Abstract
Despite recent groundbreaking advances in multiple myeloma (MM) treatment, most MM patients ultimately experience relapse, and the relapse biology is not entirely understood. To define altered gene expression in MM relapse, gene expression profiles were examined and compared among 16 MM patients grouped by 12 months progression-free survival (PFS) after autologous stem cell transplantation. To maximize the difference between prognostic groups, patients at each end of the PFS spectrum (the four with the shortest PFS and four with the longest PFS) were chosen for additional analyses. We discovered that integrin-α8 (ITGA8) is highly expressed in MM patients with early relapse. The integrin family is well known to be involved in MM progression; however, the role of integrin-α8 is largely unknown. We functionally overexpressed integrin-α8 in MM cell lines, and surprisingly, stemness features including HIF1α, VEGF, OCT4, and Nanog, as well as epithelial mesenchymal transition (EMT)-related phenotypes, including N-cadherin, Slug, Snail and CXCR4, were induced. These, consequently, enhanced migration and invasion abilities, which are crucial to MM pathogenesis. Moreover, the gain of integrin-α8 expression mediated drug resistance against melphalan and bortezomib, which are the main therapeutic agents in MM. The cBioPortal genomic database revealed that ITGA8 have significant tendency to co-occur with PDGFRA and PDGFRB and their mRNA expression were up-regulated in ITGA8 overexpressed MM cells. In summary, integrin-α8, which was up-regulated in MM of early relapse, mediates EMT-like phenotype, enhancing migration and invasion; therefore, it could serve as a potential marker of MM relapse and be a new therapeutic target.
Keywords: epithelial-mesenchymal transition, gene expression profile, integrin-α8, multiple myeloma relapse
INTRODUCTION
Multiple myeloma (MM) is characterized by a clonal proliferation of malignant plasma (Palumbo and Anderson, 2011). The recent inundation of novel chemotherapeutic agents has led to tremendous advances in the treatment and survival outcomes and extended overall survival in MM patients (Kyle and Rajkumar, 2004). Nevertheless, MM still poses a great clinical challenge as most patients experience a relapse during their disease, but the exact mechanisms of relapse remain obscure (Avet-Loiseau, 2010; Lonial, 2010; Pandit and Vesole, 2001).
Gene expression profiling (GEP) has been used to predict the prognosis of cancer and apply tailored therapy (Potti et al., 2006; Reverter et al., 2010). Several groups have reported GEPs associated with MM progression and tried to develop novel therapeutic targets (Anguiano et al., 2009; Broyl et al., 2010; Kuiper et al., 2012; Harousseau et al., 2004; Zhan et al., 2002). However, less has been done to define novel therapeutic targets due to the complexity of MM relapse, and, to the best of our knowledge, our study is the first to analyze the mechanisms of relapse after high-dose chemotherapy with stem cell rescue. Investigation of altered gene expression related to relapse will provide a better understanding of relapse biology.
Epithelial-mesenchymal transition (EMT) is physiologically involved in embryogenesis, embryo implantation, and organ development, and contributes to cancer progression, metastasis, and tumor recurrence in pathologic circumstances (Kalluri and Weinberg, 2009; Thiery et al., 2009). The acquisition of EMT by cancer cells is associated with stem cell phenotype, cell migration, invasion and metastasis, and it is a well-recognized phenomenon in solid tumor (Guarino, 2007; Guarino et al., 2007). In MM, hypoxic conditions regulate the transcription of EMT related markers, causing MM cells to disseminate and metastasize to the new bone marrow (BM) region (Azab et al., 2012). CXCR4, which is one of the potential therapeutic targets in MM, is crucial for migration and homing of MM cells into BM. EMT has previously been reported to mediate CXCR4 to enhance metastasis in both solid tumors and MM (Alsayed et al., 2007; Azab et al., 2009; 2012; Yagi et al., 2011). MM cells with migration activity continuously egress out of the original location and metastasize into new areas, which ultimately translates into the development of disease heterogeneity, drug resistance, metastasis and relapse of MM (Bianchi et al., 2015; Hideshima et al., 2002; Menu et al., 2006).
The integrin family consists of cell adhesion receptors widely expressed in various types of cancer, including MM. Integrins play a crucial role in tumor initiation, progression, and metastasis by mediating tumor cell proliferation, migration, and invasion (Damiano and Dalton, 2000; Desgrosellier and Cheresh, 2010; Rathinam and Alahari, 2010). In MM, α4β1 (VLA-4) and α5β1 (VLA-5) are recognized as major integrins mediating the interaction between MM cells and BM niches through adhesion, migration and homing (Damiano and Dalton, 2000; Damiano et al., 1999; Sanz-Rodriguez and Teixido, 2001). Integrin-β7 is also important as it enhances adhesive interaction of MM cells with BM, and increases the production of vascular endothelial growth factor (VEGF), resulting in drug resistance (Neri et al., 2011). Overall, the integrin family is regarded as an attractive and promising therapeutic target for MM.
Our study focused on the genetic events of 16 relapsed MM patients after autologous bone marrow transplantation with distinguished gene expression patterns relative to PFS duration. We hypothesized that highly expressed integrin-α8, newly discovered from GEP in early relapsed MM patients, interprets MM cells EMT-like features, causing migration, invasion and drug resistance. Therefore, integrin-α8 could be utilized as a prognostic marker and a therapeutic target.
MATERIALS AND METHODS
Reagents
Melphalan was purchased from Sigma Aldrich (USA) while bortezomib and sunitinib were obtained from Selleckchem.com.
Cell lines and cultures
The human MM cell lines, KMS-12-BM, KMS-12-PE, MOLP8, NCI-H929, LP-1 and EJM, were obtained from Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures, Germany). The U266 and RPMI-8226 cell lines were kindly provided by Dr. Dong Soon Lee (Seoul National University, College of Medicine, Korea). Those cell lines were cultured in RPMI-1640 supplemented with penicillin (100 U/ml)/streptomycin (100 μg/ml) and 10% FBS (RPMI-8226), 15% FBS (U266) or 20% FBS (KMS-12-BM, KMS-12-PE, MOLP8 and NCI-H929). The LP1 and EJM were maintained in IMDM supplemented with penicillin (100 U/ml)/streptomycin (100μg/ml) and 20% FBS. The cells were incubated at 37°C in a 5% CO2 atmosphere.
Primary bone marrow samples collection and RNA purification
Bone marrow aspirates were obtained from 16 MM patients under a protocol approved by the Seoul National University Hospital Institutional Review Board for the use of samples for research. Each total RNA sample was isolated using an RNA Blood Mini Kit (QIAGEN, USA), and the concentration was measured using a ND-1000 Spectrometer (Nano Drop Technologies, Inc., USA).
Whole-genome gene expression assay
All 16 MM patients’ BM were run in triplicate. RNA isolated from the 16 MM patients was used for gene expression analysis with the Human-6 Whole-Genome Expression BeadChip (Illumina). Biotin-labeled cRNA was produced using a linear amplification kit (Ambion, USA) with 300 ng of quality-checked total RNA as an input. Chip hybridizations, washing, Cy3-streptavidin staining, and scanning were performed on a BeadArray™ Reader (Illumina) platform using reagents and by following protocols supplied by the manufacturer. The “Detection Score” was used to determine expression. Microarray data are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-4881.
