Abstract
The development of immune responses is influenced by the interaction between environmental and genetic factors. Our previous study showed a close association between maternal and young infant's cytokine responses. The question is how this association evolves over time and the contribution of genetic polymorphisms to this association. Five cytokines in mitogen-stimulated whole blood culture were measured from pregnant mothers and their children aged 2, 5, 12, 24 and 48 months. Cytokine gene polymorphisms were determined in both mothers and children. High production of maternal interleukin (IL)-10, tumour necrosis factor-α (TNF-α) and interferon-γ (IFN-γ) was significantly associated with higher levels of the corresponding cytokines in their children at 2 months (T2), but the association decreased over time. Maternal single-nucleotide polymorphism (SNP) in IFN-γ gene, rs3181032, was found to be associated with child's IFN-γ levels at T2 only, whereas maternal IL-10 rs4579758 and child's TNF-α rs13215091 were associated with child's corresponding cytokines at later ages but not at T2. In the final models including the gene polymorphisms, maternal cytokines were still the strongest determinant of child cytokines. Maternal cytokine during pregnancy, which could be a proxy for child's environmental factors, showed its highest impact at early age, with no or little influence from genetic factors.
Introduction
There has been an increasing number of studies looking at immune responses in early life and its modulation by environmental factors, which may predispose an individual to certain diseases in later life. Several studies have shown parallels in cellular immune responses between mothers and their children.1, 2, 3, 4, 5 The mechanisms behind this have not been elucidated. Moreover, all these studies were conducted in developed countries, where infectious diseases are better controlled, while allergic and autoimmune diseases are increasingly affecting the population.6 Specifically, the relationship between in utero exposure, early immune profiles after birth and the development of immune responses in early childhood has not been examined comprehensively in populations where chronic parasitic infections are endemic. In this regard, our previous study in a helminth-endemic area found that maternal cytokines (interleukin (IL)-10 and interferon-γ (IFN-γ)) were associated with infant's cytokines at the age of 2 months, even after taking into account several environmental factors including maternal parasitic infections,7 in agreement with the findings from the studies in industrialized countries where maternal and child cytokine responses appear to be tightly linked.1, 2, 3, 4, 5
Interaction between environment and genetic factors will determine the phenotypic outcome of an individual. To our knowledge, there has been no study looking at the changing pattern of the mother–child cytokine relationship over time and whether genetic factors can modify this relationship. To investigate this, we measured child's cytokine responses to mitogen at five time points, starting around 2 months and up to 4 years of age, and examined the relationship with maternal cytokine responses during pregnancy. We genotyped single-nucleotide polymorphisms (SNPs) from mothers and children and asked whether they modified the cytokine relationship of mother and child.
Results
Characteristics of mother–child pairs
The flow diagram of the entire study is described elsewere.8 We included 119 mothers and children in the present study, with 54% (64/119) children being males. Among 107 children with data on breastfeeding, 83 children (78%) received breastfeeding exclusively for 6 months, followed by 21 (19%) children who were partially breastfed (mixed with formula milk) and 3 (3%) children who were not breastfed. Although all 119 mother and child pairs had genotype data, the number of children with cytokine data was different at each time point: 111 at T2, 95 at T5, 90 at T12, 88 at T24 and 86 at T48. The median maternal cytokine levels in response to phytohaemmaglutinin (PHA) were used to classify a mother as a high or low cytokine producer (see legend to Figure 1).
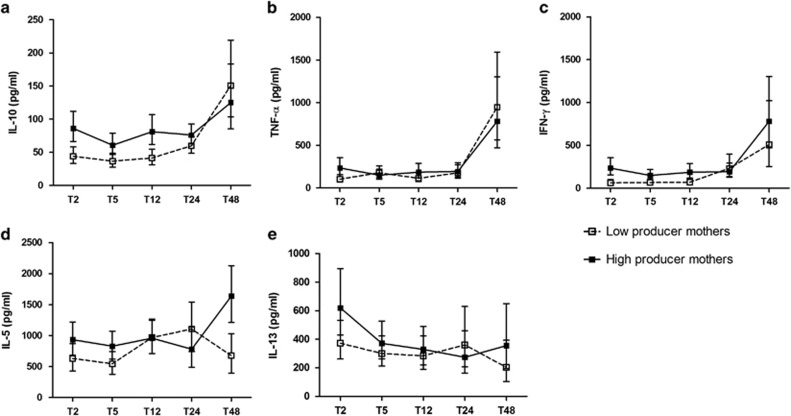
Figure 1.
