Abstract
The response to growth hormone in humans is dependent on phenotypic, genetic and environmental factors. The present study in children with growth hormone deficiency (GHD) collected worldwide characterised gene–environment interactions on growth response to recombinant human growth hormone (r-hGH). Growth responses in children are linked to latitude, and we found that a correlate of latitude, summer daylight exposure (SDE), was a key environmental factor related to growth response to r-hGH. In turn growth response was determined by an interaction between both SDE and genes known to affect growth response to r-hGH. In addition, analysis of associated networks of gene expression implicated a role for circadian clock pathways and specifically the developmental transcription factor NANOG. This work provides the first observation of gene–environment interactions in children treated with r-hGH.
Introduction
Relationships have been identified between adult stature and geographical location; in particular these concern latitude, with adult height increasing as distance from the equator increases.1, 2, 3 This phenomenon is seen in Europe: people from North Europe are taller than people from South Europe.4, 5 In addition, genetic studies have indicated that the height differences between these closely related populations can be partially explained by multiple minor genetic differences.4 This raises the possibility of a relation between genotype and geography that influences height.
There are few published data in children regarding geographical height gradients.1, 3 A latitudinal gradient has been shown for height in Japanese children from North to South Japan. This may relate to a difference in day length as this is an important predictor of height in early and late adolescence.3
Human growth is a complex process, with variations in growth rate occurring in the short-term over weeks and also seasonally.6, 7 Growth rate is greatest during the summer, suggesting a relationship with day length.8, 9, 10 However, little is known regarding growth rate in children with short stature living at different latitudes, and there are few published data analysing growth rate with respect to number of daylight hours. One report from the National Cooperative Growth Study in children with growth hormone deficiency (GHD) on recombinant human growth hormone (r-hGH) therapy, has shown that there was a ‘seasonal' variation in growth at all latitudes, with summer annualised height velocity (HV) being greater than winter HV.11 This difference was greatest in the first year of therapy but persisted in the subsequent years. This difference also increased with distance from the equator and correlated with number of daylight hours across different latitudes. Most recently, growth rate in GHD children in response to r-hGH treatment has been considered in terms of genetic background. Carriage of specific genetic markers has been associated with both high and low growth responses to r-hGH in GHD children.12
The aims of the present study were (A) to assess whether living at different latitudes with different numbers of summer daylight hours impacted on annual HV in children with GHD treated with r-hGH, (B) to investigate the possible interaction between summer daylight and growth-related genetic markers on HV12 and (C) to use the difference in gene expression profile associations to identify pathways and hence mechanisms associated with this interaction.
Subjects and methods
Study design
Children were enrolled from the PREDICT long-term follow-up prospective study, which was conducted at 41 sites in 14 countries worldwide (NCT00699855; Merck Serono SA—Geneva). In the current study, we have analysed the data on first-year response to r-hGH from the PREDICT long-term follow-up study, which uses a pharmacogenomic approach to evaluate the association of single-nucleotide polymorphisms (SNPs) in growth and metabolism genes with long-term changes in growth whilst on r-hGH therapy.12
The following data had been collected: location of the study centre, ethnicity, gender, birth weight, parental heights, peak serum GH response to stimulation testing, age, height, weight and r-hGH dose at baseline and height and weight after 1 year of treatment. Absolute latitude was assumed to be that of the study site. For each patient, annualised HV (cm per year) at the first year of r-hGH therapy was assessed. To look at mechanisms underlying the latitude effect, the average number of daylight hours during summer at each centre was obtained using data obtained from the Geographical Information System and mesh climatic data.13 The number of daylight hours depends both on date and latitude. At the sites in the Northern hemisphere, the three months with the longest days, in terms of daylight hours, were classed as summer (May 6 to August 6).11 In the sites of the Southern hemisphere, summer period was considered to extend from November 6 to February 5.
Patients were categorised into three groups based on the distribution of their locations and the corresponding summer daylight exposure (SDE) (as high (>75th percentile of the cohort: ⩾15.5 h), intermediate (between the 25th and 75th percentiles: >15.5 to <14.3 h) and low (<25th percentile: ⩽14.3 h) exposure) (Figure 1a). Basal gene expression was correlated with SDE and HV (cm per year) using rank regression. Network models were constructed from the gene expression overlap between these two regressions, and highly connected regions were assessed to define biological functions.
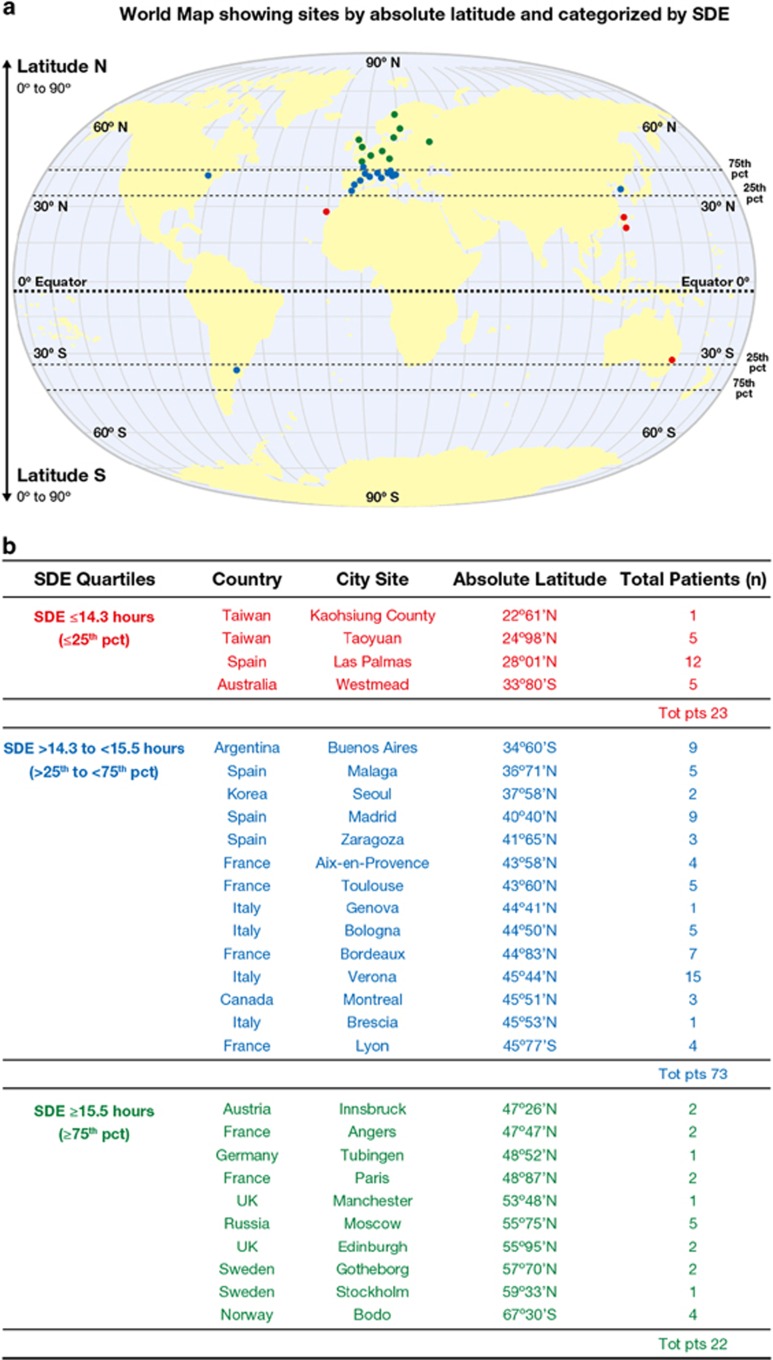
Figure 1.
(a) The absolute latitude at each study site extracted from the GIS.13 Patients were divided into three groups according to the summer daylight exposure (SDE) at their site: high (>75th percentile: ⩾15.5 h; n=22), intermediate (between the 25th and 75th percentiles: >15.5 to <14.3 h; n=73) and low (<25th percentile: ⩽14.3 h; n=23) SDE. (b) Distribution of the summer daylight exposure groups according to the latitude of the centres: four centres with lower SDE and nearest to the equator (low latitude), 10 sites had highest SDE and were farthest from the equator (high latitude) and 14 sites were in the intermediate latitude group. Number of patients at each site is shown. The three groups are coloured differently.
The growth response was then analysed both by SDE and also by carriage/non-carriage of SNPs previously associated with a high growth response.12
This study was conducted in compliance with ethical principles based on the Declaration of Helsinki, the International Conference on Harmonisation Tripartite Guideline for Good Clinical Practice, and all applicable regulatory requirements.
