Abstract
Background
Korean ginseng (Panax ginseng) is a well-known medicinal plant of Oriental medicine that is still in practice today. Until now, a total of 11 Korean ginseng cultivars with unique features to Korean ginseng have been developed based on the pure-line-selection method. Among them, a new cultivar namely G-1 with different agricultural traits related to yield and content of ginsenosides, was developed in 2012.
Methods
The aim of this study was to distinguish the new ginseng cultivar G-1 by identifying the unique single-nucleotide polymorphism (SNP) at its 45S ribosomal DNA and Panax quinquefolius region than other Korean ginseng cultivars using multiplex amplification-refractory mutation system–polymerase chain reaction (ARMS-PCR).
Results
A SNP at position of 45S ribosomal DNA region between G-1, P. quinquefolius, and the other Korean ginseng cultivars was identified. By designing modified allele-specific primers based on this site, we could specifically identified G-1 and P. quinquefolius via multiplex PCR. The unique primer for the SNP yielded an amplicon of size 449 bp in G-1 cultivar and P. quinquefolius. This study presents an effective method for the genetic identification of the G-1 cultivar and P. quinquefolius.
Conclusion
The results from our study shows that this SNP-based approach to identify the G-1 cultivar will be a good way to distinguish accurately the G-1 cultivar and P. quinquefolius from other Korean ginseng cultivars using a SNP at 45S ribosomal DNA region.
Keywords: G-1 cultivar, multiplex polymerase chain reaction, Panax ginseng, Panax quinquefolius, Single-nucleotide polymorphism
1. Introduction
Panax ginseng Meyer is a deciduous perennial herb plant that belongs to the family Araliaceae. They are native to East Asia, while two species of them are found in North America. Ginseng has been used as a medicinal plant for over 2,000 years in Korea, China, and Japan as an immunostimulant, and acts as an agent to foster resistance to fatigue and stress [1], [2], [3]. The usage of ginsenoside-based medicinal products is increasing worldwide. Among the different species of ginseng, P. ginseng and Panax quinquefolius are the most popular for consumption, as well as for medicinal purposes.
Most of the commercial cultivation of P. ginseng has been centralized to South Korea and the northeastern part of China and Japan, whereas P. quinquefolius has been cultivated in China, Canada, and the United States. In South Korea, the ideal climatic conditions to grow ginseng plants in all four seasons favor the cultivation of many species of Panax for commercial purposes, such as P. ginseng, Panax notoginseng (Chinese ginseng), Panax japonicus (Japanese ginseng), and P. quinquefolius L. [4]. Recently, there are a total of nine cultivars namely, Yunpoong, Gopoong, Sunpoong, Gumpoong, Chunpoong, Sunun, Sunone, Sunhyang, and Chungsun with features unique to Korean ginseng, which have been selected from three basic (varieties) lines (Jakyung, Chungkyung, and Hwangsook) using the pure-line-selection method [5]. A similar method was followed to develop the K-1, a new cultivar G-1 and registered with the Korea Seed & Variety Service (http://www.seed.go.kr).
Each Korean ginseng cultivar has unique features in relation to the improved agronomical properties, such as root yield, root shape, and disease resistance. Yunpoong has the highest root yield [6] and Chunpoong, Gopoong, Gumpoong have good root shapes. While considering the quality of red ginseng (steamed ginseng roots) in these cultivars, Chunpoong has the highest root yield of Chun Sam and it is considered the first-grade ginseng followed by Gumpoong, Gopoong, Yunpoong, and Sunpoong. Pertaining to the ginsenoside unit content and total content of ginsenoside in 6-year-old ginseng roots, the ginsenoside content is higher in the order of Gopoong, Yunpoong, Chunpoong, Gumpoong, and Sunpoong [7]. The Korean ginseng cultivar G-1 was developed in 2012 and the morphological characteristics of G-1 are short flower stalk, violet color on stem stronger than Chunpoong (green stem with light violet), lighter than K-1, budding later than Sunpoong and red berry color (Table 1). In addition, the G-1 root appearance, ginsenoside analysis and disease resistance were also analyzed (data not shown).
