Abstract
Rare cases of culture-negative infective endocarditis are caused by Tropheryma whipplei, the uncommon bacterium of Whipple's disease. We evaluated an 80-year-old woman with valvular heart disease but without intestinal Whipple's disease. The diagnosis of aortic valve xenograft culture-negative infection with T. whipplei was established by multiple molecular assays and by electron microscopy. First, a PCR with broad-range primers identified the complete 16S ribosomal DNA of T. whipplei in bioprosthesis tissue. Novel real-time reverse transcription-PCR assays were developed to detect mRNAs encoding recently identified proteins determined from the T. whipplei genome, specifically Whipplei surface protein (TW113) and a DNA polymerase III subunit (TW727). The positive detection of mRNAs indicated the presence of metabolically active bacteria and suggested the viability of T. whipplei. The quantification of T. whipplei genome equivalents by real-time PCR indicated a high-density bacterial colonization of the valve tissue. Additionally, an ultrastructural examination revealed numerous rod-shaped bacteria consistent in size with T. whipplei in the extracellular collagen matrix of the bioprosthesis. We conclude that extracellular growth of T. whipplei can occur in the microenvironment of biological prosthetic valve tissue and that T. whipplei endocarditis can occur in the absence of intestinal Whipple's disease.
In 1907, George H. Whipple presented a case report of a 36-year-old man with migratory polyarthritis, diarrhea and malabsorption, weight loss, fever and lymphadenopathy (31). Whipple's disease is a rare infective systemic disorder with clinical symptoms including abdominal pain, diarrhea and malabsorption, weight loss, fever, and lymphadenopathy as well as, occasionally, symptoms including involvement of the heart and central nervous system. The disease is fatal if left untreated. Since 1961, electron microscopy studies have shown small, rod-shaped bacteria in affected tissues (3, 26). Since 1992, the causative organism, T. whipplei, has been identified to have a phylogenetic position between the actinomycetes with group B peptidoglycan and the family Cellulomonadaceae (13), as determined by the bacterial 16S ribosomal DNA (rDNA) and the 16S-23S rRNA intergenic spacer sequence (21).
Despite numerous attempts, T. whipplei remained uncultured for decades. Eventually, in 2000, cocultivation with a human fibroblast cell line inoculated with infected heart valve tissue allowed in vitro replication of this organism, and the first strain, Twist, was established (18). Further strains were established from small-intestinal mucosa (19) and from cerebrospinal fluid (CSF) (14).
The availability of T. whipplei strain TW08/27 (14) permitted the sequencing and analysis of the complete genome of 925,938 bp (1). This revealed a lack of key biosynthetic pathways and a reduced capacity for energy metabolism. The genome sequence analysis identified a novel family of predicted Whipplei surface proteins (WiSP). The WiSP genes exhibit DNA sequence variations and hypervariation, leading to phase regulation in protein expression, and they are thought to be responsible for effective pathogenesis and immune system evasion (1). Large regions of noncoding repetitive DNA are located in two repeat clusters (RCs) that could be involved in the generation of gene variation by conversion events.
Until now, a total of 11 cases of PCR-based diagnosis of T. whipplei endocarditis without intestinal Whipple's disease have been reported (5, 6, 8, 17, 18, 23, 24, 27), suggesting a possible role of T. whipplei in cardiac disease which has not been considered adequately until now. However, certainty about the uncommon diagnosis in the cases reported to date has been limited to single-test PCR results which were yielded with heterogeneous PCR protocols. Herein we report the first case of T. whipplei endocarditis in the absence of intestinal Whipple's disease that is documented by more extensive molecular testing and by electron microscopy.
CASE REPORT
An 80-year-old woman was operated on in January 2001 for aortic valvular dysfunction with predominant stenosis and cardiac insufficiency. Considering her age, a porcine xenograft (Mosaic aortic bioprosthesis; Medtronic Inc., Minneapolis, Minn.) was chosen to avoid the need for anticoagulation. In October 2002, symptoms of cardiac insufficiency and findings of prosthetic valve dysfunction recurred. The patient had no fever, multiple blood cultures were negative for bacteria, and transesophageal echocardiography did not show vegetation of the prosthetic valve leaflets. Thus, there were no major Duke's criteria present to cause suspicion of infective endocarditis (12), despite the finding of a moderately elevated c-reactive protein level of 1.8 mg/dl (reference range, <0.05 mg/dl) and a white blood count of 8.0 × 109 leukocytes/liter. Other parameters for basic laboratory data were in the normal range, without indications of a bacterial infection. Prosthetic valvular degeneration was diagnosed by transesophageal and transthoracal echocardiography, and in November 2002, a second operation was performed. The porcine bioprosthesis was excised and replaced with another porcine xenograft (Mitroflow Synergy bioprosthesis; CarboMedics Inc., Richmond, British Columbia, Canada). Cardiac surgery and the postoperative course were uneventful. At no time did the patient have abdominal symptoms of intestinal Whipple's disease, but she reported myalgia over the last 2 months prior to the second cardiac surgery.
