Abstract
Background
In the present study, metabolite profiles of ginsenosides Rk1 and Rg5 from red ginseng or red notoginseng in zebrafish were qualitatively analyzed with ultraperformance liquid chromatography/quadrupole–time-of-flight MS, and the possible metabolic were pathways proposed.
Methods
After exposing to zebrafish for 24 h, we determined the metabolites of ginsenosides Rk1 and Rg5. The chromatography was accomplished on UPLC BEH C18 column using a binary gradient elution of 0.1% formic acetonitrile–0.1% formic acid water. The quasimolecular ions of compounds were analyzed in the negative mode. With reference to quasimolecular ions and MS2 spectra, by comparing with reference standards and matching the empirical molecular formula with that of known published compounds, and then the potential structures of metabolites of ginsenosides Rk1 and Rg5 were acquired.
Results
Four and seven metabolites of ginsenoside Rk1 and ginsenoside Rg5, respectively, were identified in zebrafish. The mechanisms involved were further deduced to be desugarization, glucuronidation, sulfation, and dehydroxymethylation pathways. Dehydroxylation and loss of C-17 residue were also metabolic pathways of ginsenoside Rg5 in zebrafish.
Conclusion
Loss of glucose at position C-3 and glucuronidation at position C-12 in zebrafish were regarded as the primary physiological processes of ginsenosides Rk1 and Rg5.
Keywords: ginsenoside Rg5, ginsenoside Rk1, metabolism, ultraperformance liquid chromatography/quadrupole–time-of-flight MS, zebrafish
1. Introduction
Panax ginseng (PG) has been widely used as a general tonic in traditional oriental medicine for over 2,000 y to increase disease resistance, physical fitness, and longevity, especially in elderly people [1]. There are reports recorded that steamed ginseng possesses enhances pharmacological effects compared with nonsteamed ginseng [1]. Notoginseng, the dry root and rhizome of Panax notoginseng (Burk.) F. H. Chen, has been one of the most popular Chinese herbal medicines used for removing blood stasis, stopping bleeding, relieving swelling, and alleviating pain in the past hundreds of years [2]. Steamed notoginseng nourishes the blood, enhances immune function, inhibits tumors, increases robustness, and promotes tissue regeneration [2]. These activities of ginseng and notoginseng are known to mainly originate from ginsenosides [3], [4], [5]. During the steaming process, the chemicals are transformed. Some compounds disappear, and some of the active compounds are produced. Reports show that saponin compounds including ginsenosides F4, Rg6, Rh2, Rk3, Rh4, Rk1, Rg5, and Rg3 can be isolated from steamed ginseng and notoginseng [6], [7]. Among them, ginsenosides Rk1 and Rg5 are both protopanoxadiol disaccharide glycosides [position C-3 is connected with glucose-(2-1)-glucose] whose dehydrated positions are at C-20. The only difference in structure between ginsenoside Rk1 and ginsenoside Rg5 is the C-20 double bond position. The former position is at double bond Δ20,21, while the latter position is at double bond Δ20,22. Pharmacological references published have demonstrated memory enhancing and neuroprotective effects of ginsenoside Rk1 [8]. Ginsenoside Rg5 is associated with reinforcement of cytotoxic effectiveness in human hepatoma SK-HEP-1 cells [9]. Anticoagulation, antianxiety, and induction of neuronal differentiation in neurosphere stem cells of ginsenoside Rg5 also have been reported [10], [11]. However, the metabolism of ginsenoside Rk1 and Rg5 has not been reported yet in vitro and in vivo. The metabolic study method in vitro is simple to perform but a high standard of experimental conditions is necessary, and the approach in vivo is resource costly and highly labor intensive. Many results show that studying the metabolism in vivo is more accurate than in vitro. In vitro metabolism experiments often neglect the interaction of the body, so there is the absence of consistency with metabolism in vivo. The experimental results require validation of metabolic consequences in vivo. In the overall consideration, we choose the method in vivo. In terms of the experimental animals, rats are often chosen as the object. Then we first use zebrafish as the model due to lots of advantages of genetic and physiological characteristics. Zebrafish has similar genes and complex organ system just like the mammals. In addition, zebrafish possess similar types of metabolic enzymes [12], [13], [14] and intestinal flora [15] as those observed in mammals. We used ultraperformance liquid chromatography/quadrupole–time-of-flight (UPLC-Q-TOF)/MS, which can improve the sensitivity, reduce experimental error, and contribute to the reliability of the results. The similarities and differences of metabolic behavior of ginsenoside Rk1 and Rg5 were estimated rapidly. Our work supplied data for their metabolic mechanism investigation in vivo and research on druggability during development of new drug on future.
2. Materials and methods
2.1. Chemicals
Ginsenoside Rk1 (purity > 98%, Batch No: 12052401) was purchased from Must Biological Technology Company (Chengdu, China), and ginsenoside Rg5 (purity > 98%, Batch No: 20100728) was purchased from Yong Heng Biological Technology Company (Shanghai, China). MS-grade formic acid was purchased from Nanjing Chemical Reagent Co., Ltd. (Nanjing, China). Dimethyl sulfoxide (DMSO) was purchased from Sinopharm Chemical Reagent Co. Ltd (Shanghai, China), and the other reagents and chemicals were of analytical grade. HPLC-grade acetonitrile and methanol were purchased from Merck (Darmstadt, Germany). Deionized water was purified using the Milli-Q water purification system (Millipore, Bedford, MA, USA).
