Abstract
Early B cell factor 3 (EBF3) is an atypical transcription factor that is thought to influence the laminar formation of the cerebral cortex. Here, we report that de novo mutations in EBF3 cause a complex neurodevelopmental syndrome. The mutations were identified in two large-scale sequencing projects: the UK Deciphering Developmental Disorders (DDD) study and the Canadian Clinical Assessment of the Utility of Sequencing and Evaluation as a Service (CAUSES) study. The core phenotype includes moderate to severe intellectual disability, and many individuals exhibit cerebellar ataxia, subtle facial dysmorphism, strabismus, and vesicoureteric reflux, suggesting that EBF3 has a widespread developmental role. Pathogenic de novo variants identified in EBF3 include multiple loss-of-function and missense mutations. Structural modeling suggested that the missense mutations affect DNA binding. Functional analysis of mutant proteins with missense substitutions revealed reduced transcriptional activities and abilities to form heterodimers with wild-type EBF3. We conclude that EBF3, a transcription factor previously unknown to be associated with human disease, is important for brain and other organ development and warrants further investigation.
Main Text
Neurodevelopmental disorders are increasingly found to have a genetic component by next-generation sequencing (NGS) technologies. Despite these successes, many individuals still remain undiagnosed as a result of a lack of functional data to support gene-variant pathogenicity in combination with the rarity of individual gene mutations.
Deciphering Developmental Disorders (DDD) is a large UK-wide collaborative recruitment network in which genome-wide microarrays and exome sequencing have been performed on families affected by severe undiagnosed developmental disorders.1, 2, 3 Stringent genome-wide levels of significance were developed to enable robust gene discovery,1, 4 but reducing the rate of false positives could cause previously unknown disease-associated genes to be missed as a result of lower significance levels. Methods for quickly prioritizing variants in these genes are essential as NGS becomes integrated into clinical practice and the number of variants of uncertain significance increases dramatically. Here, we report that mutations in early B cell factor 3 (EBF3 [MIM: 607407]) cause a neurodevelopmental disorder. The mutations were initially missed as a result of low significance levels but were found on reanalysis of the DDD dataset by a simplified variant-prioritization strategy combined with functional investigations. Subsequently, two additional individuals were identified in the parallel Canadian Clinical Assessment of the Utility of Sequencing and Evaluation as a Service (CAUSES) study. Mutations in EBF3 are of particular interest because this gene encodes a transcription factor that is thought to be involved in lamination of the cerebral cortex.
Individuals 1–6 (IDs 272588, 280219, 265391, 262955, 263361, and 279995, respectively) in the DDD study were identified as part of an ongoing search for ataxia-associated genes. In the first 4,293 DDD families, 343 subjects were identified to have cerebellar ataxia (HP: 0001251). Families were recruited from around the UK, and written consent was obtained from all participating families. The DDD study received UK research ethics committee (REC) approval (10/H0305/83 granted by the Cambridge South REC and GEN/284/12 granted by the Republic of Ireland REC). The exome sequencing and initial bioinformatics pipeline for DDD individuals has been reported previously.1, 3 In brief, fragmented genomic DNA was used for targeted pull-down with a custom Agilent SureSelect 55 MB Exome Plus and 75-bp paired-end reads sequenced on an Illumina HiSeq. Average sequencing depth (ASD, ratio of sequenced bases to targeted bases) was 90× across the whole targeted sequence or 93× across autosomal targets only. Alignment was performed with the Burrows-Wheeler Aligner (v.0.59), and realignment around indels was performed with the Genome Analysis Toolkit (GATK).5 Putative de novo mutations were identified from exome data with DeNovoGear software. The functional consequence of each variant was assessed according to the most severe consequence from the Ensembl Variant Effect Predictor (VEP).
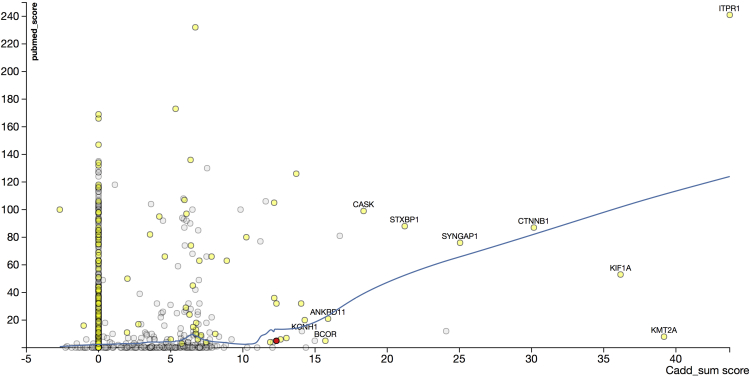
Candidate variants were identified through filtering as previously described3 according to the frequency of the alternate variant in the population (minor allele frequency < 1%) and the function of the variant (protein altering, loss of function, or change in gene dosage), but genes known to be associated with a developmental disorder were not excluded (see Gene2Phenotype in the Web Resources). In total, 2,928 variants in 2,175 genes were further prioritized according to a combination of Cadd_sum and PubMed scores. The former is calculated on the basis of the Combined Annotation Dependent Depletion (CADD) Phred score, a tool for assessing pathogenicity.6 The latter is a method for mining PubMed to identify the 50 most recent publications on the specific genes in which variants are present in the dataset under consideration by using phenotype search keywords. The program obtains a quantitative measure of the presence in the literature for each keyword. Using the relevant keywords (ataxia, cerebellar, cerebellum, cortex, intellectual, and neuron), we identified high PubMed and Cadd_sum scores for EBF3 and other genes in which de novo mutations were present (Figure 1). We validated the method by identifying numerous genes known to be associated with ataxia, including CACNA1A (MIM: 601011), SPTBN2 (MIM: 604985), and ITPR1 (MIM: 147265), and by generating comparative scatterplots with different keyword searches (Figure S1). Having identified two individuals with EBF3 de novo variants of interest in the initial DDD ataxia dataset (individual 1 [272588] with a missense mutation and individual 2 [280219] with a splice mutation), we used an iterative process of re-interrogating the entire DDD data without referring to phenotype and found four more individuals, including two with only a formal diagnosis of intellectual disability (ID). The p value for EBF3 as a gene with potentially pathogenic de novo variants within the first 4,293 families in the DDD study was 2.93 × 10−6, well above the p < 7 × 10−7 cutoff level of genome-wide significance.7 Therefore, variants in the gene had not been identified as disease causing and, prior to the current analysis, were neither published nor reported back to the referring clinicians.
Figure 1.