Patients
One hundred and eighteen patients (75 boys, mean age±s.d. scores (SDS): 8.9±3.3; 43 girls, 8.5±3.1) with GHD from a per-protocol population at 1 year were studied. Seven patients were excluded from the population: for three patients, no information on geographical location was available; for four patients, the country of origin did not match that of the study site. All patients were naïve to r-hGH therapy and pre-pubertal at the start of treatment. The diagnosis of GHD was based on two different stimulation tests with a peak GH <10 μg l−1.12 The median peak GH value was 4.1 μg l−1. Patients with GHD received r-hGH at an average dose of 0.035 mg kg−1 per day. Compliance was monitored by recall in the last month of the study and was estimated at an average of ~90% of patients. The great majority of the cohort had isolated GHD; however, a small number of children (<7%) had additional hormone deficiencies (thyroid-stimulating hormone, adrenocorticotropic hormone) and were treated with replacement therapies during the study, this group was considered too small to undertake sub-analyses. During the year of r-hGH treatment 31 patients entered puberty (Tanner stage 2).
Country of origin
Data were submitted from 28 study centres in 14 countries (Figure 1b). The population included 77% Europeans and 33% patients originating from countries across the world including Australia, Canada, Argentina, Taiwan and Korea.
Growth parameters
For each patient, annualised height gain and annualised HV (cm per year) at the first-year visit (12 ±3 month of first-year treatment anniversary) were calculated, converted to SDS14 and expressed as delta (Δ) height SDS and HV SDS. Body mass index (BMI) was calculated as the weight in kilograms divided by the square of the height in metres and converted to SDS using published reference values15 and expressed as ΔBMI SDS.
Target height (TH) was calculated as previously shown and converted to SDS.16 In addition, to take into account the patient's genetic potential for growth, distance to TH SDS, which is the difference between the child's height SDS and TH SDS,17 was calculated.
Genetic markers associated with height change in children with GHD
Analysis was performed using seven polymorphisms within five different genes previously associated with high growth response.12 These included the gene coding for the major GH-dependent carrier of insulin-like growth factor I in the circulation, IGFBP-3; a signalling molecule, GRB10; the growth factor TGF-α; the tumour suppressor TP53; and CYP19A1, a P450 cytochrome enzyme with aromatase activity. For each polymorphism, the difference in growth between alleles or genotypes was >1 cm over the first year, representing ~20% of first-year increment in growth.12 For all the SNPs, the analysis was conducted using full genotypes based on the presence or absence of the major allele (dominant model), and the presence or absence of the minor allele (recessive model).
Statistical analysis
All auxological data were expressed as median and inter-quartile ranges [median (Q1,Q3)]. Uncorrected P-values <0.05 were considered statistically significant. Statistical analysis was performed using the Statistical Package for Social Science program, version 20.0 software for Windows (SPSS, Chicago, IL, USA). Differences in continuous variables were examined for unpaired samples by the Kruskal–Wallis test, whereas differences in categorical variables were assessed by Fisher's exact test. Correlations between variables were assessed by Pearson's correlation coefficient. Partial least squares regression (PLSR) was applied to overcome multi-co-linearity between variables.18 By using PLSR, the ‘variable important for projection' coefficients were computed and a value of <0.8 was considered to be small and not contributing significantly to the prediction model.19 To examine which variables had a major impact on the prediction of HV, independent variables were used, including latitude and summer daylight, GH peak, r-hGH dose, BW SDS, baseline BMI SDS, distance to TH SDS, age and gender; HV was used as the dependent variable.
To investigate the impact of carriage of the growth-related SNPs and summer daylight on HV, a generalised linear model was used, with an interaction term for carriage/non-carriage of the growth-related SNPs and SDE, which was modelled as a fixed effect. HV (cm per year) was considered as the dependent variable with covariates for multiple other variables: gender, GH peak, r-hGH dose, BW SDS, baseline age, BMI and distance to TH SDS. Significance of each term was tested using an Anova type III.
Transcriptome analysis
Gene expression profiling was performed at baseline on whole-blood RNA extracted centrally by qLAB (Edinburgh, UK) using the PAXgene 96 blood RNA kit (Qiagen, Crawley, UK). Reduction of globin messenger RNA was undertaken using the Ambion GLOBIN Clear Human Kit (Life Technologies, Paisley, UK). Complementary RNA was generated using the Two-Cycle Eukaryotic Target Labelling Kit (Affymetrix, Santa Clara, CA, USA) and a final quality check performed before hybridisation to Affymetrix GeneChip Human Genome U133 Plus 2.0 Arrays. Arrays were then scanned on an Affymetrix GeneChip 7G scanner and assessed for quality against internal and hybridisation controls. All analyses were performed centrally by the Bioinformatics Group at Merck Serono.
Processing and normalisation of gene expression data were performed using a Robust Multi-array Average background correction modified for probe sequence with quantile normalisation and median polish (Partek Genomics Suite, version 6.3, St Louis, MO, USA). Confounding effects due to variations in cell populations and outliers were examined by cross-validation using principal component analysis and iso-map multidimensional scaling (Qlucore Omics Explorer 2.2, Qlucore, Lund, Sweden).
Correlations between basal gene expression with summer daylight and HV were assessed using rank regression with and without all the variables included in the PLSR model as confounding factors (Qlucore Omics Explorer 2.2).
Network analysis of transcriptomic data
Network analysis is directed towards the identification and prioritisation of key functional elements within interactome models. An interactome model of all known protein–protein interactions between the differentially expressed genes as ‘seeds' and their inferred immediate neighbours was calculated using BioGRID database (31.2.114).20 Network processing was performed using Cytoscape 2.8.3.21
The ModuLand plugin for Cytoscape 2.8.3 was used to determine overlapping modules within the network and to identify hierarchical structure within the model thus enabling the identification of key network elements and prioritise biological function.22, 23 Network modules were prioritised for further investigation by their centrality property and the most central set of 10 genes within each module was used to assess associated biological pathways using the geneontology.org database.24 The network structure observed with community modelling in Moduland was confirmed by cluster analysis using the ClusterOne algorithm.25 Cluster robustness was tested by random sample removal.26
Causal Network Analysis was performed within the overlap of associated gene expression between SDE and HV. Causal Network Analysis identifies upstream molecules up to three steps distant that control the expression of the genes in the data set, and thus provides insight into information flow within the network.27
Results
Clinical characteristics varied over 1 year of r-hGH therapy
Subjects were classified on the basis of SDE groups: as high, intermediate and low SDE (see Methods). At the start of treatment, the age, height and BMI SDS, peak GH and administered dose of r-hGH were not significantly different between these groups (Table 1). TH SDS and distance to TH SDS were greater at high compared with intermediate and low groups (P<0.05).
Table 1. Clinical characteristics of the study population and response to r-hGH defined by SDE groups.
| GHD (n=118) | Low SDE ⩽14.3 h | Intermediate SDE SDE >15.5 to <14.3 h | High SDE ⩾15.5 h | P-value |
|---|---|---|---|---|
| Number of patients (n) | 23 | 73 | 22 | |
| Gender (M/F) | 15/8 | 47/26 | 13/9 | 0.888 |
| BW SDS | −0.5 (−1.3, 0.8) | −0.5 (−1.3, 0.5) | −0.5 (−0.9, 0.1) | 0.883 |
| At baseline | ||||
| GH peak response (μg l−1) | 4.5 (3.0, 6.2) | 4.0 (2.3, 5.5) | 3.8 (1.2, 5.3) | 0.331 |
| Age (years) | 8.9 (6.2, 11.5) | 9.1 (7.3, 11.5) | 8.0 (5.0, 11.0) | 0.120 |
| Height SDS | −2.0 (−2.4, −1.6) | −2.3 (−2.8, −1.8) | −2.1 (−2.8, −1.6) | 0.066 |
| BMI SDS | −0.1 (−0.8, 1.6) | −0.3 (−1.1, 0.6) | −0.7 (−1.6, 0.1) | 0.090 |
| TH SDS | −1.6 (−2.3, −0.5) | −0.9 (−1.7, −0.3) | −0.1 (−1.1, 0.7) | 0.002 |
| Distance to TH SDS | −0.6 (−1.3, 0.6) | −1.3 (−2.2, −0.6) | −2.1 (−3.2, −1.3) | <0.0001 |
| Average r-hGH dose (mg kg−1) | 0.03 (0.030, 0.036) | 0.03 (0.031, 0.035) | 0.03 (0.030, 0.035) | 0.263 |
| 1-year visit | ||||
| HV (cm per year) | 8.2 (7.0, 10.3) | 8.1 (7.0, 9.8) | 9.8 (8.5, 11.4) | 0.019 |
| HV SDS | 2.0 (0.5, 4.0) | 1.8 (0.5, 3.2) | 3.0 (1.4, 6.8) | 0.024 |
| Δ Height SDS | 0.6 (0.3, 1.0) | 0.6 (0.3, 0.9) | 0.8 (0.6, 1.4) | 0.017 |
| Δ BMI SDS | 0.1 (−0.2, 0.6) | −0.1 (−0.4, 0.2) | −0.1 (−0.3, 0.2) | 0.143 |
| Puberty onset (%) | 7/23 (30%) | 20/73 (27%) | 4/22 (18%) | 0.607 |
Abbreviations: BMI, body mass index; BW, birth weight; Δ, delta; GH, growth hormone; HV; height velocity; r-hGH, recombinant human GH; SDS, s.d. score; TH, target height. Data are number or median (Q1, Q3). P-value: low vs intermediate vs high SDE.