Child's PHA-induced cytokines over time, based on maternal cytokine producer status during pregnancy. Error bars represent geometric means of (a) IL-10, (b) TNF-α, (c) IFN-γ, (d) IL-5 levels and mean of square of (e) IL-13 levels of children at the age of 2 months (T2), 5 months (T5), 12 months (T12), 24 months (T24) and 48 months (T48). Closed dots are children born to high producer mothers, whereas open dots are children born to low producer mothers. The median of maternal cytokines is used to divide high or low producer mothers: IL-10 (91.7, interquartile range (IQR): 46.9–167.1 pg ml−1), TNF-α (274.2, IQR: 54.7–915.6 pg ml−1), IFN-γ (152.5, IQR: 43.7–425.5 pg ml−1), IL-5 (859.8, IQR: 295.0–1673.8 pg ml−1) and IL-13 (243.5, IQR: 75.7–627.2 pg ml−1). All values were adjusted for mother and child's gene polymorphism, village of residence and child's gender.
Minor allele frequency and P-values for testing the null hypothesis that Hardy–Weinberg equilibrium holds for all SNPs of the mother–child pairs are shown in the Supplementary Table S1. All genotyped SNPs were in Hardy–Weinberg equilibrium with the exception of the two IL-5 gene polymorphisms, rs4143832 and rs17690122 (both in perfect linkage disequilibrium (LD)/r2=1), which slightly deviated from Hardy–Weinberg equilibrium (P=0.039). The other SNPs that showed perfect LD were rs1878672 and rs1800896 in IL-10 gene. Several SNPs were in high LD (r2>0.8), such as between rs10494879 and rs1878672 or rs1800896 in the IL-10 gene, between rs10878763 and rs10784683 in the IFN-γ gene, between rs2040704 in RAD50 gene and rs1800925 in IL-13 gene, between rs1881457 and rs1800925, and between rs1295686 and rs20541 in IL-13 gene (Supplementary Table S1). Altogether, there were six pairs of SNPs in perfect LD or high LD, which caused similar results in the analyses.
Association between maternal and child cytokines over time
Table 1 shows that high cytokine producer mothers were associated with higher child cytokine production in response to PHA at 2 months of age (T2), this is true for IL-10 (estimate: 0.31; 95% confidence interval (95% CI): 0.19, 0.43), IFN-γ (estimate: 0.26; 95% CI: 0.03, 0.49) and, to a lesser extent, for tumour necrosis factor-α (TNF-α estimate: 0.19; 95% CI: 0.02, 0.36). With increasing age, the mother–child relationship for IL-10 disappeared (interaction between maternal IL-10 with time, P=0.002), whereas for TNF-α and IFN-γ the relationship did not change significantly. The production of TH2-type cytokines, IL-5 and IL-13, were not significantly associated between mother and child at baseline or over time (Table 1).
Table 1. Effect of maternal cytokine on child's cytokine production over time.
| Estimate (95% CI)a | P-value | |
|---|---|---|
| IL10 | ||
| Maternal cytokine | 0.31 (0.19, 0.43) | <0.0001 |
| Time maternal cytokineb | −0.009 (−0.01, −0.003) | 0.002 |
| TNF-α | ||
| Maternal cytokine | 0.19 (0.02, 0.36) | 0.030 |
| Time maternal cytokineb | −0.006 (−0.01, 0.002) | 0.146 |
| IFN-γ | ||
| Maternal cytokine | 0.26 (0.03, 0.49) | 0.027 |
| Time maternal cytokineb | −0.009 (−0.02, 0.002) | 0.093 |
| IL-5 | ||
| Maternal cytokine | 1.51 (−2.58, 5.60) | 0.466 |
| Time maternal cytokineb | 0.15 (−0.04, 0.34) | 0.121 |
| IL-13 | ||
| Maternal cytokine | 0.07 (−0.08, 0.23) | 0.348 |
| Time maternal cytokineb | 0.0002 (−0.008, 0.009) | 0.954 |
Abbreviations: CI, confidence interval; IFN-γ, interferon-γ IL, interleukin; TNF-α, tumor necrosis factor-α.
The estimates shows associations between maternal and child's cytokines at 2 months of age.
Bold P-value <0.05.
Adjusted for child gender.
Interaction between time and maternal cytokine, showing the change of association between maternal and child's cytokines over time. Time units are 2, 5, 12, 24 and 48 months.
