Abstract
Early B cell factor 3 (EBF3) is a member of the highly evolutionarily conserved Collier/Olf/EBF (COE) family of transcription factors. Prior studies on invertebrate and vertebrate animals have shown that EBF3 homologs are essential for survival and that loss-of-function mutations are associated with a range of nervous system developmental defects, including perturbation of neuronal development and migration. Interestingly, aristaless-related homeobox (ARX), a homeobox-containing transcription factor critical for the regulation of nervous system development, transcriptionally represses EBF3 expression. However, human neurodevelopmental disorders related to EBF3 have not been reported. Here, we describe three individuals who are affected by global developmental delay, intellectual disability, and expressive speech disorder and carry de novo variants in EBF3. Associated features seen in these individuals include congenital hypotonia, structural CNS malformations, ataxia, and genitourinary abnormalities. The de novo variants affect a single conserved residue in a zinc finger motif crucial for DNA binding and are deleterious in a fly model. Our findings indicate that mutations in EBF3 cause a genetic neurodevelopmental syndrome and suggest that loss of EBF3 function might mediate a subset of neurologic phenotypes shared by ARX-related disorders, including intellectual disability, abnormal genitalia, and structural CNS malformations.
Keywords: Drosophila, knot, ataxia, hypotonia, intellectual disability, expressive speech disorder, COE3, transcription factor, vermian hypoplasia, inhibitory GABAergic neurons
Main Text
An estimated 7.6 million children are born annually with congenital neurodevelopmental disorders, encompassing several clinically and biologically heterogeneous conditions including intellectual disability, autism spectrum disorder (ASD), and epilepsy.1, 2, 3 The underlying disease-causing mechanism remains elusive when a genetic disorder lacks strong unique features to stratify the affected individuals for traditional phenotypically driven gene discovery. The advent of whole-exome sequencing (WES) has provided a powerful tool for discovering disease-associated genes by identifying mutations in a population of individuals presenting with rather non-specific clinical features.4, 5, 6
We have identified three individuals with a previously unrecognized genetic syndromic disorder characterized by global developmental delay (3/3), hypotonia (3/3), intellectual disability (3/3), mild facial dysmorphisms (3/3), facial weakness (3/3), expressive speech disorder (3/3), ataxia (3/3), perseverative social behaviors (1/3), motor stereotypies (1/3), decreased pain response (2/3), structural CNS malformations (2/3), and genitourinary malformations (2/3) (Figure 1 and Table S1). These three individuals all have de novo EBF3 (MIM: 607407; HGNC: 19087) missense variants that affect the same amino acid residue (Arg163) in the Zn2+ finger Collier/Olf/Ebf (COE) motif and are predicted to be putatively deleterious by SIFT and PolyPhen-2 models (Figure 1A and Table S2). The coincidental occurrence of three de novo variants affecting the same residue in individuals with similar phenotypes from a total WES cohort of 7,595 individuals is highly unlikely and remains statistically significant after correction for the size of the targeted exome (p = 2.1 × 10−3).7, 8, 9 Prior statistical models examining observed versus expected functional coding variation revealed that EBF3 undergoes selective restraint, a process where selection has reduced functional variation, suggesting that mutations are more likely to be deleterious.10, 11 Statistical analysis comparing the observed to the expected functional variation across the genome for EBF3 resulted in a Residual Variation Intolerance Score (RVIS) of −0.646, where RVIS < 0 indicates that there is less common functional variation than predicted.10 Furthermore, in a model of de novo mutations for ASD and intellectual disability, the statistical analysis identified EBF3 as one of ∼1,000 genes that significantly lack functional variation in non-ASD individuals but are enriched with de novo loss-of-function mutations in affected individuals.11 Similarly, analysis of the ExAC (Exome Aggregation Consortium) Browser revealed that EBF3 has a high probability of loss-of-function intolerance (pLI = 1.0), given that 23.2 loss-of-function variants were expected given the gene’s size and GC content but only one loss-of-function variant was observed.12 Together, these statistical findings provide strong evidence that the recurrent de novo variants in EBF3 cause the observed neurodevelopmental disorder.
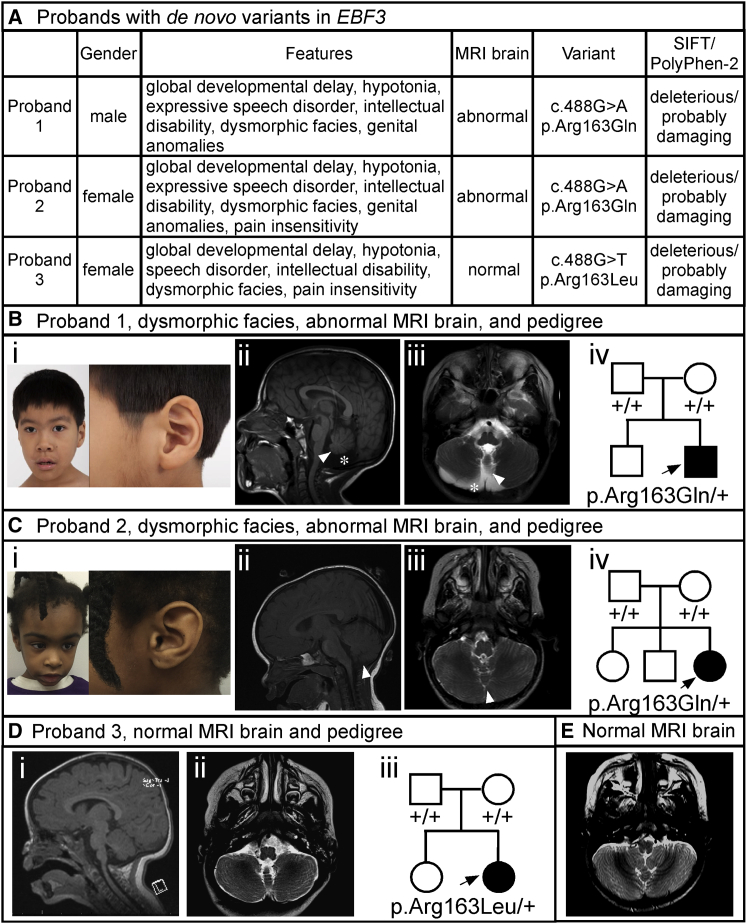
Figure 1.
Probands and Phenotypic Features
(A) Summary of phenotypic features, brain MRI findings, gene variants, and SIFT and PolyPhen-2 predictions for the three probands with the de novo EBF3 p.Arg163Gln and p.Arg163Leu variants.
(B) Proband 1. Representative images show (i) mild facial dysmorphisms including oval-shaped myopathic facies, short anteverted nostrils, and overfolding of the superior helices, (ii) mid-sagittal T1-weighted and (iii) axial T2-weighted images depicting vermian hypoplasia (white arrows) and reduced cerebellar hemispheres volume (white asterisk), and (iv) a pedigree showing the de novo p.Arg163Gln variant.
(C) Proband 2. Representative images show (i) mild facial dysmorphisms including triangular myopathic facies and overfolding of the superior helices, (ii) mid-sagittal T1-weighted and (iii) axial T2-weighted images depicting vermian hypoplasia (white arrows) with normal cerebellar hemispheres, and (iv) a pedigree showing the de novo p.Arg163Gln variant.
(D) Proband 3. Brain MRI shows (i) mid-sagittal T1-weighted and (ii) axial T2-weighted images depicting normal cerebellar vermis and hemispheres. A pedigree (iii) shows the de novo p.Arg163Leu variant.
(E) A representative axial T2-weighted image from the normal brain MRI of a 23-month-old control individual is shown for comparison. Note the typical cerebellar hemispheres and vermian structures.
Clinical data were obtained after written informed consent was provided and procedures were followed in accordance with the ethical standards of the participating institutional review boards on human research and in keeping with national standards. The NIH Undiagnosed Diseases Program (UDP) under protocol 76-HG-0238, “Diagnosis and Treatment of Patients with Inborn Errors of Metabolism or Other Genetic Disorders,” approved by the National Human Genome Research Institute Institutional Review Board, identified a de novo c.488G>A (p.Arg163Gln) missense change in EBF3 (GenBank: NM_001005463.2) for proband 1 by exome sequencing (Supplemental Note and Table S3). Sequence changes c.488G>A (p.Arg163Gln) and c.488G>T (p.Arg163Leu) were identified for probands 2 and 3, respectively, by clinically based exome sequencing performed in the Baylor Genetics Laboratory, certified by the Clinical Laboratory Improvement Amendments (Supplemental Note and Table S3). Sanger sequencing of the parental samples to confirm segregation revealed de novo variants for all three probands. Paternity was confirmed by the inheritance of rare SNPs from the parents. Sample swap was excluded. Neither of the changes is present in the ExAC Browser.12 The clinical phenotypes of these individuals and their EBF3 variants are depicted in Figure 1, described in the Supplemental Note, and summarized below.
