Abstract
Inherited retinal disease is a common cause of visual impairment and represents a highly heterogeneous group of conditions. Here, we present findings from a cohort of 722 individuals with inherited retinal disease, who have had whole-genome sequencing (n = 605), whole-exome sequencing (n = 72), or both (n = 45) performed, as part of the NIHR-BioResource Rare Diseases research study. We identified pathogenic variants (single-nucleotide variants, indels, or structural variants) for 404/722 (56%) individuals. Whole-genome sequencing gives unprecedented power to detect three categories of pathogenic variants in particular: structural variants, variants in GC-rich regions, which have significantly improved coverage compared to whole-exome sequencing, and variants in non-coding regulatory regions. In addition to previously reported pathogenic regulatory variants, we have identified a previously unreported pathogenic intronic variant in CHM in two males with choroideremia. We have also identified 19 genes not previously known to be associated with inherited retinal disease, which harbor biallelic predicted protein-truncating variants in unsolved cases. Whole-genome sequencing is an increasingly important comprehensive method with which to investigate the genetic causes of inherited retinal disease.
Keywords: retinal dystrophy, whole-genome sequence, copy-number variants, rare sequence variant
Introduction
Inherited retinal disease (IRD) describes a phenotypically heterogeneous group of conditions consequent upon dysfunction and/or degeneration of the neural retina or retinal pigment epithelium, resulting in visual impairment. IRD is the most common cause of severe visual impairment among working-age individuals in the UK.1 Determining the genetic cause of IRD in affected individuals allows accurate assessment of risk of the disease to other family members, provides useful prognostic information for affected individuals, and can provide much-needed insight and understanding of the disease for those affected. Furthermore, genetic stratification of affected individuals is increasingly being used to direct specific treatment options, including clinical trials of medicines and restoration of protein function through gene therapy trials.2
Whole-exome sequencing (WES) of large cohorts of individuals has transformed research into rare Mendelian diseases in recent years by facilitating discovery of pathogenic variants, newly described disease-associated genes, and other insights into the genetic architecture of rare diseases.3, 4, 5 Whole-genome sequencing (WGS) has thus far typically been employed on a smaller scale and has demonstrated advantages over other methods.6, 7 Large-scale projects such as the Genomics England 100,000 Genome Project are also beginning to use WGS to investigate rare diseases.
IRD represents a particularly good clinical phenotype to provide comprehensive investigation by high-throughput sequencing technologies because it exhibits a high degree of phenotypic, genetic, and allelic heterogeneity, with more than 250 genes and loci associated with autosomal-dominant, autosomal-recessive, X-linked, and mitochondrial inheritance (RetNet), and it is often difficult to predict the disease-associated gene from an individual’s phenotype alone.8 IRD is therefore among the many rare diseases that are increasingly being investigated through high-throughput sequencing.9, 10, 11, 12 These technologies are also transforming clinical practice by being incorporated into diagnostic services.13, 14 Most of these studies have used targeted pull-down of a predetermined panel of known genes, which are limited not only in that they cover only certain genes, but also because they cannot reliably detect structural variants (SVs). WES, on the other hand, is not limited to known disease-associated genes, but coverage is generally variable across the exome, negatively affecting variant detection compared to targeted gene panel testing.15 PCR-free WGS should overcome the disadvantages of WES and targeted gene panels, although the remaining challenge will be in handling the large amount of data generated and variant interpretation. Indeed, a recent study compared WGS to targeted gene panels and found that WGS improves the pathogenic variant detection rate by facilitating detection of SVs and variants in regulatory regions.16
The aims of the current study were three-fold. First, to identify pathogenic variants in a large cohort of individuals with IRD, including intractable cases in which previous investigations had not yielded a diagnosis. Second, to explore advantages and disadvantages of WGS as a tool with which to investigate IRD. Third, to identify candidate IRD-associated genes and provide new insights into phenotypes and the genetic architecture of IRD. Here we present a cohort of 722 individuals with IRD who have had high-throughput sequencing as part of the NIHR-BioResource Rare Diseases research study.
Subjects and Methods
Cohort
722 individuals with IRD were recruited to the NIHR BioResource Rare Diseases research study. All participants provided written informed consent and the study was approved by the East of England Cambridge South national institutional review board (13/EE/0325). Most of the individuals (n = 657) were recruited at the Inherited Eye Disease clinics, Moorfields Eye Hospital NHS Foundation Trust (London, UK). The remainder were recruited at Cambridge University Hospitals NHS Foundation Trust (Cambridge, UK), Great Ormond Street Hospital For Children NHS Foundation Trust (London, UK), Guy’s And St Thomas’ NHS Foundation Trust (London, UK), London North West Healthcare NHS Trust (London, UK), and University College London Hospitals NHS Foundation Trust (London, UK).
Most individuals had undergone some previous genetic testing using routine diagnostic approaches: direct Sanger sequencing of highly suggestive genes based on clinical characteristics, direct sequencing of candidate genes in gene discovery projects, and arrayed primer extension (APEX, Asper Biotech Ltd.) assays for retinitis pigmentosa (MIM: 268000) and Leber congenital amaurosis (LCA [MIM: 204000]). Individuals with no likely pathogenic variant detected by these methods were recruited to the study although systematic documentation of pre-screening of all cases was not available. 152 individuals of the cohort had no pre-screening performed.
High-Throughput Sequencing
WES and WGS were performed as previously described.17, 18 Genome build GRCh37 was used for mapping and variant calling. SVs were identified in the individuals who had WGS using two independent algorithms: Isaac Copy Number Variant Caller (Canvas, Illumina), which identifies copy number gains and deletions based on read depth, and Isaac Structural Variant Caller (Manta, Illumina), which identifies translocations, deletions, tandem duplications, insertions, and inversions based on both paired read fragment spanning and split read evidence.19
To identify the likely ethnicity of each individual, a principal-component analysis was performed on WGS data from individuals of various ethnicities in the 1000 Genomes project with 20,000 single-nucleotide variants (SNVs) using PLINK.20 The location of the center of the cluster of each ethnicity was then calculated, and a likely ethnicity assigned to each of the WGS samples based on the closest cluster.
Variant Interpretation
To facilitate variant interpretation, a list of reported IRD-associated genes was assembled, including genes associated with syndromic forms of IRD or albinism, from various sources including RetNet and literature searches. This list was manually curated according to published evidence of pathogenicity to compile a shortlist of 224 high-confidence IRD-associated genes (Table S1).21
To identify pathogenic variants, a two-step variant filtering protocol was designed, utilizing automated filtering followed by manual review. For SNVs and indels, automated filtering identified variants that fulfil the following criteria: passes standard Illumina quality filters in >80% of the whole NIHR BioResource Rare Diseases cohort (n = 6,688); predicted to be a high impact, medium impact, or splice region variant, or present in the HGMD Pro database;22 and has minor allele frequency (MAF) < 0.01 in control datasets including the NIHR BioResource Rare Diseases cohort and the Exome Aggregation Consortium (ExAC) database.23 If a variant is present in the HGMD Pro database, a higher MAF threshold of 0.1 was used. Finally, we identified just those variants that affect an IRD-associated gene. In one individual (G002628) with hypomagnesemia and retinitis pigmentosa, we interrogated known hypomagnesemia-associated genes in addition to IRD-associated genes. For SVs in the WGS data, automated filtering identified variants that fulfil the following criteria: passes standard Illumina quality filters, overlaps at least one exon, does not overlap known benign SVs in healthy cohorts,24 has MAF < 0.01 in the NIHR BioResource Rare Diseases cohort, and affects an IRD-associated gene.
Manual review of all the variants that passed the automated filtering was then performed. The variant is considered to be pathogenic if it fulfils the following criteria. First, the genotype and frequency of the variant is consistent with the expected mode of inheritance of the individual’s family (if known) and with the expected mode of inheritance of the gene. Second, the phenotype of the individual is consistent with the phenotype known to be associated with the gene. Third, the variant is predicted to result in truncation of the protein, or it is predicted to be damaging to the protein using scores such as CADD,25 or it has previously been reported as pathogenic in HGMD Pro. Fourth, the variant appears to be of good quality upon examination of the sequencing reads with Integrative Genomics Viewer (IGV).26 Fifth, the variant affects the Ensembl canonical transcript or a known retinal transcript.
Variant Confirmations
A subset of the pathogenic SNVs and indels was confirmed by Sanger sequencing using standard protocols. For SVs, Sanger sequencing was performed across the predicted breakpoints to generate a unique junction fragment sequence. Genotyping using HumanCoreExome-24 v.1.0 BeadChip (Illumina) was also performed, followed by SV identification using GenomeStudio and cnvPartition software (Illumina). Sequences of all primers are available on request.
Comparing Coverage of WGS and WES Datasets
Protein-coding regions of the Ensembl canonical transcript of each autosomal IRD-associated gene were split into 50 bp bins. The mean GC% of each bin was calculated using data available from the UCSC Genome Browser. The mean coverage of each bin was calculated across a sample of 100 individuals with IRD in the NIHR BioResource Rare Diseases cohort (WGS data) and also in the ExAC cohort, for which per-base coverage has been previously published on 10% of the cohort.23 We used the ExAC WES dataset for the comparison of the coverage of IRD-associated genes, rather than the WES data generated as part of this study, because ExAC was used as a frequency control set in this study and because it is based on >60,000 individuals.
Coverage data are presented as relative to the mean coverage of all bins (34.4× in the WGS dataset and 65.6× in ExAC). Coverage for each observed range of GC content (20%–30%, 30%–40%, 40%–50%, 50%–60%, 60%–70%, 70%–80%, and 80%–90%) was compared using one-tailed Mann-Whitney U tests. p values were corrected for testing of seven observed ranges of GC content.
Variant Analysis in CHM
Two individuals with a clinical diagnosis of choroideremia (MIM: 303100) in whom no pathogenic coding variant was found underwent intronic variant analysis of CHM ([MIM: 300390]; ENST00000357749.2, GRCh37/hg19 chrX: 85,116,185–85,302,566). Intronic variants in CHM that passed standard quality filters and had a MAF ≤ 0.01 in the UK10K WGS cohort and the NIHR BioResource Rare Diseases cohort were prioritized for analysis. Splice site prediction of the reference and alternate sequences were compared using Human Splice Finder V3.0 (HSF) and NNSPLICE0.9.27, 28 We identified a candidate variant that was confirmed by direct Sanger sequencing of genomic DNA from affected members of both families and an obligate carrier. Reverse-transcription PCR (RT-PCR) spanning CHM exons 4–5 was carried out on total RNA isolated from peripheral blood mononuclear cells.
Identification of Genes Containing Likely Biallelic Predicted Protein-Truncating Variants in Unsolved Cases
Variants (SNVs, indels, and large deletions) were identified that fulfil the following criteria: in an unsolved case where an individual has undergone WGS, are high impact and autosomal, the genotype is consistent with autosomal-recessive inheritance, have MAF < 0.001 in the control datasets described, are in the Ensembl canonical transcript, are in a gene that contains no homozygous predicted protein-truncating variants in the ExAC database, and appear to be real on examination in IGV. This analysis was not limited to IRD-associated genes, but included all protein-coding genes.
