 with its paralogs (BRPF2 and BRPF3), as well as the orthologs from zebrafish (z), Xenopus (x), sea urchin (s), Drosophila (d), and C. elegans (Lin-49). There is one ortholog per organism from the worm to Xenopus, but there are three in zebrafish. Pro370 of BRPF1 and its corresponding residues in its paralogs and orthologs are boxed in red.
(C) Pro370 is located within a loop connecting the C2HC knuckle to the second PHD finger. PHD1 recognizes the free N terminus of histone H3,21, 23, 34 whereas the C2HC zinc knuckle and PHD2 form a module for non-specific interaction with the DNA backbone of the nucleosome.23 The structural model was generated based on a published report,23 with kind help of Tatiana G. Kutateladze.
BRPF1 Loss Reduces Histone H3K23 Acetylation In Vivo
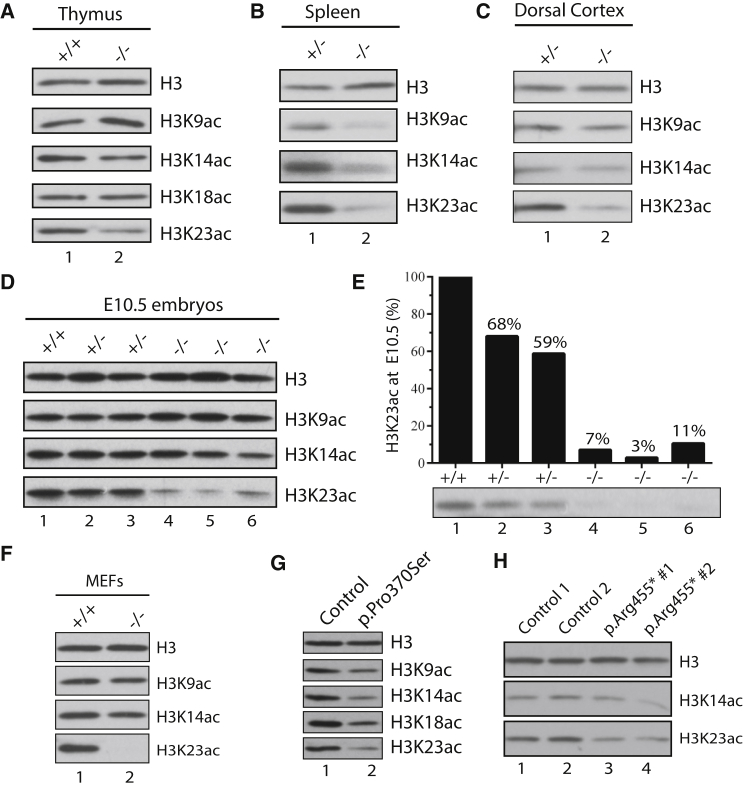
Through the EPC-like motif N-terminal to the PZP domain and a conserved region further N-terminal to this motif, BRPF1 interacts with and activates three histone acetyltransferases, KAT6A, KAT6B, and KAT7.19, 20, 21 Drosophila Enok (a fly acetyltransferase with a catalytic domain highly homologous to those of KAT6A and KAT6B) and Br140 (orthologous to BRPF1) are crucial for histone H3K23 acetylation.38, 39 We found that BRPF1 is also important for histone H3K23 acetylation in the mouse bone marrow.29 In agreement with this, H3K23 acetylation was defective in thymus and spleen protein extracts from hematopoietic-specific Brpf1- knockout pups (Figures 3A and 3B). Moreover, similar defects were present in dorsal cortex extracts of forebrain-specific Brpf1-knockout pups (Figure 3C) and protein extracts of epiblast-specific Brpf1-knockout embryos (Figure 3D, lanes 1 and 4–6). Interestingly, heterozygous mutant embryos also displayed reduced H3K23 acetylation (Figures 3D and 3E, compare lanes 1–3), which is relevant to monoallelic mutations in the ten individuals (Table 1). Furthermore, H3K23 acetylation was not detectable in Brpf1−/− mouse embryonic fibroblasts (Figure 3F). By contrast, histone H3K9 and H3K14 acetylation was less consistently affected in different Brpf1-knockout samples (Figures 3A–3F). Therefore, mouse Brpf1 governs H3K23 acetylation in different tissues and cells in vivo.
Figure 3.
BRPF1 Loss Reduces Histone H3K23 Acetylation In Vivo
(A and B) Histone H3 acetylation in thymus and spleen protein extracts from control and hematopoietic-specific Brpf1-knockout mice.
(C) Histone H3 acetylation in dorsal cortical extracts from heterozygous and forebrain-specific Brpf1 knockouts.
(D) Histone H3 acetylation in protein extracts from wild-type and epiblast-specific Brpf1-knockout embryos at embryonic day 10.5 (E10.5).
(E) Quantification of H3K23 acetylation in proteins extracts from wild-type and epiblast-specific Brpf1 knockout embryos at E10.5. The blot was from the same experiment as shown in (D), but the exposure time was shorter than the bottom image in (D). The quantification was performed with NIH ImageJ.
(F) Histone H3 acetylation in protein extracts from control and Brpf1-knockout mouse embryonic fibroblasts (MEFs). The fibroblasts were prepared from control and tamoxifen-inducible knockout embryos at E15.5.27
(G) Histone H3 acetylation in protein extracts from control and p.Pro370Ser lymphoblastoid cells.
(H) Histone H3 acetylation in protein extracts from control and p.Arg455∗ fibroblasts.