Association between maternal cytokines or genetic factors and child's cytokines over time
IL-10 and TNF-α
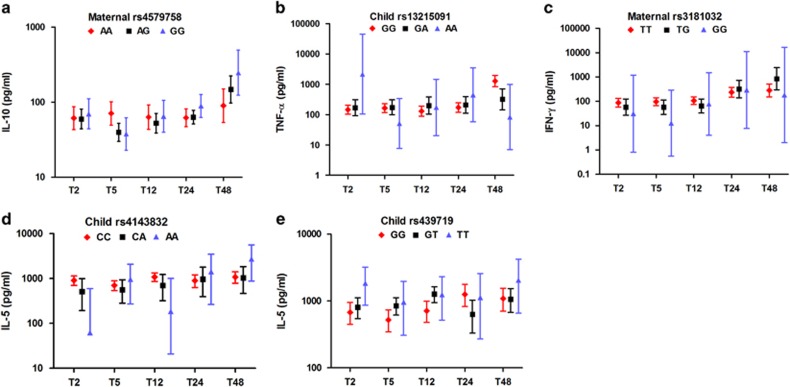
The estimated effect of maternal IL-10 and maternal or child's genotype on child's IL-10 production in response to PHA is shown in Table 2. Here, only genotypes that revealed significant associations are presented. In this model, at 2 months of age the estimates for mother–child cytokine relationship were similar to the estimates in the previous model without genetic factors (Table 2 and Figure 1a; estimate: 0.31; 95% CI: 0.18, 0.43), indicating that the variants tested here did not explain the relationship between maternal and child cytokines. In contrast to maternal cytokine status, neither maternal nor child's IL-10 genotypes were associated with child's cytokine at T2. However, at later time points we observed a tendency for children born to mothers with more G allele in rs4579758 to have significantly higher production of IL-10 over time (Table 2 and Figure 2a). When formally tested in a dominant model, the contribution of maternal genotype at this locus on the child's IL-10 levels over time was significant. However, the child's genotype had no effect on IL-10 production over time.
Table 2. Effect of maternal cytokine, maternal and child's genotype on child's PHA-induced cytokine production over time.
| Child cytokine | Variable | Estimate (95% CI)a | P-value |
|---|---|---|---|
| IL10 | Maternal cytokine | 0.31 (0.18, 0.43) | <0.0001b |
| rs4579758 | Maternal genotype | −0.05 (−0.16, 0.05) | 0.321 |
| Child's genotype | −0.009 (−0.11, 0.10) | 0.866 | |
| Time maternal cytokinec | −0.008 (−0.01, −0.003) | 0.004 | |
| Time maternal genotypec | 0.005 (0.0002, 0.01) | 0.039 | |
| Time child's genotypec | 0.001 (−0.003, 0.006) | 0.616 | |
| TNF-α | Maternal cytokine | 0.20 (0.03, 0.37) | 0.024 |
| rs13215091 | Maternal genotype | −0.005 (−0.21, 0.20) | 0.961 |
| Child's genotype | 0.17 (−0.02, 0.37) | 0.080 | |
| Time maternal cytokinec | −0.006 (−0.01, 0.001) | 0.108 | |
| Time maternal genotypec | 0.004 (−0.005, 0.01) | 0.354 | |
| Time child's genotypec | −0.01 (−0.02, −0.004) | 0.005 | |
| IFN-γ | Maternal cytokine | 0.26 (0.04, 0.48) | 0.023 |
| rs3181032 | Maternal genotype | −0.30 (−0.58, −0.01) | 0.039 |
| Child's genotype | 0.26 (−0.10, 0.61) | 0.152 | |
| Time maternal cytokinec | −0.009 (−0.02, 0.001) | 0.077 | |
| Time maternal genotypec | 0.01 (0.002, 0.03) | 0.024 | |
| Time child's genotypec | −0.0008 (−0.02, 0.01) | 0.919 | |
| IL-5 | Maternal cytokine | 1.46 (−2.59, 5.51) | 0.476 |
| Rs4143832 | Maternal genotype | 4.32 (−1.27, 9.91) | 0.129 |
| Child's genotype | −4.93 (−9.60, −0.27) | 0.038 | |
| Time maternal cytokinec | 0.17 (−0.02, 0.36) | 0.075 | |
| Time maternal genotypec | −0.24 (−0.49, 0.02) | 0.066 | |
| Time child's genotypec | 0.14 (−0.08, 0.35) | 0.210 | |
| IL-5 | Maternal cytokine | 1.02 (−3.07, 5.10) | 0.622 |
| Rs739719 | Maternal genotype | −2.10 (−5.76, 1.56) | 0.259 |
| Child's genotype | 4.17 (0.12, 8.23) | 0.044 | |
| Time maternal cytokinec | 0.14 (−0.05, 0.34) | 0.143 | |
| Time maternal genotypec | −0.02 (−0.19, 0.15) | 0.799 | |
| Time child's genotypec | −0.02 (−0.21, 0.17) | 0.849 |
Abbreviations: CI, confidence interval; IFN-γ, interferon-γ IL, interleukin; TNF-α, tumor necrosis factor-α.