2.2. Animals
Adult zebrafish (Danio rerio; age 6–10 mo, weight, 0.8–1.2 g) of mixed sex were supplied by Model Animal Research Center of Nanjing University (Nanjing, China), and acclimatized to tap water in a glass aquarium for at least 10 d preceding experimentation. Fish were kept at a temperature of 23 ± 1°C in a photoperiod of 12:12 h. The fish were fed daily during the acclimatization period, and were fasted overnight before the day of the experiment.
2.3. Instruments
Acquity UPLC system (Waters Corp., Milford, MA, USA) furnished with autosampler, a vacuum degasser, thermostatted column compartment, and quadruple pump, and Synapt Q-TOF mass spectrometer (Waters Corp.) furnished with an electrospray ionization (ESI) source were used. KQ3200DE Digital Ultrasonic Washer (Kunshan Ultrasonic Instruments Co. Ltd., Kunshan, China); XW-80A Vortex Mixer (Shanghai Huxi Scientific Instrument Factory, Shanghai, China); Mettler Toledo AB135-S Analytical Balance (Mettler Toledo, Schwerzenbach, Switzerland); Labconco Freezer Dryer (Labconco, Kansas City, MO, USA); TGL-16G Desk Centrifuge (Shanghai Anting Scientific Instrument Factory, Shanghai, China); Organomation N- EVAP 112 Nitrogen Evaporator (Organomation Associates, Inc., Berlin, MA, USA) were also used.
2.4. Drug administration and sample collection
Adult zebrafish were separated into three experimental groups of six fish each at random. After fasting for 12 h, these fish in each group were kept individually in brown glass bottles maintaining at a temperature of 23 ± 1°C in a waterbath with 30 mL solution: one blank control group was exposed to 0.5% DMSO purified water (blank zebrafish group), two groups were exposed to 30 mL solution of ginsenoside Rk1 (0.78 μg/mL) or ginsenoside Rg5 (2.54 μg/mL) in 0.5% DMSO purified water (drug zebrafish groups). In addition, the above solutions of 0.5% DMSO purified water, ginsenoside Rk1 (0.78 μg/mL) and ginsenoside Rg5 (2.54 μg/mL) without zebrafish were used as blank solvent and blank drug controls. Both zebrafish body and solution of the blank zebrafish group and drug zebrafish groups were sampled at 24 h. The zebrafish bodies of each group were combined and washed quickly with purified water three times, then weight after sacrifice and removal of fins and scales of fish and stored at −70°C prior to analysis; the solution of each group were also combined. Combined solutions of each group were sampled and also stored at −70°C prior to analysis. Solutions of blank solvent and blank drug control groups were sampled same as above at 24 h.
2.5. Sample preparation
The solution sample (30 mL) was freeze-dried to dryness, and the residue dissolved in 90% methanol (1 mL). The solution was filtered through a 0.22-μm filter and 20 μL was injected into the UPLC-Q-TOF/MS system for analysis. The zebrafish body samples (6 fish in each group) were cut with scissors, and 1 g was sampled and homogenized with physiological saline (5 mL), followed by centrifugation at 1852 g for 10 min, then the supernatant was mixed by vortexing with methanol at the ratio of 1:4 (v/v) three times, followed by centrifugation at 1852 g for 10 min. The supernatant was evaporated to dryness with nitrogen at room temperature, and the residue was dissolved in 90% methanol (1 mL). After filtering through a 0.22-μm filter, 20 μL of the solution containing 1 g fish tissue/mL was introduced into the UPLC-Q-TOF/MS system for analysis.
2.6. Analysis condition
Chromatography was accomplished using an ACQUITY UPLC system (Waters) furnished with a conditioned autosampler at 4°C. The separation was completed on an Acquity UPLC BEH C18 column (i.d., 1.7 μm; 2.1 mm × 50 mm; Waters Corp.). The column temperature was kept at 35°C. The mobile phase comprised 0.1% formic acid–water (solvent A) and 0.1% formic acid–acetonitrile (solvent B). The linear gradient elution program was as follows: 0–4 min, 10–100% B; 4–4.3 min, 100–100% B; 4.3–4.7 min, 100–10% B; 4.7–5 min, 10% B. The flow velocity was 0.2 mL/min. The injection volume was 5 μL.
The high-mass resolution experiments were carried out on a Synapt Q-TOF mass spectrometer (Waters Corp.) furnished with an electrospray ionization (ESI) source in the negative ion mode. The capillary and cone voltages were 3,000 V and 30 V, respectively. The desolvation gas (nitrogen) was set to 800 L/h at 400°C, and the source temperature was 100°C. The mass range was scanned from 100 Da to 1,500 Da and corrected during acquisition using an external reference (lock spray) comprising a solution of 600 ng/mL leucine enkephalin (m/z 566.2771) infused at a rate of 5 μL/min. The transfer collision energy (Ec) and trap Ec to acquire MS data were 4 eV and 2 eV, respectively, whereas the transfer Ec was 2 eV and the trap Ec was 6 eV to acquire MS/MS data. The raw data were collected and processed using MassLynx (version 4.1; Waters Corp.).