Heatmap analysis and functional pathway prediction
A heatmap of the differentially expressed genes based on the 12 month PFS was created using Java TreeView 1.1.6r2 version (Department of Genetics, Stanford University School of Medicine, USA: http://jtreeview.sourceforge.net) (Saldanha, 2004). To investigate the biologically relevant pathways most impacted by the gene input list, the online program Pathway-Express (Onto-Tools, Wayne State University, Detroit, MI, USA; http://vortex.cs.wayne.edu/Projects.html) was used (Draghici et al., 2007). Specific biological pathways were defined using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Kanehisa Laboratories, Kyoto, Japan; http://www.genome.jp/kegg/pathway.html). Pathway-Express selects pathways based on an impact analysis that considers not only conventional statistical analysis, but also other biologic factors, including the expression levels of input genes, their type and position in a given pathway, and protein-protein interactions. In our pathway analysis, we used the gene expression level (fold change), while corrected p-values < 0.05 were used as an empirical cutoff for retrieving altered pathways.
Reverse transcription PCR and quantitative real-time PCR
Total RNA was isolated using TRIzol reagent (Gibco BRL, USA). cDNA was synthesized by reverse-transcription PCR with Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA, USA). Quantitative real-time PCR was conducted using an iCycler iQ detection system (BioRad Laboratories, Inc., USA) with SYBR Green. All data from the samples were normalized to the 18s cDNA levels. The primers used for qRT-PCR are listed in Table 1.
Table 1.
Sequence of primer
| Gene | Forward(5′-3′) | Reverse (5′-3′) |
|---|---|---|
| ITGA8 | ACATTCTGGTGGACTGTGG | AATCCCTTGTTGTTGCGTTC |
| VEGF | CCTTGCTGCTCTACCTCCAC | ATCTGCATGGTGATGTTGGA |
| HIF1α | TGATGACCAGCAACTTGAGG | TGGGGCATGGTAAAAGAAAG |
| OCT4 | GTACTCCTCGGTCCCTTTCC | CAAAAACCCTGGCACAAACT |
| Nanog | ATGCCTCACACGGAGACTG | CTGCGTCACACCATTGCTA |
| CDH1 | TGCCCAGAAAATGAAAAAGG | GTGTATGTGGCAATGCGTTC |
| CDH2 | CCATCACTCGGCTTAATGGT | GATGATGATGCAGAGCAGGA |
| Vimentin | TGCCCTTAAAGGAACCAATG | TCCAGCCAGCTTCCTGTAAGGT |
| Twist | GTCCGCAGTCTTACGAGGAG | CCAGCTTGAGGGTCTGAATC |
| Slug | CTTTTTCTTGCCCTCACTGC | GCTTCGGAGTGAAGAAATGC |
| Snail | ACCCCACATCCTTCTCACTG | TACAAAAACCCACGCAGACA |
| CXCR4 | GGTGGTCTATGTTGGCGTCTT | TGGAGTGTGACAGCTTGGAG |
| PDGFRA | GAAGCTGTCAACCTGCATGA | CTTCCTTAGCACGGATCAGC |
| PDGFRB | GCACTTTTATCCACCCAGGA | GTACTTGGCTCAGCCTCCAG |
Western blotting
Whole cell lysates were collected using Kinexus protein lysis buffer (containing 20 nM MOPS (pH 7.0), 2 mM EGTA, 5 mM EDTA, 30 mM sodium fluoride, 60 mM β-glycerophosphate (pH 7.2), 20 mM sodium pyrophosphate, 1 mM sodium orthovanadate, 1% Triton X-100, 1 mM PMSF and 1g/mL protein inhibitor cocktail (Hoffmann-La Roche Ltd., Swiss). Cell lysates were separated on 8% to 15% SDS-PAGE, transferred onto nitrocellulose membranes and then probed overnight at 4°C using the following appropriate primary antibodies: integrin-α8 (Santa Cruz Biotechnology), N-cadherin, vimentin, slug, snail (Cell Signaling Technology), phospho-CXCR4 and total CXCR4 (Abcam). Antibody binding was detected using the enhanced chemiluminescence system reagents (Amersham Pharmacia Biotec., USA). GAPDH or α-tubulin (Cell Signaling Technology) was used as a loading control.
Plasmid transfection
Transfection with pCMV3-ITGA8-GFPSpark (Sino Biological Inc., China) or pCMV3 control vector was performed using Lipofectamine RNAiMAX reagent (Invitrogen, USA) according to the manufacturer’s instructions.
Cell adhesion assay
LP1 or RPMI-8226 cells (5 × 106 cells/ml), control or ITGA8 overexpressed, were loaded into human fibronectin (Millipore, Germany) or MM patient derived BMSCs coated 96 well plates and incubated for 2–4 h at 37°C. Non-adherent cells were washed out with serum free RPMI 1640 media three times, after which 10 μl of CCK8 (Dojindo, Japan) was added into each well and incubated for 4 h. The adhered cells were detected at 450 nm using a microplate reader (Becton Dickinson Labware, France).
Chemotaxis assay
A chemotaxis assay was measured using an 8-μ-pore transwell plate (BioVision, Inc., USA) according to the manufacturer’s protocol. Next, 2–3 × 105 serum starved MM cells (LP1 and RPMI-8226), control or ITAG8 overexpressed, suspended in serum free RPMI1640 media were added to the upper chamber in the presence of medium containing 0 or 30 nM recombinant SDF1α (ProSpec, USA) in the lower chamber. After 24 h of incubation, the cells in the lower chamber were counted using Countess II FL (Thermo Fisher Scientific, USA) or the O.D. value at 450 nm was measured using a Versamax microplate reader (Molecular Devices).
Invasion assay
MM cell invasion was measured using an 8-μ-pore transwell plate coated with gel basement membrane solution (BioVision, Inc., USA) according to the manufacturer’s protocols. Next, 2–3 × 105 serum starved MM cells (LP1 and RPMI-8226), control or ITAG8 overexpressed, suspended in serum free RPMI1640 media were added to the upper chamber in the presence of supplemented medium in the lower chamber. After 24–48 h of incubation, the cells in the lower chamber were counted using a Countess II FL (Thermo Fisher Scientific, USA) or the O.D. value was measured at 450 nm.
Cell viability assay
Cells were seeded in 96 well plates, then exposed to 50 or 100 μM melphalan, or 1 μM bortezomib for 24 h. Next, 10 μl of CCK8 (Dojindo, Japan) was added into each well and the cells were incubated for 2–4 h. The absorbance of each well was detected at 450 nm using a microplate reader (Becton Dickinson Labware, France).
Genomic data
Genetic and expression alterations data were obtained from the cBioPortal cancer genomics platform (http://www.cbioportal.org). The 877 samples of all tumors containing mRNA, CNA and sequencing data were analyzed.
Statistical analysis
The statistical significance of differences observed between the two groups was analyzed by the Student’s t-test, using the GrahpPad Prism 4 statistical software (GraphPad Software, Inc., USA). For all statistical analyses, a P value < 0.05 was considered to indicate significance.
RESULTS
Integrin-α8 is highly expressed in MM patients with early relapse
To define the gene expression aspects in MM relapse, gene expression profiles were analyzed in 16 MM relapsed patients using the Human-6 Whole-Genome Expression BeadChip (Illumina). The whole BM aspirated from MM relapsed patients was used for RNA extraction and we interpreted whole BM as a tumor burden. The range of progression free survival (PFS) duration after autologous stem cell transplantation among the 16 MM patients was 54 to 1795 days, and one of them still did not relapse (Table 2). Patients were divided into two groups on the basis of 12 months PFS for GEP analysis. After filtering based on a P ≤ 0.01, heatmapping with 529 genes revealed markedly different gene expression pattern between the PFS < 12 month group and > 12 month group (Fig. 1A). From the 12 months PFS point, 77 genes upregulated more than two-fold in the < 12 months group were defined and the altered biological pathways were investigated (Supplementary Tables 1 and 2). Pathway enrichment analysis was performed using Pathway-Express identifying KEGG pathways from the process of an input list of genes (Draghici et al., 2007). We found eight pathways that were associated with the PFS < 12 months relapsed MM group (corrected P < 0.05), focal adhesion, glioma, melanoma, regulation of actin cytoskeleton, extracellular matrix-receptor interaction, prostate cancer, Toll-like receptor signaling pathway, and cytokine-cytokine receptor interaction. Platelet-derived growth factor beta (PDGFB, 2.02 fold), insulin-like growth factor 1 (IGF1, 7.76 fold), integrin alpha 7 (ITGA7, 2.34 fold) and integrin alpha 8 (ITGA8, 7.69 fold) were most involved among these pathways.
