Abstract
New gene origination is a major source of genomic innovations that confer phenotypic changes and biological diversity. Generation of new mitochondrial genes in plants may cause cytoplasmic male sterility (CMS), which can promote outcrossing and increase fitness. However, how mitochondrial genes originate and evolve in structure and function remains unclear. The rice Wild Abortive type of CMS is conferred by the mitochondrial gene WA352c (previously named WA352) and has been widely exploited in hybrid rice breeding. Here, we reconstruct the evolutionary trajectory of WA352c by the identification and analyses of 11 mitochondrial genomic recombinant structures related to WA352c in wild and cultivated rice. We deduce that these structures arose through multiple rearrangements among conserved mitochondrial sequences in the mitochondrial genome of the wild rice Oryza rufipogon, coupled with substoichiometric shifting and sequence variation. We identify two expressed but nonfunctional protogenes among these structures, and show that they could evolve into functional CMS genes via sequence variations that could relieve the self-inhibitory potential of the proteins. These sequence changes would endow the proteins the ability to interact with the nucleus-encoded mitochondrial protein COX11, resulting in premature programmed cell death in the anther tapetum and male sterility. Furthermore, we show that the sequences that encode the COX11-interaction domains in these WA352c-related genes have experienced purifying selection during evolution. We propose a model for the formation and evolution of new CMS genes via a “multi-recombination/protogene formation/functionalization” mechanism involving gradual variations in the structure, sequence, copy number, and function.
Keywords: cytoplasmic male sterility, new gene origination, mitochondrial genome, rice
Introduction
Mitochondria are thought to be generated by endosymbiosis over one billion years ago and function as energy-producing and biological signaling centers in eukaryotes1. Unlike the small (15-18 kb) and homogenous mitochondrial genomes of animals, plant mitochondrial genomes are large (hundreds of kb or more) and complicated, with dramatic variations in structure, size, and organization2. Studies have shown that in the angiosperm mitochondrial genomes, frequent DNA recombination may result in genomic rearrangements, yielding multiple types of recombinant structures that may vary dramatically in copy number (substoichiometric shifting (SSS)); the recombinant structures may contain new open reading frames (ORFs) and some of them can cause cytoplasmic male sterility (CMS)3,4,5. CMS is a widespread phenomenon observed in more than 150 flowering plant species6 and often associated with unusual ORFs present in the mitochondrial genomes5. To date, a number of CMS genes have been cloned from various plant species, and they are usually chimeric, involving sequences homologous to mitochondrial essential genes for ATPase, cytochrome c oxidase, or ribosomal proteins4,5. However, how new mitochondrial genes including CMS genes originate, evolve, and especially how they acquire their functions still remain a mystery.
The Asian cultivated rice (Oryza sativa L.) was domesticated from the wild rice (Oryza rufipogon Griff)7. Several types of CMS systems have been identified in rice (wild rice), including the Wild Abortive (CMS-WA), Boro II (CMS-BT), and HongLian (CMS-HL); these CMS systems have been explored for hybrid rice production5. The CMS-based hybrid seed technology is a three-line system, using a CMS line, a maintainer line, and a restorer line. The CMS line has a male-sterile cytoplasm with a CMS gene and carries non-functional (recessive) nuclear restorer gene(s). The maintainer line has normal fertile cytoplasm and contains the same nuclear genome as the CMS line, thus serving as the male parent in hybridization with the CMS line for the CMS line propagation. The restorer line possesses functional (dominant) restorer gene(s), and is used as the male parent to cross with the CMS line for production of F1 hybrid seeds. In the F1 plants, the functional restorer gene(s) restore male fertility, and the combination of the divergent nuclear genomes from the CMS line and the restorer line produces hybrid vigor and higher grain yields.