Proband 1 is a 7-year-old male Pacific Islander of Chinese and Japanese descent and has a de novo c.488G>A (p.Arg163Gln) missense change in EBF3. No significant findings were revealed during previous targeted genetic testing, karyotype analysis, or chromosomal microarray analysis; SNP array analysis showed no anomalous regions of homozygosity or significant copy-number variants (CNVs). His prenatal history is significant for decreased fetal movements, and he was born at full term at 38 weeks of gestation by caesarean section with a birth weight of 3.4 kg (25th–50th percentile). His clinical features include congenital hypotonia, facial weakness, global developmental delay, expressive speech disorder, dysarthria, dysphagia, gastroesophageal reflux disease, strabismus, ataxia, dysmorphisms including myopathic facies and overfolding of the superior helices, hockey-stick palmar creases, short anteverted nostrils, micropenis, and cryptorchidism. Brain MRI obtained at 7 years of age revealed small inferior posterior cerebellar lobes and hypoplasia of the posterior vermis with mild prominence of the ventricles and sulci (Figure 1B).
Proband 2 is a 5-year-old female of African American descent and has a de novo c.488G>A (p.Arg163Gln) missense change in EBF3. Her prenatal history is significant for decreased fetal movements, and she was born at full term at 40 weeks of gestation via induced vaginal delivery for oligohydramnios with a birth weight of 3.35 kg (25th–50th percentile). Her clinical features include congenital hypotonia, facial weakness, global developmental delay, expressive speech disorder, apraxia, dysarthria, dysphagia, strabismus, ataxia, perseverative social behaviors, dysmorphisms including triangular-shaped facies and overfolding of the superior helices, abnormal palmar creases, fifth-finger clinodactyly, and mild reduction in volume of the labia majora. She is reported to have marked insensitivity to pain such that she does not cry when she falls or receives vaccinations. Brain MRI obtained at 18 months of age revealed hypoplasia of the anterior and posterior vermis (Figure 1C).
Proband 3 is a 3-year-old female of English, Irish, German, and Polish descent and has a de novo c.488G>T (p.Arg163Leu) missense change in EBF3. Her prenatal history is significant for delivery at 39 weeks of gestation by caesarean section due to breech position and a birth weight of 2.7 kg (tenth percentile). Her clinical features include congenital hypotonia, facial weakness, global developmental delay, expressive speech disorder, dysphagia, motor stereotypies, and dysmorphisms including a triangular facies, small feet, and torticollis. She has no genitourinary abnormalities. At present, she speaks only one word, achieved ambulation late, and has a pincer grasp. She is reported to have marked insensitivity to pain such that she does not cry when she falls or receives vaccinations. Brain MRI obtained at 1 year of age was normal, and follow-up at 2 years of age was also normal (Figure 1D; a normal brain MRI axial image from a 23-month-old unaffected individual is included in Figure 1E for comparison).
All three probands presented here have neurodevelopmental disorders composed of intellectual disability, global developmental delay, ataxia, and motor incoordination (Figure 1 and Figure 1, Figure 2, Figure 3, Figure 4). Consistent neurological abnormalities in these probands include congenital hypotonia, facial weakness, dysphagia, and pronounced expressive speech disorder with dysarthria. Two probands also have decreased pain sensitivity. Brain anomalies are present in two of the three probands and include vermian hypoplasia with or without reduced cerebellar hemispheres. No other structural brain anomalies have been noted. Both probands with brain anomalies (1 and 2) have the de novo p.Arg163Gln variant. In contrast, proband 3 has the de novo p.Arg163Leu variant and lacks brain anomalies. Probands 1 and 2 have genitourinary defects, but proband 3 does not. A variety of dysmorphic features were also observed in the three probands (Figure 1 and Supplemental Note).
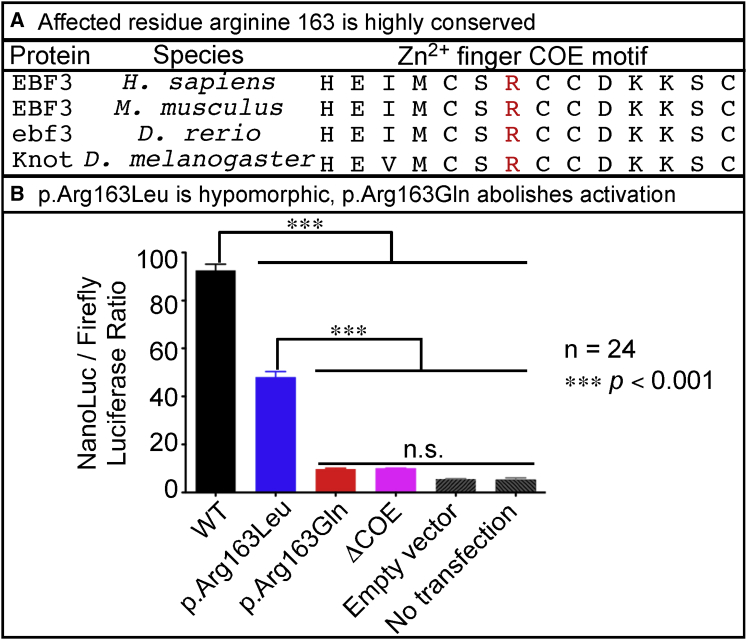
Figure 2.
EBF3 p.Arg163Gln and p.Arg163Leu Impair Transcriptional Activation
(A) Affected residue Arg163 in the Zn2+ finger COE motif is highly conserved across vertebrates and invertebrates.
(B) Activation of reporter-gene expression in HEK293 cells was assessed as the ratio of NanoLuc to firefly luciferase according to the Promega NanoGlo Dual Reporter protocol and was measured on the TD 20/20 Luminometer. The cDNAs encoding WT EBF3 and the EBF3 p.Arg163Gln and p.Arg163Leu variants had a stop codon to ensure that the proteins were untagged to minimize off-target effects from a protein tag. The cDNAs were subcloned into the mammalian expression vector, pcDNA-DEST40. Two synthetic oligonucleotides containing the imperfect palindromic COE binding sequence were used to generate six concatamerized COE binding sites in the NanoLuc vector, pNL3.1. The pGL4.53 firefly luciferase vector was used as an internal transfection control. Additional experimental controls included EBF3 with deletion of the Zn2+ finger COE motif (denoted as ΔCOE). As transfection background controls, we either transfected only the pNL3.1 with six COE binding sites and pGL4.53 but no cDNA expression vectors (denoted as “empty vector”) or did not transfect any vectors (denoted as “no transfection”). A 92-fold induction was observed with WT EBF3 (black). However, EBF3 p.Arg163Leu caused only a 45-fold induction (blue), indicating a partial loss of transcriptional activation. EBF3 p.Arg163Gln showed a very poor induction of transcription (red) similar in level to the transfection background (gray) and that of EBF3 ΔCOE (magenta) controls, indicating severe loss of activity. Data represent the mean ± SEM. n = 24 (six replicates per four separate transfections per experimental condition); ∗∗∗p < 0.001 via one-way ANOVA with Tukey’s post hoc analysis; n.s., no significant difference.
