Abstract
Three isolates collected from human mycetomas and showing an unusual brownish purple pigmentation on Bennett agar plates were analyzed by a polyphasic taxonomic approach, including morphological, biochemical, physiological, and chemotaxonomic properties coupled with genomic and phylogenetic analysis. It clearly appeared that these microorganisms were distinct from their closest phenotypic and genetic match, the most related species according to 16S rRNA gene sequence analysis being Nocardia pseudobrasiliensis. The data obtained indicated that the three clinical strains should be recognized as a new species for which the name Nocardia mexicana sp. nov. is proposed.
The Nocardia genus consists of gram-positive, variably acid fast, strictly aerobic bacteria that form filamentous, branched cells that fragment into pleomorphic, rod-shaped, or coccoid elements (1). It is widely distributed in the environment and causes a variety of suppurative and granulomatous infections of humans and animals, including cutaneous, subcutaneous, lymphocutaneous, pulmonary, cerebral, or disseminated nocardiosis. These bacteria are increasingly recognized as a cause of opportunistic infections (28).
The identification of new species was based for a long time on investigation of epidemiology, clinical disease spectrum, and/or drug susceptibility combined to culture features and biochemical or chemotaxonomic characteristics. Such a phenotypic approach was probably responsible for the low number (n = 12) of species described between the first description of the genus by Nocard in 1888 (32) and 1995. In the last decade, the introduction of genomic methods based on the analysis of sequences and polymorphism of the 16S rRNA gene or of the hsp65 gene has been of value for the description of many species. Thus, 17 new species had been validated from 1995 to 2003, leading to a major taxonomic revolution for this genus. It seems that it was just the beginning of the story and that the Nocardia genus will undergo the same dramatic taxonomic changes as Mycobacterium genus, for which almost a hundred species have now been described. This promoted a radical reappraisal of nocardial systematics.
According to this new taxonomic background, we decided to reevaluate the taxonomic status of the strains deposited in the culture collection of the French Observatory for Nocardiosis (OFN; Lyon, France). We observed that three of them revealed atypical characteristics. These strains originated from mycetoma samples collected from Mexican patients. The strains were initially identified as Nocardia brasiliensis but showed morphological differences compared to this species. The nocardial systematic position of these three strains was investigated extensively according to an integrated use of phenotypic, chemotaxonomic, and molecular methods. The data obtained indicated that these strains should be recognized as a new species, for which the name Nocardia mexicana sp. nov. is proposed.
MATERIALS AND METHODS
Strains and culture conditions.
The analysis of a large number of nocardial isolates from the collection of the OFN enabled us to select three isolates with unusual brownish purple pigmentation on Bennett agar plates.
The three strains (CIP 108295T, OFN 704.62, and OFN 1325.82) had been initially isolated from pus samples of Mexican patients with mycetomas at the Instituto de Salubridad y Enfermedades Tropicales in Mexico city. Each isolate was studied and compared to reference strains of the Nocardia genus. Routine cultivation was performed on Bennett agar at 37°C for 7 days. The strains were maintained as glycerol suspensions (20% [vol/vol]) at −20°C.
Growth and morphology.
Strains CIP 108295T, OFN 704.62, and OFN 1325.82 were grown on Bennett agar medium at 37°C for 1 week and were examined for pigmentation, production of aerial hyphae, and morphological characteristics. Prior to their incubation, sterile slides were placed in an angle of 45° in the medium. They were removed after 5 days and fixed by heat. The Gram stain was performed on these samples. This simplified technique allowed us to observe very well preserved cellular morphology and stain affinities of the cells. Observations were made with an Olympus BX50 microscope, and the image was captured by a charge-coupled device camera and Image-Pro Plus software (Medi Cybernetics, Carlsbad, Calif.). Acid fastness was observed by use of a modified Ziehl-Neelsen method (1% acid decoloration) (2). In addition, the ability to 25, 37, and 45°C was determined after 2 weeks on Bennett agar.
Physiological and biochemical characteristics.
The methods described by Boiron et al. (2), Goodfellow (11, 12) and Goodfellow and Lechevalier (10) were used (i) to determine decomposition of adenine, casein, hypoxanthine, testosterone, tyrosine, uric acid, and xanthine; (ii) to determine the utilization of substrates (l-arabinose, d-fructose, d-fucose, d-galactose, d-glucose, maltose, d-mannitol, mannose, raffinose, l-rhamnose, d-ribose, saccharose, sorbitol, and d-xylose) as the sole carbon source; and (iii) to determine the production of urease, arylsulfatase, β-galactosidase, and catalase. β-Lactamase production was determined by using the chromogenic cephalosporin disk method (Cefinase; bioMérieux, Marcy l'Etoile, France) with a loopful of colonies growing in Muller-Hinton agar and on the edge of the inhibition zone around a paper disk containing amoxicillin (Bio-Rad, Marnes-la-Coquette, France). Colonies were crushed on the disks placed in empty petri dishes, followed by a drop of distilled water. The result was read after a 1-h incubation period at 37°C.