After 1 year of r-hGH therapy, GHD patients from locations with higher SDE had a greater 1-year growth response than those from locations with intermediate and lower SDE (P=0.019 for HV (cm per year); P=0.024 for HV SDS; P=0.017 for Δ height SDS) (Table 1).
Relationships between summer daylight and growth rate
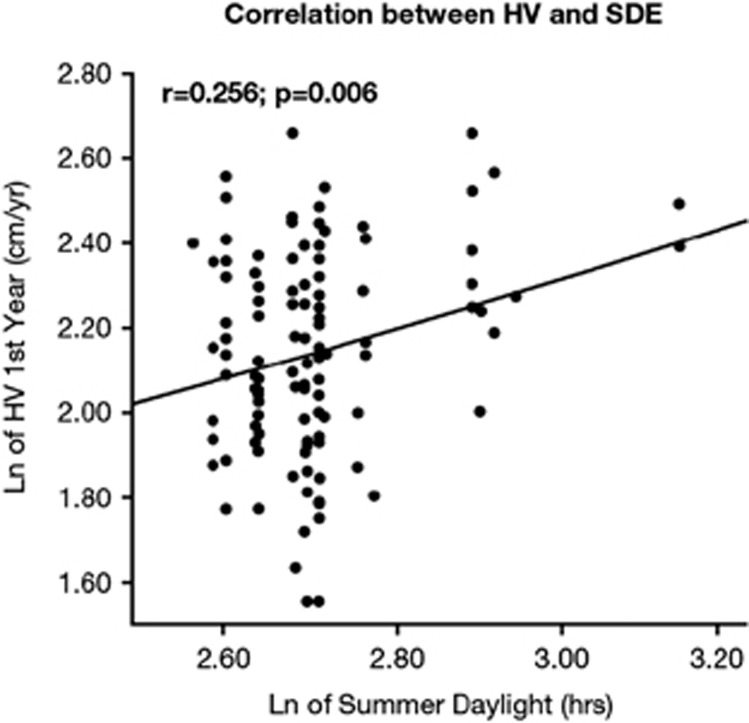
When the relationship between HV and summer daylight was evaluated, there was a significant correlation between HV (cm per year) and summer daylight (r=0.256, P=0.006; Figure 2).
Figure 2.
Correlation between HV (cm per year) and summer daylight. The variables are expressed as natural logarithm (Ln).
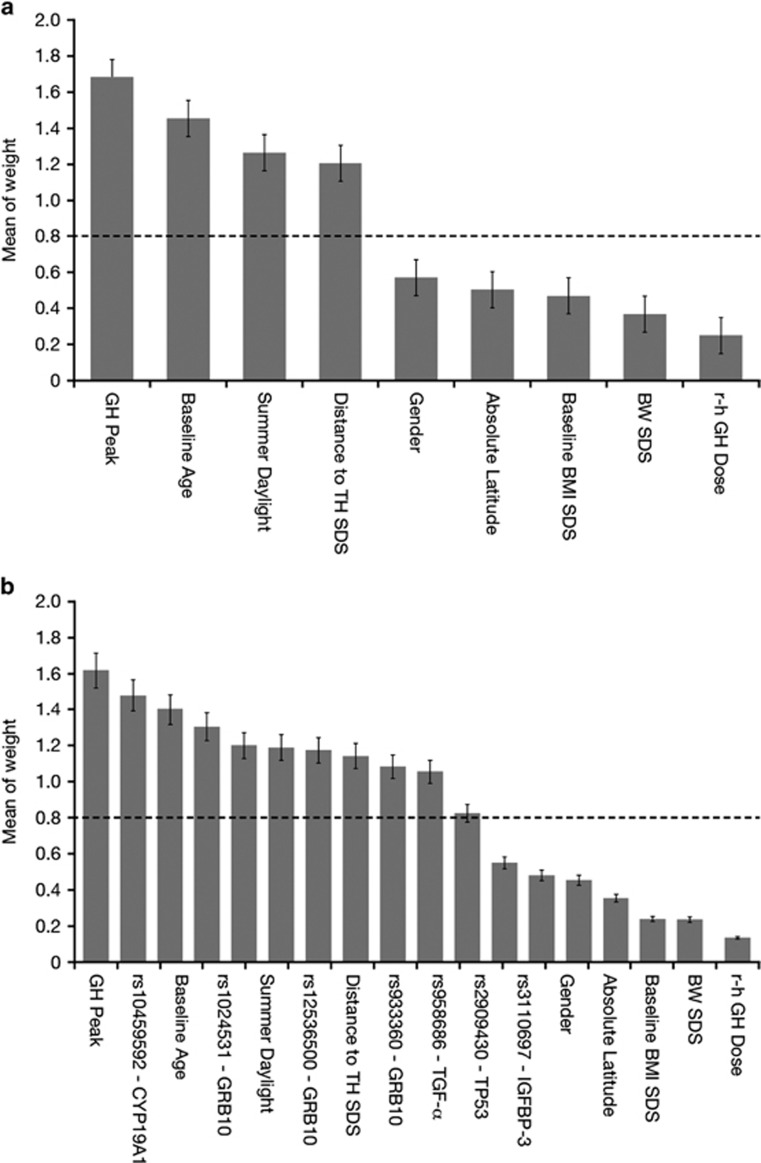
To identify which variables (baseline characteristics, latitude and SDE) had the greatest effect on HV, a PLSR analysis was used, which accounts for multi-co-linearity between variables. Four variables had a variable important for projection value >0.8 – in order: GH peak, baseline age, summer daylight and distance to TH (Figure 3a). The effect of the other variables, including latitude, was smaller and not considered significant. Inclusion of SNP data into the PLSR analysis confirmed that genotypes as well as summer daylight had a significant impact on HV (Figure 3b).
Figure 3.
The importance of each variable to the prediction of HV (cm per year) was assessed by partial least squares regression (PLSR). Variables were plotted according to their importance in the prediction of HV. A cutoff (dashed line) of 0.8 has been used to identify ‘important‘ variables. High values indicate that the variable has high impact in the prediction of HV. Panel (a) shows the effect of latitude, summer daylight and the main clinical variables; and panel (b) includes the effect of genotypes.
Interactions between carriage/non-carriage of growth-related SNPs and SDE: gene–environment interaction
To test the relationship between genetic and environmental factors, a generalised linear model was fitted. The effect of genotype (carriage vs non-carriage of the SNP) and group (high vs intermediate vs low SDE) on 1-year HV (cm per year) was investigated (Table 2). There was no difference in genotype frequency between the three summer daylight groups. HV was affected by a significant interaction between the carriage of a high growth response SNP and summer daylight for SNPs within GRB10, IGFBP-3, TGF-α, CYP19A1 and TP53 (interaction P-value <0.05 for each gene) (Table 2). HV SDS was also tested as a dependent variable in the generalised linear model and results were similar (data not shown).
Table 2. Generalised linear models showing differences in HV (cm per year) by genotype and SDE group.
| Gene | SNP ID | Model | Genotype | HV (cm per year) SDE ⩽14.3 h (n of patients=23) | HV (cm per year) SDE >15.5 to <14.3 h (n of patients=73) | HV (cm per year) SDE ⩾15.5 h (n of patients=22) | SDE P-value | Genotype P-value | Interaction P-value |
|---|---|---|---|---|---|---|---|---|---|
| GRB10 | rs1024531 | Allele* | AA | (13) | (36) | (12) | 0.019 | 0.001 | <0.0001† |
| 9.7 (8.7, 10.8) | 9.0 (8.4, 9.6) | 10.0 (8.7, 11.0) | |||||||
| GG & GA | (10) | (37) | (10) | ||||||
| 8.0 (6.8, 9.3) | 7.7 (7.0, 8.3) | 9.8 (8.6, 11.1) | |||||||
| rs12536500 | Allele* | CC | (16) | (43) | (12) | 0.015 | 0.001 | <0.0001† | |
| 9.5 (8.5, 10.5) | 9.0 (8.3, 9.5) | 10.0 (8.7, 11.0) | |||||||
| TT & TC | (7) | (30) | (10) | ||||||
| 7.8 (6.4, 9.3) | 7.5 (6.8, 8.3) | 9.8 (8.6, 11.1) | |||||||
| rs933360 | Allele* | TT | (14) | (46) | (15) | 0.014 | 0.044 | 0.008† | |
| 9.8 (8.8, 10.9) | 8.5 (8.0, 9.1) | 10.5 (9.5, 11.6) | |||||||
| TC & CC | (9) | (27) | (7) | ||||||
| 7.7 (6.4, 9.0) | 8.0 (7.2, 8.8) | 8.7 (7.2, 10.1) | |||||||
| IGFBP-3 | rs3110697 | Allele** | GG & GA | (20) | (60) | (18) | |||
| 9.1 (8.2, 10.0) | 8.6 (8.1, 9.1) | 10.0 (9.1, 11.0) | 0.031 | 0.025 | 0.009† | ||||
| AA | (3) | (13) | (4) | ||||||
| 8.4 (6.1, 10.6) | 7.3 (6.1, 8.4) | 8.4 (5.7, 11.1) | |||||||
| TGF-α | rs958686 | Allele* | CC | (3) | (10) | (2) | |||
| 9.3 (7.0, 11.7) | 8.5 (8.1, 8.9) | 11.4 (8.0, 14.8) | 0.027 | 0.152 | 0.015† | ||||
| GG & GC | (20) | (63) | (20) | ||||||
| 9.3 (8.5, 10.1) | 7.5 (6.3, 8.5) | 9.0 (8.2, 9.9) | |||||||
| CYP19A1 | rs10459592 | Allele* | GG | (7) | (14) | (7) | |||
| 10.7 (9.2, 12.1) | 9.4 (8.4, 10.4) | 10.8 (9.3, 12.1) | 0.041 | 0.039 | 0.026† | ||||
| TT & TG | (16) | (59) | (15) | ||||||
| 8.3 (7.3, 9.2) | 8.1 (7.6, 8.6) | 9.6 (8.5, 10.6) | |||||||
| TP53 | rs2909430 | Allele* | TT | (18) | (48) | (18) | |||
| 9.1 (8.2, 10.0) | 8.6 (8.1, 9.2) | 10.3 (9.3, 11.3) | 0.028 | 0.128 | 0.033† | ||||
| TC & CC | (5) | (25) | (4) | ||||||
| 8.8 (7.0, 10.5) | 7.8 (7.0, 8.6) | 8.1 (6.1, 10.1) |
Abbreviations: CI, confidence intervals; HV, height velocity; n, number of patients; SDE, summer daylight exposure. SDE had a significant effect on HV in all genotype groups, whereas genotypes for GRB10, IGFBP-3 and CYP19A1, but not TGF-α and TP53 had an effect on HV across the SDE groups. However, a significant interaction between the genotype and SDE was present for all the SNPs (†).