Table 2.
The characteristics of 16 MM patients
| Relapsed after auto BMT | <12 month (n = 7) | >12 month (n = 9) |
|---|---|---|
| PFS, days | ||
| Average | 182.1 | 663.4 |
| Range | 54–343 | 406–1795 |
| Age, years | ||
| Average | 58.5 | 55.7 |
| Range | 54.2–63.9 | 49.4–66.4 |
| Sex, n | ||
| Male | 5 | 7 |
| Female | 2 | 2 |
Fig. 1.
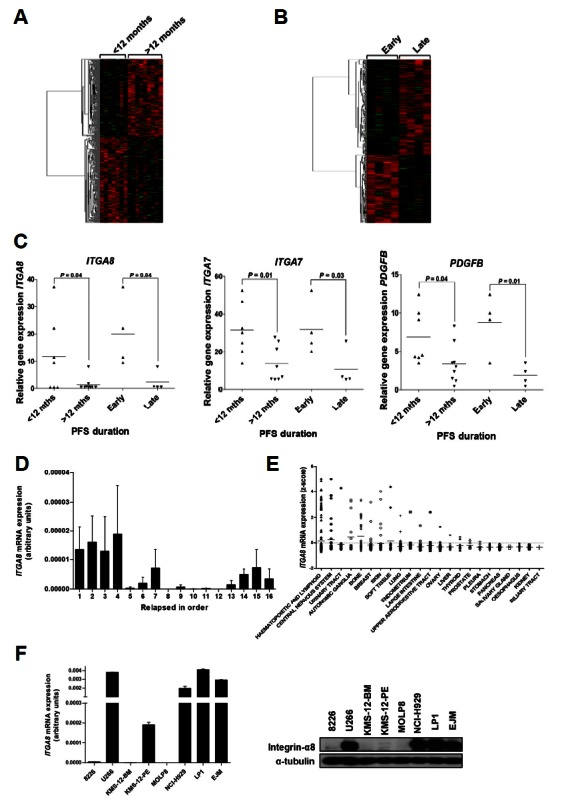
Integrin-α8 is highly expressed in MM patients with early relapse. (A) Heatmap of 529 genes differentially expressed between the PFS duration < 12 months group (n=7) and > 12 months group (n = 9) (P < 0.05). (B) Heatmap of 1018 genes differentially expressed between the early group (n = 4) and late group (n = 4) (P < 0.05). (C) Relative expression level based on microarray analysis of the group of PFS 12-month criterion (< 12 months or > 12 months) and extreme PFS (early or late): ITGA8, ITGA7, and PDGFB. (D) ITGA8 mRNA level in 16 relapsed patients was validated by real time PCR and the bar graph indicates the order of PFS duration. The mRNA expression was normalized to 18s transcript and data shown are the mean ± SD from three independent experiments (arbitrary units). (E) ITGA8 mRNA expression among the cancer type was obtained from cBioPortal (http://www.cbioportal.org). (F) Integrin-α8 mRNA and protein expression were screened in 8 MM cell lines (RPMI-8226, U266, KMS-12-BM, KMS-12-PE, MOLP8, NCI-H929, LP1 and EJM) by qRT-PCR and western blotting. The mRNA expression data shown are the means ± SD from three independent experiments (arbitrary units).
To maximize the difference between different prognostic groups, patients at each end of the PFS spectrum (the four with the shortest PFS and the four with the longest PFS) were selected for analyses and each group was defined as an ‘early group’ or ‘late group’ (Table 3). Heatmapping with a total of 1018 genes filtered at P ≤ 0.01, also showed a distinguished expression pattern between two groups (Fig. 1B). The 163 genes that showed expression upregulated by more than two-fold were defined based on comparison to the late group (Supplementary Table 3). These genes were involved in six KEGG pathways: melanogenesis, insulin signaling pathway, ECM-receptor interaction, focal adhesion, regulation of actin cytoskeleton, and basal cell carcinoma (corrected P < 0.05) (Supplementary Table 4). The genes most involved among the pathways were PDGFB (4.54 fold), FZD2 (8.99 fold), ITGA7 (2.95 fold) and ITGA8 (8.18 fold). We were particularly interested that up-regulated in the expression of ITGA8 and ITGA7, PDGFB (Fig. 1C) and their associated pathways, focal adhesion, ECM-receptor interaction, and regulation of actin cytoskeleton, were commonly identified with previous analysis. To validate the significance of ITGA8, ITGA7 and PDGFB expression in MM relapse, cDNA was synthesized from the RNA extracted from 16 MM patients and qRT-PCR was performed. The ITGA8 expression level matched microarray data that showed higher expression in early relapsed patients, and the four patients with the shortest PFS duration showed the highest ITGA8 expression (Fig. 1D). However, PDGFB and ITGA7 was not validated by qRT-PCR (data not shown).
Table 3.
The characteristics of 8 MM patients
| Relapsed after auto BMT | Early group (n = 4) | Late group (n = 4) |
|---|---|---|
| PFS, days | ||
| Average | 101.5 | 924.25 |
| Range | 54–169 | 517–1795 |
| Age, years | ||
| Average | 55.4 | 55.2 |
| Range | 45.25–63.94 | 49.36–62.68 |
| Sex, n | ||
| Male | 2 | 3 |
| Female | 2 | 1 |
The ITGA8 mRNA expression among cancer types was investigated using the cBioPortal genomic database. Interestingly, the ITGA8 level was highest in the hematopoietic and lymphoid system (Fig. 1E). These results indicate that ITGA8 expression may have a critical role in hematologic malignancies. We next screened the basal expression of integrin-α8 in mRNA and protein in the panel of MM cell lines (RPMI-8226, U266, KMS-12-BM, KMS-12-PE, MOLP8, HCI-H929, LP1, and EJM) by qRT-PCR and western blotting. As shown in Fig. 1F, among the MM cell lines, U266, NCI-H929, LP1, and EJM showed higher expression of integrin-α8 in both mRNA and protein compared to the others. Notably, the NCI-H929 cell line was established at the relapse state, and the U266, LP1, and EJM cell lines were established at the terminal refractory stage. The results provided further information regarding the relationship between integrin-α8 and MM relapse/refractory.
Integrin-α8 has not been thoroughly investigated in cancer, but it is expressed in mammary tumors undergoing metastasis, where it acts as a tumor cell receptor (Scherberich et al., 2005). Moreover, in many studies not related to cancer, integrin-α8 has been found to be localized on the extracellular domain in mesenchymal cells, such as fibroblast cells, mesangial cells and vascular smooth muscle cells, where it serves as a receptor for fibronectin, vitronectin, and osteopontin (Brandenberger et al., 2001; Denda et al., 1998; Muller et al., 1995). We conducted immunofluorescence staining to determine where integrin-a8 localizes in MM cells. As shown in Supplementary Fig. 1, integrin-α8 appears to be expressed in cytoplasm, but is primarily found in the membrane.