The preliminary diagnosis of culture-negative endocarditis prompted further examinations, as detailed below. When the diagnosis of T. whipplei endocarditis was reached, some further diagnostic tests were performed which are now common as staging examinations for patients with Whipple's disease (15, 29). Upper endoscopy was performed, and multiple biopsies were obtained from the grossly normal distal duodenum. They were subjected to histologic examinations, which included serial sections and periodic acid-Schiff (PAS) staining (30), and to PCR analysis by means of an established and validated PCR assay (28). A lumbar puncture was performed to obtain CSF, without pathological results, and a PCR analysis of the CSF was performed (by means of the assay described below). Informed consent was provided by the patient. The patient was treated with ceftriaxone (intravenously) for 2 weeks and cotrimoxazole (orally) for 1 year, according to the protocol of the Study of Initial Therapy in Whipple's Disease (www.whipplesdisease.net).
MATERIALS AND METHODS
Nucleic acid isolation.
Total DNAs were extracted from 30 mg of porcine valvular tissue, peripheral blood, duodenal mucosa, and CSF by use of a QIAamp DNA blood kit (Qiagen, Hilden, Germany) according to the manufacturer's protocols. DNAs were eluted from the column with 60 μl of AE buffer.
Total RNAs were extracted from 50 mg of tissue by the use of Trizol reagent (Invitrogen GmbH, Karlsruhe, Germany) according to the manufacturer's instructions. Extracted RNA samples were digested with 10 U of RQ1 DNase (Promega GmbH, Mannheim, Germany)/ml for 15 min at 37°C, and clean-up of the samples was performed by use of an RNeasy mini kit (Qiagen).
Broad-range bacterial 16S rDNA.
As is common for cases of culture-negative infective endocarditis, we performed a 16S rDNA broad-range PCR to detect culture-negative bacteria. DNAs were prepared from the native bioprosthetic valve and then screened for bacterial 16S rDNA. A broad-range PCR with primers B4 and B5 (11) was applied that amplifies a 511-bp fragment of the 5′ region of the 16S rRNA gene (positions 9 to 520).
Purified PCR products were sequenced by the BigDye dideoxy extension method (ABI Prism Dye Terminator cycle sequencing ready reaction kit, v. 2.0; Applied Biosystems). The sequencing products were analyzed with an ABI Prism 310 DNA sequencer (Applied Biosystems). Partial 16S rDNA sequences were compared to entries in the GenBank and EMBL databases.
hsp65 PCR.
T. whipplei-specific heat shock protein 65 gene (hsp65) primers (whipp-frw1, whipp-frw2, and whipp-rev) were used in a seminested PCR as described previously (16). A sample from another infected heart valve from a patient with intestinal Whipple's Disease (provided from the Whipple's Disease Registry, Heidelberg, Germany [www.whipplesdisease.net]) was used as a positive control. The PCR products were directly sequenced, and the DNA sequences of the amplicons were compared to entries in the GenBank and EMBL databases.
16S rDNA and 16S-23S rDNA internal transcribed spacer analysis.
Amplification of the major parts of the T. whipplei 16S rDNA (positions 22 to 1541) was performed by use of the universal bacterial primers 41f (5′-GCTCAGATTGAACGCTGGCG-3′) and BSR1541 (5′-AGGAGGTGATCCAGCCGCA-3′). The PCR products were cloned into Escherichia coli by use of the pCR-TOPO cloning system (Invitrogen GmbH), and sequencing of different clones was performed with standard primers as described previously (32).
Amplification and direct sequencing of the 16S-23S rDNA spacer region were performed with primers tws3-f and tws4-r as described previously (9).
Real-time reverse transcription-PCR (RT-PCR).
New PCR primers were designed to amplify gene sequences that were recently identified within the genome of T. whipplei (1). The oligonucleotides TW221U (5′-CCCCAGTCTCTTATCTTGTCT-3′) and TW394L (5′-CTGTAACCGGTGCTGTAAAT-3′) amplify a 192-bp fragment of TW113, which encodes a specific Whipplei surface protein (ID CAD66796.1 [1]). Amplification of a 257-bp fragment of the noncoding degenerate repeat cluster region RC1 was done with primers TW235U (5′-GGCCACACTGTTAGGATTATC-3′) and TW476L (5′-GACGCCCTCTGTTTTTGTTC-3′). In addition, a PCR was performed with primers TW116U (5′-CAGAACAGCCTCCCCCGTAA-3′) and TW259L (5′-TGTCAGGAAAGCCCATTGATG-3′), generating a 163-bp fragment of the TW727 gene encoding the DNA polymerase III gamma and tau subunits (protein ID CAD67386.1 [1]).