PubMed Score versus Cadd_sum Score
Shown are x-y plots to prioritize genes associated with ataxia. The Cadd_sum score is on the x axis, the PubMed score is on the y axis, and the Loess regression line is shown in blue. Known genes are highlighted in yellow, and a selected few known ataxia-related genes are labeled with gene symbols. EBF3 is shaded red. Only de novo variants are presented in this analysis. The PubMed score was calculated as follows: all genes on the variant list were searched in PubMed for keywords “[gene name] AND (ataxia [title/abstract] OR cerebellar [title/abstract] OR cerebellum [Title/Abstract] OR cortex [title/abstract] OR intellectual [title/abstract] OR neuron [title/abstract]).” The returned top 50 articles were searched for the occurrence of the searched keywords (ataxia, cerebellar, cerebellum, cortex, intellectual, and neuron), and the PubMed score was calculated as the sum of the occurrence. The assumption is that the PubMed score captures the relevance between the phenotype and a given gene as supported by the literature. The Cadd_sum was calculated as follows: all variants on the list were assigned a CADD score through the CADD web service.6 For de novo variants, only those where an alternative allele was called in the child but in neither parent were counted (the collection is denoted as denovo_v), and the Cadd_sum of a given gene was calculated on a max per-individual level. (We believe only the most damaging de novo variant of a given gene contributes to the observed phenotype. In fact, for any individual in the cohort, no two or more de novo variants were found on the same gene.) If we have M individuals [1… m … M], G genes [1 … g … G], and V variants [1…v… V], the Cadd_sum of a gene g for de novo cases is given by the following formula: . The method has been validated with other keyword searches (Figure S1) and demonstrates that for Cadd_sum scores greater than 10, there is a linear increase in the PubMed score. The correlation between the PubMed score and the Cadd_sum score, which is observed only when relevant phenotype terms are used in the PubMed search, as well as the successful annotation of known ataxia-related genes with high specificity, demonstrates the method’s efficacy in the discovery of pathogenic genes in the cohort.
Subsequently, two additional siblings (individuals 7 [67-1] and 8 [67-4]) out of a total of 65 probands with ID were identified in an independent exome sequencing study, the Canadian CAUSES project (see Web Resources). Individuals were enrolled with written informed consent of their caregivers under two institutional-review-board-approved protocols (H15-00092 and H09-01228, University of British Columbia). All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation.
In brief, the exome capture for each family member of the trio was performed with peripheral-blood DNA and the Agilent All Exon V5+UTR, and sequencing was performed on the Illumina HiSeq 2500. A customized bioinformatics pipeline, coupled with visual validation (Integrative Genomics Viewer) and prioritization, identified variants of interest: Bowtie 28 was used for fast gapped-read alignment to the reference human genome (UCSC Genome Browser hg19), Picard was used to sort and mark duplicate reads, GATK was applied for indel realignment, SAMtools and BCFtools were used to call and filter SNVs and indels, and snpEff9 annotated Ensembl 75 transcripts. Functional variants were then filtered against the public databases dbSNP, NHLBI Exome Sequencing Project Exome Variant Server, and Exome Aggregation Consortium (ExAC) Browser, and a list of rare functional variants compatible with Mendelian inheritance patterns was annotated with the use of custom scripts, ANNOVAR, Ensembl VEP, OMIM, Human Gene Mutation Database, ClinVar, DECIPHER, Leiden Open Variation Database, NCBI Gene, Residual Variation Intolerance Score,10 dbNSFP, and the Loss-of-Function Transcript Effect Estimator (see Web Resources). The variant found in individuals 7 and 8 is absent from the ExAC Browser, and importantly, no other plausible variants of interest are shared by the siblings.
The neurodevelopmental phenotype in the eight individuals ranges from mild to severe ID (Table 1), such that some children require special education and others remain in mainstream school; behavioral disturbances have been reported in some. Six have obvious ataxia, but cerebellar abnormalities were revealed by brain imaging in only individuals 1, 4, and 5 (Figure S2; Table 1). The individuals with splice mutations were not submitted to DDD with a diagnosis of ataxia, but on individual review, both were noted to have poor balance. Most have mild facial dysmorphism (e.g., wide nasal bridge, hypertelorism, synophrys, and deep-set eyes), but there does not appear to be an obvious facial “gestalt” (Figure S3). Additional features that occur in more than one individual include extra-ocular abnormalities (strabismus in four individuals and one Duane abnormality) and renal dysfunction (vesicoureteric reflux and recurrent urinary-tract infections in three individuals). Four children have proportionate short stature.
Table 1.
Summary of Clinical and Molecular Findings in Individuals with EBF3 Mutations
|
DDD Study |
CAUSES Study |
|||||||
|---|---|---|---|---|---|---|---|---|
| Individual 1 (272588) | Individual 2 (280219) | Individual 3 (265391) | Individual 4 (262955) | Individual 5 (263361) | Individual 6 (279995) | Individual 7 (67-1) | Individual 8 (67-4) | |
| cDNA | c.488G>C | c.530C>T | c.355+1G>C | c.579G>T | c.280_283del | c.554+1G>A | c.616C>T | c.616C>T |
| Protein | p.Arg163Pro | p.Pro177Leu | p.? | p.Lys193Asn | p.Glu94Lysfs∗37 | p.? | p.Arg206∗ | p.Arg206∗ |
| Background | ||||||||
| Gender | male | male | male | female | female | female | male | female |
| Age at most recent assessment | 13 years | 7 years | 8 years, 4 months | not available | 8 years, 5 months | 4 years, 8 months | 14 years | 9 years |
| Ethnicity | white British | mixed race (white British and Caribbean) | white British | Irish | English | Irish | Indian | Indian |
| General | ||||||||
| Age at onset | neonate | 9 months | neonate | neonate | neonate | neonate | within first months | within first year |
| Presenting features | developmental delay | developmental delay, hypermobile, ataxia | developmental delay, hypotonia | developmental delay, hypotonia, poor feeding | hypotonia, poor feeding | hypotonia | hypotonia, gross motor delays | hypotonia, delayed gross motor skills |
| Family history | none | none | none | none | none | none | affected sibling | affected sibling |
| Mother’s age at conception (years) | 21 | 38 | 18 | 32 | not available | 32 | 32 | 37 |
| Father’s age at conception (years) | 29 | 42 | 18 | 40 | not available | 33 | 33 | 38 |
| Consanguinity | no | no | no | no | no | no | no | no |
| Stature | short stature 3 SDs below mean, proportionate | normal, weight 75th percentile, height 50th–70th percentile | mild proportionate short stature, height and weight below 0.4th percentile, no hormone investigations | short stature, proportionate | height, weight, and OFC all below 0.3rd percentile | normal | normal | normal |