Susceptibility testing (broth microdilution).
The different drugs tested (see Table 2) were provided by each of the manufacturers. The three strains (CIP 108295T, OFN 704.62, and OFN 1325.82) were tested by using a broth microdilution method according to the National Committee for Clinical Laboratory Standards M24-A guidelines for dilutional susceptibility tests for aerobic actinomycetes (31). Escherichia coli ATCC 25922 was inoculated as a control. The inoculum was standardized to obtain a solution with a final concentration of 1 × 104 to 5 × 105 CFU per well in 0.5 ml. This solution was added to each tubes containing an equal volume of broth with serial dilutions of the drugs to be tested (see Table 2). As a growth control, we inoculated in the same way a well containing cation-adjusted Muller-Hinton broth without drug. After 3 days (1 day for E. coli) of incubation at 37°C, the tubes were read, and the MIC was the lowest concentration of drug at which no visible growth was detected by visual inspection.
TABLE 2.
MICs determined by broth microdilution method for three strains (CIP 108295T, OFN 704.62, and OFN 1325.82) of N. mexicana sp. nov.
| Druga | MIC (μg/ml) for N. mexicana sp. nov. strain:
|
|||
|---|---|---|---|---|
| Range | CIP 108295T | OFN 704.62 | OFN 1325.82 | |
| Amikacin | 128-0.06 | 32 | 32 | 32 |
| Amoxicillin | 128-0.06 | >128 | >128 | >128 |
| Ampicillin | 128-0.06 | >128 | >128 | >128 |
| Cefotaxime | 256-0.12 | 8 | 8 | 8 |
| Ceftriaxone | 256-0.12 | 8 | 8 | 8 |
| Ciprofloxacin | 128-0.06 | 2 | 2 | 2 |
| Gentamicin | 128-0.06 | 128 | 128 | 128 |
| Imipenem | 128-0.06 | 16 | 16 | 16 |
| Smx | 512-0.25 | 128 | 128 | 128 |
| Tmp/Smx | 12.8/256-0.12/0.006 | 3.2/64 | 3.2/64 | 3.2/64 |
| Tetracycline | 128-0.06 | 64 | 64 | 64 |
Abbreviations: Smx, sulfamethoxazole; Tmp/Smx, trimethoprim-sulfamethoxazole.
Chemotaxonomic studies.
The isomeric of diaminopimelic acid was determined by thin-layer chromatography of whole-organism hydrolysates according to the procedure described by Boiron et al. (2). The fatty acids and mycolic acids composition was determined by the Deutsche Sammlung von Mikroorganismen und Zellkulturen by using the standard Microbial Identification System (MIDI) for automated GC analyses as described previously (21). Isoprenoid quinones were extracted from freeze-dried biomass of strain CIP 108295T by using the small-scale procedure of Minnikin et al. (29, 30), separated by high-pressure liquid chromatography and analyzed as described by Kroppenstedt (23, 24).
PRA identification.
16S amplification (used for genus identification) and hsp65 gene polymorphism restriction analysis (PRA) (used for species identification) were performed as previously described (25, 36).
16S phylogeny.
For each strain, a nearly complete 16S rRNA gene sequence (1,330-nucleotide fragment) was determined by using amplification primers SQ1 (5′-AGAGTTGATCMTGGCTCAG-3′) and SQ6 (5′-CGGTGTGTACAAGGCCC-3′). After 35 cycles consisting of denaturation at 98°C for 30 s, primer annealing at 58°C for 30 s, and primer extension at 72°C for 30 s, direct sequencing of amplicon was achieved with the following primer sets: SQ1-SQ5 (5′-CGCGGCTGCTGGCACG-3′), SQ4 (5′-CGTGCCAGCAGCCGCG-3′)-SQ3 (5′-CCCGTCAATYCTTTGAGTTT-3′), and SQ2 (5′-AAACTCAAAGRATTGACGGG-3′)-SQ6 by using a Taq DyeDeoxy terminator cycle sequencing kit (Applied Biosystems) and an Applied Biosystems model 373A DNA sequencer. Overlapping DNA segments from both the forward and the reverse strands were analyzed to determine a consensus sequence.
For phylogenetic analysis, the 16S ribosomal DNA (rDNA) nucleotide sequences were aligned with corresponding sequences of representative Nocardia species from the GenBank database by using the multiple sequence alignment program CLUSTAL W (37). According to Phylo_win software (9), evolutionary trees were inferred according to three treeing algorithms, namely, by maximum-likelihood (8), maximum-parsimony (22), and neighbor-joining (35) methods by using the Kimura two-parameter model (20). The robustness of this tree was assessed by bootstrap resampling (1,000 replicates each).