These results prompted us to investigate whether histone H3K23 acetylation is affected in cells from individuals with BRPF1 mutations. Lymphoblastoid cells from individual P4 (p.Pro370Ser; Table 1) showed reduced H3K23 acetylation (∼50% of the control; Figure 3G). In comparison, qRT-PCR revealed no decrease in the amount of BRPF1 mRNA in mutant cells (Figure S3A). We also obtained a skin biopsy from individual P5 (p.Arg455∗; Table 1) for preparation of fibroblasts. As shown in Figure 3H, H3K23 acetylation in the fibroblasts also decreased to ∼50% of the control. These results indicate that two mutations lead to decreased H3K23 acetylation in individuals P4 and P5 (Table 1 and Figures 3G and 3H). Consistent with this, RNAi knockdown showed that BRPF1 is important for H3K23 acetylation in human U2OS cells.40
NMD degradates a premature stop codon-containing transcript expressed in Brpf1-knockout mice.26, 28, 29 The mutation in individual P5 introduces a stop codon in a middle exon (Figure 2A) and might thus trigger NMD. To investigate this, we first performed qRT-PCR to analyze BRPF1 mRNA amounts in the control and mutant fibroblasts. One primer set, but not the other, detected some decrease of BRPF1 mRNA in the mutant cells (Figure S3B). To resolve this difference, cDNA from the mutant fibroblasts was prepared and sequenced. The results revealed that the wild-type and mutant alleles were similarly expressed (Figure S1C), indicating that NMD is not at play with the mutant transcript in fibroblasts from individual P5. To substantiate this, we compared lymphoblastoid cells from this individual and her parents. qRT-PCR analysis of BRPF1 mRNA detected no difference between the mutant and control cells (Figure S3C). Furthermore, we analyzed lymphoblastoid cells from individual P2, also carrying a premature stop codon in the mutant allele (Table 1). qRT-PCR analysis of BRPF1 mRNA detected no difference between the mutant and control cells (Figure S3D). Together, these results suggest that NMD does not apply to the mutant transcripts in individuals P2 and P5. Notably, a similar conclusion has been reached for KAT6A and KAT6B mutations.9, 12
Impact of BRPF1 Mutations on Histone H3K23 Acetylation In Vitro
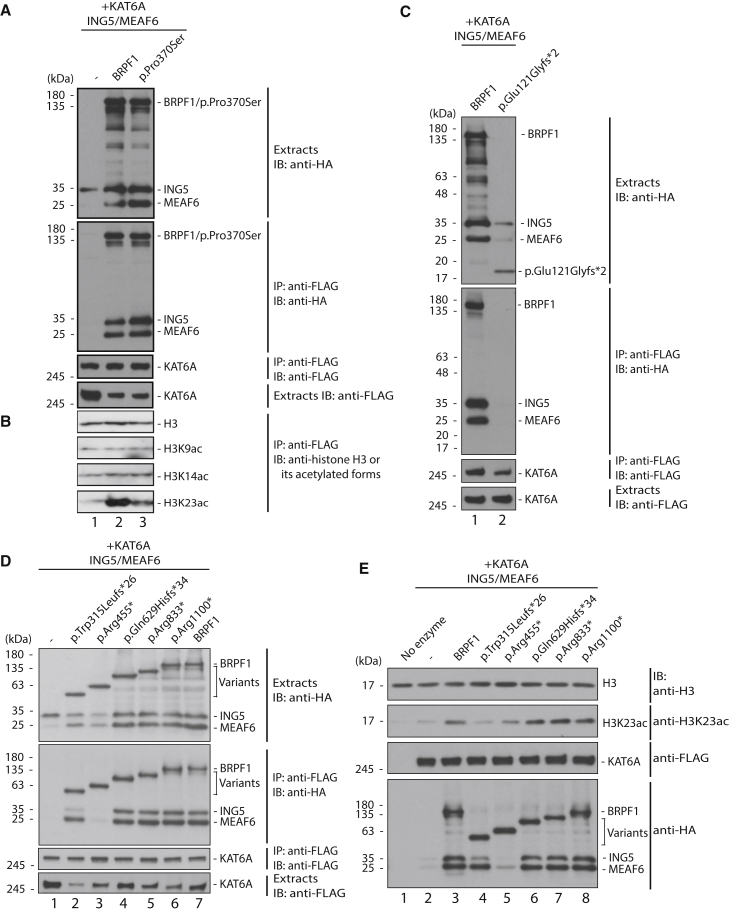
To delineate further the molecular mechanisms by which the BRPF1 mutations affect H3K23 acetylation, we engineered seven variants encoded by the mutations and analyzed the impact on formation of tetrameric complexes with KAT6A (Figure 4). The variants were produced transiently in HEK293 cells with KAT6A, ING5, and MEAF6 for co-immunoprecipitation. BRPF1 serves as a scaffold protein to bridge interaction of KAT6A and KAT6B with ING5 and MEAF6.19, 20 Exogenous BRPF1 promoted production of ING5 and MEAF6 (Figure 4A, top panel, lanes 1 and 2). We observed similar effects with p.Pro370Ser, p.Gln629Hisfs∗34, p.Arg833∗, and p.Arg1100∗ (Figures 4A, 4C, and 4D, top panels). Moreover, like wild-type BRPF1, these four variants form tetrameric complexes with KAT6A, ING5, and MEAF6 (Figures 4A, lanes 2 and 3, and 4D, lanes 4–7). By contrast, p.Glu121Glyfs∗2, p.Trp315Leufs∗26, and p.Arg455∗ failed to promote expression of ING5 and MEAF6 (Figures 4C and 4D, top panel). p.Glu121Glyfs∗2 was not produced as well as the wild-type (Figure 4C, top panel) and failed to interact with KAT6A to form a tetrameric complex with ING5 and MEAF6 (Figure 4C). The remaining two variants interacted with KAT6A similarly to wild-type BRPF1 (Figure 4D, lanes 1–3 and 7). As expected, given its lack of the ING5- and MEAF6-interacting domain (Figure 2A), p.Arg455∗ failed to mediate the interaction of KAT6A with ING5 and MEAF6 (Figure 4D, lanes 3 and 7). Unexpectedly, pTrp315Leufs∗26 was still able to interact with MEAF6 (compare lanes 2 and 7). Whether this is due to the extra 26 residues introduced by the reading frameshift remains unclear. These results support that the nine variants (Table 1 and Figure 2A) form different groups according to the ability to form tetrameric complexes with KAT6A, ING5, and MEAF6.
Figure 4.
Functional Characterization of BRPF1 Variants In Vitro
(A) Interaction of BRPF1 and the variants with KAT6A, ING5, and MEAF6. KAT6A was produced in HEK293 cells as a FLAG-tagged fusion protein along with HA-tagged BRPF1 (or the p.Pro370Ser variant), ING5, and MEAF6 as indicated. Soluble protein extracts were prepared for affinity purification on anti-FLAG agarose, and bound proteins were eluted with the FLAG peptide for immunoblotting with anti-FLAG and -HA antibodies.
(B) Histone acetylation assays. HeLa oligonucleosomes were used as substrates for acetylation by proteins affinity purified in (A). Acetylation of histone H3 was detected with antibodies recognizing histone H3 and its acetylated forms as indicated.