The CMS-WA system, which is most widely used for hybrid rice production since 1970s, was developed by successive backcrossing to introduce the CMS cytoplasm of a male sterile O. rufipogon plant into the nuclear backgrounds of rice cultivars8,9. We previously identified the CMS-WA gene WA352 (here renamed WA352c, see below) and revealed that WA352c causes CMS by interaction with the nucleus-encoded, highly conserved mitochondrial protein COX11, leading to premature programmed cell death in the anther tapetum and defective pollen development. Male fertility of WA352c-carrying hybrids can be restored by the dominant restorer genes Rf4 and Rf3, which suppress WA352c function at the mRNA and protein levels, respectively, in diploid anther-wall cells of the hybrids (i.e., in a sporophytic manner)10,11. We noted that the WA352c-containing structure in the mitochondrial genome (including the upstream and downstream flanking regions) consists of multiple segments that are homologous to different sequences of the mitochondrial genomes in wild rice species, suggesting a complicated origin through DNA recombination and rearrangement. However, the source of WA352c sequences and the routes of its evolution, how it exerts its effect on adaptive evolution of rice, and how it co-evolves with the nuclear effector (COX11) and the restorer genes (Rf3 and Rf4), remain obscure.
In this study, we surveyed the mitochondrial genomes of 808 wild and cultivated rice lines, and identified 11 recombinant structures containing chimeric ORFs related to WA352c in the mitochondrial genomes. We deduced that these structures likely have arisen from complex evolutionary routes through multiple recombination events among conserved mitochondrial sequences of unknown function, coupled with SSS and sequence variation. We further identified two transitional, non-CMS protogenes from these structures and showed that they likely evolved into functional CMS genes via sequence variations that unlocked the self-inhibitory COX11-interacting function. We provide evidence that these sequence variations were under purifying selection, suggesting that origination of new CMS genes may play a role in promoting out-crosses and improving the adaptive fitness of wild rice under natural conditions.
Results
Identification of CMS-related recombinant structures
The Oryza genus has 20 wild and two cultivated species12. To investigate the origin and evolutionary history of WA352c, we screened for possible recombinant structures related to WA352c in the mitochondrial genomes of a wide range of wild and cultivated rice. We first analyzed the sequence of the WA352c-containing recombinant structure (named S352c for convenience) by searching GenBank (http://www.ncbi.nlm.nih.gov/), and re-annotated the sequence composition of S352c (including the ORF and its upstream and downstream flanking sequences) in an elite indica rice (O. sativa L. ssp. indica) CMS-WA line Zhenshan 97A (ZS97A) (Figure 1A and 1B). The WA352c ORF is located downstream of rpl5, and consists of four segments: 284s, cs3 (previously defined as unknown origin and orf224-homologous sequence10), cs2 and cs1 (previously defined as an orf288-homologous sequence10). The 284s segment is identical to the promoter/5′-ORF of a putative mitochondrial ORF orf284, and cs1-cs3 and the downstream sequences cs4-cs6 are conserved in the mitochondrial genomes of the Oryza species and other plants (such as maize, sorghum, wheat, and soybean) (see below). The downstream flanking sequences also include an atp6-containing segment, and a segment (ch12) homologous to a chromosome 12 region. These WA352c sequences have no similarity to any known functional cytoplasmic or nuclear genes.