Hybridization DNA-DNA.
Levels of genomic relatedness between strain CIP 108295T, N. pseudobrasiliensis DSM 44290, and N. asteroides ATCC 19247T were determined by DNA-DNA hybridization experiments. DNA was isolated by using a French pressure cell (Thermo Spectronic) and was purified by chromatography on hydroxyapatite as described by Cashion et al. (5). DNA-DNA hybridization was carried out as described by De Ley et al. (6), with the modifications described by Huss et al. (16) and Escara and Hutton (7) with a model 2600 spectrophotometer equipped with a model 2527-R thermoprogrammer and plotter (Gilford Instrument Laboratories). Renaturation rates were computed with the TRANSFER.BAS program (17, 18).
Nucleotide sequence accession number.
The 16S rDNA nucleotide sequences that we determined have been deposited in GenBank under accession numbers AY555577 (N. mexicana sp. nov. CIP 108295T), AY560656 (N. mexicana sp. nov. OFN 704.62), and AY560655 (N. mexicana sp. nov. OFN 1325.82).
RESULTS AND DISCUSSION
Mycetoma is endemic in tropical and subtropical regions. It is a chronic granulomatous, progressive inflammatory disease that involves the subcutaneous tissue and sometimes bone after a minor trauma when walking barefoot, such as a cut with a thorn or splinter contaminated with soil containing the etiologic agent. The characteristic triad of a painless subcutaneous mass, sinuses, and the discharge of grains with microorganisms is pathogenomonic of mycetoma. The main localizations of this disease are the lower extremities (28). It may be caused by true fungi (eumycetes) or by higher bacteria (actinomycetes), and therefore it is classified into eumycetoma and actinomycetoma, respectively (4, 28).
Around 60% of the mycetoma cases worldwide are caused by aerobic actinomycetes (39), but the causative agents of mycetoma vary from region to region. N. brasiliensis, N. asteroides, N. otitidiscaviarum, N. transvalensis, and newly recognized species such as N. veterana, N. africana, and N. abscessus have been reported to cause human mycetomas (1, 14, 15, 19, 27). However, the major causal agent remains N. brasiliensis. A survey carried out in Mexico to determine the incidence and epidemiological characteristics of mycetoma from a total of 2,105 cases throughout a 30 year period (1956 to 1985) showed that eumycetoma (2.2%) was due to Madurella grisea and Madurella mycetomatis in most cases and that the predominant etiologic agents found corresponded in 97.8% of the cases to actinomycetes, of which Actinomadura madurae (10.2%) and Nocardia brasiliensis (86.6%) showed the greatest prevalence (26). Identification of this last species was often based on simple characteristics (e.g., culture, microscopy, and limited biochemical tests) which can explain the low diversity of Nocardia species reported. In this context, the three isolates included in our study were initially identified as N. brasiliensis according to phenotypic characteristics. They all originated from Mexico and were collected from mycetoma without available additional clinical history.
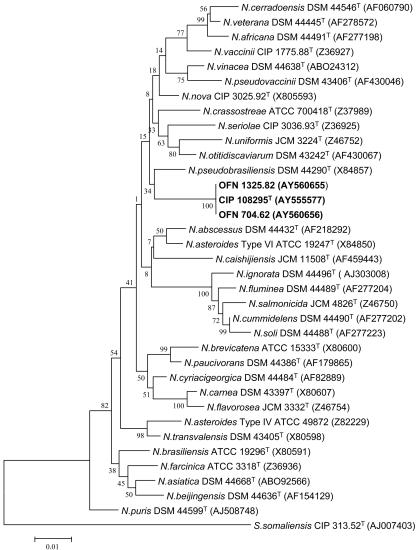
The almost complete 16S rRNA genes (1,330 nucleotides, positions 46 to 1400 based on E. coli numbering) (3) of strains CIP 108295T, OFN 704.62, and OFN 1325.82 were analyzed. The degree of similarity was found to be 100%. These sequences were aligned by CLUSTAL W (37) with reference sequences of species belonging to the genus Nocardia (sequences extracted from GenBank). It clearly appears that the three strains are affiliated with the genus Nocardia. According to the phylogenetic trees (Fig. 1), the three strains are clearly individualized among Nocardia species, and they form a monophyletic clade. The most closely related species was N. pseudobrasiliensis, but the bootstrap value (equal to 34) in the analysis based on the neighbor-joining method was low. The 16S rRNA sequence similarity between strain CIP 108295T and N. pseudobrasiliensis DSM 44290 is 97%, a value that corresponds to 40 nucleotide differences out of 1,330 nucleotide positions. Interestingly, the sequence AF 430064 originally deposited by Moore et al. as Nocardia transvalensis and then reclassified by Roth et al. (33) as Nocardia sp. (DSM46067) revealed 99.5% (7 nucleotide differences/1,330 nucleotides) homology with N. mexicana sp. nov. CIP 108295T. It clearly appears that this clinical isolate from a mycetoma belonged to the new species described here.