An aliquot of 5 μl of the RNA was added to 15 μl of a reaction mixture containing 3.0 mM Mn(OAc)2, a 0.4 μM concentration of each primer, and 1× LightCycler-RNA Master SYBR Green I (from a kit for one-step RT-PCR; Roche Diagnostics GmbH, Mannheim, Germany). Reaction capillaries were loaded, centrifuged, and placed into the carousel of a LightCycler instrument (Roche Diagnostics GmbH). The experimental RT-PCR protocol was performed under the following conditions: reverse transcription for 20 min at 60°C and denaturation for 2 min at 95°C, followed by 40 cycles of 2 s of denaturation at 95°C, 10 s of annealing at 60°C, and 12 s of extension at 72°C. Temperature transitions were set at 20°C/s. The fluorescence was sampled once per cycle in the channel F1/F2 at 80°C after the end of the extension cycle to generate amplification plots. A melting point analysis measuring the fluorescence used was performed after PCR to determine that the correct products had been amplified. The temperature was slowly raised 0.1°C/s, from 72 to 95°C, while the fluorescence emissions were continually sampled. The melting temperatures of the T. whipplei-specific PCR products were 79.5 ± 1.0°C for TW113, 80.8 ± 1.0°C for RC2, and 79.2 ± 1.0°C for TW727.
Construction of T. whipplei-specific plasmid standard.
PCR products of 357 bp flanked by primers whipp-frw2 and whipp-rev (16) were cloned into E. coli by use of the plasmid vector pCRII-TOPO, which was supplied by a TA-TOPO cloning kit (Invitrogen GmbH), according to the protocol provided by the supplier. An analysis of the recombinant plasmids was performed by extracting plasmid DNAs from small-volume cultures grown overnight at 37°C by use of a QIAprep Spin plasmid kit (Qiagen). Plasmid concentrations were determined by a fluorometric analysis with the PicoGreen DNA quantitation reagent (Molecular Probes, Leiden, Netherlands), and defined plasmid copies in the range from 2 to 2.3 × 107 copies per PCR were used as standards in real-time PCR assays.
Quantitative PCR.
PCRs for T. whipplei-specific hsp65 were carried out on a LightCycler instrument (Roche Diagnostics GmbH) in capillaries containing 15 μl of reaction mix and 5 μl of nucleic acid extract or plasmid standard. The reaction mixture for LightCycler PCR included the following reagents: 1× LC-FastStart DNA Master SYBR Green I (Roche Diagnostics), 3 mM MgCl2, a 500 nM concentration (each) of primers whipp-frw2 and whipp-rev (16), and 0.01 U of uracil DNA glycosidase (Roche Diagnostics)/μl. The reaction conditions were 10 min at 30°C and 10 min at 95°C, followed by 40 cycles of 0 s at 95°C, 10 s at 60°C, and 15 s at 72°C. The double-stranded PCR product was measured after the 72°C elongation step by detection of the fluorescence associated with the binding of the SYBR Green dye to the DNA. The specificity of the obtained fluorescence signal was checked by a melting curve analysis after each run. This analysis was initiated at a temperature of 60°C for 30 s, and the temperature was gradually raised by 0.1°C/s up to 95°C. During this process, the fluorescence signal was continuously monitored. The melting temperature of the T. whipplei-specific PCR product was 86.0 ± 1.0°C.
Electron microscopy.
Xenograft tissue kept frozen in sterile brain-heart solution was thawed in 3% buffered glutaraldehyde and postfixed in 1.0% osmium tetroxide. Ultrathin sections were stained with uranyl acetate and lead citrate for examination by transmission electron microscopy (Zeiss EM 902).
Nucleotide sequence accession numbers.
The sequence of the hsp65 DNA has been included in the GenBank and EMBL databases under accession number AJ551322. The sequence of the 16S-23S rDNA spacer region has been included in the GenBank and EMBL databases under accession number AJ551273.
RESULTS
16S rDNA sequencing.
The PCR with primers B4 and B5 yielded a 511-bp product. Automatic sequencing revealed 100% homology with T. whipplei (positions 22 to 1541; GenBank accession number AJ551273). In this screening test, no positive controls for the bacterium were used, so false-positive results by cross-contamination were precluded. In order to confirm the screening result, we further amplified major parts of 16S rRNA showing 100% homology over 1,841 nucleotides, covering the 16S rRNA of T. whipplei and its intergenic 16S-23S rRNA spacer (X99636 [13]). This patient's T. whipplei isolate was characterized as having 16S-23S rRNA spacer type 1 (9).