HV (cm per year) is presented as mean and 95% CIs in parentheses. Allele**=major allele dominant, Allele*=major allele recessive.
Directional differences in growth response
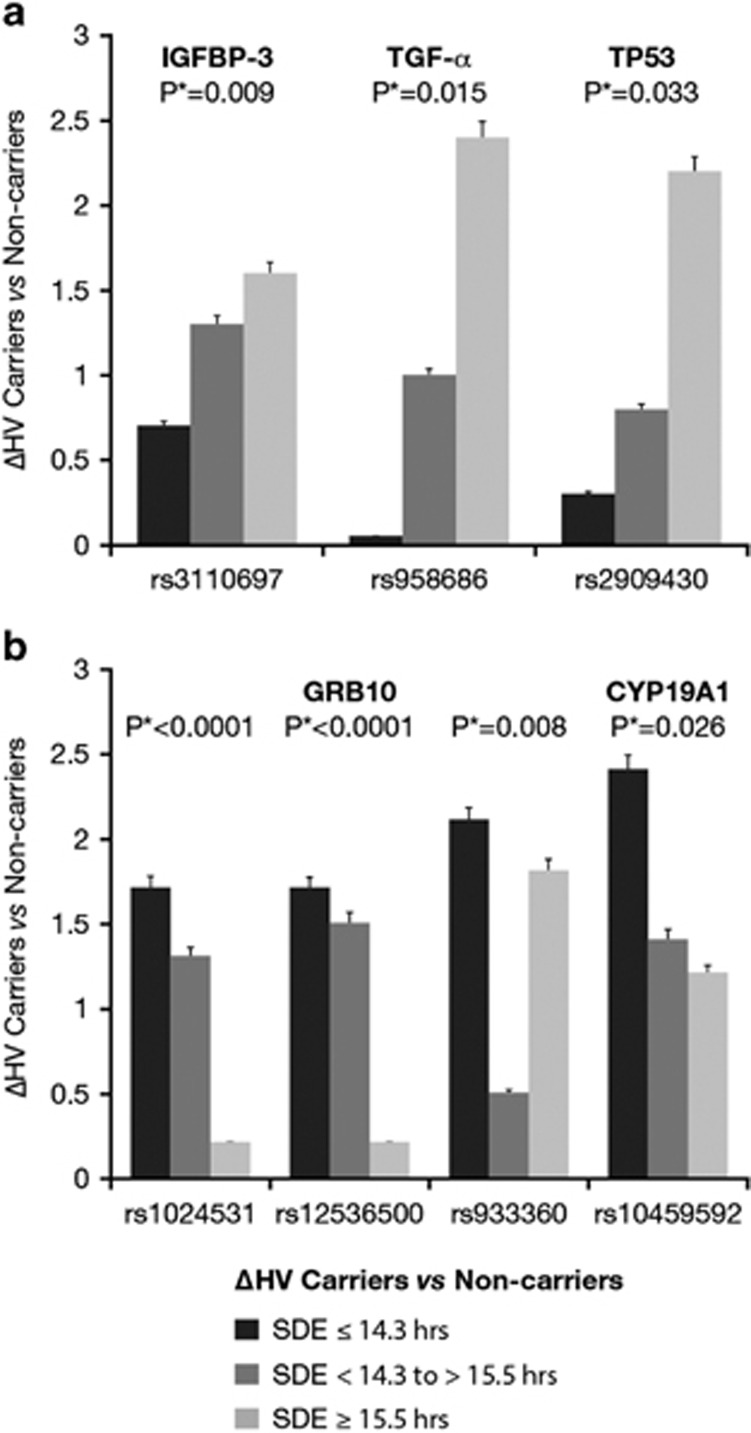
For each SNP, the direction of the impact on growth response in each SDE group was analysed (Figure 4). The difference in HV (in centimetres) between carriers and non-carriers for IGFBP-3, TGF-α and TP53 SNPs was greatest in those exposed to the highest number of summer daylight hours, which corresponds to the higher latitudes (Figure 4a). In contrast, for GRB10 and CYP19A1, the difference in HV between carriers and non-carriers was higher in those exposed to the lowest number of summer daylight hours, corresponding to lower latitudes (Figure 4b).
Figure 4.
Delta HV (ΔHV, cm per year) between carriers and non-carriers for each SNP by summer daylight group is shown. This relates to the results from the generalised linear model, evaluating a carriage (carriage vs non-carriage of the SNP) and group (high vs intermediate vs low summer daylight exposure (SDE)) effect on 1-year growth velocity (cm per year). P*=significant (<0.05). In (a) the ΔHV is greatest in the group with the highest number of SDE hours (for IGFBP-3, TGF-α and TP53). The difference in HV varies among SDE groups, ranging from 2.4 to 1.6 cm per year at higher to 0.0–0.7 cm per year at lower SDE. In (b) the ΔHV is greatest in the group with the lowest number of SDE hours (for GRB10 and CYP19A1). The difference in HV ranged from 2.4 to 1.7 cm per year at lower to 0.2 to 1.8 cm per year at higher SDE.
Network analysis of transcriptomic data
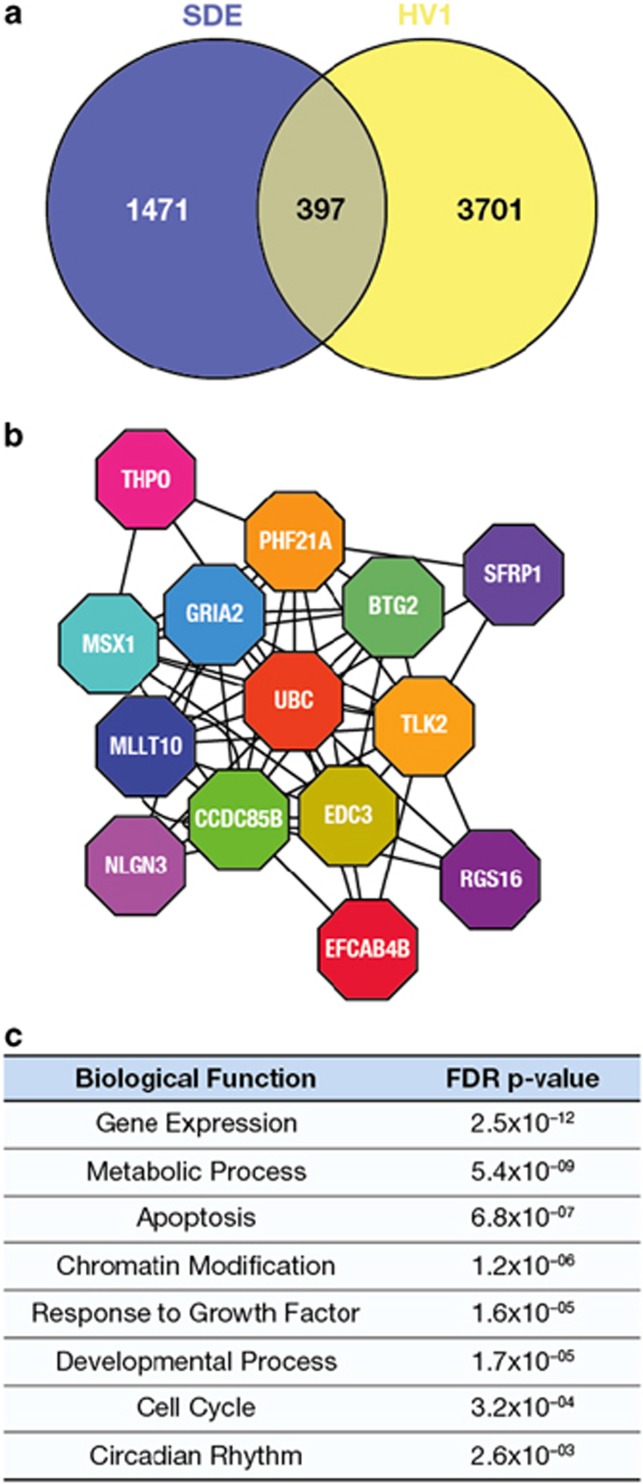
The expression of 1868 and 4098 genes was correlated with SDE and HV, respectively, with no covariates; 397 genes were present in both data sets (overlap, P=0.0015) (Figure 5a) [n=60, Gene Expression Omnibus GSE72439].