(C and D) Same as (A) except that different BRPF1 variants were analyzed. Unexpectedly, the variant p.Trp315Leufs∗26 still showed strong interaction with MEAF6; whether this is due to the extra 26 residues introduced after the reading frameshift remains unclear.
(E) Histone acetylation assays. Proteins affinity-purified in (D) were used for the enzymatic assays as in (B).
We next investigated whether the BRPF1 variants stimulate the acetyltransferase activity of KAT6A. As shown in Figure 4B (lanes 1 and 2), wild-type BRPF1 promoted H3K23 acetylation by KAT6A when oligonucleosomes were used as the substrate. By contrast, p.Pro370Ser failed to do so (Figure 4B, lanes 1–3). This is consistent with reduced H3K23 acetylation in lymphoblastoid cells prepared from individual P4 (Figure 3G). These results attest to the importance of the DNA-binding loop of BRPF1 in regulating the acetyltransferase activity of KAT6A. Like this mutant, the variant pTrp315Leufs∗26 was inactive in stimulating the acetyltransferase activity of KAT6A (Figure 4E, lanes 1–4). The variant p.Arg455∗ showed reduced ability to stimulate the acetyltransferase activity of KAT6A (compare lanes 1–3 and 5). The impaired ability of the variants pTrp315Leufs∗26 and p.Arg455∗ is likely due to the loss of the domain crucial for ING5 and EAF6 interaction (Figure 1A).20 By contrast, p.Gln629Hisfs∗34, p.Arg833∗, and p.Arg1100∗ were as active as wild-type BRPF1 in stimulating the acetyltransferase activity of KAT6A (Figure 4E, lanes 6–8), in agreement with their ability to form tetrameric complexes with KAT6A, ING5, and MEAF6, similarly to wild-type BRPF1 (Figure 4D, lanes 4–7). The variants p.Arg833∗ and p.Arg1100∗ lack the PWWP domain, which binds to trimethylated histone H3 in vitro.36, 37 Moreover, a mutant lacking this domain causes developmental defects in zebrafish.24 Thus, the findings on these two variants reiterate the importance of the PWWP domain in vivo.
We also analyzed subcellular localization of BRPF1 and its mutants by fluorescence microscopy. As reported for NIH 3T3 fibroblasts,20 when produced alone in HEK293 cells, BRPF1 formed cytoplasmic dots but became more nuclear with co-production of KAT6A, ING5, and MEAF6 (Figure S4). By themselves, p.Glu121Glyfs∗2 and p.Trp315Leufs∗26 presented uniform cytoplasmic distribution, whereas p.Arg455∗ and p.Gln629Hisfs∗34 were mainly nuclear (Figure S4). In the presence of exogenous KAT6A, ING5, and MEAF6, these four variants were all nuclear (Figure S4). The variant p.Arg833∗ formed large aggregates in the cytoplasm but became nuclear upon co-production of KAT6A, ING5, and MEAF6 (Figure S4). By itself, the missense variant p.Pro370Ser showed cytoplasmic distribution different from the wild-type but translocated to the nucleus upon co-production of KAT6A, ING5, and MEAF6 (Figure S4). Thus, the missense and truncation variants all behaved differently from wild-type BRPF1 (Figure S4). Together, results from determination of enzymatic activity (Figures 3 and 4) and subcellular localization (Figure S4) support that the BRPF1 mutations lead to pathogenicity through different mechanisms.
Distribution of Histone H3K23 Acetylation in the Mouse Brain
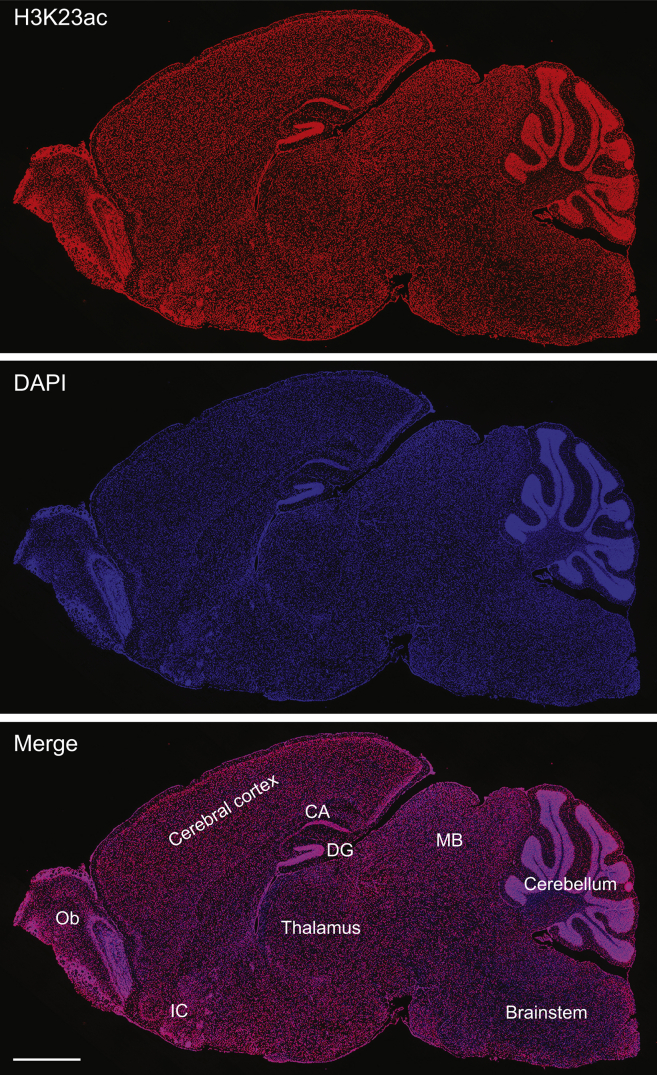
Intellectual disability in the individuals with BRPF1 mutations (Tables 1, 2, and S1) suggests the importance of BRPF1 in human brain development. In support of this, BRPF1 was highly expressed in the brain,25 and forebrain-specific inactivation of mouse Brpf1 leads to defects in the cerebrum and hippocampus.26, 28 In addition, mouse Kat6b is important for cerebral development.41 We thus analyzed distribution of H3K23 acetylation in the mouse brain. For this, indirect immunofluorescence microscopy was performed on mouse brain sections. As shown in Figure 5, fluorescence signals for H3K23 acetylation were detectable in the cerebral cortex but enhanced in the hippocampus and cerebellum. In the hippocampus, the signal was enriched within Cornu Ammonis areas and the dentate gyrus (Figure 5). In addition, the signals were high at islands of Calleja located within the olfactory tubercle (Figure 5). The cerebrum, hippocampus, and olfactory tubercle are all important for learning and memory. Moreover, the H3K23ac distribution is very similar to the expression pattern of mouse Brpf1 in the brain, as determined with a knockin reporter.25 These results further support that BRPF1 acts through H3K23 acetylation to regulate brain functions.