Heat shock protein Hsp65.
The detection of T. whipplei DNA in prosthetic valve tissue was confirmed further by a second DNA extraction and nested PCR assays targeting the T. whipplei-specific hsp65 gene (Fig. 1).
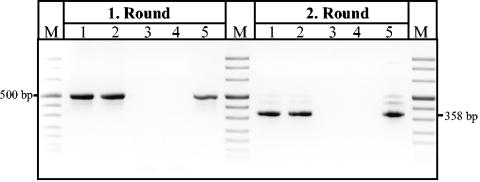
FIG. 1.
Direct detection of T. whipplei with nested hsp65 PCR in prosthetic valve tissue samples. Products of specific nested amplifications (first and second round) were analyzed in an ethidium bromide-stained agarose gel. Marker (M), pUC mix marker (pUC19 and pUC57 DNA digested with MspI, DraI, and HindIII); lanes 1 and 2, two samples of the patient's bioprosthetic heart valve (BPHV) that were positive for T. whipplei; lanes 3 and 4, water control; lane 5, sample from a patient who was positive for T. whipplei (control).
Quantification of T. whipplei DNA.
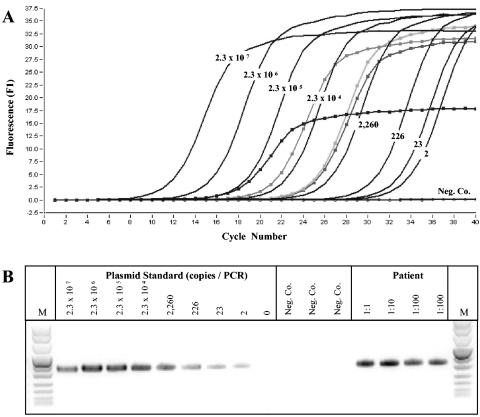
Because of the difficulty of long-term cocultivation with human fibroblasts (14, 18), we performed quantitative real-time PCR with T. whipplei-specific primers and an external plasmid standard. We used the single-copy Hsp65 gene for this purpose. With DNA preparations of valve tissue, we determined that there were 1.5 × 106 copies of the gene per PCR, which corresponds to 1.7 × 107 copies per μg of total DNA (Fig. 2). This suggests a high-density colonization of the valve tissue with T. whipplei.
FIG. 2.
Quantification of T. whipplei DNA by LightCycler real-time hsp65 PCR. (Top) Amplification plot of T. whipplei hsp65 PCR. Serial 10-fold dilutions of standard T. whipplei DNA (plasmid DNA) from 2.3 × 100 to 2.3 × 107 copies per PCR were prepared. The fluorescence (F1) was plotted against each cycle number. The numbers of copies of each standard DNA are shown. DNA samples from the bioprosthetic heart valve are marked (▪). (Bottom) Agarose gel electrophoresis analysis of PCR products after quantitative detection of T. whipplei hsp65 DNA. M, pUC mix marker (see legend to Fig. 1); lane 1, 2.3 × 107 copies; lane 2, 2.3 × 106 copies; lane 3, 2.3 × 105 copies; lane 4, 2.3 × 104 copies; lane 5, 2,260 copies; lane 6, 266 copies; lane 7, 23 copies; lane 8, 2 copies; lane 9, 0 copies; lanes 10 to 12, water control; lanes 13 to 16, dilutions of DNA preparation samples from the patient's bioprosthetic heart valve that were positive for T. whipplei.
Bacterial viability.
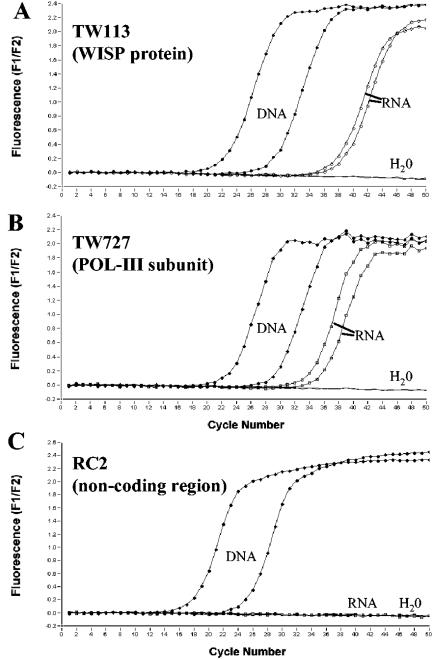
Conventional culture methods to control the titer and viability of T. whipplei are not available so far. In order to detect viable bacteria at a molecular level, we developed a real-time RT-PCR assay using a LightCycler instrument based on SYBR Green detection and melting curve analysis. The ability to distinguish between viable and nonviable bacteria is crucial. For this purpose, we used RT-PCR assays targeting two genes, one encoding a T. whipplei-specific member of the Whipplei surface protein (WiSP) family (TW113) and one encoding a DNA polymerase III subunit (TW727) (1). The mRNAs of these regions were detected by one-step real-time RT-PCR. In order to gain information about the viability of the bacteria, we used a third primer system to amplify a part of the repeat cluster region RC1, which is thought to not be transcribed in T. whipplei (1). This approach was used as a control for the effectiveness of a DNase treatment of RNA samples. RT-PCR was performed on different nucleic acid preparations from valve samples. Total nucleic acids and DNA-free RNAs were isolated in parallel and were subjected to RT-PCR. For both primer sets for the TW113 and TW727 targets, T. whipplei-specific products were detectable from RNA and DNA samples (Fig. 3A and B). RNase treatment of the RNA samples led to the loss of amplification products (data not shown). In contrast, the PCR products of the noncoding region RC1 were only amplified with DNA probes from tissue, while RNA preparations yielded no detectable PCR product. The existence of mRNA was interpreted as a hint for the viability of T. whipplei in the prosthetic valve tissue.