Figure 5.
Overlap of gene expression associated with both HV and SDE and subsequent network analysis of the common genes [n=60 GHD patients]. (a) Venn diagram of overlap (397, P=0.0015 hypergeometric test) between genes correlated with SDE (1868) and HV (4098) (no covariates, rank regression P<0.05). (b) Overlapping gene set correlated with SDE and HV using the same covariates as used for the partial least square regression (gender, GH peak, r-hGH dose, BW SDS, baseline age, BMI and distance to TH SDS) (Supplementary Table S1) was used to generate an interactome model. Clusters of related genes were identified within the interactome model using the Moduland algorithm and a network of the cluster modules was generated (shown) where the different coloured octagons represent clusters and the gene name is the most central gene element within that cluster (Supplementary Table S2). (c) Biological pathways associated with the overlap between the clusters were identified using the Geneontology.org database (hypergeometric test with a Benjamini–Hochberg false discovery rate (FDR) modified P-value).
The subset of common genes (143 unique genes with GH peak, r-hGH dose, BW SDS, baseline BMI SDS, distance to TH SDS, age and gender as covariates (Supplementary Table S1)) was used to generate an interactome model on which network analysis was performed (Figure 5b).
Network clusters were identified as markers of biological function23, 28 and robustness was confirmed using random sample removal (Supplementary Figure S1). The biological functions associated with the clusters were centred on chromatin remodelling (PHF21A), signal transduction (TLK2) and transcriptional regulation (MLLT10 and MSX1) (Supplementary Table S2 and Figure 5b). Biological functions represented by the network clusters included ‘gene expression' (P=2.5x10−12), ‘metabolic process' (P=5.4 × 10−09), ‘circadian rhythm' (P=2.6 × 10−03; Supplementary Table S2 and Figure 5c).
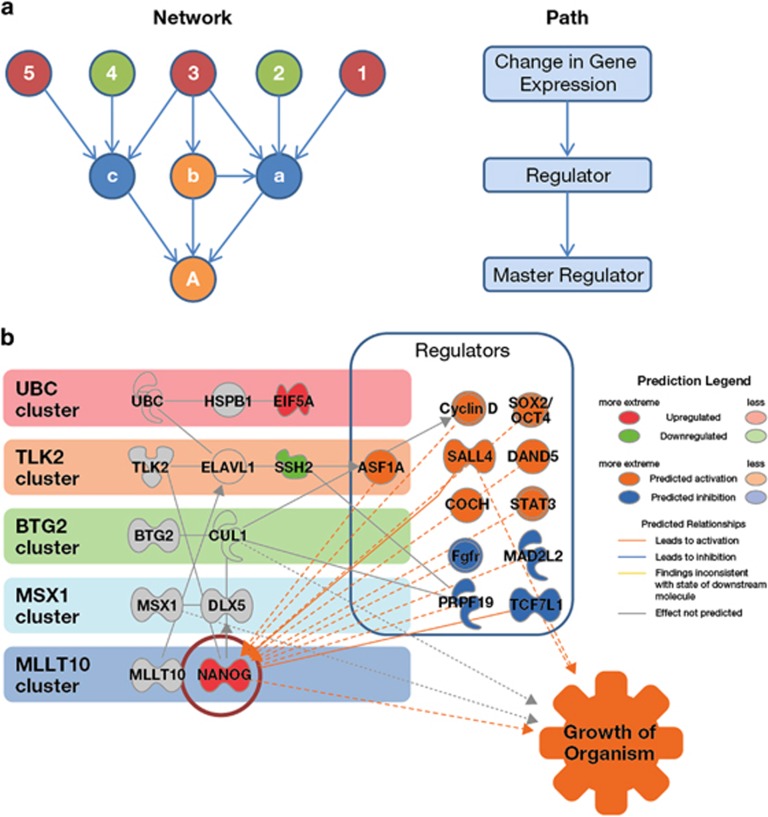
The causal network analysis (Figure 6a) of upstream regulation identified a set of transcriptional regulators with similar action in relation to both summer daylight and HV (using hierarchical clustering, Supplementary Table S3). The transcription factor NANOG had positively correlated expression with both SDE and HV and was the primary target of the regulators within the causal networks (Figure 6b).
Figure 6.
Causal analysis and mechanistic modelling of the subset of genes common to both HV and SDE. (a) Causal network analysis takes the genes with altered expression (examples numbered 1–5, green (low expression) and red (high expression)) and identifies upstream molecules up to three steps distant. This approach provides insight into information flow within the network using the known literature to identify network edges linking to upstream regulators (a–c) and master regulators (A), for which there is statistical evidence (Fisher's exact test) to support a corresponding causal relationship (within Ingenuity Pathway Analysis software). The most significant causal edges between regulators are then used to construct networks downstream of a ‘master' regulator to indicate possible mechanisms. (b) Regulators of gene expression with matched action in both HV and SDE were identified by causal network analysis and hierarchical clustering of results (Supplementary Table S3). These data were mapped onto the clusters identified within the network model of the overlap of gene expression and implicated NANOG as a prime target of regulation. Grey=opposing correlated expression with HV and SDE, green=negatively correlated with both HV and SDE, red=positively correlated with both HV and SDE, uncoloured=inferred interaction, orange=predicted activated regulator, blue=predicted inhibited regulator.
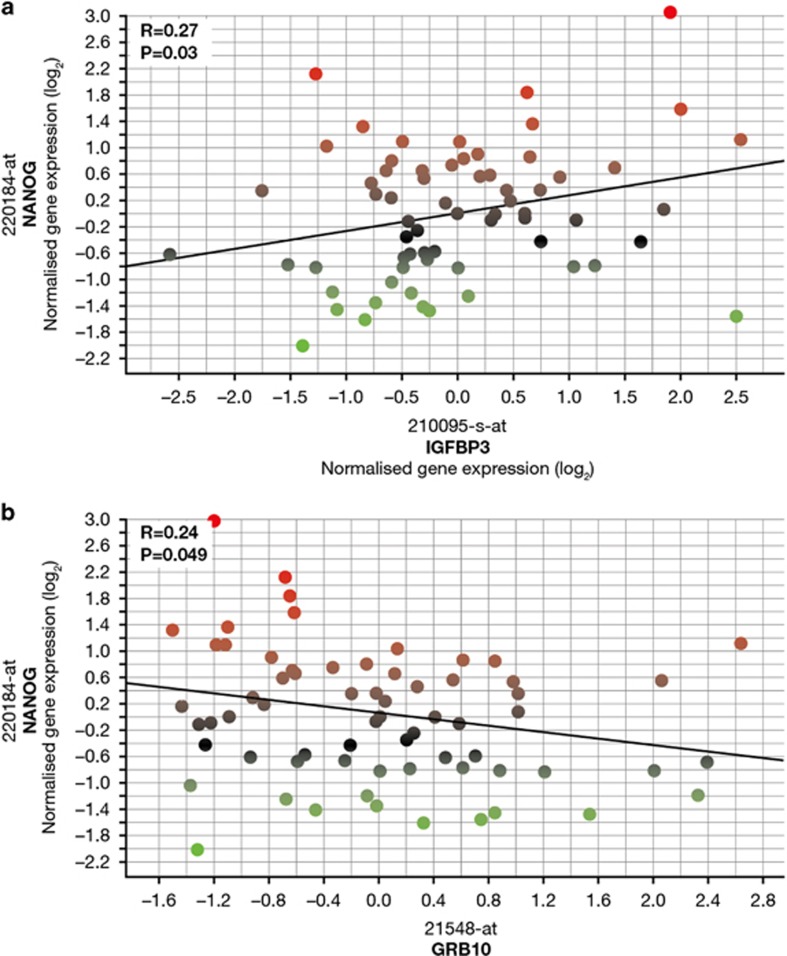
The expression of NANOG correlated positively (R=0.27, P=0.03) with the expression of IGFBP-3, a gene where the rs3110697 SNP has the greatest effect on HV at higher levels of SDE (Figures 4a and 7a). The expression of NANOG also correlated negatively (R=–0.24, P=0.05) with the expression of GRB10, a gene where the rs1024531, rs12536500 and rs933360 SNPs have the greatest effect on HV at lower levels of SDE (Figures 4b and 7b).
