Abstract
Polyploid rice hybrids have a powerful biological and yield potential that may become a new way for rice breeding; however, low fertility is major hindrance in commercial utilization. Here, we developed a neo-tetraploid rice that could overcome the sterility of autotetraploid rice and produce high heterosis. Transcriptome analysis of F1 hybrid developed by crossing neo-tetraploid with autotetraploid rice displayed 807, 663 and 866 differentially expressed genes that uniquely associated with F1 and specific to (DEGFu-sp) anther, ovary and leaf, respectively. Of the DEGFu-sp, 1224 genes displayed nonadditive expression; 44 and 10 genes were annotated as TFs and methyltransferase or hydroxymethyltransferase, respectively. Gene ontology enrichment and co-expression analysis revealed specific differential gene expressions in the DEGFu-sp to leaf, anther and ovary, such as genes related to photosynthesis, metabolic process and transport, and co-expression network including fertility, resistance and epigenetic elements. Of the DEGFu-sp to anther, 42 meiosis stage-specific genes, eight meiosis-related genes, such as RAD51 and SMC2, were identified. We identified 38 miRNAs from DEGFu-sp to anther, and their targets were associated with pollen fertility and retrotransposon protein. Our study provides new germplasm for polyploid rice breeding, and revealed complex regulatory mechanisms that might be associated with heterosis and fertility.
Rice is one of the main food crops of the world’s population. Heterosis or hybrid vigor is a phenomenon which refers to the superior performance of a hybrid relative to one or both parental phenotypes. Heterosis had been significantly increased the yield of rice crop in the world during last fifty years1. However, the rice yield is stagnant from the last few years. So, it is of utmost importance for the rice breeders to breed new rice varieties with high yield and resistant against various biotic and abiotic stresses. Rice polyploidization can improve the heterozygosity of “allogeneic”, and increases the probability of combination and interaction of elite genes, thereby, increases the adaptability to adverse environmental conditions2,3,4,5.
Polyploidy plant does not only provide a large number of material basis for mutants, evolutionary adaptation and selection, but also has become one of the important ways to promote evolution and plant breeding6. Intersubspecific hybrids (indica × japonica) of autotetraploid rice have a powerful biological and yield potential, and it is expected to become a new way to breed rice in the future. Therefore, polyploidy rice has attracted the attention of rice scientist3,4,5,7,8,9.
However, autotetraploid rice has many unfavorable traits, just like other newly synthesized polyploids. Among these unfavorable traits, low fertility has the largest negative impact on rice breeding, and it is difficult to take out the advantages of gigantic features of autotetraploid rice, so autotetraploid rice hardly applied in commercial production10,11. Recently, we revealed that polyploidy strengthen F1 pollen sterility multi-loci interactions, which cause meiosis abnormalities and resulted in high pollen sterility in autotetraploid rice hybrids10. Consequently, how to create new tetraploid rice lines with normal fertility and to overcome the sterility of F1 hybrids is a key step to use the tetraploid rice. To solve this problem, after 20 years of unremitting efforts, our research group has successfully bred a number of new tetraploid rice lines with high seed setting (>80%). Our research group has registered two new lines of tetraploid rice for PVP (Protection for New Varieties of Plants) in China4. These new tetraploid rice lines with high fertility were designated as neo-tetraploid rice according to neo-tetraploid Arabidopsis6.
RNA-seq-based transcriptome data provide very helpful platform for investigating the molecular aspects of heterosis in plants. The complexity of gene expression profiles associated with heterosis and differential gene expressions in different tissues had been revealed using transcriptome in rice12,13, maize14,15 and wheat16. Therefore, we concentrated on the transcriptomic data to analyze the molecular aspects of heterosis and fertility of neo-tetraploid rice in the present study. We reported the breeding procedure of Huaduo 3, a neo-tetraploid rice line, and heterosis and transcriptomic analysis of hybrids developed by crossing Huaduo 3 with different autotetraploid rice lines. Our study provides new germplasm for polyploid rice breeding and reveals some genes that may associate with the fertility and heterosis in neo-tetraploid rice.
Results
The breeding procedure of neo-tetraploid rice, Huaduo 3
In this study, an autotetraploid rice line, Jackson-4x, and another autotetraploid rice line, 96025, were self-crossed for more than 10 years at our farm. The combination of Jackson-4x ×96025 was generated in 2005. The F1 hybrid was harvested and continuously self-crossed until F4, and one plant with more than 80% seed setting was found in F5. We planted more than 60 plants with high fertility from the F5 plant, and continuously planted in the field from F7 to F11. These lines maintained high seed setting in next successive generations (F10 to F13), and named as “Huaduo 3” in 2012 (Supplementary Fig. S1). Huaduo 3 (H3) exhibited “high fertility”, and has an excellent plant type and yield-related traits (Fig. 1). Cytogenetic observations revealed more than 90% normal pollen mother cells in meiosis (Supplementary Fig. S2).
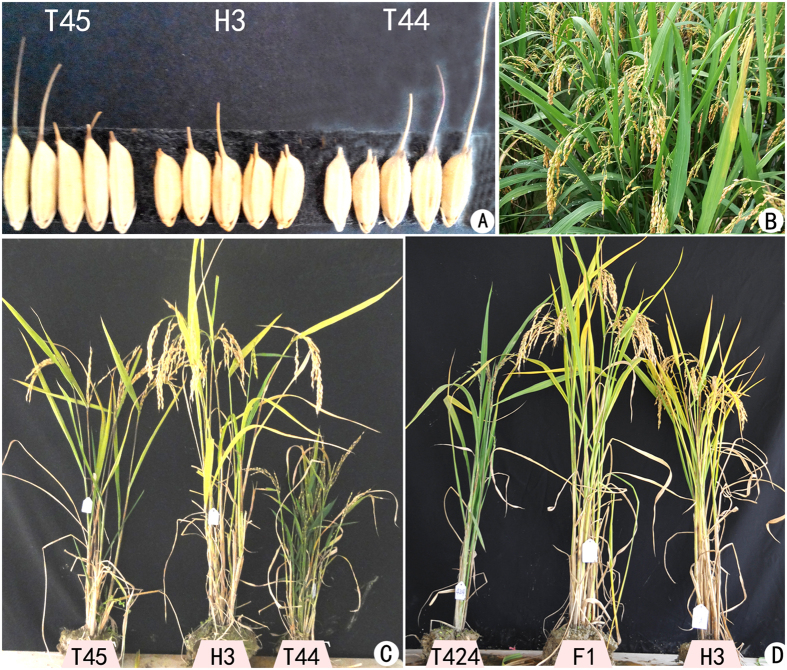
Figure 1. Morphological characteristics of F1 hybrid and their parents.
(A) Grains of Jackson-4x (T45), Huaduo 3 (H3) and 96025 (T44). H3 was developed from T45 and T44; (B) Huaduo 3 at grain filling stage in a paddy field; (C) Plant appearance of 96025 (T44), Huaduo 3 (H3), and Jackson-4x (T45); (D) Plant structure of Shennong15-4x (T424), Huaduo 3 and their F1 hybrid.