Results
In a Highly Heterogeneous Cohort of Individuals with Inherited Retinal Disease, High-Throughput Sequencing Achieves a Pathogenic Variant Detection Rate of 56%
High-throughput sequencing was performed on a cohort of 722 individuals with IRD, as part of the NIHR-BioResource Rare Diseases research study. 605 of the individuals had WGS, 72 had WES, and an additional 45 had both. For WGS average coverage was 37× (SD = 2.7), with a minimum of 95% of each genome covered to at least 15×. For WES average target coverage was 43× (SD = 14.9), with 83% of target bases covered to at least 15×.
The cohort is phenotypically heterogeneous, and the most frequent phenotypes are retinitis pigmentosa (n = 311), retinal dystrophy (n = 101), cone-rod dystrophy (n = 53), Stargardt disease (n = 45), macular dystrophy (n = 37), and Usher syndrome (n = 37). The majority of individuals are unrelated probands (714/722), and there are three parent-offspring pairs and one pair of siblings. The majority of the cohort had a negative result for a genetic test, such as single-gene Sanger sequencing or targeted gene panel sequencing, prior to enrolment in the current study.
To identify the pathogenic variants, rare, coding, and high-quality variants, including large deletions, in 224 genes that are known to be associated with IRD were considered. The likely cause of IRD was identified in 404/722 individuals, corresponding to a pathogenic variant detection rate of 56% (Tables 1 and S2). A further 5% of individuals (36/722) have been classified as partially solved. These individuals either have a single likely pathogenic variant in a known gene associated with recessive IRD and in keeping with the individual’s phenotype, more than two heterozygous variants in a gene associated with recessive IRD, or a variant that explains only part of the phenotype (Table S2). The remaining 282/722 (39%) of individuals remain without a complete or partial molecular diagnosis.
Table 1.
Pathogenic Variant Detection Rates by Sequencing Technology
| Sequencing Method | Total Cases | Cases Solved | Cases Partially Solved | Cases Unsolved |
|---|---|---|---|---|
| WGS | 605 | 331 (55%) | 31 (5%) | 243 (40%) |
| WES | 72 | 59 (82%) | 3 (4%) | 10 (14%) |
| WES and WGS | 45 | 14 (31%) | 2 (4%) | 29 (64%) |
| TOTAL | 722 | 404 (56%) | 36 (5%) | 282 (39%) |
WES alone solved 59/117 (50%) cases. Subsequently, 45 of the 58 cases that are unsolved by WES also underwent WGS. Partially solved cases have either one likely pathogenic variant in a gene with biallelic inheritance or more than two heterozygous variants in a gene with biallelic inheritance, or a variant that appears to explain only part of the phenotype.
By using WES alone we were able to identify pathogenic variants in 59/117 (50%) individuals. Subsequently, 45 of the 58 individuals who were unsolved by WES underwent WGS, and we identified or confirmed pathogenic variants in a further 14 (Table 1). For three individuals the bait for the variant location was absent from the capture kit (Roche NimbleGen, SeqCap EZ Exome v.3), for two individuals each had a large deletion not called by WES, and in one individual, a large indel was also not called by WES. For three more individuals, the variant was called by WES but the quality was poor. For the remaining five individuals, although the variants were identified by WES, they were not the expected mode of inheritance for the family, and therefore WGS was performed to exclude more plausible cause of disease by including analysis of copy-number variants and exons of high GC content.
The pathogenic variant detection rate also varies depending on the phenotype of the individual (Table 2). For example, 168/311 (54%) of individuals with retinitis pigmentosa have been solved, which is similar to the overall rate; 31/37 (84%) of individuals with Usher syndrome have been solved but only 6/21 (29%) of individuals with cone dystrophy have been solved. Also, individuals who had not had any genetic test prior to this study had a notably higher pathogenic variant detection rate than the overall rate. Of 152 individuals who had no pre-screening, including individuals with a diagnosis of retinitis pigmentosa, cone dystrophy, or congenital stationary night blindness, 96 were solved (63%), suggesting that our cohort is enriched for intractable cases.
Table 2.
Pathogenic Variant Detection Rate by Phenotype
| Phenotype | Total Cases | Cases Solved | Cases Partially Solved | Cases Unsolved |
|---|---|---|---|---|
| RP | 311 | 168 (54%) | 11 (4%) | 132 (42%) |
| RD | 101 | 55 (54%) | 5 (5%) | 41 (41%) |
| CRD | 53 | 29 (55%) | 3 (6%) | 21 (40%) |
| Stargardt | 45 | 27 (60%) | 10 (22%) | 8 (18%) |
| Macular dystrophy | 37 | 18 (49%) | 1 (3%) | 18 (49%) |
| Usher | 37 | 31 (84%) | 2 (5%) | 4 (11%) |
| Other | 27 | 10 (37%) | 0 (0%) | 17 (63%) |
| CSNB | 26 | 23 (88%) | 0 (0%) | 3 (12%) |
| Cone dystrophy | 21 | 6 (29%) | 1 (5%) | 14 (67%) |
| Multiple | 21 | 6 (29%) | 3 (14%) | 12 (57%) |
| LCA | 18 | 16 (89%) | 0 (0%) | 2 (11%) |
| Achromatopsia | 9 | 6 (67%) | 0 (0%) | 3 (33%) |
| Albinism | 8 | 6 (75%) | 0 (0%) | 2 (25%) |
| FEVR | 8 | 3 (38%) | 0 (0%) | 5 (62%) |
| TOTAL | 722 | 404 (56%) | 36 (5%) | 282 (39%) |
Abbreviations: RP, retinitis pigmentosa; RD, retinal dystrophy; CRD, cone-rod dystrophy; other, any phenotype with frequency of less than eight; CSNB, congenital stationary night blindness; multiple, more than one phenotype including syndromic cases; LCA, Leber congenital amaurosis; FEVR, familial exudative vitreoretinopathy.
The pathogenic variant detection rate within our cohort also varies depending on the ethnicity of the individual (Table 3). Likely ethnicity was estimated from the WGS data. Only 13/43 (30%) of individuals of African ancestry were solved, compared to 259/467 (55%) of individuals of European ancestry and 70/123 (57%) of individuals of South Asian ancestry. Higher genetic diversity in African populations, combined with underrepresentation of non-European populations in control datasets, result in an excess of rare and apparently rare variation in these individuals, rendering variant interpretation more challenging.23, 29 There was a median of 12 rare coding variants in IRD-associated genes for manual review in individuals of European ancestry and 30 in individuals of African ancestry. Interestingly, in individuals of South Asian ancestry, 59/89 pathogenic variants (66%) were homozygous, compared to only 82/446 (18%) in individuals of European ancestry. This is likely due to increased rates of consanguinity in South Asian populations, and probably explains why they have comparable pathogenic variant detection rates to individuals of European ancestry in this study, despite also being underrepresented in control databases of allele frequencies.
Table 3.
Pathogenic Variant Detection Rate by Ethnicity
| Likely Ethnicity | Total Cases | Cases Solved | Cases Partially Solved | Cases Unsolved |
|---|---|---|---|---|
| EUR | 467 | 259 (55%) | 23 (5%) | 185 (40%) |
| SAS | 123 | 70 (57%) | 4 (3%) | 49 (40%) |
| AFR | 43 | 13 (30%) | 4 (9%) | 26 (60%) |
| EAS | 13 | 1 (8%) | 1 (8%) | 11 (85%) |
| AMR | 4 | 2 (50%) | 1 (25%) | 1 (25%) |
| TOTAL | 650 | 345 (53%) | 33 (5%) | 272 (42%) |
Likely ethnicity estimated from WGS data using principal component analysis. Table includes individuals who had WGS only. Abbreviations: EUR, European; SAS, South Asian; AFR, African; EAS, East Asian; AMR, Ad Mixed American.
The genes in which pathogenic variants are most frequently found are summarized in Table 4. ABCA4 (MIM: 601691) is implicated in 73/440 solved or partially solved cases (17%). In total, 796 pathogenic alleles were identified in 95 different known IRD-associated genes. Of these alleles, 687 (86.3%) are autosomal recessive, 72 (9%) are autosomal dominant, 35 (4.4%) are X-linked hemizygous, and 2 (0.3%) are X-linked monoallelic. Some of the pathogenic alleles are homozygous, and some occur in more than one individual in the cohort, thus there are 537 different pathogenic alleles. Of these, 499 (92.9%) are exonic SNVs or indels, 30 (5.6%) are large deletions overlapping at least one exon, 6 (1.1%) are SNVs or indels that are synonymous or in regulatory regions, and 2 (0.4%) are tandem duplications overlapping at least one exon. 291/537 (54.2%) are previously unreported in publically available databases emphasizing the allelic heterogeneity of IRD, and the rest have been previously reported and are in the HGMD Pro database.22
Table 4.
Number of Solved and Partially Solved Cases by Gene
| Gene | Total Cases | Cases Solved | Cases Partially Solved |
|---|---|---|---|
| ABCA4 | 73 | 57 | 16 |
| USH2A | 61 | 50 | 11 |
| EYS | 16 | 15 | 1 |
| RP1 | 16 | 16 | 0 |
| CACNA1F | 13 | 13 | 0 |
| RPGR | 13 | 13 | 0 |
| CRB1 | 12 | 11 | 1 |
| CNGB1 | 9 | 8 | 1 |
| MYO7A | 8 | 8 | 0 |
| PDE6B | 8 | 8 | 0 |
| BBS1 | 7 | 7 | 0 |
| CERKL | 7 | 7 | 0 |
| CNGB3 | 7 | 7 | 0 |
| PROM1 | 7 | 7 | 0 |
| RHO | 7 | 7 | 0 |
| CDHR1 | 6 | 6 | 0 |
| CLN3 | 6 | 6 | 0 |
| PRPF31 | 6 | 6 | 0 |
| PRPH2 | 6 | 6 | 0 |
| RP2 | 5 | 5 | 0 |
| TRPM1 | 5 | 5 | 0 |
Only genes with pathogenic variants in five or more individuals are shown.
We performed confirmatory Sanger sequencing and segregation in families in a subset of variants. Of 177 alleles Sanger sequenced, all were confirmed. Of these, 52 were tested in at least one additional informative family member, and segregated as expected. In five of the individuals whose case is unsolved, a pathogenic variant was identified by a different method concurrent to this study. Four of these are variants in highly repetitive regions requiring specific optimized Sanger sequencing protocols, for example RPGR (MIM: 312610) exon ORF15, and one is a heteroplasmic variant in the mitochondrial genome, which was apparent retrospectively in the WGS data but was not identified by the variant caller.