FIG. 3.
Real-time RT-PCR for T. whipplei-specific targets on a LightCycler instrument. Fluorescence curves for SYBR Green I detection are shown for PCR systems specific for TW113, TW727, and the RC2 noncoding region (A, B, and C, respectively). Data for nucleic acid preparations of the patient's bioprosthetic heart valve are shown. ▪, DNA samples; □, RNA (DNA-free); −, negative control.
Electron microscopy.
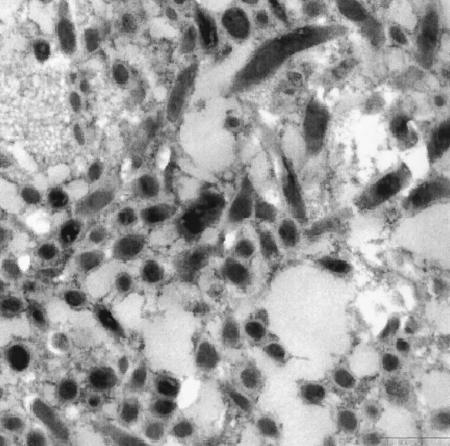
An ultrastructural examination revealed the presence of numerous rod-shaped bacteria within an acellular matrix of collagen fibers (which characterizes any cardiac valve bioprosthesis from cadaveric sources). The bacteria had a fairly constant diameter of 0.21 to 0.25 μm and a broad electron-dense outer membrane, which identifies T. whipplei (Fig. 4). The bacterial cytoplasm was electron-dense, indicating numerous ribosomes, which characterize vital bacteria. Taken together, the electron microscopy findings, the diverse molecular test results, the elevated level of c-reactive protein, and the normal white blood cell count led us to diagnose infective endocarditis of the aortic xenograft with T. whipplei.
FIG. 4.
Electron micrograph of porcine valve prosthesis. Numerous rod-shaped bacteria are present in between the acellular collagen matrix. Virtually all of the bacteria have an electron-dense cytoplasm indicating numerous ribosomes, which suggests metabolic activity. Bar = 500 nm.
Staging.
The diagnosis of T. whipplei endocarditis led to some further diagnostic tests which are now common as staging examinations for patients with Whipple's disease (15, 29). Upper endoscopy was performed, and multiple biopsies were obtained from the grossly normal distal duodenum. Histologic examinations, which included serial sections and PAS staining, did not reveal any PAS-positive macrophages (30). PCR analysis (by means of an established and validated PCR assay) (28) was negative for DNA of T. whipplei. A lumbar puncture was performed to obtain CSF for PCR analysis. Broad-range 16S rDNA- and T. whipplei-specific hsp65 PCRs were performed. No DNA of T. whipplei was detected in the CSF. Preantibiotic blood and serum were subjected to T. whipplei-specific and broad-range bacterial PCR assays, but no bacteria were identified.
The patient was treated with antibiotics according to the protocol of the Study of Initial Therapy in Whipple's Disease. The patient had no complaints or clinical symptoms of Whipple's disease in 1 year of follow-up.
