Abstract
Antimicrobial susceptibility of 120 Helicobacter pylori isolates to metronidazole, tetracycline, clarithromycin, and amoxicillin was determined, and 77.5, 15, 10, and 6.6% of the isolates, respectively, were resistant. Only rdxA inactivation and both rdxA and frxA inactivation were responsible for metronidazole resistance in 66% (8 of 12) and 33% (4 of 12) of the isolates, respectively.
Eradication of Helicobacter pylori infection by treatment with two antimicrobial agents (clarithromycin and amoxicillin or metronidazole) and a proton pump inhibitor is recommended by various consensus groups (10, 16, 20). Antimicrobial resistance in H. pylori is a growing problem as it is the most important factor in determining treatment outcome. The prevalence of antimicrobial resistance varies with geographical regions (3, 25). Metronidazole resistance in H. pylori has been shown to be due to mutation in rdxA; mutation in frxA has also been shown to be associated with metronidazole resistance (11, 12, 23). In Bangladesh, the prevalences of H. pylori infection among infants, children, and adults are 61, 84, and 92%, respectively (1, 21, 22); however, information on antimicrobial susceptibility to commonly used drugs in H. pylori treatment is lacking. This study was conducted to evaluate (i) the prevalence of primary antibiotic resistance to commonly used antimicrobial agents and (ii) the genetic basis for metronidazole resistance in H. pylori isolates from Bangladesh.
Consecutive patients attending the Gastroenterology Department of Dhaka Medical College Hospital for upper gastrointestinal endoscopy were enrolled during 1999 to 2001. Diagnosis of peptic ulcer (PU) and non-ulcer dyspepsia (NUD) or gastritis was based on endoscopic examination of the stomach and duodenum. Biopsy samples were taken from each patient for culture.
Bacteria were grown in brain heart infusion agar with 7% sheep blood and incubated at 37°C in 5% O2, 10% CO2, and 85% N2 for 3 to 6 days. The MICs of amoxicillin, clarithromycin, metronidazole, and tetracycline for the isolates were determined by the agar dilution method as described elsewhere (18, 19). All tests were repeated twice, and H. pylori 26695 was used as a control. β-Lactamase production was tested by the chromogenic cephalosporin method (6). The molecular mechanism of susceptibility and resistance to metronidazole was studied in 12 isolates. Metronidazole-susceptible (Mtzs) isolates were further studied (by inactivation of rdxA alone or rdxA and frxA for conversion into an Mtzr phenotype) by transformation of Mtzs isolates with plasmids pBS-rdxA-cam (rdxA::cat) and pBS-frxA-kan (frxA::kan) as described earlier (11, 12).
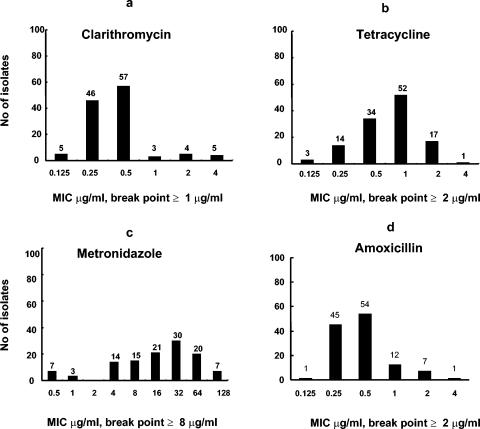
A total of 278 consecutive patients between 15 and 78 years of age were enrolled, and among them, 72.7% (202 patients) were male and 27.3% (76 patients) were female. Among the patients, 162 had PU and 116 had NUD and 62.6% (174 of 278) were culture positive for H. pylori. Among the culture-positive patients, 121 (69.5%) were male and 53 (30.4%) were female and 112 (64.3%) had PU and 62 (35.6%) had NUD. Of the 174 isolates, a total of 120 were available for antimicrobial susceptibility testing and 73.3% (88 of 120) and 26.6% (32 of 120) were from PU and NUD patients, respectively. Among the isolates, 77.5% (93 of 120), 15% (18 of 120), 10% (12 of 120), and 6.6% (8 of 120) were resistant to metronidazole, tetracycline, clarithromycin, and amoxicillin, respectively (Table 1). The range and distribution of MICs for the isolates are shown in Fig. 1. All amoxicillin-resistant isolates were β-lactamase negative. Antimicrobial susceptibilities of the isolates collected from patients with PU and NUD and males and females were compared, and no significant difference (P ≤ 0.05) in antimicrobial resistance was observed among these groups (Table 1).
