Abstract
Genome-wide association studies (GWASs) have identified loci for erythrocyte traits in primarily European ancestry populations. We conducted GWAS meta-analyses of six erythrocyte traits in 71,638 individuals from European, East Asian, and African ancestries using a Bayesian approach to account for heterogeneity in allelic effects and variation in the structure of linkage disequilibrium between ethnicities. We identified seven loci for erythrocyte traits including a locus (RBPMS/GTF2E2) associated with mean corpuscular hemoglobin and mean corpuscular volume. Statistical fine-mapping at this locus pointed to RBPMS at this locus and excluded nearby GTF2E2. Using zebrafish morpholino to evaluate loss of function, we observed a strong in vivo erythropoietic effect for RBPMS but not for GTF2E2, supporting the statistical fine-mapping at this locus and demonstrating that RBPMS is a regulator of erythropoiesis. Our findings show the utility of trans-ethnic GWASs for discovery and characterization of genetic loci influencing hematologic traits.
Introduction
Erythrocyte disorders are common worldwide, contributing to substantial morbidity and mortality.1 Erythrocyte counts and indices are heritable (estimated h2 = 0.40–0.902, 3, 4), exhibit different patterns across ethnic groups, and have been influenced by selection in various ethnic groups, most notably for protection against infection by parasites such as those that cause malaria.5, 6, 7 Erythrocyte traits have been studied most extensively in European ancestry populations,8, 9, 10 with smaller studies in non-European populations, and have shown both shared and distinct genetic loci influencing erythrocyte traits.11, 12
Trans-ethnic meta-analysis of genome-wide association studies (GWASs) offers improved signal detection in a combined meta-analysis when heterogeneity of allelic effects, allele frequencies, and differences in linkage disequilibrium (LD) between ethnicities are accounted for. Trans-ethnic meta-analysis can also enable fine-mapping of association intervals by evaluating differences in LD structure between diverse populations, thereby enhancing the detection of causal variants.13
We conducted trans-ethnic GWAS meta-analyses with the goal of elucidating the genetic architecture of erythrocyte traits and to evaluate (1) whether combining data across populations of diverse ancestry may improve power to detect associations for erythrocyte traits and (2) whether differences in LD structure can be exploited to identify causal variants driving the observed associations with common SNPs. In this study, we analyzed GWAS summary statistics from 71,638 individuals from three diverse populations of European (EUR), East Asian (EAS), and African (AFR) ancestry. We conducted replication analyses in independent samples and performed functional testing to support our approach to fine-mapping.
Subjects and Methods
Study Samples
We aggregated HapMap-imputed GWAS results from 71,638 individuals represented in 23 cohorts embedded in the CHARGE Consortium (40,258 individuals of EUR ancestry), the RIKEN/BioBank Japan Project and AGEN cohorts (15,252 individuals of EAS ancestry), and the COGENT Consortium (16,128 individuals of AFR ancestry). Phenotypic information on all participating cohorts is provided in Table S1 and has been reported previously.8, 11, 12, 14, 15 We conducted replication analyses of the identified trait-loci associations in six independent studies: the Gutenberg Health Study (GHS cohorts 1 and 2, both EUR ancestry), the Genes and Blood-Clotting Study (GBC, EUR ancestry), the NEO study (EUR ancestry), the JUPITER trial (EUR ancestry), and the HANDLS study (AFR ancestry)16, 17, 18, 19, 20, 21 (total replication size N = 16,389).
Erythrocyte Phenotype Modeling
We analyzed six erythrocyte traits: hemoglobin concentration (Hb, g/dL), hematocrit (Hct, percentage), mean corpuscular hemoglobin (MCH, picograms), mean corpuscular hemoglobin concentration (MCHC, g/dL), mean corpuscular volume (MCV, femtoliters), and red blood cell count (RBC, 1M cells/cm3). Trait units were harmonized across all studies. MCH, MCHC, MCV, and RBC were transformed to obtain normal distributions. We excluded samples deviating more than 3 SD from the ethnic- and trait-specific mean within each contributing study, because we focused on determinants of variation in the general population rather than on specific hematological diseases that are overrepresented at the extremes of the trait distribution (Table S2).
Genotyping
In brief, the cohorts comprise unrelated individuals, except for the Framingham Heart Study (related individuals of European ancestry) and GeneSTAR (related individuals of European or African ancestry). SNPs with a minor allele frequency < 1%, missingness > 5, or HWE p < 10−7 were excluded. Genotypes were imputed to approximately 2.5 million SNPs using HapMap Phase II CEU. The RIKEN and the BioBank Japan Project and AGEN cohorts comprise unrelated individuals of East Asian ancestry (EAS). SNPs with a minor allele frequency < 0.01, missingness > 1%, or HWE p < 10−7 were excluded. Individuals with a call rate < 98% were excluded as well. Genotypes were imputed to approximately 2.5 million SNPs using HapMap Phase II JPT and CHB. The COGENT consortium cohorts comprise individuals of African American ancestry (AFR). SNPs with a minor allele frequency < 1% or missingness > 10% were excluded. Genotypes were imputed to approximately 2.5 million SNPs using HapMap Phase II CEU and YRI.
Cohort-Specific GWASs
For the initial GWA analyses, each cohort used linear regression to assess the association of all SNPs meeting the quality control criteria with each of the six traits separately. An additive genetic model was used and the regressions were adjusted for age, sex, and study site (if applicable). The Framingham Heart Study and the GeneSTAR study used linear mixed effects models to account for relatedness, and these models included adjustment for principal components.
Ethnic-Specific GWAS Meta-analyses
GWAS results of SNPs with a minor allele frequency (MAF) ≥ 1% and an imputation quality > 30% were analyzed in a fixed-effect meta-analysis (METAL software22) within each ancestry group, with genomic control (GC) correction of the individual GWAS results of each contributing cohort and the final meta-analysis results.23
Trans-ethnic Meta-analyses
For the trans-ethnic meta-analyses, the three sets of the ethnic-specific meta-analysis summary statistics were then combined with three approaches. First, we performed for each trait a trans-ethnic fixed-effect inverse variance-weighted meta-analysis of the EUR, EAS, and AFR GWAS summary statistics using METAL. Second, the ethnic-specific GWAS summary statistics were also combined using the MANTRA (Meta-Analysis of Trans-ethnic Association Studies) package, a meta-analysis software tool allowing for heterogeneity in allelic effects due to differences in LD structure in different ancestry clusters.24 MANTRA results are reported as log10 Bayes’s factors (log10BF). Finally, the three sets of ethnic-specific results were analyzed by means of the Han and Eskin RE2 model, a meta-analysis method developed for higher statistical power under heterogeneity.25 We used the METASOFT 3.0c tool as developed by the Buhm Han laboratories (Web Resources). For the fixed-effects and the RE2 models, we applied a genome-wide significance threshold adjusted for multiple testing, as we analyzed six traits in our study. Given that the traits under investigation are correlated (Table S10), we used eigenvalues to assess the effective number of independent traits according to Ji and Li,26 and we estimated this number at 4.0549 using the Matrix Spectral Decomposition tool (Web Resources). We therefore considered p values smaller than 1.25 × 10−8 (i.e., 5 × 10−8 / 4.0549) as genome-wide significant. For the MANTRA discovery analyses, a log10BF > 6.1 was considered as a genome-wide significant threshold value.27
Replication in Human Cohorts
The six independent replication studies—the Gutenberg Health Study (GHS cohorts 1 and 2, both EUR ancestry), the Genes and Blood-Clotting Study (GBC, EUR ancestry), the NEO study (EUR ancestry), the JUPITER trial (EUR ancestry), and the HANDLS study (AFR ancestry)16, 17, 18, 19, 20, 21 (total replication size N = 16,389)—provided linear regression results for the nine trait-locus combinations. Their results were meta-analyzed with a fixed effects inverse variance weighted method (METAL) and the RE2 methodology. Additionally, we meta-analyzed replication results with the discovery data using fixed-effects, MANTRA, and RE2 methods. For the replication analyses of the nine individual trait-locus combinations, we applied a threshold of p < 0.05/9. Additional human replication findings are provided in Supplemental Data.