Figure 5.
Distribution of Histone H3K23 Acetylation in the Mouse Brain
Adult mouse brain sections were used for indirect immunofluorescence microscopy with the anti-H3K23ac antibody (upper), with nuclei counterstained with DAPI (middle). H3K23ac signals are enriched in the hippocampus, cerebellum, olfactory bulb (OB), and islands of Calleja (IC). Within the hippocampus, the fluorescence signals were high in Cornu Ammonis (CA) areas and the dentate gyrus (DG). Images of one representative parasagittal brain section are shown here. MB, mid-brain. Scale bar, 1 mm.
BRPF1 Governs H3K23 Acetylation through KAT6A and KAT6B
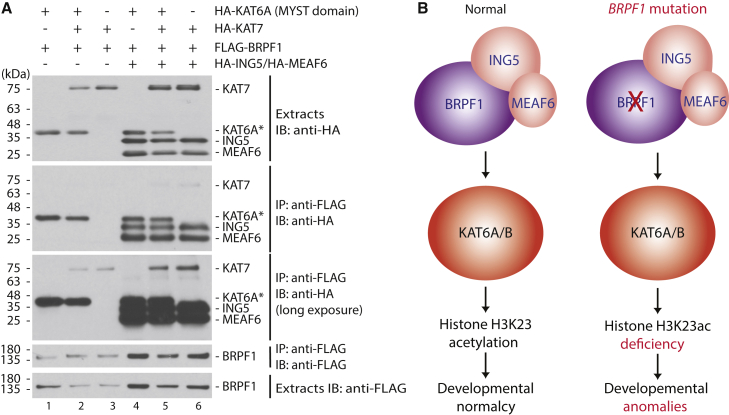
Via the EPC-like motif N-terminal to the PZP domain and a conserved region further N-terminal to this motif (Figure 2A), mammalian BRPF1 interacts with three histone acetyltransferases, KAT6A, KAT6B, and KAT7.19, 20, 21 Although Drosophila Br140 co-purified efficiently with Enok (homologous to KAT6A and KAT6B), Chameau (orthologous to KAT7) was not detectable.38, 39, 42 An interesting question is how KAT6A, KAT6B, and KAT7 contribute to BRPF1 functions in mammals. The binding sites for these KATs are all located within the N-terminal domain of BRPF1,19, 20, 21 so we investigated its competitive interaction with KAT6A and KAT7. For this, BRPF1 was produced in HEK293 cells as a FLAG-tagged fusion protein along with HA-tagged KAT6A (or KAT7), ING5, and MEAF6, as indicated (Figure 6A). Soluble protein extracts were prepared for co-immunoprecipitation. As shown in Figure 6A, the MYST domain of KAT6A formed an almost stoichiometric complex with ING5 and MEAF6, whereas the amount of KAT7 co-precipitated with BRPF1 was much lower (compare lanes 4 and 6). When both acetyltransferases were produced together (lane 5), competition was not obvious, supporting that they utilize distinct binding sites. Related to this, KAT7 binds to the BN domain of BRPF1,21 whereas KAT6A binds to a region more C-terminal to the BN domain (Figure 4).20 Based on the amount of KAT7 recovered from co-immunoprecipitation (Figure 6A), its binding to BRPF1 appeared to be much weaker than that of KAT6A. KAT6A is paralogous to KAT6B,20, 43 so these results suggest that BRPF1 mainly acts through KAT6A and KAT6B to control H3K23 acetylation in vivo (Figure 6B, left). These results also imply that through altering H3K23 acetylation by these two acetyltransferases, BRPF1 mutations cause developmental anomalies (Figure 6B, right).
Figure 6.
BRPF1 Governs H3K23 Acetylation through KAT6A and KAT6B
(A) Competitive interaction of BRPF1 with KAT6A and KAT7. BRPF1 was produced in HEK293 cells as a FLAG-tagged fusion protein along with HA-tagged KAT6A (or KAT7), ING5, and MEAF6 as indicated. For KAT6A, only its MYST domain was produced (labeled as KAT6A∗). Soluble protein extracts were prepared for affinity purification on anti-FLAG agarose, and bound proteins were eluted with the FLAG peptide for immunoblotting with anti-FLAG and -HA antibodies.
(B) Cartoon showing how BRPF1 mutations affect histone H3K23 acetylation and cause developmental abnormalities. In individuals without BRPF1 mutations (left), BRPF1 forms a trimeric complex with ING5 and EAF6 to control histone H3K23 acetylation by KAT6A and KAT6B, which in turn regulate developmental programs. In individuals with BRPF1 mutations (right), BRPF1 variants fail to exert proper control on histone H3K23 acetylation, thereby deregulating normal gene expression and developmental programs. For simplicity, the cartoon does not illustrate additional partners that BRPF1 might have in vivo.
Discussion
Chromatin structure and function constitute an integral component of epigenetic regulation. To interpret different chromatin modification states, the human genome encodes several hundred chromatin readers. Some of them have been investigated at the molecular level and in model organisms, but very few have been associated with Mendelian disorders in humans. Results presented herein show that BRPF1 mutations cause a developmental disorder in ten individuals with an intellectual disability syndrome (Figures 1 and 2 and Table 2). Developmental delay, intellectual disability, and language impairment are consistent with recent knockout studies showing that mouse Brpf1 is critical for embryo survival and forebrain development.26, 27, 28 The individuals are heterozygous for the mutations (Figures 1 and 2A). The mutated alleles are not functional in individuals P1–P5, P7, and P8 (Figures 3, 4, and S4), and perhaps also in individuals P6, P9, and P10 (Tables 1), so haploinsufficiency might be the pathological mechanism, as observed in several developmental disorders due to KAT6A mutations or KAT6B mutations.7, 8, 9, 10, 11, 12, 13 Some clinical features such as intellectual disability and developmental delay reported herein are common to those in persons with KAT6A or KAT6B mutations,7, 8, 9, 10, 11, 12, 13 but craniofacial dysmorphisms are less striking in individuals with BRPF1 mutations (Figure 1 and Table 2). Although the number of patients is not large enough to draw broad conclusions, most individuals with BRPF1 variants do not exhibit microcephaly, as seen in persons with KAT6A variants,12, 13 or possess genital and patellar abnormalities observed in genitopatellar syndrome (MIM: 606170) due to KAT6B mutations.9, 10 However, clinical variability has been observed in persons with KAT6B variants, which can present as genitopatellar syndrome or the Say-Barber-Biesecker-Young-Simpson variant of Ohdo syndrome (MIM: 603736).7, 8, 9, 10, 11 Notably, a majority of KAT6A and KAT6B mutations remove the serine- and methionine-rich transcriptional activation domain but leave the acetyltransferase domain intact,7, 8, 9, 10, 11, 12, 13 so they are not expected to affect global histone H3K23 acetylation per se. This is different from the major impact of BRPF1 mutations on histone H3K23 acetylation (Figure 2, Figure 3, Figure 4), raising a therapeutic prospect whereby it might be beneficial to correct this deficiency with deacetylase inhibitors.