Heterosis analysis of neo-tetraploid rice
The agronomic traits of 33 control autotetraploid hybrids and 40 other hybrids generated by crossing with various autotetraploid lines (i.e., H3 crossed with 26 indica and 14 japonica autotetraploid rice lines) improved significantly compared to their parents, especially the average values of the traits like filled grains per plant, seed setting and grain yield per plant for the hybrids prepared by H3. Of these traits, filled grains per plant were more than 662, the seed setting was more than 76%, and grain yield per plant was more than 22 g for all hybrids. The average values of filled grains per plant, seed setting and grain yield per plant for the hybrids were in the following order: hybrids of H3 and indica autotetraploid lines > hybrids of H3 and japonica autotetraploid lines > hybrids of indica and japonica autotetraploid lines (Supplementary Table S1). It is worth mentioning that the seed setting of the hybrids from H3 was significantly higher than those hybrids generated from other autotetraploid lines.
For the 26 hybrid combinations between H3 and indica, the mid-parent heterosis values were positive for all the traits except width of ten grains. A total of 15 hybrids showed positive high-parent heterosis for six traits, except length and width of 10-grains (Supplementary Fig. S3, Table S2). For 14 combinations generated by crossing H3 with japonica autotetraploid lines, the average value of high-parent heterosis for five agronomic traits, including plant height, effective number of panicles per plant, filled grains per plant, total grains per plant and grain yield per plant, were positive. The highest high-parent heterosis was detected for the total grains per plant (i.e. 68.12%), followed by the effective number of panicles per plant > filled grains per plant > grain yield per plant > plant height (Supplementary Fig. S3, Table S3).
For further analysis, we selected one hybrid, Huajingxian 74-4x (T452) ×H3, to analyze F1 heterosis and to investigate the transcriptome of hybrid and its parents. T452 is an autotetraploid rice line with low fertility. The hybrid showed positive high-parent heterosis for all the traits except grain length and width of 10-grains, and the values for high parent heterosis were very high for effective number of panicles per plant and grain yield per plant (Table 1). All the traits showed positive mid-parent heterosis values except for the width of 10-grains.
Table 1. Heterosis analysis of the F1 hybrid (Huajingxian 74-4x ×Huaduo 3) and its parents.
| Traits | H3 | T452 | F1 hybrid | HPH (%) | MPH (%) |
|---|---|---|---|---|---|
| PH (cm) | 121.67±3.51 | 92.00±1.73 | 126.67±0.58 | 4.11 | 18.56 |
| EP | 4.00±0.32 | 4.33±2.08 | 8.33±2.52 | 92.31 | 100.00 |
| FG | 123.25±16.88 | 107.91±19.43 | 131.78±11.16 | 6.92 | 14.02 |
| SS (%) | 83.93±6.41 | 11.84±0.48 | 87.86±5.09 | 4.69 | 83.49 |
| GYP (g) | 16.03±0.23 | 1.64±0.51 | 37.07±16.67 | 131.27 | 319.70 |
| GL (cm) | 8.90±0.32 | 10.00±0.35 | 9.50±0.01 | −5.00 | 0.53 |
| GW (cm) | 4.00±0.36 | 3.10±0.21 | 3.13±0.15 | −21.67 | −11.74 |
H3: Huaduo 3; T452: Huajingxian 74-4x; F1 hybrid: (Huajingxian 74-4x ×Huaduo 3); HPH: High-parent heterosis; MPH: Mid-parent heterosis.
PH: Plant height; EP: Effective number of panicles per plant; FG: Filled grains per panicle; SS: Seed setting; GYP: Grain yield per plant; GL: Grain length (10-grains); GW: Grain width (10-grains).
Whole genome transcriptome profiles of F1 and its two parental lines are of satisfactory quality
Since we found high yield-related traits heterosis in F1 compared to its parents, and higher F1 fertility than T452, so the transcriptome profiles of F1 and its two parental lines were analyzed in three tissues, including anthers and ovaries at meiosis stage, and flag leaves during flowering. In total, we detected more than 548 million clean reads from the F1 and its parents by sequencing the transcriptomes of three tissues. By aligning the reads against the Nipponbare reference genome, an average of 89.06% annotated transcripts of the reference genome was obtained in our material, which was much greater than the minimum value (70%). The average percentage of unique mapped reads was 93.45%. There was a significant correlation between three biological replicates of transcriptome data in hybrid and its parents, and correlation coefficient was more than 0.8. The qRT-PCR was employed to validate the transcriptomic data by using 12 primers and the results showed that the expression profiles of 12 differentially expressed genes (DEGs) were consistent with the transcriptome data (Supplementary Fig. S4).
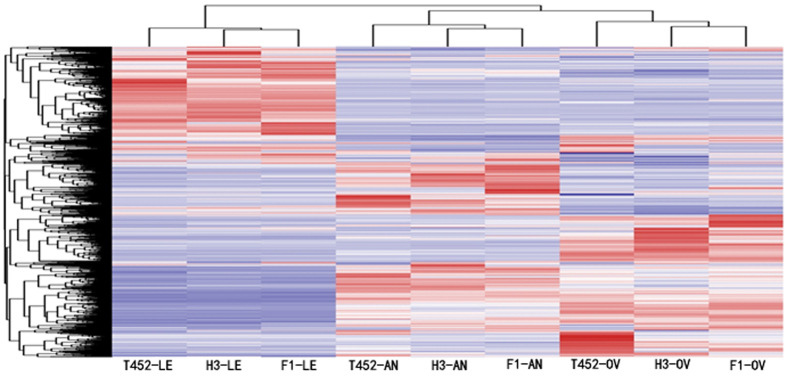
Hierarchical Cluster analysis was employed to investigate the correlations between differentially expressed genes in F1 and its two parents using transcriptomic data at different stages, and the results revealed that the three tissues, anther, ovary and leaf, from the F1 and its parents always assembled into primary groups at the same stage (Fig. 2). Moreover, we also found that most of the expressed genes in F1 were more similar to those in H3 (Supplementary Fig. S5), suggesting that the transcriptome profiles were consistent with the phenotypic data of F1 and its two parents.
Figure 2. Hierarchical clustering analysis of all expressed genes based on transcriptome data.
T452: Huajingxian 74-4x; H3: Huaduo 3; F1: (Huajingxian 74-4x ×Huaduo 3); AN: Anther; OV: Ovary; LE: Leaf.
Identification of differentially expressed genes (DEGs) and functional annotation of DEGs associated with anther, ovary and leaf in F1 and its parents
A total of 17877 DEGs were detected in three tissues, anther, ovary and leaf, of F1 and its parents, and ranged from 1521 to 2498 DEGs between F1 compared to its parents and between two parents (Table 2). DEGs between the hybrid and its parents are designated as DEGF, and the DEGs between the parents are called as DEGP. The DEGF could be divided into two groups, i.e. one shared by DEGP and DEGF, and another uniquely belonging to F1 compared to parents, which was designated as DEGFu. The DEGFu could explain the phenotypic differences between F1 and its two parents12; therefore, we specifically focused on the DEGFu to find out the genes associated with the heterosis and fertility. Of the 17877 DEGs, we obtained 1150, 1014 and 1122 DEGFu associated with anther, ovary and leaf between F1 and its parents, respectively. Among DEGFu, 807, 663 and 866 were specifically associated with anther, ovary and leaf, respectively (hereafter referred as DEGFu-sp). Among the 2336 DEGFu-sp, 10 genes encoding methyltransferase or hydroxymethyltransferase were detected (Supplementary Table S4).