FIG. 1.
Unrooted phylogenetic tree showing the positions of strains CIP 108295T, OFN 704.62, and OFN 1325.82 within the radiation of species of the genus Nocardia. The tree, constructed by using the neighbor-joining method was based on a comparison of 1,300 nucleotides. Bootstrap values, expressed as a percentage of 1,000 replications. The scale bar represents 0.01 substitutions per nucleotide position.
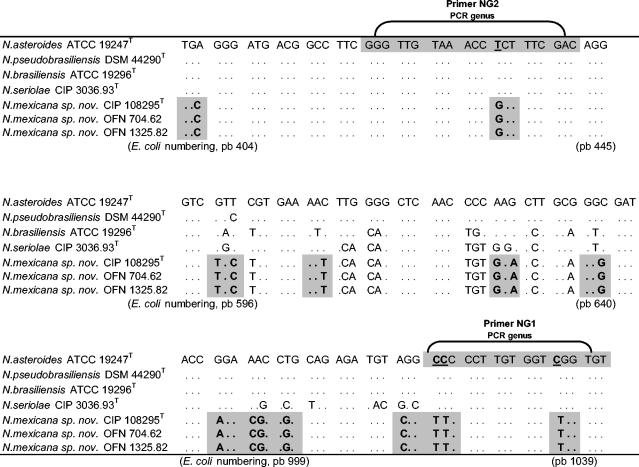
The level of relatedness between strain CIP 108295T, N. pseudobrasiliensis DSM 44290, or N. asteroides ATCC 19247T was low, with DNA-DNA reassociation levels from 32.1 and 26.3%, respectively. Such results were previously reported for Nocardia (42). Thus, strain CIP 108295T is not clearly related at the species level to the two tested strains when the threshold value of 70% for the definition of bacterial species according to Wayne et al. (40) is considered. The 16S rRNA gene between closely related nocardial species revealed a low interspecies heterogeneity (33). For example, the highest 16S rDNA similarity value between N. paucivorans and N. brevicatena is 99.6%, whereas DNA-DNA relatedness value was <70% (41). These data confirmed the novel species status of this strain. Finally, for these strains (CIP 108295T, OFN 704.62, and OFN 1325.82), three hypervariable regions in the 16S rRNA sequence were located from nucleotide positions 404 to 445, 596 to 640, and 999 to 1039 (E. coli numbering) (Fig. 2). These regions showed specific “sequences signatures” for the three studied strains that distinguish them from genotypically related species.
FIG. 2.
Alignment of hypervariable regions of the 16S rRNA genes of N. mexicana sp. nov. and genotypically related species. Nucleotides that differ from N. asteroides are shown. Base positions, in parentheses, correspond to positions on the 16S rRNA gene of E. coli. The sequences used for comparison, determined in our laboratory, were N. mexicana sp. nov. CIP 108295T, OFN 704.62, and OFN 1325.82. The sequences obtained from GenBank were N. asteroides ATCC 19247T (Z82218), N. brasiliensis ATCC 19296T (X80591), N. pseudobrasiliensis DSM 44290T (X84857), and N. seriolae CIP 3036.93 (Z36925)T.
These molecular data were supported by phenotypic and chemotaxonomic analysis. The three isolates studied have well-developed substrate mycelium with poor aerial mycelium and no detectable presence of spores. They produced unusual brownish purple pigmentation on Bennett agar plates. Bacteria are gram-positive cells with extensively branched hyphae and a tendency to fragmentation, even in early stages of growth. Branching is generally located near septa, and branches are almost at a right angle. The presence of inclusions, probably polyphosphate granules or lipid globules, give a beaded appearance to the hyphae. The filamentous are acid and alcohol fast.
Standard biochemical reactions were homogeneous for the three strains and allowed us to easily differentiate them from representatives of the validated species belonging to Nocardia genus (Table 1). In particular, they were able to grow on l-arabinose and sorbitol as the sole sources of carbon and to decompose adenine, hypoxanthine, and uric acid. The phenotypic pattern observed did not match any of closest related Nocardia species (Table 1).
TABLE 1.