Table 1.
Main characteristics of aerial parts of 4-year-old ginseng cultivars
| Line | Cultivars | Color of stem | Color of berry | Leaf type | Registered date |
|---|---|---|---|---|---|
| Jakyung | Yunpoong | Light violet | Red | Having stipule | 1998 |
| Gopoong | Violet | Red | Long oval | 2000 | |
| Sunpoong | Violet | Red | Long oval | 2000 | |
| Sunun | Violet | Red | Long oval | 2004 | |
| Sunone | Violet | Red | Long oval | 2004 | |
| Sunhyang | Violet | Red | Long oval, occurrence of stipule | 2007 | |
| K-1 | Violet | Red | Long oval, tipple | 2012 | |
| G-1 | Violet | Red | Occurrence of stipule | 2013 | |
| Chungkyung | Chunpoong | Green and violet spot in green | Orange yellow | Narrow elliptical | 1998 |
| Chungsun | Green | Red | Long oval | 2005 | |
| Hwangsook | Gumpoong | Green | Yellow | Long oval | 2000 |
These cultivars are grown in mixed ginseng fields, and are also sold mixed with other Panax species in the market. Therefore, the development of a valid authentication method is necessary for the preservation of these varieties, and to protect the rights of farmers and consumers. Although the medicinal components and efficacy of P. ginseng have been widely explored [8], [9], [10], [11], there is little information available on the genome of P. ginseng, making the molecular identification of different cultivars difficult. However, with the development of robust molecular markers, such as polymerase chain reaction (PCR)–restriction fragment length polymorphism [12], single-strand conformation polymorphism [13], randomly amplified polymorphic DNA [14], sequence-characterized amplified region [15], intersimple sequence repeat-derived sequence-characterized amplified region [16], amplification-refractory mutation system (ARMS) [17], amplified fragment length polymorphism, and directed amplification of minisatellite region DNA [18] for the Korean ginseng cultivars, this difficulty is prevailed.
Traditional methods based on phenotypic observations have been used to identify the G-1 cultivar from the rest of the Korean ginseng cultivars, but morphological characteristics are often affected by environmental and developmental factors. Due to very similar phenotypical characteristics of these cultivars, the identification and authentication of G-1 becomes difficult especially during the seed-development and seedling stages. Thus, it is advantageous to use molecular methods to differentiate the ginseng cultivars.
In this study, we investigated the possibility of using a single-nucleotide polymorphism (SNP) in 45S ribosomal DNA (rDNA) to differentiate ginseng cultivars. The nucleolar organizing regions (NORs) are cytologically observed as a secondary constriction containing many tandem repeats of 45S ribosomal ribonucleic acid genes [19]. The 45S rDNA sites were observed to be restricted to the NORs, although in some species, smaller or less active sites have also been detected outside the NORs [20]. Based on the SNP sites found for G-1, other Korean ginseng cultivars, and American ginseng, specific primers were designed and multiplex ARMS–PCR was conducted to authenticate these plants. This method based on DNA analysis is widely accepted as a means of identifying medicinal plants, because it is not affected by the growth stage and environmental conditions.
2. Materials and methods
2.1. Plant materials
Eleven ginseng samples (Table 2) were provided by the Ginseng Resource Bank. All voucher specimens were morphologically identified by Professor Woo-Saeng Kwon (Department of Oriental Medicinal Biotechnology, College of Life Sciences, Kyung Hee University).
Table 2.