Table 2. Differentially expressed genes (DEGs) associated with anther, ovary and leaf.
| Name of combination | Total DEGs |
Up-DEGs |
Down-DEGs |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Total No | Known DEG | New gene | No of Up DEG | Known DEG | New gene | No of down DEG | Known DEG | New gene | |
| H3vsT452-AN* | 2189 | 1273 | 916 | 1318 | 826 | 492 | 871 | 447 | 424 |
| F1vsT452- AN | 2498 | 1405 | 1093 | 1781 | 991 | 790 | 717 | 414 | 303 |
| F1vsH3- AN | 1693 | 1131 | 562 | 1068 | 594 | 474 | 625 | 537 | 88 |
| H3vsT452-OV | 2286 | 1319 | 967 | 1349 | 835 | 514 | 937 | 484 | 453 |
| F1vsT452- OV | 2424 | 1354 | 1070 | 1674 | 942 | 732 | 750 | 412 | 338 |
| F1vsH3- OV | 1521 | 973 | 548 | 1030 | 569 | 461 | 491 | 404 | 87 |
| H3vsT452-LE | 2166 | 1824 | 342 | 840 | 693 | 147 | 1326 | 1131 | 195 |
| F1vsT452-LE | 1543 | 1362 | 181 | 1012 | 851 | 161 | 531 | 511 | 20 |
| F1vsH3-LE | 1557 | 1389 | 168 | 725 | 598 | 127 | 832 | 791 | 41 |
| Total | 17877 | 12030 | 5847 | 10797 | 6899 | 3898 | 7080 | 5131 | 1949 |
*H3: Huaduo 3; T452: Huajingxian 74-4x; F1: (Huajingxian 74-4x × Huaduo 3); AN: Anther; OV: Ovary; LE: Leaf.
To understand the functions of DEGFu-sp genes in different tissues, we investigated the transcription factors (TFs), and found that 44 TFs, including 16 in anther, 10 in ovary and 18 in leaf, exhibited different levels of expressions in F1 compared to its parents. Meanwhile, we also used DEGFu-sp genes to identify protein-protein interactions, and the results revealed significant interactions in different tissues. Eight subnetworks, and 14 significant pathways, such as metabolic pathways, starch and sucrose metabolism, were detected in anther. A total of 17 subnetworks, and three significant pathways, such as Diterpenoid biosynthesis and biosynthesis of secondary metabolites, were identified in ovary. Moreover, we detected seven subnetworks and 32 significant pathways related to DEGFu-sp genes in leaf (Supplementary Fig. S6a–c and Table S4).
Then, gene ontology (GO) enrichment analysis was employed for the functional categorization of genes, and the results revealed that differential gene expressions might be associated with fertility and heterosis in anther, ovary and leaf. We identified 19 prominent categories associated with DEGFu-sp to anther in biological process category that were mainly related to photosynthesis, regulation of metabolic process, regulation of biosynthetic process and regulation of transcription (Supplementary Fig. S7). Six pathways were involved in the DEGFu-sp to anther, such as photosynthesis and metabolic pathways. Here, 13 genes showed high levels of expressions in F1 compared to T452. Of 807 DEGFu-sp to anther, 15 genes exhibited co-expression with 46 other important genes (Supplementary Table S4).
The DEGFu-sp to ovary could be divided into 38 clusters, which showed 13 prominent functional gene groups that associated with metabolic and biosynthetic processes in the biological process category, and microtubule associated complex (Supplementary Fig. S8a,b). Among 663 DEGFu-sp to ovary, 26 genes showed co-expression with 79 other genes (Supplementary Table S4).
The 866 DEGFu-sp to leaf were mainly categorized into 62 clusters, and 32 prominent genes groups were detected, including 81 genes related to transport (Fig. 3), localization and small molecule metabolic process (Supplementary Fig. S9). We identified 87 genes associated with metabolic pathways (Supplementary Table S4), among them, five genes displayed high levels of expressions in F1 compared to T452. They were annotated as aldehyde dehydrogenase (LOC_Os02g43280), aminotransferase (LOC_Os03g07570), cytochrome P450 (LOC_Os03g12660), GHMP kinases ATP-binding protein (LOC_Os06g48940) and molybdenum cofactor biosynthesis protein 1 (LOC_Os12g32230). Since H3 is an excellent neo-tetraploid rice line with high fertility and its leaves color changed with the passage of time during flowering, and T452 is an autotetraploid rice line with low fertility and leaves stay-green during flowering. Therefore, we compared the up-regulated genes between F1 compared to T452 and H3 compared to T452, and 74.01% up-regulated genes were similar in both comparisons. Interestingly, we detected two main prominent functional gene classes in leaf, including programmed cell death and defense response. Among 866 DEGFu in leaf, 19 genes displayed co-expression with 20 other important genes (Supplementary Table S4).
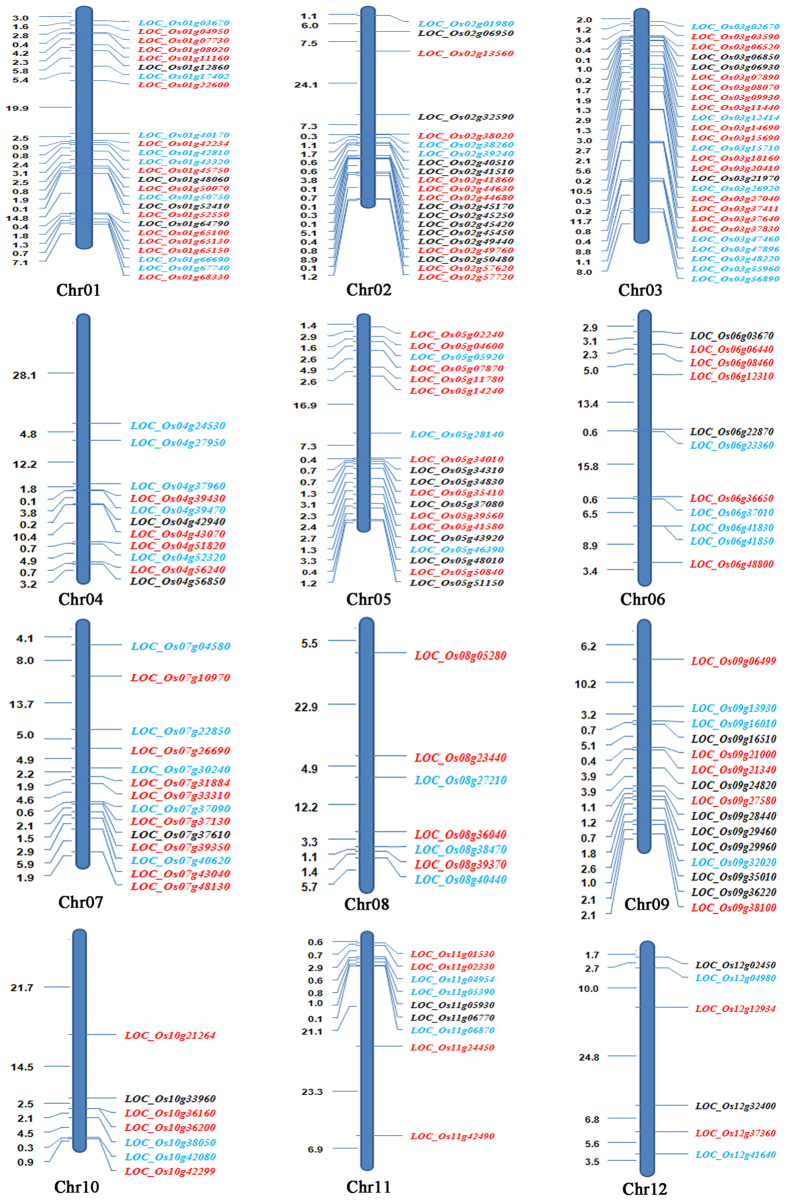
Figure 3. Location of genes associated with fertility and heterosis, and specific to leaf, anther and ovary.