Molecular analyses showed that the mutations impair H3K23 acetylation (Figure 3) due to defects in activating the acetyltransferases KAT6A and KAT6B (Figure 4) and in maintaining proper subcellular distribution (Figure S4). The latter is especially important for those variants lacking the PWWP domain only (Figure 2A). Moreover, it has been shown that the PWWP domain is important both in vitro and in vivo.24, 36, 37 For H3K23 acetylation, BRPF1 acts predominantly through KAT6A and KAT6B (Figure 6A). These molecular results are consistent with two recent reports showing that BRPF1 is important for histone H3K23 acetylation in mouse hematopoietic stem cells and that KAT6B is a histone H3K23 acetyltransferase in lung cancer cells.29, 44 Moreover, Drosophila Enok and Br140 are crucial for histone H3K23 acetylation.38, 39 Together, these studies suggest that BRPF1 acts through KAT6A and KAT6B to govern H3K23 acetylation and that mutations in BRPF1 might deregulate related epigenetic and developmental programs (Figure 6B).
Compared to other histone acetylation sites (such as H3K9, H3K14, H3K27, and H4K16), H3K23 is not as well characterized. However, three recent studies have unexpectedly revealed that acetylation at this site is highly abundant from C. elegans to humans,45, 46, 47 suggesting evolutionarily conserved importance in different organisms. At the mechanistic level, H3K23 acetylation serves as a docking site for the chromatin reader TRIM24 in breast cancer.48 This epigenetic mark also competes with H3K23 methylation and ubiquitination, both of which have recently emerged as two important epigenetic marks.49, 50 H3K23 acetylation might even interplay with H3K18 acetylation and H3K27 methylation, given the physical proximity to these two modification sites. Furthermore, a recent study has linked H3K23 acetylation to DNA replication and perhaps also DNA repair in Drosophila.39 Thus, defective H3K23 acetylation in individuals with BRPF1 mutations might exert polytrophic effects on molecular and cellular processes underlying the developmental anomalies.
BRPF1 is conserved from worms to humans.51 Lin-49, a related protein in C. elegans, regulates neuron asymmetry, hindgut development, and fecundity.52 Drosophila Br140 is highly homologous to mammalian BRPF1 and forms a tetrameric complex with Enok (homologous to KAT6A and KAT6B) to target histone H3K23 acetylation.39, 42 Enok is critical for neuroblast proliferation in the fly brain.53 Both Br140 and Enok are important for neuronal wiring of the fly visual system too.54 Although their functions in the nervous system remain unclear, both Brpf1 and Kat6a are important for maintaining pharyngeal segmental identity and skeletal development in zebrafish.24, 55 Both Brpf1 and Kat6b are critical for mouse brain development.26, 28, 41 Consistent with this, intellectual disability is a major clinical feature in individuals with mutations in BRPF1 (Table 2) or KAT6B.7, 8, 9, 10, 11 Although the individuals described herein carry monoallelic mutations, no obvious phenotypes have been observed for heterozygous Brpf1 mice.24, 25, 26, 27, 28, 29 Similarly, although monoallelic mutations in KAT6A or KAT6B cause developmental anomalies, no obvious phenotypes have been reported for heterozygous Kat6a or Kat6b mice.41, 56, 57 We have recently found that Brpf1 is critical for mouse hematopoietic stem cells and that its inactivation leads to pre-weaning lethality as a result of acute bone-marrow failure,29 but no hematological abnormalities have been observed in the individuals with BRPF1 mutations (Tables 2 and S1). Similarly, mouse Kat6a is critical for hematopoietic stem cells,56, 57 but no hematological abnormalities have been discovered in patients with the KAT6A mutations. One major difference is that both alleles are inactivated in knockout mice but only one allele is affected in individuals with BRPF1 or KAT6A mutations. In addition, over 100 missense and truncating somatic mutations in BRPF1 have been identified in different types of cancer.23, 51 About one third of these mutations are predicted to inactivate BRPF1 functions, but no indications of cancer predisposition are present in the ten individuals with BRPF1 mutations, although the oldest is now only 17 years old (Tables 2 and S1). Thus, further studies are needed to investigate these interesting issues.
In summary, we provide evidence that heterozygous BRPF1 mutations cause intellectual disability and other anomalies. The mutations affect physical and functional interactions of BRPF1 with KAT6A and KAT6B, resulting in deficient histone H3K23 acetylation (Figure 6B). Our findings provide a basis for phenotypic and molecular diagnosis of the developmental disorder with BRPF1 mutations, highlight the importance of this chromatin regulator in human development, and shed light on functions of its orthologs in different organisms. The findings also suggest potential value to using Brpf1 mutant mice as preclinical models for exploring treatment options. This study on BRPF1 mutations and the recent reports7, 8, 9, 10, 11, 12, 13 on KAT6A and KAT6B mutations uncover an emerging group of intellectual disability disorders caused by aberrant histone H3 acetylation.
Consortia
The members of the CAUSES Study consortium are Shelin Adam, Christele Du Souich, Jane Gillis, Alison Elliott, Anna Lehman, Jill Mwenifumbo, Tanya Nelson, Clara Van Karnebeek, and Jan Friedman.