Ginseng plant samples used in this study
| Ginseng sample | Voucher | Location | GenBank accession number of 45S |
|---|---|---|---|
| Chunpoong | GB001 | Kochang, Korea | KF727964 |
| Yunpoong | GB002 | Kochang, Korea | KF727965 |
| Gopoong | GB003 | Kochang, Korea | KF727966 |
| Sunpoong | GB004 | Kochang, Korea | KF727967 |
| Gumpoong | GB005 | Kochang, Korea | KF727968 |
| Sunun | GBD048 | Daejeon, Korea | KF727969 |
| Chungsun | GBD073 | Daejeon, Korea | KF727970 |
| Sunone | GBD043 | Daejeon, Korea | KF727971 |
| Sunhyang | GBD058 | Daejeon, Korea | KF727972 |
| K-1 | GBD201 | Kochang, Korea | KF727973 |
| G-1 | GBD101 | Kochang, Korea | KF727974 |
| G-1 | GBD102 | Kochang, Korea | |
| G-1 | GBD103 | Kochang, Korea | |
| Panax quinquefolius | GBD099 | USA | KF727975 |
| P. quinquefolius | GBD100 | USA | |
| P. quinquefolius | GBD101 | USA |
2.2. DNA extraction and PCR amplification of ribosomal 45S region
The collected leaf samples were frozen in liquid nitrogen and ground to a fine powder. Genomic DNA was isolated and purified using G Spin Kit for plants (iNtRON, Seongnam, Korea), as per the manufacturer's instructions. The primer pairs used for amplification of the 45S region were 45SF (5′-GCG AGA ATT CCA CTG AAC CT-3′) and 45SR (5′-ACG AAT TCC CTC CGC TTA TTG ATA TGC TTA-3′). PCR amplification was performed in a total volume of 20 μL, and the reaction mixture consisted of each of the primers at a concentration of 0.5μM, 20 ng of template DNA, and 10 μL of 2× PCR premix (Genotech, Daejeon, Korea). The amplification profile consisted of one predenaturation cycle of 4 min at 94°C, followed by 35 cycles of 30 s at 94°C, 30 s at 60°C, 2 min at 72°C, and a final extension at 72°C for 7 min. The PCR products were separated by 1.0% agarose gel electrophoresis and detected by ethidium-bromide staining under UV.
2.3. Sequencing and DNA sequence analysis
The PCR products were purified by PCRquick-spin (iNtRON) according to the manufacturer's instructions, and the purified products were sequenced by Genotech. The DNA sequences of the ribosomal 45S region obtained in the sequencing experiments were compiled using SeqMan software (SeqMan 2, DNASTAR, Inc., 3801, Regent St, Madison, WI, 53705, USA) and edited using the BioEdit program (BioEdit 7.2.5, Ibis Biosciences, 2251 Faraday Avenue, Carlsbad, CA 92008, USA) [21]. Multiple sequence alignments were performed using the online ClustalW2 program (http://www.ebi.ac.uk/Tools/clustalw2/).
2.4. Design of specific primers
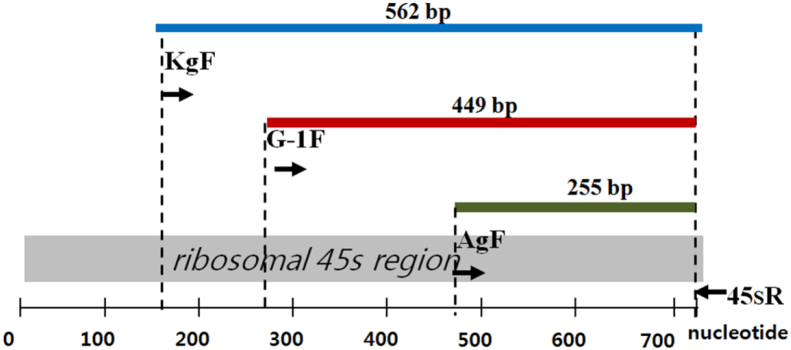
Three specific primers were designed based on the SNP site: KgF (a positive control specific to all cultivars of Korean ginseng), G-1F (specific to G-1 and P. quinquefolius), and AgF (specific to P. quinquefolius) (Fig. 1). We introduced an additional mismatch into KgF and G-1F through the substitution of T for G and A for C at the third base from the 3′ end. The reverse primer used in combination with these primers was 45SR. The sequences and orientations of common and specific primers are shown in Table 3 and Fig. 2, respectively.
Fig. 1.