Time units are 2, 5, 12, 24 and 48 months. Bold P-values <0.05.
The estimates shows associations between maternal cytokines or genotypes and child's genotypes at with child's cytokine production at 2 months of age.
Adjusted for child gender.
Significant after Bonferroni's correction (P<0.0028).
Interaction between time and maternal cytokine/genotype or child's genotype, showing the change of association between maternal cytokine/genotype or child's genotype with child's cytokine production over time.
Figure 2.
Child's PHA-induced cytokines over time, based on maternal or child genotypes. Only SNPs that showed significant associations in the multivariate models are shown. Error bars represent geometric means of (a) IL-10, (b) TNF-α, (c) IFN-γ and (d and e) IL-5 levels of children at the age of 2 months (T2), 5 months (T5), 12 months (T12), 24 months (T24) and 48 months (T48). Red dots/interrupted lines indicate the children having or born to mothers with major homozygote alleles, whereas black dots/solid lines for heterozygotes and blue dots and solid lines for minor homozygotes. All values were adjusted for maternal cytokines, village of residence and child's gender.
In a similar manner to IL-10 but weaker, the production of the child's PHA-induced TNF-α at 2 months of age was associated with maternal TNF-α production (Table 2 and Figure 1b; estimate: 0.20, 95% CI: 0.03, 0.37) and this association decreased over time. Although at the beginning there was a positive trend for association between polymorphism of child's rs13215091 and TNF-α levels (estimate: 0.17; 95% CI: −0.02, 0.37), the direction of association reversed over time with significantly stronger effect exerted by child's decreasing number of minor allele (estimate: −0.01; 95% CI: −0.02, −0.004; Table 2 and Figure 2b). For the other three SNPs of TNF-α, maternal and child's genotype did not show significant associations with child's cytokine at baseline and over time (data not shown).
IFN-γ, IL-5 and IL-13
The production of TH1-type responses (IFN-γ) in response to PHA in children after adjustment for genotypes was associated with maternal cytokine producer status at baseline (Table 2 and Figure 1c; estimate: 0.26; 95% CI: 0.04, 0.48) and this association did not change over time (estimate: −0.009; 95% CI: −0.02, 0.001). Among six IFN-γ polymorphisms, the only genotype found to be significantly associated with child's cytokine was rs3181032 of mother, which is an IFN-γ gene polymorphism located in the promoter region. Mothers with increasing number of this SNP's minor allele were more likely to have children producing lower IFN-γ levels at T2 (Table 2 and Figure 2c; estimate: −0.30; 95% CI: −0.58, −0.01); however, over time the direction of the association reversed (estimate: 0.01; 95% CI: 0.002, 0.03).
Among all genotyped IL-5 SNPs, only two of child's IL-5 polymorphisms, rs4143832 and rs739719, were associated with child's cytokine production at T2 (Table 2 and Figure 2d and e; estimate: −4.93; 95% CI: −9.60, −0.27 and estimate: 4.17; 95% CI: 0.12, 8,23, respectively) with no significant difference of slopes with time. None of IL-13 or RAD50 gene polymorphisms were associated with child's IL-13 or with IL-5 and IL-13 levels, respectively (data not shown).
Discussion
Our study is the first to examine longitudinally, at five different time points from 6 weeks to 4 years of age, the association between cytokine production of a pregnant mother and her child in Indonesia, a developing country. In the present study, we show that a number of variants tested at cytokine candidate genes do not explain the strong association between maternal cytokine production during pregnancy with child's cytokine production in the first year of life. We also found that most of the mother–child cytokine relationships became weaker over time (IL-10, IFN-γ and TNF-α). These findings indicate that the strong association between cytokine responses of a pregnant mother and her child is not directly due to genetic factors but could likely result from similar immune conditioning during gestational period extending into early childhood. This was different for IL-5. The association between maternal and child IL-5 production, which was not significant in the first 2 years of age, became significant when the child reached 4 years of age.