Acknowledgments
This project was supported in part by operating grants from the Canadian Institutes of Health Research (RN324373 and RN315908 to P.M.C. and MOP-142252 X.J.Y.) and the Natural Sciences and Engineering Research Council of Canada (342146-12 to X.J.Y.). Funding for the CAUSES Study was from Mining for Miracles, British Columbia Hospital Foundation. The DDD Study presents independent research commissioned by the Health Innovation Challenge Fund (grant no. HICF-1009-003), a parallel funding partnership between the Wellcome Trust and the UK Department of Health, and the Wellcome Trust Sanger Institute (grant no. WT098051). The views expressed in this publication are those of the author(s) and not necessarily those of the Wellcome Trust or the Department of Health. The study has UK Research Ethics Committee (REC) approval (10/H0305/83, granted by the Cambridge South REC, and GEN/284/12, granted by the Republic of Ireland REC). The research team acknowledges the support of the National Institute for Health Research, through the Comprehensive Clinical Research Network. M.G., B.B. and M.T.C. are employees of GeneDx.
Published: December 8, 2016
Footnotes
Supplemental Data include four figures and one table and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.11.011.
Contributor Information
Philippe M. Campeau, Email: p.campeau@umontreal.ca.
Xiang-Jiao Yang, Email: xiang-jiao.yang@mcgill.ca.
Web Resources
cBioPortal for Cancer Genomics, http://www.cbioportal.org/index.do
DECIPHER, http://decipher.sanger.ac.uk/
ExAC Browser, http://exac.broadinstitute.org/
GenBank, http://www.ncbi.nlm.nih.gov/genbank/
GeneMatcher, https://genematcher.org/
OMIM, http://www.omim.org/
UCSC Genome Browser, http://genome.ucsc.edu
Supplemental Data
References
- 1.Li B., Carey M., Workman J.L. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 2.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Allis C.D., Jenuwein T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016;17:487–500. doi: 10.1038/nrg.2016.59. [DOI] [PubMed] [Google Scholar]
- 4.Berdasco M., Esteller M. Genetic syndromes caused by mutations in epigenetic genes. Hum. Genet. 2013;132:359–383. doi: 10.1007/s00439-013-1271-x. [DOI] [PubMed] [Google Scholar]
- 5.Bernier R., Golzio C., Xiong B., Stessman H.A., Coe B.P., Penn O., Witherspoon K., Gerdts J., Baker C., Vulto-van Silfhout A.T. Disruptive CHD8 mutations define a subtype of autism early in development. Cell. 2014;158:263–276. doi: 10.1016/j.cell.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weiss K., Terhal P.A., Cohen L., Bruccoleri M., Irving M., Martinez A.F., Rosenfeld J.A., Machol K., Yang Y., Liu P., DDD Study De Novo Mutations in CHD4, an ATP-Dependent Chromatin Remodeler Gene, Cause an Intellectual Disability Syndrome with Distinctive Dysmorphisms. Am. J. Hum. Genet. 2016;99:934–941. doi: 10.1016/j.ajhg.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kraft M., Cirstea I.C., Voss A.K., Thomas T., Goehring I., Sheikh B.N., Gordon L., Scott H., Smyth G.K., Ahmadian M.R. Disruption of the histone acetyltransferase MYST4 leads to a Noonan syndrome-like phenotype and hyperactivated MAPK signaling in humans and mice. J. Clin. Invest. 2011;121:3479–3491. doi: 10.1172/JCI43428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clayton-Smith J., O’Sullivan J., Daly S., Bhaskar S., Day R., Anderson B., Voss A.K., Thomas T., Biesecker L.G., Smith P. Whole-exome-sequencing identifies mutations in histone acetyltransferase gene KAT6B in individuals with the Say-Barber-Biesecker variant of Ohdo syndrome. Am. J. Hum. Genet. 2011;89:675–681. doi: 10.1016/j.ajhg.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simpson M.A., Deshpande C., Dafou D., Vissers L.E., Woollard W.J., Holder S.E., Gillessen-Kaesbach G., Derks R., White S.M., Cohen-Snuijf R. De novo mutations of the gene encoding the histone acetyltransferase KAT6B cause Genitopatellar syndrome. Am. J. Hum. Genet. 2012;90:290–294. doi: 10.1016/j.ajhg.2011.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campeau P.M., Kim J.C., Lu J.T., Schwartzentruber J.A., Abdul-Rahman O.A., Schlaubitz S., Murdock D.M., Jiang M.M., Lammer E.J., Enns G.M. Mutations in KAT6B, encoding a histone acetyltransferase, cause Genitopatellar syndrome. Am. J. Hum. Genet. 2012;90:282–289. doi: 10.1016/j.ajhg.2011.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu H.C., Geiger E.A., Medne L., Zackai E.H., Shaikh T.H. An individual with blepharophimosis-ptosis-epicanthus inversus syndrome (BPES) and additional features expands the phenotype associated with mutations in KAT6B. Am. J. Med. Genet. A. 2014;164A:950–957. doi: 10.1002/ajmg.a.36379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arboleda V.A., Lee H., Dorrani N., Zadeh N., Willis M., Macmurdo C.F., Manning M.A., Kwan A., Hudgins L., Barthelemy F., UCLA Clinical Genomics Center De novo nonsense mutations in KAT6A, a lysine acetyl-transferase gene, cause a syndrome including microcephaly and global developmental delay. Am. J. Hum. Genet. 2015;96:498–506. doi: 10.1016/j.ajhg.2015.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tham E., Lindstrand A., Santani A., Malmgren H., Nesbitt A., Dubbs H.A., Zackai E.H., Parker M.J., Millan F., Rosenbaum K. Dominant mutations in KAT6A cause intellectual disability with recognizable syndromic features. Am. J. Hum. Genet. 2015;96:507–513. doi: 10.1016/j.ajhg.2015.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vega H., Waisfisz Q., Gordillo M., Sakai N., Yanagihara I., Yamada M., van Gosliga D., Kayserili H., Xu C., Ozono K. Roberts syndrome is caused by mutations in ESCO2, a human homolog of yeast ECO1 that is essential for the establishment of sister chromatid cohesion. Nat. Genet. 2005;37:468–470. doi: 10.1038/ng1548. [DOI] [PubMed] [Google Scholar]