Pathogenic Structural Variants Are an Important Cause of IRD, and Their Detection Is Facilitated by WGS
There are 33 SVs (31 deletions and 2 tandem duplications) that are pathogenic in 31 individuals (Table S2). Each SV was further investigated using either SNP genotyping array, direct Sanger sequencing of a unique PCR amplified product spanning the predicted breakpoints, or both techniques. 23 (70%) have been confirmed by Sanger sequencing of the breakpoint, 6 (18%) have been confirmed by both Sanger sequencing the breakpoint and SNP genotyping array, 2 (6%) have been confirmed by genotyping only, and 2 (6%) have not yet been confirmed by an alternative method, although visual inspection of the IGV plots predict a deletion. In seven SVs, confirmatory Sanger sequencing of PCR generated products over the breakpoint also revealed an insertion (1–51 bps) at the breakpoint of the variants predicted by the algorithms.
The Manta algorithm, which identifies structural variants based on both paired read fragment spanning and split read evidence, predicts the correct breakpoints with 100% accuracy in 13/26 SVs that were called by Manta and have been confirmed by Sanger sequencing. It predicts the correct breakpoints to within 3 bp in 21/26 SVs. Interestingly, we have identified one identical SV in two unrelated individuals. G001296 and W000139 both have 15: 89,750,128–89,757,489del, which overlaps exons 7–9 of RLBP1 (MIM: 180090). In G001296 the deletion is heterozygous and occurs in conjunction with a second pathogenic variant, and in W000139 it is homozygous. It does not occur in any other individual in the NIHR BioResource Rare Diseases cohort.
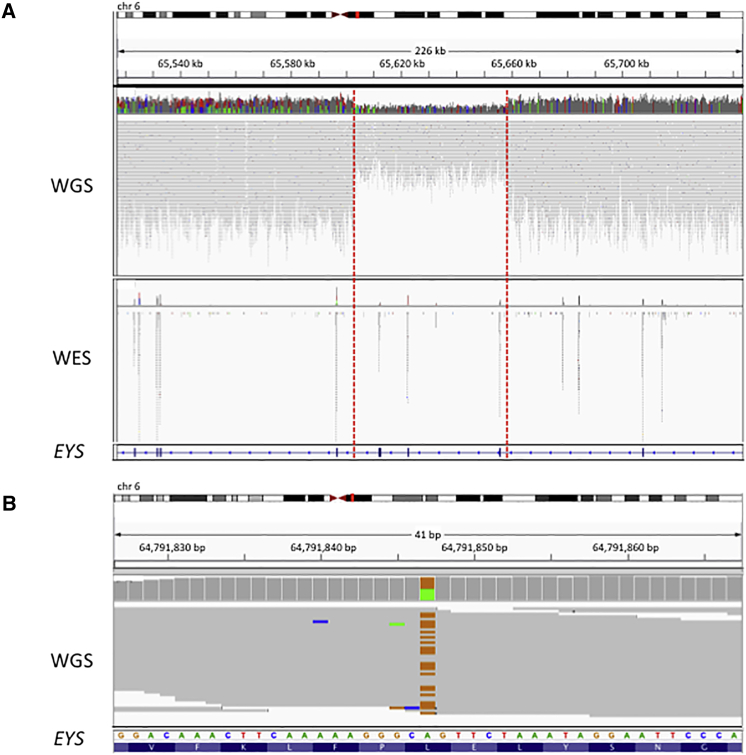
Case study one illustrates the power of WGS to detect SVs. Two likely pathogenic heterozygous variants were identified in EYS (MIM: 612424) in individual W000325, who presented with typical retinitis pigmentosa. The variants are a missense variant ENST00000503581.1; c.6473T>C (p.Leu2158Pro) that is predicted to be damaging by CADD (phred-scaled CADD score = 23.5) and has not been reported before in public databases, and a 55 kb deletion (chr6: 65,602,819–65,658,187del), confirmed by Sanger sequencing, which overlaps exons 15–18 (c.2260-2380_2847-6084del). If transcribed, this large deletion is predicted to lead to a frameshift and premature termination in exon 19 (p.Arg949Ilefs∗4). This individual had undergone WES analysis prior to WGS analysis, from which it was not possible to identify the pathogenic deletion (Figure 1).
Figure 1.
Case Study 1: WGS, Compared to WES, Increases Power to Detect SVs
(A) One individual (W000325) with retinitis pigmentosa has a pathogenic heterozygous deletion within EYS (6:656,02,819–65,658,187). IGV plots showing the deleted region in both WGS and WES data in the individual. The deletion was identified by WGS due to the drop in coverage and increased distance between mate-paired reads. However, it was not identified from WES data.
(B) The individual also has a pathogenic missense variant ENST00000503581.1; c.6473T>C (p.Leu2158Pro).
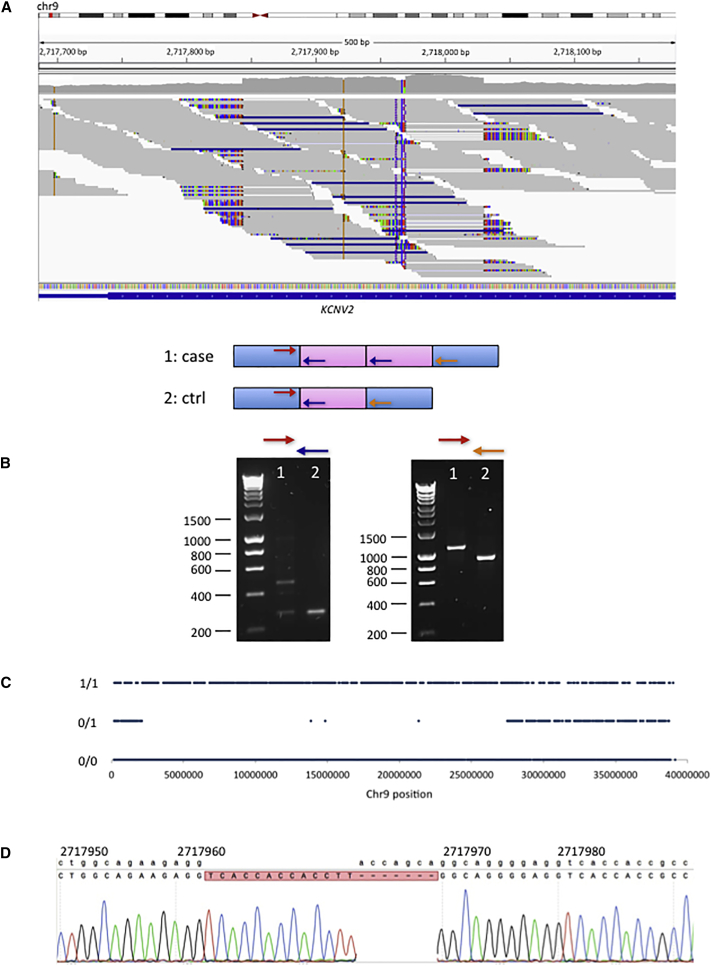
Case study two is an interesting example of likely uniparental isodisomy discovered through analysis of SVs. Individual W000170 presented with an atypical early-onset form of retinal dystrophy. A homozygous in-frame combination indel in KCNV2 (MIM: 607604) was first identified: ENST00000382082.3; c.222_232delGGACCAGCAGGinsGGTCACCACCACCTTGG (ENSP00000371514.3; p.Asp75_Gln77delinsValThrThrThrLeu). The mother of the individual is heterozygous for this indel, and the father is homozygous for the reference allele. By visual inspection of the surrounding region using the IGV software,26 a homozygous tandem duplication was identified (chr9: 2,717,844–2,718,030dup), flanking the indel (Figure 2), which was confirmed by Sanger sequencing. Further investigation revealed a long run of homozygosity with approximate coordinates chr9: 2,100,000–27,400,000. Taken together, we consider that the most likely explanation for these observations is homozygosity for the KCNV2 disease allele due to partial maternal uniparental isodisomy of chromosome 9.
Figure 2.
Case Study 2: Identification by WGS of Pathogenic Tandem Duplication and Likely UPD
One individual (W000170) with atypical, early-onset retinal dystrophy, has a homozygous pathogenic tandem duplication within KCNV2, likely due to partial maternal uniparental isodisomy of chromosome 9.
(A) IGV plot showing the 184 bp tandem duplication of 9: 2,717,844–2,718,028 in WGS data.
(B) The tandem duplication was confirmed by PCR and Sanger sequencing. The arrows represent the positions of the primers.
(C) There is a ∼25 Mb region of homozygosity in chromosome 9 in this individual, which encompasses KCNV2. Approximate coordinates of this region of homozygosity are 9:2,100,000–27,400,000.
(D) Sanger sequencing also confirms a homozygous combined in-frame insertion/deletion in KCNV2, within the tandem duplication: ENST00000382082.3; c.222_232delGGACCAGCAGGinsGGTCACCACCACCTTGG (ENSP00000371514.3; p.Asp75_Gln77delinsValThrThrThrLeu). The mother of the affected individual is heterozygous for this variant, but the father is homozygous for the reference allele (not shown).
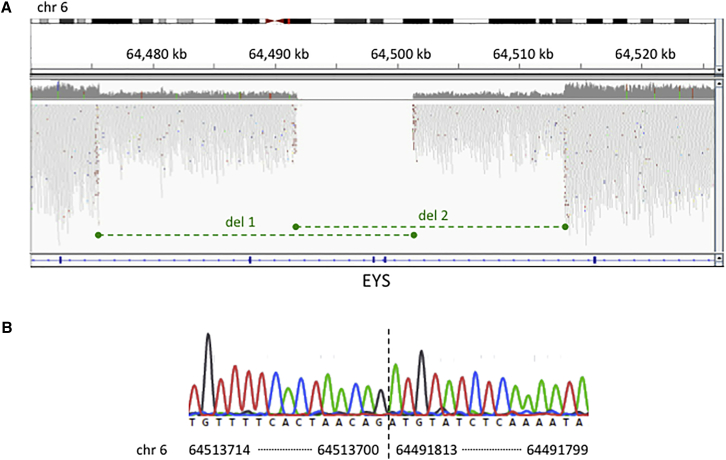
Case study three is an interesting example of overlapping compound heterozygous deletions. Individual W000164 presented with typical retinitis pigmentosa. Two likely pathogenic overlapping heterozygous deletions were identified by WGS. The first (chr6: 64,475,599–64,501,270del) encompasses exons 38–40 of EYS and the second (chr6: 64,491,812–64,513,698del) encompasses exons 38–39 of EYS (Figure 3). Sanger sequencing confirmed both deletions. This example illustrates the value of algorithms that use paired read fragment spanning evidence such as Manta. Only these are able to provide characterization of breakpoints to a single base pair resolution. WES or indeed read depth analysis alone of WGS (e.g., the Canvas algorithm) would fail to characterize the breakpoints precisely, rendering Sanger sequencing confirmation or family analysis more difficult.
Figure 3.
Case Study 3: Identification and Characterization of Compound Heterozygous Deletions in EYS by WGS
(A) One individual (W000164) with retinitis pigmentosa has compound heterozygous deletions in EYS, shown by the green dashed lines.
(B) Sanger sequencing confirmed both deletions. Sequencing across the breakpoint (vertical dashed line) of deletion 2 is shown.