Genes in red, blue and black letters associated with transport (leaf), meiosis (anther) and metabolic process (ovary), respectively.
Differentially expressed anther specific genes were associated with meiosis stage-specific genes between F1 and its parents
Since we used transcriptome to analyze the DEGF in neo-tetraploid hybrid during pollen mother cell (PMC) meiosis, and T452 is an autotetraploid rice line with low fertility, we specifically investigated meiosis stage-specific genes associated with the differentially up-regulated genes in F1 compared to T452 to identify genes related to high fertility of F1. Firstly, we compared the up-regulated genes in F1 compared to T452 with those up-regulated genes in H3 compared to T452, and 643 specifically expressed genes were detected (Supplementary Table S5). AgriGo analysis of the 643 genes displayed six prominent functional gene groups associated with defense response, photosynthesis, cell death and programmed cell death. Among these genes, seven genes were related to photosynthesis (GO:0015979). A total of 41 genes exhibited down-regulation in autotetraploid hybrid with low fertility compared to diploid rice (Supplementary Table S5)10. Of these 41 genes, four genes, including LOC_Os04g33830, LOC_Os12g08770, LOC_Os06g21590 and LOC_Os07g25430, were associated with photosynthesis. AgriGo analysis of the anther specific up-regulated genes revealed that there were no significant GO terms in F1 compared to H3.
Secondly, we compared 643 anther specific up-regulated genes in F1 compared to T452 with microarray data of wild type rice anther meiosis stage-specific expression17,18, and meiosis-related expression18,19,20. We identified nine overlapping genes that showed meiosis stage-specific expression from zygotene to tetrad (Supplementary Table S5).
Thirdly, we compared DEGFu-sp to anther with the microarray data of wild type rice meiosis stage-specific genes17,18 and meiosis-related genes18,19,20, and 42 meiosis stage-specific and eight meiosis-related overlapping genes were detected (Fig. 3). Of the eight meiosis-related genes, two genes, including LOC_Os11g04954 and LOC_Os12g04980, encoded DNA repair (RAD51) family protein, and one gene, LOC_Os01g67740, was annotated as structural maintenance of chromosomes protein 2 (SMC2) (Supplementary Table S5). Moreover, four genes associated with photosynthesis, including LOC_Os04g33830, LOC_Os12g08770, LOC_Os06g21590 and LOC_Os07g25430, exhibited down-regulation in autotetraploid hybrid with low fertility compared to diploid rice10.
Since transcriptomes may also be regulated through epigenetic mechanisms21, we also analyzed micro-RNA (miRNA). A total of 288 differentially expressed miRNAs (DERs), including 13 novel miRNAs, were detected in anther. Of these DERs, 38 were uniquely belonging to F1 compared to its parents. Together, 397 target genes were predicted by the 38 DERFu (Differentially expressed miRNA uniquely belonging to F1 compared to its parents) (Table 3 and Supplementary Table S6). Gene ontology (GO) enrichment analysis of the 397 targets showed seven prominent functional gene groups related to cell death, defense response, programmed cell death, apoptosis, DNA integration and response to stress. Most of the targets were the same as those detected by a comparison between the up-regulated genes in F1 compared to T452 and up-regulated genes in H3 compared to T452. Of the 397 targets, five genes were annotated as ubiquitin-conjugating enzyme or ubiquitin family protein, three genes encoded BURP-domain containing protein, four genes enriched in ATP binding protein, one gene was involved in chlorophyll a-b binding protein 1, seventy seven genes were associated with retrotransposon and nine genes encoded transposon protein or transposable element protein (Supplementary Table S6). Then we compared 38 DERs uniquely belonging to F1 compared to its parents with previously identified differentially expressed miRNAs. We detected one miRNA, osa-miR5072_L-2_1ss3AG, which was down-regulated in meiosis between Taichung65-2x and Taichung65-4x during pollen development, and predicted a target, LOC_Os02g24960, which was annotated as retrotransposon protein22.
Table 3. Identification of differentially expressed miRNAs (DERs) in anther.
| Name of combination | Total DER |
Up-DER |
Down-DER |
|||
|---|---|---|---|---|---|---|
| Total No | Novel miRNA | No of Up DER | Novel miRNA | No of down DER | Novel miRNA | |
| H3vsT452-AN* | 122 | 35 | 82 | 33 | 40 | 2 |
| F1vsT452-AN | 83 | 21 | 34 | 9 | 49 | 12 |
| F1vsH3- AN | 93 | 20 | 50 | 19 | 43 | 1 |
| Total | 298 | 76 | 166 | 61 | 132 | 15 |
| DERFu- AN | 38 | 13 | 24 | 12 | 14 | 1 |
*H3: Huaduo 3; T452: Huajingxian 74-4x; F1: (Huajingxian 74-4x × Huaduo 3); AN: Anther.
Nonadditive gene expression in anther, ovary and flag leaf of F1 hybrid
The gene expression in F1 could be divided into two types, the first was designated as additive expression that contributed by each allele from both parents, and another was nonadditive expression that deviates from the mid-parent value12. A total of 1224 nonadditive differentially expressed genes (NDEGs) were detected in the three tissues, including 314, 578 and 332 in anther, ovary and leaf, respectively (Supplementary Table S7). Of the 1224 NDEGs, 895 and 329 genes were found to be up- and down-regulated, respectively (Table 4). We compared the NDEGs in anther with the microarray data of rice anther meiosis specific genes17,18,19,20, and found 53 overlapping genes between anther and microarray data in meiosis. Among 53 genes, seven genes, including LOC_Os07g40620, LOC_Os02g38260, LOC_Os01g65630, LOC_Os03g21090, LOC_Os05g41120, LOC_Os04g49260 and LOC_Os05g38350, displayed down-regulation in autotetraploid hybrid compared to diploid rice with low fertility10. These seven genes were annotated as putative uncharacterized protein.