Fine-Mapping
We used the MANTRA results to fine-map the regions of trait-associated index SNPs. We defined regions by identifying variants within a 1 Mb window around each index SNP (500 kb upstream and 500 kb downstream). For each SNP in a region, the posterior probability that this SNP is driving the region’s association signal was calculated by dividing the SNP’s BF by the summation of the BFs of all SNPs in the region. Credible sets (CSs) were subsequently created by sorting the SNPs in each region in descending order based on their BF (starting with the index SNP since this SNP has the region’s largest BF by definition). Going down the sorted list, the SNPs’ posterior probabilities were summed until the cumulative value exceeded 99% of the total cumulative posterior probability for all SNPs in the region. The length of a CS was expressed in base pairs. We compared 99% CSs for the trans-ethnic results and the results of a EUR-only MANTRA analysis.13, 24, 28 For the MANTRA fine-mapping analyses, a less stringent threshold value of log10BF > 5 was applied, because we wanted to include previously identified regions that may not have showed up in the more stringent MANTRA discovery analyses.
Heterogeneity Analysis
Heterogeneity of the associations across the different ethnicities was assessed by the I2 and Cochran’s Q statistics as reported by METAL22 and the posterior probability of heterogeneity as reported by MANTRA.24
ENCODE Annotation
We evaluated the SNPs identified in the discovery analyses against the ENCODE Project Consortium’s database of functional elements in the K562 erythroleukemic line.29
Experiments in Zebrafish
To substantiate the fine mapping of the RBPMS/GTF2E2 region biologically, we tested the effect of morpholino knockdown in zebrafish for both RBPMS and GTF2E2 orthologous genes, followed by assays of erythrocyte development.
Zebrafish rbpms, rbpms2, and gtf2e2 were identified and confirmed by peptide sequence homology study and gene synteny analysis. For rbpms, we relied solely on peptide homology comparison and domain structure since no syntenic region was previously annotated and found by this study.
For each morpholino (MO), its design incorporated information about gene structure and translational initiation sites (Gene-Tool Inc.). MOs targeting each transcript were injected into single-cell embryos at 1, 3, and 5 ng/embryo to find an optimal dose at which there was minimal non-specific toxicity. The stepwise doses also give a range of phenotypes from a hypomorph to a near complete knockdown for most transcripts, which were used to assess the additive model of genetic association. After injection, embryos were collected at specified time points, 16–18 ss, 22–26 hpf, and 48 hpf using both standard morphological features of the whole embryo and hours post-fertilization (hpf) to minimize differences in embryonic development staging caused by the MO injection.30, 31 The embryos were then assayed for hematopoietic development by whole-mount in situ hybridization and benzidine staining. We conducted two assays simultaneously for globin transcription and hemoglobin formation. For the globin transcription, developing erythrocytes in the intermediate cell mass of the embryos were assayed by embryonic β-globin 3 expression at the 16 somite stage, or 16–18 hpf.31 Benzidine staining phenotype was categorized from subtle decrease to complete absence of staining, which was categorized as mild, intermediate, or strong effect. Morphologically normal morphants with decreased blood formation were scored for hematopoietic effect.
In zebrafish, rbpms was not annotated in the known EST and cDNA databases, although a genomic sequence in the telomeric region on chromosome 7 predicting a coding sequence (80% peptide sequence similarity) was identified. In addition, the synteny between human RBPMS and GTF2E2 is not conserved in zebrafish where rbpms and gtf2e2 are located on two separate chromosomes, chromosomes 7 and 1, respectively. rbpms2 was annotated with two paralogs on chromosome 7 (26 Mb away from and centromeric to the true rbpms) and chromosome 25 of the zebrafish genome. This orthology mapping was confirmed again by this research based on gene synteny and 88% and 91% sequence similarity, respectively, for rbpms2b and rbpms2a to human RBPMS2. These two zebrafish RBPMS2 orthologs have a higher overall sequence similarity to human RBPMS than the true zebrafish rbpms, but both have a RBPMS2-signature stretch of alanine in the C terminus of the protein. Therefore, to confirm our rbpms orthology study and to confirm functional conservation of rbpms in zebrafish, MO individual knockdown of both rbpms2a and rbpms2b was also performed in independent experiments, showing much less or no effect by rbpms2a knock-down and moderate effect by rbpms2b impact on erythropoiesis, suggesting functional compensation of the genes in the rbpms family in zebrafish during embryonic erythropoiesis.
Chromatin Immunoprecipitation and Assay for Transposase Accessible Chromatin in Human CD34+ Cell Lines
For ChIP-seq experiments, the following antibodies were used: Gata1 (Santa Cruz cat# sc265X), Gata2 (Santa Cruz cat# sc9008X), and H3K27ac (Abcam cat# ab4729; RRID: AB_2118291). ChIP experiments were performed as previously described with slight modifications.32, 33 In brief, 20–30 million cells for each ChIP were crosslinked by the addition of 1/10 volume 11% fresh formaldehyde for 10 min at room temperature. The crosslinking was quenched by the addition of 1/20 volume 2.5 M glycine. Cells were washed twice with ice-cold PBS and the pellet was flash-frozen in liquid nitrogen. Cells were kept at −80°C until the experiments were performed. Cells were lysed in 10 mL of lysis buffer 1 (50 mM HEPES-KOH [pH 7.5], 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100, and protease inhibitors) for 10 min at 4°C. After centrifugation, cells were resuspended in 10 mL of lysis buffer 2 (10 mM Tris-HCl [pH 8.0], 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, and protease inhibitors) for 10 min at room temperature. Cells were pelleted and resuspended in 3 mL of sonication buffer for K562 and U937 and 1 mL for other cells used (10 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% Na-Deoxycholate, 0.05% Nlauroylsarcosine, and protease inhibitors) and sonicated in a Bioruptor sonicator for 24–40 cycles of 30 s followed by 1 min resting intervals. Samples were centrifuged for 10 min at 18,000 × g and 1% of TritonX was added to the supernatant. Prior to the immunoprecipitation, 50 mL of protein G beads (Invitrogen 100-04D) for each reaction were washed twice with PBS, 0.5% BSA. Finally, the beads were resuspended in 250 mL of PBS, 0.5% BSA, and 5 mg of each antibody. Beads were rotated for at least 6 hr at 40°C and then washed twice with PBS, 0.5% BSA. Cell lysates were added to the beads and incubated at 40°C overnight. Beads were washed 1× with 20 mM Tris-HCl (pH 8), 150 mM NaCl, 2 mM EDTA, 0.1% SDS, 1% Triton X-100, 1× with 20 mM Tris-HCl (pH 8), 500 mM NaCl, 2 mM EDTA, 0.1% SDS, 1% Triton X-100, 1× with 10 mM Tris-HCl (pH 8), 250 nM LiCl, 2 mM EDTA, 1% NP40, and 1× with TE and finally resuspended in 200 mL elution buffer (50 mM Tris-HCl [pH 8.0], 10 mM EDTA, and 0.5%–1% SDS). 50 μL of cell lysates prior to addition to the beads was kept as input. Crosslinking was reversed by incubating samples at 65°C for at least 6 hr. Afterward the cells were treated with RNase and proteinase K and the DNA was extracted by phenol/chloroform extraction.
ChIP-seq libraries were prepared using the following protocol. End repair of immunoprecipitated DNA was performed using the End-It DNA End-Repair kit (Epicenter, ER81050) and incubating the samples at 25°C for 45 min. End-repaired DNA was purified using AMPure XP Beads (1.8× the reaction volume) (Agencourt AMPure XP – PCR purification Beads, BeckmanCoulter, A63881) and separating beads using DynaMag-96 Side Skirted Magnet (Life Technologies, 12027). A tail was added to the end-repaired DNA using NEB Klenow Fragment Enzyme (3′-5′ exo, M0212L), 1× NEB buffer 2, and 0.2 mM dATP (Invitrogen, 18252-015) and incubating the reaction mix at 37°C for 30 min. A-tailed DNA was cleaned up using AMPure beads (1.8× reaction volume). Subsequently, cleaned-up dA-tailed DNA went through Adaptor ligation reaction using Quick Ligation Kit (NEB, M2200L) according to the manufacturer’s protocol. Adaptor-ligated DNA was first cleaned up using AMPure beads (1.8× of reaction volume), eluted in 100 μL and then size-selected using AMPure beads (0.9× of the final supernatant volume, 90 μL). Adaptor ligated DNA fragments of proper size were enriched with PCR reaction using Fusion High-Fidelity PCR Master Mix kit (NEB, M0531S) and specific index primers supplied in NEBNext Multiplex Oligo Kit for Illumina (Index Primer Set 1, NEB, E7335L). Conditions for PCR used are as follows: 98°C, 30 s; (98°C, 10 s; 65°C, 30 s; 72°C, 30 s) × 15 to 18 cycles; 72°C, 5 min; hold at 4°C. PCR-enriched fragments were further size selected by running the PCR reaction mix in 2% low-molecular-weight agarose gel (Bio-Rad, 161-3107) and subsequently purifying them using QIAquick Gel Extraction Kit (28704). Libraries were eluted in 25 μL elution buffer. After measuring concentration in Qubit, all the libraries went through quality-control analysis using an Agilent Bioanalyzer. Samples with proper size (250–300 bp) were selected for next generation sequencing using Illumina Hiseq 2000 or 2500 platform.