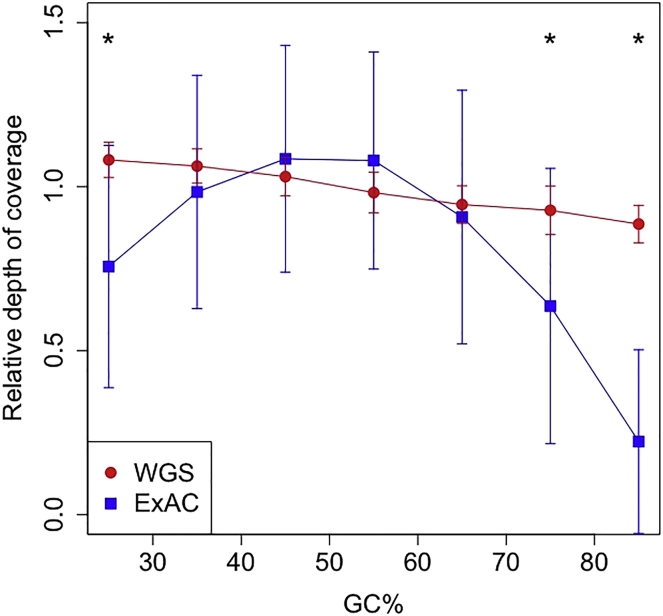
WGS Improves Coverage of GC-Rich Regions Compared to WES
In the current study, we have observed increased power to detect variants in GC-rich regions. To assess the possible impact of this, we calculated the average coverage of exons of known IRD-associated genes, split into 50 bp bins, in our WGS dataset and in the ExAC WES dataset. For exons of IRD-associated genes with a GC content of 45%–65%, the average coverage by read depth of sequence using WES is higher than for WGS. The calling of SNV exonic variants in this range is not significantly different for WES and WGS. Comparing WES from ExAC, we find significantly higher coverage in our WGS dataset in bins with GC < 30% or > 70% (Figure 4). Additionally, the variability of coverage in our WGS data is much less than that of the ExAC dataset. This uniformity of coverage is one of the main factors that make WGS particularly powerful for SV detection. Calling SVs in WES where the variance of read depth is so great is unreliable (Figure 1A).
Figure 4.
Regions of IRD-Associated Genes that Are Low or High in GC Content Have Significantly Higher Coverage in WGS Data than in the ExAC WES Project
Coverage was calculated across protein-coding regions of autosomal IRD-associated genes (Ensembl canonical transcript), split into 50 bp bins. Coverage shown is relative to the average coverage of each dataset. Error bars show standard deviation. ∗p < 1 × 10−15.
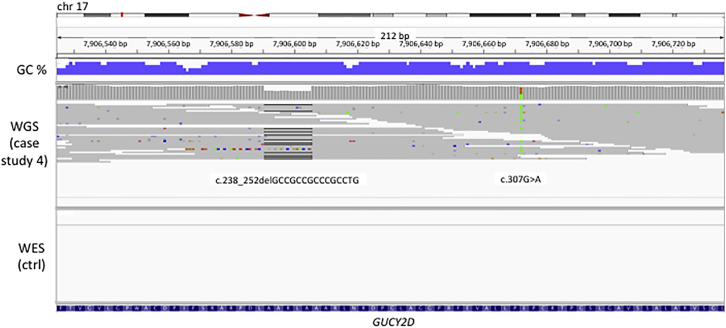
Case study four illustrates the clinical relevance of this improvement in coverage. Individual G004991 presented with Leber congenital amaurosis. Pathogenic compound heterozygous variants were identified in the first coding exon of GUCY2D (MIM: 600179), which is 76% GC (Figure 5). The variants are an in-frame deletion (ENST00000254854.4; c.238_252delGCCGCCGCCCGCCTG [p.Ala80_Leu84del]) not previously reported in publically available databases and a previously reported missense variant (ENST00000254854.4; c.307G>A [p.Glu103Lys]).30 This exon is not covered in the capture kit used for WES, and had WES been used rather than WGS to investigate this individual, neither pathogenic variant would have been identified. Furthermore, visual inspection of the sequencing reads using IGV demonstrates biallelic inheritance of the two variants, since they occur within 70 nucleotides and are never observed on the same 150 bp read.
Figure 5.
Case Study 4: Identification of Pathogenic Variants by WGS in GC-Rich Regions Not Covered by WES
One individual (G004991) with Leber congenital amaurosis has pathogenic compound heterozygous variants ENST00000254854.4; c.238_252delGCCGCCGCCCGCCTG (p.Ala80_Leu84del) and ENST00000254854.4; c.307G>A (p.Glu103Lys) in exon 1 of GUCY2D, which is 76% GC-rich. This exon is not covered by WES in our cohort, as demonstrated by the WES data of a control sample shown here. It is also not well covered in ExAC.
WGS Allows Identification of Pathogenic Variants in Non-coding Regions
In our cohort, three different pathogenic non-coding SNVs were identified. For example, in G008165, who presented with Stargardt disease, one intronic SNV was identified in ABCA4 in trans with a previously reported synonymous SNV, 1:94466602C>T (ENST00000370225.3; c.6342G>A), that results in a premature donor splice site that truncates exon 46.31 The heterozygous intronic variant was 1:94476951A>G (ENST00000370225.3; c.5461−10T>C), which has also been previously reported and causes aberrant splicing.32 This variant was found in 16 individuals, all of whom presented with typical features of ABCA4 retinopathy, and thus represents a significant portion of disease alleles. In two of these individuals this variant is homozygous, in a further nine individuals it occurs in conjunction with a second heterozygous likely pathogenic allele, and in five, we have identified no second likely pathogenic allele and the individuals remain partially solved. This variant is rare in both ExAC (MAF = 2.2 × 10−4, no homozygotes) and the whole NIHR BioResource Rare Diseases cohort (MAF = 1.8 × 10−3, no homozygotes other than the two described here).
In another example, individual G001035, who presented with Usher syndrome, the deep intronic USH2A (MIM: 608400) variant 1:216064540T>C (ENST00000307340.3; c.7595−2144A>G) was identified. This is heterozygous and occurs in conjunction with a second heterozygous likely pathogenic allele. It has been previously reported, and results in the retention of a pseudoexon, which causes a frameshift and premature truncation of the protein.33
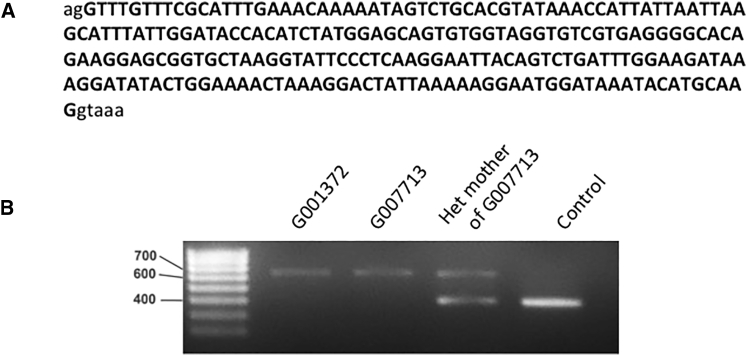
Finally, in two unrelated males with a clinical diagnosis of choroideremia (G001372 and G007713), we identified a deep intronic variant in CHM that has not been previously reported. Both individuals had previously undergone CHM screens, which were negative, and no convincing pathogenic coding variant was found in any IRD-associated gene in either individual upon analysis of the WGS data. Given the unequivocal diagnosis of choroideremia in these individuals and the single genetic cause of choroideremia, the sequence of these case subjects was selected for interrogation of rare non-coding variants in CHM (chrX: 85,116,185–85,302,566). We hypothesized that a deep intronic variant may act as a null variant by altering splicing as previously described.34
A rare (MAF ≤ 0.01) deep intronic hemizygous variant was identified in G001372 in the genomic region of CHM: chrX: 85,220,593T>C (ENST00000357749.2; c.315–1536A>G). Individual G007713 had three rare intronic variants, including the same chrX: 85,220,593T>C variant. This variant is absent in the UK10K genome project dataset and the 1000 Genomes dataset.29, 35 The variant was confirmed in both individuals by direct Sanger sequencing, and it was also confirmed in an affected male cousin of G007713 and in the heterozygous state in the mother of G007713.
Splice prediction analysis using HSF27 and NNSPLICE0.928 demonstrated the likely introduction of a cryptic splice acceptor site by this variant (HSF consensus value: 92.66, NNSPLICE0.9 score 0.94). The presence of a strong donor site (HSF consensus value: 84.71, NNSPLICE0.9 score: 0.89, sequence atgcaaggtaaactg) 224 bp downstream of the cryptic acceptor site suggested that a cryptic exon could arise from this variant (Figure 6).
Figure 6.
The Deep Intronic CHM Variant c.315-1536A>G Results in the Inclusion of a Cryptic Exon
(A) Sequence of the cryptic exon included by the deep intronic CHM variant chrX:85,220,593T>C (ENST00000357749.2; c.315-1536A>G).
(B) RT-PCR showing increased size of fragment due to inclusion of the cryptic exon.
To confirm this, we used RT-PCR amplification spanning exons 4–5 of CHM from G001372, G007713, and the heterozygous mother of G007713. This demonstrated a fragment approximately 200 bp larger than expected (2 fragments in the heterozygous mother). Upon sequencing of the fragments, the predicted cryptic exon was confirmed. The variant is predicted to lead to a premature termination codon after 9 altered amino acid residues (p.Ser105Argfs∗10).
Monoallelic Variants in Genes Associated with a Recessive Mode of Inheritance
Interpreting single heterozygous variants in recessive IRD-associated genes presents a challenge, as it is typically difficult to distinguish between a case in which a second pathogenic allele in the same gene has been missed from a case in which the individual is just a carrier of the single heterozygous variant and the real cause of disease lies elsewhere. In our cohort there are only 16 individuals (2%) who have a single heterozygous likely pathogenic variant in a recessive IRD-associated gene. Of these, 12 are in ABCA4 and one is in each of CEP290 (MIM: 610142), CNGB1 (MIM: 600724), GUCY2D, and CRB1 (MIM: 604210). These individuals all have phenotypes that are strongly indicative of variants in the gene in question, and the identified variant in each case is either a predicted protein-truncating variant or a missense previously reported as being pathogenic. We excluded the possibility that these individuals have an additional previously reported pathogenic deep intronic variant. The probability is high that at least some of these 16 partially solved individuals harbor a second pathogenic variant in the same gene, perhaps in a regulatory region, that remains elusive. This cohort is under continuing investigation to identify further variants, including variants in regulatory regions. The proportion of individuals who have a single likely pathogenic variant in an autosomal-recessive gene in this study is lower than previously reported (2%).16 This may reflect a combination of detailed specialist phenotyping to reduce phenocopies and the comprehensive exon and non-coding coverage achieved using WGS.