DISCUSSION
Infective endocarditis (IE) is a life-threatening disease which is sometimes difficult to diagnose. Blood cultures are regarded as the most important laboratory test, as they allow the identification of the etiologic agent. However, for a significant proportion (up to 40%) of patients, blood cultures remain sterile due to the presence of slow-growing, fastidious bacteria or the administration of antibiotics (2). Nevertheless, a diagnosis of the causative agent of IE is mandatory for appropriate antimicrobial therapy. Common clinical criteria for the diagnosis of IE, such as the Duke criteria (12), do not take into account that noncultivable causative organisms may be present in patients with IE. Several molecular approaches have been assessed for detecting and identifying pathogens in a wide variety of infectious diseases. Among these, PCR has been the most widely used method in recent years and is a powerful aid to microbiological diagnosis. Various investigators have explored the ability of PCR to detect the nucleic acids of fastidious and noncultivable agents in blood and other samples, such as heart valves or xenografts, from patients with IE, especially culture-negative endocarditis (7, 10). Among these, Mycobacteria sp., Bartonella sp., Coxsiella sp., Ehrlichia sp., and T. whipplei have been detected by broad-range bacterial PCR (2, 7).
PCR has been widely used for the diagnosis of intestinal Whipple's disease (5) and is consistently negative when histology is negative for Whipple's disease (15). Therefore, positive PCR results should be validated by histology. In this case, PCR tests established the diagnosis of a culture-negative bacterial endocarditis, and this molecular diagnosis was corroborated by electron microscopy.
Since the advent of PCR testing, a total of 11 cases have been reported of T. whipplei endocarditis without (overt) intestinal Whipple's disease (6, 8, 17, 18, 23, 24, 27), including one case report of a xenograft infection with T. whipplei (5). Notably, the majority of patients reported did not fulfill the common criteria for a diagnosis of Whipple's disease, and it has to be questioned whether a diagnosis of isolated cardiac Whipple's disease is adequate. Alternatively, these patients may be designated as suffering from T. whipplei endocarditis. The source of infection of the porcine xenograft is unknown. Therefore, further studies of T. whipplei in a porcine animal model should be conducted.
Our patient had the second reported case of cardiac valvular endocarditis due to T. whipplei which was studied by electron microscopy. As in the first case, which was reported 20 years ago (20), numerous rod-shaped Whipple's bacteria were detected. The observation of crowds of viable bacteria within an extracellular matrix of collagen fibers is interesting, as this suggests that T. whipplei lives and grows extracellularly, but without a visible host cell. This finding is in line with previous experimental work (4) but is puzzling against the background of recent knowledge that T. whipplei has a small genome which lacks key metabolic pathways (1).
At present, a diagnostic laboratory culture of T. whipplei is not available (14, 18, 19, 22). Furthermore, the identification of slow-growing bacteria to the species level is far easier with molecular biological approaches. Another molecular biological approach is the quantification of bacteria with the help of quantitative PCR, especially for slow-growing bacteria such as T. whipplei, for which the usual quantification by microbiological means is very difficult to perform.
In order to test the viability and metabolic activity of the T. whipplei bacteria, we evaluated a new molecular genetic RT-PCR assay. A previous in situ hybridization study discussed the use of rRNA probes to assess metabolic activity in this bacterium (4). Although the rRNA content of bacteria is correlated with metabolic activity in fluorescence in situ hybridization analyses, this marker is unsuitable for the differentiation of dead and viable cells in RT-PCR assays using total nucleic acid preparations. The high stability of rRNA due to its protection from degradation in the ribosomal complex leads to a long half-life for the molecule (25), and differentiation between DNA and rRNA is difficult. In the infected valve tissue, we detected mRNAs of two T. whipplei-specific genes, while the untranscribed control target RC2 was only detectable in DNA preparations from the valve tissue. These results suggest the viability of the previously diagnosed T. whipplei bacteria. So far nothing is known about the stability and half-lives of these mRNAs. We believe that these transcripts, which are needed for replication and growth, are expressed during active phases of the bacterial cell cycle. Further studies need to be performed to prove the usefulness of these markers of viability for the diagnosis of IE caused by T. whipplei.
In conclusion, our case report illustrates the potential of new molecular genetic approaches for the diagnosis of T. whipplei endocarditis. The PCR assays for quantitation and viability control of T. whipplei need to be tested in more extensive trials to elucidate the possible involvement of this bacterium in cardiac disease.
Acknowledgments
We thank Peter Rieger, Heidelberg, Germany, for help with electron microscopy, G. Seitz, Bamberg, Germany, for part of the histologic studies of intestinal biopsies, and G. E. Feurle, Neuwied, Germany, for therapeutic recommendations. We thank Sarah Kirkby for linguistic advice.