TABLE 1.
Antibiotic susceptibility of 120 H. pylori strains isolated during 1999 to 2001 from Dhaka, Bangladesh
| Antimicrobial agent | No. (%) of resistant isolates in patients:
|
||||
|---|---|---|---|---|---|
| All patients (n = 120) | PU (n = 88) | NUD (n = 32) | Male (n = 88) | Female (n = 32) | |
| Amoxicillin | 8 (6.6) | 7 (8.0) | 1 (3.1) | 4 (4.5) | 4 (12.5) |
| Clarithromycin | 12 (10) | 8 (9.1) | 4 (12.5) | 7 (8.0) | 5 (15.6) |
| Metronidazole | 93 (77.5) | 67 (76.1) | 26 (81.2) | 68 (77.3) | 25 (78.1) |
| Tetracycline | 18 (15) | 12 (13.6) | 6 (18.7) | 14 (16.0) | 4 (12.5) |
FIG. 1.
Distribution of MICs of clarithromycin (a), tetracycline (b), metronidazole (c), and amoxicillin (d) for 120 H. pylori isolates. The numbers above the bars represent the numbers of the isolates for which the particular MIC applies.
Inactivation of only rdxA was sufficient to confer the Mtzr phenotype in 66% (8 of 12) of isolates, 33% (4 of 12) of isolates were Mtzs, and inactivation of only frxA had little effect on Mtzs of all 12 isolates. Subsequent frxA inactivation of all rdxA-deficient strains increased the MIC of metronidazole from 16 μg/ml to 32 μg/ml for the eight strains which became resistant after only rdxA inactivation, and four strains which were sensitive after only rdxA inactivation reverted to the Mtzr phenotype (MIC, 32 μg/ml).
Resistance to metronidazole was the most common type of resistance, with worldwide rates of 10 to 90% (3, 25). The high prevalence (77%) of metronidazole resistance in Bangladesh might be due to frequent use of metronidazole for other intestinal and gynecological problems. Previous use of metronidazole has been shown to be associated with H. pylori resistance to this antimicrobial agent (17). Two types of Mtzs H. pylori were isolated in the present study: type I, requiring only inactivation of rdxA to become resistant; and type II, requiring inactivation of both rdxA and frxA to become resistant. Only frxA inactivation did not have any role in metronidazole resistance, as only subsequent inactivation of frxA in rdxA-inactivated isolates reverted from the Mtzs phenotype to the Mtzr phenotype and increased the MIC for the Mtzr phenotype. This is in contrast to the findings of Kwon et al., who interpreted that the resistant phenotype can obtained by inactivation either of frxA or rdxA (13). Thus, resistance to metronidazole in H. pylori is mainly due to mutation in the rdxA gene and results from de novo mutation in the resident rdxA gene, rather than lateral transfer of a mutant rdxA gene.
The reported prevalence of primary resistance to clarithromycin ranges between 0 and 15% in most countries (3, 25). Around 10% of the isolates in the present study were clarithromycin resistant. In Bangladesh, clarithromycin was introduced in the late 1990s, and it has been widely used for eradication of H. pylori. Previous use of macrolides has been shown to be associated with H. pylori resistance to clarithromycin (17).
Amoxicillin resistance was not considered important until recently identified in the United States, Canada, and Italy (7, 8). Amoxicillin is one of the most commonly used antimicrobial agents in Bangladesh in recent years. Although 6.6% of the isolates were resistant, none was positive for β-lactamase. Amoxicillin resistance develops due to structural alterations in one of the penicillin-binding proteins (4, 5, 9) or changes in other proteins involved in cell wall synthesis (2, 15, 26), and the resistant phenotype may be lost due to freezing or storage. All isolates tested in the present study were frozen at least once, and the low prevalence of the resistance phenotype may be due to loss during storage. Primary resistance to tetracycline ranges between 5 and 59% in Asian countries (14, 24, 27). Around 15% of the isolates in the present study were tetracycline resistant, which is in agreement with an earlier finding from this region.
Therefore, it is reasonable to conclude that in our geographical area, antibiotic resistance is an emerging problem for the treatment of H. pylori-infected patients. The present study also demonstrates the need for continuous monitoring of the antimicrobial susceptibility in H. pylori for determination of optimal treatment regimens.
Acknowledgments
This study was conducted at the ICDDR, B: Centre for Health and Population Research with the support of SIDA agreement no 2001-3970, component 75000255/A/7500260.