Comparison of the 45S ribosmal DNA sequences of 11 Korean ginseng cultivars and Panax quinquefolius. The specific primers designed in ribosomal DNA 45S region. KgF: universal primer to Korean ginseng; G-1F: positive primer specific to G-1 cultivar and Panax quinquefolius; AgF: positive primer specific to P. quinquefolius.
Table 3.
Oligonucleotide sequences of primers used in this study
| Primer name | Nucleotide sequence (5′→3′) | Position in 45S region |
|---|---|---|
| 45SF | GCGAGAATTCCACTGAACC | [12] |
| 45SR | ACGAATTCCCTCCGCTTATTG ATATGCTTA | [12] |
| KgF | GACCACCCTTGGGTTGA (G→T) | 170–186 |
| G-1F | TCTAAAACACAAACGACTCTAGG (C→A) | 283–305 |
| AgF | TCACTCCTTTGCGGGAATC | 477–495 |
Note. Bold underlined nucleotide is the additional mismatch introduced via substitution of G for T and C for A.
Fig. 2.
Schematic diagram of the ribosomal 45S region and the positions of the specific primers used for multiplex polymerase chain reaction. G-1, the other Korean ginseng cultivars, and Panax quinquefolius yielded the universal band of 562 bp amplified by primers KgF and 45SR from the ribosomal 45S region.
2.5. ARMS–PCR
Based on the SNP site detected in the ribosomal 45S region, the primer pairs KgF and 45SR were used as the universal primers for authentication of G-1 and P. ginseng. G-1F and AgF were designed for specific authentication of G-1 and P. quinquefolius, respectively. ARMS–PCR was conducted with all primer pairs for simultaneous molecular authentication of G-1, the other Korean ginseng cultivars, and P. quinquefolius. The 20-μL reaction mixture consisted of 20 ng of template DNA, 10 μL of 2 × PreMix DNA polymerase (Genotech), and the four primers (KgF, G-1F, AgF, and 45SR) at concentrations of 0.13 μM, 0.13 μM, 0.13 μM, and 0.13 μM, respectively. The amplification profile consisted of 5 min at 94°C, followed by 35 cycles of 30 s at 94°C, 30 s at 62.5°C, 40 s at 72°C, and a final 7-min extension at 72°C.
3. Results and discussion
Molecular markers have become a popular tool for the identification and authentication of plant and animal species at the DNA level.
Recently, a new class of biomarkers has become the method of choice for genomic identification and authentication. SNPs are the simplest form of sequence variation that can occur between different genomes. It essentially involves substitution of a nucleotide at one position to another one at a specific location [22], [23], which is advantageous over previous methods of variation assessment. In plants, SNP research is still in its infancy, and SNPs have been rigorously documented only in a few species, including several major crops, such as barley [24], rice, maize, wheat, sugar beet, Arabidopsis thaliana, Panax species, and Acanthopanacis cortex [25].
To develop a simple and reliable method for identifying different ginseng cultivars and P. quinquefolius, SNP genotyping with allele-specific PCR was used in this study. The 45S rDNA region of 11 ginseng cultivars and P. quinquefolius was amplified using specific primers, and was found to be 731 bp in length. The results from sequence alignment showed that the 45S rDNA region of the 11 cultivars was identical, except for an SNP specific for G-1 and P. quinquefolius. These sequences were deposited in GenBank (Table 2). The SNP is located at the 305th nucleotide position of G-1 and P. quinquefolius. It contains a guanine (G) residue at the 305th position, whereas the other cultivars substituted with adenine (A) at the same site. Based on this SNP region, KgF, G-1F, AgF, and 45SR (Fig. 2) were designed to authenticate G-1 from the other Korean ginseng cultivars and P. quinquefolius.