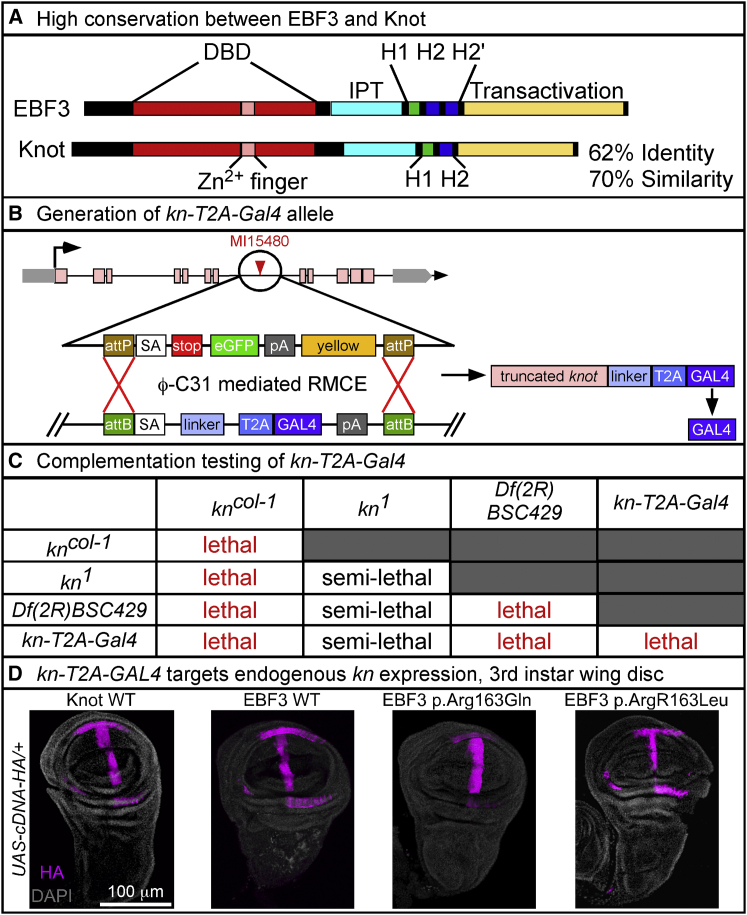
Figure 3.
Generation and Characterization of the Fly kn-T2A-GAL4 Allele
(A) High conservation of protein structure and amino acid sequence between human EBF3 and fly Knot with 62% identity and 70% similarity. Conserved domains include the DNA-binding domain (DBD, red), Zn2+ finger COE motif (pink), Ig-like/plexins/transcription factors (IPT) domain (cyan), helix-loop-helix dimerization motif with α helices H1 (green) and H2 or H2′ (blue), and C-terminal transactivation domain (yellow).
(B) Conversion of the MI15480 line with a MiMIC transposable-element insertion in the fourth coding intron of knot via ϕC31-mediated RMCE for generation of the kn-T2A-GAL4 allele, which expresses GAL4 transactivator in the pattern of kn. This allele also creates a loss-of-function allele of knot by prematurely truncating the transcript by a ribosomal skipping signal (T2A) and a premature polyadenylation signal (pA).
(C) Complementation testing shows that kn-T2A-GAL4 with both the amorphic kncol-1 and genomic deficiency Df(2R)BSC429, encompassing the entire kn locus, fails to complement the lethality. Complementation with hypomorphic kn1 is semi-lethal with <10% viability.
(D) For expression analysis, yw/y; kn-T2A-GAL4/CyO, Kr-GAL4, UAS-GFP males were crossed with yw; UAS-cDNA/TM3 Sb, Kr-GAL4, UAS-GFP virgin females, and double heterozygotes were selected by loss of GFP expression. Images were acquired on a Leica Sp8 laser-scanning confocal microscope. The same settings for laser power and detector gain were used for all genotypes. Images were acquired as a z stack with a z-step of 1 μm and line average of 4 at 400 Hz with a 25× water objective at 1024 × 1024 pixel resolution. Maximum intensity projections were created from the stack in ImageJ. Immunolabeling revealed that kn-T2A-GAL4 recapitulates the endogenous knot expression pattern in third instar wing disc, as shown with the HA-tagged UAS fly lines for WT Knot, WT EBF3, and the EBF3 p.Arg163Gln and p.Arg163Leu variants. Images show HA (magenta) and DAPI (gray). The scale bar represents 100 μm.
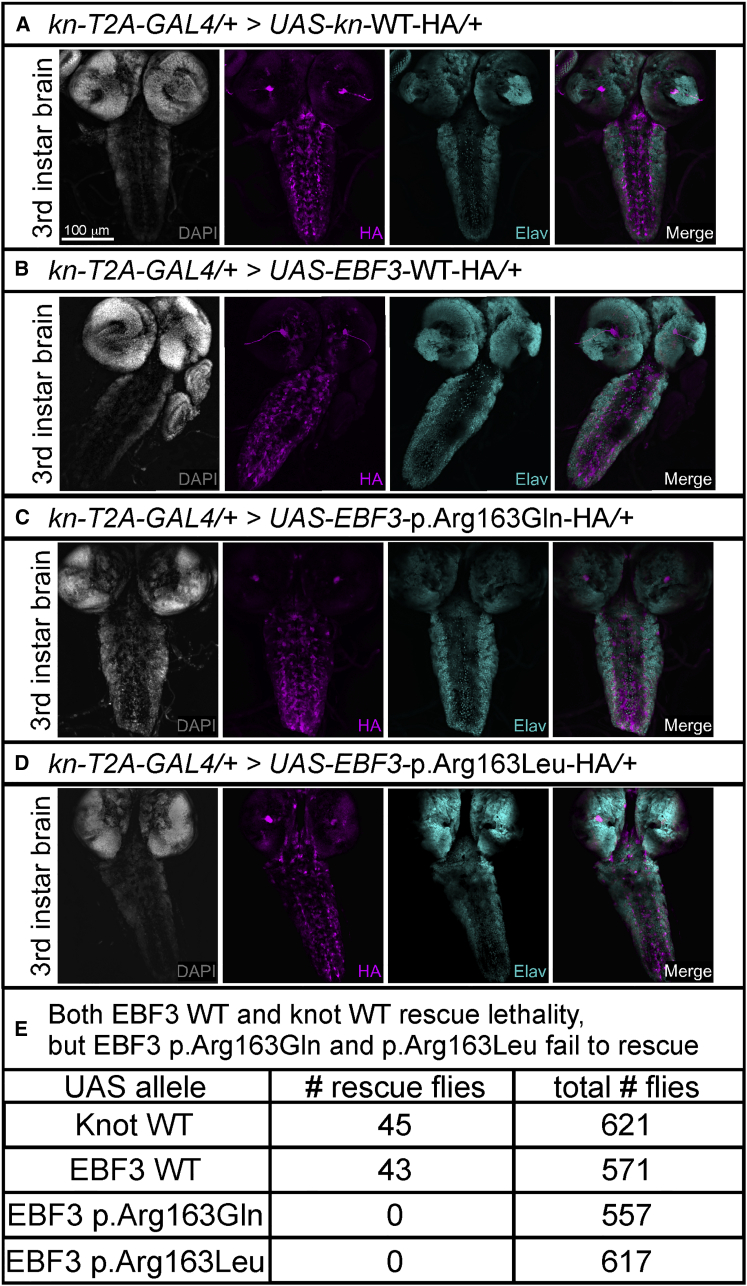
Figure 4.
WT EBF3 and knot Rescue Lethality, but EBF3 p.Arg163Gln and p.Arg163Leu Fail to Rescue Lethality in Flies
(A–D) For expression analysis of third instar brain and ventral nerve cord, yw/y; kn-T2A-GAL4/CyO, Kr-GAL4, UAS-GFP males were crossed with yw; UAS-cDNA/TM3 Sb, Kr-GAL4, UAS-GFP virgin females, and double heterozygotes were selected by loss of GFP expression in the wandering third instar larval stage. Images were acquired on a Leica Sp8 laser-scanning confocal microscope. The same settings for laser power and detector gain were used for all genotypes. Third instar larval brain images were acquired as a z stack with a z-step of 1.51 μm and line average of 4 at 400 Hz with a 20× objective at 1024 × 1024 pixel resolution. Maximum intensity projections were created from the stack in ImageJ. Immunolabeling revealed that kn-T2A-GAL4 drives expression of HA-tagged UAS fly lines for WT Knot (A), WT EBF3 (B), EBF3 p.Arg163Gln (C), and EBF3 p.Arg163Leu (D) in the third instar brain and ventral nerve cord (pan-neuronal marker Elav in cyan; nuclei labeled with DAPI in gray). The scale bar represents 100 μm.
(E) Fly in vivo rescue analysis using the UAS-GAL4 system. For generating the rescue flies, w1118/y; Df(2R)BSC429/Sp; UAS-cDNA-WT-HA/+ males were crossed with yw; kn-T2A-GAL4/SM6a virgin females to produce rescue animals with Knot or EBF3 produced solely from the UAS allele under the control of the kn-T2A-GAL4 driver. The genotypes of the rescued flies are yw/y; Df(2R)BSC429/kn-T2A-GAL4; UAS-cDNA-HA/+ males and w1118/yw; ; Df(2R)BSC429/kn-T2A-GAL4; UAS-cDNA-HA/+ females. For each UAS-cDNA line, >550 adult flies were scored; data represent the number of observed rescue flies and the total number of flies scored. UAS fly lines expressing WT Knot or WT EBF3 rescued embryonic lethality in viable adults. UAS fly lines expressing EBF3 variant p.Arg163Gln or p.Arg163Leu completely failed to rescue the lethality such that no rescue animals were observed as adults or pupae.