Integrin-α8 overexpression in MM cells leads to stem cell properties and EMT features
To understand the role of integrin-α8 expression in the MM relapse mechanism, we transiently transfected pCMV3-ITGA8-GFPSpark into MM cell lines. To accomplish this, we used RPMI-8226 and LP1 cells, which showed different basal expression of integrin-α8, to investigate whether the biological effects of integrin-α8 would come out the same, regardless of the expression level. The transfection results were confirmed based on the mRNA and protein level of integrin-α8 (Fig. 2A).
Fig. 2.
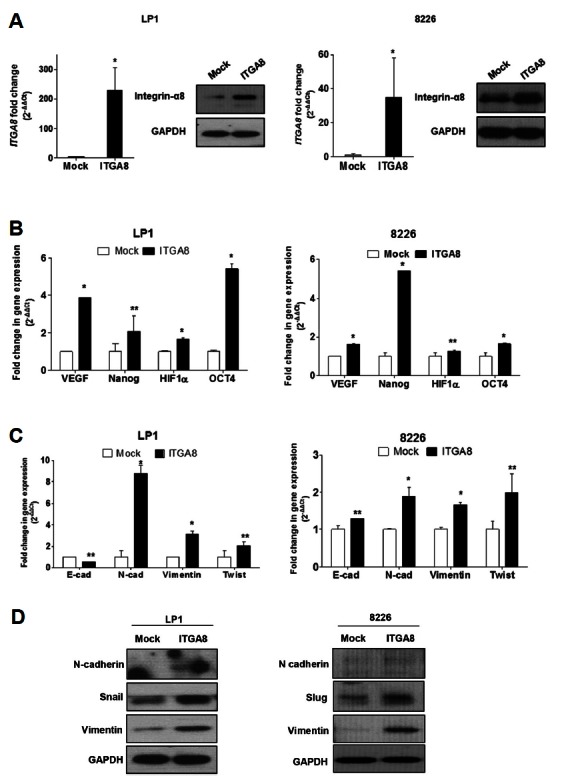
Gain of integrin-α8 expression regulates the markers of stem cell and EMT in MM cells. (A) Transfection of pCMV3-ITGA8-GFPSpark in LP1 and RPMI 8226 was confirmed based on the mRNA and protein level by qRT-PCR and western blotting, respectively (*P < 0.05). The 10 μg protein sample of LP1 and 15 μg protein sample of 8226 was loaded for western blot analyses. (B) Stemness phenotype including VEGF, NANOG, HIF1A, and OCT4 mRNA expression in integrin-α8 overexpressed LP1 and 8226 cells was confirmed by qRT-PCR and the data shown are the mean relative fold changes ± SD of three independent experiments compared to the control (*P < 0.05, **P > 0.2). Gene expression values in the samples were divided by those in the controls (i.e. each gene in the control is ‘1’). (C) The mRNA expression of EMT markers including E-cadherin, N-cadherin, vimentin and twist were measured in integrin-α8 overexpressed LP1 and 8226 cells. The data shown are the mean relative fold changes ± SD of three independent experiments (*P < 0.05, **P > 0.15). Gene expression values in the samples were divided by those in the controls (i.e. each gene in the control is ‘1’). (D) The protein level of EMT markers including N-cadherin, vimentin, slug, and snail was confirmed by Western blot analysis, and GAPDH was used as a loading control.
It is well known that cancer recurrence occurs due to the cancer stem cells (CSCs) having self-renewal ability, chemo-resistance, potential to metastasize, etc. (Lobo et al., 2007). Some studies have revealed that cancer cells acquire stem cell features and therefore behave like CSCs and contribute to malignancy of the tumor as well as cancer relapse (Blagosklonny, 2006; Mitra et al., 2015). We investigated whether integrin-α8 overexpression induces stemness properties. The mRNA level of stem cell markers including VEGF, HIF-1α, Nanog, and OCT4 was quantified by qRT-PCR, and compared to that of the control. Integrin-α8 overexpression in LP1 and RPMI-8226 cells induced the mRNA expression of the stemness factor (Fig. 2B), indicating that integrin-α8 mediates the stem cell-like features in MM cells.
In solid tumors, a subpopulation of stem cell like-cancer cells regulate EMT phenotype. Similarly, clonogenic MM cells also show a stem cell like-phenotype and consequently acquire EMT like-features (Blagosklonny, 2006; Matsui et al., 2008; Mitra et al., 2015). We demonstrated whether integrin-α8 overexpression can mediate the EMT phenomenon. To accomplish this, we screened the transcriptional level of EMT markers including E-cadherin, N-cadherin, vimentin and twist in overexpressed integrin-α8 LP1 and RPMI-8226 cells (Fig. 2C). It is well known that down-regulation of E-cadherin and upregulation of N-cadherin mediates EMT (Thiery et al., 2009). Our results demonstrate that increasing integrin-α8 expression increased N-cadherin level, as well as that of vimentin and twist, by more than 2 fold compared to the control. However, E-cadherin expression showed no significant changes. Protein levels also showed upregulation of N-cadherin, snail, slug and vimentin in integrin-α8 overexpressed cells (Fig. 2D), whereas there were no differences in E-cadherin protein expression (data not shown). We also checked the transcriptional level of MMP2 and MMP9, critical molecules involved in cancer metastasis (Gialeli et al., 2011), by qRT-PCR; however, MMP2 was not detected in integrin-α8 overexpressed LP1 because of its low expression. Moreover, there was no significant change in MMP9 compared to the control. Integrin-α8 overexpressed 8226 cells showed slight induction in both MMP2 and MMP9, but we found that it did not differ significantly from the control (Supplementary Fig. 2).
Overexpression of integrin-α8 mediates MM cell invasion and migration
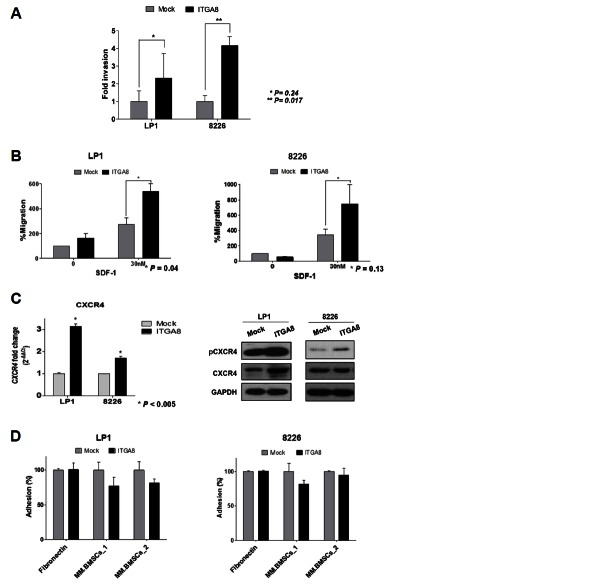
The interaction between MM cells and BM niches through invasion, migration, and adhesion is crucial to disease progression through which MM cells acquire proliferation and survival advantages, as well as drug resistance (Bianchi et al., 2015; Damiano et al., 1999; Pagnucco et al., 2004). Regarding the previous results that showed upregulated integrin-α8 induces stemness features and EMT markers, we next investigated the invasive ability of integrin-α8 overexpressed cells. As shown in Fig. 3A, a gain of integrin-α8 expression significantly increased RPMI-8226 and LP1 cell invasion that those cells penetrated the basement membrane coated chamber more actively than the control. These results show that integrin-α8 is one of the key factors of MM relapse biology, and that it functions by mediating the EMT-like features.