Alignment and visualization ChIP-seq reads were aligned to the human reference genome (hg19) using bowtie with parameters -k 2 -m 2 -S.34 WIG files for display were created using MACS35 with parameters -w -S–space = 50–nomodel–shiftsize = 200 and were displayed in IGV.36, 37
High-confidence peaks of ChIP-seq signal were identified using MACS with parameters–keepdup = auto -p 1e-9 and corresponding input control. Bound genes are RefSeq genes that contact a MACS-defined peak between −10,000 bp from the TSS and +5,000 bp from the TES.
For the assay for transposase accessible chromatin (ATAC-seq), CD34+ cells were expanded and differentiated using the protocol mentioned above. Before collection, cells were treated with 25 ng/mL hrBMP4 for 2 hr. 5 × 104 cells per differentiation stage were harvested by spinning at 500 × g for 5 min, 4°C. Cells were washed once with 50 μL of cold 1× PBS and spun down at 500 × g for 5 min, 4°C. After discarding supernatant, cells were lysed using 50 μL cold lysis buffer (10 mM Tris-HCl [pH 7.4], 10 mM NaCl, 3 mM MgCl2, 0.1% IGEPAL CA-360) and spun down immediately at 500 × g for 10 min, 4°C. The cells were then precipitated and kept on ice and subsequently resuspended in 25 μL 2X TD Buffer (Illumina Nextera kit), 2.5 μL transposase enzyme (Illumina Nextera kit, 15028252), and 22.5 μL nuclease-free water in a total of 50 μL reaction for 1 hr at 37°C. DNA was then purified using QIAGEN MinElute PCR purification kit (28004) in a final volume of 10 μL. Libraries were constructed according to Illumina protocol using the DNA treated with transposase, NEB PCR master mix, Sybr green, and universal and library-specific Nextera index primers. The first round of PCR was performed under the following conditions: 72°C, 5 min; 98°C, 30 s; (98°C, 10 s; 63°C, 30 s; 72°C, 1 min) × 5 cycles; hold at 4°C. Reactions were kept on ice and, using a 5 μL reaction aliquot, the appropriate number of additional cycles required for further amplification was determined in a side qPCR reaction: 98°C, 30 s; (98°C, 10 s; 63°C, 30 s; 72°C, 1 min) × 20 cycles; hold at 4°C. Upon determining the additional number of PCR cycles required further for each sample, library amplification was conducted using the following conditions: 98°C, 30 s; (98°C,10 s; 63°C, 30 s; 72°C, 1 min) × appropriate number of cycles; hold at 4°C. Libraries prepared went through quality-control analysis using an Agilent Bioanalyzer. Samples with appropriate nucleosomal laddering profiles were selected for next generation sequencing using Illumina Hiseq 2500 platform.
All human ChIP-seq datasets were aligned to build version NCBI37/HG19 of the human genome using Bowtie2 (v.2.2.1)34 with the following parameters:–end-to-end, -N0, -L20. We used the MACS2 v.2.1.035 peak finding algorithm to identify regions of ATAC-seq peaks, with the following parameter: –nomodel–shift −100–extsize 200. A q-value threshold of enrichment of 0.05 was used for all datasets.
Evaluation in Mouse Crosses
To further affirm the trait loci we identified, and in an attempt to further fine-map the intervals identified in our discovery analyses through cross-species comparisons, we evaluated the new loci in syntenic regions in 12 inter-strain mouse QTL crosses.38
In brief, mice from 12 different strains were inter-crossed38 and the same erythrocyte traits we have studied by GWAS were measured in peripheral blood. The Jackson Laboratory Animal Care and Use Committee approved all protocols. The number of markers genotyped per cross varied by the platform used, and the total number per cross is provided in Table S9. QTL analysis was performed for each erythrocyte trait using R/qtl v1.07-12 (Web Resources).39 Genetic map positions of all markers used were updated to the new mouse genetic map using online mouse map converter tool (Web Resources).40 All phenotypic data were ranked-Z transformed to approximate the normal distribution prior to analysis. The QTL analysis was performed as a genome-wide scan with sex as an additive covariate. Permutation testing (1,000 permutations) was used to determine significance, and LOD scores greater than the 95th percentile (p < 0.05) were considered significant. QTL confidence intervals were determined by the posterior probability.41, 42 For each candidate region in the mouse, the coordinates were obtained from the Mouse Genome Database, which is part of Mouse Genome Informatics (MGI), using the “Genes and Markers” query (Web Resources). Protein coding genes, non-coding RNA genes, and unclassified genes were queried.
Results
In this study we analyzed the association of genetic variation in 71,638 individuals and 6 clinically relevant erythrocyte traits which are commonly measured, accounting for the diverse ethnic background of the participants.
We identified 44 previously reported loci7, 8, 9, 10, 11, 12, 43, 44, 45, 46, 47 (Table S3) and 9 other significant trait-locus associations at 7 loci (p < 5 × 10−8 or log10BF > 6.1, Table 1). SHROOM3 was simultaneously identified in an exome chip analysis by our group in overlapping samples.48 Ethnic-specific results are presented in Table S4. Regional association plots are shown for each region in Figure S1, showing ethnic-specific results, the trans-ethnic meta-analysis, and plots of pairwise LD across the regions for EUR, EAS, and AFR ancestry.
Table 1.
Findings from the METAL and MANTRA Trans-ethnic Analyses
| Trait | SNP | Chr | Gene | c/nc | N |
METAL |
MANTRA |
RE2 |
||
|---|---|---|---|---|---|---|---|---|---|---|
| Effect (SE) | p | Log10BF | posthg | p | ||||||
| Hb | rs2299433 | 7 | MET | T/C | 63,091 | 0.041 (0.008) | 6.16 × 10−8 | 6.195 | 0.027 | 1.20 × 10−7 |
| Hct | rs6430549 | 2 | TMEM163 / ACMSD | A/G | 71,647 | 0.103 (0.018) | 4.96 × 10−9 | 7.408 | 0.120 | 8.46 × 10−9 |
| Hct | rs2299433 | 7 | MET | T/C | 63,532 | 0.102 (0.019) | 5.66 × 10−8 | 6.199 | 0.099 | 9.87 × 10−8 |
| MCH | rs2060597 | 3 | PLCL2 | T/C | 38,836 | 0.006 (0.001) | 4.18 × 10−10 | 8.178 | 0.009 | 9.75 × 10−10 |
| MCH | rs2979489 | 8 | RBPMS | A/G | 37,531 | −0.002 (0.001) | 8.89 × 10−5 | 9.723 | 1.000 | 1.19 × 10−12 |
| MCV | rs10929547 | 2 | ID2 | A/C | 50,870 | −0.002 (0.0003) | 2.50 × 10−9 | 7.977 | 0.007 | 2.14 × 10−9 |
| MCV | rs9821630 | 3 | PLCL2 | A/G | 48,697 | −0.002 (0.0004) | 6.86 × 10−9 | 7.864 | 0.004 | 2.44 × 10−9 |
| MCV | rs2979489 | 8 | RBPMS | A/G | 48,697 | −0.002 (0.0004) | 7.24 × 10−9 | 7.961 | 0.003 | 1.65 × 10−9 |
| MCV | rs6121246 | 20 | FOXS1 | T/C | 49,896 | 0.003 (0.001) | 4.05 × 10−7 | 6.296 | 0.003 | 8.31 × 10−8 |
Abbreviations are as follows: chr, chromosome number; c/nc, coding/non-coding allele; n, number of participants; SE, standard error; p, p value; log10BF, logarithm of Bayes Factor; posthg, posterior probability of heterogeneity.