ICDDR, B acknowledges with gratitude the commitment of SIDA to the Centre's research efforts.
REFERENCES
- 1.Ahmad, M. M., M. Rahman, A. K. Rumi, S. Islam, F. Huq, M. F. Chowdhury, F. Jinnah, M. G. Morshed, M. S. Hassan, A. K. Khan, and M. Hasan. 1997. Prevalence of Helicobacter pylori in asymptomatic population—a pilot serological study in Bangladesh. J. Epidemiol. 7:251-254. [DOI] [PubMed] [Google Scholar]
- 2.Costa, C. S., and D. N. Anton. 1993. Round-cell mutants of Salmonella typhimurium produced by transposition mutagenesis: lethality of rodA and mre mutations. Mol. Gen. Genet. 236:387-394. [DOI] [PubMed] [Google Scholar]
- 3.Debets-Ossenkopp, Y. J., A. J. Herscheid, R. G. Pot, E. J. Kuipers, J. G. Kusters, and C. M. Vandenbrouck-Grauls. 1999. Prevalence of Helicobacter pylori resistance to metronidazole, clarithromycin, amoxycillin, tetracycline and trovafloxacin in The Netherlands. J. Antimicrob. Chemother. 43:511-515. [DOI] [PubMed] [Google Scholar]
- 4.DeLoney, C. R., and N. L. Schiller. 2000. Characterization of an in vitro-selected amoxicillin-resistant strain of Helicobacter pylori. Antimicrob. Agents Chemother. 44:3368-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dore, M. P., D. Y. Graham, and A. R. Sepulveda. 1999. Different penicillin-binding protein profiles in amoxicillin-resistant Helicobacter pylori. Helicobacter 4:154-161. [DOI] [PubMed] [Google Scholar]
- 6.Dore, M. P., M. S. Osato, G. Realdi, I. Mura, D. Y. Graham, and A. R. Sepulveda. 1999. Amoxycillin tolerance in Helicobacter pylori. J. Antimicrob. Chemother. 43:47-54. [DOI] [PubMed] [Google Scholar]
- 7.Dore, M. P., A. R. Sepulveda, I. Mura, G. Realdi, M. S. Osato, and D. Y. Graham. 1997. Explanation for variability of omeprazole amoxycillin therapy? Tolerance of H. pylori to amoxycillin. Gastroenterology 112:A105. [Google Scholar]
- 8.Fedorak, R., A. Archambault, R. Flamm, M. Osato, and D. Stamle. 1997. Antimicrobial susceptibility of H. pylori in Canada to three key antibiotics: metronidazole, clarithromycin, and amoxicillin. Gastroenterology 112:A115. [Google Scholar]
- 9.Gerrits, M. M., D. Schuijffel, A. A. van Zwet, E. J. Kuipers, C. M. Vandenbroucke-Grauls, and J. G. Kusters. 2002. Alterations in penicillin-binding protein 1A confer resistance to β-lactam antibiotics in Helicobacter pylori. Antimicrob. Agents Chemother. 46:2229-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graham, D. Y., W. A. de Boer, and G. N. Tytgat. 1996. Choosing the best anti-Helicobacter pylori therapy: effect of antimicrobial resistance. Am. J. Gastroenterol. 91:1072-1076. [PubMed] [Google Scholar]
- 11.Jeong, J.-Y., A. K. Mukhopadhyay, D. Dailidiene, Y. Wang, B. Velapatino, R. H. Gilman, A. J. Parkinson, G. B. Nair, B. C. Wong, S. K. Lam, R. Mistry, I. Segal, Y. Yuan, H. Gao, T. Alarcon, M. L. Brea, Y. Ito, D. Kersulyte, H. K. Lee, Y. Gong, A. Goodwin, P. S. Hoffman, and D. E. Berg. 2000. Sequential inactivation of rdxA (HP0954) and frxA (HP0642) nitroreductase genes causes moderate and high-level metronidazole resistance in Helicobacter pylori. J. Bacteriol. 182:5082-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeong, J.-Y., A. K. Mukhopadhyay, J. K. Akada, D. Dailidiene, P. S. Hoffman, and D. E. Berg. 2001. Roles of FrxA and RdxA nitroreductases of Helicobacter pylori in susceptibility and resistance to metronidazole. J. Bacteriol. 183:5155-5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwon, D. H., F. A. El-Zaatari, M. Kato, M. S. Osato, R. Reddy, Y. Yamaoka, and D. Y. Graham. 2000. Analysis of rdxA and involvement of additional genes encoding NAD(P)H flavin oxidoreductase (FrxA) and ferredoxin-like protein (FdxB) in metronidazole resistance of Helicobacter pylori. Antimicrob. Agents Chemother. 