| Facial dysmorphism | dolichocephaly, prominent forehead and occiput, deep-set eyes | hypertelorism | broad forehead, straight eyebrows, tubular-shaped nose, broad nasal tip, small mouth with a slightly thin upper lip, and low-set, posteriorly rotated ears | broad deep forehead, synophrys, hypertelorism, upslanting palpebral fissures, irregular dentition, downturned mouth, short neck, minimal facial expression | deep-set eyes | normal | deep-set eyes, thick eyebrows, | deep-set eyes, thick eyebrows |
| Other abnormalities | left cryptorchidism, pectus excavatum, tapering fingers, pes planus, shortened great toes | no | fullness on backs of hands, short tapering fingers, clinodactyly of fifth finger, short toes | bilateral moderately severe vesicoureteric reflux and fixed talipes equinovarus, severe FTT, severe cyclical vomiting from 9 weeks | vesicoureteric reflux, recurrent urinary-tract infections, neurogenic bladder, constipation | vesicoureteric reflux and renal dysplasia, recurrent urinary-tract infections | no | no |
| Neurology | ||||||||
| Gait ataxia | yes | yes | poor balance | difficult to assess because of talipes | yes | yes (4 years) | yes | yes |
| Truncal ataxia | yes | yes, but not prominent | no | not apparent | no | yes | yes | yes |
| Hypotonia | yes | yes | yes | yes | yes | yes | yes | yes |
| Dysarthria | yes | mild | no | no | yes | no | no | no |
| Tremor (postural, intention) | no | not noted | no | no | no | no | intention tremor | no |
| Seizures or abnormal EEG and details | no | no | abnormal movements, EEG inconclusive | EEG: intermittent slow activity with occipital sharp features, nil epileptiform | single febrile seizure | recent episodes suggestive of seizures are being analyzed by video telemetry | no | no |
| Microcephaly | 75th–91st percentile | 50th–75th percentile | yes | no | no | progressive microcephaly | no | no |
| OFC at birth | not available | not available | not available | not available | not available | 40 cm | not available | not available |
| OFC at latest assessment (age) | 56.2 cm (13 years); 50th–75th percentile | 53.5 cm (5 years) | 44.8 cm (7 years, 7 months); 0.4th percentile | 52.5 cm (8 years) | 49 cm (5 years, 2 months) | 49 cm (5 years) | 54.5 cm (12.5 years) | 49.5 cm (4 years) |
| Additional features | high-pitched voice | high pain threshold | no | minimal facial expression | no | no | no | no |
| Eye Signs | ||||||||
| Nystagmus | no | no | intermittent on upgaze | no | no | yes | no | no |
| Saccadic abnormalities | no | slow eye movements | no | no | no | no | no | no |
| Jerky eye movements | no | no | no | no | no | no | no | no |
| Additional features | strabismus (esotropia) | strabismus | left convergent squint | blue sclerae, absent tears, left convergent strabismus | strabismus | Duane anomaly | no | no |
| Development | ||||||||
| Head control | not available | not available | not available | 2 years | not available | 18 months | not available | not available |
| Sat independently | not available | 9 months | unknown | 30 months | 12 months | 2 years | 8 months | 8–9 months |
| Stood with support | 2 years | 19 months | 2 years | 4 years | unknown | no | not available | not available |
| Walked independently | 5 years, 8 months | 24 months | >4 years | no | 32 months | no | 18 months | 16 months |
| Speech | 6 years, 6 months | 20 months | 3 years, slightly slurred, needs speech and language therapy | first single words by 7 years, <20 single words by 10.5 years | 40 single words by 3 years | no speech | first word at 19 months | first word after 2 years, 10 words by 2.5 years |
| Current speech ability | 50 words, just putting two words together | mild delay | mild delay | <20 single words | sentences, some echolalia | not applicable | normal | normal |
| School | special (IQ 71) | mainstream | mainstream with 1:1 support | special | special | special | mainstream | mainstream |
| Other | ||||||||
| Behavior | no problems reported | mild emotional and behavioral difficulties and attention deficit | behavioral difficulties | very placid | no | no | attentional and mild behavioral challenges | attentional challenges |
| Imaging | described as cerebellar cleft or absent vermis, cerebellar atrophy suggested from comparison of scans at ages 1 and 5 years, atrophy of pontine tegmentum | normal MRI | first MRI non-specific features and delayed global myelination, repeat normal | cerebellar arachnoid cyst considered unlikely to be of clinical significance | subtle dysplasia cerebellar cortex | normal MRI | normal MRI | normal MRI |
Abbreviations are as follows: EEG, electroencephalography; and OFC, occipital frontal circumference.
EBF3 is an extremely interesting candidate for a role in ID and ataxia. The ortholog in Xenopus, xebf3, is thought to regulate neuronal differentiation during primary neurogenesis,11 and in mice, Ebf3 is highly expressed in Cajal-Retzius (C-R) cells12 during corticogenesis. C-R cells are thought to influence both laminar and areal specification of the developing cerebral cortex, and both EBF3 and the highly homologous EBF2 are implicated in the migration of C-R cells;13 moreover, EBF3 has been shown to heterodimerize with EBF2.14 There is good evidence that EBF2 plays a role in Purkinje cell migration and cerebellar patterning,15 but there are no specific data on the role of EBF3 in cortical or cerebellar development, although it is expressed throughout the brain from at least 11.5 days after conception in mouse embryos.16 EBF3 is also highly expressed in the olfactory epithelium.17 Data from CORTECON, an expression atlas from neural stem cells, show a bi-phasic pattern of EBF3 expression.18 Data on the peripheral expression of EBF3 are limited, but it appears to be low, at least in adults. EBF3 is a direct target of the transcription factor ARX,19, 20 which has been implicated in neurodevelopmental disorders both with and without structural brain anomalies, as well as in genital abnormalities.21, 22 In addition, genome-wide association studies on late-onset dementia have implicated EBF3 as a risk factor in Alzheimer disease.23 Somatic mutations in EBF3 have been identified in glioblastoma and pancreatic cancer, suggesting that it might also function as a tumor suppressor.24
EBF3 is one of a family of highly homologous transcription factors. The first of the encoding gene family to be analyzed in detail (and the earliest from an evolutionary perspective) was early B cell factor 1 (EBF1 [MIM: 164343]), whose expression and function have been studied extensively in the B cell lineage. EBF1 has been shown to be an atypical transcription factor that binds to a palindromic site within the early-B-cell-specific Cd79a promoter and drives expression of Ig-α, a transmembrane protein that is essential for display of the pre-B cell receptor (pre-BCR) and the BCR on the B cell plasma membrane.25, 26 EBF1 contains a DNA-binding domain (DBD) (Figure 2A) whose function is dependent on the coordination of a zinc ion by a single histidine and three cysteines within a 14-residue motif termed the “zinc knuckle,” which is required for DNA binding.27, 28
Figure 2.
Schematic Representations of EBF3 and EBF3 Structure
(A) Structure of EBF3, including the position of the zinc knuckle within the DBD. Numbering refers to amino acids.
(B) Exon structure of EBF3. Numbering at the top refers to amino acid residues. Mutations identified in this study are shown in red with arrows.