ARMS has become a standard technique that was first described by Newton and colleagues in 1989 [26] it allows the discrimination of alleles that differ by as little as 1 bp [27]. The molecular authentication of G-1 was performed using ARMS–PCR using KgF, G-1F, AgF, and 45SR primers based on the G-1-specific SNP site. All of the Korean ginseng cultivars and G-1 yielded an amplicon of 562 bp amplified by the combination of KgF and 45SR from the 45S rDNA region. G-1 and P. quinquefolius generated an amplicon of 449 bp amplified by G-1F and 45SR primers, which flank the unique SNP site, whereas only P. quinquefolius yielded an amplicon of 255 bp with AgF and 45SR primer pairs (Fig. 3). The experiments were conducted several times with a large pool of ginseng samples, and the reproducibility of the ARMS results was validated. Thus, we can conclude that G-1 and P. quinquefolius can be differentiated from the other ginseng cultivars by simultaneous detection of the specific 45S rDNA region.
Fig. 3.
Products of multiplex amplification-refractory mutation system–polymerase chain reaction using primers KgF, G-1F, AgF, and 45SR. Lane M: 1,000-bp DNA ladder; lane 1: Chunpoong; lane 2: Yunpoong; lane 3: Gopoong; lane 4: Sunpoong; lane 5: Gumpoong; lane 6: Sunun; lane 7: Chungsun; lane 8: Sunone; lane 9: Sunhyang; lane 10: K-1; lanes 11–13: G-1; lanes 14–16: Panax quinquefolius. G-1 and P. quinquefolius yielded 449 bp and 255 bp for G-1F and AgF, respectively. The experiments were conducted 10 times with a large number of ginseng samples, and the reproducibility of the amplification-refractory-mutation-system results were validated.
The critical part of ARMS–PCR depends on the designing of the primer. In general, a primer binds with high specificity to a normal allele, and has a 3′ mismatch with a nonspecific allele; this hinders the efficient amplification by DNA polymerase. In many cases, only one mismatch at the 3′ end is not sufficient for the proper differentiation between two alleles. To enhance the amplification of specific alleles, an extra mismatch is introduced in the primers at the 3′ end; this reduces any nonspecific amplification [23], [26]. The expected product sizes for G-1 and P. quinquefolius were 449 bp and 255 bp with primers G-1F and AgF, respectively. Multiplex ARMS–PCR was successfully conducted using these specific primers for molecular identification of G-1 and P. quinquefolius.
In this study, we present an effective method for the simultaneous identification of G-1, other Korean ginseng cultivars, and P. quinquefolius using ARMS–PCR. The method described in this study requires neither restriction digestion nor sequence analysis of PCR products, and the detection of the results employs a simple gel-based assay, which is accessible in any molecular-biology laboratory. In the case of multiplex PCR, other factors, such as primer dimers and hairpins, should also be avoided as far as possible to minimize false-positive results due to false priming. Specific primers based on the 45S rDNA region were designed by the introduction of additional mismatches to ensure absolute specificity. The DNA analysis-based method utilized in this study is widely accepted for the identification of medicinal plants because it is not influenced by growth stages and environmental conditions.
Therefore, a simple and reliable method for the simultaneous authentication of G-1, other Korean ginseng cultivars, and American ginseng was established using the ARMS assay. Our data indicate that G-1 and P. quinquefolius can be differentiated from other ginseng cultivars by the simultaneous detection of the specific 45S rDNA region. We believe that this method will be a useful tool for the authentication of the new Korean ginseng cultivar G-1 and American ginseng.
Conflicts of interest
All authors declare no conflicts of interest.
Acknowledgments
This research was supported by KIPET (113014-03-3-SB010); the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries, Republic of Korea; a grant from YOUNG SCIENTIST (YS) personnel expenditure (490009-1297) from the National Research Council of Science & Technology; and the grant K15060 awarded to the Korea Institute of Oriental Medicine from the Ministry of Education, Science and Technology, Korea.
Contributor Information
Min-Kyeoung Kim, Email: kyeoung0207@khu.ac.kr.
Deok-Chun Yang, Email: dcyang@khu.ac.kr.