Earlier studies had found associations in cytokine production between mother (pre- or postpartum) and child at one or two time points, such as at birth in cord blood,1, 2, 5 at a time when infant was 3 months,3 1 year5, 9 or 2 years of age.4 The mother–child cytokine relationship was not always apparent directly after birth. For example, the birth cohort study by Halonen et al.3 showed no correlation of mitogen-stimulated IFN-γ and IL-13 between a pregnant mother and fetus, but instead with the child at 3 months of age. Similarly, in another study with the lipopolysaccharide-induced IL-10 and TNF-α production, there was a significant association between pregnant mother and the child at the age of 1 year but not with cord blood.9 Our results are in agreement with the previous two studies in that maternal IL-10 or IFN-γ production during pregnancy was positively associated with child's corresponding cytokines up to 1 year of age but with longer observation, the association was no longer present (Figure 1). This particular finding regarding IL-10 or IFN-γ may reflect intra uterine and nursing effect on child's developing immune system. Although no trans-placental transfer of maternal cytokines in humans are believed to occur,10 the components of uterine microenvironment may modulate the fetal naive immune cells. This might explain why maternal IL-10 during pregnancy showed the strongest association with child's IL-10. After birth, the maturating immune system of infants is believed to get compensations from breast milk, which contains maternal humoral and cellular immune components including cytokines, chemokines and immune cells. As the majority (97%) of the infants/children in our study was breastfed, breast milk may also contribute to the transfer of maternal immunological information to infants in this population. Both IL-10 and IFN-γ are present in breast milk11, 12 In the case of IL-10 this is thought to be a continuation of immune regulation during gestation, to avoid rejection of fetal allograft, whereas for IFN-γ the nursing can be the best way to complement the infant's immune system whose capacity to produce TH1-type responses are less than in adults.13 Interestingly, high levels of TNF-α are present in early milk but become almost undetectable after 1 month, whereas IL-10, IFN-γ, IL-5 and IL-13 tend to maintain certain levels in mature milk (reviewed in Agarwal et al.12). This might explain the weak association we observed in TNF-α production between mother and child in the first year of life.
The pattern of mother–child relationship in IL-5 responses was different from other cytokines, in that the significant association was found only when the children reached 4 years of age. There are no other studies examining IL-5 in this context and one possibility to explain our observation might be the fact that the study area is endemic for helminth infections. As children grow older they increasingly become infected with helminths,14 which drive TH2 responses, and therefore the closer association between maternal and child IL-5 at 4 years of age might reflect the sharing of helminth-infected environment with the mother.
It appears that at early age, the capacity of infant's immune cells to produce pro- and anti-inflammatory cytokines (TNF and IL10, respectively) is more influenced by maternal cytokines than by the genetic variation tested, which seem to have more influence at later age. On the other hand, we found that the production of TH1-type cytokine at early age was independently associated with maternal cytokine and gene polymorphisms. Previous studies in adult twins showed that cytokines can have a low to high proportion of heritability, ranging from 30 to 75% for IL-10,15, 16, 17 40 to 85% for IFN-γ15, 18 and 17 to 80% for TNF-α.15, 16, 17, 18, 19 The participants of these studies were adults. Our cohort study is unique in showing the effect of maternal cytokines on child cytokines in early age may overrule or mask the genetic effect during the maturation of child immune responses. This notion is supported by the finding in a twin study, which showed that the genetic effect on serum TNF-α increased with age.17 In the case of IL-10, at the later age (T48), when the association with maternal cytokine weakened, the pattern of IL-10 production from children born to rs4579758-AA mothers did not increase as much as those born to AG/GG mothers. It is known that a child inherits half maternal genes; therefore, it is not necessary that the child will have the same alleles as the mother. On the other hand, there were several studies showing that maternal gene polymorphisms may affect child's response through non-genetic factor, but epigenetic programming via exposure of child in utero or early life to maternal cytokines. The function of child's cytokine gene polymorphism may appear later in life, which may overcome the weakening impact of maternal cytokine. The fact that in the present study maternal genotype contributed more to child's IL-10 production than the child's own genotype may indicate that maternal genetic factors could have their effect on child's cytokines not necessarily through genetic inheritance but through interaction with the environmental factors (epigenetic programming), via in utero effects, and/or breastfeeding. Indeed, previous studies found such interactions between maternal genotype and environment on child's immune outcomes.20
Regarding the IL-10 SNP of mother (rs4579758) that showed significant association with child's cytokine, so far no other studies have shown any functional consequences of this SNP in association with cytokine production or disease. Nevertheless, this SNP was found in high LD (r2=0.97 for Asian) with another SNP, rs6692511. The latter SNP is located in the site of DNase I hypersensitivity clusters and transcription factors, which are associated with regulation of gene expression (http://genome-euro.ucsc.edu) and therefore could contribute to our observation regarding IL-10.