Physiological characteristics of CIP 108295T, OFN 704.62, OFN 1325.82 and reference Nocardia strains
| Test | Characteristics of straina
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
| Growth on carbon sources (% [wt/vol]) | ||||||||||
| l-Arabinose (1.0) | + | + | + | − | − | − | − | − | − | + |
| d-Fructose (1.0) | + | + | + | + | + | + | + | + | + | + |
| d-Fucose (1.0) | − | − | − | − | − | − | − | − | − | − |
| d-Galactose (1.0) | + | + | + | − | − | − | − | + | + | + |
| d-Glucose (1.0) | + | + | + | + | + | + | + | + | + | + |
| Maltose (1.0) | − | − | − | + | − | − | − | + | + | + |
| d-Mannitol (1.0) | + | + | + | W | − | − | − | − | W | + |
| Mannose (1.0) | + | + | + | + | + | + | + | + | + | + |
| Raffinose (1.0) | − | − | − | − | − | − | − | − | − | − |
| l-Rhamnose (1.0) | + | + | + | − | − | + | − | + | − | − |
| d-Ribose (1.0) | + | + | + | + | + | + | + | − | + | + |
| Saccharose (1.0) | − | − | − | + | + | − | + | + | + | + |
| Sorbitol (1.0) | + | + | + | − | − | − | − | − | − | − |
| d-Xylose (1.0) | − | − | − | − | − | − | − | − | − | − |
| Growth on Bennett agar at: | ||||||||||
| 25°C | + | + | + | + | + | + | + | + | + | + |
| 37°C | + | + | + | + | + | + | + | + | + | + |
| 45°C | − | − | − | − | − | − | − | + | − | − |
| Arylsulfatase production at: | ||||||||||
| 3 days | − | − | − | − | − | − | − | − | − | − |
| 14 days | − | − | − | − | − | − | − | − | − | − |
| Catalase | + | + | + | + | + | + | + | + | + | + |
| Urease | + | + | + | + | + | + | + | + | + | + |
| β-Galactosidase | + | + | W | + | W | + | + | − | + | W |
| β-Lactamase | + | + | + | + | − | − | W | + | + | + |
| Decomposition of (% [wt/vol]): | ||||||||||
| Adenine (0.4) | + | + | + | − | − | − | − | − | − | + |
| Casein (1.0) | − | − | − | − | − | − | − | − | − | + |
| Hypoxanthine (0.4) | + | + | + | − | − | − | − | − | + | + |
| Testosterone (0.1) | − | − | − | + | − | − | + | + | − | − |
| Tyrosine (0.5) | W | − | − | − | − | − | − | − | + | + |
| Uric acid (0.5) | + | + | + | − | − | + | − | − | − | − |
| Xanthine (0.4) | − | − | − | − | − | − | − | − | − | − |
Strains: 1, CIP 108295T; 2, OFN 704.62; 3, OFN 1325.82; 4, N. ignorata DSM 44496T; 5, N. soli DSM 44488T; 6, N. cummidelens DSM 44490T; 7, N. asteroides ATCC 19247T; 8, N. farcinica DSM 43578T; 9, N. brasiliensis ATCC 19296T; 10, N. pseudobrasiliensis DSM 44290T. Reactions: −, negative; +, positive; W, weak.
Whole-cell hydrolysates of strain CIP 108295T contained meso-diaminopimelic acid as the only diamino acid of the peptidoglycan. The analyses of parietal composition revealed mycolic acids that showed a chain length from 54 to 60 carbon atoms (the principal mycolic acids have a chain length of 56 carbons). This pattern was in the range of the mycolic acids expected for Nocardia species (C50 to C62) (28). The fatty acid pattern is composed of C13:1 (1.7%), C14:0 (1.5%), C15:1 (1.5%), C15:0 (0.7%), C16:0 (42.4%), C16:1 (22.3%), C17:1 (1.4%), C18:1 (12.7%), and C18:0 (15.5%). The most common fatty acids consisted in palmitic acid (C16:0 [40.29%]), palmitoleic acid (C16:1cis [20.9%]), and tuberculostearic acid (10 methyl branched C18:0 [15.5%]). The Nocardia-specific quinone, MK-8(H4cycl.), represents 84% of the menaquinones. This menaquinone is observed only in members of the genera Nocardia and Skermania (13). Small amounts of MK-8(H2) (12%) and MK-8(H4) (4%) were also detected.
Strains CIP 108295T, OFN 704.62, and OFN 1325.82 had homogeneous susceptibility profiles (Table 2). A β-lactamase activity (cefinase disk) was observed for the three strains. For these strains the amoxicillin, ampicillin, imipenem, amikacin, gentamicin, and sulfamethoxazole MICs were high; the cefotaxime and ceftriaxone MICs were low, and the ciprofloxacin and sulfamethoxazole-trimethoprim MICs were intermediate.
It is important to note that trimethoprim-sulfamethoxazole combination is one of the treatments of choice for actinomycetoma. However, Vera-Cabrera et al. reported resistance to trimethoprim-sulfamethoxazole in the treatment in clinical cases (39). According to the MIC results, the combination of the two drugs revealed a low in vitro activity against N. mexicana sp. nov. that questioned its therapeutic use. Our results include only three representatives of N. mexicana sp. nov., and it is too early to establish a definitive susceptibility pattern for this new species. For instance, Van Gelderen de Komaid et al. (38) demonstrated that the majority of N. brasiliensis isolates obtained from soils showed higher susceptibilities to antibiotics than the strains isolated from human mycetomas.