Fig. 3.
Gain of integrin-α8 expression mediates MM cell invasion and migration. (A) Integrin-α8 overexpressed cells that invaded through a basement membrane coated 8 μM pore transwell were used for this invasion assay. Integrin-α8 overexpressed LP1 and RPMI 8226 cells were starved for 24 h, after which cells that invaded to the bottom were counted. (B) An 8 μM pore transwell migration assay was used. Integrin-α8 overexpressed MM cells were starved for 24 h, and then allowed to migrate to the bottom well RPMI/IMDM serum free media supplemented with SDF-1a (0 or 30 nM). (C) The CXCR4 mRNA and protein level in integrin-α8 overexpressed MM cells were confirmed by qRT-PCR and Western blot, respectively. The mRNA data shown are the mean fold changes ± SD of three independent experiments compared to the control. Gene expression values in the samples were divided by those in the controls (i.e. each gene in the control is ‘1’). (D) Adhesion of overexpressed integrin-α8 LP1 and RPMI 8266 cells to FN or MM patient isolated BMSCs coated 96 well microplates. Unattached cells were washed out, after which adherent cells were determined based on the O.D. value.
Migration activity was also evaluated by a transwell migration assay, which revealed that 0 or 30 nM SDF-1α induced MM cells to migrate. Overexpression of integrin-α8 significantly induced chemotaxis of LP1 and RPMI-8226 cells compared to the control (Fig. 3B). There is growing evidence that the acquisition of EMT-like features accompanied the upregulated CXCR4 expression during MM cell dissemination (Roccaro et al., 2015). The CXCR4/SDF-1α axis plays a major role in chemotaxis in MM cells to BM, and we evaluated CXCR4 expression (Alsayed et al., 2007). As shown in Fig. 3C, the mRNA level of CXCR4 and protein level of pCXCR4 were upregulated in overexpressed integrin-α8 MM cells. These findings indicate that integrin-α8 may regulate CXCR4/SDF-1α signaling, causing MM cells to migrate actively. We next examined the integrin-α8 effect on MM cells adhesion to fibronectin and BMSCs isolated from MM patients. Compared to control, we could not find any significant effect of integrin-α8 in adhesive activity to BM elements (Fig. 3D). Our results are contradictory to the well-known finding that integrin is one of the major molecules involved in regulation of cell adhesion. However, the knockdown conditions of integrin-α8 decreased adhesive interaction on BM (Supplementary Fig. 3). Further study is needed to confirm the function of integrin-α8 in cell adhesion according to its expression. Taken together, these findings indicate that, because integrin-α8 induced MM cell chemotaxis and invasion, cells may have motility to egress in and out from the BM rather than adhere to the BM elements.
MM cells with overexpressed integrin-α8 enhances drug resistance to melphalan and bortezomib
The alkylating agent, melphalan, and the proteasome inhibitor, bortezomib, are the most widely used treatment agents in MM patients, and failure of these chemotherapeutic strategies leads to MM recurrence. Therefore, we investigated whether integrin-α8 overexpression affected the sensitivity of melphalan and bortezomib. As shown in Fig. 4, integrin-α8 overexpressed LP1, while RPMI 8226 cells showed lower sensitivity in the presence of 50 μM and100 μM of melphalan as well as high doses of bortezomib after 24 h. These results indicate that induction of EMT-like features mediated by integrin-α8 may contribute to drug resistance, which can lead MM disease progression.
Fig. 4.
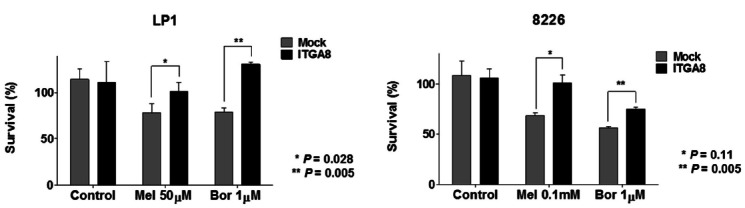
Gain of integrin-α8 expression shows drug resistance in MM cells. Drug treated integrin-α8 overexpressed LP1 (50 μM melphalan, 1 μM bortezomib) and 8226 (0.1 mM melphalan, 1 μM bortezomib) were incubated for 24 h. Cell viability was measured using CCK8 solution. The data shown are the means ± SD from three independent experiments.
Integrin-α8 may crosstalk with PDGFRA and PDGFRB
It is well known that the crosstalk between integrin and growth factor receptor regulates migration and adhesion of cancer cells, consequently mediating cancer initiation, drug resistance, and metastasis (Eliceiri, 2001). We obtained evidence from cBioPortal cancer genomic platforms that integrin-α8 mRNA upregulation is correlated with 42.2% of PDGFRA and 33.3% of PDGFRB mRNA upregulation (Fig. 5A). The co-occurrence of ITGA8 and PDGFRA or ITGA8 and PDGFRB was significant (Fig. 5B). To clarify these results, we investigated PDGFRA and PDGFRB mRNA expression in integrin-α8 overexpressed LP1 and RPMI-8226 cells. Both PDGF receptors mRNA expression were co-upregulated following integrin-α8 overexpression (Fig. 5C). PDGF receptor signaling is an important pathway for tumor progression. Taken together, these findings indicate that the crosstalk between integrin-α8 and PDGF receptor signaling may mediate MM pathogenesis; however, further study is needed to confirm this.
Fig. 5.
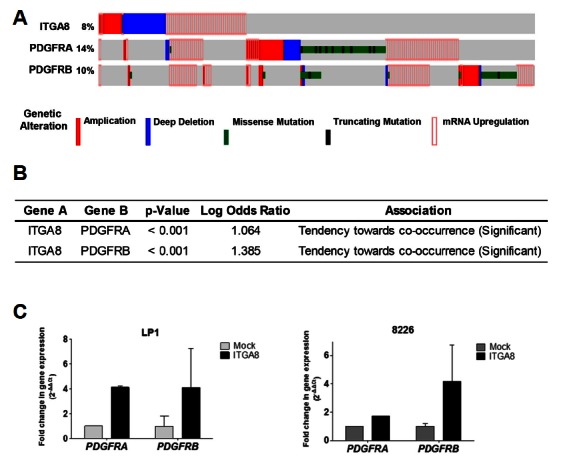
Crosstalk between ITGA8 and PDGFR signaling. (A) Data for genetic and expression alterations of ITGA8 were obtained from the cBioPortal cancer genomics platform (http://www.cbioportal.org). The 877 samples of all tumors having mRNA, CNA and sequencing data were analyzed. The % on the left represents the percentage of cases with alteration. (B) The tendency of co-occurrence between ITGA8 and PDGFRA or ITGA8 and PDGFRB. (C) The relative levels of PDGFRA and PDGFRB mRNA expression were confirmed using qRT-PCR in integrin-α8 overexpressed LP1 and 8226 cells and normalized to 18s transcript. The data shown are the mean relative fold changes ± SD of three independent experiments compared to the control. Gene expression values in the samples were divided by those in the controls (i.e. each gene in the control is ‘1’).
DISCUSSION
Despite remarkable recent advances in MM treatment, MM remains an incurable disease with unidentified mechanisms of relapse (Kyle and Rajkumar, 2004; Richardson et al., 2005). In this study, to address the unresolved questions regarding MM relapse mechanisms, we focused on identifying the key molecules responsible for controlling adhesion, migration and invasion of MM cells. A recent study showed that MM cells disseminate into peripheral circulation through acquisition of EMT-like features under hypoxic conditions to migrate and invade into a new region of BM. These behaviors are a similar to those observed in solid tumor metastasis, in which MM cells become more malignant to survive, proliferate, escape and become resistant against chemotherapy, eventually leading to MM recurrence (Azab et al., 2012; Muz et al., 2014; Thompson and Haviv, 2011). Similarly, in the present study, GEP revealed that integrin-α8 is constantly highly expressed in MM with early relapse, and that integrin-α8 induces EMT-like features in these patients, which could play a pivotal role in relapse.