Table 4. Nonadditive differentially expressed genes (NDEGs) and number of differentially expressed genes uniquely associated with F1 and specific to (DEGFu-sp) anther, ovary, and leaf in F1 hybrid.
| Tissues | No. of DEGFu |
No. of NDEGs |
Total | %* | ||
|---|---|---|---|---|---|---|
| Total | Specific | Up | Down | |||
| Anther | 1150 | 807 | 197 | 117 | 314 | 0.09 |
| Ovary | 1014 | 663 | 386 | 192 | 578 | 1.61 |
| Leaf | 1122 | 866 | 312 | 20 | 332 | 1.5 |
| Total | 3286 | 2336 | 895 | 329 | 1224 | 3.2 |
*Indicate the percent ratio of NDEGs to total expressed genes in F1.
The Plant GeneSet Enrichment Analysis Toolkit (PlantGSEA) was employed to analyze the gene set enrichment of 578 NDEGs of ovary, and it showed a prominent category comprising of 74 genes that associated with a primary metabolic process (GO:0044238) in biological process. Moreover, we detected a microtubule associated complex (GO:0005875) that related to four genes in cellular component. These four genes encoded kinesin motor domain containing protein. Among the 332 NDEGs of leaf, nine prominent enrichment gene groups were detected, which mainly related to response to stress (GO: 0006950), defense response (GO: 0006952), programmed cell death (GO:0012501) (Supplementary Table S7).
Association between DNA sequence variations and differentially expressed genes (DEGs) in F1 compared to its parents
DNA re-sequencing revealed a huge difference in genome sequence of maternal, T452, and paternal, H3. A total of 2351039 SNPs were detected in T452 and H3, among them 1192412 and 374460 SNPs were specifically expressed in T452 and H3, respectively. A total number of 499448 Indels were observed in T452 and H3, of these 248194 and 84196 Indels showed specific variation in T452 and H3, respectively (Supplementary Table S8). The polymorphism rates of SNPs and Indels were 66.77% and 69.97% between T452 and H3, respectively (Supplementary Table S9). Of those polymorphic loci, about 65% of SNPs and 72% of Indels were in genic regions, 47.67% SNPs and 53.37% Indels were found to be in up or down regulatory regions that might be associated with the differentially expressed genes (DEGs). Moreover, we detected 58908 SNPs and 5561 Indels in coding regions (Supplementary Fig. S10 and Table S10).
Since many DNA sequences variation were detected between T452 and H3, we detected genes variation in DEGFu-sp, and a total of 330, 291 and 459 genes showed DNA sequence variation (SNP+Indel) in anther, ovary and leaf, respectively (Supplementary Table S7). The genes of three groups displayed differential expressions in anther, ovary and leaf of F1 compared to its parents. Of the 330 genes in anther, we detected 15 prominent categories in biological process, mainly related to photosynthesis, regulation of cellular metabolic process, regulation of transcription and biosynthetic process. We identified a prominent enrichment category associated with metabolic process (GO:0008152), consisting of 98 genes in biological process of ovary. Eight prominent enrichment genes, mainly related to cellular nitrogen compound metabolic process, establishment of localization and transport, were found in biological process category of leaf. These main enrichment categories were nearly consistent with the DEGFu-sp to anther, ovary and leaf (Supplementary Table S7).
Moreover, 67, 133 and 87 genes involved in SNP and Indel variations between two parents were detected in nonadditive differentially expressed genes (NDEGs) of anther, ovary and leaf, respectively. We detected 25 and 39 metabolic process related genes in anther and ovary, respectively. Notably, we found 25 genes related to TFs involved in SNP and Indel variations between parents (Supplementary Table S7).
Discussion
Both hybridity and induced polyploidy are the potent sources to increase the biomass, yield and resistance against various biotic and abiotic stresses. Agronomic traits, such as larger grain size, stronger stem, and longer panicles of autotetraploid rice than their corresponding parents, are of special interest for rice breeders2,8,9,10,11,23. However, low seed set has been a major obstacle in the use of autotetraploid rice and cannot be used directly in the breeding programs10,11. Autotetraploid lines with high seed setting might be developed through artificial selection of plants generated by the crossing of indica and japonica autotetraploid rice lines24,25. Here, we reported a neo-tetraploid rice, Huaduo 3, with high fertility (>80%) that was developed in 2012 by our research group. We developed 40 tetraploid rice hybrids by crossing H3 with different autotetraploid rice lines to observe the fertility and heterosis, and found encouraging results.
Firstly, our results revealed that H3 has the ability to improve the seed setting of different autotetraploid rice lines by hybridization. For example, a hybrid between H3 and autotetraploid rice line, with a seed setting of 8.43%, exhibited a normal seed set (67.08%). Another hybrid between H3 and autotetraploid rice line with seed setting of 42.97%, yielded high levels of seed set (84.73%). Overall, all the hybrids generated by H3 showed high seed setting (>80%), though some parental lines had very low seed setting.
Secondly, we detected positive and higher levels of heterosis for the yield and yield-related traits, such as effective number of panicles per plant, filled grains per plant, total number of grains per plant and grain yield per plant in the hybrids generated by H3. These results are consistent with the previous studies who found that autotetraploid hybrids had high-parent heterosis for filled grains per panicle, seed setting and yield7,8,9. However, these studies were limited to few lines or/and hybrids. Together, our work strongly suggests that H3 is not only producing hybrids with high fertility, but also exhibited powerful hybrid vigor for yield and yield-related traits compared to other autotetraploid lines.
In the present study, length and width of 10-grains showed no positive high-parent heterosis. These results are in agreement with the previous studies, who also found negative high-parent heterosis for grain length and width in autotetraploid rice8,9. Guo et al.25 investigated several autotetraploid rice hybrids, and the results showed that the mid-parent heterosis for grain length and width was significant, but high-parent heterosis exhibited opposite trends, i.e., the former was negative, and the latter was positive. We inferred that a complex genetic mechanism controlled the heterosis for length and width of 10-grains in autotetraploid rice.
The complexity of gene expression profiles associated with heterosis and differential gene expressions in different tissues had been revealed using transcriptome in diploid rice12,13. For example, genes associated with energy metabolism and transport enriched in DGHP (differentially expressed genes between the hybrid and its parents) rather than in DGPP (differentially expressed genes between the parental lines), and the genes involved in starch synthesis had much higher expression in the panicle of F1 than sterile parent at grain filling stage12. A transcriptome analysis of the root heterosis showed that DGHP were significantly enriched in carbohydrate metabolism and plant hormone signal transduction26. RNA-seq data of two rice genotypes and their reciprocal hybrids showed that DEGs tend to have tissue-specific expression patterns27. Moreover, RNA-seq-based transcriptome profiling analysis has revealed that polyploidy level is greatly associated with genome-wide disruption of gene expression and ultimately altered phenotypes in the polyploid rice hybrids (indica × japonica)28. Transcriptome analysis revealed that about 16% of the 16,112 expressed genes showed nonadditive expressions in leaf tissue of an interspecific F1 triploid hybrid29. The differential gene expressions for cell growth and functional secondary metabolites were found in induced autotetraploid of Chinese Woad (Isatis indigotica Fort.) by transcriptomic analysis30.