Figure 7.
Correlation of NANOG expression with gene expression from genes with single-nucleotide polymorphisms associated with both height velocity and summer daylight exposure. NANOG gene expression probeset 220184_at correlated against: (a) IGFBP-3, a gene where the rs3110697 SNP has a greater effect on height velocity at higher levels of SDE (Figure 5), using gene expression probeset 210095_s_at and (b) GRB10, a gene where the rs1024531, rs12536500 and rs933360 SNPs have a reduced effect on height velocity at higher levels of SDE (Figure 5), using gene expression probeset 215248_at. Analysis performed with the same covariates as used for the PLSR (gender, GH peak, r-hGH dose, BW SDS, baseline age, BMI and distance to TH SDS). Red (high) to green (low) gradation of colour represents level of expression of NANOG.
Distance from light-modulated pathways
To address the relationship of light modulation to gene expression within the overlap data set we considered three pathways (i) circadian rhythm, (ii) melatonin and (iii) vitamin D signalling.
To assess how close these pathways were to the network of genes identified, we calculated the ‘shortest path' between all primary network regulators identified by causal network analysis and the melatonin, vitamin D and circadian rhythm pathways (Supplementary Table S4). The melatonin pathway was significantly closer to the network model of the overlap of SDE (Supplementary Figure S2A) and HV (Supplementary Figure S2B) gene expression (P<0.01) than either vitamin D or circadian rhythm pathways.
Discussion
In the present study we observed that growth response is influenced by both the genetic background and geographical location. This is the first study, to the authors' knowledge, which has shown an interaction between growth-related polymorphisms and SDE in influencing growth response in patients treated with r-hGH therapy.
First-year growth velocity was significantly greater at locations with longer SDE, which correspond to higher latitudes (Table 1). It is important to consider whether there are any confounding factors influencing this relationship. First, those patients in locations with longer SDE had a lower baseline and ΔBMI SDS over the year, but neither was significant. If BMI was assumed to be an index of food intake and used as a marker of nutritional status, as previously shown,29 then a lower baseline and ΔBMI SDS should be associated with a poorer growth rate rather than a higher rate.30
Second, GH peak levels in GH stimulation tests were used in this study to define GHD severity. Such tests are recognised to have low-diagnostic specificity for GHD.31 However, all patients underwent two different stimulation tests, and both tests needed to have generated a peak GH level <10 μg l−1 for the child to be included in the study. It is possible that at a later date some of these patients may retest with a normal peak GH level but that at the time of the study they were all ‘biochemically' GHD. Importantly, across the SDE groups GH peak levels were not significantly different and therefore any inadequacy in the GH testing would apply equally across the groups (Table 1).
Third, another potential bias was the onset of puberty. Thirty-one patients entered puberty (Tanner stage 2) over the year of r-hGH treatment. However, there was no difference in the proportion of patients entering puberty across the three SDE groups (Table 1), indicating that any pubertal influence on growth would occur in all three groups. In addition, the pubertal acceleration in growth does not occur at the start of puberty in boys and does not reach maximal impact until Tanner breast stage 3 in girls. Therefore, puberty will have had a minimal impact on growth rate in the first year of treatment.32 In fact children living at higher latitudes/higher SDE had the lowest percentage of patients entering puberty, yet the highest growth rate (Table 1). Overall this indicates that differences in nutritional intake, severity of GH deficiency and puberty onset in our cohort of GHD children cannot account for the geographical gradient in the growth patterns and that other factors are involved.
In the examination of mechanisms related to the latitude effect, a positive correlation was found between HV and summer daylight and this was further supported by the PLSR analysis showing that summer daylight, but not latitude had a significant linear effect on growth rate. Several potential mechanisms explaining the association between daylight and height in children in Northern vs Southern Japan have been described by Yokoya et al.3 Climatic variables of temperature, solar radiation and day length were analysed. It was shown that day length, but not other climate variables, was the primary predictor for the geographical gradient in body height. Differences in melatonin secretion due to variation in day length were proposed to explain the geographical variation in height,33 inhibiting sexual and skeletal maturation.34 The link between day length and melatonin secretion resides in the eye, a light-sensitive organ whose function is to maintain circadian and seasonal rhythms,35 as facilitated by retinal communication via neural tracts with the pineal gland.35, 36 A circadian clock has been found to be primarily mediated by melanopsin-containing retinal ganglion cells,36, 37, 38 which are intrinsically blue-light sensitive.39 A number of studies have also provided support for a link between seasonal changes in daylight and physiological alterations, including human growth,40, 41 with the existence of a seasonal variability in growth patterns in normal children. Growth appears to speed up during times of greatest daylight exposure and slow down during periods of darkness.7, 8, 9, 11 The same phenomenon has been demonstrated in impaired growth with the exogenous administration of r-hGH. A report from the National Cooperative Growth Study has shown the existence of a seasonal variability in children with GHD on r-hGH therapy. Growth rate was greatest during summer and correlated with different numbers of daylight hours.11
Melatonin is related to the glucocorticoid receptor (GR) pathway and there is a well-established link between the circadian cortisol cycle and growth rate.42, 43 Therefore, the GR pathway modulation is a possible mechanism by which daylight could modulate growth response. We previously found that the expression of several genes involved in the GR pathway were correlated with both 1 month insulin-like growth factor I generation44 and 1 year growth response to r-hGH.12 We also showed that variation in the gene expression of GR pathway members relates to phase of growth in normal children.45 It is likely that a complex interaction between melatonin, GR pathways and growth mechanisms is generating these seasonal differences.
Gene expression (transcriptomic) data were studied on whole blood to explain biological mechanisms related to the interaction between SDE and HV. The transcriptome of whole blood has a substantial overlap with other human tissues.45, 46 A number of studies have characterised mononuclear cells in whole blood as a growth-responsive tissue and an appropriate model to study GH action.44, 47, 48 There is also a significant overlap between the regulation of lymphoid cell function and the classic growth pathways, as mononuclear blood cells have been shown to share important growth-related genes involved in both T/B cell proliferation and the regulation of bone development.49, 50, 51, 52 We are, therefore, confident that analysis of GE from whole blood gives us an opportunity to define potential mechanims for the SDE effect. The analysis of correlated GE profiles between SDE and HV (Figure 5c) has suggested a role of the circadian clock pathway. Mapping gene expression correlated with SDE and HV onto the canonical circadian rhythm pathway showed gene expression clustering around CREB activity (Supplementary Figure S2). In relation to light-sensitive biological pathways further analysis of network properties showed that the melatonin pathway was closer to SDE and HV correlated gene expression than the vitamin D and circadian rhythm pathways, but no distinct network modules were identified to support a direct involvement of these pathways.
Causal network analysis identified NANOG as a primary target for both SDE- and HV-related regulatory pathways. NANOG has been implicated in the development of circadian oscillator action,53 shown to reduce adipogenesis54 and enhance bone growth.55 MSX1 was the most central gene in a transcription factor related network cluster (Figure 6b) and has been recently found to induce light-controlled cell growth and tail development in vivo in vertebrate models.56 Both NANOG and MSX1 are associated with the regulation of growth-related pathways, including the Wnt, MAP/ERK and CREB pathways. Mutations in the MAP/ERK signalling pathways have been implicated in the aetiology of human RASopathies, dysmorphic syndromes presenting with short stature57 and the CREB pathway is involved in somatic growth and bone development,58 along with the hormonal hypothalamic-pituitary regulation. CREB-mutant mice have been found to have reduced postnatal growth consistent with dwarfism caused by GHD, owing to a reduction of GHRH expression.59 This network analysis has established a mechanistic hypothesis for the role of NANOG in differential growth response at different latitudes.
Some limitations of the present study exist, such as the lack of 25-hydroxyvitamin D measurement as a reflection of ultraviolet light exposure and the diversity of children enrolled at any one site. However, to reduce any bias, all the analyses were corrected for distance to TH and BMI and only ethnicity consistent with the country of origin was considered.
It is unlikely that only environmental variables can explain the geographical gradient in growth as its inter-individual variation depends also on the child's genetic constitution. The relationship between mid-parental height and the final height of offspring has been shown to explain 40% of the sex- and age-adjusted height variance in normal growth.60 We found greater TH at higher latitudes, reflecting the fact that body size and adult stature are associated with latitude.2, 5, 61, 62 To assess whether the environmental or genetic factors had the greatest impact on growth response, a PLSR analysis was performed that accounts for co-linearity between the data and allows distinct contributions to the variance within the data to be identified. SDE and distance to TH were demonstrated to have similar effects in the PLSR analysis (Figure 3a), which included variables that were utilised in previous GHD-specific models of growth.63 These previous models have incorporated surrogate genetic markers, such as parental heights and, although using identified clinical or biochemical factors that influence growth response to GH, explain only 40–61% of GH responsiveness over the first year of therapy.63 These models have not incorporated any primary genetic information. This implies that further parameters, such as specific genetic markers, could be included to improve current prediction models.