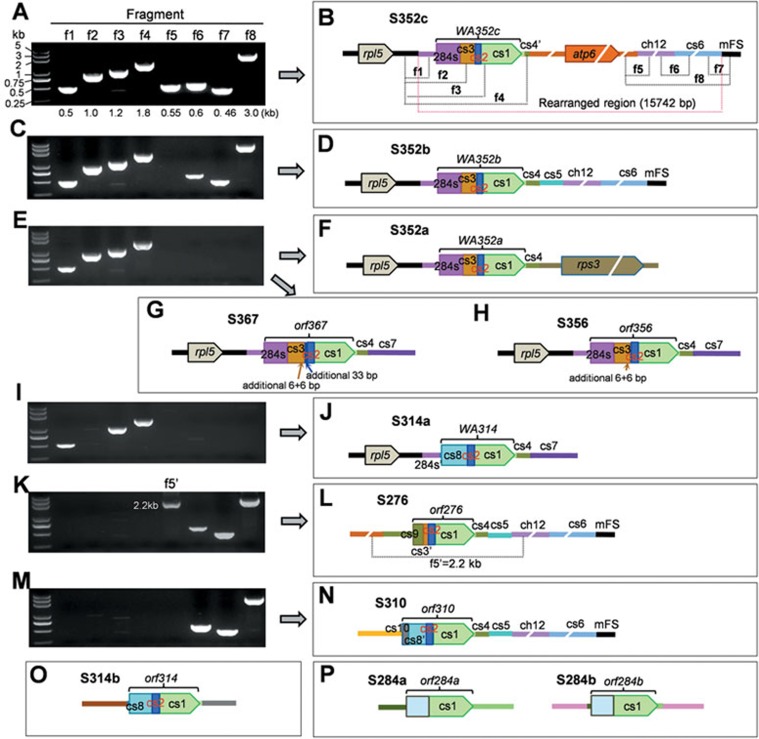
Figure 1.
Detection and annotation of the CMS-WA mitochondrial genomic structure S352c and its related variant structures in wild and cultivated rice. (A-N) Based on the sequence of S352c (including the upstream and downstream regions), primer sets were prepared (Supplementary information, Table S1) for amplification of 8 fragments (f1-f8) in the Oryza species (Table 1). The obtained PCR products were sequenced. For some structures, of which some fragments in one side (S352a, S367, S356, S314a, and S352b) or both sides of cs1 (S284a, S284b, and S310) were not amplified, hiTAIL-PCR was used to recover their unknown flanking sequences followed by sequencing. The whole-recombinant structures (including the ORF-flanking sequences) are named S###, where “###” indicates the number of amino acids (aa) encoded by the putative ORFs. The ORFs verified as functional CMS genes are named “WA###”, and those of non-CMS and functionally uncharacterized ones named “orf###”. The cs1 to cs10 sequences are conserved in the mitochondrial genomes of Oryza and other plant species (Supplementary information, Figure S4). The sequence ch12 is homologous to rice chromosome 12. The homologous sequences among these recombinant structures are indicated with the same colors. For simplification, the short (277 bp) pseudogene sequence of rps14 at the immediate downstream of rpl5 is not shown. (O) S314b (orf314) was detected in some accessions of O. rufipogon and many rice cultivars (Table 1), including the determined mitochondrial genomic sequences of 3 rice cultivars (DQ167400.1, DQ167807.1, and DQ167399.1), in which the same orf314 ORF was previously annotated as orf288 (GenBank BA000029.3). (P) S284a and S284b have similar putative ORFs but different flanking sequences. Note that orf284a and orf284b are distinct from the orf284 sequence locating upstream of cox1 (see Figure 4B)
Next, we examined a total of 808 lines of 15 Oryza species, including 478 rice cultivars, 292 O. rufipogon accessions, and 38 accessions of 13 other Oryza wild species (Table 1). By PCR using primers designed for amplification of 8 recombinant segments of S352c (Supplementary information, Table S1), all the 8 segments were amplified from S352c in ZS97A and several lines of O. rufipogon and O. sativa. However, in some other lines, only some of these segments were amplified (Figure 1A-1P), indicating the presence of S352c-related structure variants. From these lines we then amplified and sequenced the unknown sequences flanking these known sequences using hiTAIL-PCR13. As a result, we identified eleven mitochondrial recombinant structures (mitotypes) related to S352c, which contain WA352c-related putative ORFs and the same or different upstream and downstream flanking sequences (Table 1, Figure 1). However, many (391) wild rice accessions and cultivated rice lines do not carry any one of these types recombinant structures (in high copy-number state) (Table 1). These recombinant structures including the ORFs and their flanking sequences, of which cs1 to cs10 are conserved in the mitochondrial genomes of Oryza and other plant species, were named S###. The ORFs that were verified as functional CMS genes in this study (see below) and a previous report14 were named “WA###”, while the putative ORFs with no CMS function (see below) and those functionally uncharacterized were named “orf###”, where “###” indicates the number of amino acids (aa) encoded by the ORFs. For the homologous ORFs of the same aa number but with different flanking sequences, lower case letters (a, b, c, etc) were added at the end.