3. Results
The full scan mass spectrum of fragment ions of the biosamples acquired from zebrafish treated with ginsenosides Rk1 and Rg5 was compared with that of biosamples acquired from blank zebrafish and blank solvent to identify the possible metabolites of ginsenosides Rk1 and Rg5. Two parent compounds and their metabolites showed their quasimolecular ions [M-H]− or [M+HCOO]− in negative mode, and were confirmed by a general analysis of the MS and MS/MS fragmentation behaviors and retention times. The structures of most metabolites were also extrapolated. The major product ions of the metabolites, along with their UPLC retention times are shown in Table 1, Table 2.
Table 1.
Accurate mass measurement for the ionized molecules of metabolites of ginsenoside Rk1 in zebrafish
| No. | Formula | tR (min) | [M−H]− |
[M+HCOO]− (mg/L error) | Fragment ions of [M−H]− or [M+HCOO]−m/z |
Part | ||
|---|---|---|---|---|---|---|---|---|
| calcd m/z | exptl m/z | mg/L error | ||||||
| Parent | C42H70O12 | 4.01 | 765.4795 | 765.4792 | 0.4 | 811.4858 (1.7) | — | — |
| M1 | C36H60O10S | 3.26 | 683.3834 | 683.3824 | −0.8 | — | MS2[527.3030] | Zebrafish body |
| M2 | C42H68O13 | 3.19 | 779.4587 | 779.4609 | 3.5 | — | MS2[515.3047] | Zebrafish body |
| M3 | C47H76O17 | 3.00 | 911.5010 | 911.4960 | −4.9 | — | MS2[545.3144] | Zebrafish body |
| M4 | C48H78O18 | 3.03 | 941.5115 | 941.5114 | 0.4 | — | MS2[501.2884], [573.3090] | Zebrafish body |
| M5 | C42H68O13 | 3.62 | 779.4587 | 779.4595 | 1.7 | — | MS2[515.3047] | Solution sample of zebrafish |
| M6 | C47H76O17 | 3.00 | 911.5010 | 911.5001 | −0.4 | — | MS2[545.3144] | Solution sample of zebrafish |
Table 2.
Accurate mass measurement for the ionized molecules of metabolites of ginsenoside Rg5 in zebrafish
| No. | Formula | tR(min) | [M−H]− |
[M+HCOO]− (mg/L error) | Fragment ions of [M−H]− or [M+HCOO]−m/z |
Part | ||
|---|---|---|---|---|---|---|---|---|
| calcd m /z | exptl m /z | mg/L error | ||||||
| Parent | C42H70O12 | 4.03 | 765.4795 | 765.4811 | 2.8 | 811.4856 (0.3) | — | — |
| N1 | C28H48O9S | 3.03 | 559.2946 | 559.2946 | 0.9 | — | MS2[513.2891] | Zebrafish body |
| N2 | C48H78O18 | 2.91 | 941.5115 | 941.5109 | −0.1 | — | MS2[599.3195], [615.3898] | Zebrafish body |
| N3 | C28H48O9S | 2.39 | 559.2946 | 559.2934 | −1.3 | — | MS2[513.2891] | Solution sample of zebrafish |
| N4 | C34H56O12 | 2.54 | 655.3699 | 655.3684 | −1.5 | — | MS2[475.2688] | Solution sample of zebrafish |
| N5 | C34H56O13 | 2.10 | 671.3648 | 671.3629 | −2.1 | — | MS2[539.3220] | Solution sample of zebrafish |
| N6 | C34H58O12 | 2.54 | — | — | — | 703.3906 (0.1) | MS2[657.3842] | Solution sample of zebrafish |
| N7 | C42H68O13 | 3.10 | 779.4587 | 779.4568 | −1.8 | 825.4640 (0.4) | MS2[599.3204] | Solution sample of zebrafish |
| N8 | C47H76O17 | 3.00 | 911.5010 | 911.5022 | 1.9 | — | MS2[545.3146], [599.3203] | Solution sample of zebrafish |