The recurrent de novo variant at the same nucleotide (c.488G>A [p.Arg163Gln]) is potentially due to the location of nucleotide 488 in a CpG-dinucleotide island. CpG-dinucleotide islands are mutational hotspots underlying over one-third of de novo missense variants associated with human diseases.13, 14 Given that paternal age was >40 years old for both probands with the p.Arg163Gln variant, other potential mechanisms are the selfish spermatogonial selection process proposed to underlie the association between advanced paternal age and neurodevelopmental disorders,15 unrecognized side effects of the mutation resulting in recurrent de novo variants, or a selection bias for consistent neurodevelopmental phenotypes.
EBF3 is a downstream transcriptional target of aristaless-related homeobox (ARX) and is thought to be transcriptionally repressed by ARX. Gene-expression analysis performed on mouse Arx-mutant medial ganglionic eminence (MGE) showed that Ebf3 had the highest increase in expression levels with the deletion of Arx. Furthermore, in vitro assays confirmed that Arx directly represses Ebf3 expression, indicating that ARX and EBF3 share a strong molecular interaction in regulating γ-aminobutyric acid (GABA)ergic interneuron development and migration.16, 17
Various mutations in ARX (MIM: 300382; HGNC: 18060) cause a diverse range of neurodevelopmental disorders ranging from structural CNS malformations and genitourinary abnormalities associated with premature truncation mutations (MIM: 300004 and 300215) to infantile spasms and epileptic encephalopathies associated with polyalanine repeats (MIM: 308350, 309510, and 300419) to isolated intellectual disability associated with missense mutations or ARX duplications (MIM: 300419).16, 18, 19, 20, 21, 22, 23, 24 Although the exact pathogenic nature of these ARX mutations remains to be fully elucidated, the types of mutations and CNVs suggest that haploinsufficient, gain-of-function, and dominant-negative mechanisms exist for ARX-related disorders.18, 21, 23, 25 Recent advances in sequencing technologies have revealed a growing number of putative ARX transcriptional targets identified by gene-expression analysis in Arx mouse models. At least three of the genes regulated by ARX, including MAGEL2 (MIM: 605283 and 615547; HGNC: 6814),26, 27 FOXP1 (MIM: 605515 and 613670; HGNC: 3823),28, 29 and SOX8 (MIM: 605923 and 141750; HGNC: 11203),30, 31 are associated with neurodevelopmental disorders. These findings suggest that the transcriptional cascade mediated by ARX represents a pathway enriched with disease-associated genes in which mutations cause neurodevelopmental syndromes with phenotypic features overlapping ARX-related disorders, such as EBF3.
Prior studies have shown that Ebf3 haploinsufficiency in mice results in abnormal GABAergic interneuron migration and projection,32 indicating that EBF3 is a critical regulator of inhibitory GABAergic neuronal development. EBF3 and other members of the COE family are also transiently expressed in Cajal-Retzius cells during corticogenesis, where these cells play an essential role in regulating laminar and areal specification.33, 34, 35, 36, 37 Furthermore, extensive loss-of-function manipulations in well-conserved homologs of EBF3 in worms (unc-3, also known as CeO/E), flies (knot, also known as collier), frogs (Xcoe2), and mice (Ebf3) have consistently been shown to impair survival and be deleterious to neuronal development, migration, and function32, 38, 39, 40, 41, 42
The EBF3 Arg163 residue affected in all three probands is a highly conserved arginine across vertebrate and invertebrate species and is located in the Zn2+ finger COE motif (H-X3C-X2-C-X5-C) present in the amino-terminal DNA-binding domain (Figure 2A). A prior mutational study of the paralogous EBF1 showed that disrupting highly conserved cysteine and histidine residues in the 14-aa COE motif abolishes DNA-binding activity by destabilizing the conformation of the Zn2+ finger COE motif.43 In the same study, EBF1 p.Arg163Ala also affected DNA binding.43 To assess the functional consequences of p.Arg163Gln and p.Arg163Leu on EBF3 activation of gene expression, we performed a luciferase activity assay by utilizing previously identified COE transcription factor binding sites.44, 45 To generate the EBF3 variants, we utilized the human full-length cDNA clone for the most abundant splicing isoform of EBF3 (full-length ORF [GenBank: HQ258299] and mRNA [GenBank: BC130479.1]). Site-directed mutagenesis was performed for EBF3 in the pENTR223 Gateway compatible donor vector via either the Agilent QuikChange Lightning or the NEB Q5 site-directed mutagenesis protocol.
We compared the transcriptional activity of the EBF3 variants to that of two controls: wild-type (WT) EBF3 and EBF3 with deletion of the Zn2+ finger COE motif (ΔCOE). HEK293 cells were co-transfected with each individual EBF3 cDNA expression construct in combination with nanoluciferase (NanoLuc) and firefly luciferase vectors. Activation of reporter-gene expression in vitro was assessed as the ratio of NanoLuc to firefly luciferase. A 92-fold induction was observed with WT EBF3. However, EBF3 p.Arg163Leu had only a 45-fold induction, suggesting that the variant is a hypomorphic alteration. EBF3 p.Arg163Gln showed a very poor induction similar in level to the transfection background and that of EBF3 ΔCOE controls, indicating that it is a severe loss-of-function variant (Figure 2B).
To decipher the functional significance of the observed de novo variants affecting residue Arg163, we turned to a recently proposed method to test their pathogenicity in vivo in flies.46 EBF3 is one of four COE homologs in mammals, but Drosophila melanogaster has only one COE transcription factor, knot, also known as collier (gene symbol: kn [FlyBase: FBgn0001319]), which has 62% identity with and 70% similarity to EBF3 (Figure 3A). Prior studies in the fly have shown that loss of knot function is detrimental to nervous system development and that homozygous-null alleles of knot are embryonically lethal.39, 40, 47 We utilized rescue of the homozygous embryonically lethal phenotype and the availability of sophisticated genetic tools to assess the human EBF3 variants by taking advantage of a Minos-mediated integration cassette (MiMIC) transposon inserted in the fourth coding intron of knot (Figure 3B).48, 49 Utilizing recombinase-mediated cassette exchange (RMCE), we created a novel knot allele that truncates the knot transcript and expresses the yeast transactivator gene, GAL4, under the control of the endogenous regulatory elements of knot (Figure 3B).50, 51 Heterozygous kn-T2A-GAL4 flies are viable and do not show any obvious phenotype. Complementation testing showed that the kn-T2A-GAL4 is a severe loss-of-function or null allele causing homozygous embryonic lethality. kn-T2A-GAL4 also fails to complement the lethality of the previously well-characterized amorphic kncol-1 allele,40 the hypomorphic kn1 allele,47 and a molecularly defined deficiency allele, Df(2R)BSC429, which includes the kn locus52 (Figure 3C), confirming the specificity of the kn-T2A-GAL4 mutation.
To assess the functional significance of the EBF3 p.Arg163Gln and p.Arg163Leu variants in flies, we generated several transgenic fly alleles by utilizing the pUASg-HA-attB vector53 to express the human WT EBF3 and variant cDNAs with a C-terminal hemagglutinin (HA) tag under the control of upstream activating system (UAS) elements. As an internal control, we assessed rescue with the WT knot fly cDNA with a C-terminal HA tag under the control of UAS. In conjunction with the kn-T2A-GAL4 allele, the UAS-GAL4 system allowed us to express the human EBF3 and fly knot cDNAs in the same endogenous spatiotemporal pattern as knot. We determined that kn-T2A-GAL4 recapitulates the previously characterized endogenous expression pattern of Knot in the wing imaginal disc of third instar larvae for the UAS lines encoding WT Knot, WT EBF3, and the EBF3 p.Arg163Gln and p.Arg163Leu variants when probed with anti-HA (magenta) (Figure 3D).47
We also determined that kn-T2A-GAL4 drives expression of WT Knot, WT EBF3, and the EBF3 p.Arg163Gln and p.Arg163Leu variants when probed with anti-HA (magenta) in a similar subset of neurons labeled with the pan-neuronal marker Elav (cyan) in the third instar larval brain and ventral nerve cord (Figures 4A–4D). These data suggest that the GAL4 produced by the fusion transcript is expressed in the proper spatial and temporal expression pattern.