In the present study, the differentially expressed genes that uniquely associated with F1 and specific to (DEGFu-sp) anther were mainly enriched in four pathways, including photosynthesis, regulation of metabolic process, regulation of biosynthetic process and regulation of transcription. The DEGFu-sp genes to ovary were involved in metabolic process, biosynthetic process and response to stimulus. Interestingly, a prominent functional gene group associated with microtubule complex was detected in ovary. In higher plants, microtubule associated proteins are involved in the regulation of microtubules assembly and disassembly, and also responsible for the organization of microtubule structures and functions31. Therefore, we speculate that DEGFu-sp to microtubule-associated complex may involve in the embryo sac fertility.
Most of the leaf DEGFu-sp genes expressed in F1 were similar to that in Huaduo 3, and consisted of significantly enriched genes in transport, localization, and small molecule metabolic process. We also detected genes related to metabolic pathways. Moreover, more than 70% up-regulated genes in F1 were also found to be up-regulated in H3 compared to T452. The genes associated with leaf displayed two main prominent functional gene classes, including programmed cell death and defense response. These genes may be associated with energy transfer and heterosis, which inherited from the neo-tetraploid rice, Huaduo 3.
The dosage effect of transcription factors (TFs) had been considered as an important factor to affect phenotypes of hybrids in rice12. In the present study, we obtained 44 TFs that showed different expressions in DEGFu-sp, and the number of TFs was less than those (187 TFs) detected in diploid rice hybrids. Since previous study had revealed that DNA methylation and activity of class II transposable elements (TEs) involved in gene expression in autotetraploid rice32, this may be associated with the decrease in the number of TFs in F1 compared to its parents in our study. Meanwhile, we detected 25 TFs involved in SNP and Indel variations between the parents, T452 and H3. These TFs may alter the gene expression and is a possible molecular mechanism underlying heterosis and fertility in neo-tetraploid rice12. Among DEGFu-sp, we found 10 genes annotated as methyltransferase or hydroxymethyltransferase, which were related to epigenetic genes. Moreover, about 3% of total genes displayed nonadditive expressions, which might be regulated by trans-acting factors33. These results are consistent with the previous studies, who also found nonadditive genes in rice12,14,29. All of these results demonstrated complex regulatory mechanisms associated with heterosis, and differential gene expression was involved in fertility of neo-tetraploid rice hybrids.
Meiosis is a conserved process associated with many key genes in rice reproduction. More than 400 meiosis-specific or stage-specific genes have been identified in rice anther, and 28 genes were characterized in rice meiosis17,18,19,20,34. Since there are four chromosomes in autotetraploid rice, meiosis played more crucial role in fertility. A total of 55 meiosis-related or meiosis-stage-specific genes involved in pollen sterility loci interactions in autotetraploid rice hybrids were detected in our previous study10. In the present study, 42 meiosis stage-specific and eight meiosis-related genes revealed differential expressions in DEGFu-sp to anther. Of the 42 meiosis stage-specific genes, 26 genes were found to be down-regulated in autotetraploid hybrid with low fertility, and most of them were key genes for pollen fertility, such as Os09g0493500 that encoded ubiquitin fusion degradation protein and Os09g0329000 that was annotated as BURP domain containing protein10. These results suggested that fertility could be improved by suppressing alternation of the expression profiles of important meiosis-related genes, and by reducing epistatic interactions between alleles of pollen sterility loci in neo-tetraploid rice.
miRNA, an important epigenetic factor, regulates the changes in transcriptomes associated with heterosis and fertility21,22. Here, 38 miRNAs, uniquely belonging to F1 compared to its parents, were identified, and they predicted 397 target genes. Of these targets, 13 targets were found to be involved in pollen fertility, and 86 target genes were related to retrotransposon or transposon protein or transposable element protein. Moreover, one miRNA, osa-miR5072_L-2_1ss3AG, which predicted a target, LOC_Os02g24960, and annotated as retrotransposon protein, was detected in anther meiosis. Since DNA methylation variation of transposable elements are widespread and had a remarkable effect on gene expression in autotetraploid rice22,32, the changes in rich retrotransposon, transposon and transposable elements may act as a “super-genome shock” response factor for creating adaptability and to produce high fertility and heterosis in neo-tetraploid hybrids.
Plant Material
A neo-tetraploid rice, Huaduo (H3), and 40 different autotetraploid rice lines were used for developing hybrids in the present study (Supplementary Table S11). A total of 73 hybrids were developed, including 33 hybrids (control, CK) were generated by crossing different autotetraploid rice lines with each other (Supplementary Table S12), and 40 hybrids were developed by crossing H3 with different autotetraploid rice lines (Supplementary Table S13).
Methods
Evaluation of agronomic traits and data analysis
A total of 15 plants of each parent and F1 hybrid were harvested from the field at maturity. Agronomic traits, including plant height, effective number of panicles per plant, filled grains per plant, empty grains per plant, total grains per plant, seed setting ratio, grain yield per plant, length and width of ten grains of the selected plants, were measured. These agronomic traits were selected and investigated or measured according to the protocols of People’s Republic of China for the registration of a new plant variety DUS (Distinctness, Uniformity and Stability) test guidelines of rice (Oryza sativa L) (Guidelines for the DUS test in China, 2012)4.
The single factor variance analysis of each trait (different combinations) was done by SPSS 16.0. Multiple comparison was done by Duncan’s New Multiple-Range test (DMRT), using α = 0.05 significant level. High-parent heterosis (HPH) and mid-parent heterosis (MPH) were estimated by the following formula: HPH = (F1 − HP)/HP × 100%, and MPH = (F1 − MP)/MP × 100%, where F1 is the performance of first filial generation (hybrid), HP is the performance of the best parent, and MP is the average performance of two parents.
Whole-genome re-sequencing analysis
Young Leaves of Huaduo 3 and Huajingxian 74-4x were collected and stored at −80 °C for DNA isolation. Genomic DNA (gDNA) was extracted from each young leaf tissue by using a modified CTAB method35. The task of whole-genome re-sequencing was performed on an Illumina HiSeq2000™ by Biomarker Technologies (Beijing, China). The procedure was performed in accordance with the standard Illumina protocol as described previously36. The detection of SNPs and Indels were mainly performed by GATK software tools (https://www.broadinstitute.org/gatk/guide/best-practices.php). The SNPs and Indels annotations were performed based on the reference genome data by using SnpEff software.