| N9 | C48H78O18 | 2.91 | 941.5115 | 941.5109 | −0.1 | — | MS2[599.3195], [615.3898] | Solution sample of zebrafish |
3.1. Metabolites of ginsenoside Rk1 detected in zebrafish body samples and the solution sample of zebrafish
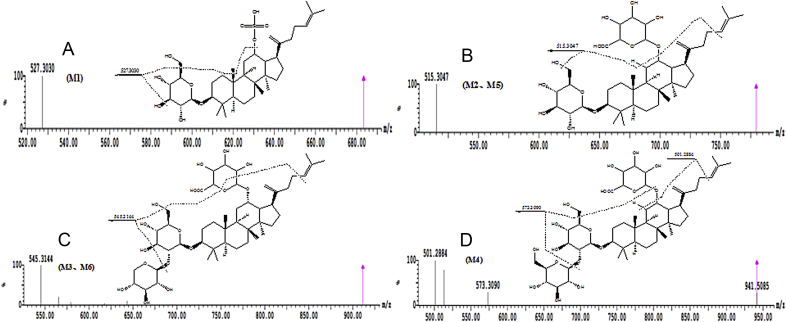
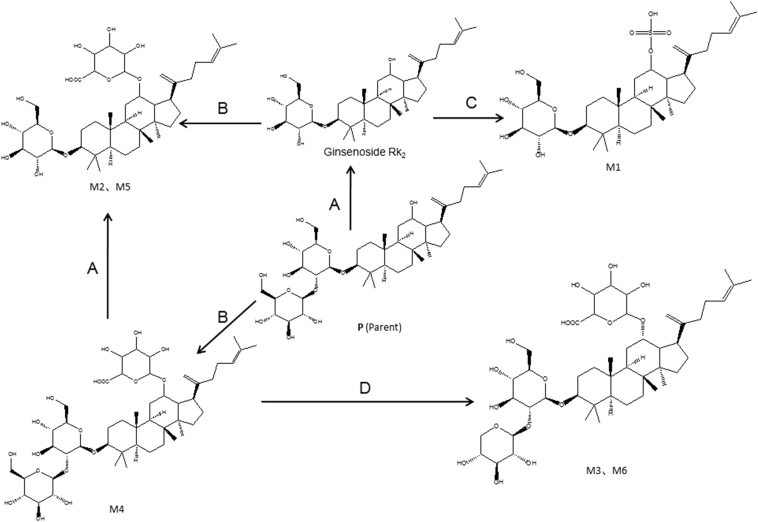
In all, four metabolites (M1–M4) and two metabolites (M5, M6) were tentatively characterized by MS and MS/MS in the zebrafish body and the solution sample of zebrafish, respectively. (Table 1). Of these, four—M1, M2(M5), M3(M6), M4—are, to our knowledge, reported for the first time. The TOF-MS spectra of these metabolites and the proposed major metabolic pathway of ginsenoside Rk1 in zebrafish are shown in Fig. 1, Fig. 2, respectively.
Fig. 1.
Mass spectra of metabolites of ginsenoside Rk1 in zebrafish. (A) 23-de-2-methyl propenyl ginsenoside Rk2-12-O-sulfate, M1; (B) ginsenoside Rk2-12-O-glucuronate, M2 and M5; (C) 5″-dehydroxymethyl ginsenoside Rk1-12-O-glucuronate, M3 and M6; and (D) ginsenoside Rk1-12-O-glucuronate, M4.
Fig. 2.
Proposed major metabolic pathway of ginsenoside Rk1 in zebrafish. (A) Desugarization; (B) glucuronidation; (C) sulfation; and (D) dehydroxymethylation.
The molecular ions of M4 (m/z 941.5115) were produced by the glucuronated conjugation of 176 Da, corresponding to the glucuronidation from the parent compound (P). Therefore, M4 (Fig. 1) was thought to be the glucuronated product of ginsenoside Rk1 due to glucuronidation at position C-12. According to the structure of ginsenoside Rk1, M4 was called ginsenoside Rk1-12-O-glucuronate.
The molecular ions at m/z 779.4587 (M2 and M5) and their product ions at m/z 515.3047 and m/z 507.2781 (Fig. 1B) was 162 Da less than m/z 941.5115 (M4) and its product ions at m/z 573.3090 and m/z 501.2884. Therefore, M2 and M5 were identified as the losing one molecular glucose products of M4, and their positions of loss of glucose were thought to be at C-1′′. According to the structure of M4, M2 and M5 were called ginsenoside Rk2-12-O-glucuronate.
The molecular ions at m/z 911.5010 (M3 and M6) and their product ions at m/z 545.3144 and m/z 545.3145 (Fig. 1C) was 30 Da less than m/z 941.5115 (M4) and its product ions at m/z 573.3090 and m/z 501.2884. Therefore, M3 and M6 were identified as the losing one molecular hydroxymethyl products of M4, and their positions of loss of hydroxymethyl were thought to be at C-5′ of glucose. According to the structure of M4, M3 and M6 were called 5″-dehydroxymethyl ginsenoside Rk1-12-O-glucuronate.
The molecular ions at m/z 683.3834 (M1) and its product ions at m/z 527.3030 (Fig. 1A) was 80 Da more than that of ginsenoside Rk2. Therefore, M1 was identified as the sulfated product of ginsenoside Rk2, and the sulfated position was thought to be at C-12. According to the structure of ginsenoside Rk2, M1 was called ginsenoside Rk2-12-O-sulfate.