To determine the in vivo functional consequences of the EBF3 p.Arg163Gln and p.Arg163Leu variants, we utilized the UAS-GAL4 system with the kn-T2A-GAL4 allele to assess whether WT Knot, WT EBF3, and EBF3 p.Arg163Gln and p.Arg163Leu are capable of rescuing the embryonic lethality observed with complete loss of endogenous Knot. We scored >550 adult flies per cDNA variant and found that both WT EBF3 and WT Knot rescued the embryonic lethality at near Mendelian expectations but that both the EBF3 p.Arg163Gln and p.Arg163Leu variants failed to rescue the lethality, and no adults or pupae were observed (Figure 4E).
Together, the in vitro luciferase reporter-gene activation assay and in vivo fly functional assessment demonstrate that our probands’ de novo EBF3 variants result in a loss of EBF3 function in activating transcription, which is consistent with prior in vitro findings showing that altering the charged Arg163 to neutral alanine abolishes DNA binding.43 Pathogenic mechanisms for these variants could potentially be gain of function, haploinsufficiency loss of function, and dominant negative. A gain-of-function mechanism is less likely given the luciferase data and the near complete loss of activation of reporter-gene expression. The normal development of heterozygous kncol-1 flies and the subtle morphological phenotypes displayed in mice with Ebf3 haploinsufficiency suggest that haploinsufficiency might not solely contribute to the observed phenotypes in our probands. However, the prior findings in fly and mouse models are limited in the assessment of the in vivo consequences of knot or Ebf3 haploinsufficiency to neural network activity and behaviors, and we therefore cannot exclude haploinsufficiency on the basis of these results from prior animal models.
Deletions of at least part of the EBF3 locus have been observed in 59 of 21,770 samples in DECIPHER; they range in size from 120 kb to 14.49 Mb and are associated with syndromic neurodevelopmental features including cerebellar vermian hypoplasia, behavioral abnormalities, motor stereotypies, intellectual disability, delayed speech and language, motor delay, seizures, and genitourinary abnormalities, which were observed in our three probands.54 The majority of these deletion CNVs were reported as de novo in DECIPHER, but some were also inherited from a reportedly unaffected parent, suggesting that the parent might have somatic mosaicism for the CNV or genetic modifiers of phenotypic severity. The smallest deletion encompassing the entire EBF3 locus was 656 kb and only included two genes in addition to EBF3.
In contrast, the Database of Genomic Variants (DGV) reports no CNVs with deletion of the entire EBF3 locus in 54,946 samples from the general population.55 However, the DGV does report two individuals with deletion CNVs (1.25 and 1.34 Mb) containing a breakpoint within the EBF3 locus. Given that the full phenotypic range of EBF3 loss of function has not been elucidated, it remains to be determined whether these reportedly unaffected individuals carrying EBF3 deletion CNVs might represent the milder presentation of EBF3 loss of function or are unaffected carriers. Relative to those in reportedly unaffected individuals in the general population, the enrichment of EBF3 deletion CNVs reported in affected individuals with a range of neurodevelopmental phenotypes, together with the phenotypic overlap between our probands and de novo loss-of-function variants in EBF3, suggests that haploinsufficiency of EBF3 is potentially pathogenic. Given that EBF3 transcription factors function as homodimers that bind to each other via their basic helix-loop-helix domain, another potential mechanism for the loss of function observed with variants p.Arg163Gln and p.Arg163Leu could be that they correspond to dominant-negative mutations that poison the function of the WT protein. The autosomal-dominant mode of inheritance of these variants is therefore most likely due to EBF3 loss of function via either a dominant-negative or haploinsufficient effect.
Interestingly, EBF3 is a well-characterized transcriptional target of ARX, and loss of ARX repression of EBF3 causes disrupted inhibitory GABAergic neuronal migration in Arx-deletion mouse models. Our findings suggest that EBF3 loss of function might mediate a subset of features seen in ARX-related disorders associated with ARX gain of function, specifically with regard to the shared features of intellectual disability, abnormal genitalia, and structural CNS anomalies. Additionally, the phenotypes of the three probands are clearly similar and consistent with the functional results for the EBF3 p.Arg163Gln and p.Arg163Leu variants. We show that EBF3 p.Arg163Gln is a severe loss-of-function variant and that p.Arg163Leu is a partial loss-of-function variant. Determining possible genotype-phenotype correlations related to residual activity of the variant protein is limited by the small sample size. However, it is interesting that probands 1 and 2 (who have the p.Arg163Gln variant and minimal transcriptional activity) have structural anomalies of the cerebellum, whereas proband 3 (who has the p.Arg163Leu variant and partial transcriptional activity) has no structural brain or genitourinary abnormalities. We also note that Harms et al.56 (in this issue of The Journal) identify EBF3 mutations in individuals with similar neurological phenotypes. A larger sample size and longitudinal monitoring of neurodevelopment would be beneficial to determining the genotype-phenotype correlations between the EBF3 variants identified in the two studies. Our findings further suggest that ARX target genes might represent an enriched population of disease-associated genes for neurodevelopmental disorders given the diversity and heterogeneity of ARX-related disorders.
Consortia
Members of the Undiagnosed Diseases Network (UDN) include David R. Adams, Christopher J. Adams, Mercedes E. Alejandro, Patrick Allard, Euan A. Ashley, Carlos A. Bacino, Ashok Balasubramanyam, Hayk Barseghyan, Alan H. Beggs, Hugo J. Bellen, Jonathan A. Bernstein, David P. Bick, Camille L. Birch, Braden E. Boone, Lauren C. Briere, Donna M. Brown, Matthew Brush, Lindsay C. Burrage, Katherine R. Chao, Gary D. Clark, Joy D. Cogan, Cynthia M. Cooper, William J. Craigen, Mariska Davids, Jyoti G. Dayal, Esteban C. Dell’Angelica, Shweta U. Dhar, Katrina M. Dipple, Laurel A. Donnell-Fink, Naghmeh Dorrani, Dan C. Dorset, David D. Draper, Annika M. Dries, David J. Eckstein, Lisa T. Emrick, Christine M. Eng, Cecilia Esteves, Tyra Estwick, Paul G. Fisher, Trevor S. Frisby, Kate Frost, William A. Gahl, Valerie Gartner, Rena A. Godfrey, Mitchell Goheen, Gretchen A. Golas, David B. Goldstein, Mary “Gracie” G. Gordon, Sarah E. Gould, Jean-Philippe F. Gourdine, Brett H. Graham, Catherine A. Groden, Andrea L. Gropman, Mary E. Hackbarth, Melissa Haendel, Rizwan Hamid, Neil A. Hanchard, Lori H. Handley, Isabel Hardee, Matthew R. Herzog, Ingrid A. Holm, Ellen M. Howerton, Howard J. Jacob, Mahim Jain, Yong-hui Jiang, Jean M. Johnston, Angela L. Jones, Alanna E. Koehler, David M. Koeller, Isaac S. Kohane, Jennefer N. Kohler, Donna M. Krasnewich, Elizabeth L. Krieg, Joel B. Krier, Jennifer E. Kyle, Seema R. Lalani, Lea Latham, Yvonne L. Latour, C. Christopher Lau, Jozef Lazar, Brendan H. Lee, Hane Lee, Paul R. Lee, Shawn E. Levy, Denise J. Levy, Richard A. Lewis, Adam P. Liebendorder, Sharyn A. Lincoln, Carson R. Loomis, Joseph Loscalzo, Richard L. Maas, Ellen F. Macnamara, Calum A. MacRae, Valerie V. Maduro, May Christine V. Malicdan, Laura A. Mamounas, Teri A. Manolio, Thomas C. Markello, Azamian S. Mashid, Paul Mazur, Alexandra J. McCarty, Allyn McConkie-Rosell, Alexa T. McCray, Thomas O. Metz, Matthew Might, Paolo M. Moretti, John J. Mulvihill, Jennifer L. Murphy, Donna M. Muzny, Michele E. Nehrebecky, Stan F. Nelson, J. Scott Newberry, John H. Newman, Sarah K. Nicholas, Donna Novacic, Jordan S. Orange, J. Carl Pallais, Christina G.S. Palmer, Jeanette C. Papp, Loren D.M. Pena, John A. Phillips III, Jennifer E. Posey, John H. Postlethwait, Lorraine Potocki, Barbara N. Pusey, Rachel B. Ramoni, Lance H. Rodan, Sarah Sadozai, Katherine E. Schaffer, Kelly Schoch, Molly C. Schroeder, Daryl A. Scott, Prashant Sharma, Vandana Shashi, Edwin K. Silverman, Janet S. Sinsheimer, Ariane G. Soldatos, Rebecca C. Spillmann, Kimberly Splinter, Joan M. Stoler, Nicholas Stong, Kimberly A. Strong, Jennifer A. Sullivan, David A. Sweetser, Sara P. Thomas, Cynthia J. Tift, Nathanial J. Tolman, Camilo Toro, Alyssa A. Tran, Zaheer M. Valivullah, Eric Vilain, Daryl M. Waggott, Colleen E. Wahl, Nicole M. Walley, Chris A. Walsh, Michael F. Wangler, Mike Warburton, Patricia A. Ward, Katrina M. Waters, Bobbie-Jo M. Webb-Robertson, Alec A. Weech, Monte Westerfield, Matt T. Wheeler, Anastasia L. Wise, Lynne A. Worthe, Elizabeth A. Worthey, Shinya Yamamoto, Yaping Yang, Guoyun Yu, and Patricia A. Zornio.