For the first part of our study, gene expression of 16 relapsed MM patients undergoing autologous stem cell transplantation was analyzed. Whole BM from relapsed MM patients was used for RNA extraction that we interpreted whole BM as a tumor burden. Furthermore, genes resistant to bone marrow transplantation or chemotherapy may not have any impact on their expression level (Batist et al., 2011; Nooter and Herweijer, 1991), so the acquired date of BM from patients was not considered. Grouping with PFS duration at the point of 12 months or grouping at each end of the PFS spectrum (the four patients with the shortest PFS and the four patients with the longest PFS) were compared and found to have considerably different patterns of gene expression. In both analyses, three genes, ITGA8, ITGA7 and PDGFB, were identified and regarded as candidate genes that may be involved in and mediate MM relapse; however, only ITGA8 was confirmed to play such a role.
The integrin family plays an important role in mediating MM pathogenesis and drug resistance by promoting adhesion, migration and homing to BM niche (Damiano and Dalton, 2000; Neri et al., 2011; Sanz-Rodriguez et al., 1999). However, integrin-α8, which we newly discovered from GEP of relapsed MM, has never previously been reported in MM. In this study, we investigated whether the induction of integrin-α8 may potentially be involved in early MM relapse by regulating the interaction with the BM niche through adhesion, migration, and invasion. The cBioPortal database revealed the highest frequency of ITGA8 mRNA expression in the hematopoietic and lymphoid system among cancer. In addition, highly expressed integrin-α8 was observed in approximately 50% of the MM cell lines that were established from the relapse/refractory stage. These results indicate integrin-α8 is primarily associated with hematologic malignancies and partially support our hypothesis that highly expressed integrin-α8 is related to MM relapse/refractory.
Since integrin-α8 was shown to be upregulated in MM early relapse, we overexpressed integrin-α8 in LP1 and RPMI-8226 to investigate its functional role. We screened stem cell markers in integrin-α8 overexpressed MM cells and found that integrin-α8 upregulation may mediate the stemness feature of MM cells. There is growing evidence that cancer cells share the signaling of cancer stem cells, which express the markers of normal stem cells, tumor initiation ability, drug resistance, and acquisition of the EMT phenotype. The subpopulation of MM cells was also found to have cancer stem cell properties and play important roles in MM progression and relapse (Lobo et al., 2007; Matsui et al., 2008; Mitra et al., 2015; Muz et al., 2014). Regarding these results, we investigated the EMT phenomenon in which acquired stemness features mediated by integrin-α8 might induce a transcriptional level of EMT markers. Consistent with these results, an invasion assay demonstrated that integrin-α8 overexpression enhanced cell invasive ability. E-cadherin, which is a marker of mesenchymal-epithelial transition (MET), showed a slight reduction or no change of mRNA level in integrin-α8 overexpressed MM cells and the protein level was not detectable.
CXCR4, which is one of the potential therapeutic targets in MM, is essential to migration and homing of MM cells into BM (Alsayed et al., 2007; Azab et al., 2009). Circulating MM cells in patients have been reported to predict poor survival outcomes and to be related to MM bone disease, which is a major symptom of MM (Nowakowski et al., 2005; Ooi and Dunstan, 2009). Our results showed that increased integrin-α8 expression significantly induced migration ability mediated by upregulated CXCR4 mRNA and protein. Unlike previous studies, overexpressed integrin-α8 cells showed no change or little reduction of adhesion to MM patients derived BMSCs or FN when compared to the control. Considering these data, integrin-α8 mediates the acquisition of EMT properties and induction of CXCR4 expression, which eventually promotes MM cells to migrate and invade to new regions of BM rather than adhere and be retained by the BM microenvironment. Previous observations support our finding that the induction of EMT features enhances CXCR4 expression in epithelial tumors, and that acquisition of EMT-like phenotype with the upregulation of CXCR4 expression develops the extra-medullary in MM, resulting in MM cell dissemination and metastasis (Muz et al., 2014; Roccaro et al., 2015).
Failure to respond to chemotherapy leads to spread of cancer, and these two processes of drug resistance development and cancer metastasis are intricately related (Mitra et al., 2015; Singh and Settleman, 2010; Shang et al., 2013). Arumugam et al. showed that EMT mediates induction of Zeb1, an EMT marker, causing drug resistance to gemcitabine, 5-fluorouracil, and cisplatin in pancreatic cancer cells (Arumugam et al., 2009). In wild-type EGFR non-small cell lung cancer (NSCLC) cells, the sensitivities of EGFR inhibition were determined by the degree of EMT. Moreover, the NSCLC cell line, which expresses E-cadherin, a negative regulator of EMT, showed greater response to EGFR inhibition (Nurwidya et al., 2012). In agreement with these previous reports, our results also show that acquisition of the EMT-like phenotype mediated by integrin-a8 lowered the sensitivity of melphalan and bortezomib in MM cells. Thus, targeting EMT may be essential for reverse drug resistance, metastasis, and tumor recurrence.
The crosstalk between integrin and growth factor receptor activates intercellular signaling of cancer cell proliferation, survival, invasion, and migration (Eliceiri, 2001). We investigated whether PDGFRA and PDGFRB has significant co-occurrence with ITGA8 using the cBioPortal genome database, and the results were consistent with our finding that increasing integrin-α8 expression induced PDGFRA and PDGFRB mRNA expression. PDGFR signaling has been thoroughly investigated in cancer, including MM supporting tumor growth and angiogenesis, and clinically used as a therapeutic target (Tsirakis et al., 2012; Yu et al., 2003). The PDGFR inhibitor, sunitinib, is currently in a phase II trial to test its effects on MM relapse; specifically, to determine if it can inhibit growth of cancer cells by blocking enzymes related to cancer cell proliferation and blood flow to cancer (https://clinicaltrials.gov). However, further study is needed to identify the crosstalk between integrin-α8 and PDGF receptors and the synergistic effects of PDGFR inhibitor with other MM therapeutic strategies.
In conclusion, we present the first demonstration of highly expressed integrin-α8 inducing EMT-like phenotype in MM cells, which in turn mediates migration, invasion, and drug resistance acquisition. We suggest integrin-α8 as a potential marker of early relapse. Further research should be conducted to establish integrin-α8 as a therapeutic target to reach complete remission of MM.
Supplementary Figures
ACKNOWLEDGMENTS
This work was supported by the Seoul National University research grant in 2015 (800-20150219) and a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI14C2399).