Recently, the main genes involved in the determination of human height have been identified in normal individuals.64 Analysis of 183,727 individuals by the Genetic Investigation of ANthropometric Traits (GIANT) consortium has identified 180 loci that influence adult height, which could also be potential candidates for growth and markers of GH responsiveness. The PREDICT long-term follow-up study has also highlighted genes involved in growth response and has revealed that carriage of a range of genetic markers is associated with change in insulin-like growth factor I over the first month of r-hGH treatment44 and HV over the first year of treatment.12 The inclusion of these genetic markers into the PLSR analysis confirmed that they had a significant impact on HV, along with distance to TH, implying that genetic contribution and carriage of specific growth-related SNPs is important in determination of growth response in children treated with r-hGH (Figure 4).
This study also showed an interaction between the carriage of high growth response SNPs and SDE within five genes involved in growth pathways and also known to affect adult height.64 These genes include GRB10, IGFBP-3, TGF-α and TP53 involved in IGF-1 system and cell growth and CYP19A1 in oestrogen synthesis. To assess the latitudinal effect we used different SDE groups, showing a gene–environmental interaction, which leads to differential growth response for children carrying the same SNP at different latitudes. Specifically, for IGFBP-3, TGF-α and TP53 the difference in HV between carriers and non-carriers was increased at longer summer daylight hours. In contrast, for GRB10 and CYP19A1 we showed the converse. Although in several conditions a gene–environment interaction has been shown causing variation in the phenotypic effect,28 in human growth it has only been hypothesised65 and this study suggests the existence of an interaction between environmental and genetic factors.
In conclusion, the present report suggests that growth response in GHD children involves a complex gene–environment interaction. The growth response to r-hGH appears to be related to both daylight exposure and to gene polymorphisms, with the magnitude and direction of the interaction being gene dependent. In addition, the gene expression data suggest that pathways related to the circadian clock and in particular the transcriptional regulator NANOG may contribute to mechanisms that link this gene–environment interaction.
Acknowledgments
The PREDICT study was supported by Merck Serono SA—Geneva, Switzerland. Chiara De Leonibus was supported by ESPE Research Fellowship, sponsored by Novo Nordisk A/S. We also thank Ekaterina Koledova, Olivier Morteau, John Raelson, Jerome Wojcik and Benoit Destenaves (supported by Merck Serono SA—Geneva, Switzerland) for their assistance in the preparation of the manuscript and Esther Dodge and Simon Lancaster of inScience Communications, UK (supported by Merck Serono S.A—Geneva, Switzerland) for assistance with editing and formatting of the manuscript. This work was also supported by the Manchester Biomedical Research centre. The PREDICT investigator group includes: Argentina: A Belgorosky, Buenos Aires. Australia: G Ambler, Westmead. Austria: K Kapelari, Innsbruck. Canada: C Deal, Montreal; J Hamilton, Toronto. Finland: J Jääskeläinen, Kuopio. France: Y Brusquet, Puyricard (Aix-en-Provence); P Chatelain, Lyon; M Colle, Bordeaux; R Coutant, Angers; Y Le Bouc, Paris; R Reynaud, Marseille; J-P Salles, Toulouse; J Weill, Lille. Germany: R Pfäffle, Leipzig; M Ranke, Tübingen; G Binder, Tübingen. Italy: M Bozzola, Pavia; F Buzi, Brescia; M Cappa, Rome; A Cicognani, Bologna; M Maghnie, Genova; L Tato, Verona; F Antoniazzi, Verona. Korea: DH Kim, Seoul; SW Yang, Seoul; HW Yoo, Seoul. Norway: E Vangsøy Hansen, Bergen; D Veimo, Bodø. Russia: E Bashnina, St Petersburg; V Peterkova, Moscow; J Skorodok, St Petersburg; L Sultanova, Kazan. Spain: A Carrascosa, Barcelona; A Ferrandez Longas, Zaragoza; R Gracia Bouthellier, Madrid; JP Lopez Siguero, Malaga; S Quinteiro, Las Palmas de Gran Canaria; MD Rodriguez-Arnao, Madrid; A Rodriguez Sanchez, Madrid. Sweden: J Dahlgren, Göteborg; L Hagenäs, Stockholm. Taiwan: JW Hou, Taoyuan; TJ Wang, Kaohsiung County. UK: P Clayton, Manchester; C Kelnar, Edinburgh.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the The Pharmacogenomics Journal website (http://www.nature.com/tpj)
DATA DEPOSITION
The transcriptomic data described in this manuscript is available from Gene Expression Omnibus (GEO) - GSE72439.
Supplementary Material
References
- Endo A, Omoe K, Ishikawa H. Ecological factors affecting body size of Japanese adolescents. Am J Phys Anthropol 1993; 91: 299–303. [DOI] [PubMed] [Google Scholar]
- Gustafsson A, Lindenfors P. Latitudinal patterns in human stature and sexual stature dimorphism. Ann Hum Biol 2009; 36: 74–87. [DOI] [PubMed] [Google Scholar]
- Yokoya M, Shimizu H, Higuchi Y. Geographical distribution of adolescent body height with respect to effective day length in Japan: an ecological analysis. PLoS One 2012; 7: e50994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavelaars AE, Kunst AE, Geurts JJ, Crialesi R, Grotvedt L, Helmert U et al. Persistent variations in average height between countries and between socio-economic groups: an overview of 10 European countries. Ann Hum Biol 2000; 27: 407–421. [DOI] [PubMed] [Google Scholar]
- Turchin MC, Chiang CW, Palmer CD, Sankararaman S, Reich D et al. Evidence of widespread selection on standing variation in Europe at height-associated SNPs. Nat Genet 2012; 44: 1015–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalange NK, Foster PJ, Gill MS, Price DA, Clayton PE. Model of normal prepubertal growth. Arch Dis Child 1996; 75: 427–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillmann V, Thalange NK, Foster PJ, Gill MS, Price DA, Clayton PE. The relationship between stature, growth, and short-term changes in height and weight in normal prepubertal children. Pediatr Res 1998; 44: 882–886. [DOI] [PubMed] [Google Scholar]
- Hermanussen M, Geiger-Benoit K, Burmeister J, Sippell WG. Periodical changes of short term growth velocity ('mini growth spurts') in human growth. Ann Hum Biol 1988; 15: 103–109. [DOI] [PubMed] [Google Scholar]
- Lampl M, Veldhuis JD, Johnson ML. Saltation and stasis: a model of human growth. Science 1992; 258: 801–803. [DOI] [PubMed] [Google Scholar]
- Land C, Blum WF, Stabrey A, Schoenau E. Seasonality of growth response to GH therapy in prepubertal children with idiopathic growth hormone deficiency. Eur J Endocrinol 2005; 152: 727–733. [DOI] [PubMed] [Google Scholar]
- Shulman DI, Frane J, Lippe B. Is there ‘seasonal' variation in height velocity in children treated with growth hormone? Data from the National Cooperative Growth Study. Int J Pediatr Endocrinol 2013; 2013: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton P, Chatelain P, Tato L, Yoo HW, Ambler GR, Belgorosky A et al. A pharmacogenomic approach to the treatment of children with GH deficiency or Turner syndrome. Eur J Endocrinol 2013; 169: 277–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuckols JR, Ward MH, Jarup L. Using geographic information systems for exposure assessment in environmental epidemiology studies. Environ Health Perspect 2004; 112: 1007–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole TJ, Freeman JV, Preece MA. British 1990 growth reference centiles for weight, height, body mass index and head circumference fitted by maximum penalized likelihood. Stat Med 1998; 17: 407–429. [PubMed] [Google Scholar]
- Cole TJ, Freeman JV, Preece MA. Body mass index reference curves for the UK, 1990. Arch Dis Child 1995; 73: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole TJ. Galton's midparent height revisited. Ann Hum Biol 2000; 27: 401–405. [DOI] [PubMed] [Google Scholar]
- Radetti G, D'Addato G, Gatti D, Bozzola M, Adami S. Influence of two different GH dosage regimens on final height, bone geometry and bone strength in GH-deficient children. Eur J Endocrinol 2006; 154: 479–482. [DOI] [PubMed] [Google Scholar]
- Abdi H, Williams LJ. Principal component analysis. WIREs Comp Stat 2010; 2: 433–459. [Google Scholar]
- Wold S. Exponentially weighted moving principal components analysis and projections to latent structures. Chemometr Intell Lab Syst 1994; 23: 149–161. [Google Scholar]
- Chatr-Aryamontri A, Breitkreutz BJ, Heinicke S, Boucher L, Winter A, Stark C et al. The BioGRID interaction database: 2013 update. Nucleic Acids Res 2013; 41 (Database issue): D816–D823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 2011; 27: 431–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens A, De Leonibus C, Hanson D, Dowsey AW, Whatmore A, Meyer S et al. Network analysis: a new approach to study endocrine disorders. J Mol Endocrinol 2014; 52: R79–R93. [DOI] [PubMed] [Google Scholar]
- Szalay-Beko M, Palotai R, Szappanos B, Kovacs IA, Papp B, Csermely P. ModuLand plug-in for Cytoscape: determination of hierarchical layers of overlapping network modules and community centrality. Bioinformatics 2012; 28: 2202–2204. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepusz T, Yu H, Paccanaro A. Detecting overlapping protein complexes in protein-protein interaction networks. Nat Methods 2012; 9: 471–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimand J, Bader G, Margolin A, Gonzalez-Perez A, Tamborero D, Lopez-Bigas N et al. TCGA pan-cancer project, thread 2: Network Models. Network models. Nat Genet 2013; doi:10.1038/ng.2787.