Five of the discovered trait loci showed a significant association in the fixed-effects trans-ethnic METAL analyses, in the Bayesian MANTRA analyses, and in the RE2 analyses; these were TMEM163/ACMSD for Hct, PLCL2:rs2060597 for MCH, and ID2, PLCL2:rs9821630, and RBPMS for MCV. Two loci (MET and FOXS1) showed a borderline significant effect in METAL and RE2 and a strong significant effect in MANTRA for HB and MCV, respectively. The association of rs2979489 (RBPMS) further showed a strong association with MCH in the multi-ethnic Bayesian meta-analysis and in the RE2 model but was not detected in the multi-ethnic fixed-effects meta-analysis, nor in any of the ethnic-specific meta-analyses for this trait. Interestingly, MCH and MCV are correlated traits, yet strong heterogeneity of effect was observed for this SNP’s association with MCH only, as indicated by both METAL (I2 statistic 94%, p value Cochran’s Q statistic of heterogeneity 6.48 × 10−8) and MANTRA (posterior probability of heterogeneity = 1) (Table 1). Inspection of the discovery datasets showed that one of the African American cohorts supplied data for MCV but not for MCH, which resulted in a stronger positive association of rs2979489 with MCH than with MCV in the AFR meta-analyses. This phenomenon was accompanied by greater evidence of heterogeneity for MCH in the trans-ethnic meta-analyses because the EUR and EAS associations were in the opposite direction to that observed in the AFR meta-analysis. The MANTRA and RE2 analyses were able to account for this heterogeneity and thus yield a stronger result as compared to METAL for this trait locus.
Replication Analyses
In the meta-analyses of the replication cohorts, the trait-SNP combinations HT-TMEM163/ACMSD and MCH-RBPMS achieved a Bonferroni-corrected significance threshold with both fixed effects and RE2 methods (p < 0.05/9). ID2 was Bonferroni-significant in the fixed-effects model and nominally significant in the RE2 model. Furthermore, we found nominal significance for MCV-RBPMS (fixed-effects analyses) and FOXS1 (fixed-effects and RE2) (Table S5).
When we compared the discovery and replication combined meta-analyses with the discovery analyses alone, we observed stronger associations for Hct-TMEM163/ACMSD, MCH-PLCL2, MCV-ID2, and MCV-RBPMS in all three models (fixed-effects, MANTRA, and RE2). For MCH-RBPMS, we found a stronger association in the fixed-effects analysis (Table S6).
Statistical Fine-Mapping
We found that 31 trait-specific trans-ethnic 99% CSs showed a decrease in length of at least 50% as compared to their EUR-only CS counterparts (26 unique loci across the 6 erythrocyte traits) (Table S7).
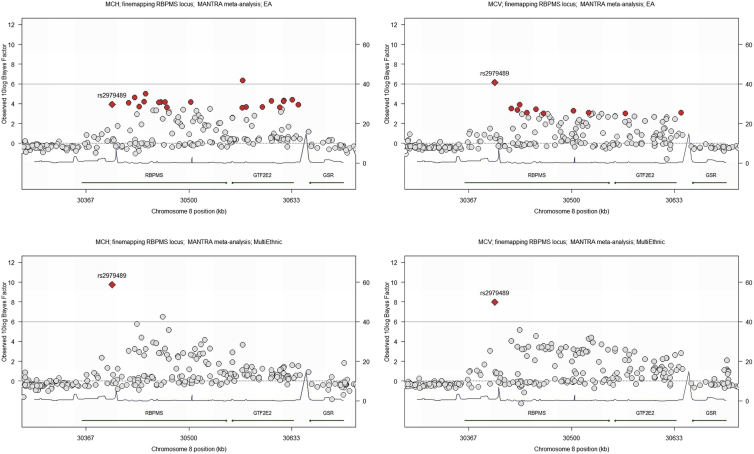
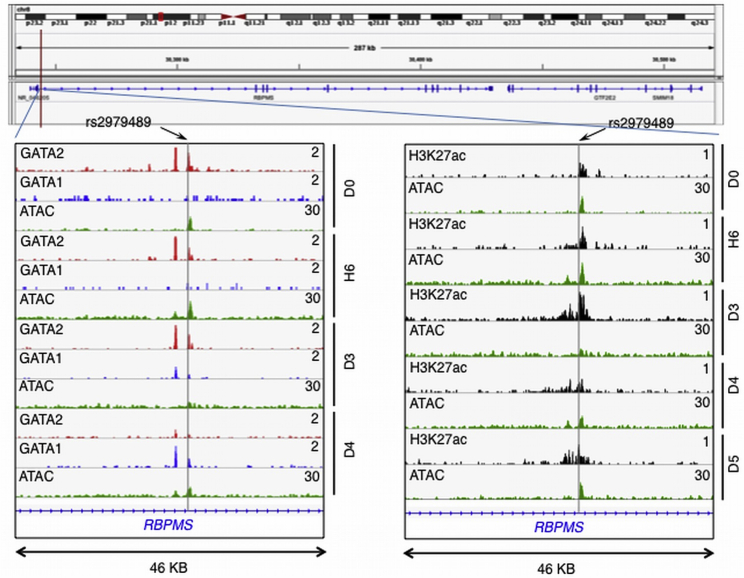
Among the loci identified in this study, the chromosome 8 RBPMS locus showed fine-mapping according to this criterion (Table 2, Figure 1). For MCH, the EUR credible set spanned 204,200 bp, encompassing RBPMS and GTF2E2. The multi-ethnic credible set comprised just one SNP, rs2979489, within the first intron of RBPMS (Figure 1). Remarkably, this associated SNP rs2979489 is located adjacent to a GATA-motif where a gradual switch of binding from GATA2 to GATA1 takes place during commitment of human CD34 progenitors toward erythroid lineage (Figure 2, bottom left). Moreover, an assay for chromatin accessibility sites (ATAC-seq) and H3K27a ChIP-seq clearly identify that the genomic region proximal to this SNP is actively regulated during human erythroid differentiation (Figure 2, bottom right).
Table 2.
Fine Mapping of a Chromosome 8 Locus Identified in European Ancestry Meta-analysis by MANTRA Trans-ethnic Analysis
| Trait | Chr | Gene |
EUR |
Multi-ethnic |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| topSNP | log10BF | n_SNPs | width (bp) | topSNP | log10BF | n_SNPs | width (bp) | |||
| MCH | 8 | RBPMS | rs2979502 | 6.32982 | 21 | 241480 | rs2979489 | 9.72267 | 1 | 1 |
| MCV | 8 | RBPMS | rs2979489 | 6.13733 | 11 | 241480 | rs2979489 | 7.96132 | 1 | 1 |
Abbreviations are as follows: chr, chromosome number; log10BF, logarithm of Bayes Factor; n_SNPs, number of SNPs in the region.
Figure 1.
Fine Mapping of the Chromosome 8 RBPMS/GTF2E2 Locus
99% credible sets (red dots) around the top hit rs2979489 (red diamond). European Ancestry MANTRA analyses (top) for MCH (left) and MCV (right) are shown, compared to 99% credible sets of the trans-ethnic MANTRA analyses (bottom, MCH on the left and MCV on the right).
Figure 2.
rs2979489 Is Localized to a Potential Regulatory Site that Involves Transition Binding of GATA2 to GATA1 during Erythrocyte Differentiation
Top shows gene-track view of rs2979489 location in the RBPMS/GTF2E2 gene region. Bottom left: gene track of RBPMS gene showing overlap of GATA2, GATA1, and ATAC-seq peaks (red, blue, and green, respectively) during human erythroid differentiation. Bottom right: overlap of ATAC-seq (green) and H3K27ac ChIP-seq (black) during differentiation at the region proximal to the SNP rs2979489. The gray horizontal line indicates the position of SNP rs 2979489. D0, day 0; H6, hour 6; D3, day 3; D4, day 4; and D5, day 5 of erythroid differentiation time-course post-induction of differentiation.
Among the known loci, fine mapping narrowed signals as shown in Table S7.
Interestingly, trans-ethnic fine-mapping of the XRN1 locus (MCH) led us to the rs6791816 polymorphism. Van der Harst et al. identified the same SNP in their exploration of nucleosome-depleted regions (NDRs, representing active regulatory elements for erythropoeisis) in a follow-up analysis of their GWAS results.10 By means of subsequent formaldehyde-assisted isolation of regulatory elements followed by next-generation sequencing (FAIRE-seq), they pinpointed rs6791816 as an NDR SNP in LD with their initial index SNP for MCH and MCV.
Furthermore, fine-mapping of both the MPND locus (MCH) and SH3GL1 locus (MCV) pointed to the rs8887 SNP within the 3′ UTR of PLIN4. The rs8887 SNP minor allele has been shown experimentally to create a novel seed site for miR-522, resulting in decreased PLIN4 expression.49 miR-522 is expressed in circulating blood,50 and these data suggest that an allele-specific miR-522 regulation of PLIN4 by rs8887 could serve as a functional mechanism underlying the identified association.