(C) Structure of mouse EBF1 (PDB: 3MLP) in complex with DNA (the dimer chain is hidden for clarity); it was created with CCP4mg. Mouse EBF1 has 89% sequence identity with human EBF3 and 100% in the Zinc knuckle. The protein is shown as a blue-gray ribbon, and the DNA is green. The five EBF3 missense variants are labeled and displayed as space-filling models.
(D) Close up of DNA-binding interactions of EBF3 missense variants (generated with PyMol). The protein is shown in green, and the DNA is shown in orange and magenta. Depicted are the numerous interactions involving Pro177 and Lys193.
In vertebrates, there is 100% amino acid conservation between EBF1 and EBF3 across the zinc knuckle and very high homology (95% identity) across the rest of the DBD (Figure S4A), strongly suggesting that the two factors have similar DNA-binding properties and transcriptional activity. EBF family proteins typically form homodimers, which are essential for the recognition of palindromic DNA-binding sites. Homodimerization requires the DBD, the adjacent transcription factor-like/immunoglobulin-like (TIG) domain (also known as the Ig-like/plexins/transcription factors [IPT] domain), and the atypical helix-loop-helix (HLH) domain. The C-terminal domain of EBF1 is required for transcriptional activation of some, but not all, target genes.29
The identified EBF3 variants (GenBank: NM_001005463.2; GI: 347658909) are all de novo (neither parent showed evidence of the alternative allele on Sanger sequencing), although in two individuals, detailed inspection of the read depth of the parental samples showed very low levels of alternative reads, suggesting germline mosaicism (Table S1). The variants include missense, splice, frameshift, and stop-gain mutations (Figure 2B; Table 1). None are present in the ExAC Browser. The missense Z score for EBF3 is high (4.89), indicating intolerance to variation; the pLI score is 1.0, indicating very poor tolerance for heterozygous loss-of-function alleles.30
The mechanism of pathogenicity of the frameshift mutation (c.280_283del [p.Glu94Lysfs∗37]) is almost certainly loss of function because it is predicted to cause nonsense-mediated decay (NMD). The stop-gain and splice mutations are also highly likely to cause heterozygous loss of function (haploinsufficiency), consistent with the bioinformatic evidence. In individual 3 (c.355+1G>C) and individual 6 (c.554+1G>A), formal proof that abnormal splicing occurs has not been possible given the negligible expression in peripheral lymphocytes (data not shown). However, both splicing mutations alter the invariant +1G donor splice site and are therefore predicted to drastically affect splicing according to four splice prediction programs (Table S2). c.355+1G>C is predicted to cause skipping of exon 3, which would result in a frameshift, p.Pro99Serfs∗12, and most likely NMD. However, it is also possible that alternative donor sites could be used, resulting in intronic inclusion. A potential donor site adjacent to position c.355+202 was identified; use of this cryptic site would result in the inclusion of 202 nucleotides of intron 3, again resulting in a frameshift at the protein level and NMD. c.554+1G>A is predicted to cause skipping of exon 6, which would result in an in-frame deletion of 22 amino acids, p.Ser162_Asp184del, across the zinc knuckle and again would lead to loss of DNA-binding activity (Figure 2A). A potential donor site was identified adjacent to position c.554+155, and use of this cryptic site would result in the inclusion of 155 nucleotides of intron 6, resulting in a frameshift at the protein level and, again, most likely NMD.
The bioinformatic data also strongly support the pathogenicity of the EBF3 missense mutations. These mutations are located in regions that encode highly conserved residues within, or near, the zinc knuckle in both vertebrates and invertebrates (Figure S4B; Figure 2C). They are predicted to affect DNA binding and possibly dimerization, and most are predicted to be pathogenic by standard pathogenicity prediction programs (Table S3). Similarly, experimentally induced missense mutations affecting the zinc knuckle of EBF1 have previously been shown to markedly reduce DNA binding.28 Additionally, the p.Arg163Pro substitution, which changes a key DNA contact residue of the zinc knuckle,27, 31 is predicted to disrupt coordination of the zinc ion and DNA binding. The crystal structure of the EBF-DNA complex shows that Pro177 forms van der Waals interactions with Phe191 and a hydrogen bond with the main chain of Arg173, capping off the C terminus of an α helix in the DBD. A more flexible backbone (as in p.Pro177Leu) might destabilize this region and alter DNA binding. Lys193 is involved in the electrostatic network of interactions with the DNA phosphate backbone and the carbonyl oxygen of Ser65. The neutral short residue in the mutant (in p.Lys193Asn) is unlikely to compensate for loss of the positive charge of the lysine (Figure 2D).
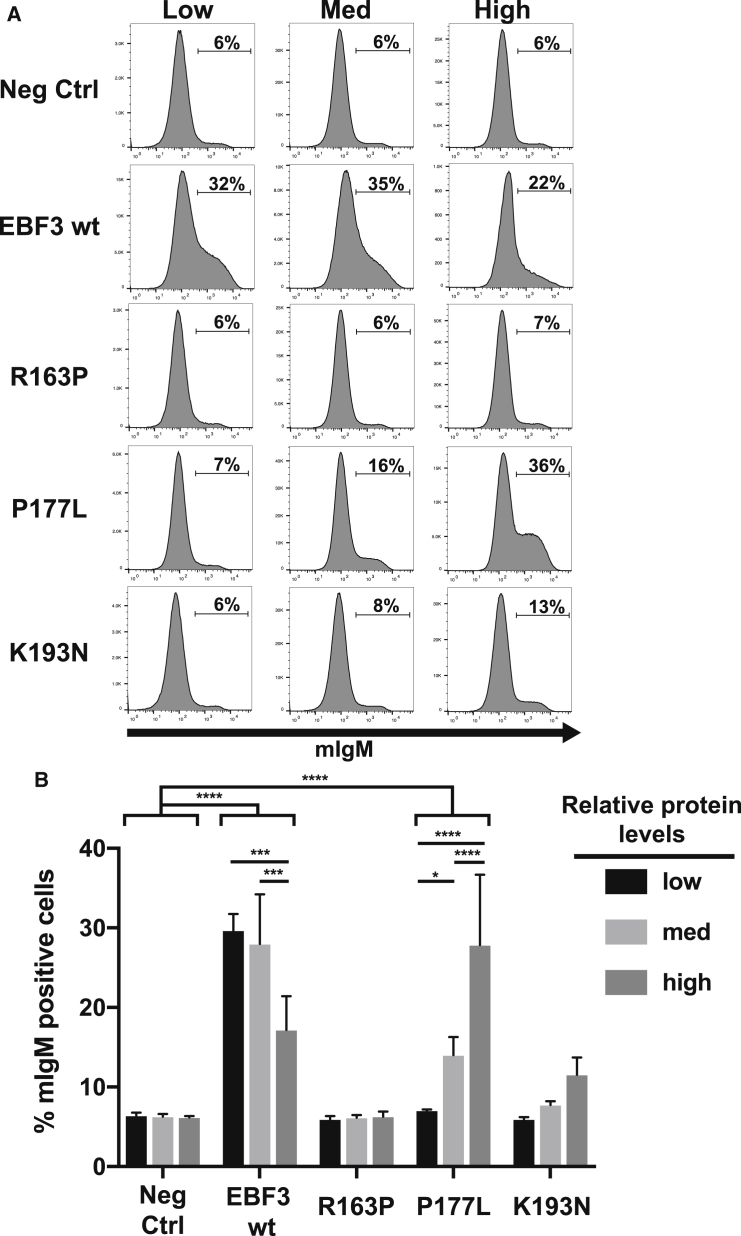