It is important to note that the nonsignificant associations between child cytokine production with SNPs detected in this study might also be caused by other unmeasured SNPs that were located in a larger distance but still in LD. We realize that using single-nucleotide analysis in this study may not entirely represent the genetic effect on child's cytokine responses, as there are unmeasured genetic variations besides the measured SNPs, which may reveal a cytokine relationship between mother and child, such as copy number variation, haplotype, microsatellite alleles as well as epigenetic processes (DNA methylation, histone modifications and small non-coding RNA). Nevertheless, using the tagging SNPs we expected to limit the number of SNPs to be tested by covering those not genotyped, which were in high LD with the tagging SNPs in the same gene.21
As the aim of the study was more focused on mother–child cytokine relationship, we consider the SNP analysis in relation to the child' cytokine responses exploratory. Therefore, the weak associations (P<0.05) found between genotypes and cytokine production before correction for multiple testing were not considered immediately as not significant. The confirmation of the gene association results would need replication with larger sample size in similar population/race. It would also be interesting to investigate whether the effect of gene polymorphism seen at 4 years of age will be maintained at older age; for example, the children born to mothers with AA genotype of rs4579758 will continue to have a stabile lower IL-10 production compared with those born to AG/GG mothers or will they eventually catch up.
In conclusion, the close relationship of mother and child cytokine production, especially IL-10 and IFN-γ, was prominent in early life, before 1 year of age. This immunological relationship appeared to be independent of cytokine gene polymorphisms, suggesting that infant's cytokine responses were more influenced by the environment shared with the mother during intra uterine and breastfeeding period. Different cytokines can have different interaction with maternal cytokine and maternal/child genetic factors at certain time points, depending on the maturation of child immune responses and the challenges from the environment. Furthermore, whether the cytokine profile of children born to high or low producer mother is associated with clinical outcomes or only reflects physiological variation needs to be investigated.
Subjects and methods
Study population
This study is part of the longitudinal study of children living in a peri-urban area in Bekasi Distric, West Java province, Indonesia. Children were followed up at five time points: 2 (T2), 5 (T5), 12 (T12), 24 (T24) and 48 (T48) months of age. Among all participants, there were 126 pairs of pregnant mothers in second or third trimester and their children who had both cytokine measurements and genotyping data. All mothers provided written informed consent for themselves and their children. This study was approved by the Ethical Committee Faculty of Medicine Universitas Indonesia.
Whole-blood culture and cytokine measurement
Whole-blood culture and cytokine measurement were performed as described previously.7 Briefly, heparinized venous blood was diluted 1:10 with RPMI-1640 medium (added with 1 mM pyruvate and 2 mM glutamate), followed by incubation with PHA (2 μg ml−1; Wellcome Diagnostics, Dartford, UK) in 37 °C and 5% CO2. The concentrations of IL-10 and TNF-α were measured in day 1 supernatant, whereas IL-5, IL-13 and IFN-γ were measured in day 6 supernatant. Supernatants were kept frozen at −20 °C and later on were thawed for the measurement with in-house multiplex bead-based assay (Luminex IS 100, Luminexcorp, Austin, TX, USA) for IL-10, TNF-α, IL-13 and IFN-γ, whereas enzyme-linked imminosorbent assay was performed for measurement of IL-5. The detection limits for IL-10, TNF-α, IL-13, IFN-γ and IL-5 were 6.5, 1.7, 12.5, 3.6 and 2 pg ml−1, respectively. All cytokine levels below detection limit were given half of the threshold value.
DNA purification and genotyping
Genomic DNA was purified from 200 μl of whole-blood samples from pregnant mothers and their children, which was kept frozen at −20 °C, using QIAamp DNA Mini kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. The purified DNA was quantified using NanoDrop 1000 Spectrophotometer (Thermo Scientific, Wilmington, DE, USA).
The source of cytokine gene polymorphisms for Indonesians living in West Java were derived from Malays population in Singapore Variation Genome Project database (http://www.statgen.nus.edu.sg/~SGVP),22 with some additional SNPs, which were not found in the database but were associated with phenotypes in Asian or other populations. A set of gene polymorphisms spanning from 20 kb upstream and 10 kb downstream from each cytokine gene region were included in the genotyping. Pairwise tagging SNPs were obtained from Haploview's Tagger program (http://www.broad.mit.edu/mpg/tagger), with selection based on minor allele frequency ⩾5% and r2 threshold=0.8. All SNPs selected with this method were included in the analysis, including those SNPs that have not been found to be functional or associated with diseases.