Table 1. Detection of the WA352c-related recombinant structures in wild and cultivated rice.
| Species (genome) | No. accession | S352c | S352b | S352a | S367 | S356 | S314a | S314b | S276 | S310 | S284a | S284b |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| O. rufipogon (AA) | 292 (200) | 2 | 11 | 1 | 10 | 4 | 3 | 10 | 73 | 81 | 0 | 5 |
| O. sativa (AA) | 478 (211) | 6 | 0 | 0 | 0 | 0 | 0 | 142 | 18 | 45 | 0 | 0 |
| O. officinalis (CC) | 4 (3) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
| O. rhizomatis (CC) | 1 (1) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| O. eichingeri (CC) | 2 (1) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| O. minuta (BBCC) | 6 (1) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Other 9 species | 25 (0) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 808 (417) | 8 | 11 | 1 | 10 | 4 | 3 | 152 | 91 | 126 | 3 | 8 |
Note: in the investigated 808 accessions, 417 accessions (in parentheses) carry predominant forms (in high copy numbers) of the structures that were detected by PCR with 28 cycles. The recombinant structures with very low copy numbers that co-exist with the predominant structures in some accessions (see Figure 5) are not included here. The nine Oryza species (no. accessions analyzed) from which no these structures were detected are: O. australiensis (2), O. barthii (4), O. brachyantha (3), O. glumaepatula (2), O. grandiglumis (2), O. latifolia (3), O. longistaminata (4), O. meridionalis (2), and O. punctata (3).
Four structures (S352a, S352b, S367, and S356) show strong structural similarity to S352c, but contain different downstream flanking sequences (Figure 1C-1H). The five ORFs (WA352a, WA352b, WA352c, orf367, and orf356) in these structures share the same segments (284s, cs3, cs2, and cs1). WA352a is the same as orf352 of the functional CMS gene reported in CMS-RT102 rice14. WA352b and WA352c have identical sequences, but differ from WA352a by five single-nucleotide polymorphisms (SNPs). Compared with WA352a, WA352b, and WA352c (WA352a/b/c), orf367 and orf356 have two 6-bp segments and several SNPs within cs3, and orf367 has a further 33-bp segment within cs2 (Figure 1G). WA314 and orf314 in S314a and S314b, respectively, have the same structural composition (cs8/cs2/cs1) with nineteen SNPs between them, but they have completely different upstream and downstream sequences (Figure 1I, 1J, 1O). S276 and S310 have the same downstream sequences (cs4/cs5/ch12/cs6/mFS) as in S352b, but different upstream sequences (Figure 1K-1N). The CMS-like orf276 and orf310 in S276 and S310 are the same as the ORFs identified in the CMS-RT98 and CMS-Lead rice lines, respectively15,16. A recent report showed that in the CMS-Lead rice, the CMS-BT homologous gene orf79, but not orf310, confers the CMS trait17, suggesting that orf310 is not a CMS factor. The simplest cs1-containing structures, S284a and S284b, have the same putative ORF (orf284a and orf284b, which are distinct from the orf284 sequence located upstream of cox1), but their upstream and downstream flanking sequences differ from each other and from those of the other structures (Figure 1P).