References
- 1.Attele A.S., Wu J.A., Yuan C.S. Ginseng pharmacology: multiple constituents and multiple actions. Biochem Pharmacol. 1999;58:1685–1693. doi: 10.1016/s0006-2952(99)00212-9. [DOI] [PubMed] [Google Scholar]
- 2.Chong S.K., Oberholzer V.G. Ginseng—is there a use in clinical medicine? Postgrad Med J. 1988;64:841–846. doi: 10.1136/pgmj.64.757.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park H.J., Kim D.H., Park S.J., Kim J.M., Ryu J.H. Ginseng in traditional herbal prescriptions. J Ginseng Res. 2012;36:225–241. doi: 10.5142/jgr.2012.36.3.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jung J., Kim K.H., Yang K.W., Bang K.H., Yang T.J. Practical application of DNA markers for high-throughput authentication of Panax ginseng and Panax quinquefolius from commercial ginseng products. J Ginseng Res. 2014;38:123–129. doi: 10.1016/j.jgr.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwon W.S., Chung C.M., Kim Y.T., Lee M.G., Choi K.T. Breeding process and characteristics of KG101, a superior line of Panax ginseng C. A. Meyer. Korean J Ginseng Sci. 1998;22:11–17. [Google Scholar]
- 6.Kwon W.S., Lee M.G., Choi K.T. Breeding process and characteristics of Yunpoong, a new variety of Panax ginseng C.A. Meyer. J Ginseng Res. 2000;24:1–7. [Google Scholar]
- 7.Lee J.H., Lee J.S., Kwon W.S., Kang J.Y., Lee D.Y., In J.G., Kim Y.S., Seo J., Baeg I.H., Chang I.M. Characteristics of Korean ginseng varieties of Gumpoong, Sunun, Sunpoong, Sunone, Cheongsun, and Sunhyang. J Ginseng Res. 2015;39:94–104. doi: 10.1016/j.jgr.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen C.F., Chiou W.F., Zhang J.T. Comparison of the pharmacological effects of Panax ginseng and Panax quinquefolius. Acta Pharmacol Sin. 2008;29:1103–1108. doi: 10.1111/j.1745-7254.2008.00868.x. [DOI] [PubMed] [Google Scholar]
- 9.Choi K.T. Botanical characteristics, pharmacological effects and medicinal components of Korean Panax ginseng C A Meyer. Acta Pharmacol Sin. 2008;29:1109–1118. doi: 10.1111/j.1745-7254.2008.00869.x. [DOI] [PubMed] [Google Scholar]
- 10.Mathiyalagan R., Subramaniyam S., Kim Y.J., Kim Y.C., Yang D.C. Ginsenoside compound K-bearing glycol chitosan conjugates: synthesis, physicochemical characterization and in vitro biological studies. Carbohydr Polym. 2014;112:359–366. doi: 10.1016/j.carbpol.2014.05.098. [DOI] [PubMed] [Google Scholar]
- 11.Mathiyalagan R., Subramaniyam S., Kim Y.J., Natarajan S., Min J.W., Kim S.Y., Yang D.C. Synthesis and pharmacokinetic characterization of a pH-sensitive polyethylene glycol ginsenoside CK (PEG-CK) conjugate. Biosci Biotechnol Biochem. 2014;78:466–468. doi: 10.1080/09168451.2014.885827. [DOI] [PubMed] [Google Scholar]
- 12.Ngan F., Shaw P., But P., Wang J. Molecular authentication of Panax species. Phytochemistry. 1999;50:787–791. doi: 10.1016/s0031-9422(98)00606-2. [DOI] [PubMed] [Google Scholar]
- 13.Zhan X.J., Tian C., Zhang Y., Liu C.S. PCR–SSCP molecular identification of Panax ginseng and P. quinquefolius based on ITS2 bar coding SNPs. Zhongguo Zhong yao za zhi. 2012;37:3748–3751. [PubMed] [Google Scholar]