According to hsp65 PRA proposed for species identification by Steingrube et al. (36), the three strains showed a specific profile (BstEII [440 pb, no cut], MspI [70, 130, and 155 bp], and HinfI [190 and 250 pb] enzymes), which is different from Nocardia species previously tested by Steingrube et al. (36). Conversely, amplifications with 16S Nocardia-genus specific primers previously described (25) were negative, demonstrating (for the first time) a lack of specificity for this procedure. This is related to nucleotide substitutions observed in positions 1024 (C to T) and 1025 (C to T) (E. coli numbering) that correspond to the 3′ extremity of primer NG1 (Fig. 2).
Biochemical, chemotaxonomic, and molecular data indicate that the three studied strains formerly listed in our collection as N. brasiliensis were misclassified and form a specific group compared to the type strains of all current validated species of Nocardia. On the basis of our results, we propose that strains CIP 108295T, OFN 704.62, and OFN 1325.82 should be assigned as a new species within the Nocardia genus. The name Nocardia mexicana sp. nov. is proposed, and the strain CIP 108295T is chosen as the type strain.
N. brasiliensis was previously considered to constitute a homogeneous population. However, in 1996 Ruimy et al. (34) described a new taxon, named N. pseudobrasiliensis, for some N. brasiliensis strains on the basis of taxonomic criteria, as well as clinical criteria. However, in most clinical laboratories, both species were not delineated. Members of this new taxon generally induce noncutaneous nocadioses, whereas N. brasiliensis sensu stricto is mainly isolated from cutaneous sites (34). In fact, the same observation is achieved today among these cutaneous sites with the description of N. mexicana sp. nov. Further investigations are required to establish the true incidence of this new species in mycetomas and more generally in the other forms of nocardiosis (pulmonary, cerebral, etc.).
Description of Nocardia mexicana sp. nov.
(me.xi.ca′na. M.L. fem. adj. mexicana referring to Mexico city, the geographical area from which isolates were collected).
Cells are strictly aerobic and gram positive, with extensively branched hyphae and tendency to fragmentation, even in early stages of growth. The filaments are acid and alcohol fast. Growth on Bennett agar is observed at 25 and 37°C but not at 45°C.
Colonies are rough and pigmented to brownish purple and range in size from 1 to 3 mm in diameter on Bennett agar plates. The bacteria are able to utilize l-arabinose, d-fructose, d-galactose, d-glucose, d-mannitol, mannose, l-rhamnose, d-ribose, and sorbitol as the sole carbon source. Adenine, hypoxanthine, and uric acid are decomposed but not casein, testosterone, tyrosine, and xanthine. Urease, β-galactosidase, catalase, and β-lactamase are detected, but it is arylsulfatase negative. The most common fatty acids consisted included the following: palmitic acid (C16:0), 40.29%; palmitoleic acid (C16:1cis), 20.9%; and tuberculostearic acid (10 methyl branched C18:0), 15.5%. Mycolic acids are 54 to 60 carbon atoms in length. The organism contains meso-diaminopimelic acid and MK-8(H4cycl.) menaquinones. MICs were estimated to be as follows: amikacin, 32 μg ml−1; amoxicillin, 128 μg ml−1; ampicillin, 128 μg ml−1; cefotaxime, 8 μg ml−1; ceftriaxone, 8 μg ml−1; ciprofloxacin, 2 μg ml−1; gentamicin, 128 μg ml−1; imipenem, 16 μg ml−1; sulfamethoxazole, 128 μg ml−1; and trimethoprim-sulfamethoxazole, 3.2 and 64 μg ml−1. The strains studied were isolated from human mycetomas. The type strain of Nocardia mexicana is CIP 108295T.
Acknowledgments
We thank the Deutsche Sammlung von Mikroorganismen und Zellkulturen Gmb for technical contributions. We thank Gregory Devulder for considerable assistance with this project.
Verónica Rodríguez-Nava is grateful to the Cosejo Nacional de Ciencia y Tecnologia, México City, Mexico, and to the Société Française d'Exportation des Ressources Educatives, Paris, France, for financial support.
REFERENCES
- 1.Beaman, B. L., and L. Beaman. 1994. Nocardia species: host-parasite relationships. Clin. Microbiol. Rev. 7:213-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boiron, P., F. Provost, and B. Dupont. 1993. Technical protocols, p. 107-126. In Methodes de laboratoire pour le diagnostic de la nocardiose. Institut Pasteur, Paris, France.