Except for S284a (found in O. rhizomatis, O. minuta, and O. eichingeri that possess the CC-type or CCBB-type nuclear genome) and S284b (detected in O. officinalis with the CC nuclear genome and O. rufipogon with the AA nuclear genome), these recombinant structures were detected only in certain O. rufipogon accessions and some rice cultivars but not in the few accessions of the other nine Oryza species sampled here (Table 1). It is proposed that the Oryza species with the BB-genome and CC-genome are phylogenetically close to O. rufipogon18. Although there is the possibility that some relative ancestral structures, such as S367 and S356 (see below), might have arisen in the ancestor of O. rufipogon, the results suggest that most of these mitochondrial structures were generated in O. rufipogon, which diverged from other AA-genome-containing Oryza species ca. 2 million years ago19. Thus, these genes (ORFs) are evolutionarily young, and they are not fixed in O. rufipogon and O. sativa, i.e., not spreading to all individuals of the species.
Functional test of the candidate CMS genes
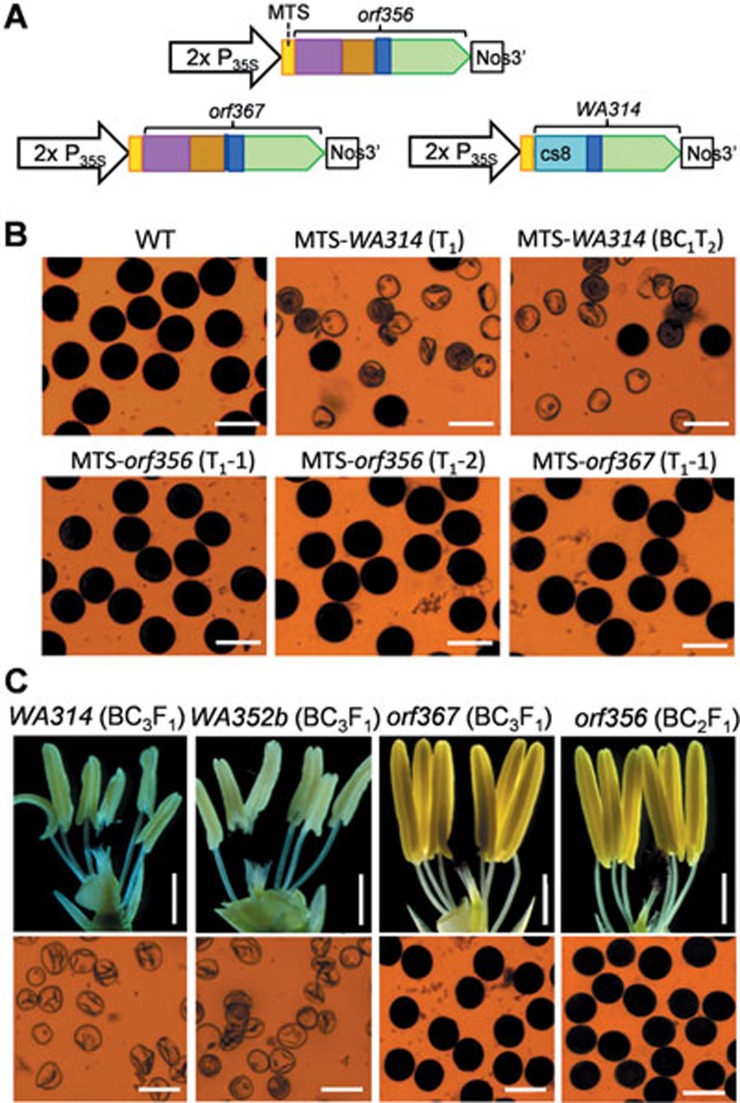
Previous studies proved that WA352c and WA352a have CMS function10,14. As orf356, orf367 and WA314 have structures similar to WA352a and WA352c, we analyzed whether they also have CMS function. We prepared three binary constructs with MTS-orf356 (MTS, mitochondrial transit sequence), MTS-orf367, and MTS-WA314 (Figure 2A) and transferred them into a CMS-WA maintainer line ZS97B with normal cytoplasm and the recessive restorer genes rf3 and rf4-i, an indica type rf4 allele11. The MTS fusion localizes the protein to mitochondria, thus allowing the test of CMS function using nuclear transformation10,20. Four T0 transgenic rice plants with MTS-WA314 showed partial pollen sterility. The transgenic progenies (T1) were obtained from self-pollination of the T0 plants (with 25%-38% seed set) and BC1T1 plants were produced by backcrossing of the T0 plants with ZS97B. These segregants carrying MTS-WA314 exhibited partial male sterility while those without the transgene were male fertile (Figure 2B and Supplementary information, Figure S1A). We also tested the WA314 construct in Arabidopsis thaliana and observed partial male sterility (with very few seeds) in 22 T1 plants carrying MTS-WA314; the partial sterility and fertility in the T2 plants co-segregated with the presence and absence of the transgene (Supplementary information, Figures S1A and S2A-S2C). However, all T0 and T1 rice plants with either the MTS-orf367 or the MTS-orf356 construct, had normal male fertility, even though the transgenes were expressed at high levels (Figure 2B and Supplementary information, Figure S1A, S1B).