- 15.Williams S.R., Aldred M.A., Der Kaloustian V.M., Halal F., Gowans G., McLeod D.R., Zondag S., Toriello H.V., Magenis R.E., Elsea S.H. Haploinsufficiency of HDAC4 causes brachydactyly mental retardation syndrome, with brachydactyly type E, developmental delays, and behavioral problems. Am. J. Hum. Genet. 2010;87:219–228. doi: 10.1016/j.ajhg.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deardorff M.A., Bando M., Nakato R., Watrin E., Itoh T., Minamino M., Saitoh K., Komata M., Katou Y., Clark D. HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle. Nature. 2012;489:313–317. doi: 10.1038/nature11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taverna S.D., Li H., Ruthenburg A.J., Allis C.D., Patel D.J. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Musselman C.A., Lalonde M.E., Côté J., Kutateladze T.G. Perceiving the epigenetic landscape through histone readers. Nat. Struct. Mol. Biol. 2012;19:1218–1227. doi: 10.1038/nsmb.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doyon Y., Cayrou C., Ullah M., Landry A.J., Côté V., Selleck W., Lane W.S., Tan S., Yang X.J., Côté J. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol. Cell. 2006;21:51–64. doi: 10.1016/j.molcel.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 20.Ullah M., Pelletier N., Xiao L., Zhao S.P., Wang K., Degerny C., Tahmasebi S., Cayrou C., Doyon Y., Goh S.L. Molecular architecture of quartet MOZ/MORF histone acetyltransferase complexes. Mol. Cell. Biol. 2008;28:6828–6843. doi: 10.1128/MCB.01297-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lalonde M.E., Avvakumov N., Glass K.C., Joncas F.H., Saksouk N., Holliday M., Paquet E., Yan K., Tong Q., Klein B.J. Exchange of associated factors directs a switch in HBO1 acetyltransferase histone tail specificity. Genes Dev. 2013;27:2009–2024. doi: 10.1101/gad.223396.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu L., Qin S., Zhang J., Ji P., Shi Y., Wu J. Solution structure of an atypical PHD finger in BRPF2 and its interaction with DNA. J. Struct. Biol. 2012;180:165–173. doi: 10.1016/j.jsb.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 23.Klein B.J., Muthurajan U.M., Lalonde M.E., Gibson M.D., Andrews F.H., Hepler M., Machida S., Yan K., Kurumizaka H., Poirier M.G. Bivalent interaction of the PZP domain of BRPF1 with the nucleosome impacts chromatin dynamics and acetylation. Nucleic Acids Res. 2016;44:472–484. doi: 10.1093/nar/gkv1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laue K., Daujat S., Crump J.G., Plaster N., Roehl H.H., Kimmel C.B., Schneider R., Hammerschmidt M., Tübingen 2000 Screen Consortium The multidomain protein Brpf1 binds histones and is required for Hox gene expression and segmental identity. Development. 2008;135:1935–1946. doi: 10.1242/dev.017160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.You L., Chen L., Penney J., Miao D., Yang X.J. Expression atlas of the multivalent epigenetic regulator Brpf1 and its requirement for survival of mouse embryos. Epigenetics. 2014;9:860–872. doi: 10.4161/epi.28530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.You L., Yan K., Zou J., Zhao H., Bertos N.R., Park M., Wang E., Yang X.J. The lysine acetyltransferase activator Brpf1 governs dentate gyrus development through neural stem cells and progenitors. PLoS Genet. 2015;11:e1005034. doi: 10.1371/journal.pgen.1005034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.You L., Yan K., Zou J., Zhao H., Bertos N.R., Park M., Wang E., Yang X.J. The chromatin regulator Brpf1 regulates embryo development and cell proliferation. J. Biol. Chem. 2015;290:11349–11364. doi: 10.1074/jbc.M115.643189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.You L., Zou J., Zhao H., Bertos N.R., Park M., Wang E., Yang X.J. Deficiency of the chromatin regulator BRPF1 causes abnormal brain development. J. Biol. Chem. 2015;290:7114–7129. doi: 10.1074/jbc.M114.635250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.You L., Li L., Zou J., Yan K., Belle J., Nijnik A., Wang E., Yang X.J. BRPF1 is essential for development of fetal hematopoietic stem cells. J. Clin. Invest. 2016;126:3247–3262. doi: 10.1172/JCI80711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ling P.D., Huls H.M. Isolation and immortalization of lymphocytes. Curr. Protoc. Mol. Biol. 2005;Chapter 28 doi: 10.1002/0471142727.mb2802s70. unit 28.2. [DOI] [PubMed] [Google Scholar]
- 31.Yan K., You L., Degerny C., Ghorbani M., Liu X., Chen L., Li L., Miao D., Yang X.J. The chromatin regulator BRPF3 preferentially activates the HBO1 acetyltransferase but is dispensable for mouse development and survival. J. Biol. Chem. 2016;291:2647–2663. doi: 10.1074/jbc.M115.703041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Firth H.V., Richards S.M., Bevan A.P., Clayton S., Corpas M., Rajan D., Van Vooren S., Moreau Y., Pettett R.M., Carter N.P. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am. J. Hum. Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qin S., Jin L., Zhang J., Liu L., Ji P., Wu M., Wu J., Shi Y. Recognition of unmodified histone H3 by the first PHD finger of bromodomain-PHD finger protein 2 provides insights into the regulation of histone acetyltransferases monocytic leukemic zinc-finger protein (MOZ) and MOZ-related factor (MORF) J. Biol. Chem. 2011;286:36944–36955. doi: 10.1074/jbc.M111.244400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poplawski A., Hu K., Lee W., Natesan S., Peng D., Carlson S., Shi X., Balaz S., Markley J.L., Glass K.C. Molecular insights into the recognition of N-terminal histone modifications by the BRPF1 bromodomain. J. Mol. Biol. 2014;426:1661–1676. doi: 10.1016/j.jmb.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vezzoli A., Bonadies N., Allen M.D., Freund S.M., Santiveri C.M., Kvinlaug B.T., Huntly B.J., Göttgens B., Bycroft M. Molecular basis of histone H3K36me3 recognition by the PWWP domain of Brpf1. Nat. Struct. Mol. Biol. 2010;17:617–619. doi: 10.1038/nsmb.1797. [DOI] [PubMed] [Google Scholar]