- 3.Brosius, J., M. L. Palmer, P. J. Kennedy, and H. F. Noller. 1978. Complete nucleotide sequence of the 16S rRNA gene from Escherichia coli. Proc. Natl. Acad. Sci. USA 75:4801-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buot, G., P. Lavalle, F. Mariat, and P. Suchil. 1987. Epidemiologic study of mycetomas in Mexico: a propos of 502 cases. Bull. Soc. Pathol. Exot. 3:329-339. [PubMed] [Google Scholar]
- 5.Cashion, P., M. A. Hodler-Franklin, J. McCully, and M. Franklin. 1977. A rapid method for base ratio determination of bacterial DNA. Anal. Biochem. 81:461-466. [DOI] [PubMed] [Google Scholar]
- 6.De Ley, J., H. Cattoir, and A. Reynaerts. 1970. The quantitative measurement of DNA hybridization from renaturation rates. Eur. J. Biochem. 12:133-142. [DOI] [PubMed] [Google Scholar]
- 7.Escara, J. F., and J. R. Hutton. 1980. Thermal stability and renaturation of DNA in dimethyl sulphoxide solutions: acceleration of renaturation rate. Biopolymers 19:1315-1327. [DOI] [PubMed] [Google Scholar]
- 8.Felsenstein, J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17:368-376. [DOI] [PubMed] [Google Scholar]
- 9.Galtier, N., M. Gouy, and C. Gautier. 1996. SeaView and Phylo_win, two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 12:543-548. [DOI] [PubMed] [Google Scholar]
- 10.Goodfellow, M., and M. P. Lechevalier. 1989. Genus Nocardia Trevisan, p. 2350-2361. In S. T. Williams, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 4. The Williams & Wilkins Co., Baltimore, Md.
- 11.Goodfellow, M. 1992. The family Nocardiaceae, p. 1188-1213. In A. Balows, H. G. Truper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes, 2nd ed. Springer, New York, N.Y.
- 12.Goodfellow, M. 1998. The genus Nocardia Trevisan 1889, p. 464-489. In A. Balows and B. I. Duerden (ed.), Topley and Wilson's microbiology and microbial infections, vol. 2. Edward Arnold, London, United Kingdom.
- 13.Hamid, M. E., L. Maldonado, G. S. Sharaf Eldin, M. F. Mohamed, N. S. Saeed, and M. Goodfellow. 2001. Nocardia africana sp. nov., a new pathogen isolated from patients with pulmonary infections. J. Clin. Microbiol. 39:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hattori, Y., R. Kano, Y. Kunitani, T. Yanai, and A. Hasegawa. 2003. Nocardia africana isolated from a feline mycetoma. J. Clin. Microbiol. 2:908-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horre, R., G. Schumacher, G. Marklein, H. Stratmann, E. Wardelmann, S. Gilges, G. S. De Hoog, and K. P. Schaal. 2002. Mycetoma due to Pseudallescheria boydii and co-isolation of Nocardia abscessus in a patient injured in road accident. Med. Mycol. 5:525-527. [DOI] [PubMed] [Google Scholar]
- 16.Huss, V. A. R., H. Festl, and K. H. Schleifer. 1983. Studies on the spectrophotometric determination of DNA hybridization from renaturation rates. Syst. Appl. Microbiol. 4:184-192. [DOI] [PubMed] [Google Scholar]
- 17.Jahnke, K. D., and G. Bahnweg. 1986. Assessing natural relationships in the Basidiomycetes by DNA analysis. Trans. Br. Mycol. Soc. 87:175-191. [Google Scholar]
- 18.Jahnke, K. D. 1992. Basic computer program for evaluation of spectroscopic DNA renaturation data from GILFORD System 2600 spectrometer on a PC/XT/AT type personal computer. J. Microbiol. Methods 15:61-73. [Google Scholar]
- 19.Kano, R., Y. Hattori, N. Murakami, N. Mine, M. Kashima, R. Kroppenstedt, M. Mizoguchi, and A. Hasegawa. 2002. The first isolation of Nocardia veterana from a human mycetoma. Microbiol. Immunol. 46:409-412. [DOI] [PubMed] [Google Scholar]
- 20.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 21.Klatte, S., F. A. Rainey, and M. Kroppenstedt. 1994. Transfer of Rhodococcus aichiensis Tsukamura 1982 and Nocardia amarae Lechevalier and Lechevalier 1974 to the genus Gordona as Gordona aichiensis comb. nov. and Gordona amarae comb. nov. Int. J. Syst. Bacteriol. 44:769-773. [DOI] [PubMed] [Google Scholar]
- 22.Kluge, A. G., and F. S. Farris. 1969. Quantitative phyletics and the evolution of anurans. Syst. Zool. 18:1-32. [Google Scholar]
- 23.Kroppenstedt, R. M. 1982. Separation of bacterial menaquinones by HPLC using reverse phase (RP-18) and a silver loaded ion exchanger. J. Liquid Chromatogr. 5:2359-2367. [Google Scholar]
- 24.Kroppenstedt, M. 1985. Fatty acids and menaquinone analysis of actinomycetes and related organisms, p. 173-189. In M. Goodfellow and D. E. Minnikin (ed.), Chemical methods in bacterial systematics. Academic Press, London, England.