To determine the functional consequences of the EBF3 missense mutations, we took advantage of the μM2.21 cell system, which has proven insightful for mutations in the closely related transcription factor EBF1 (Figure S5A).27, 32, 33 This assay provides a measure of the relative transactivation abilities of mutant and wild-type EBF3 proteins across graded levels of expression. In brief, the μM2.21 assay quantitatively measures the percentage of surface mIgM expression as a direct readout of EBF3 function. Structural similarities between EBF1 and EBF3 suggest that they can substitute for each other (at least partially) in transduced cells. We confirmed this by detecting similar amounts of mIgM on the surface of μM2.21 cells after infection with retroviruses expressing either human EBF1 or EBF3 (Figures S5B and S5C). To assess the relative transcriptional activities of FLAG-epitope-tagged wild-type and mutant EBF3 proteins, we transduced them into μM2.21 cells by using bicistronic retroviruses that also express GFP. After 72 hr, cells were stained, and GFP+ populations were examined for mIgM expression by flow cytometry. Because EBF1 is known to have dose-dependent transcriptional effects on its target genes,34 we wanted to determine whether EBF3 function is similarly dose dependent. To examine functional differences based on EBF3 dosage, we gated transduced μM2.21 cells on similar but non-overlapping low, medium, or high expression of GFP (each increasing by approximately 10-fold) and assessed mIgM on each of these populations (Figures 3A and 3B; Figure S6). Wild-type EBF3 activated mIgM similarly at low and medium expression (over a 10-fold range). However, mIgM expression was significantly reduced at the highest levels of GFP, suggesting dose-dependent inhibitory effects of EBF3 function at high concentrations, consistent with a lack of tolerance of high-level expression of the wild-type. In contrast to the wild-type, the p.Arg163Pro substitution was inactive across all amounts of GFP expression, suggesting that disruption of the zinc knuckle motif ablates EBF3 function regardless of its expression level (Figures 3A and 3B). Both p.Pro177Leu and p.Lys193Asn mutant proteins activated Cd79a transcription and mIgM display at high dosages. This was especially true for p.Pro177Leu, which activated Cd79a expression to near wild-type levels at the highest dosage measured (Figures 3 and 4). These data suggest that neither p.Pro177Leu nor p.Lys193Asn completely abolishes binding to the CD79a promoter but instead probably reduces the affinity and therefore requires larger amounts of protein to activate transcription.
Figure 3.
Dose-Dependent Effects of EBF3 on Function
Flow cytometric analysis of mIgM on plasmacytoma cells in response to increasing amounts of wild-type or mutant EBF3 (each by itself). The retroviral vector for expression of FLAG-tagged EBF3 was generated in two steps. First, primer sequences encoding the FLAG tag, a Gly-Ala-Leu-Thr spacer, and a linker SpeI site were ligated into BS-KS(+) to produce 5′-GTCGACCATGGATTACAAGGACGACGACGATAAAGGTGCTCTGACTAGT-3′. EBF3 was amplified with the EBF3 cDNA clone (I.M.A.G.E clone IRCMp5012D0321D, Source Bioscience), Pfu Ultra II Fusion HS DNA polymerase (Agilent Technologies), and primers 1 and 2 (Table S4). The amplified fragment was digested with SpeI and NotI for ligations into a similarly digested BSK-FLAG vector to make BSK-FLAG-EBF3(wt). FLAG-EBF3 was excised as a SalI-NotI fragment and subcloned into the retroviral vector MSCV-IRES-MCFP (S.J.W. and J.H., manuscript in preparation), which was digested with XhoI and NotI. Mutations were introduced into EBF3 according to a protocol based on that in Fitzsimmons et al.35 In brief, each mutant sense or antisense primer was used together with primer 1 or 2 to amplify EBF3 from BSK-FLAG-EBF3. Fragments were gel purified, and 5′ and 3′ fragments were combined and amplified with primers 1 and 2 alone. PCR fragments were digested with BglII and BstEII for ligation into the similarly digested BSK-FLAG-EBF3. Inserts were excised with SalI and NotI for ligation into MSCV-IRES-MCFP. The T7 epitope tag was added upstream of wild-type EBF3 via subcloning of the SpeI-NotI fragment of BSK-FLAG-EBF3 into NheI-NotI-digested BSK-T73-CHD4.36 The SalI-NotI fragment including T7-EBF3 was subcloned into XhoI-NotI-digested MSCV-IRES-GFP2α (provided by P. Marrack). All plasmids were sequenced for confirmation of the correct cloning and mutagenesis. Culture of the μM2.21 plasmacytoma cell line, infection with the retroviruses, and detection of mIgM by flow cytometry were described previously.33 Whole-cell extracts were obtained via sorting of GFP+, mCFP+, or double-positive populations, washing once in cold PBS (pH 7.5), and then cell lysing for 15 min on ice in a mixture of radioimmunoprecipitation assay buffer (25 mM Tris-HCL [pH 7.6], 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, and 1% SDS) containing 1× HALT and protease inhibitors (Thermo Fisher) and an additional 1 mM phenylmethylsulfonyl fluoride, 1 mM Na3VO4, 5 mM NaF, 5 mM DTT, and 1 μg/mL Pepstatin A prepared fresh. Total protein concentrations were determined by Bradford assay. For mIgM expression, p values comparing column mean differences for individual constructs across their expression dosages were obtained via two-way ANOVA with Tukey’s correction for multiple comparisons. Similar analysis was performed to compare main effects across the mean of each construct. ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗p < 0.05. Significance was set to p < 0.05.
(A) Cells were sorted for low, medium, or high levels of GFP expression (a correlate for EBF protein level) across non-overlapping decades representing 1-, 10-, and 100-fold expression. Histograms represent three independent experiments.
(B) Quantification and statistical analysis of mIgM expression on plasmacytoma cells in response to wild-type or mutant EBF3 across increasing levels of expression. Error bars represent mean values ± SD of three independent experiments.
Figure 4.