Transcriptome analysis
Anthers and ovaries at meiosis stage, and flag leaves during flowering were collected from Huaduo 3, Huajingxian 74-4x and their F1 hybrid. All samples were collected in three biological replicates and stored at −80 °C for RNA isolation. The total RNA from each sample was extracted from the anthers, ovaries and leaves according to the manual instruction of the TRlzol Reagent (Life technologies, California, USA). The samples from anther and ovary were mixed from three biological replicates for RNA extraction. The quantity and quality of each RNA sample were assessed using 1% agarose gel and examined with a Nanodrop 1000 spectrophotometer (Nanodrop, Wilmington, DE, USA). RNA integrity number and concentration were checked using an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA). The mRNA was isolated by NEBNext Poly (A) mRNA Magnetic Isolation Module (NEB, E7490). The enriched and purified mRNA was broken into approximately 200 nt short RNA inserts, which were used to synthesize the first-strand cDNA and the second cDNA. The double-stranded cDNA were performed end-repair/dA-tail and adaptor ligation. The suitable fragments were isolated by AgencourtAMPure XP beads (Beckman Coulter, Inc.), and enriched by PCR amplification. Finally, the constructed cDNA libraries of the samples were sequenced on a flow cell using an Illumina HiSeq™ 2500 sequencing platform.
Transcriptome analysis was done using reference genome-based reads mapping. Low quality reads, such as adaptor sequences, unknown nucleotides>5%, or Q20 <20% (percentage of sequences with sequencing error rates <1%), were removed by perl script. The clean reads, which were filtered from the raw reads, were mapped to Nipponbare (IRGSP-1.0 pseudomolecule/MSU7) reference genome using Bowtie2 and Tophat2 software37. The aligned records from the aligners in BAM/SAM format were further examined to remove potential duplicate molecules. Gene expression levels were estimated using FPKM values (Fragments Per Kilobase of transcript per Million fragments mapped) by the Cufflinks software38.
DESeq was employed to evaluate the differential gene expression between F1 and its parents. After that, gene abundance differences between those samples were calculated based on the ratio of the FPKM values. The false discovery rate (FDR) control method was used to identify the threshold of the P-value in multiple tests in order to compute the significance of the differences. Here, only genes with an absolute value of Fold Change c and FDR significance score <0.01 were used for subsequent analysis.
GO analysis was performed for the functional categorization of differentially expressed genes using David analysis tools (http://david.abcc.ncifcrf.gov/home.jsp), the Plant GeneSet Enrichment Analysis Toolkit (http://structuralbiology.cau.edu.cn/PlantGSEA/) and AgriGO tool (http://bioinfo.cau.edu.cn/agriGO/). Cluster analysis was performed using Cluster 3.0 software. Pathway analysis was performed using PlantGSEA software. Predicted protein-protein interactions were analyzed using STRING software (http://www.string-db.org/). Annotations for differentially expressed genes were retrieved from the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/). Gene co-expression analysis was performed by using Gene Co-expression Networks in the website (http://bioinfo.sibs.ac.cn/carmo/Gene_Annotation.php). Venny software was used to identify the overlapped differentially expressed genes in different samples and tissues (http://bioinfogp.cnb.csic.es/tools/venny/)10.
miRNA analysis
Anthers at meiosis stage were collected from Huaduo 3, Huajingxian 74-4x and their hybrid. Total RNA was isolated from the anthers by using Trizol reagent (Invitrogen, CA, USA) according to the manufacturer’s protocol. The quantity and purity of total RNA were analyzed by RNA 6000 Nano LabChip Kit (Agilent, CA, USA) and Bioanalyzer 2100 with RIN number >7.0. Small RNA library was prepared by using about 1 μg of RNA according to the protocol of Illumina’s TruSeq small RNA sample preparation Kits (San Diego, USA). Single-end sequencing (36 bp) was executed on an Illumina Hiseq2500 at the LC-BIO (Hangzhou, China) following the manufacturer’s protocol. After sequencing, the data was further analyzed with an in-house program, ACGT101-miR, to remove common RNA families (snRNA, snoRNA, rRNA and tRNA), low complexity, adapter dimers, junk and repeats. Then, the unique sequences with 18–25 nt length were BLASTed to rice precursors in miRBase 20.0 (ftp://mirbase.org/pub/mirbase/CURRENT/) to detect known miRNAs. RNAfold software (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi) was employed to identify the novel predicted miRNAs. The miRNAs were considered as differentially expressed according to the normalized deep-sequencing levels (with the exclusion of 10 RPM) in F1 and its two parents. Chi-square (X2) test and Fisher exact test were used to estimate P-value. miRNAs with P-value < 0.05 and log2 (fold change ratio) >1 were considered as differentially expressed miRNAs. Targeted genes of differentially expressed miRNAs were predicted by using psRNATarget (http://plantgrn.noble.org/psRNATarget/)22,39.
Whole-genome re-sequencing, transcriptome and small RNA sequencing data have been deposited to NCBI SRA database (SRA accession: SRP078960).
qRT-PCR
A set of 12 selected DEGs from the transcriptome analysis were validated by qRT-PCR using the same RNA samples that used for transcriptome analysis. The sequences of corresponding target genes were obtained from the National Center for Biotechnology Information (NCBI). The target gene sequences were used to design the 12 gene-specific primer pairs using Primer Premier5.0 and Oligo6.0 software (Supplementary Table S14). About 1 μg of isolated RNA was used to synthesize first-strand complementary DNA (cDNA) using the Transcriptor First Strand cDNA Synthesis Kit (Roche). qRT-PCR was performed using SYBR Green (Bio-Rad) in a LightCycler480 System (Roche). The final reaction volume for qRT-PCR was 20 μL, and each reaction contained 2 μL of cDNA, 10 μM of forward and reverse primers, and 10 μL of universal SYBR Green supermix (Bio-RAD). The following amplification program was used: denaturation at 95 °C for 30 s and 40 cycles of amplification (95 °C for 5 s followed by 58 °C for 20 s). Melting curve analysis was performed from 65 °C to 95 °C, with 5-s increments of 0.5 °C. The rice Ubiquitin gene was used as an internal control. The relative expression levels were calculated as 2−(ΔCt of treatment−ΔCt) of control40, where Ct represents threshold cycle. Each PCR reaction, including the control reaction, was performed in triplicate10.
Additional Information
How to cite this article: Guo, H. et al. Transcriptome analysis of neo-tetraploid rice reveals specific differential gene expressions associated with fertility and heterosis. Sci. Rep. 7, 40139; doi: 10.1038/srep40139 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
The authors thank Prof. Yuanqing Li and Yuhua Zhang for donating Jackson-4x and 96025. We thank our group members for technical assistance. This work was supported by the National Natural Science Foundation of China Grant 31571625 (to X.D.L.), the Guangdong Provincial Science and Technique Project Grant 2014A030304055 (to X.D.L.) and Characteristic Innovation Project of Guangdong Provincial Key Platform of University Grant Yue–JK2014 [65] (to X.D.L.).
Footnotes
Author Contributions X.D.L. and M.Q.S. designed research. X.D.L., M.Q.S., H.G., M.J.N. and L.X. wrote the paper. H.G., M.J.N., L.X., J.D., Z.L. and M.Q.S. performed research. X.D.L., H.G., L.X., J.W., X.L. and M.Q.S. analyzed data. X.D.L. bred neo-tetraploid rice and contributed reagents/materials/analysis tools. All authors reviewed the manuscript.