We additionally showed fine mapping in several other intervals (Table S7) with fine-mapped genes about which less is known about their potential biologic role in erythropoeisis or red blood cell function. These regions are of interest for further hypothesis generation based upon the GWAS findings.
ENCODE Analyses
We further evaluated the SNPs from the chromosome 8 RBPMS region against the ENCODE Project Consortium’s database of numerous functional elements in the K562 erythroleukemic line.29 The lone SNP that was fine mapped at the locus, rs2979489, was found in a strong enhancer element as defined by Segway, supporting a functional role for this SNP and RBPMS. The other SNPs in the RBPMS region, excluded by the statistical fine-mapping exercise, were not annotated as regulatory in the ENCODE data (Table S8).
Experiments in Zebrafish
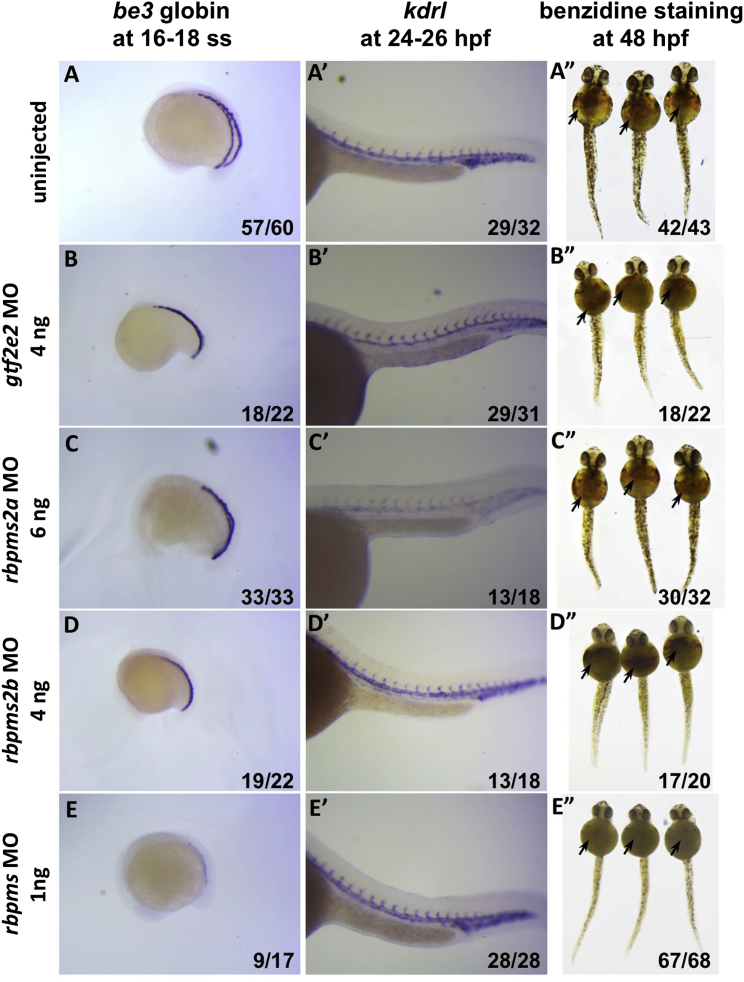
We identified a erythropoietic effect for the zebrafish rbpms. Both embryonic globin expression at 16 ss and o-dianisidine/benzidine staining at 48 hpf significantly decreased in morphants, indicating a decrease in both globin transcription and Hb levels (Figure 3). This loss-of-function finding is consistent with a decreased mean erythrocyte Hb content observed in our human association results. In zebrafish, the rbpms orthology mapping included rbpms2a, rbpms2b, and rbpms, and loss-of-function phenotypes of all orthologs were tested experimentally. The results suggested a clear erythropoietic effect with limited functional compensation of the genes in the rbpms family in zebrafish during embryonic erythropoiesis. On the other hand, morpholino knockdown experiments with the zebrafish ortholog of GTF2E2 did not show an apparent erythropoietic effect.
Figure 3.
Loss-of-Function Analysis of the RBPMS, RBPMS2, and GTF2E2 Orthologs in Zebrafish
After injection of 0–3 ng ATG and splicing morpholinos (MOs) against the RBPMS zebrafish ortholog (row E), both the o-dianisidine/benzidine staining (arrows) in embryos at 48 hpf (right) and the embryonic βe3 globin expression in embryos at 16–18 ss (left) are obviously decreased, indicating a dose-dependent disruption in erythropoiesis in the experimentally treated embryos as compared to uninjected and gtf2e2-, rbpms2a-, and rbpms2b-MO-injected controls (rows A–D). Representative results are shown for the embryos injected with MOs against the RBPMS ortholog in (E) as well as for the embryos injected with MOs against rbpms2a (C) and rbpms2b (D) at higher doses. Injections of MO against the zebrafish GTF2E2 ortholog (B) also at a higher dose show no obvious effect on βe3 globin expression at 16–18 ss and o-dianisidine/benzidine staining at 48 hpf. Expression pattern of vascular marker gene kdrl (A–E, middle) is relatively normal in all MO-injected embryos at 24–26 hpf, suggesting grossly normal development of cells in other organs. The numbers on the lower right corner of each image indicate the number of embryos with phenotypes similar to the ones shown on each of the images over the total number of embryos examined in each of the experimental groups.
Review of the human association results showed no evidence of pleiotropy across the RBPMS family of genes and denote that the human association is specific to RBPMS (Supplemental Data). This review was conducted because the orthology in the fish led to inclusion of rbpms2 in the zebrafish analyses as well. These findings indicate that the statistical fine-mapping was useful to home in on RPBMS as a causal gene influencing erythropoiesis.
Evaluation in Mouse Crosses
In the eight regions from our discovery analysis, six had evidence of cross-species validation by evidence of syntenic gene within the linkage peak in the mouse QTL results (Table 3). However, the human GWAS intervals were not narrowed by the mouse QTL results for any of these loci (Table S9).
Table 3.
Mouse QTL Validation of the Findings from MANTRA Trans-ethnic Analyses
| Trait | Chr | Gene |
Human (hg18/Build 36) |
Mouse (37 mm9) |
Significant and Suggestive Mouse QTLa |
LOD |
|---|---|---|---|---|---|---|
| (Chromosome:Position) | (Chromosome:Position) | Peak (95% CI) (Mb) | ||||
| Hct | 2 | TMEM163/ACMSD | chr2: 135,196,450–135,438,613 | chr1: 129,581,372–129,711,586b | 141.0 (54.8–158.9)∗ | 3.72∗ |
| Hct | 4 | SHROOM3 | chr4: 77,586,311–77,629,342 | chr5: 93,112,461–93,394,344 | 46.0 (19.6–106.5) | 2.34 |
| Hct | 7 | MET | chr7: 116,118,114–116,131,947 | chr6: 17,432,318–17,447,418b | 37.6 (6.6–127.9) | 2.75 |
| MCH | 8 | RBPMS | chr8: 30,400,375–30,400,375 | chr8: 34,893,115–35,040,335 | 78.9 (28.0–96.1)∗ | 3.98∗ |
| MCV | 3 | PLCL2 | chr3: 16,860,239–16,945,942 | chr17: 50,604,848–50,698,773b | 46.0 (28.6–55.3)∗ | 5.46∗ |
| MCV | 20 | FOXS1 | chr20: 29,684,484–29,897,013 | chr2: 152,576,419–152,758,874b | 170.1 (147.6–179.3)∗ | 4.69∗ |
Gene found in a significant (indicated with asterisk) or suggestive 95% CI mouse QTL, not corresponding to the human interval.
Within the corresponding human interval (±250 kb).
Discussion
We conducted GWASs and meta-analyses of six erythrocyte traits (Hb, Hct, MCH, MCHC, MCV, and RBC) in 71,638 individuals from European, Asian, and African American ancestry. While prior genome-wide association studies have identified loci associated with erythrocyte traits through the analysis of ancestrally homogeneous cohorts and consortia, largely biased toward European ancestry studies, trans-ethnic analysis has not previously been performed while accounting for differences in genetic architecture in ethnically diverse groups.
We identified seven loci for erythrocyte traits (nine locus-trait combinations) and replicated 44 previously identified loci. We fine-mapped several known and new loci. One fine-mapped locus led us to a region on chromosome 8 associated with MCH and MCV.