In total, there were 10 SNPs for IL-10, 4 SNPs for TNF-α, 6 SNPs for IFN-γ, 6 SNPs for IL-5 and 8 SNPs for IL-13. In addition to these cytokine genes, 4 SNPs of RAD50 genes were added, as they occupy the region between IL-4, IL-5 and IL-13 genes (TH2 cytokine locus) in chromosome 5q31. RAD50 gene encodes a DNA repair enzyme; it was recently found to have locus control region at its 3′-end and is shown to be associated with asthma and eczema.23, 24 Genotyping of all SNPs were performed using Sequenom MassARRAY iPLEX Platform (Sequenom, Inc., CA, USA). Quality control was performed by including ±10% of successfully genotyped samples and positive controls in the repeated measurement of failed samples. At the end, all SNPs were genotyped successfully with call rates ⩾90%. After exclusion of two mother–child pairs, who had call rates <95%, and five pairs with Mendelian inconsistencies, data were available for a total of 119 pairs.
Statistical analysis
Cytokine levels displayed skewed distributions, therefore log (base 10) transformation was used for all cytokines, except for IL-5, for which a square-root transformation was used. To investigate the association between mother and child's cytokine productions, we divided maternal cytokines based on median levels into high or low producer mothers7 and by using this category we compared child cytokine levels at each time point. Minor allele frequency, deviations from Hardy–Weinberg equilibrium and pairwise LD were calculated for each mother or child's SNP using Haploview software (http://www.broadinstitute.org).
First, we modeled the association between maternal cytokines and child cytokines. Next, we modeled the maternal cytokines together with maternal or child genotype. We used linear mixed models to study the effect of maternal cytokines and maternal or child genotypes over time on child cytokines. Using a likelihood ratio test, we tested whether the simpler first-order autoregressive heterogenous (for IL-10 and TNF-α) structrure could be used to model correlation over time instead of the unstructured (for IFN-γ, IL-5 and IL-13) covariance structure. Each genotype was coded 0, 1 and 2 for the increasing number of minor alleles and all SNPs were tested using an additive genetic model. First, we used model with main effects and one interaction term between time and SNP (that is, linear change over time) for all SNPs. For significant SNPs, we continued with a larger model where time was included as a categorical variable to obtain more insight of the effect of the SNP over time. Bonferroni corrections were done for multiple testing, where a P-value was considered significant if <0.0028. All models were adjusted for child gender as an a priori factor for child's cytokine responses.15 The statistical analyses were performed using IBM SPSS version 20 (International Business Machines Corporation, Chicago, IL, USA).
Acknowledgments
We are very grateful to all families for their participation in the study and local health staff of Puskesmas Jati Sampurna for their assistance in blood collections. We also thank venipuncturists Sudirman and Heru, Erlinda Marthias for cytokine measurement and Dennis Kremer for his expertise and help in genotyping. This study was supported by The Netherlands Foundation for the Advancement of Tropical Research (W93-364 and W93-468), European Commission, Sixth Framework Programme (MEST-CT-2005-020524-GALTRAIN), Royal Netherlands Academy of Arts and Sciences (Ref.KNAW-05-PP-35) and Directorate of Research and Community Services University of Indonesia (Hibah Kompetensi untuk Publikasi di Jurnal Internasional Terindeks Universitas Indonesia No.2527/UN2.R12/HKP.05.00/2016).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Genes and Immunity website (http://www.nature.com/gene)
Supplementary Material
References
- Prescott SL, Taylor A, Roper J, Wahdan A, Noakes P, Thornton C et al. Maternal reactivity to fetal alloantigens is related to newborn immune responses and subsequent allergic disease. Clin Exp Allergy 2005; 35: 417–425. [DOI] [PubMed] [Google Scholar]
- Tse DB, Young BK. Co-ordinate expression of Th1/Th2 phenotypes in maternal and fetal blood: evidence for a transplacental nexus. J Perinat Med 2012; 40: 165–170. [DOI] [PubMed] [Google Scholar]