Acknowledgments
We thank the families and clinical staff at the Undiagnosed Diseases Program (UDP), Baylor College of Medicine (BCM), and New York University for participating in this study. This work was supported in part by NIH grants U54NS093793, R24OD022005, and R01GM067858 to H.J.B, by the Intramural Research Program of the National Human Genome Research Institute, and by the Common Fund of the NIH Office of the Director. H.J.B. is an investigator of the Howard Hughes Medical Institute (HHMI). F.X. and J.A.R. are supported by NIH grant U01HG007709. The Department of Molecular and Human Genetics at BCM derives revenue from the clinical exome sequencing offered at Baylor Genetics. We are thankful for the technical assistance provided by Y. Huang (NIH UDP Translational Laboratory) for Sanger sequencing; A. Weech (NIH UDP Translational Laboratory) for input on protein modeling; H. Pan (HHMI, BCM) for fly embryo injections; S. Nagarkar-Jaiswal, P. Lee, Y. He, J. Li, Z. Wang, Q. Gao, and L. Wang (BCM) for creating the 15,000 MiMIC insertion stocks; and W. Lin (BCM) for T2A-GAL4 conversion. We thank the Bloomington Drosophila Stock Center, Drosophila Genomics and Genetic Resources, and FlyORF for numerous stocks and the Developmental Studies Hybridoma Bank for antibodies. This study made use of data generated by the DECIPHER community. A full list of centers that contributed to the generation of the data is available at http://decipher.sanger.ac.uk and via email at decipher@sanger.ac.uk. Funding for the project was provided by the Wellcome Trust.
Published: December 22, 2016
Footnotes
Supplemental Data include a Supplemental Note and six tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.11.018.
Contributor Information
Michael F. Wangler, Email: mw147467@bcm.edu.
May Christine V. Malicdan, Email: maychristine.malicdan@nih.gov.
Undiagnosed Diseases Network:
David R. Adams, Christopher J. Adams, Mercedes E. Alejandro, Patrick Allard, Euan A. Ashley, Carlos A. Bacino, Ashok Balasubramanyam, Hayk Barseghyan, Alan H. Beggs, Hugo J. Bellen, Jonathan A. Bernstein, David P. Bick, Camille L. Birch, Braden E. Boone, Lauren C. Briere, Donna M. Brown, Matthew Brush, Lindsay C. Burrage, Katherine R. Chao, Gary D. Clark, Joy D. Cogan, Cynthia M. Cooper, William J. Craigen, Mariska Davids, Jyoti G. Dayal, Esteban C. Dell'Angelica, Shweta U. Dhar, Katrina M. Dipple, Laurel A. Donnell-Fink, Naghmeh Dorrani, Dan C. Dorset, David D. Draper, Annika M. Dries, David J. Eckstein, Lisa T. Emrick, Christine M. Eng, Cecilia Esteves, Tyra Estwick, Paul G. Fisher, Trevor S. Frisby, Kate Frost, William A. Gahl, Valerie Gartner, Rena A. Godfrey, Mitchell Goheen, Gretchen A. Golas, David B. Goldstein, Mary “Gracie” G. Gordon, Sarah E. Gould, Jean-Philippe F. Gourdine, Brett H. Graham, Catherine A. Groden, Andrea L. Gropman, Mary E. Hackbarth, Melissa Haendel, Rizwan Hamid, Neil A. Hanchard, Lori H. Handley, Isabel Hardee, Matthew R. Herzog, Ingrid A. Holm, Ellen M. Howerton, Howard J. Jacob, Mahim Jain, Yong-hui Jiang, Jean M. Johnston, Angela L. Jones, Alanna E. Koehler, David M. Koeller, Isaac S. Kohane, Jennefer N. Kohler, Donna M. Krasnewich, Elizabeth L. Krieg, Joel B. Krier, Jennifer E. Kyle, Seema R. Lalani, Lea Latham, Yvonne L. Latour, C. Christopher Lau, Jozef Lazar, Brendan H. Lee, Hane Lee, Paul R. Lee, Shawn E. Levy, Denise J. Levy, Richard A. Lewis, Adam P. Liebendorder, Sharyn A. Lincoln, Carson R. Loomis, Joseph Loscalzo, Richard L. Maas, Ellen F. Macnamara, Calum A. MacRae, Valerie V. Maduro, May Christine V. Malicdan, Laura A. Mamounas, Teri A. Manolio, Thomas C. Markello, Azamian S. Mashid, Paul Mazur, Alexandra J. McCarty, Allyn McConkie-Rosell, Alexa T. McCray, Thomas O. Metz, Matthew Might, Paolo M. Moretti, John J. Mulvihill, Jennifer L. Murphy, Donna M. Muzny, Michele E. Nehrebecky, Stan F. Nelson, J. Scott Newberry, John H. Newman, Sarah K. Nicholas, Donna Novacic, Jordan S. Orange, J. Carl Pallais, Christina G.S. Palmer, Jeanette C. Papp, Loren D.M. Pena, John A. Phillips, III, Jennifer E. Posey, John H. Postlethwait, Lorraine Potocki, Barbara N. Pusey, Rachel B. Ramoni, Lance H. Rodan, Sarah Sadozai, Katherine E. Schaffer, Kelly Schoch, Molly C. Schroeder, Daryl A. Scott, Prashant Sharma, Vandana Shashi, Edwin K. Silverman, Janet S. Sinsheimer, Ariane G. Soldatos, Rebecca C. Spillmann, Kimberly Splinter, Joan M. Stoler, Nicholas Stong, Kimberly A. Strong, Jennifer A. Sullivan, David A. Sweetser, Sara P. Thomas, Cynthia J. Tift, Nathanial J. Tolman, Camilo Toro, Alyssa A. Tran, Zaheer M. Valivullah, Eric Vilain, Daryl M. Waggott, Colleen E. Wahl, Nicole M. Walley, Chris A. Walsh, Michael F. Wangler, Mike Warburton, Patricia A. Ward, Katrina M. Waters, Bobbie-Jo M. Webb-Robertson, Alec A. Weech, Monte Westerfield, Matt T. Wheeler, Anastasia L. Wise, Lynne A. Worthe, Elizabeth A. Worthey, Shinya Yamamoto, Yaping Yang, Guoyun Yu, and Patricia A. Zornio
Accession Numbers
The accession numbers for the c.488G>A (p.Arg163Gln) and c.488G>T (p.Arg163Leu) sequences reported in this paper are ClinVar: SCV000328550 and SCV000328551, respectively.