- Kramer A, Green J, Pollard J Jr., Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014; 30: 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickels S, Truong T, Hein R, Stevens K, Buck K, Behrens S et al. Evidence of gene-environment interactions between common breast cancer susceptibility loci and established environmental risk factors. PLoS Genet 2013; 9: e1003284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd C, Senerat A, Chambers E, Keller KL. PROP taster status interacts with the built environment to influence children's food acceptance and body weight status. Obesity 2013; 21: 786–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogol AD, Clark PA, Roemmich JN. Growth and pubertal development in children and adolescents: effects of diet and physical activity. Am J Clin Nutr 2000; 72 (2 Suppl): 521S–528S. [DOI] [PubMed] [Google Scholar]
- Shalet SM, Toogood A, Rahim A, Brennan BM. The diagnosis of growth hormone deficiency in children and adults. Endocr Rev 1998; 19: 203–223. [DOI] [PubMed] [Google Scholar]
- Rogol AD, Roemmich JN, Clark PA. Growth at puberty. J Adolesc Health 2002; 31 (6 Suppl): 192–200. [DOI] [PubMed] [Google Scholar]
- Perreau-Lenz S, Kalsbeek A, Garidou ML, Wortel J, van der Vliet J, van Heijningen C et al. Suprachiasmatic control of melatonin synthesis in rats: inhibitory and stimulatory mechanisms. Eur J Neurosci 2003; 17: 221–228. [DOI] [PubMed] [Google Scholar]
- Reiter RJ. Melatonin and human reproduction. Ann Med 1998; 30: 103–108. [DOI] [PubMed] [Google Scholar]
- Lamb TD. Evolution of the eye. Scientists now have a clear vision of how our notoriously complex eye came to be. Sci Am 2011; 305: 64–69. [PubMed] [Google Scholar]
- Young IM, Francis PL, Leone AM, Stovell P, Silman RE. Constant pineal output and increasing body mass account for declining melatonin levels during human growth and sexual maturation. J Pineal Res 1988; 5: 71–85. [DOI] [PubMed] [Google Scholar]
- Ikegami K, Iigo M, Yoshimura T. Circadian clock gene Per2 is not necessary for the photoperiodic response in mice. PLoS One 2013; 8: e58482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuo S, Yoshimura T, Ebihara S, Korf HW. Melatonin transmits photoperiodic signals through the MT1 melatonin receptor. J Neurosci 2009; 29: 2885–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooley JJ, Rajaratnam SM, Brainard GC, Kronauer RE, Czeisler CA, Lockley SW. Spectral responses of the human circadian system depend on the irradiance and duration of exposure to light. Sci Transl Med 2010; 2: 31ra33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggs JE, Hogenesch JB. Genomics and systems approaches in the mammalian circadian clock. Curr Opin Genet Dev 2010; 20: 581–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Ramsey KM, Marcheva B, Bass J. Circadian rhythms, sleep, and metabolism. J Clin Invest 2011; 121: 2133–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell GM, Henley DE, Leendertz J, Douthwaite JA, Wood SA, Stevens A et al. Rapid glucocorticoid receptor-mediated inhibition of hypothalamic-pituitary-adrenal ultradian activity in healthy males. J Neurosci 2010; 30: 6106–6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiga F, Waite EJ, Liu Y, Kershaw YM, Aguilera G, Lightman SL. ACTH-dependent ultradian rhythm of corticosterone secretion. Endocrinology 2011; 152: 1448–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens A, Clayton P, Tato L, Yoo HW, Rodriguez-Arnao MD, Skorodok J et al. Pharmacogenomics of insulin-like growth factor-I generation during GH treatment in children with GH deficiency or Turner syndrome. Pharmacogenomics J 2014; 14: 54–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens A, Hanson D, Whatmore A, Destenaves B, Chatelain P, Clayton P. Human growth is associated with distinct patterns of gene expression in evolutionarily conserved networks. BMC Genomics 2013; 14: 547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai I, Benjamin H, Shmoish M, Chalifa-Caspi V, Shklar M, Ophir R et al. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics 2005; 21: 650–659. [DOI] [PubMed] [Google Scholar]
- Whatmore AJ, Patel L, Clayton PE. A pilot study to evaluate gene expression profiles in peripheral blood mononuclear cells (PBMCs) from children with GH deficiency and Turner syndrome in response to GH treatment. Clin Endocrinol 2009; 70: 429–434. [DOI] [PubMed] [Google Scholar]
- Mitchell CJ, Nelson AE, Cowley MJ, Kaplan W, Stone G, Sutton SK et al. Detection of growth hormone doping by gene expression profiling of peripheral blood. J Clin Endocrinol Metab 2009; 94: 4703–4709. [DOI] [PubMed] [Google Scholar]
- Romanyukha AA, Yashin AI. Age related changes in population of peripheral T cells: towards a model of immunosenescence. Mech Ageing Dev 2003; 124: 433–443. [DOI] [PubMed] [Google Scholar]
- Weksler ME, Szabo P. The effect of age on the B-cell repertoire. J Clin Immunol 2000; 20: 240–249. [DOI] [PubMed] [Google Scholar]
- Horowitz MC, Lorenzo JA. Immunologic regulation of bone development. Adv Exp Med Biol 2007; 602: 47–56. [DOI] [PubMed] [Google Scholar]
- Finkielstain GP, Forcinito P, Lui JC, Barnes KM, Marino R, Makaroun S et al. An extensive genetic program occurring during postnatal growth in multiple tissues. Endocrinology 2009; 150: 1791–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagita K, Horie K, Koinuma S, Nakamura W, Yamanaka I, Urasaki A et al. Development of the circadian oscillator during differentiation of mouse embryonic stem cells in vitro. Proc Natl Acad Sci USA 2010; 107: 3846–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TM, Wu YN, Guo XM, Hui JH, Lee EH, Lim B. Effects of ectopic Nanog and Oct4 overexpression on mesenchymal stem cells. Stem Cells Dev 2009; 18: 1013–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bais MV, Shabin ZM, Young M, Einhorn TA, Kotton DN, Gerstnefeld LC. Role of Nanog in the maintenance of marrow stromal stem cells during post natal bone regeneration. Biochem Biophys Res Commun 2012; 417: 211–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams DS, Tseng AS, Levin M. Light-activation of the Archaerhodopsin H(+)-pump reverses age-dependent loss of vertebrate regeneration: sparking system-level controls in vivo. Biol Open 2013; 2: 306–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tidyman WE, Rauen KA. The RASopathies: developmental syndromes of Ras/MAPK pathway dysregulation. Curr Opin Genet Dev 2009; 19: 230–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long F, Schipani E, Asahara H, Kronenberg H, Montminy M. The CREB family of activators is required for endochondral bone development. Development 2001; 128: 541–550. [DOI] [PubMed] [Google Scholar]
- Mantamadiotis T, Kretz O, Ridder S, Bleckmann SC, Bock D, Grone HJ et al. Hypothalamic 3',5'-cyclic adenosine monophosphate response element-binding protein loss causes anterior pituitary hypoplasia and dwarfism in mice. Mol Endocrinol 2006; 20: 204–211. [DOI] [PubMed] [Google Scholar]
- Aulchenko YS, Struchalin MV, Belonogova NM, Axenovich TI, Weedon MN, Hofman A et al. Predicting human height by Victorian and genomic methods. Eur J Hum Genet 2009; 17: 1070–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashton KG, Tracy MC, de Queiroz A. Is Bergmann's rule valid for mammals? Am Naturalist 2000; 156: 390–415. [DOI] [PubMed] [Google Scholar]
- Blackburn TM, Gaston KJ, Loder N. Geographic gradients in body size: a clarification of Bergmann's rule. Divers Distrib 1999; 5: 165–174. [Google Scholar]
- Ranke MB, Lindberg A, Chatelain P, Wilton P, Cutfield W, Albertsson-Wikland K et al. Derivation and validation of a mathematical model for predicting the response to exogenous recombinant human growth hormone (GH) in prepubertal children with idiopathic GH deficiency. KIGS International Board. Kabi Pharmacia International Growth Study. J Clin Endocrinol Metab 1999; 84: 1174–1183. [DOI] [PubMed] [Google Scholar]
- Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 2010; 467: 832–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois L, Ohm Kyvik K, Girard M, Tatone-Tokuda F, Perusse D, Hjelmborg J et al. Genetic and environmental contributions to weight, height, and BMI from birth to 19 years of age: an international study of over 12 000 twin pairs. PLoS One 2012; 7: e30153. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.