3.2. Metabolites of ginsenoside Rg5 detected in zebrafish body samples and the solution sample of zebrafish
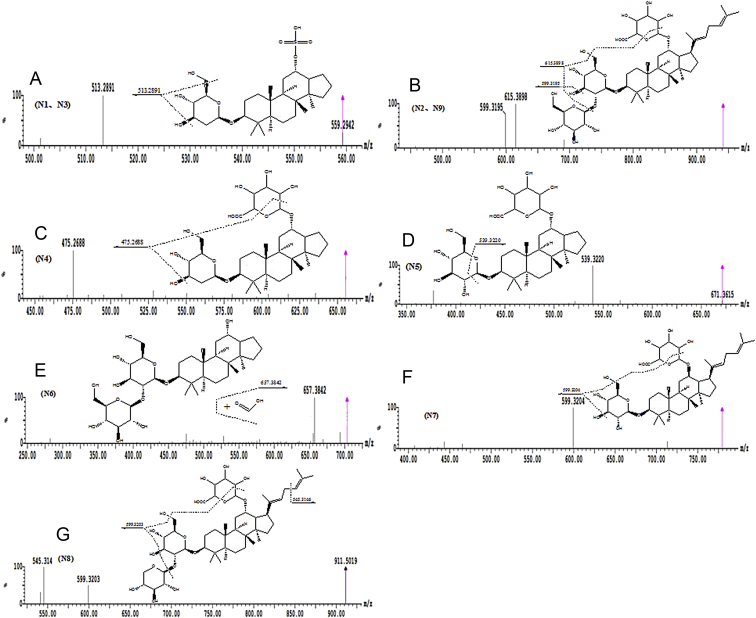
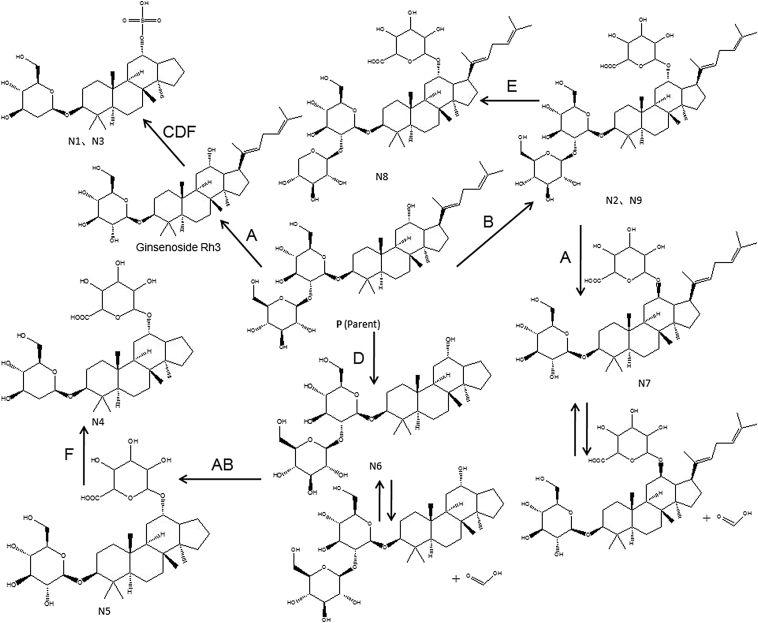
In all, two metabolites (N1, N2) and eight metabolites (N3–N9) were tentatively characterized by MS and MS/MS in the zebrafish body and the solution sample of zebrafish, respectively (Table 2). Of these, seven—N1(N3), N2(N9), N4–N8—are, to our knowledge, reported for the first time. The TOF-MS spectra of these metabolites and the proposed major metabolic pathway of ginsenoside Rg5 in zebrafish are shown in Fig. 3, Fig. 4, respectively.
Fig. 3.
Mass spectra of metabolites of ginsenoside Rg5 in zebrafish. (A) 2′-dehydroxyl, 17-de-1,5-dimethyl-1,4-hexadienyl ginsenoside Rh3-12-O- sulfate, N1 and N3; (B) ginsenoside Rg5-12-O-glucuronate, N2 and N9; (C) 2′-dehydroxyl, 17-de-1,5-dimethyl-1,4-hexadienyl ginsenoside Rh3-12-O-glucuronate, N4; (D) 17-de-1,5-dimethyl-1,4-hexadienyl ginsenoside Rh3-12-O-glucuronate, N5; (E) 17-de-1,5-dimethyl-1,4-hexadienyl ginsenoside Rg5, N6; (F) ginsenoside Rh3-12-O-glucuronate, N7; and (G) 5″-dehydroxymethyl ginsenoside Rg5-12-O-glucuronate, N8.
Fig. 4.
Proposed major metabolic pathway of ginsenoside Rg5 in zebrafish. (A) Desugarization; (B) glucuronidation; (C) sulfation; (D) loss of C-17 residue (de-1,5-dimethyl-1,4-hexadienylation); (E) dehydroxymethylation; and (F) dihydroxylation.
The molecular ions of N1 and N3 (m/z 559.2946) were produced by the addition of 206 Da, corresponding to the desugarization, sulfation, de-1,5-dimethyl-1,4-hexadienyl, dehydroxylation of the parent compound. These results indicate that the ions of the parent compound were first desugarized at positions C-3 and the ions of ginsenoside Rh3, which then sulfated, losing 1,5-dimethyl-1,4-hexadienyl, dehydroxy was formed. Therefore, N1 and N3 (Fig. 2A) were thought to be the sulfated, losing 1,5-dimethyl-1,4-hexadienyl, dehydroxy products of ginsenoside Rh3. The positions of sulfation, losses of 1,5-dimethyl-1,4-hexadienyl and dehydroxylation are C-12,C-17 and C-2′ of glucose, respectively. According to the structure of ginsenoside Rh3, N1 and N3 were called 2′-dehydroxyl, 17-de-1,5-dimethyl-1,4-hexadienyl ginsenoside Rh3-12-O- sulfate.