Figure 2.
Functional analysis of the putative CMS genes. (A) Binary constructs for testing CMS function of the putative CMS genes by nuclear transformation (2× P35S, doubled CaMV35S promoter; MTS, sequence encoding a mitochondrial transit signal; Nos3′, transcriptional termination sequence). (B) Pollen phenotype of the rice plants carrying the transgenes. The BC1T1 plants with MTS-WA314 were obtained by backcrossing with ZS97B. WT is a negative segregant without the transgene (MTS-WA314) selected from a T1 family. The transgenic plants carrying MTS-WA314 exhibited partial male sterility (deflated pollen grains), but those with MTS-orf356 and MTS-orf367 produced fertile pollen. Pollen grains were stained with 1% potassium iodide. Scale bars, 50 μm. (C) Anther (top) and pollen (bottom) phenotypes of the backcrossed plants with ZS97B as the recurrent parent and wild rice accessions containing the putative CMS genes as cytoplasm donors. The plants with WA314 and WA352b showed aberrant (pale-white) anthers with sterile pollen. Scale bars, 1 mm for anthers and 50 μm for pollen.
To further study the CMS effect of orf356, orf367, WA352b, and WA314 in a rice line with recessive rf3 and rf4, we introduced the cytoplasm with these ORFs of the O. rufipogon accessions into the ZS97B nuclear background by successive backcrossing. All BC2F1, BC3F1, and BC4F1 plants with WA314 or WA352b (and homozygous rf3 and rf4-i) had aberrant anthers with sterile pollen, similar to the WA352c-carrying CMS-WA lines10; however, all the backcrossed plants with orf367 (BC1F1 to BC3F1) and orf356 (BC1F1 and BC2F1) were male fertile (Figure 2C and Supplementary information, Figure S3). These results demonstrate that both WA314 and WA352b can confer CMS, whereas orf356 and orf367 lack CMS activity.