Identification of Genes Containing Likely Biallelic Predicted Protein-Truncating Variants in Unsolved Cases
We performed further investigation of 247 individuals, in whom no pathogenic variants were detected in the known IRD-associated genes, and whose family history is not inconsistent with recessive disease. These were screened for any gene containing ≥2 predicted protein-truncating alleles, including SVs. This yielded a list of 19 genes in 16 individuals (Tables 5 and S3). None occurred in more than one individual, but three individuals have variants in two genes. Segregation has not been performed on the double heterozygous variants. None of these genes contain any homozygous predicted protein-truncating variants in the ExAC dataset, suggesting that they may not tolerate biallelic loss-of-function variation.
Table 5.
High-Impact, Likely Biallelic Variants in 19 Genes in Unsolved Cases
| Individual | Phenotype | Sex | Ethnicity | Gene | Variant genomic | Variant HGVSc | Variant HGVSp | GT | Consequence | Description |
|---|---|---|---|---|---|---|---|---|---|---|
| G001284 | multiple | F | SAS | SCAPER | 15:76998312G>A | ENST00000563290.1; c.2179C>T | p.Arg727Ter | 0/1 | stop_gained | S-phase cyclin A-associated protein in the ER |
| G001284 | multiple | F | SAS | SCAPER | 15:77064214CA>C | ENST00000563290.1; c.1116delT | p.Val373SerfsTer21 | 0/1 | frameshift | S-phase cyclin A-associated protein in the ER |
| G001298 | RD | F | SAS | FUT5 | 19:5867071G>T | ENST00000252675.5; c.666C>A | p.Tyr222Ter | 1/1 | stop_gained | fucosyltransferase 5 (alpha (1,3) fucosyltransferase) |
| G001298 | RD | F | SAS | PODNL1 | 19:14046820C>CAGCT | ENST00000339560.5; c.374_377dupAGCT | p.Gln127AlafsTer119 | 1/1 | frameshift | podocan-like protein 1-like |
| G001411 | RP | M | AFR | NAALADL1 | 11:64812774G>GC | ENST00000358658.3; c.2191dupG | p.Ala731GlyfsTer9 | 0/1 | frameshift | N-acetylated alpha-linked acidic dipeptidase-like 1 |
| G001411 | RP | M | AFR | NAALADL1 | 11:64822078G>A | ENST00000358658.3; c.736C>T | p.Arg246Ter | 0/1 | stop_gained | N-acetylated alpha-linked acidic dipeptidase-like 1 |
| G005002 | CRD | M | SAS | WASF3 | 13:27216381GTGTTTTCAATTTTCAGATTGTGAACCA>G | ENST00000335327.5; c.−10-14_3delTTTTCAATTTTCAGATTGTGAACCATG | NA | 1/1 | splice_acceptor | WAS protein family, member 3 |
| G005019 | Usher | F | SAS | PLD4 | 14:105395186GC>G | ENST00000392593.4; c.388delC | p.Gln130ArgfsTer108 | 0/1 | frameshift | phospholipase D family, member 4 |
| G005019 | Usher | F | SAS | PLD4 | 14:105398102CTGTCCCCA>C | ENST00000392593.4; c.937_944delTGTCCCCA | p.Cys313GlyfsTer167 | 0/1 | frameshift | phospholipase D family, member 4 |
| G005203 | RP | F | SAS | FAM71A | 1:212799206T>TGCAG | ENST00000294829.3; c.991_994dupGGCA | p.Thr332ArgfsTer88 | 1/1 | frameshift | family with sequence similarity 71, member A |
| G005251 | cone dystrophy | M | SAS | POMZP3 | 7:76240807T>TG | ENST00000310842.4; c.538dupC | p.Gln180ProfsTer14 | 1/1 | frameshift | POM121 and ZP3 fusion |
| G005492 | RP | F | EUR | IRX5 | 16:54967694CTAAAG>C | ENST00000394636.4; c.1362_1366delTAAAG | p.Lys455ProfsTer19 | 1/1 | frameshift | iroquois homeobox 5 |
| G005513 | Stargardt | M | EUR | ITIH2 | 10:7776934CT>C | ENST00000358415.4; c.1838delT | p.Leu613ArgfsTer5 | 0/1 | frameshift | inter-alpha-trypsin inhibitor heavy chain 2 |
| G005513 | Stargardt | M | EUR | ITIH2 | 10:7780709CAT>C | ENST00000358415.4; c.2084_2085delAT | p.His695ArgfsTer5 | 0/1 | frameshift | inter-alpha-trypsin inhibitor heavy chain 2 |
| G005514 | RP | M | AFR | SLC37A3 | 7:140064249G>A | ENST00000326232.9; c.334C>T | p.Arg112Ter | 1/1 | stop_gained | solute carrier family 37, member 3 |
| G007696 | CRD | M | SAS | NUMB | 14:73746066G>GC | ENST00000355058.3; c.1162dupG | p.Ala388GlyfsTer6 | 1/1 | frameshift | numb homolog (Drosophila) |
| G007696 | CRD | M | SAS | FAM57B | 16:30038139GC>G | ENST00000380495.4; c.234delG | p.Gln79AsnfsTer43 | 1/1 | frameshift | family with sequence similarity 57, member B |
| G007723 | RP | M | SAS | FOXI2 | 10:129536034AC>A | ENST00000388920.4; c.498delC | p.Asp166GlufsTer87 | 1/1 | frameshift | forkhead box I2 |
| G008152 | RP | M | EUR | CROCC | 1:17292217G>A | ENST00000375541.5; c.4406−1G>A | NA | 1/1 | splice_acceptor | ciliary rootlet coiled-coil, rootletin |
| W000146 | RP | F | EUR | CCZ1B | 7:6844600C>A | ENST00000316731.8; c.1075G>T ENST00000375541.5; c.4406−1G>A |
p.Glu359Ter | 1/1 | stop_gained | CCZ1 vacuolar protein trafficking and biogenesis associated homolog B (S. cerevisiae) |
| W000278 | other | F | SAS | OR2M7 | 1:248487484AG>A | ENST00000317965.2; c.386delC | p.Pro129LeufsTer2 | 1/1 | frameshift | olfactory receptor, family 2, subfamily M, member 7 |
| W000375 | RP | M | SAS | PRTFDC1 | 10:25231367T>C | ENST00000320152.6; c.49−2A>G | NA | 1/1 | splice_acceptor | phosphoribosyl transferase domain containing 1 |
| W000375 | RP | M | SAS | UBAP1L | 15:65395024T>G | ENST00000559089.1; c.121−2A>C | NA | 1/1 | splice_acceptor | ubiquitin associated protein 1-like |
Genomic coordinates refer to genome build GRCh37. Analysis is limited to cases that underwent WGS in whom pathogenic variants in known genes were not detected and whose family history is not inconsistent with recessive inheritance of disease. Abbreviations: RP, retinitis pigmentosa; RD, retinal dystrophy; CRD, cone-rod dystrophy; other, any phenotype with frequency of fewer than eight individuals; multiple, more than one phenotype including syndromic cases.
Discussion
Overall, we have identified pathogenic variants for 404/722 individuals with IRD in the NIHR-BioResource Rare Diseases study, which is a pathogenic variant detection rate of 56%. Factors that influence this rate within our cohort include the phenotype of the individual, for example individuals with Usher syndrome are substantially more likely to receive a molecular diagnosis than individuals with retinitis pigmentosa. There are several possible reasons for this. First, it may be that a higher proportion of Usher syndrome genes overall have been identified than retinitis pigmentosa genes. Second, more specific phenotypes can suggest a smaller number of candidate genes; retinitis pigmentosa is far more genetically heterogeneous than Usher syndrome. Third, it is more difficult to distinguish pathogenic monoallelic variants from the many rare benign inherited monoallelic variants than it is to distinguish pathogenic biallelic variation; therefore, phenotypes that are predominantly recessive tend to have higher pathogenic variant detection rates. Also, it may be that individuals with some phenotypes were more likely to undergo some pre-screening prior to enrolment in the project, which would exclude individuals with variants in known genes and likely reduce the pathogenic variant detection rate.
Our pathogenic variant detection rate of 56% with WGS is comparable to those previously reported in other similar studies of IRD, which ranges between 39% and 70%.9, 10, 11, 12, 13, 14, 16 Factors that influence differences between these rates include the technology used and the degree of pre-screening performed on the cohort, as well as the phenotypes included and the ethnic distribution of the cohort as discussed. Our detection rate of 50% using WES in 117 individuals is perhaps lower than expected due to the WES depth of coverage of 43× used compared to diagnostic laboratory median coverage of >80× that is recommended.36 Our observation that the subset of our cohort who had no pre-screening had a higher pathogenic variant detection rate than the overall rate suggests that our cohort is enriched for intractable cases and that the overall rate of 56% is an underestimate compared with what would be expected if WGS was used as the first-line test.
We have observed that using primarily WGS instead of WES or targeted gene panels has improved our power to detect three categories of variants in particular: SVs, variants in GC-rich regions, and variants in regulatory regions. Previous studies have also observed that WGS is superior to WES for the detection of SVs, particularly small deletions (e.g., single exon deletions).6, 37, 38 This is due to high uniformity of coverage achieved with WGS and the very high probability that the breakpoints of a SV will be covered by WGS reads. With WGS, therefore, precise characterization of the SV to single base pair resolution is often possible, without any further investigation.
Our study also supports previous reports that PCR-free sequencing protocols, such as WGS, capture genomic regions that are particularly high or low in GC content much more effectively than methods such as WES and targeted gene panels, which require a PCR amplification step.36, 37 GC-rich regions tend to be poorly covered by methods that require a PCR amplification step due to their high stability and consequent resistance to standard denaturation protocols, and variant calling suffers as a consequence.39, 40, 41
The WGS analysis identified three intronic variants that would not have been identified by standard WES without specific prior knowledge. The interpretation of these variants required the availability of sequence from a large number of internal controls (∼3,000 alleles from the NIHR BioResource Rare Diseases consortium) and external WGS controls (∼8,000 alleles from UK10K). Frequency data on non-coding variants comparable to the ExAC dataset for coding variants is needed and will transform rare disease non-coding variant interpretation. Clinical WES platforms can be designed to target known pathogenic variants in regulatory regions in addition to the exome footprint.42 However, they require regular redesign to capture newly reported non-coding pathogenic variants, and they do not give the option of identifying pathogenic deep intronic variants that have not been previously reported.