EBF3 Forms Multimers with EBF3 Mutants In Vitro
Western blotting (WB, top four lanes) and co-immunoprecipitation (co-IP, bottom four lanes) of EBF3 in retrovirally infected plasmacytoma cells. For western blots, 20 μg of total protein was mixed with 5% 2-mercaptoethanol and 1× Laemmli buffer and resolved on a mini-PROTEAN or Criterion 4%–20% TGX gel (Bio-Rad) at 80–120 V for 1–2 hr. Proteins were then wet transferred onto a 0.45 μm Amersham Protran nitrocellulose membrane (GE Healthcare Life Sciences) for 2.5 hr at 4°C, blocked for 2 hr in 5% milk at room temperature, and stained overnight with primary antibodies in a 5% milk solution in 1× PBS and 0.1% TWEEN. The next day, membranes were washed three times with 1× PBS and 0.1% Tween20, stained with horseradish peroxidase (HRP)-conjugated secondary antibody for 1 hr at room temperature, washed three times with 1× PBS and 0.1% TWEEN, and then washed four times with 1× PBS. Membranes were then incubated with ECL-Plus WB substrate (Thermo Scientific) for 5 min and imaged on a Typhoon FLA9500 (GE Healthcare Life Sciences). For co-IP, μM2.21 cells were infected with listed constructs plus empty-vector control or co-infected with retroviruses expressing N-terminal T7-epitope-tagged EBF3 together with N-terminal FLAG-EBF3, -(R163P), -(P177L), or -(K193N). For all conditions, double-positive GFP+mCFP+ cells were sorted and whole-cell extracts were prepared as described for WB. For each condition, 100 μg of total protein was incubated at 4°C overnight with 1 μL of antibody (anti-FLAG, Rockland Rb-600-401-383; anti-T7, Novagen 69522-3). The next day, 25 μL of Protein A/G magnetic beads (Thermo Scientific) was washed and added to each sample, incubated for 3 hr at 4°C, washed twice with lysis buffer and inhibitors, and then eluted with 1× Laemmli buffer and 5% 2-ME for 10 min at 90°C. Samples were loaded onto mini-PROTEAN gels (Bio-Rad). WB was performed as described previously. Primary antibodies used were anti-EBF1(Abnova H00001879-M01), anti-β-actin (Abcam ab8227), anti-T7-TAG HRP (Novagen 69048-3), and anti-FLAG M2 peroxidase conjugate (Sigma A8592). Secondary antibodies used were anti-mouse HRP conjugate (Promega w402b) and anti-rabbit HRP conjugate (Promega w401b). All primary antibodies were used at 1:1,000, and secondary antibodies were used at 1:10,000. WB and co-IP images were generated from the same sample and represent two independent experiments.
Binding of EBFs to DNA requires the formation of homodimers. Because the genetic changes identified are heterozygous, we wanted to determine whether wild-type EBF3 is capable of forming heterodimers with the mutant EBF3 constructs. To test this, we co-expressed wild-type EBF3 together with each mutated protein (Figure 4). To discriminate between wild-type and mutant proteins, we tagged wild-type EBF3 with the bacteriophage T7 epitope tag in a bicistronic retrovirus that also expresses M-Cherry fluorescent protein (MCFP). μM2.21 cells were simultaneously co-infected with T7-EBF3 (MCFP+) along with wild-type or mutated EBF3 (GFP+). Double-positive cells were sorted and used for western blotting and co-immunoprecipitation. As expected, double-positive wild-type EBF3 co-immunoprecipitated. Notably, wild-type EBF3 also co-immunoprecipitated with each mutant protein. On the basis of previous literature demonstrating the formation of wild-type dimers, our co-immunoprecipitation experiments suggest that heterodimers composed of wild-type and mutant proteins are also formed and that the pathogenicity of the missense substitutions might be due, in part, to dominant-negative effects similar to those previously reported for mutant Xenopus EBF2.37
Overall, the evidence from the bioinformatic, structural, and functional data suggests that the phenotype of EBF3 mutations is caused by heterozygous loss of function. Evidence in support of this interpretation includes the observation of partially overlapping phenotypes in individuals with heterozygous whole-gene deletions across EBF3 (Patricia Maciel et al., personal communication) and a high likelihood of haploinsufficiency as measured by the “haploinsufficiency index.”38 Moreover, haploinsufficiency of the B cell master regulator EBF1 results in reduced or delayed expression of normal B cell developmental markers, an inability to repress non-B lineage genes during developmental progression, and increased DNA damage and cell death (or tumorigenesis).39, 40, 41 Therefore, it is likely that, similar to those reported for EBF1, the human mutations result in reduced EBF3 activity, albeit in different cell populations (i.e., cerebral cortex, cerebellum, and possibly elsewhere). Nevertheless, alternative or additional pathogenic mechanisms—including the formation of heterodimeric EBF3 complexes (composed of wild-type and mutant EBF3), changes in DNA recognition, or abnormal EBF3-EBF2 heterodimerization leading to dominant-negative impairment of function—should be considered.
Recently, in an unpublished manuscript, Harms et al.42 have reported a series of different individuals who have the same phenotype and carry EBF3 nonsense, missense, and splice variants, including an identical mutation (encoding p.Pro177Leu). Interestingly, using a reporter assay in HEK293T cells, the authors found that several of the mutants caused a significant reduction of reporter activity, suggesting dominant-negative effects. However, they also note that the nonsense variants are likely to cause NMD, suggesting heterozygous loss of function.
In conclusion, we present here an ID syndrome in which other key findings are ataxia, facial dysmorphism, and vesicoureteric reflux. The mechanism of action is most likely related to a heterozygous loss of function, which is likely to lead to a reduction in transcriptional activation of EBF3 early in development. Dominant-negative effects could also occur. The effects on brain development remain unknown, but EBF3 is highly expressed in C-R cells, which are known to influence cortical lamination. There is no imaging evidence of cortical abnormalities in the individuals presented here, which would suggest that effects on cortical development are subtle. However, some individuals had some imaging evidence of cerebellar abnormalities, suggesting differing effects of EBF3 on cortical and cerebellar development. The identification of variants in EBF3 presents an opportunity to investigate the role of this transcription factor in neuronal development in much more detail. In this regard, both cellular and animal models of EBF3 mutations will provide novel insights into the mechanisms that perturb these complex systems.
Acknowledgments
The authors would like to thank Gregory Downey and Philippa Marrack for helpful suggestions and support, Desiree Straign for excellent technical assistance, and the National Jewish Health Flow Cytometry Core and Josh Loomis for their support and technical assistance. This work was funded by Action Medical Research and the Henry Smith Charity (A.H.N. and H.S.), RP Fighting Blindness and Fight for Sight (J.Y.), the Wendy Siegel Fund for Leukemia and Cancer Research (J.H.), and the Victor W. Bolie and Earleen D. Bolie Graduate Scholarship Fund (S.J.W.). The DDD study presents independent research commissioned by the Health Innovation Challenge Fund (grant HICF-1009-003), a parallel funding partnership between the Wellcome Trust and the Department of Health, and the Wellcome Trust Sanger Institute (grant WT098051). The views expressed in this publication are those of the author(s) and not necessarily those of the Wellcome Trust or the Department of Health. The research team acknowledges the support of the National Institute for Health Research through the Comprehensive Clinical Research Network. The CAUSES Study is funded by Mining for Miracles, British Columbia Children’s Hospital Foundation, and Genome British Columbia.