In the chromosome 8 RBPMS/GTF2E2 locus, the index variant rs2979489, which was associated with MCV and MCH and highlighted in the trans-ethnic fine-mapping analyses, is located within the first intron of RBPMS (RNA binding protein with multiple splicing), notably at an open chromatin site at which a switch of GATA1/2 binding occurs during erythroid differentiation. The RBPMS protein product regulates a variety of RNA processes, including pre-mRNA splicing, RNA transport, localization, translation, and stability.51, 52 RBPMS is expressed at relatively low levels in mammalian erythroblasts and the protein product has not been detected in mature human erythrocytes.53, 54
The rs2979489 polymorphism showed remarkably high heterogeneity in effect on the MCH trait across the different ethnicities, with different directions of effect for the AFR meta-analysis results compared to the EUR and ASN findings. If the variant is causal, this pattern of association could reflect gene-environment interaction. In this case, different exposures in AFR compared to EUR/ASN populations may lead to a marginal effect of the SNP in opposing directions by different selection pressures. If, however, rs2979489 is not causal, but rather a marker in LD with the causal variant, then the opposing direction of effects could reflect very different LD structures in the different populations, also indicating selection, or theoretically it could even reflect different causal variants in AFR and EUR/EAS—and rs2979489 being just in strong LD with both causal variants.
The SNP rs2979489 is located adjacent to a GATA-motif where a gradual switch of binding from GATA2 to GATA1 takes place during commitment of human CD34 progenitors toward erythroid lineage. These observations suggest that rs2979489 localizes at a potential regulatory site where a modulation of erythroid cell differentiation occurs and the presence of rs2979489 may lead to observed red cell trait alterations in human populations, possibly through regulation of RBPMS expression timing, level, and/or splicing variation. Although RBPMS previously had no known role in hematopoiesis or more specifically in erythropoiesis, RBPMS has been previously shown to be upregulated in transcriptional profiles of murine and human hematopoietic stem cells.55, 56, 57 Its role may be at much earlier stages during the differentiation of erythrocytes from erythroblasts and/or hematopoietic stem cells. RBPMS is known to physically interact with Smad2, Smad3, and Smad4 and stimulate smad-mediated transactivation through enhanced Smad2 and Smad3 phosphorylation and associated promotion of nuclear accumulation of Smad proteins.58 These Smad proteins are known to regulate the TGF-β-mediated regulation of hematopoietic cell fate and erythroid differentiation.59 RBPMS has four annotated transcript isoforms, and further delineation of the tissue specificity, timing of expression, and function of these transcripts in the context of the genetic variant we identified warrants further study.
Among the additional six loci, we identified two loci in which the index SNP was located within annotated genes, rs6430549 in ACMSD (aminocarboxymuconate semi aldehyde decarboxylase, intronic) and rs2299433 in MET (mesenchymal epithelial transition factor, intronic). No previous hematologic role has been described for either region. Variants in the chromosome 2q21.3 ACMSD region have previously been associated with blood metabolite levels, obesity, and Parkinson disease.60, 61, 62 A genetic variant in the first intron of MET was significantly associated with both Hb and Hct; however, association was not observed in replication samples, possibly due to lower power in the replication experiment. Three additional loci were intergenic but close to a coding gene (rs10929547 near ID2 [inhibitor of DNA binding 2, dominant-negative helix-loop-helix protein], rs6121246 near FOXS1 [forkhead box S1], and rs2060597 approximately 40 kbp upstream of PLCL2 [phospholipase C-like 2]). The roles of variants in these regions in determining erythrocyte traits are unknown.53, 63
In the statistical fine-mapping analyses, the trans-ethnic meta-analysis approach resulted in smaller 99% credible intervals in all of the loci identified in this study. Since these loci were identified in analyses that accounted for heterogeneity in allelic effects between ethnic groups, in which the heterogeneity may be due to variation in LD patterns, we examined the LD patterns in these loci. Not surprisingly, we noted that the consistent decrease in the size of 99% credible interval across all loci is likely due to the inclusion of cohorts of African ancestry, an ethnic group with generally smaller LD blocks throughout the genome. The loss-of-function screens in zebrafish for the chromosome 8 signal suggested that these analyses successfully identified a single gene (RBPMS) with erythropoietic effect within one of the fine-mapped intervals. We also fine-mapped previously known regions such as the chromosome 6p21.1 region associated with RBC count and highlighted CCND3, which has been experimentally shown to regulate RBC count experimentally in a knock-out mouse model.64 These examples suggest that attempts to refine association signals using these types of approaches in existing samples may yield functional candidates for further mechanistic hypothesis testing, which is a major goal of GWASs.
Trans-ethnic genome-wide meta-analyses of common variants have aided in the characterization of genetic loci for various complex traits.13, 65, 66, 67 Our data demonstrate the benefits of trans-ethnic genome-wide meta-analysis in identifying and fine-mapping genetic loci of erythrocyte traits. By exploiting the differences in genetic architecture of the associations within these loci in various ethnic groups, we may identify causal genes influencing clinically relevant hematologic traits. Use of a similar approach for other complex traits is likely to provide deeper insights into the biological mechanisms underlying human traits.
Acknowledgments
B.M.P. serves on the DSMB of a clinical trial funded by the manufacturer (Zoll LifeCor) and on the Steering Committee of the Yale Open Data Access project funded by Johnson & Johnson.
Published: December 22, 2016
Footnotes
Supplemental data include Supplemental Acknowledgments, individual study methods and cohort descriptions, pleiotropy analysis, 10 tables, and a figure with 123 panels.
Accession Numbers
Summary data have been deposited in the database of Genotypes and Phenotypes (dbGaP) under CHARGE (Cohorts for Heart and Aging Research in Genomic Epidemiology) Consortium Summary Results from Genomic Studies. The dbGaP study accession number is phs000930.
Web Resources
Center for Genome Dynamics, http://cgd.jax.org
Matrix Spectral Decomposition, http://neurogenetics.qimrberghofer.edu.au/matSpD/
METASOFT 3.0c, http://www.buhmhan.com/software
MGI Genes and Markers Query, http://www.informatics.jax.org/marker
R/qtl v1.07-12, http://www.rqtl.org
Supplemental Data
References
- 1.Koury M.J. Abnormal erythropoiesis and the pathophysiology of chronic anemia. Blood Rev. 2014;28:49–66. doi: 10.1016/j.blre.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 2.Whitfield J.B., Martin N.G. Genetic and environmental influences on the size and number of cells in the blood. Genet. Epidemiol. 1985;2:133–144. doi: 10.1002/gepi.1370020204. [DOI] [PubMed] [Google Scholar]
- 3.Evans D.M., Frazer I.H., Martin N.G. Genetic and environmental causes of variation in basal levels of blood cells. Twin Res. 1999;2:250–257. doi: 10.1375/136905299320565735. [DOI] [PubMed] [Google Scholar]
- 4.Lin J.-P., O’Donnell C.J., Jin L., Fox C., Yang Q., Cupples L.A. Evidence for linkage of red blood cell size and count: genome-wide scans in the Framingham Heart Study. Am. J. Hematol. 2007;82:605–610. doi: 10.1002/ajh.20868. [DOI] [PubMed] [Google Scholar]
- 5.Guindo A., Fairhurst R.M., Doumbo O.K., Wellems T.E., Diallo D.A. X-linked G6PD deficiency protects hemizygous males but not heterozygous females against severe malaria. PLoS Med. 2007;4:e66. doi: 10.1371/journal.pmed.0040066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tishkoff S.A., Varkonyi R., Cahinhinan N., Abbes S., Argyropoulos G., Destro-Bisol G., Drousiotou A., Dangerfield B., Lefranc G., Loiselet J. Haplotype diversity and linkage disequilibrium at human G6PD: recent origin of alleles that confer malarial resistance. Science. 2001;293:455–462. doi: 10.1126/science.1061573. [DOI] [PubMed] [Google Scholar]