44:2133-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwon, D. H., J. J. Kim, M. Lee, Y. Yamaoka, M. Kato, M. S. Osato, F. A. El-Zaatari, and D. Y. Graham. 2000. Isolation and characterization of tetracycline-resistant clinical isolates of Helicobacter pylori. Antimicrob. Agents Chemother. 44:3203-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maki, H., and K. Murakami. 1997. Formation of potent hybrid promoters of the mutant llm gene by IS256 transposition in methicillin-resistant Staphylococcus aureus. J. Bacteriol. 179:6944-6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malfertheiner, P., F. Megraud, C. O'Morain, A. P. Hungin, R. Jones, A. Axon, D. Y. Graham, G. Tytgat et al. 2002. Current concepts in the management of Helicobacter pylori infection—the Maastricht 2-2000 consensus report. Aliment. Pharmacol. Ther. 16:167-180. [DOI] [PubMed] [Google Scholar]
- 17.McMahon, B. J., T. W. Hennessy, J. M. Bensler, D. L. Bruden, A. J. Parkinson, J. M. Morris, A. L. Reasonover, D. A. Hurlburt, M. G. Bruce, F. Sarco, and J. C. Butler. 2003. The relationship among previous antimicrobial use, antimicrobial resistance, and treatment outcomes for Helicobacter pylori infections. Ann. Intern. Med. 139:463-469. [DOI] [PubMed] [Google Scholar]
- 18.National Committee for Clinical Laboratory Standards. 1997. Methods for dilution antimicrobial susceptibility test for bacteria that grow aerobically. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 19.National Committee for Clinical Laboratory Standards. 1999. Performance standard antimicrobial susceptibility testing. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 20.NIH Consensus Development Panel on Helicobacter pylori in Peptic Ulcer Disease. 1994. NIH, Consensus, and Conference: Helicobacter pylori in peptic ulcer disease. JAMA 272:65-69. [PubMed] [Google Scholar]
- 21.Sarker, S. A., D. Mahalanabis, P. Hildebrand, M. M. Rahaman, P. K. Bardhan, G. Fuchs, C. Beglinger, and K. Gyr. 1997. Helicobacter pylori: prevalence, transmission, and serum pepsinogen II concentrations in children of a poor periurban community in Bangladesh. Clin. Infect. Dis. 25:990-995. [DOI] [PubMed] [Google Scholar]
- 22.Sarker, S. A., M. M. Rahman, D. Mahalanabis, P. K. Bardhan, P. Hildebrand, C. Beglinger, and K. Gyr. 1995. Prevalence of Helicobacter pylori infection in infants and family contacts in a poor Bangladesh community. Dig. Dis. Sci. 40:2669-2672. [DOI] [PubMed] [Google Scholar]
- 23.Sisson, G., J. Y. Jeong, A. Goodwin, L. Bryden, N. Rossler, S. Lim-Morrison, A. Raudonikiene, D. E. Berg, and P. S. Hoffman. 2000. Metronidazole activation is mutagenic and causes DNA fragmentation in Helicobacter pylori and in Escherichia coli containing a cloned H. pylori rdxA+ (nitroreductase) gene. J. Bacteriol. 182:5091-5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thyagarajan, S. P., et al. 2003. Geographical difference in antimicrobial resistance pattern of Helicobacter pylori clinical isolates from Indian patients: multicentric study. J. Gastroenterol. Hepatol. 18:1373-1378. [DOI] [PubMed] [Google Scholar]
- 25.Toracchio, S., and L. Marzio. 2003. Primary and secondary antibiotic resistance of Helicobacter pylori strains isolated in central Italy during the years 1998-2002. Dig. Liver Dis. 35:541-545. [DOI] [PubMed] [Google Scholar]
- 26.Wosten, M. M., E. E. Ishiguro, and B. A. van der Zeijst. 1997. Cloning and characterization of the lytB gene of Campylobacter jejuni. FEMS Microbiol. Lett. 157:117-121. [DOI] [PubMed] [Google Scholar]
- 27.Wu, H., X. D. Shi, H. T. Wang, and J. X. Liu. 2000. Resistance of Helicobacter pylori to metronidazole, tetracycline and amoxycillin. J. Antimicrob. Chemother. 46:121-123. [DOI] [PubMed] [Google Scholar]