REFERENCES
- 1.Bentley, S. D., M. Maiwald, L. D. Murphy, M. J. Pallen, C. A. Yeats, L. G. Dover, H. T. Norbertczak, G. S. Besra, M. A. Quail, D. E. Harris, A. von Herbay, A. Goble, S. Rutter, R. Squares, S. Squares, B. G. Barrell, J. Parkhill, and D. A. Relman. 2003. Sequencing and analysis of the genome of the Whipple's disease bacterium Tropheryma whipplei. Lancet 361:637-644. [DOI] [PubMed] [Google Scholar]
- 2.Brouqui, P., and D. Raoult. 2001. Endocarditis due to rare and fastidious bacteria. Clin. Microbiol. Rev. 14:177-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chears, W. C., Jr., and C. T. Ashworth. 1961. Electron microscopic study of the intestinal mucosa in Whipple's disease. Demonstration of encapsulated bacilliform bodies in the lesion. Gastroenterology 41:129-138. [PubMed] [Google Scholar]
- 4.Fredricks, D. N., and D. A. Relman. 2001. Localization of Tropheryma whippelii rRNA in tissues from patients with Whipple's disease. J. Infect. Dis. 183:1229-1237. [DOI] [PubMed] [Google Scholar]
- 5.Geissdorfer, W., I. Wittmann, M. Rollinghoff, C. Schoerner, and C. Bogdan. 2001. Detection of a new 16S-23S rRNA spacer sequence variant (type 7) of Tropheryma whippelii in a patient with prosthetic aortic valve endocarditis. Eur. J. Clin. Microbiol. Infect. Dis. 20:762-763. [DOI] [PubMed] [Google Scholar]
- 6.Geissdorfer, W., I. Wittmann, G. Seitz, R. Cesnjevar, M. Rollinghoff, C. Schoerner, and C. Bogdan. 2001. A case of aortic valve disease associated with Tropheryma whippelii infection in the absence of other signs of Whipple's disease. Infection 29:44-47. [DOI] [PubMed] [Google Scholar]
- 7.Goldenberger, D., A. Kunzli, P. Vogt, R. Zbinden, and M. Altwegg. 1997. Molecular diagnosis of bacterial endocarditis by broad-range PCR amplification and direct sequencing. J. Clin. Microbiol. 35:2733-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gubler, J. G., M. Kuster, F. Dutly, F. Bannwart, M. Krause, H. P. Vogelin, G. Garzoli, and M. Altwegg. 1999. Whipple endocarditis without overt gastrointestinal disease: report of four cases. Ann. Intern. Med. 131:112-116. [DOI] [PubMed] [Google Scholar]
- 9.Hinrikson, H. P., F. Dutly, and M. Altwegg. 2000. Evaluation of a specific nested PCR targeting domain III of the 23S rRNA gene of “Tropheryma whippelii” and proposal of a classification system for its molecular variants. J. Clin. Microbiol. 38:595-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamas, C. C., and S. J. Eykyn. 1997. Suggested modifications to the Duke criteria for the diagnosis of native valve and prosthetic valve disease. Clin. Infect. Dis. 25:713-719. [DOI] [PubMed] [Google Scholar]
- 11.Ley, B. E., C. J. Linton, D. M. Bennett, H. Jalal, A. B. Foot, and M. R. Millar. 1998. Detection of bacteraemia in patients with fever and neutropenia using 16S rRNA gene amplification by polymerase chain reaction. Eur. J. Clin. Microbiol. Infect. Dis. 17:247-253. [DOI] [PubMed] [Google Scholar]
- 12.Li, J. S., D. J. Sexton, and N. Mick. 2000. Proposed modifications to the Duke criteria for the diagnosis of infective endocarditis. Clin. Infect. Dis. 30:633-638. [DOI] [PubMed] [Google Scholar]
- 13.Maiwald, M., H. J. Ditton, A. von Herbay, F. A. Rainey, and E. Stackebrandt. 1996. Reassessment of the phylogenetic position of the bacterium associated with Whipple's disease and determination of the 16S-23S ribosomal intergenic spacer sequence. Int. J. Syst. Bacteriol. 46:1078-1082. [DOI] [PubMed] [Google Scholar]
- 14.Maiwald, M., A. von Herbay, D. N. Fredricks, C. C. Ouverney, J. C. Kosek, and D. A. Relman. 2003. Cultivation of Tropheryma whipplei from cerebrospinal fluid. J. Infect. Dis. 188:801-808. [DOI] [PubMed] [Google Scholar]
- 15.Maiwald, M., A. von Herbay, D. H. Persing, P. P. Mitchell, M. F. Abdelmalek, J. N. Thorvilson, D. N. Fredricks, and D. A. Relman. 2001. Tropheryma whippelii DNA is rare in the intestinal mucosa of patients without other evidence of Whipple disease. Ann. Intern. Med. 134:115-119. [DOI] [PubMed] [Google Scholar]