Web Resources
Database of Genomic Variation (DGV), http://dgv.tcag.ca/dgv/app/home/
DECIPHER, http://decipher.sanger.ac.uk/
ENSEMBL Variant Effect Predictor, http://useast.ensembl.org/info/docs/tools/vep/index.html
Exome Aggregation Consortium (ExAC) Browser, http://exac.broadinstitute.org/
FlyBase, http://flybase.org/
MutationTaster, http://www.mutationtaster.org
OMIM, http://www.omim.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
Supplemental Data
References
- 1.Alwan A., Modell B. Recommendations for introducing genetics services in developing countries. Nat. Rev. Genet. 2003;4:61–68. doi: 10.1038/nrg978. [DOI] [PubMed] [Google Scholar]
- 2.American Psychiatric Association, editor. Diagnostic and statistical manual of mental disorders. Fifth Edition. American Psychiatric Association Publishing; 2013. [Google Scholar]
- 3.Boivin M.J., Kakooza A.M., Warf B.C., Davidson L.L., Grigorenko E.L. Reducing neurodevelopmental disorders and disability through research and interventions. Nature. 2015;527:S155–S160. doi: 10.1038/nature16029. [DOI] [PubMed] [Google Scholar]
- 4.Yang Y., Muzny D.M., Reid J.G., Bainbridge M.N., Willis A., Ward P.A., Braxton A., Beuten J., Xia F., Niu Z. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N. Engl. J. Med. 2013;369:1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu X., Petrovski S., Xie P., Ruzzo E.K., Lu Y.F., McSweeney K.M., Ben-Zeev B., Nissenkorn A., Anikster Y., Oz-Levi D. Whole-exome sequencing in undiagnosed genetic diseases: interpreting 119 trios. Genet. Med. 2015;17:774–781. doi: 10.1038/gim.2014.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Ligt J., Willemsen M.H., van Bon B.W., Kleefstra T., Yntema H.G., Kroes T., Vulto-van Silfhout A.T., Koolen D.A., de Vries P., Gilissen C. Diagnostic exome sequencing in persons with severe intellectual disability. N. Engl. J. Med. 2012;367:1921–1929. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 7.Neale B.M., Kou Y., Liu L., Ma’ayan A., Samocha K.E., Sabo A., Lin C.F., Stevens C., Wang L.S., Makarov V. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bainbridge M.N., Hu H., Muzny D.M., Musante L., Lupski J.R., Graham B.H., Chen W., Gripp K.W., Jenny K., Wienker T.F. De novo truncating mutations in ASXL3 are associated with a novel clinical phenotype with similarities to Bohring-Opitz syndrome. Genome Med. 2013;5:11. doi: 10.1186/gm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allen A.S., Berkovic S.F., Cossette P., Delanty N., Dlugos D., Eichler E.E., Epstein M.P., Glauser T., Goldstein D.B., Han Y., Epi4K Consortium. Epilepsy Phenome/Genome Project De novo mutations in epileptic encephalopathies. Nature. 2013;501:217–221. doi: 10.1038/nature12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petrovski S., Wang Q., Heinzen E.L., Allen A.S., Goldstein D.B. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Samocha K.E., Robinson E.B., Sanders S.J., Stevens C., Sabo A., McGrath L.M., Kosmicki J.A., Rehnström K., Mallick S., Kirby A. A framework for the interpretation of de novo mutation in human disease. Nat. Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Magewu A.N., Jones P.A. Ubiquitous and tenacious methylation of the CpG site in codon 248 of the p53 gene may explain its frequent appearance as a mutational hot spot in human cancer. Mol. Cell. Biol. 1994;14:4225–4232. doi: 10.1128/mcb.14.6.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mancini D., Singh S., Ainsworth P., Rodenhiser D. Constitutively methylated CpG dinucleotides as mutation hot spots in the retinoblastoma gene (RB1) Am. J. Hum. Genet. 1997;61:80–87. doi: 10.1086/513898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goriely A., McGrath J.J., Hultman C.M., Wilkie A.O., Malaspina D. “Selfish spermatogonial selection”: a novel mechanism for the association between advanced paternal age and neurodevelopmental disorders. Am. J. Psychiatry. 2013;170:599–608. doi: 10.1176/appi.ajp.2013.12101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colasante G., Sessa A., Crispi S., Calogero R., Mansouri A., Collombat P., Broccoli V. Arx acts as a regional key selector gene in the ventral telencephalon mainly through its transcriptional repression activity. Dev. Biol. 2009;334:59–71. doi: 10.1016/j.ydbio.2009.07.014. [DOI] [PubMed] [Google Scholar]
- 17.Fulp C.T., Cho G., Marsh E.D., Nasrallah I.M., Labosky P.A., Golden J.A. Identification of Arx transcriptional targets in the developing basal forebrain. Hum. Mol. Genet. 2008;17:3740–3760. doi: 10.1093/hmg/ddn271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strømme P., Mangelsdorf M.E., Shaw M.A., Lower K.M., Lewis S.M., Bruyere H., Lütcherath V., Gedeon A.K., Wallace R.H., Scheffer I.E. Mutations in the human ortholog of Aristaless cause X-linked mental retardation and epilepsy. Nat. Genet. 2002;30:441–445. doi: 10.1038/ng862. [DOI] [PubMed] [Google Scholar]
- 19.Bienvenu T., Poirier K., Friocourt G., Bahi N., Beaumont D., Fauchereau F., Ben Jeema L., Zemni R., Vinet M.C., Francis F. ARX, a novel Prd-class-homeobox gene highly expressed in the telencephalon, is mutated in X-linked mental retardation. Hum. Mol. Genet. 2002;11:981–991. doi: 10.1093/hmg/11.8.981. [DOI] [PubMed] [Google Scholar]
- 20.Kato M., Das S., Petras K., Kitamura K., Morohashi K., Abuelo D.N., Barr M., Bonneau D., Brady A.F., Carpenter N.J. Mutations of ARX are associated with striking pleiotropy and consistent genotype-phenotype correlation. Hum. Mutat. 2004;23:147–159. doi: 10.1002/humu.10310. [DOI] [PubMed] [Google Scholar]
- 21.Kato M., Saitoh S., Kamei A., Shiraishi H., Ueda Y., Akasaka M., Tohyama J., Akasaka N., Hayasaka K. A longer polyalanine expansion mutation in the ARX gene causes early infantile epileptic encephalopathy with suppression-burst pattern (Ohtahara syndrome) Am. J. Hum. Genet. 2007;81:361–366. doi: 10.1086/518903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kitamura K., Yanazawa M., Sugiyama N., Miura H., Iizuka-Kogo A., Kusaka M., Omichi K., Suzuki R., Kato-Fukui Y., Kamiirisa K. Mutation of ARX causes abnormal development of forebrain and testes in mice and X-linked lissencephaly with abnormal genitalia in humans. Nat. Genet. 2002;32:359–369. doi: 10.1038/ng1009. [DOI] [PubMed] [Google Scholar]
- 23.Ishibashi M., Manning E., Shoubridge C., Krecsmarik M., Hawkins T.A., Giacomotto J., Zhao T., Mueller T., Bader P.I., Cheung S.W. Copy number variants in patients with intellectual disability affect the regulation of ARX transcription factor gene. Hum. Genet. 2015;134:1163–1182. doi: 10.1007/s00439-015-1594-x. [DOI] [PubMed] [Google Scholar]
- 24.Turner G., Partington M., Kerr B., Mangelsdorf M., Gecz J. Variable expression of mental retardation, autism, seizures, and dystonic hand movements in two families with an identical ARX gene mutation. Am. J. Med. Genet. 2002;112:405–411. doi: 10.1002/ajmg.10714. [DOI] [PubMed] [Google Scholar]
- 25.Okazaki S., Ohsawa M., Kuki I., Kawawaki H., Koriyama T., Ri S., Ichiba H., Hai E., Inoue T., Nakamura H. Aristaless-related homeobox gene disruption leads to abnormal distribution of GABAergic interneurons in human neocortex: evidence based on a case of X-linked lissencephaly with abnormal genitalia (XLAG) Acta Neuropathol. 2008;116:453–462. doi: 10.1007/s00401-008-0382-2. [DOI] [PubMed] [Google Scholar]
- 26.Schaaf C.P., Gonzalez-Garay M.L., Xia F., Potocki L., Gripp K.W., Zhang B., Peters B.A., McElwain M.A., Drmanac R., Beaudet A.L. Truncating mutations of MAGEL2 cause Prader-Willi phenotypes and autism. Nat. Genet. 2013;45:1405–1408. doi: 10.1038/ng.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fountain M.D., Aten E., Cho M.T., Juusola J., Walkiewicz M.A., Ray J.W., Xia F., Yang Y., Graham B.H., Bacino C.A. The phenotypic spectrum of Schaaf-Yang syndrome: 18 new affected individuals from 14 families. Genet. Med. 2016 doi: 10.1038/gim.2016.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Fevre A.K., Taylor S., Malek N.H., Horn D., Carr C.W., Abdul-Rahman O.A., O’Donnell S., Burgess T., Shaw M., Gecz J. FOXP1 mutations cause intellectual disability and a recognizable phenotype. Am. J. Med. Genet. A. 2013;161A:3166–3175. doi: 10.1002/ajmg.a.36174. [DOI] [PubMed] [Google Scholar]