Published: December 22, 2016
Footnotes
Supplemental Data include a Supplemental Note, six figures, and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.11.020.
Contributor Information
James Hagman, Email: hagmanj@njhealth.org.
Andrea H. Németh, Email: andrea.nemeth@ndcn.ox.ac.uk.
Web Resources
CAUSES Clinic, http://www.causes.clinic/
CCP4 Molecular Graphics, http://www.ccp4.ac.uk/MG/
Combined Annotation Dependent Depletion (CADD), http://cadd.gs.washington.edu/score
DECIPHER, http://decipher.sanger.ac.uk/
Deciphering Developmental Disorders (DDD) Project, http://www.ddduk.org/
DeNovoGear, https://sourceforge.net/projects/denovogear/files/
Ensembl, http://www.ensembl.org
Ensembl Variant Effect Predictor, http://www.ensembl.org/vep
ExAC Browser, http://exac.broadinstitute.org/
Gene2Phenotype, http://www.ebi.ac.uk/gene2phenotype/
Human Gene Mutation Database (HGMD), http://www.hgmd.cf.ac.uk/
Human Phenotype Ontology, http://compbio.charite.de/hpoweb/showterm?id=HP:0000118
LOFTEE (Loss-Of-Function Transcript Effect Estimator), https://github.com/konradjk/loftee
LOVD (Leiden Open Variation Database) 3.0, http://www.LOVD.nl/
MutationTaster, http://www.mutationtaster.org/
NCBI Gene, https://www.ncbi.nlm.nih.gov/gene/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
Picard, http://picard.sourceforge.net
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
PyMOL, http://www.pymol.org/
SAMtools, http://samtools.sourceforge.net
SIFT, http://sift.jcvi.org/
UCSC Genome Browser, https://genome.ucsc.edu/
Supplemental Data
References
- 1.Deciphering Developmental Disorders Study Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519:223–228. doi: 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Firth H.V., Wright C.F., DDD Study The Deciphering Developmental Disorders (DDD) study. Dev. Med. Child Neurol. 2011;53:702–703. doi: 10.1111/j.1469-8749.2011.04032.x. [DOI] [PubMed] [Google Scholar]
- 3.Wright C.F., Fitzgerald T.W., Jones W.D., Clayton S., McRae J.F., van Kogelenberg M., King D.A., Ambridge K., Barrett D.M., Bayzetinova T., DDD study Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet. 2015;385:1305–1314. doi: 10.1016/S0140-6736(14)61705-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Samocha K.E., Robinson E.B., Sanders S.J., Stevens C., Sabo A., McGrath L.M., Kosmicki J.A., Rehnström K., Mallick S., Kirby A. A framework for the interpretation of de novo mutation in human disease. Nat. Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McRae J.F., Clayton S., Fitzgerald T.W., Kaplanis J., Prigmore E., Rajan D., Sifrim A., Aitken S., Akawi N., Alvi M. Prevalence, phenotype and architecture of developmental disorders caused by de novo mutation. bioRxiv. 2016 [Google Scholar]
- 8.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cingolani P., Platts A., Wang L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petrovski S., Wang Q., Heinzen E.L., Allen A.S., Goldstein D.B. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pozzoli O., Bosetti A., Croci L., Consalez G.G., Vetter M.L. Xebf3 is a regulator of neuronal differentiation during primary neurogenesis in Xenopus. Dev. Biol. 2001;233:495–512. doi: 10.1006/dbio.2001.0230. [DOI] [PubMed] [Google Scholar]
- 12.Yamazaki H., Sekiguchi M., Takamatsu M., Tanabe Y., Nakanishi S. Distinct ontogenic and regional expressions of newly identified Cajal-Retzius cell-specific genes during neocorticogenesis. Proc. Natl. Acad. Sci. USA. 2004;101:14509–14514. doi: 10.1073/pnas.0406295101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiara F., Badaloni A., Croci L., Yeh M.L., Cariboni A., Hoerder-Suabedissen A., Consalez G.G., Eickholt B., Shimogori T., Parnavelas J.G., Rakić S. Early B-cell factors 2 and 3 (EBF2/3) regulate early migration of Cajal-Retzius cells from the cortical hem. Dev. Biol. 2012;365:277–289. doi: 10.1016/j.ydbio.2012.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang S.S., Tsai R.Y., Reed R.R. The characterization of the Olf-1/EBF-like HLH transcription factor family: implications in olfactory gene regulation and neuronal development. J. Neurosci. 1997;17:4149–4158. doi: 10.1523/JNEUROSCI.17-11-04149.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Croci L., Chung S.H., Masserdotti G., Gianola S., Bizzoca A., Gennarini G., Corradi A., Rossi F., Hawkes R., Consalez G.G. A key role for the HLH transcription factor EBF2COE2,O/E-3 in Purkinje neuron migration and cerebellar cortical topography. Development. 2006;133:2719–2729. doi: 10.1242/dev.02437. [DOI] [PubMed] [Google Scholar]
- 16.Garel S., Marín F., Mattéi M.G., Vesque C., Vincent A., Charnay P. Family of Ebf/Olf-1-related genes potentially involved in neuronal differentiation and regional specification in the central nervous system. Dev. Dyn. 1997;210:191–205. doi: 10.1002/(SICI)1097-0177(199711)210:3<191::AID-AJA1>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 17.Wang S.S., Lewcock J.W., Feinstein P., Mombaerts P., Reed R.R. Genetic disruptions of O/E2 and O/E3 genes reveal involvement in olfactory receptor neuron projection. Development. 2004;131:1377–1388. doi: 10.1242/dev.01009. [DOI] [PubMed] [Google Scholar]
- 18.van de Leemput J., Boles N.C., Kiehl T.R., Corneo B., Lederman P., Menon V., Lee C., Martinez R.A., Levi B.P., Thompson C.L. CORTECON: a temporal transcriptome analysis of in vitro human cerebral cortex development from human embryonic stem cells. Neuron. 2014;83:51–68. doi: 10.1016/j.neuron.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 19.Fulp C.T., Cho G., Marsh E.D., Nasrallah I.M., Labosky P.A., Golden J.A. Identification of Arx transcriptional targets in the developing basal forebrain. Hum. Mol. Genet. 2008;17:3740–3760. doi: 10.1093/hmg/ddn271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friocourt G., Parnavelas J.G. Identification of Arx targets unveils new candidates for controlling cortical interneuron migration and differentiation. Front. Cell. Neurosci. 2011;5:28. doi: 10.3389/fncel.2011.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kitamura K., Yanazawa M., Sugiyama N., Miura H., Iizuka-Kogo A., Kusaka M., Omichi K., Suzuki R., Kato-Fukui Y., Kamiirisa K. Mutation of ARX causes abnormal development of forebrain and testes in mice and X-linked lissencephaly with abnormal genitalia in humans. Nat. Genet. 2002;32:359–369. doi: 10.1038/ng1009. [DOI] [PubMed] [Google Scholar]