- 7.Lo K.S., Wilson J.G., Lange L.A., Folsom A.R., Galarneau G., Ganesh S.K., Grant S.F.A., Keating B.J., McCarroll S.A., Mohler E.R., 3rd Genetic association analysis highlights new loci that modulate hematological trait variation in Caucasians and African Americans. Hum. Genet. 2011;129:307–317. doi: 10.1007/s00439-010-0925-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ganesh S.K., Zakai N.A., van Rooij F.J.A., Soranzo N., Smith A.V., Nalls M.A., Chen M.-H., Kottgen A., Glazer N.L., Dehghan A. Multiple loci influence erythrocyte phenotypes in the CHARGE Consortium. Nat. Genet. 2009;41:1191–1198. doi: 10.1038/ng.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soranzo N., Spector T.D., Mangino M., Kühnel B., Rendon A., Teumer A., Willenborg C., Wright B., Chen L., Li M. A genome-wide meta-analysis identifies 22 loci associated with eight hematological parameters in the HaemGen consortium. Nat. Genet. 2009;41:1182–1190. doi: 10.1038/ng.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Harst P., Zhang W., Mateo Leach I., Rendon A., Verweij N., Sehmi J., Paul D.S., Elling U., Allayee H., Li X. Seventy-five genetic loci influencing the human red blood cell. Nature. 2012;492:369–375. doi: 10.1038/nature11677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamatani Y., Matsuda K., Okada Y., Kubo M., Hosono N., Daigo Y., Nakamura Y., Kamatani N. Genome-wide association study of hematological and biochemical traits in a Japanese population. Nat. Genet. 2010;42:210–215. doi: 10.1038/ng.531. [DOI] [PubMed] [Google Scholar]
- 12.Chen Z., Tang H., Qayyum R., Schick U.M., Nalls M.A., Handsaker R., Li J., Lu Y., Yanek L.R., Keating B., BioBank Japan Project. CHARGE Consortium Genome-wide association analysis of red blood cell traits in African Americans: the COGENT Network. Hum. Mol. Genet. 2013;22:2529–2538. doi: 10.1093/hmg/ddt087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franceschini N., van Rooij F.J.A., Prins B.P., Feitosa M.F., Karakas M., Eckfeldt J.H., Folsom A.R., Kopp J., Vaez A., Andrews J.S., LifeLines Cohort Study Discovery and fine mapping of serum protein loci through transethnic meta-analysis. Am. J. Hum. Genet. 2012;91:744–753. doi: 10.1016/j.ajhg.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nalls M.A., Couper D.J., Tanaka T., van Rooij F.J.A., Chen M.-H., Smith A.V., Toniolo D., Zakai N.A., Yang Q., Greinacher A. Multiple loci are associated with white blood cell phenotypes. PLoS Genet. 2011;7:e1002113. doi: 10.1371/journal.pgen.1002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen P., Takeuchi F., Lee J.-Y., Li H., Wu J.-Y., Liang J., Long J., Tabara Y., Goodarzi M.O., Pereira M.A., CHARGE Hematology Working Group Multiple nonglycemic genomic loci are newly associated with blood level of glycated hemoglobin in East Asians. Diabetes. 2014;63:2551–2562. doi: 10.2337/db13-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wild P.S., Zeller T., Beutel M., Blettner M., Dugi K.A., Lackner K.J., Pfeiffer N., Münzel T., Blankenberg S. Die Gutenberg Gesundheitsstudie. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2012;55:824–829. doi: 10.1007/s00103-012-1502-7. [DOI] [PubMed] [Google Scholar]
- 17.Desch K.C., Ozel A.B., Siemieniak D., Kalish Y., Shavit J.A., Thornburg C.D., Sharathkumar A.A., McHugh C.P., Laurie C.C., Crenshaw A. Linkage analysis identifies a locus for plasma von Willebrand factor undetected by genome-wide association. Proc. Natl. Acad. Sci. USA. 2013;110:588–593. doi: 10.1073/pnas.1219885110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Mutsert R., den Heijer M., Rabelink T.J., Smit J.W.A., Romijn J.A., Jukema J.W., de Roos A., Cobbaert C.M., Kloppenburg M., le Cessie S. The Netherlands Epidemiology of Obesity (NEO) study: study design and data collection. Eur. J. Epidemiol. 2013;28:513–523. doi: 10.1007/s10654-013-9801-3. [DOI] [PubMed] [Google Scholar]
- 19.Ridker P.M., JUPITER Study Group Rosuvastatin in the primary prevention of cardiovascular disease among patients with low levels of low-density lipoprotein cholesterol and elevated high-sensitivity C-reactive protein: rationale and design of the JUPITER trial. Circulation. 2003;108:2292–2297. doi: 10.1161/01.CIR.0000100688.17280.E6. [DOI] [PubMed] [Google Scholar]
- 20.Qayyum R., Snively B.M., Ziv E., Nalls M.A., Liu Y., Tang W., Yanek L.R., Lange L., Evans M.K., Ganesh S. A meta-analysis and genome-wide association study of platelet count and mean platelet volume in african americans. PLoS Genet. 2012;8:e1002491. doi: 10.1371/journal.pgen.1002491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reiner A.P., Lettre G., Nalls M.A., Ganesh S.K., Mathias R., Austin M.A., Dean E., Arepalli S., Britton A., Chen Z. Genome-wide association study of white blood cell count in 16,388 African Americans: the continental origins and genetic epidemiology network (COGENT) PLoS Genet. 2011;7:e1002108. doi: 10.1371/journal.pgen.1002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Devlin B., Roeder K., Wasserman L. Genomic control, a new approach to genetic-based association studies. Theor. Popul. Biol. 2001;60:155–166. doi: 10.1006/tpbi.2001.1542. [DOI] [PubMed] [Google Scholar]
- 24.Morris A.P. Transethnic meta-analysis of genomewide association studies. Genet. Epidemiol. 2011;35:809–822. doi: 10.1002/gepi.20630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han B., Eskin E. Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. Am. J. Hum. Genet. 2011;88:586–598. doi: 10.1016/j.ajhg.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J., Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity (Edinb) 2005;95:221–227. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- 27.Wang X., Chua H.-X., Chen P., Ong R.T.-H., Sim X., Zhang W., Takeuchi F., Liu X., Khor C.-C., Tay W.-T. Comparing methods for performing trans-ethnic meta-analysis of genome-wide association studies. Hum. Mol. Genet. 2013;22:2303–2311. doi: 10.1093/hmg/ddt064. [DOI] [PubMed] [Google Scholar]
- 28.Maller J.B., McVean G., Byrnes J., Vukcevic D., Palin K., Su Z., Howson J.M., Auton A., Myers S., Morris A., Wellcome Trust Case Control Consortium Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat. Genet. 2012;44:1294–1301. doi: 10.1038/ng.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.The ENCODE Project Consortium A user’s guide to the Encyclopedia of DNA Elements (ENCODE) PLoS Biol. 2011 doi: 10.1371/journal.pbio.1001046. Published online April 19, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 31.Huang H.-T., Kathrein K.L., Barton A., Gitlin Z., Huang Y.-H., Ward T.P., Hofmann O., Dibiase A., Song A., Tyekucheva S. A network of epigenetic regulators guides developmental haematopoiesis in vivo. Nat. Cell Biol. 2013;15:1516–1525. doi: 10.1038/ncb2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee T.I., Johnstone S.E., Young R.A. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trompouki E., Bowman T.V., Lawton L.N., Fan Z.P., Wu D.-C., DiBiase A., Martin C.S., Cech J.N., Sessa A.K., Leblanc J.L. Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell. 2011;147:577–589. doi: 10.1016/j.cell.2011.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thorvaldsdóttir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peters L.L., Shavit J.A., Lambert A.J., Tsaih S.-W., Li Q., Su Z., Leduc M.S., Paigen B., Churchill G.A., Ginsburg D., Brugnara C. Sequence variation at multiple loci influences red cell hemoglobin concentration. Blood. 2010;116:e139–e149. doi: 10.1182/blood-2010-05-283879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Broman K.W., Wu H., Sen S., Churchill G.A. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- 40.Cox A., Ackert-Bicknell C.L., Dumont B.L., Ding Y., Bell J.T., Brockmann G.A., Wergedal J.E., Bult C., Paigen B., Flint J. A new standard genetic map for the laboratory mouse. Genetics. 2009;182:1335–1344. doi: 10.1534/genetics.109.105486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Churchill G.A., Doerge R.W. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sen S., Churchill G.A. A statistical framework for quantitative trait mapping. Genetics. 2001;159:371–387. doi: 10.1093/genetics/159.1.