The molecular ions at m/z 941.5115 (N2 and N9) and their product ions at m/z 615.3898 and m/z 599.3195 (Fig. 2B) was 176 Da more than the molecular ions of ginsenoside Rg5. Therefore, N2 and N9 were identified as the glucuronated products of ginsenoside Rg5, and the glucuronated position was thought to be at C-12. According to the structure of ginsenoside Rg5, N2 and N10 were called ginsenoside Rg5-12-O-glucuronate.
The molecular ions at m/z 779.4587 (N7) whose formic acid association ion was at m/z 825.4642 and its product ion was at m/z 599.3204 (Fig. 2F) was 162 Da less than m/z 941.5115 (N2 and N9). Therefore, N7 was identified as the losing one molecular glucose product of N2 and N9 due to loss of glucose at position C-2′. According to the chemical structures of N2 and N9, N7 was called ginsenoside Rh3-12-O-glucuronate.
The molecular ions at m/z 911.5010 (N8) and their product ions at m/z 599.3203 and m/z 545.3146 (Fig. 2G) was 30 Da less than m/z 941.5115 (N2 and N9) and their product ions at m/z 615.3898 and m/z 599.3195. Therefore, N8 was identified as the losing hydroxymethyl product of N2 and N9, and the position of loss of hydroxymethyl was thought to be at C-5″ of glucose. According to the structures of N2 and N9, N8 was called 5″-dehydroxymethyl ginsenoside Rg5-12-O-glucuronate.
The molecular ions of formic acid of association of N6 was at m/z 703.3910, and its product ions at m/z 657.3842 (Fig. 2E) was 108 Da less than the parent compound (P). Therefore, N6 was identified as the losing 1,5-dimethyl-1,4-hexadienyl product of ginsenoside Rg5 due to loss of 1,5-dimethyl-1,4-hexadienyl at position C-17. According to the chemical structure of ginsenoside Rg5, N6 was called 17-de-1,5-dimethyl-1,4-hexadienyl ginsenoside Rg5.
The molecular ions at m/z 671.3648 (N5) and its product ions at m/z 539.3220 (Fig. 2D) was 14 Da more than that of N6 whose formic acid association ions was at m/z 703.3910 and its product ions at m/z 657.3842. Therefore, N5 was identified as the losing one molecular glucose and the glucuronated product of N6, due to the losses of glucose at position C-1″ of glucose and the glucuronated position at C-12. According to the structures of N6, N5 was called 17-de-1,5-dimethyl-1,4-hexadienyl ginsenoside Rh3-12-O-glucuronate.
The molecular ions at m/z 655.3699 (N4) and its product ions at m/z 475.2688 (Fig. 2C) was 16 Da less than m/z 671.3648 (N5) and its product ions at m/z 539.3220. Therefore, N4 was identified as the dehydroxy products of N5, and the dehydroxy position was thought to be at C′-2 of glucose. According to the structures of N5, N4 was called 2′-dehydroxyl, 17-de-1,5-dimethyl-1,4-hexadienyl ginsenoside Rh3-12-O-glucuronate.
4. Discussion
The zebrafish, a well-characterized vertebrate model, is being applied to the fields of molecular genetics and developmental biology, and is used as an increasingly popular preclinical testing model organism in drug toxicology and screening, as its genes and complex organs are similar to those found in mammals [16], [17], [18], [19]. The conversion of drugs is related to their activation and inactivation in vivo. So we should pay more attention to applying zebrafish model for pharmaceutical metabolism [20], [21], [22], [23]. In biology, zebrafish and humans have certain similarities, most human genes and have zebrafish gene orthologs. Zebrafish whole-genome sequencing has been completed, the similarity of the zebrafish gene with human genes is 87%, and some studies have shown that drug reactions in zebrafish and humans have a very high similarity.
We here used zebrafish as a model to study the metabolism of ginsenosides Rk1 and Rg5 from steamed notoginseng. Zebrafish are exposed to ginsenosides Rk1 and Rg5 solutions, and they can absorb compounds independently and continuously from the solution through the skin and cavity (mouth). Metabolites of the drug are continuously discharged to water along with the waste of zebrafish, so we can determine part of the drug metabolic information by analyzing the liquid composition changes. This method is simple and feasible, which has overcomed the defects of big difference between the results of metabolism in vitro and that of metabolism in vivo, prediction difficulty and high labor intensity in vivo.