- 29.Horn D., Kapeller J., Rivera-Brugués N., Moog U., Lorenz-Depiereux B., Eck S., Hempel M., Wagenstaller J., Gawthrope A., Monaco A.P. Identification of FOXP1 deletions in three unrelated patients with mental retardation and significant speech and language deficits. Hum. Mutat. 2010;31:E1851–E1860. doi: 10.1002/humu.21362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pfeifer D., Poulat F., Holinski-Feder E., Kooy F., Scherer G. The SOX8 gene is located within 700 kb of the tip of chromosome 16p and is deleted in a patient with ATR-16 syndrome. Genomics. 2000;63:108–116. doi: 10.1006/geno.1999.6060. [DOI] [PubMed] [Google Scholar]
- 31.Harteveld C.L., Kriek M., Bijlsma E.K., Erjavec Z., Balak D., Phylipsen M., Voskamp A., di Capua E., White S.J., Giordano P.C. Refinement of the genetic cause of ATR-16. Hum. Genet. 2007;122:283–292. doi: 10.1007/s00439-007-0399-y. [DOI] [PubMed] [Google Scholar]
- 32.Wang S.S., Lewcock J.W., Feinstein P., Mombaerts P., Reed R.R. Genetic disruptions of O/E2 and O/E3 genes reveal involvement in olfactory receptor neuron projection. Development. 2004;131:1377–1388. doi: 10.1242/dev.01009. [DOI] [PubMed] [Google Scholar]
- 33.Chiara F., Badaloni A., Croci L., Yeh M.L., Cariboni A., Hoerder-Suabedissen A., Consalez G.G., Eickholt B., Shimogori T., Parnavelas J.G., Rakić S. Early B-cell factors 2 and 3 (EBF2/3) regulate early migration of Cajal-Retzius cells from the cortical hem. Dev. Biol. 2012;365:277–289. doi: 10.1016/j.ydbio.2012.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cooper J.A. A mechanism for inside-out lamination in the neocortex. Trends Neurosci. 2008;31:113–119. doi: 10.1016/j.tins.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 35.Garel S., Marín F., Mattéi M.G., Vesque C., Vincent A., Charnay P. Family of Ebf/Olf-1-related genes potentially involved in neuronal differentiation and regional specification in the central nervous system. Dev. Dyn. 1997;210:191–205. doi: 10.1002/(SICI)1097-0177(199711)210:3<191::AID-AJA1>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 36.Yamazaki H., Sekiguchi M., Takamatsu M., Tanabe Y., Nakanishi S. Distinct ontogenic and regional expressions of newly identified Cajal-Retzius cell-specific genes during neocorticogenesis. Proc. Natl. Acad. Sci. USA. 2004;101:14509–14514. doi: 10.1073/pnas.0406295101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chowdhury T.G., Jimenez J.C., Bomar J.M., Cruz-Martin A., Cantle J.P., Portera-Cailliau C. Fate of cajal-retzius neurons in the postnatal mouse neocortex. Front. Neuroanat. 2010;4:10. doi: 10.3389/neuro.05.010.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liberg D., Sigvardsson M., Akerblad P. The EBF/Olf/Collier family of transcription factors: regulators of differentiation in cells originating from all three embryonal germ layers. Mol. Cell. Biol. 2002;22:8389–8397. doi: 10.1128/MCB.22.24.8389-8397.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crozatier M., Valle D., Dubois L., Ibnsouda S., Vincent A. Collier, a novel regulator of Drosophila head development, is expressed in a single mitotic domain. Curr. Biol. 1996;6:707–718. doi: 10.1016/s0960-9822(09)00452-7. [DOI] [PubMed] [Google Scholar]
- 40.Crozatier M., Valle D., Dubois L., Ibnsouda S., Vincent A. Head versus trunk patterning in the Drosophila embryo; collier requirement for formation of the intercalary segment. Development. 1999;126:4385–4394. doi: 10.1242/dev.126.19.4385. [DOI] [PubMed] [Google Scholar]
- 41.Pozzoli O., Bosetti A., Croci L., Consalez G.G., Vetter M.L. Xebf3 is a regulator of neuronal differentiation during primary neurogenesis in Xenopus. Dev. Biol. 2001;233:495–512. doi: 10.1006/dbio.2001.0230. [DOI] [PubMed] [Google Scholar]
- 42.Prasad B.C., Ye B., Zackhary R., Schrader K., Seydoux G., Reed R.R. unc-3, a gene required for axonal guidance in Caenorhabditis elegans, encodes a member of the O/E family of transcription factors. Development. 1998;125:1561–1568. doi: 10.1242/dev.125.8.1561. [DOI] [PubMed] [Google Scholar]
- 43.Hagman J., Gutch M.J., Lin H., Grosschedl R. EBF contains a novel zinc coordination motif and multiple dimerization and transcriptional activation domains. EMBO J. 1995;14:2907–2916. doi: 10.1002/j.1460-2075.1995.tb07290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang S.S., Tsai R.Y., Reed R.R. The characterization of the Olf-1/EBF-like HLH transcription factor family: implications in olfactory gene regulation and neuronal development. J. Neurosci. 1997;17:4149–4158. doi: 10.1523/JNEUROSCI.17-11-04149.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang M.M., Tsai R.Y., Schrader K.A., Reed R.R. Genes encoding components of the olfactory signal transduction cascade contain a DNA binding site that may direct neuronal expression. Mol. Cell. Biol. 1993;13:5805–5813. doi: 10.1128/mcb.13.9.5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bellen H.J., Yamamoto S. Morgan’s legacy: fruit flies and the functional annotation of conserved genes. Cell. 2015;163:12–14. doi: 10.1016/j.cell.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vervoort M., Crozatier M., Valle D., Vincent A. The COE transcription factor Collier is a mediator of short-range Hedgehog-induced patterning of the Drosophila wing. Curr. Biol. 1999;9:632–639. doi: 10.1016/s0960-9822(99)80285-1. [DOI] [PubMed] [Google Scholar]
- 48.Nagarkar-Jaiswal S., Lee P.T., Campbell M.E., Chen K., Anguiano-Zarate S., Gutierrez M.C., Busby T., Lin W.W., He Y., Schulze K.L. A library of MiMICs allows tagging of genes and reversible, spatial and temporal knockdown of proteins in Drosophila. eLife. 2015;4 doi: 10.7554/eLife.05338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Venken K.J., Schulze K.L., Haelterman N.A., Pan H., He Y., Evans-Holm M., Carlson J.W., Levis R.W., Spradling A.C., Hoskins R.A., Bellen H.J. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat. Methods. 2011;8:737–743. doi: 10.1038/nmeth.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Diao F., Ironfield H., Luan H., Diao F., Shropshire W.C., Ewer J., Marr E., Potter C.J., Landgraf M., White B.H. Plug-and-play genetic access to drosophila cell types using exchangeable exon cassettes. Cell Rep. 2015;10:1410–1421. doi: 10.1016/j.celrep.2015.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gnerer J.P., Venken K.J., Dierick H.A. Gene-specific cell labeling using MiMIC transposons. Nucleic Acids Res. 2015;43:e56. doi: 10.1093/nar/gkv113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cook R.K., Christensen S.J., Deal J.A., Coburn R.A., Deal M.E., Gresens J.M., Kaufman T.C., Cook K.R. The generation of chromosomal deletions to provide extensive coverage and subdivision of the Drosophila melanogaster genome. Genome Biol. 2012;13:R21. doi: 10.1186/gb-2012-13-3-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bischof J., Sheils E.M., Björklund M., Basler K. Generation of a transgenic ORFeome library in Drosophila. Nat. Protoc. 2014;9:1607–1620. doi: 10.1038/nprot.2014.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Firth H.V., Richards S.M., Bevan A.P., Clayton S., Corpas M., Rajan D., Van Vooren S., Moreau Y., Pettett R.M., Carter N.P. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am. J. Hum. Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.MacDonald J.R., Ziman R., Yuen R.K., Feuk L., Scherer S.W. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 2014;42:D986–D992. doi: 10.1093/nar/gkt958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harms F.L., Girisha K.M., Hardigan A.A., Kortüm F., Shukla A., Alawi M., Dalal A., Brady L., Tarnopolsky M., Bird L.M. Mutations in EBF3 Disturb Transcriptional Profiles and Cause Intellectual Disability, Ataxia, and Facial Dysmorphism. Am. J. Hum. Genet. 2016;100:117–127. doi: 10.1016/j.ajhg.2016.11.012. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.