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chambers J.C., Zhang W., Li Y., Sehmi J., Wass M.N., Zabaneh D., Hoggart C., Bayele H., McCarthy M.I., Peltonen L. Genome-wide association study identifies variants in TMPRSS6 associated with hemoglobin levels. Nat. Genet. 2009;41:1170–1172. doi: 10.1038/ng.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ding K., de Andrade M., Manolio T.A., Crawford D.C., Rasmussen-Torvik L.J., Ritchie M.D., Denny J.C., Masys D.R., Jouni H., Pachecho J.A. Genetic variants that confer resistance to malaria are associated with red blood cell traits in African-Americans: an electronic medical record-based genome-wide association study. G3 (Bethesda) 2013;3:1061–1068. doi: 10.1534/g3.113.006452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kullo I.J., Ding K., Jouni H., Smith C.Y., Chute C.G. A genome-wide association study of red blood cell traits using the electronic medical record. PLoS ONE. 2010;5:e13011. doi: 10.1371/journal.pone.0013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li J., Glessner J.T., Zhang H., Hou C., Wei Z., Bradfield J.P., Mentch F.D., Guo Y., Kim C., Xia Q. GWAS of blood cell traits identifies novel associated loci and epistatic interactions in Caucasian and African-American children. Hum. Mol. Genet. 2013;22:1457–1464. doi: 10.1093/hmg/dds534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pistis G., Okonkwo S.U., Traglia M., Sala C., Shin S.-Y., Masciullo C., Buetti I., Massacane R., Mangino M., Thein S.-L., CHARGE Consortium Hematology Working Genome wide association analysis of a founder population identified TAF3 as a gene for MCHC in humans. PLoS ONE. 2013;8:e69206. doi: 10.1371/journal.pone.0069206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.CHARGE Consortium Hematology Working Group Meta-analysis of rare and common exome chip variants identifies S1PR4 and other loci influencing blood cell traits. Nat. Genet. 2016;48:867–876. doi: 10.1038/ng.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Richardson K., Louie-Gao Q., Arnett D.K., Parnell L.D., Lai C.-Q., Davalos A., Fox C.S., Demissie S., Cupples L.A., Fernandez-Hernando C., Ordovas J.M. The PLIN4 variant rs8887 modulates obesity related phenotypes in humans through creation of a novel miR-522 seed site. PLoS ONE. 2011;6:e17944. doi: 10.1371/journal.pone.0017944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Williams Z., Ben-Dov I.Z., Elias R., Mihailovic A., Brown M., Rosenwaks Z., Tuschl T. Comprehensive profiling of circulating microRNA via small RNA sequencing of cDNA libraries reveals biomarker potential and limitations. Proc. Natl. Acad. Sci. USA. 2013;110:4255–4260. doi: 10.1073/pnas.1214046110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shimamoto A., Kitao S., Ichikawa K., Suzuki N., Yamabe Y., Imamura O., Tokutake Y., Satoh M., Matsumoto T., Kuromitsu J. A unique human gene that spans over 230 kb in the human chromosome 8p11-12 and codes multiple family proteins sharing RNA-binding motifs. Proc. Natl. Acad. Sci. USA. 1996;93:10913–10917. doi: 10.1073/pnas.93.20.10913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ascano M., Hafner M., Cekan P., Gerstberger S., Tuschl T. Identification of RNA-protein interaction networks using PAR-CLIP. Wiley Interdiscip. Rev. RNA. 2012;3:159–177. doi: 10.1002/wrna.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trakarnsanga K., Wilson M.C., Griffiths R.E., Toye A.M., Carpenter L., Heesom K.J., Parsons S.F., Anstee D.J., Frayne J. Qualitative and quantitative comparison of the proteome of erythroid cells differentiated from human iPSCs and adult erythroid cells by multiplex TMT labelling and nanoLC-MS/MS. PLoS ONE. 2014;9:e100874. doi: 10.1371/journal.pone.0100874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kingsley P.D., Greenfest-Allen E., Frame J.M., Bushnell T.P., Malik J., McGrath K.E., Stoeckert C.J., Palis J. Ontogeny of erythroid gene expression. Blood. 2013;121:e5–e13. doi: 10.1182/blood-2012-04-422394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ramalho-Santos M., Yoon S., Matsuzaki Y., Mulligan R.C., Melton D.A. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- 56.Georgantas R.W., 3rd, Tanadve V., Malehorn M., Heimfeld S., Chen C., Carr L., Martinez-Murillo F., Riggins G., Kowalski J., Civin C.I. Microarray and serial analysis of gene expression analyses identify known and novel transcripts overexpressed in hematopoietic stem cells. Cancer Res. 2004;64:4434–4441. doi: 10.1158/0008-5472.CAN-03-3247. [DOI] [PubMed] [Google Scholar]
- 57.Wagner W., Ansorge A., Wirkner U., Eckstein V., Schwager C., Blake J., Miesala K., Selig J., Saffrich R., Ansorge W., Ho A.D. Molecular evidence for stem cell function of the slow-dividing fraction among human hematopoietic progenitor cells by genome-wide analysis. Blood. 2004;104:675–686. doi: 10.1182/blood-2003-10-3423. [DOI] [PubMed] [Google Scholar]
- 58.Sun Y., Ding L., Zhang H., Han J., Yang X., Yan J., Zhu Y., Li J., Song H., Ye Q. Potentiation of Smad-mediated transcriptional activation by the RNA-binding protein RBPMS. Nucleic Acids Res. 2006;34:6314–6326. doi: 10.1093/nar/gkl914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He W., Dorn D.C., Erdjument-Bromage H., Tempst P., Moore M.A.S., Massagué J. Hematopoiesis controlled by distinct TIF1γ and Smad4 branches of the TGFbeta pathway. Cell. 2006;125:929–941. doi: 10.1016/j.cell.2006.03.045. [DOI] [PubMed] [Google Scholar]
- 60.Shin S.-Y., Fauman E.B., Petersen A.-K., Krumsiek J., Santos R., Huang J., Arnold M., Erte I., Forgetta V., Yang T.-P., Multiple Tissue Human Expression Resource (MuTHER) Consortium An atlas of genetic influences on human blood metabolites. Nat. Genet. 2014;46:543–550. doi: 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Comuzzie A.G., Cole S.A., Laston S.L., Voruganti V.S., Haack K., Gibbs R.A., Butte N.F. Novel genetic loci identified for the pathophysiology of childhood obesity in the Hispanic population. PLoS ONE. 2012;7:e51954. doi: 10.1371/journal.pone.0051954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nalls M.A., Plagnol V., Hernandez D.G., Sharma M., Sheerin U.M., Saad M., Simón-Sánchez J., Schulte C., Lesage S., Sveinbjörnsdóttir S., International Parkinson Disease Genomics Consortium Imputation of sequence variants for identification of genetic risks for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet. 2011;377:641–649. doi: 10.1016/S0140-6736(10)62345-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Otsuki M., Fukami K., Kohno T., Yokota J., Takenawa T. Identification and characterization of a new phospholipase C-like protein, PLC-L(2) Biochem. Biophys. Res. Commun. 1999;266:97–103. doi: 10.1006/bbrc.1999.1784. [DOI] [PubMed] [Google Scholar]
- 64.Sankaran V.G., Ludwig L.S., Sicinska E., Xu J., Bauer D.E., Eng J.C., Patterson H.C., Metcalf R.A., Natkunam Y., Orkin S.H. Cyclin D3 coordinates the cell cycle during differentiation to regulate erythrocyte size and number. Genes Dev. 2012;26:2075–2087. doi: 10.1101/gad.197020.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Keller M.F., Reiner A.P., Okada Y., van Rooij F.J.A., Johnson A.D., Chen M.-H., Smith A.V., Morris A.P., Tanaka T., Ferrucci L., CHARGE Hematology. COGENT. BioBank Japan Project (RIKEN) Working Groups Trans-ethnic meta-analysis of white blood cell phenotypes. Hum. Mol. Genet. 2014;23:6944–6960. doi: 10.1093/hmg/ddu401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dastani Z., Hivert M.-F., Timpson N., Perry J.R.B., Yuan X., Scott R.A., Henneman P., Heid I.M., Kizer J.R., Lyytikäinen L.-P., DIAGRAM+ Consortium. MAGIC Consortium. GLGC Investigators. MuTHER Consortium. DIAGRAM Consortium. GIANT Consortium. Global B Pgen Consortium. Procardis Consortium. MAGIC investigators. GLGC Consortium Novel loci for adiponectin levels and their influence on type 2 diabetes and metabolic traits: a multi-ethnic meta-analysis of 45,891 individuals. PLoS Genet. 2012;8:e1002607. doi: 10.1371/journal.pgen.1002607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu C.-T., Buchkovich M.L., Winkler T.W., Heid I.M., Borecki I.B., Fox C.S., Mohlke K.L., North K.E., Adrienne Cupples L., African Ancestry Anthropometry Genetics Consortium. GIANT Consortium Multi-ethnic fine-mapping of 14 central adiposity loci. Hum. Mol. Genet. 2014;23:4738–4744. doi: 10.1093/hmg/ddu183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.