- 37.Wu H., Zeng H., Lam R., Tempel W., Amaya M.F., Xu C., Dombrovski L., Qiu W., Wang Y., Min J. Structural and histone binding ability characterizations of human PWWP domains. PLoS ONE. 2011;6:e18919. doi: 10.1371/journal.pone.0018919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang F., Paulson A., Dutta A., Venkatesh S., Smolle M., Abmayr S.M., Workman J.L. Histone acetyltransferase Enok regulates oocyte polarization by promoting expression of the actin nucleation factor spire. Genes Dev. 2014;28:2750–2763. doi: 10.1101/gad.249730.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang F., Saraf A., Florens L., Kusch T., Swanson S.K., Szerszen L.T., Li G., Dutta A., Washburn M.P., Abmayr S.M., Workman J.L. The Enok acetyltransferase complex interacts with Elg1 and negatively regulates PCNA unloading to promote the G1/S transition. Genes Dev. 2016;30:1198–1210. doi: 10.1101/gad.271429.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feng Y., Vlassis A., Roques C., Lalonde M.E., González-Aguilera C., Lambert J.P., Lee S.B., Zhao X., Alabert C., Johansen J.V. BRPF3-HBO1 regulates replication origin activation and histone H3K14 acetylation. EMBO J. 2016;35:176–192. doi: 10.15252/embj.201591293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas T., Voss A.K., Chowdhury K., Gruss P. Querkopf, a MYST family histone acetyltransferase, is required for normal cerebral cortex development. Development. 2000;127:2537–2548. doi: 10.1242/dev.127.12.2537. [DOI] [PubMed] [Google Scholar]
- 42.Strübbe G., Popp C., Schmidt A., Pauli A., Ringrose L., Beisel C., Paro R. Polycomb purification by in vivo biotinylation tagging reveals cohesin and Trithorax group proteins as interaction partners. Proc. Natl. Acad. Sci. USA. 2011;108:5572–5577. doi: 10.1073/pnas.1007916108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Champagne N., Bertos N.R., Pelletier N., Wang A.H., Vezmar M., Yang Y., Heng H.H., Yang X.J. Identification of a human histone acetyltransferase related to monocytic leukemia zinc finger protein. J. Biol. Chem. 1999;274:28528–28536. doi: 10.1074/jbc.274.40.28528. [DOI] [PubMed] [Google Scholar]
- 44.Simó-Riudalbas L., Pérez-Salvia M., Setien F., Villanueva A., Moutinho C., Martínez-Cardús A., Moran S., Berdasco M., Gomez A., Vidal E. KAT6B Is a Tumor Suppressor Histone H3 Lysine 23 Acetyltransferase Undergoing Genomic Loss in Small Cell Lung Cancer. Cancer Res. 2015;75:3936–3945. doi: 10.1158/0008-5472.CAN-14-3702. [DOI] [PubMed] [Google Scholar]
- 45.Vandamme J., Sidoli S., Mariani L., Friis C., Christensen J., Helin K., Jensen O.N., Salcini A.E. H3K23me2 is a new heterochromatic mark in Caenorhabditis elegans. Nucleic Acids Res. 2015;43:9694–9710. doi: 10.1093/nar/gkv1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen S., Yang Z., Wilkinson A.W., Deshpande A.J., Sidoli S., Krajewski K., Strahl B.D., Garcia B.A., Armstrong S.A., Patel D.J., Gozani O. The PZP Domain of AF10 Senses Unmodified H3K27 to Regulate DOT1L-Mediated Methylation of H3K79. Mol. Cell. 2015;60:319–327. doi: 10.1016/j.molcel.2015.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou T., Chung Y.H., Chen J., Chen Y. Site-Specific Identification of Lysine Acetylation Stoichiometries in Mammalian Cells. J. Proteome Res. 2016;15:1103–1113. doi: 10.1021/acs.jproteome.5b01097. [DOI] [PubMed] [Google Scholar]
- 48.Tsai W.W., Wang Z., Yiu T.T., Akdemir K.C., Xia W., Winter S., Tsai C.Y., Shi X., Schwarzer D., Plunkett W. TRIM24 links a non-canonical histone signature to breast cancer. Nature. 2010;468:927–932. doi: 10.1038/nature09542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nishiyama A., Yamaguchi L., Sharif J., Johmura Y., Kawamura T., Nakanishi K., Shimamura S., Arita K., Kodama T., Ishikawa F. Uhrf1-dependent H3K23 ubiquitylation couples maintenance DNA methylation and replication. Nature. 2013;502:249–253. doi: 10.1038/nature12488. [DOI] [PubMed] [Google Scholar]
- 50.Papazyan R., Voronina E., Chapman J.R., Luperchio T.R., Gilbert T.M., Meier E., Mackintosh S.G., Shabanowitz J., Tackett A.J., Reddy K.L. Methylation of histone H3K23 blocks DNA damage in pericentric heterochromatin during meiosis. eLife. 2014;3:e02996. doi: 10.7554/eLife.02996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang X.J. MOZ and MORF acetyltransferases: Molecular interaction, animal development and human disease. Biochim. Biophys. Acta. 2015;1853:1818–1826. doi: 10.1016/j.bbamcr.2015.04.014. [DOI] [PubMed] [Google Scholar]
- 52.O’Meara M.M., Zhang F., Hobert O. Maintenance of neuronal laterality in Caenorhabditis elegans through MYST histone acetyltransferase complex components LSY-12, LSY-13 and LIN-49. Genetics. 2010;186:1497–1502. doi: 10.1534/genetics.110.123661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scott E.K., Lee T., Luo L. enok encodes a Drosophila putative histone acetyltransferase required for mushroom body neuroblast proliferation. Curr. Biol. 2001;11:99–104. doi: 10.1016/s0960-9822(01)00020-3. [DOI] [PubMed] [Google Scholar]
- 54.Berger J., Senti K.A., Senti G., Newsome T.P., Asling B., Dickson B.J., Suzuki T. Systematic identification of genes that regulate neuronal wiring in the Drosophila visual system. PLoS Genet. 2008;4:e1000085. doi: 10.1371/journal.pgen.1000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miller C.T., Maves L., Kimmel C.B. moz regulates Hox expression and pharyngeal segmental identity in zebrafish. Development. 2004;131:2443–2461. doi: 10.1242/dev.01134. [DOI] [PubMed] [Google Scholar]
- 56.Katsumoto T., Aikawa Y., Iwama A., Ueda S., Ichikawa H., Ochiya T., Kitabayashi I. MOZ is essential for maintenance of hematopoietic stem cells. Genes Dev. 2006;20:1321–1330. doi: 10.1101/gad.1393106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomas T., Corcoran L.M., Gugasyan R., Dixon M.P., Brodnicki T., Nutt S.L., Metcalf D., Voss A.K. Monocytic leukemia zinc finger protein is essential for the development of long-term reconstituting hematopoietic stem cells. Genes Dev. 2006;20:1175–1186. doi: 10.1101/gad.1382606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.