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Alsayed Y., Ngo H., Runnels J., Leleu X., Singha U.K., Pitsillides C.M., Spencer J.A., Kimlinger T., Ghobrial J.M., Jia X., et al. Mechanisms of regulation of CXCR4/SDF-1 (CXCL12)-dependent migration and homing in multiple myeloma. Blood. 2007;109:2708–2717. doi: 10.1182/blood-2006-07-035857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anguiano A., Tuchman S.A., Acharya C., Salter K., Gasparetto C., Zhan F., Dhodapkar M., Nevins J., Barlogie B., Shaughnessy J.D., Jr, et al. Gene expression profiles of tumor biology provide a novel approach to prognosis and may guide the selection of therapeutic targets in multiple myeloma. J Clin Oncol. 2009;27:4197–4203. doi: 10.1200/JCO.2008.19.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam T., Ramachandran V., Fournier K.F., Wang H., Marquis L., Abbruzzese J.L., Gallick G.E., Logsdon C.D., McConkey D.J., Choi W. Epithelial to mesenchymal transition contributes to drug resistance in pancreatic cancer. Cancer Res. 2009;69:5820–5828. doi: 10.1158/0008-5472.CAN-08-2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avet-Loiseau H. Ultra high-risk myeloma. Hematology/the Education Program of the American Society of Hematology. American Society of Hematology. Education Program. 2010;2010:489–493. doi: 10.1182/asheducation-2010.1.489. [DOI] [PubMed] [Google Scholar]
- Azab A.K., Runnels J.M., Pitsillides C., Moreau A.S., Azab F., Leleu X., Jia X., Wright R., Ospina B., Carlson A.L., et al. CXCR4 inhibitor AMD3100 disrupts the interaction of multiple myeloma cells with the bone marrow microenvironment and enhances their sensitivity to therapy. Blood. 2009;113:4341–4351. doi: 10.1182/blood-2008-10-186668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azab A.K., Hu J., Quang P., Azab F., Pitsillides C., Awwad R., Thompson B., Maiso P., Sun J.D., Hart C.P., et al. Hypoxia promotes dissemination of multiple myeloma through acquisition of epithelial to mesenchymal transition-like features. Blood. 2012;119:5782–5794. doi: 10.1182/blood-2011-09-380410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batist G., Wu J.H., Spatz A., Miller W.H., Jr, Cocolakis E., Rousseau C., Diaz Z., Ferrario C., Basik M. Resistance to cancer treatment: the role of somatic genetic events and the challenges for targeted therapies. Front Pharmacol. 2011;2:59. doi: 10.3389/fphar.2011.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi G., Richardson P.G., Anderson K.C. Promising therapies in multiple myeloma. Blood. 2015;126:300–310. doi: 10.1182/blood-2015-03-575365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagosklonny M.V. Target for cancer therapy: proliferating cells or stem cells. Leukemia. 2006;20:385–391. doi: 10.1038/sj.leu.2404075. [DOI] [PubMed] [Google Scholar]
- Brandenberger R., Schmidt A., Linton J., Wang D., Backus C., Denda S., Muller U., Reichardt L.F. Identification and characterization of a novel extracellular matrix protein nephronectin that is associated with integrin alpha8beta1 in the embryonic kidney. J Cell Biol. 2001;154:447–458. doi: 10.1083/jcb.200103069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broyl A., Hose D., Lokhorst H., de Knegt Y., Peeters J., Jauch A., Bertsch U., Buijs A., Stevens-Kroef M., Beverloo H.B., et al. Gene expression profiling for molecular classification of multiple myeloma in newly diagnosed patients. Blood. 2010;116:2543–2553. doi: 10.1182/blood-2009-12-261032. [DOI] [PubMed] [Google Scholar]
- Damiano J.S., Dalton W.S. Integrin-mediated drug resistance in multiple myeloma. Leuk Lymphoma. 2000;38:71–81. doi: 10.3109/10428190009060320. [DOI] [PubMed] [Google Scholar]
- Damiano J.S., Cress A.E., Hazlehurst L.A., Shtil A.A., Dalton W.S. Cell adhesion mediated drug resistance (CAM-DR): Role of integrins and resistance to apoptosis in human myeloma cell lines. Blood. 1999;93:1658–1667. [PMC free article] [PubMed] [Google Scholar]
- Denda S., Muller U., Crossin K.L., Erickson H.P., Reichardt L.F. Utilization of a soluble integrin-alkaline phosphatase chimera to characterize integrin alpha 8 beta 1 receptor interactions with tenascin: murine alpha 8 beta 1 binds to the RGD site in tenascin-C fragments, but not to native tenascin-C. Biochemistry. 1998;37:5464–5474. doi: 10.1021/bi9727489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desgrosellier J.S., Cheresh D.A. Integrins in cancer: biological implications and therapeutic opportunities. Nat Rev Cancer. 2010;10:9–22. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draghici S., Khatri P., Tarca A.L., Amin K., Done A., Voichita C., Georgescu C., Romero R. A systems biology approach for pathway level analysis. Genome Res. 2007;17:1537–1545. doi: 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliceiri B.P. Integrin and growth factor receptor crosstalk. Circulation Res. 2001;89:1104–1110. doi: 10.1161/hh2401.101084. [DOI] [PubMed] [Google Scholar]
- Gialeli C., Theocharis A.D., Karamanos N.K. Roles of matrix metalloproteinases in cancer progression and their pharmacological targeting. FEBS J. 2011;278:16–27. doi: 10.1111/j.1742-4658.2010.07919.x. [DOI] [PubMed] [Google Scholar]
- Guarino M. Epithelial-mesenchymal transition and tumour invasion. Int J Biochem Cell Biol. 2007;39:2153–2160. doi: 10.1016/j.biocel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Guarino M., Rubino B., Ballabio G. The role of epithelial-mesenchymal transition in cancer pathology. Pathology. 2007;39:305–318. doi: 10.1080/00313020701329914. [DOI] [PubMed] [Google Scholar]
- Harousseau J.L., Shaughnessy J., Jr, Richardson P. Multiple myeloma. Hematology/the Education Program of the American Society of Hematology. American Society of Hematology Education Program. 2004:237–256. doi: 10.1182/asheducation-2004.1.237. [DOI] [PubMed] [Google Scholar]
- Hideshima T., Chauhan D., Hayashi T., Podar K., Akiyama M., Gupta D., Richardson P., Munshi N., Anderson K.C. The biological sequelae of stromal cell-derived factor-1alpha in multiple myeloma. Mol Cancer Ther. 2002;1:539–544. [PubMed] [Google Scholar]
- Kalluri R., Weinberg R.A. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper R., Broyl A., de Knegt Y., van Vliet M.H., van Beers E.H., van der Holt B., el Jarari L., Mulligan G., Gregory W., Morgan G., et al. A gene expression signature for high-risk multiple myeloma. Leukemia. 2012;26:2406–2413. doi: 10.1038/leu.2012.127. [DOI] [PubMed] [Google Scholar]
- Kyle R.A., Rajkumar S.V. Multiple myeloma. The New Engl J Med. 2004;351:1860–1873. doi: 10.1056/NEJMra041875. [DOI] [PubMed] [Google Scholar]
- Lobo N.A., Shimono Y., Qian D., Clarke M.F. The biology of cancer stem cells. Ann Rev Cell Dev Biol. 2007;23:675–699. doi: 10.1146/annurev.cellbio.22.010305.104154. [DOI] [PubMed] [Google Scholar]
- Lonial S. Relapsed multiple myeloma. Hematology/the Education Program of the American Society of Hematology. American Society of Hematology. Education Program. 2010;2010:303–309. doi: 10.1182/asheducation-2010.1.303. [DOI] [PubMed] [Google Scholar]