References
- Cheng S., Zhuang J., Fan Y., Du J. & Cao L. Progress in research and development on hybrid rice: a super-domesticate in china. Ann. Bot. 100, 959–966 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D., Yuan L. & Lu X. A. New strategy of rice breeding in the 21st century II. searching a new pathway of rice breeding by utilization of double heterosis of wide cross and polyploidization. Acta.Agron. Sin. 27, 110–116 (2001). [Google Scholar]
- Shahid M. Q. et al. Yield and yield components in autotetraploid and diploid rice genotypes (indica and japonica) sown in early and late seasons. Aust. J. Crop. Sci. 7, 632–641 (2013). [Google Scholar]
- Guo H. et al. The research on autotetraploid rice. Guangzhou, South China University of Technology Press, China, pp 90–92 (2014). [Google Scholar]
- Wu J. et al. Comparative cytological and transcriptomic analysis of pollen development in autotetraploid and diploid rice. Plant Reprod. 27, 181–196 (2014). [DOI] [PubMed] [Google Scholar]
- Yu Z., Haage K., Streit V. E., Gierl A. & Torres Ruiz R. A. A large number of tetraploid Arabidopsis thaliana lines, generated by a rapid strategy, reveal high stability of neo-tetraploids during consecutive generations. Theor. Appl. Genet. 118, 1107–1119 (2009). [DOI] [PubMed] [Google Scholar]
- Tu S. et al. Production and heterosis analysis of rice autotetraploid hybrids. Crop Sci. 47, 2356–2363 (2007). [Google Scholar]
- Shahid M. Q., Liu G., Li J., Naeem M. & Liu X. Heterosis and gene action study of agronomic traits in diploid and autotetraploid rice. Acta Agri. Scand. B-Soil & Plant Sci. 61, 23–32 (2011). [Google Scholar]
- Shahid M. Q. et al. Genetic analysis and hybrid vigor study of grain yield and other quantitative traits in autotetraploid rice. Pak. J. Bot. 44, 237–246 (2012). [Google Scholar]
- Wu J. W. et al. Polyploidy enhances F1 pollen sterility loci interactions that increase meiosis abnormalities and pollen sterility in autotetraploid rice. Plant Physiol. 169, 2700–2717 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahid M. Q., Sun J., Wei C., Zhang P. & Liu X. Studies on the abnormality of embryo sac and pollen fertility in autotetraploid rice during different growing seasons. Pak. J. Bot. 42, 7–19 (2010). [Google Scholar]
- Wei G., Tao Y., Liu G., Chen C. & Luo R. A transcriptomic analysis of superhybrid rice LYP9 and its parents. Proc. Natl. Acad. Sci. USA. 106, 7695–7701 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankar R., Bhattacharjee A. & Jain M. Transcriptome analysis in different rice cultivars provides novel insights into desiccation and salinity stress responses. Sci. Rep-UK. 6, 23719 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson-Wagner R. A. et al. All possible modes of gene action are observed in a global comparison of gene expression in a maize F1 hybrid and its inbred parents. Proc. Natl. Acad. Sci. USA. 103, 6805–6810 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H. et al. Heterosis in early maize ear inflorescence development: a genome-wide transcription analysis for two maize inbred lines and their hybrid. Int. J. Mol. Sci. 15, 13892–13915 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A. et al. mRNA and small RNA transcriptomes reveal insights into dynamic homoeolog regulation of allopolyploid heterosis in nascent hexaploid wheat. The Plant Cell. 26, 1878–1900 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveshwar P., Bovill W. D., Sharma R., Able J. A. & Kapoor S. Analysis of anther transcriptomes to identify genes contributing to meiosis and male gametophyte development in rice. BMC Plant Biol. 11, 78 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M. et al. Rice expression atlas in reproductive development. Plant Cell Physiol. 51, 2060–2081 (2010). [DOI] [PubMed] [Google Scholar]
- Yant L. et al. Meiotic adaptation to genome duplication in Arabidopsis arenosa. Curr. Biol. 23, 2151–2156 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright K. M. et al. Selection on meiosis genes in diploid and tetraploid Arabidopsis arenosa. Mol. Biol. Evol. 32, 944–955 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z. et al. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature. 457, 327–331 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. et al. Comparative small RNA analysis of pollen development in autotetraploid and diploid rice. Int. J. Mol. Sci. 17, 499 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. W. et al. Analysis on genetic diversification and heterosis in autotetraploid rice. Springerplus. 2, 439 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W. & Yan Y. A preliminary report on investigations of auto-polyploids and amphidiploids in some cereal crops. Bulletin Bot. 5, 297–316 (1956). [Google Scholar]
- Guo H. et al. Agronomic traits and cytogenetic evaluation of newly developed autotetraploid rice line. Pak. J. Agri. Sci., 53, 291–301 (2016). [Google Scholar]
- Zhai R. et al. Transcriptome analysis of rice root heterosis by transcriptome analysis of rice root heterosis by RNA-Seq. BMC Genomics. 14, 19 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- E Z., Huang S., Zhang Y., Ge L. & Wang L. Genome-wide transcriptome profiles of rice hybrids and their parents. Int. J. Mol. Sci. 15, 20833–20845 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C. et al. Genome-wide disruption of gene expression in allopolyploids but not hybrids of rice subspecies. Mol. Biol. Evol. 31, 1066–1076 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y. et al. Transcriptome shock in an interspecific F1 triploid hybrid of Oryza revealed by RNA sequencing. J. Integr. Plant Biol. 58, 150–164 (2016). [DOI] [PubMed] [Google Scholar]
- Zhou Y. et al. Transcriptomic analysis reveals differential gene expressions for cell growth and functional secondary metabolites in induced autotetraploid of Chinese woad (Isatis indigotica Fort.). Plos One. 10, e116392 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada T. Microtubule-associated proteins in higher plants. J. Plant Res. 120, 79–98 (2007). [DOI] [PubMed] [Google Scholar]
- Zhang J. et al. Autotetraploid rice methylome analysis reveals methylation variation of transposable elements and their effects on gene expression. Proc. Natl. Acad. Sci. USA. 112, E7022–E7029 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupar R. M. & Springer N. M. Cis-transcriptional variation in maize inbred lines B73 and Mo17 leads to additive expression patterns in the F1 hybrid. Genetics. 173, 2199–2210 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Q., Li Y., Shen Y. & Cheng Z. Ten years of gene discovery for meiotic event control in rice. J. Genet. Genomics. 41, 125–137 (2014). [DOI] [PubMed] [Google Scholar]
- Cota-Sanchez J. H., Remarchuk K. & Ubayasena K. Ready-to-use DNA extracted with a CTAB method adapted for herbarium specimens and mucilaginous plant tissue. Plant Mol. Biol. Rep. 24, 161–167 (2006). [Google Scholar]
- Bai H. et al. Identifying the genome-wide sequence variations and developing new molecular markers for genetics research by re-sequencing a landrace cultivar of Foxtail Millet. Plos One. 8, e73514 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X. & Zhao P. X. PsRNATarget: A plant small RNA Target analysis server. Nucleic Acids res. 39, W155–W159 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.