- 22.Strømme P., Mangelsdorf M.E., Shaw M.A., Lower K.M., Lewis S.M., Bruyere H., Lütcherath V., Gedeon A.K., Wallace R.H., Scheffer I.E. Mutations in the human ortholog of Aristaless cause X-linked mental retardation and epilepsy. Nat. Genet. 2002;30:441–445. doi: 10.1038/ng862. [DOI] [PubMed] [Google Scholar]
- 23.Belbin O., Carrasquillo M.M., Crump M., Culley O.J., Hunter T.A., Ma L., Bisceglio G., Zou F., Allen M., Dickson D.W. Investigation of 15 of the top candidate genes for late-onset Alzheimer’s disease. Hum. Genet. 2011;129:273–282. doi: 10.1007/s00439-010-0924-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao D. Emerging roles of the EBF family of transcription factors in tumor suppression. Mol. Cancer Res. 2009;7:1893–1901. doi: 10.1158/1541-7786.MCR-09-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feldhaus A.L., Mbangkollo D., Arvin K.L., Klug C.A., Singh H. BLyF, a novel cell-type- and stage-specific regulator of the B-lymphocyte gene mb-1. Mol. Cell. Biol. 1992;12:1126–1133. doi: 10.1128/mcb.12.3.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hagman J., Travis A., Grosschedl R. A novel lineage-specific nuclear factor regulates mb-1 gene transcription at the early stages of B cell differentiation. EMBO J. 1991;10:3409–3417. doi: 10.1002/j.1460-2075.1991.tb04905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fields S., Ternyak K., Gao H., Ostraat R., Akerlund J., Hagman J. The ‘zinc knuckle’ motif of Early B cell Factor is required for transcriptional activation of B cell-specific genes. Mol. Immunol. 2008;45:3786–3796. doi: 10.1016/j.molimm.2008.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hagman J., Gutch M.J., Lin H., Grosschedl R. EBF contains a novel zinc coordination motif and multiple dimerization and transcriptional activation domains. EMBO J. 1995;14:2907–2916. doi: 10.1002/j.1460-2075.1995.tb07290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boller S., Ramamoorthy S., Akbas D., Nechanitzky R., Burger L., Murr R., Schübeler D., Grosschedl R. Pioneering Activity of the C-Terminal Domain of EBF1 Shapes the Chromatin Landscape for B Cell Programming. Immunity. 2016;44:527–541. doi: 10.1016/j.immuni.2016.02.021. [DOI] [PubMed] [Google Scholar]
- 30.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Treiber N., Treiber T., Zocher G., Grosschedl R. Structure of an Ebf1:DNA complex reveals unusual DNA recognition and structural homology with Rel proteins. Genes Dev. 2010;24:2270–2275. doi: 10.1101/gad.1976610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao H., Lukin K., Ramírez J., Fields S., Lopez D., Hagman J. Opposing effects of SWI/SNF and Mi-2/NuRD chromatin remodeling complexes on epigenetic reprogramming by EBF and Pax5. Proc. Natl. Acad. Sci. USA. 2009;106:11258–11263. doi: 10.1073/pnas.0809485106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maier H., Ostraat R., Gao H., Fields S., Shinton S.A., Medina K.L., Ikawa T., Murre C., Singh H., Hardy R.R., Hagman J. Early B cell factor cooperates with Runx1 and mediates epigenetic changes associated with mb-1 transcription. Nat. Immunol. 2004;5:1069–1077. doi: 10.1038/ni1119. [DOI] [PubMed] [Google Scholar]
- 34.Lukin K., Fields S., Guerrettaz L., Straign D., Rodriguez V., Zandi S., Månsson R., Cambier J.C., Sigvardsson M., Hagman J. A dose-dependent role for EBF1 in repressing non-B-cell-specific genes. Eur. J. Immunol. 2011;41:1787–1793. doi: 10.1002/eji.201041137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fitzsimmons D., Lutz R., Wheat W., Chamberlin H.M., Hagman J. Highly conserved amino acids in Pax and Ets proteins are required for DNA binding and ternary complex assembly. Nucleic Acids Res. 2001;29:4154–4165. doi: 10.1093/nar/29.20.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Musselman C.A., Ramírez J., Sims J.K., Mansfield R.E., Oliver S.S., Denu J.M., Mackay J.P., Wade P.A., Hagman J., Kutateladze T.G. Bivalent recognition of nucleosomes by the tandem PHD fingers of the CHD4 ATPase is required for CHD4-mediated repression. Proc. Natl. Acad. Sci. USA. 2012;109:787–792. doi: 10.1073/pnas.1113655109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dubois L., Bally-Cuif L., Crozatier M., Moreau J., Paquereau L., Vincent A. XCoe2, a transcription factor of the Col/Olf-1/EBF family involved in the specification of primary neurons in Xenopus. Curr. Biol. 1998;8:199–209. doi: 10.1016/s0960-9822(98)70084-3. [DOI] [PubMed] [Google Scholar]
- 38.Huang N., Lee I., Marcotte E.M., Hurles M.E. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 2010;6:e1001154. doi: 10.1371/journal.pgen.1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heckl D., Schwarzer A., Haemmerle R., Steinemann D., Rudolph C., Skawran B., Knoess S., Krause J., Li Z., Schlegelberger B. Lentiviral vector induced insertional haploinsufficiency of Ebf1 causes murine leukemia. Mol. Ther. 2012;20:1187–1195. doi: 10.1038/mt.2012.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lukin K., Fields S., Lopez D., Cherrier M., Ternyak K., Ramírez J., Feeney A.J., Hagman J. Compound haploinsufficiencies of Ebf1 and Runx1 genes impede B cell lineage progression. Proc. Natl. Acad. Sci. USA. 2010;107:7869–7874. doi: 10.1073/pnas.1003525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prasad M.A., Ungerbäck J., Åhsberg J., Somasundaram R., Strid T., Larsson M., Månsson R., De Paepe A., Lilljebjörn H., Fioretos T. Ebf1 heterozygosity results in increased DNA damage in pro-B cells and their synergistic transformation by Pax5 haploinsufficiency. Blood. 2015;125:4052–4059. doi: 10.1182/blood-2014-12-617282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harms F.L., Girisha K.M., Hardigan A.A., Kortüm F., Shukla A., Alawi M., Dalal A., Brady L., Tarnopolsky M., Bird L.M. Mutations in EBF3 disturb transcriptional profiles and cause intellectual disability, ataxia, and facial dysmorphism. Am. J. Hum. Genet. 2016;100:117–127. doi: 10.1016/j.ajhg.2016.11.012. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.