Zebrafish have a variety of drug-metabolizing enzymes, such as cytochrome P450 isoforms or conjugation enzymes including glutathione-S-transferase, uridine diphosphoglucuronyl transferases, and sulfotransferases, which are similar to those of mammals. The gene or amino acid sequence of metabolic enzymes of zebrafish is more than half identical to human [12], [13]. Experimental results suggest that ginsenosides Rk1 and Rg5 were metabolized in zebrafish by reaction of desugarization, dihydroxylation, and dehydroxymethylation mediated by intestinal bacteria and P450s enzymes and that of glucuronidation and sulfation mediated by uridine diphosphoglucuronyl transferases and sulfotransferases. There are three glucuronidation, two sulfation, and one dehydroxymethylation products in metabolites of ginsenoside Rk1. There are five glucuronidation, one sulfation, two dihydroxylation, and one dehydroxymethylation products in metabolite of ginsenoside Rg5. There are only two and three metabolites that were not deglycosylated in metabolites of ginsenosides Rk1 and Rg5, respectively. The information above can provide for research on the metabolic mechanism of ginsenosides Rk1 and Rg5 in mammals. The foundation can also provide for further verifying and clarifying metabolism law of ginsenosides Rk1 and Rg5 in vivo and play an important role on their effects.
After 24 h metabolism in zebrafish, the chromatography and mass spectrum signal of product of loss of glucose at position C-3 and glucuronidation at position C-12 were powerful. The result indexes that loss of glucose and glucuronidation is the main physiological process of metabolism of ginsenosides Rk1 and Rg5 in zebrafish. In addition, content estimate of ginsenoside Rk1 in solution sample of zebrafish is more than that of ginsenoside Rg5, which indicates that the metabolic rate of ginsenosides Rk1 is slower than that of ginsenoside Rg5. The possible reason may be that the position of C-20 double bond is altered, which leads to distinction of solubility and permeability between ginsenoside Rk1 and ginsenoside Rg5.
In conclusion, UPLC-Q-TOF/MS was applied to the complicated metabolites of ginsenosides Rk1 and Rg5 in zebrafish, which provided an accurate detection method. For the first time, totals of four and seven metabolites of ginsenosides Rk1 and Rg5 were detected and tentatively identified from the zebrafish body and solution samples of zebrafish, respectively. From the experimental results, we found that the main Phase II metabolism of ginsenosides Rk1 and Rg5 in zebrafish was glucuronidation reactions. The possible metabolic pathways of ginsenosides Rk1 and Rg5 were subsequently offered. We speculated on the percent of mass peak area that ginsenoside Rk1-12-O-glucuronate was the predominant metabolite of ginsenoside Rk1, and ginsenoside Rg5-12-O-glucuronate was the predominant metabolite of ginsenoside Rg5. Although the structures of metabolites cannot be measured conclusively by UPLC-Q-TOF/MS alone, the current method is always very worthy and reliable for the further investigation of the metabolism of ginsenosides Rk1 and Rg5. In addition, we discovered that the metabolic velocity of ginsenosides Rg5 is faster than that of ginsenoside Rk1 on account of the difference of C-20 double bond. This research offers a new model of exploring the drug metabolism, and also provides a detection method for identifying the metabolites.
Conflicts of interest
The authors declare no competing financial interest.
Acknowledgments
This work was supported by the Natural Science Foundation of China (Nos. 81102812, and 30973978) and the Natural Science Foundation of Jiangsu Province of China (BK2011866), and Jiangsu Key Laboratory of New Drug Research and Clinical Pharmacy opening topic fund (KF-XY201506).
References
- 1.Lee M.R., Yun B.S., Sung C.K. Comparative study of white and steamed black Panax ginseng, P. quinquefolium, and P. notoginseng on cholinesterase inhibitory and antioxidative activity. J Ginseng Res. 2012;36:93–101. doi: 10.5142/jgr.2012.36.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong T.T., Cui X.M., Song Z.H., Zhao K.J., Ji Z.N., Lo C.K., Tsim K.W. Chemical assessment of roots of Panax notoginseng in China: regional and seasonal variations in its active constituents. J Agric Food Chem. 2003;51:4617–4623. doi: 10.1021/jf034229k. [DOI] [PubMed] [Google Scholar]
- 3.Chan E.C., Yap S.L., Lau A.J., Leow P.C., Toh D.F., Koh H.L. Ultra-performance liquid chromatography/time-of-flight mass spectrometry based metabolomics of raw and steamed Panax notoginseng. Rapid Commun Mass Spectrom. 2007;21:519–528. doi: 10.1002/rcm.2864. [DOI] [PubMed] [Google Scholar]