- Halonen M, Lohman IC, Stern DA, Spangenberg A, Anderson D, Mobley S et al. Th1/Th2 patterns and balance in cytokine production in the parents and infants of a large birth cohort. J Immunol 2009; 182: 3285–3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson AK, Nilsson C, Hoglind A, Sverremark-Ekstrom E, Lilja G, Troye-Blomberg M. Relationship between maternal and child cytokine responses to allergen and phytohaemagglutinin 2 years after delivery. Clin Exp Immunol 2006; 144: 401–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen M, Roponen M, Pekkanen J, Huttunen K, Hirvonen MR. Maturation of cytokine-producing capacity from birth to 1 yr of age. Pediatr Allergy Immunol 2009; 20: 714–725. [DOI] [PubMed] [Google Scholar]
- Bach JF. The effect of infections on susceptibility to autoimmune and allergic diseases. N Engl J Med 2002; 347: 911–920. [DOI] [PubMed] [Google Scholar]
- Djuardi Y, Wibowo H, Supali T, Ariawan I, Bredius RG, Yazdanbakhsh M et al. Determinants of the relationship between cytokine production in pregnant women and their infants. PLoS One 2009; 4: e7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djuardi Y, Supali T, Wibowo H, Kruize YC, Versteeg SA, van Ree R et al. The development of TH2 responses from infancy to 4 years of age and atopic sensitization in areas endemic for helminth infections. Allergy Asthma Clin Immunol 2013; 9: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herberth G, Hinz D, Roder S, Schlink U, Sack U, Diez U et al. Maternal immune status in pregnancy is related to offspring's immune responses and atopy risk. Allergy 2011; 66: 1065–1074. [DOI] [PubMed] [Google Scholar]
- Aaltonen R, Heikkinen T, Hakala K, Laine K, Alanen A. Transfer of proinflammatory cytokines across term placenta. Obstet Gynecol 2005; 106: 802–807. [DOI] [PubMed] [Google Scholar]
- Garofalo R, Chheda S, Mei F, Palkowetz KH, Rudloff HE, Schmalstieg FC et al. Interleukin-10 in human milk. Pediatr Res 1995; 37 (4 Pt 1): 444–449. [DOI] [PubMed] [Google Scholar]
- Agarwal S, Karmaus W, Davis S, Gangur V. Immune markers in breast milk and fetal and maternal body fluids: a systematic review of perinatal concentrations. J Hum Lact 2011; 27: 171–186. [DOI] [PubMed] [Google Scholar]
- Labbok MH, Clark D, Goldman AS. Breastfeeding: maintaining an irreplaceable immunological resource. Nat Rev Immunol 2004; 4: 565–572. [DOI] [PubMed] [Google Scholar]
- Galvani AP. Age-dependent epidemiological patterns and strain diversity in helminth parasites. J Parasitol 2005; 91: 24–30. [DOI] [PubMed] [Google Scholar]
- Hohler T, Reuss E, Adams P, Bartsch B, Weigmann B, Worns M et al. A genetic basis for IFN-gamma production and T-bet expression in humans. J Immunol 2005; 175: 5457–5462. [DOI] [PubMed] [Google Scholar]
- Westendorp RG, Langermans JA, Huizinga TW, Elouali AH, Verweij CL, Boomsma DI et al. Genetic influence on cytokine production and fatal meningococcal disease. Lancet 1997; 349: 170–173. [DOI] [PubMed] [Google Scholar]
- Sas AA, Jamshidi Y, Zheng D, Wu T, Korf J, Alizadeh BZ et al. The age-dependency of genetic and environmental influences on serum cytokine levels: a twin study. Cytokine 2012; 60: 108–113. [DOI] [PubMed] [Google Scholar]
- Stein CM, Guwatudde D, Nakakeeto M, Peters P, Elston RC, Tiwari HK et al. Heritability analysis of cytokines as intermediate phenotypes of tuberculosis. J Infect Dis 2003; 187: 1679–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantsulaia I, Trofimov S, Kobyliansky E, Livshits G. Genetic and environmental influences on IL-6 and TNF-alpha plasma levels in apparently healthy general population. Cytokine 2002; 19: 138–146. [DOI] [PubMed] [Google Scholar]
- Zhang G, Khoo SK, Makela MJ, Candelaria P, Hayden CM, von Hertzen L et al. Maternal genetic variants of IL4/IL13 pathway genes on IgE with “Western or Eastern Environments/Lifestyles”. Allergy Asthma Immunol Res 2014; 6: 350–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie BN, Carlson CS, Rieder MJ, Nickerson DA. Efficient selection of tagging single-nucleotide polymorphisms in multiple populations. Hum Genet 2006; 120: 58–68. [DOI] [PubMed] [Google Scholar]
- Teo YY, Sim X, Ong RT, Tan AK, Chen J, Tantoso E et al. Singapore Genome Variation Project: a haplotype map of three Southeast Asian populations. Genome Res 2009; 19: 2154–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidinger S, Gieger C, Rodriguez E, Baurecht H, Mempel M, Klopp N et al. Genome-wide scan on total serum IgE levels identifies FCER1A as novel susceptibility locus. PLoS Genet 2008; 4: e1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Howard TD, Zheng SL, Haselkorn T, Peters SP, Meyers DA et al. Genome-wide association study of asthma identifies RAD50-IL13 and HLA-DR/DQ regions. J Allergy Clin Immunol 2010; 125: 328–335.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.