- 16.Morgenegg, S., F. Dutly, and M. Altwegg. 2000. Cloning and sequencing of a part of the heat shock protein 65 gene (hsp65) of “Tropheryma whippelii” and its use for detection of “T. whippelii” in clinical specimens by PCR. J. Clin. Microbiol. 38:2248-2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naegeli, B., F. Bannwart, and O. Bertel. 2000. An uncommon cause of recurrent strokes: Tropheryma whippelii endocarditis. Stroke 31:2002-2003. [DOI] [PubMed] [Google Scholar]
- 18.Raoult, D., M. L. Birg, B. La Scola, P. E. Fournier, M. Enea, H. Lepidi, V. Roux, J. C. Piette, F. Vandenesch, D. Vital-Durand, and T. J. Marrie. 2000. Cultivation of the bacillus of Whipple's disease. N. Engl. J. Med. 342:620-625. [DOI] [PubMed] [Google Scholar]
- 19.Raoult, D., B. La Scola, P. Lecocq, H. Lepidi, and P. E. Fournier. 2001. Culture and immunological detection of Tropheryma whippelii from the duodenum of a patient with Whipple disease. JAMA 285:1039-1043. [DOI] [PubMed] [Google Scholar]
- 20.Ratliff, N. B., J. T. McMahon, T. J. Naab, and D. M. Cosgrove. 1984. Whipple's disease in the porcine leaflets of a Carpentier-Edwards prosthetic mitral valve. N. Engl. J. Med. 311:902-903. [DOI] [PubMed] [Google Scholar]
- 21.Relman, D. A., T. M. Schmidt, R. P. MacDermott, and S. Falkow. 1992. Identification of the uncultured bacillus of Whipple's disease. N. Engl. J. Med. 327:293-301. [DOI] [PubMed] [Google Scholar]
- 22.Renesto, P., N. Crapoulet, H. Ogata, B. La Scola, G. Vestris, J. M. Claverie, and D. Raoult. 2003. Genome-based design of a cell-free culture medium for Tropheryma whipplei. Lancet 362:447-449. [DOI] [PubMed] [Google Scholar]
- 23.Richardson, D. C., L. L. Burrows, B. Korithoski, I. E. Salit, J. Butany, T. E. David, and J. M. Conly. 2003. Tropheryma whippelii as a cause of afebrile culture-negative endocarditis: the evolving spectrum of Whipple's disease. J. Infect. 47:170-173. [DOI] [PubMed] [Google Scholar]
- 24.Schneider, T., M. Salamon-Looijen, A. von Herbay, H. Schwerdt, S. Weg-Remers, A. Stallmach, and M. Zeitz. 1998. Whipple's disease with aortic regurgitation requiring aortic valve replacement. Infection 26:178-180. [DOI] [PubMed] [Google Scholar]
- 25.Sheridan, G. E. C., C. L. Masters, J. A. Shallcross, and B. M. Mackey. 1998. Detection of mRNA by reverse transcription-PCR as an indicator of viability in Escherichia coli cells. Appl. Environ. Microbiol. 64:1313-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silva, M. T., P. M. Macedo, and J. F. Moura Nunes. 1985. Ultrastructure of bacilli and the bacilli origin of the macrophagic inclusions in Whipple's disease. J. Gen. Microbiol. 131:1001-1013. [DOI] [PubMed] [Google Scholar]
- 27.Smith, M. A. 2000. Whipple endocarditis without gastrointestinal disease. Ann. Intern. Med. 132:595. [DOI] [PubMed] [Google Scholar]
- 28.von Herbay, A., H. J. Ditton, and M. Maiwald. 1996. Diagnostic application of a polymerase chain reaction assay for the Whipple's disease bacterium to intestinal biopsies. Gastroenterology 110:1735-1743. [DOI] [PubMed] [Google Scholar]
- 29.von Herbay, A., H. J. Ditton, F. Schuhmacher, and M. Maiwald. 1997. Whipple's disease: staging and monitoring by cytology and polymerase chain reaction analysis of cerebrospinal fluid. Gastroenterology 113:434-441. [DOI] [PubMed] [Google Scholar]
- 30.von Herbay, A., M. Maiwald, H. J. Ditton, and H. F. Otto. 1996. Histology of intestinal Whipple's disease revisited. A study of 48 patients. Virchows Arch. 29:335-343. [DOI] [PubMed] [Google Scholar]
- 31.Whipple, G. H. 1907. A hitherto undescribed disease characterized anatomically by deposits of fat and fatty acids in the intestinal and mesenteric lymphatic tissues. Bull. Johns Hopkins Hosp. 18:382-391. [Google Scholar]
- 32.Wilmotte, A., G. Van der Auwera, and R. De Wachter. 1993. Structure of the 16 S ribosomal RNA of the thermophilic cyanobacterium Chlorogloeopsis HTF (“Mastigocladus laminosus HTF”) strain PCC7518, and phylogenetic analysis. FEBS Lett. 317:96-100. [DOI] [PubMed] [Google Scholar]