- 14.Um J.Y., Chung H.S., Kim M.S., Na H.J., Kwon H.J., Kim J.J., Lee K.M., Lee S.J., Lim J.P., Do K.R. Molecular authentication of Panax ginseng species by RAPD analysis and PCR–RFLP. Biol Pharm Bull. 2001;24:872–875. doi: 10.1248/bpb.24.872. [DOI] [PubMed] [Google Scholar]
- 15.Wang J., Ha W.Y., Ngan F.N., But P.P., Shaw P.C. Application of sequence characterized amplified region (SCAR) analysis to authenticate Panax species and their adulterants. Planta Med. 2001;67:781–783. doi: 10.1055/s-2001-18340. [DOI] [PubMed] [Google Scholar]
- 16.Lee J.W., Kim Y.C., Jo I.H., Seo A.Y., Lee J.H., Kim O.T., Hyun D.Y., Cha S.W., Bang K.H., Cho J.H. Development of an ISSR-derived SCAR marker in Korean ginseng cultivars (Panax ginseng C. A. Meyer) J Ginseng Res. 2011;35:52–59. [Google Scholar]
- 17.In J.G., Kim M.K., Lee O.R., Kim Y.J., Lee B.S., Kim S.Y., Kwon W.S., Yang D.C. Molecular identification of Korean mountain ginseng using an amplification refractory mutation system (ARMS) J Ginseng Res. 2010;34:41–46. [Google Scholar]
- 18.Ha W.Y., Shaw P.C., Liu J., Yau F.C., Wang J. Authentication of Panax ginseng and Panax quinquefolius using amplified fragment length polymorphism (AFLP) and directed amplification of minisatellite region DNA (DAMD) J Agric Food Chem. 2002;50:1871–1875. doi: 10.1021/jf011365l. [DOI] [PubMed] [Google Scholar]
- 19.Sousa A., Barros e Silva A.E., Cuadrado A., Loarce Y., Alves M.V., Guerra M. Distribution of 5S and 45S rDNA sites in plants with holokinetic chromosomes and the “chromosome field” hypothesis. Micron. 2011;42:625–631. doi: 10.1016/j.micron.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 20.Guerra M., Kenton A., Bennett M.D. rDNA sites in mitotic and polytene chromosomes of Vigna unguiculata (L.) Walp. and Phaseolus coccineus L. revealed by in situ hybridization. Ann Bot. 1996;78:157–161. [Google Scholar]
- 21.Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 22.Landegren U., Nilsson M., Kwok P.Y. Reading bits of genetic information: methods for single-nucleotide polymorphism analysis. Genome Res. 1998;8:769–776. doi: 10.1101/gr.8.8.769. [DOI] [PubMed] [Google Scholar]
- 23.Medrano R.F., de Oliveira C.A. Guidelines for the tetra-primer ARMS–PCR technique development. Mol Biotechnol. 2014;56:599–608. doi: 10.1007/s12033-014-9734-4. [DOI] [PubMed] [Google Scholar]
- 24.Kanazin V., Talbert H., See D., DeCamp P., Nevo E., Blake T. Discovery and assay of single-nucleotide polymorphisms in barley (Hordeum vulgare) Plant Mol Bio. 2002;48:529–537. doi: 10.1023/a:1014859031781. [DOI] [PubMed] [Google Scholar]
- 25.Kim M.K., Jang G.H., Yang D.C., Lee S.H., Lee H.N., Jin C.G. Molecular authentication of Acanthopanacis cortex by multiplex-PCR analysis tools. Korean J Plant Res. 2014;27:680–686. [Google Scholar]
- 26.Newton C.R., Graham A., Heptinstall L.E., Powell S.J., Summers C., Kalsheker N., Smith J.C., Markham A.F. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS) Nucleic Acids Res. 1989;17(7):2503–2516. doi: 10.1093/nar/17.7.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H.T., Hua S., Kwon W.S., Jin H.Z., Yang D.C. A PCR-based SNP marker for specific authentication of Korean ginseng (Panax ginseng) cultivar “Chunpoong”. Mol Biol Rep. 2010;37:1053–1057. doi: 10.1007/s11033-009-9827-5. [DOI] [PubMed] [Google Scholar]