- Matsui W., Wang Q., Barber J.P., Brennan S., Smith B.D., Borrello I., McNiece I., Lin L., Ambinder R.F., Peacock C., et al. Clonogenic multiple myeloma progenitors, stem cell properties, and drug resistance. Cancer Res. 2008;68:190–197. doi: 10.1158/0008-5472.CAN-07-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menu E., Asosingh K., Indraccolo S., De Raeve H., Van Riet I., Van Valckenborgh E., Vande Broek I., Fujii N., Tamamura H., Van Camp B., et al. The involvement of stromal derived factor 1alpha in homing and progression of multiple myeloma in the 5TMM model. Haematologica. 2006;91:605–612. [PubMed] [Google Scholar]
- Mitra A., Mishra L., Li S.L. EMT, CTCs and CSCs in tumor relapse and drug-resistance. Oncotarget. 2015;6:10697–10711. doi: 10.18632/oncotarget.4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller U., Bossy B., Venstrom K., Reichardt L.F. Integrin alpha 8 beta 1 promotes attachment, cell spreading, and neurite outgrowth on fibronectin. Mol Biol Cell. 1995;6:433–448. doi: 10.1091/mbc.6.4.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muz B., de la Puente P., Azab F., Luderer M., Azab A.K. Hypoxia promotes stem cell-like phenotype in multiple myeloma cells. Blood Cancer J. 2014;4:e262. doi: 10.1038/bcj.2014.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri P., Ren L., Azab A.K., Brentnall M., Gratton K., Klimowicz A.C., Lin C., Duggan P., Tassone P., Mansoor A., et al. Integrin beta7-mediated regulation of multiple myeloma cell adhesion, migration, and invasion. Blood. 2011;117:6202–6213. doi: 10.1182/blood-2010-06-292243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nooter K., Herweijer H. Multidrug resistance (mdr) genes in human cancer. Brit J Cancer. 1991;63:663–669. doi: 10.1038/bjc.1991.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowski G.S., Witzig T.E., Dingli D., Tracz M.J., Gertz M.A., Lacy M.Q., Lust J.A., Dispenzieri A., Greipp P.R., Kyle R.A., et al. Circulating plasma cells detected by flow cytometry as a predictor of survival in 302 patients with newly diagnosed multiple myeloma. Blood. 2005;106:2276–2279. doi: 10.1182/blood-2005-05-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurwidya F., Takahashi F., Murakami A., Takahashi K. Epithelial mesenchymal transition in drug resistance and metastasis of lung cancer. Cancer Res Treat. 2012;44:151–156. doi: 10.4143/crt.2012.44.3.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi L.L., Dunstan C.R. CXCL12/CXCR4 axis in tissue targeting and bone destruction in cancer and multiple myeloma. J Bone Miner Res. 2009;24:1147–1149. doi: 10.1359/jbmr.090503. [DOI] [PubMed] [Google Scholar]
- Pagnucco G., Cardinale G., Gervasi F. Targeting multiple myeloma cells and their bone marrow microenvironment. Ann N Y Acad Sci. 2004;1028:390–399. doi: 10.1196/annals.1322.047. [DOI] [PubMed] [Google Scholar]
- Palumbo A., Anderson K. Multiple Myeloma REPLY. New Engl J Med. 2011;364:2364–2364. doi: 10.1056/NEJMra1011442. [DOI] [PubMed] [Google Scholar]
- Pandit S., Vesole D.H. Relapsed multiple myeloma. Curr Treat Options Oncol. 2001;2:261–269. doi: 10.1007/s11864-001-0040-6. [DOI] [PubMed] [Google Scholar]
- Potti A., Dressman H.K., Bild A., Riedel R.F., Chan G., Sayer R., Cragun J., Cottrill H., Kelley M.J., Petersen R., et al. Genomic signatures to guide the use of chemotherapeutics. Nat Med. 2006;12:1294–1300. doi: 10.1038/nm1491. [DOI] [PubMed] [Google Scholar]
- Rathinam R., Alahari S.K. Important role of integrins in the cancer biology. Cancer Metastsis Rev. 2010;29:223–237. doi: 10.1007/s10555-010-9211-x. [DOI] [PubMed] [Google Scholar]
- Reverter F., Vegas E., Sanchez P. Mining gene expression profiles: an integrated implementation of kernel principal component analysis and singular value decomposition. Genom Proteom Bioinform. 2010;8:200–210. doi: 10.1016/S1672-0229(10)60022-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson P.G., Mitsiades C.S., Hideshima T., Anderson K.C. Novel biological therapies for the treatment of multiple myeloma. Best Pract Res Clin Haematol. 2005;18:619–634. doi: 10.1016/j.beha.2005.01.010. [DOI] [PubMed] [Google Scholar]
- Roccaro A.M., Mishima Y., Sacco A., Moschetta M., Tai Y.T., Shi J., Zhang Y., Reagan M.R., Huynh D., Kawano Y., et al. CXCR4 regulates extra-medullary myeloma through epithelial-mesenchymal-transition-like transcriptional activation. Cell Rep. 2015;12:622–635. doi: 10.1016/j.celrep.2015.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha A.J. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Sanz-Rodriguez F., Teixido J. VLA-4-dependent myeloma cell adhesion. Leuk Lymphoma. 2001;41:239–245. doi: 10.3109/10428190109057979. [DOI] [PubMed] [Google Scholar]
- Sanz-Rodriguez F., Ruiz-Velasco N., Pascual-Salcedo D., Teixido J. Characterization of VLA-4-dependent myeloma cell adhesion to fibronectin and VCAM-1. Brit J Haematol. 1999;107:825–834. doi: 10.1046/j.1365-2141.1999.01762.x. [DOI] [PubMed] [Google Scholar]
- Scherberich A., Tucker R.P., Degen M., Brown-Luedi M., Andres A.C., Chiquet-Ehrismann R. Tenascin-W is found in malignant mammary tumors, promotes alpha8 integrin-dependent motility and requires p38MAPK activity for BMP-2 and TNF-alpha induced expression in vitro. Oncogene. 2005;24:1525–1532. doi: 10.1038/sj.onc.1208342. [DOI] [PubMed] [Google Scholar]
- Shang Y., Cai X., Fan D. Roles of epithelial-mesenchymal transition in cancer drug resistance. Cur Cancer Drug Targets. 2013;13:915–929. doi: 10.2174/15680096113136660097. [DOI] [PubMed] [Google Scholar]
- Singh A., Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29:4741–4751. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery J.P., Acloque H., Huang R.Y., Nieto M.A. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Thompson E.W., Haviv I. The social aspects of EMT-MET plasticity. Nat Med. 2011;17:1048–1049. doi: 10.1038/nm.2437. [DOI] [PubMed] [Google Scholar]
- Tsirakis G., Pappa C.A., Kanellou P., Stratinaki M.A., Xekalou A., Psarakis F.E., Sakellaris G., Alegakis A., Stathopoulos E.N., Alexandrakis M.G. Role of platelet-derived growth factor-AB in tumour growth and angiogenesis in relation with other angiogenic cytokines in multiple myeloma. Hematol Oncol. 2012;30:131–136. doi: 10.1002/hon.1014. [DOI] [PubMed] [Google Scholar]
- Yagi H., Tan W., Dillenburg-Pilla P., Armando S., Amornphimoltham P., Simaan M., Weigert R., Molinolo A.A., Bouvier M., Gutkind J.S. A synthetic biology approach reveals a CXCR4-G13-Rho signaling axis driving transendothelial migration of metastatic breast cancer cells. Sci Signal. 2011;4:ra60. doi: 10.1126/scisignal.2002221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Ustach C., Kim H.R. Platelet-derived growth factor signaling and human cancer. J Biochem Mol Biol. 2003;36:49–59. doi: 10.5483/bmbrep.2003.36.1.049. [DOI] [PubMed] [Google Scholar]
- Zhan F., Hardin J., Kordsmeier B., Bumm K., Zheng M., Tian E., Sanderson R., Yang Y., Wilson C., Zangari M., et al. Global gene expression profiling of multiple myeloma, monoclonal gammopathy of undetermined significance, and normal bone marrow plasma cells. Blood. 2002;99:1745–1757. doi: 10.1182/blood.v99.5.1745. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.