Regarding limitations of WGS, highly repetitive regions generally still remain poorly covered in our WGS data, due to the difficulty of uniquely mapping the reads in these regions to the genome. This has previously been reported16 and is also a problem for targeted gene panels and WES.12, 13 This is an important issue for IRD, due to the existence of clinically important repetitive regions such as RPGR ORF15, which is highly repetitive and is a mutational hotspot that constitutes one of the most common causes of X-linked retinitis pigmentosa.43 Although in the future increasing read lengths and improved read mapping algorithms are likely to result in improved coverage over these regions, currently they must still be sequenced separately using optimized PCR amplification and Sanger sequencing protocols to exclude the possibility of them harboring pathogenic variants. WGS also remains costlier than targeted gene panels or WES, and so currently costs more per diagnosis.44 With the cost of WGS continuing to fall and the emergence of large-scale WGS projects such as the Genomics England 100,000 Genome Project, WGS is an increasingly important tool for investigating genetic causes of rare diseases, including IRD, in clinical practice as well as research, and is likely to become the preferred first-line test in the future.45
This project has already contributed to novel insights into the genetic architecture and phenotypic spectrum of IRD. We have identified a distinctive electroretinogram phenotype, predominantly involving the cone pathways, in two individuals with variants in CACNA2D4 (MIM: 608171).46 Additionally, an individual from this study with biallelic IFT140 (MIM: 614620) variants, along with additional case subjects, expanded the known phenotypic spectrum of IFT140-associated disease, which was originally reported as a severe syndromic ciliopathy and is now known to also cause non-syndromic retinitis pigmentosa.18 Several other individuals in our cohort with variants in both RGR (MIM: 600342) and CDHR1 (MIM: 609502) led to the discovery that a recessive retinal disorder previously associated with a homozygous variant in RGR is more likely to be caused by the variant in CDHR1, which is in complete linkage disequilibrium with the originally reported variant.47
There are many possible reasons why 282/722 (39%) of our cohort remain as yet unsolved. Some may have pathogenic variants that have not been called by the variant calling software because, for example, they are in regions of poor coverage such as repetitive regions. Others may have pathogenic variants that were called but not manually reviewed because they did not pass one of our filters. Some are likely to have pathogenic variants in genes associated with IRD that are not on our list of IRD-associated genes. For some unsolved individuals, the cause of their disease may be more complex than Mendelian inheritance and may be oligogenic or influenced by environmental factors. Finally, it is likely that a proportion of unsolved individuals have pathogenic variants in regulatory regions, rather than in coding exons. Regulatory variation is known to be an important cause of IRD.11, 31, 33, 48, 49 Having used WGS for the majority of our cohort, we are well placed to investigate this class of variation. However, interpretation of regulatory variation remains a significant challenge. Experimental verification of specific variants is particularly difficult as many IRD-associated genes are restricted to expression in the retina, making functional confirmation of the effects of any candidate regulatory variation challenging. Our investigation of potentially pathogenic regulatory variation is ongoing.
We have identified 19 genes that contain likely biallelic, predicted protein-truncating variants in unsolved individuals with IRD, including CROCC (MIM: 615776), IRX5 (MIM: 606195), and NUMB (MIM: 603728) that have known roles in retinal development and function.50, 51 These may represent strong candidates for previously unreported recessive IRD-associated genes (Table S3). Identification of at least two additional individuals with likely pathogenic variants is necessary to provide sufficient supportive evidence of any of these genes being associated with IRD.
In conclusion, we present here a large cohort of individuals with IRD who have had WGS, and we have achieved a pathogenic variant detection rate of 56%. We have identified three categories of pathogenic variant that WGS substantially improves the detection of: SVs, variants in GC-rich regions, and variants in regulatory regions. Studying a cohort of this size using WGS provides new insights into phenotypes and the genetic architecture of IRD. In the future, WGS is likely to become the preferred choice of test with which to investigate the genetic causes of IRD.
Consortia
Members of the NIHR-BioResource Rare Diseases Consortium are Timothy Aitman, Hana Alachkar, Sonia Ali, Louise Allen, David Allsup, Gautum Ambegaonkar, Julie Anderson, Richard Antrobus, Ruth Armstrong, Gavin Arno, Gururaj Arumugakani, Sofie Ashford, William Astle, Antony Attwood, Steve Austin, Chiara Bacchelli, Tamam Bakchoul, Tadbir K. Bariana, Helen Baxendale, David Bennett, Claire Bethune, Shahnaz Bibi, Maria Bitner-Glindzicz, Marta Bleda, Harm Boggard, Paula Bolton-Maggs, Claire Booth, John R. Bradley, Angie Brady, Matthew Brown, Michael Browning, Christine Bryson, Siobhan Burns, Paul Calleja, Natalie Canham, Jenny Carmichael, Keren Carss, Mark Caulfield, Elizabeth Chalmers, Anita Chandra, Patrick Chinnery, Manali Chitre, Colin Church, Emma Clement, Naomi Clements-Brod, Virginia Clowes, Gerry Coghlan, Peter Collins, Nichola Cooper, Amanda Creaser-Myers, Rosa DaCosta, Louise Daugherty, Sophie Davies, John Davis, Minka De Vries, Patrick Deegan, Sri V.V. Deevi, Charu Deshpande, Lisa Devlin, Eleanor Dewhurst, Rainer Doffinger, Natalie Dormand, Elizabeth Drewe, David Edgar, William Egner, Wendy N. Erber, Marie Erwood, Tamara Everington, Remi Favier, Helen Firth, Debra Fletcher, Frances Flinter, James C. Fox, Amy Frary, Kathleen Freson, Bruce Furie, Abigail Furnell, Daniel Gale, Alice Gardham, Michael Gattens, Neeti Ghali, Pavandeep K. Ghataorhe, Rohit Ghurye, Simon Gibbs, Kimberley Gilmour, Paul Gissen, Sarah Goddard, Keith Gomez, Pavel Gordins, Stefan Gräf, Daniel Greene, Alan Greenhalgh, Andreas Greinacher, Sofia Grigoriadou, Detelina Grozeva, Scott Hackett, Charaka Hadinnapola, Rosie Hague, Matthias Haimel, Csaba Halmagyi, Tracey Hammerton, Daniel Hart, Grant Hayman, Johan W.M. Heemskerk, Robert Henderson, Anke Hensiek, Yvonne Henskens, Archana Herwadkar, Simon Holden, Muriel Holder, Susan Holder, Fengyuan Hu, Aarnoud Huissoon, Marc Humbert, Jane Hurst, Roger James, Stephen Jolles, Dragana Josifova, Rashid Kazmi, David Keeling, Peter Kelleher, Anne M. Kelly, Fiona Kennedy, David Kiely, Nathalie Kingston, Ania Koziell, Deepa Krishnakumar, Taco W. Kuijpers, Dinakantha Kumararatne, Manju Kurian, Michael A. Laffan, Michele P. Lambert, Hana Lango Allen, Allan Lawrie, Sara Lear, Melissa Lees, Claire Lentaigne, Ri Liesner, Rachel Linger, Hilary Longhurst, Lorena Lorenzo, Rajiv Machado, Rob Mackenzie, Robert MacLaren, Eamonn Maher, Jesmeen Maimaris, Sarah Mangles, Ania Manson, Rutendo Mapeta, Hugh S. Markus, Jennifer Martin, Larahmie Masati, Mary Mathias, Vera Matser, Anna Maw, Elizabeth McDermott, Coleen McJannet, Stuart Meacham, Sharon Meehan, Karyn Megy, Sarju Mehta, Michel Michaelides, Carolyn M. Millar, Shahin Moledina, Anthony Moore, Nicholas Morrell, Andrew Mumford, Sai Murng, Elaine Murphy, Sergey Nejentsev, Sadia Noorani, Paquita Nurden, Eric Oksenhendler, Willem H. Ouwehand, Sofia Papadia, Soo-Mi Park, Alasdair Parker, John Pasi, Chris Patch, Joan Paterson, Jeanette Payne, Andrew Peacock, Kathelijne Peerlinck, Christopher J. Penkett, Joanna Pepke-Zaba, David J. Perry, Val Pollock, Gary Polwarth, Mark Ponsford, Waseem Qasim, Isabella Quinti, Stuart Rankin, Julia Rankin, F. Lucy Raymond, Karola Rehnstrom, Evan Reid, Christopher J. Rhodes, Michael Richards, Sylvia Richardson, Alex Richter, Irene Roberts, Matthew Rondina, Elisabeth Rosser, Catherine Roughley, Kevin Rue-Albrecht, Crina Samarghitean, Alba Sanchis-Juan, Richard Sandford, Saikat Santra, Ravishankar Sargur, Sinisa Savic, Sol Schulman, Harald Schulze, Richard Scott, Marie Scully, Suranjith Seneviratne, Carrock Sewell, Olga Shamardina, Debbie Shipley, Ilenia Simeoni, Suthesh Sivapalaratnam, Kenneth Smith, Aman Sohal, Laura Southgate, Simon Staines, Emily Staples, Hans Stauss, Penelope Stein, Jonathan Stephens, Kathleen Stirrups, Sophie Stock, Jay Suntharalingam, R. Campbell Tait, Kate Talks, Yvonne Tan, Jecko Thachil, James Thaventhiran, Ellen Thomas, Moira Thomas, Dorothy Thompson, Adrian Thrasher, Marc Tischkowitz, Catherine Titterton, Cheng-Hock Toh, Mark Toshner, Carmen Treacy, Richard Trembath, Salih Tuna, Wojciech Turek, Ernest Turro, Chris Van Geet, Marijke Veltman, Julie Vogt, Julie von Ziegenweldt, Anton Vonk Noordegraaf, Emma Wakeling, Ivy Wanjiku, Timothy Q. Warner, Evangeline Wassmer, Hugh Watkins, Andrew Webster, Steve Welch, Sarah Westbury, John Wharton, Deborah Whitehorn, Martin Wilkins, Lisa Willcocks, Catherine Williamson, Geoffrey Woods, John Wort, Nigel Yeatman, Patrick Yong, Tim Young, and Ping Yu.
Acknowledgments
This work was supported by The National Institute for Health Research England (NIHR) for the NIHR BioResource – Rare Diseases project (grant number RG65966). The Moorfields Eye Hospital cohort of patients and clinical and imaging data were ascertained and collected with the support of grants from the National Institute for Health Research Biomedical Research Centre at Moorfields Eye Hospital, National Health Service Foundation Trust, and UCL Institute of Ophthalmology, Moorfields Eye Hospital Special Trustees, Moorfields Eye Charity, the Foundation Fighting Blindness (USA), and Retinitis Pigmentosa Fighting Blindness. M.M. is a recipient of an FFB Career Development Award. E.M. is supported by UCLH/UCL NIHR Biomedical Research Centre. F.L.R. and D.G. are supported by Cambridge NIHR Biomedical Research Centre.
Published: December 29, 2016
Footnotes
Supplemental Data include three tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.12.003.