- 4.Chen B., Cai T., Jia X.B. Simultaneous determination of ten active ginsenosides in steamed notoginseng by UPLC. Chin J Chin Material Media. 2014;39:1614–1619. [PubMed] [Google Scholar]
- 5.Sun S., Wang C.Z., Tong R., Li X.L., Fishbein A., Wang Q., He T.C., Du W., Yuan C.S. Effects of steaming the root of Panax notoginseng on chemical composition and anticancer activities. Food Chem. 2010;118:307–314. [Google Scholar]
- 6.Toh D.F., New L.S., Koh H.L., Chan E.C. Ultra-high performance liquid chromatography/time-of-flight mass spectrometry (UHPLC/TOFMS) for time-dependent profiling of raw and steamed Panax notoginseng. J Pharm Biomed Anal. 2010;52:43–50. doi: 10.1016/j.jpba.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 7.Wang D., Liao P.Y., Zhu H.T., Chen K.K., Xu M., Zhang Y.J., Yang C.R. The processing of Panax notoginseng and the transformation of its saponin components. Food Chem. 2012;132:1808–1813. [Google Scholar]
- 8.Bao H.Y., Zhang J., Yeo S.J., Myung C.S., Kim H.M., Kim J.M., Park J.H., Cho J., Kang J.S. Memory enhancing and neuroprotective effects of selected ginsenosides. Arch Pharm Res. 2005;28:335–342. doi: 10.1007/BF02977802. [DOI] [PubMed] [Google Scholar]
- 9.Lee K.Y., Lee Y.H., Kim S.I., Park J.H., Lee S.K. Ginsenoside-Rg5 suppresses cyclin E-dependent protein kinase activity via up-regulating p21Cip/WAF1 and down-regulating cyclin E in SK-HEP-1 cells. Anticancer Res. 1997;17:1067–1072. [PubMed] [Google Scholar]
- 10.Lee J.G., Lee Y.Y., Kim S.Y., Pyo J.S., Yun-Choi H.S., Park J.H. Platelet antiaggregating activity of ginsenosides isolated from processed ginseng. Pharmazie. 2009;64:602–604. [PubMed] [Google Scholar]
- 11.Liu J.W., Tian S.J., de Barry J., Luu B. Panaxadiol glycosides that induce neuronal differentiation in neurosphere stem cells. J Nat Prod. 2007;70:1329–1334. doi: 10.1021/np070135j. [DOI] [PubMed] [Google Scholar]
- 12.Almeida D.V., Nornberg B.F., Geracitano L.A., Barros D.M., Monserrat J.M., Marins L.F. Induction of phase II enzymes and hsp70 genes by copper sulfate through the electrophile-responsive element (EpRE): insights obtained from a transgenic zebrafish model carrying an orthologous EpRE sequence of mammalian origin. Fish Physiol Biochem. 2010;36:347–353. doi: 10.1007/s10695-008-9299-x. [DOI] [PubMed] [Google Scholar]
- 13.Bresolin T., de Freitas Rebelo M. Celso Dias Bainy A. Expression of PXR, CYP3A and MDR1 genes in liver of zebrafish. Comp Biochem Physiol C Toxicol Pharmacol. 2005;140:403–407. doi: 10.1016/j.cca.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 14.Thompson E.D., Burwinkel K.E., Chava A.K., Notch E.G., Mayer G.D. Activity of Phase I and Phase II enzymes of the benzo[a]pyrene transformation pathway in zebrafish (Danio rerio) following waterborne exposure to arsenite. Comp Biochem Physiol C Toxicol Pharmacol. 2010;152:371–378. doi: 10.1016/j.cbpc.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 15.Lü A.J., Yang Z.X., Liu H., Hu X.C., Zhang Y.H., Cheng C. Isolation of intestinal bacteria from zebrafish (Danio rerio) and their physiological and biochemical characteristics. Chin Agric Sci Bull. 2010;26:412–415. [Google Scholar]
- 16.Goldsmith P. Zebrafish as a pharmacological tool: the how, why and when. Curr Opin Pharmacol. 2004;4:504–512. doi: 10.1016/j.coph.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 17.Liang A. Zebrafish—useful model for pharmacodynamics and toxicity screening of traditional Chinese medicine. Chin J Chin Material Media. 2009;34:2839–2842. [PubMed] [Google Scholar]
- 18.Sukardi H., Chng H.T., Chan E.C., Gong Z., Lam S.H. Zebrafish for drug toxicity screening: bridging the in vitro cell-based models and in vivo mammalian models. Expert Opin Drug Metab Toxicol. 2011;7:579–589. doi: 10.1517/17425255.2011.562197. [DOI] [PubMed] [Google Scholar]
- 19.Kitambi S.S., Nilsson E.S., Sekyrova P., Ibarra C., Tekeoh G.N., Andang M., Ernfors P., Uhlen P. Small molecule screening platform for assessment of cardiovascular toxicity on adult zebrafish heart. BMC Physiol. 2012;12:3–9. doi: 10.1186/1472-6793-12-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Q.X., Zeng S. Research progress of zebrafish used in drug metabolism. Acta Pharm Sin. 2011;46:1026–1031. [PubMed] [Google Scholar]
- 21.Wei Y.J., Jia X.B., Zhan Y., Wang C.M., Chen B. Metabolism study of Chrysin by zebrafish. Chin Pharm J. 2013;48:565–568. [Google Scholar]
- 22.Wei Y.J., Wang C.M., Zhan Y., Chen B., Sun E., Jia X.B. Reasonability of 7-hydroxyflavone metabolism in Zebrafish. Chin New Drugs J. 2013;22:1078–1082. [Google Scholar]
- 23.Wei Y.J., Zhan Y., Wang C.M., Chen B., Jia X.B. Metabolism of 5-hydroxyflavone in model organism zebrafish. Chin JMAP. 2013;30:461–465. [Google Scholar]