- 25.Laurent, F., F. Provost, and P. Boiron. 1999. Rapid identification of clinically relevant Nocardia species to genus level by 16S rRNA gene PCR. J. Clin. Microbiol. 37:99-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopez-Martinez, R., L. J. Mendez-Tovar, P. Lavalle, O. Welsh, A. Saul, and E. Macotela-Ruiz. 1992. Epidemiology of mycetoma in Mexico: study of 2105 cases. Gac. Med. Mex. 128:477-481. [PubMed] [Google Scholar]
- 27.Lum, C. A., and M. S. Vadmal. 2003. Case report: Nocardia asteroides mycetoma. Ann. Clin. Lab. Sci. 33:329-333. [PubMed] [Google Scholar]
- 28.McNeil, M. M., and J. M. Brown. 1994. The medically important aerobic actinomycetes: epidemiology and microbiology. Clin. Microbiol. Rev. 7:357-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Minnikin, D. E., L. Alshamaony, and M. Goodfellow. 1975. Differentiation of Mycobacterium, Nocardia, and related taxa by thin-layer chromatographic analyses of whole-cell methanolysates. J. Gen. Microbiol. 88:200-204. [DOI] [PubMed] [Google Scholar]
- 30.Minnikin, D. E., A. G. O'Donnell, M. Goodfellow, G. Alderson, M. Athalye, A. Schaal, and J. H. Parlett. 1984. An integrated procedure for the extraction of isoprenoid quinones and polar lipids. J. Microbiol. Methods 2:233-241. [Google Scholar]
- 31.National Committee for Clinical Laboratory Standards. 2000. Susceptibility testing of Mycobacteria, Nocardia, and other aerobic actinomycetes. Tentative standard M24-T2, 2nd ed. National Committee for Clinical Laboratory Standards, Wayne, Pa. [PubMed]
- 32.Nocard, M. E. 1888. Note sur la maladie des boeufs de la guadeloupe connue sous le nom de farcin. Ann. Inst. Pasteur 2:293-302. [Google Scholar]
- 33.Roth, A., S. Andrees, R. M. Kroppenstedt, D. Harmsen, and H. Mauch. 2003. Phylogeny of the genus Nocardia based on reassessed 16S rRNA gene sequences reveals underspeciation and division of strains classified as Nocardia asteroides into three established species and two unnamed taxons. J. Clin. Microbiol. 41:851-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruimy, R., P. Riegel, A. Carlotti, P. Boiron, G. Bernardin, H. Monteil, R. J. Wallace, Jr., and R. Christen. 1996. Nocardia pseudobrasiliensis sp. nov., a new species of Nocardia which groups bacterial strains previously identified as Nocardia brasiliensis and associated with invasive diseases. Int. J. Syst. Bacteriol. 46:259-264. [DOI] [PubMed] [Google Scholar]
- 35.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 36.Steingrube, V. A., R. W. Wilson, B. A. Brown, K. Jost, J. L. Gibson, J. Brown, Z. Blacklock, J. L. Gibson, and R. J. Wallace, Jr. 1997. Rapid identification of clinically significant species and taxa of aerobic actinomycetes, including Actinomadura, Gordona, Nocardia, Rhodococcus, Stretomyces, and Tsukamurella isolates, by DNA amplification and restriction endonuclease analysis. J. Clin. Microbiol. 35:817-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Gelderen de Komaid, A. A., and E. L. Duran. 1998. Antimicrobial susceptibilities of strains of Nocardia brasiliensis isolated from soil of Tucuman. Mycopathologia 141:115-121. [DOI] [PubMed] [Google Scholar]
- 39.Vera-Cabrera, L., E. Gonzalez, S. H. Choi, and O. Welsh. 2004. In vitro activities of new antimicrobials against Nocardia brasiliensis. Antimicrob. Agents Chemother. 2:602-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wayne, L. G., D. J. Brenner, R. R. Colwell, P. A. D. Grimont, O. Kandler, M. I. Krichevski, L. H. Moore, W. E. C. Moore, R. G. E. Murray, E. Stackebrandt, M. P. Starr, and H. G. Trüper. 1987. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37:463-464. [Google Scholar]
- 41.Yassin, A. F., F. A. Rainey, J. Burghardt, H. Brzezinka, M. Mauch, and K. P. Schaal. 2000. Nocardia paucivorans sp. nov. Int. J. Syst. Evol. Microbiol. 2:803-809. [DOI] [PubMed] [Google Scholar]
- 42.Yassin, A. F., F. A. Rainey, U. Mendrock, H. Brzezinka, and K. P. Schaal. 2000. Nocardia abscessus sp. nov. Int. J. Syst. Evol. Microbiol. 4:1487-1493. [DOI] [PubMed] [Google Scholar]