Contributor Information
F. Lucy Raymond, Email: flr24@cam.ac.uk.
NIHR-BioResource Rare Diseases Consortium:
Timothy Aitman, Hana Alachkar, Sonia Ali, Louise Allen, David Allsup, Gautum Ambegaonkar, Julie Anderson, Richard Antrobus, Ruth Armstrong, Gavin Arno, Gururaj Arumugakani, Sofie Ashford, William Astle, Antony Attwood, Steve Austin, Chiara Bacchelli, Tamam Bakchoul, Tadbir K. Bariana, Helen Baxendale, David Bennett, Claire Bethune, Shahnaz Bibi, Maria Bitner-Glindzicz, Marta Bleda, Harm Boggard, Paula Bolton-Maggs, Claire Booth, John R. Bradley, Angie Brady, Matthew Brown, Michael Browning, Christine Bryson, Siobhan Burns, Paul Calleja, Natalie Canham, Jenny Carmichael, Keren Carss, Mark Caulfield, Elizabeth Chalmers, Anita Chandra, Patrick Chinnery, Manali Chitre, Colin Church, Emma Clement, Naomi Clements-Brod, Virginia Clowes, Gerry Coghlan, Peter Collins, Nichola Cooper, Amanda Creaser-Myers, Rosa DaCosta, Louise Daugherty, Sophie Davies, John Davis, Minka De Vries, Patrick Deegan, Sri V.V. Deevi, Charu Deshpande, Lisa Devlin, Eleanor Dewhurst, Rainer Doffinger, Natalie Dormand, Elizabeth Drewe, David Edgar, William Egner, Wendy N. Erber, Marie Erwood, Tamara Everington, Remi Favier, Helen Firth, Debra Fletcher, Frances Flinter, James C. Fox, Amy Frary, Kathleen Freson, Bruce Furie, Abigail Furnell, Daniel Gale, Alice Gardham, Michael Gattens, Neeti Ghali, Pavandeep K. Ghataorhe, Rohit Ghurye, Simon Gibbs, Kimberley Gilmour, Paul Gissen, Sarah Goddard, Keith Gomez, Pavel Gordins, Stefan Gräf, Daniel Greene, Alan Greenhalgh, Andreas Greinacher, Sofia Grigoriadou, Detelina Grozeva, Scott Hackett, Charaka Hadinnapola, Rosie Hague, Matthias Haimel, Csaba Halmagyi, Tracey Hammerton, Daniel Hart, Grant Hayman, Johan W.M. Heemskerk, Robert Henderson, Anke Hensiek, Yvonne Henskens, Archana Herwadkar, Simon Holden, Muriel Holder, Susan Holder, Fengyuan Hu, Aarnoud Huissoon, Marc Humbert, Jane Hurst, Roger James, Stephen Jolles, Dragana Josifova, Rashid Kazmi, David Keeling, Peter Kelleher, Anne M. Kelly, Fiona Kennedy, David Kiely, Nathalie Kingston, Ania Koziell, Deepa Krishnakumar, Taco W. Kuijpers, Dinakantha Kumararatne, Manju Kurian, Michael A. Laffan, Michele P. Lambert, Hana Lango Allen, Allan Lawrie, Sara Lear, Melissa Lees, Claire Lentaigne, Ri Liesner, Rachel Linger, Hilary Longhurst, Lorena Lorenzo, Rajiv Machado, Rob Mackenzie, Robert MacLaren, Eamonn Maher, Jesmeen Maimaris, Sarah Mangles, Ania Manson, Rutendo Mapeta, Hugh S. Markus, Jennifer Martin, Larahmie Masati, Mary Mathias, Vera Matser, Anna Maw, Elizabeth McDermott, Coleen McJannet, Stuart Meacham, Sharon Meehan, Karyn Megy, Sarju Mehta, Michel Michaelides, Carolyn M. Millar, Shahin Moledina, Anthony Moore, Nicholas Morrell, Andrew Mumford, Sai Murng, Elaine Murphy, Sergey Nejentsev, Sadia Noorani, Paquita Nurden, Eric Oksenhendler, Willem H. Ouwehand, Sofia Papadia, Soo-Mi Park, Alasdair Parker, John Pasi, Chris Patch, Joan Paterson, Jeanette Payne, Andrew Peacock, Kathelijne Peerlinck, Christopher J. Penkett, Joanna Pepke-Zaba, David J. Perry, Val Pollock, Gary Polwarth, Mark Ponsford, Waseem Qasim, Isabella Quinti, Stuart Rankin, Julia Rankin, F. Lucy Raymond, Karola Rehnstrom, Evan Reid, Christopher J. Rhodes, Michael Richards, Sylvia Richardson, Alex Richter, Irene Roberts, Matthew Rondina, Elisabeth Rosser, Catherine Roughley, Kevin Rue-Albrecht, Crina Samarghitean, Alba Sanchis-Juan, Richard Sandford, Saikat Santra, Ravishankar Sargur, Sinisa Savic, Sol Schulman, Harald Schulze, Richard Scott, Marie Scully, Suranjith Seneviratne, Carrock Sewell, Olga Shamardina, Debbie Shipley, Ilenia Simeoni, Suthesh Sivapalaratnam, Kenneth Smith, Aman Sohal, Laura Southgate, Simon Staines, Emily Staples, Hans Stauss, Penelope Stein, Jonathan Stephens, Kathleen Stirrups, Sophie Stock, Jay Suntharalingam, R. Campbell Tait, Kate Talks, Yvonne Tan, Jecko Thachil, James Thaventhiran, Ellen Thomas, Moira Thomas, Dorothy Thompson, Adrian Thrasher, Marc Tischkowitz, Catherine Titterton, Cheng-Hock Toh, Mark Toshner, Carmen Treacy, Richard Trembath, Salih Tuna, Wojciech Turek, Ernest Turro, Chris Van Geet, Marijke Veltman, Julie Vogt, Julie von Ziegenweldt, Anton Vonk Noordegraaf, Emma Wakeling, Ivy Wanjiku, Timothy Q. Warner, Evangeline Wassmer, Hugh Watkins, Andrew Webster, Steve Welch, Sarah Westbury, John Wharton, Deborah Whitehorn, Martin Wilkins, Lisa Willcocks, Catherine Williamson, Geoffrey Woods, John Wort, Nigel Yeatman, Patrick Yong, Tim Young, and Ping Yu
Accession Numbers
The accession number for the high-throughput sequencing data reported in this paper is EGAD00001002656 as a subset of the NIHR BioResource Rare Disease Consortium Data.
Web Resources
Ensembl Genome Browser, http://www.ensembl.org/index.html
European Genome-phenome Archive (EGA), https://www.ebi.ac.uk/ega
ExAC Browser, http://exac.broadinstitute.org/
Ocular Genomics Institute, https://oculargenomics.meei.harvard.edu/index.php/ret-trans/110-human-retinal-transcriptome
OMIM, http://www.omim.org/
RetNet – Retinal Information Network, https://sph.uth.edu/retnet/home.htm
The 100,000 Genomes Project, https://www.genomicsengland.co.uk/the-100000-genomes-project
UCSC Genome Browser, http://genome.ucsc.edu
UK10K Consortium, http://www.uk10k.org/
Supplemental Data
References
- 1.Liew G., Michaelides M., Bunce C. A comparison of the causes of blindness certifications in England and Wales in working age adults (16-64 years), 1999-2000 with 2009-2010. BMJ Open. 2014;4:e004015. doi: 10.1136/bmjopen-2013-004015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ellingford J.M., Sergouniotis P.I., Lennon R., Bhaskar S., Williams S.G., Hillman K.A., O’Sullivan J., Hall G., Ramsden S.C., Lloyd I.C. Pinpointing clinical diagnosis through whole exome sequencing to direct patient care: a case of Senior-Loken syndrome. Lancet. 2015;385:1916. doi: 10.1016/S0140-6736(15)60496-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fitzgerald T.W., Gerety S.S., Jones W.D., van Kogelenberg M., King D.A., McRae J., Morley K.I., Parthiban V., Al-Turki S., Ambridge K., Deciphering Developmental Disorders Study Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519:223–228. doi: 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang Y., Muzny D.M., Reid J.G., Bainbridge M.N., Willis A., Ward P.A., Braxton A., Beuten J., Xia F., Niu Z. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N. Engl. J. Med. 2013;369:1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beaulieu C.L., Majewski J., Schwartzentruber J., Samuels M.E., Fernandez B.A., Bernier F.P., Brudno M., Knoppers B., Marcadier J., Dyment D., FORGE Canada Consortium FORGE Canada Consortium: outcomes of a 2-year national rare-disease gene-discovery project. Am. J. Hum. Genet. 2014;94:809–817. doi: 10.1016/j.ajhg.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilissen C., Hehir-Kwa J.Y., Thung D.T., van de Vorst M., van Bon B.W.M., Willemsen M.H., Kwint M., Janssen I.M., Hoischen A., Schenck A. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344–347. doi: 10.1038/nature13394. [DOI] [PubMed] [Google Scholar]
- 7.Stavropoulos D.J., Merico D., Jobling R., Bowdin S., Monfared N., Thiruvahindrapuram B., Nalpathamkalam T., Pellecchia G., Yuen R.K.C., Szego M.J. Whole-genome sequencing expands diagnostic utility and improves clinical management in paediatric medicine. Nat. Genomic Med. 2016;1:15012. doi: 10.1038/npjgenmed.2015.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berger W., Kloeckener-Gruissem B., Neidhardt J. The molecular basis of human retinal and vitreoretinal diseases. Prog. Retin. Eye Res. 2010;29:335–375. doi: 10.1016/j.preteyeres.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 9.Huang X.-F., Huang F., Wu K.-C., Wu J., Chen J., Pang C.-P., Lu F., Qu J., Jin Z.-B. Genotype-phenotype correlation and mutation spectrum in a large cohort of patients with inherited retinal dystrophy revealed by next-generation sequencing. Genet. Med. 2015;17:271–278. doi: 10.1038/gim.2014.138. [DOI] [PubMed] [Google Scholar]
- 10.Patel N., Aldahmesh M.A., Alkuraya H., Anazi S., Alsharif H., Khan A.O., Sunker A., Al-Mohsen S., Abboud E.B., Nowilaty S.R. Expanding the clinical, allelic, and locus heterogeneity of retinal dystrophies. Genet. Med. 2016;18:554–562. doi: 10.1038/gim.2015.127. [DOI] [PubMed] [Google Scholar]
- 11.Eisenberger T., Neuhaus C., Khan A.O., Decker C., Preising M.N., Friedburg C., Bieg A., Gliem M., Charbel Issa P., Holz F.G. Increasing the yield in targeted next-generation sequencing by implicating CNV analysis, non-coding exons and the overall variant load: the example of retinal dystrophies. PLoS ONE. 2013;8:e78496. doi: 10.1371/journal.pone.0078496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tiwari A., Bahr A., Bähr L., Fleischhauer J., Zinkernagel M.S., Winkler N., Barthelmes D., Berger L., Gerth-Kahlert C., Neidhardt J., Berger W. Next generation sequencing based identification of disease-associated mutations in Swiss patients with retinal dystrophies. Sci. Rep. 2016;6:28755. doi: 10.1038/srep28755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Sullivan J., Mullaney B.G., Bhaskar S.S., Dickerson J.E., Hall G., O’Grady A., Webster A., Ramsden S.C., Black G.C. A paradigm shift in the delivery of services for diagnosis of inherited retinal disease. J. Med. Genet. 2012;49:322–326. doi: 10.1136/jmedgenet-2012-100847. [DOI] [PubMed] [Google Scholar]
- 14.Khan K.N., Chana R., Ali N., Wright G., Webster A.R., Moore A.T., Michaelides M. Advanced diagnostic genetic testing in inherited retinal disease: experience from a single tertiary referral centre in the UK National Health Service. Clin. Genet. 2016 doi: 10.1111/cge.12798. Published online May 10, 2016. [DOI] [PubMed] [Google Scholar]
- 15.Consugar M.B., Navarro-Gomez D., Place E.M., Bujakowska K.M., Sousa M.E., Fonseca-Kelly Z.D.Z.D., Taub D.G., Janessian M., Wang D.Y., Au E.D. Panel-based genetic diagnostic testing for inherited eye diseases is highly accurate and reproducible, and more sensitive for variant detection, than exome sequencing. Genet. Med. 2015;17:253–261. doi: 10.1038/gim.2014.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellingford J.M., Barton S., Bhaskar S., Williams S.G., Sergouniotis P.I., O’Sullivan J., Lamb J.A., Perveen R., Hall G., Newman W.G. Whole genome sequencing increases molecular diagnostic yield compared with current diagnostic testing for inherited retinal disease. Ophthalmology. 2016;123:1143–1150. doi: 10.1016/j.ophtha.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Westbury S.K., Turro E., Greene D., Lentaigne C., Kelly A.M., Bariana T.K., Simeoni I., Pillois X., Attwood A., Austin S., BRIDGE-BPD Consortium Human phenotype ontology annotation and cluster analysis to unravel genetic defects in 707 cases with unexplained bleeding and platelet disorders. Genome Med. 2015;7:36. doi: 10.1186/s13073-015-0151-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hull S., Owen N., Islam F., Tracey-White D., Plagnol V., Holder G.E., Michaelides M., Carss K., Raymond F.L., Rozet J.-M. Nonsyndromic retinal dystrophy due to bi-allelic mutations in the ciliary transport gene IFT140. Invest. Ophthalmol. Vis. Sci. 2016;57:1053–1062. doi: 10.1167/iovs.15-17976. [DOI] [PubMed] [Google Scholar]
- 19.Chen X., Schulz-Trieglaff O., Shaw R., Barnes B., Schlesinger F., Källberg M., Cox A.J., Kruglyak S., Saunders C.T. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32:1220–1222. doi: 10.1093/bioinformatics/btv710. [DOI] [PubMed] [Google Scholar]
- 20.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wright C.F., Fitzgerald T.W., Jones W.D., Clayton S., McRae J.F., van Kogelenberg M., King D.A., Ambridge K., Barrett D.M., Bayzetinova T., DDD study Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet. 2014;385:1305–1314. doi: 10.1016/S0140-6736(14)61705-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stenson P.D., Mort M., Ball E.V., Shaw K., Phillips A., Cooper D.N. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum. Genet. 2014;133:1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zarrei M., MacDonald J.R., Merico D., Scherer S.W. A copy number variation map of the human genome. Nat. Rev. Genet. 2015;16:172–183. doi: 10.1038/nrg3871. [DOI] [PubMed] [Google Scholar]
- 25.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thorvaldsdóttir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Desmet F.O., Hamroun D., Lalande M., Collod-Béroud G., Claustres M., Béroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reese M.G., Eeckman F.H., Kulp D., Haussler D. Improved splice site detection in Genie. J. Comput. Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 29.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henderson R.H., Waseem N., Searle R., van der Spuy J., Russell-Eggitt I., Bhattacharya S.S., Thompson D.A., Holder G.E., Cheetham M.E., Webster A.R., Moore A.T. An assessment of the apex microarray technology in genotyping patients with Leber congenital amaurosis and early-onset severe retinal dystrophy. Invest. Ophthalmol. Vis. Sci. 2007;48:5684–5689. doi: 10.1167/iovs.07-0207. [DOI] [PubMed] [Google Scholar]
- 31.Braun T.A., Mullins R.F., Wagner A.H., Andorf J.L., Johnston R.M., Bakall B.B., Deluca A.P., Fishman G.A., Lam B.L., Weleber R.G. Non-exomic and synonymous variants in ABCA4 are an important cause of Stargardt disease. Hum. Mol. Genet. 2013;22:5136–5145. doi: 10.1093/hmg/ddt367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sangermano R., Bax N.M., Bauwens M., van den Born L.I., De Baere E., Garanto A., Collin R.W.J., Goercharn-Ramlal A.S.A., den Engelsman-van Dijk A.H.A., Rohrschneider K. Photoreceptor progenitor mRNA analysis reveals exon skipping resulting from the ABCA4 c.5461-10T→C mutation in Stargardt disease. Ophthalmology. 2016;123:1375–1385. doi: 10.1016/j.ophtha.2016.01.053. [DOI] [PubMed] [Google Scholar]
- 33.Vaché C., Besnard T., le Berre P., García-García G., Baux D., Larrieu L., Abadie C., Blanchet C., Bolz H.J., Millan J. Usher syndrome type 2 caused by activation of an USH2A pseudoexon: implications for diagnosis and therapy. Hum. Mutat. 2012;33:104–108. doi: 10.1002/humu.21634. [DOI] [PubMed] [Google Scholar]
- 34.van den Hurk J.A.J.M., van de Pol D.J.R., Wissinger B., van Driel M.A., Hoefsloot L.H., de Wijs I.J., van den Born L.I., Heckenlively J.R., Brunner H.G., Zrenner E. Novel types of mutation in the choroideremia ( CHM) gene: a full-length L1 insertion and an intronic mutation activating a cryptic exon. Hum. Genet. 2003;113:268–275. doi: 10.1007/s00439-003-0970-0. [DOI] [PubMed] [Google Scholar]
- 35.Walter K., Min J.L., Huang J., Crooks L., Memari Y., McCarthy S., Perry J.R.B., Xu C., Futema M., Lawson D., UK10K Consortium The UK10K project identifies rare variants in health and disease. Nature. 2015;526:82–90. doi: 10.1038/nature14962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lelieveld S.H., Spielmann M., Mundlos S., Veltman J.A., Gilissen C. Comparison of exome and genome sequencing technologies for the complete capture of protein-coding regions. Hum. Mutat. 2015;36:815–822. doi: 10.1002/humu.22813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turner T.N., Hormozdiari F., Duyzend M.H., McClymont S.A., Hook P.W., Iossifov I., Raja A., Baker C., Hoekzema K., Stessman H.A. Genome sequencing of autism-affected families reveals disruption of putative noncoding regulatory DNA. Am. J. Hum. Genet. 2016;98:58–74. doi: 10.1016/j.ajhg.2015.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Belkadi A., Bolze A., Itan Y., Cobat A., Vincent Q.B., Antipenko A., Shang L., Boisson B., Casanova J.-L., Abel L. Whole-genome sequencing is more powerful than whole-exome sequencing for detecting exome variants. Proc. Natl. Acad. Sci. USA. 2015;112:5473–5478. doi: 10.1073/pnas.1418631112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoischen A., Gilissen C., Arts P., Wieskamp N., van der Vliet W., Vermeer S., Steehouwer M., de Vries P., Meijer R., Seiqueros J. Massively parallel sequencing of ataxia genes after array-based enrichment. Hum. Mutat. 2010;31:494–499. doi: 10.1002/humu.21221. [DOI] [PubMed] [Google Scholar]
- 40.Benjamini Y., Speed T.P. Summarizing and correcting the GC content bias in high-throughput sequencing. Nucleic Acids Res. 2012;40:e72. doi: 10.1093/nar/gks001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chilamakuri C.S.R., Lorenz S., Madoui M.-A., Vodák D., Sun J., Hovig E., Myklebost O., Meza-Zepeda L.A. Performance comparison of four exome capture systems for deep sequencing. BMC Genomics. 2014;15:449. doi: 10.1186/1471-2164-15-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patwardhan A., Harris J., Leng N., Bartha G., Church D.M., Luo S., Haudenschild C., Pratt M., Zook J., Salit M. Achieving high-sensitivity for clinical applications using augmented exome sequencing. Genome Med. 2015;7:71. doi: 10.1186/s13073-015-0197-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pusch C.M., Broghammer M., Jurklies B., Besch D., Jacobi F.K. Ten novel ORF15 mutations confirm mutational hot spot in the RPGR gene in European patients with X-linked retinitis pigmentosa. Hum. Mutat. 2002;20:405. doi: 10.1002/humu.9072. [DOI] [PubMed] [Google Scholar]
- 44.McRae J.F., Clayton S., Fitzgerald T.W., Kaplanis J., Prigmore E., Rajan D., Sifrim A., Aitken S., Akawi N., Alvi M. Prevalence, phenotype and architecture of developmental disorders caused by de novo mutation. bioRxiv. 2016 [Google Scholar]
- 45.Chiang J.P., Lamey T., McLaren T., Thompson J.A., Montgomery H., De Roach J. Progress and prospects of next-generation sequencing testing for inherited retinal dystrophy. Expert Rev. Mol. Diagn. 2015;15:1269–1275. doi: 10.1586/14737159.2015.1081057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ba-Abbad R., Arno G., Carss K., Stirrups K., Penkett C.J., Moore A.T., Michaelides M., Raymond F.L., Webster A.R., Holder G.E. Mutations in CACNA2D4 cause distinctive retinal dysfunction in humans. Ophthalmology. 2016;123 doi: 10.1016/j.ophtha.2015.09.045. 668–71.e2. [DOI] [PubMed] [Google Scholar]
- 47.Arno G., Hull S., Carss K., Dev-Borman A., Chakarova C., Bujakowska K., van den Born I., Robson A.G., Holder G.E., Michaelides M. Reevaluation of the retinal dystrophy due to recessive alleles of RGR with the discovery of a cis-acting mutation in CDHR1. Invest. Ophthalmol. Vis. Sci. 2016;57:4806–4813. doi: 10.1167/iovs.16-19687. [DOI] [PubMed] [Google Scholar]
- 48.Mayer A.K., Rohrschneider K., Strom T.M., Glöckle N., Kohl S., Wissinger B., Weisschuh N. Homozygosity mapping and whole-genome sequencing reveals a deep intronic PROM1 mutation causing cone-rod dystrophy by pseudoexon activation. Eur. J. Hum. Genet. 2016;24:459–462. doi: 10.1038/ejhg.2015.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.den Hollander A.I., Koenekoop R.K., Yzer S., Lopez I., Arends M.L., Voesenek K.E.J., Zonneveld M.N., Strom T.M., Meitinger T., Brunner H.G. Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am. J. Hum. Genet. 2006;79:556–561. doi: 10.1086/507318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang J., Liu X., Yue G., Adamian M., Bulgakov O., Li T. Rootletin, a novel coiled-coil protein, is a structural component of the ciliary rootlet. J. Cell Biol. 2002;159:431–440. doi: 10.1083/jcb.200207153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheng C.W., Chow R.L., Lebel M., Sakuma R., Cheung H.O.L., Thanabalasingham V., Zhang X., Bruneau B.G., Birch D.G., Hui C.C. The Iroquois homeobox gene, Irx5, is required for retinal cone bipolar cell development. Dev. Biol. 2005;287:48–60. doi: 10.1016/j.ydbio.2005.08.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.