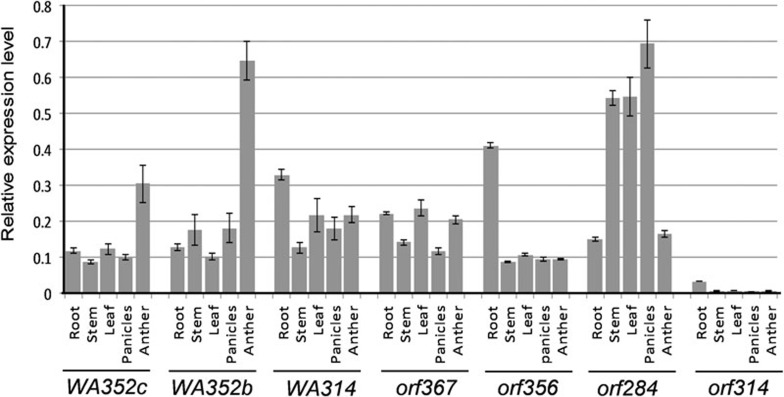
We analyzed the expression patterns of the CMS and CMS-like genes in these backcrossing lines or a japonica cultivar (O. sativa L. ssp. japonica) Nipponbare (Nip) with the recessive rf3 and rf4-j (japonica type rf4), because that the expression of the genes is not affected in the absence of the functional restorer genes10,11. Quantitative reverse transcription PCR (qRT-PCR) showed that WA352b, WA352c, WA314, orf367, and orf356 were expressed in various tissues (root, stem, leaf, panicle, and anthers) of the corresponding backcrossing lines (shown in Supplementary information, Figure S3) or Nip (for orf284 and orf314 expression) (Figure 3). However, orf314, which is present in many rice cultivars (Table 1), was not expressed in anthers and other tissues of Nip, except for its low-level expression in seedling roots (Figure 3). This result could explain why orf314, which was previously annotated as orf28821 due to a sequencing error that resulted in a mis-identification of the start codon, does not cause CMS even though its ORF sequence is similar to that of WA314 (there are 13 aa differences between the encoded proteins) and can interact with COX11 (see below). Mitochondrial genes are often transcribed as multiple mRNA species with different transcription initiation sites driven by distinct promoters22; for example, WA352c has three transcripts, one co-transcribed with rpl5 (producing a dicistronic transcript with the rpl5 and WA352c ORFs) and two transcribed independently (producing monocistronic transcripts with the WA352c ORF only)10. The putative ORF orf284, whose promoter-5′ ORF segment was involved in the formation of the CMS-related ORFs, also was expressed at quite high levels (Figure 3). Therefore, in addition to the co-utilization of the local promoter of rpl5, the transfer of the orf284-derived promoter-containing segments (284s, 284s') to the site downstream of rpl5 (see below) provided an additional active promoter to these CMS and CMS-like genes. By contrast, although orf367 and orf356 have the CMS-like structures and expression ability, their lack of CMS function may be due to other reasons.
Figure 3.
Expression analysis of the putative CMS genes and related genes by qRT-PCR. The CMS-WA line ZS97A was used for WA352c expression, BC3F1 (male fertile) carrying S367 for orf367 expression, BC2F1 (male fertile) carrying S356 for orf356 expression, BC3F1 (male sterile) carrying S352b for WA352b expression, BC3F1 (male sterile) carrying S314a for WA314 expression, and a japonica cultivar Nipponbare for expression of orf314 and orf284 (orf284 is located downstream of cox1 and it provided the promoter/5′-ORF sequence into the recombinant structures, see Figure 4B). Anthers were at the meiosis to microspore stages; roots were from seedlings; young panicles were 5-8 cm in length. Third stems and flag leaves at young panicle stage were used. The relative expression levels were normalized with the atp6 expression (i.e., ratios of target genes to atp6) and shown as mean ± SD (3 biological replicates).
Tracing the likely evolutionary routes of the CMS-related structures
We next examined how these structures could have originated. Repetitive sequences in plant mitochondrial genomes can mediate DNA recombination4. We identified short and intermediate-length repeats (8 to 84 bp in length, denoted by A-H) in these structures and their candidate source sequences (Figure 4 and Supplementary information, Figures S5 and S6). These repeats might mediate homologous recombination (HR) between the source sequences to create the described structures. However, some of these repeat sequences (B', C, D, G, H with 8, 12, 13, 10, and 19 bp, respectively) were much shorter than those previously reported to be involved in recombination events, which are generally > 50 bp4, suggesting that very short repeats can also mediate the recombination events, similar to the microhomology-mediated end joining (MMEJ) process also called alternative non-homologous end joining (altNHEJ) for repairing double-strand break (DSB) in nuclear genomes23,24,25. However, all these structures also include recombination joining sites without repeated sequences. These observations suggest that not only HR, MMEJ, and classical NHEJ (cNHEJ) mechanisms, which are common in the nuclear genomes23,24,25, might also contribute to the plant mitochondrial genomic rearrangements.
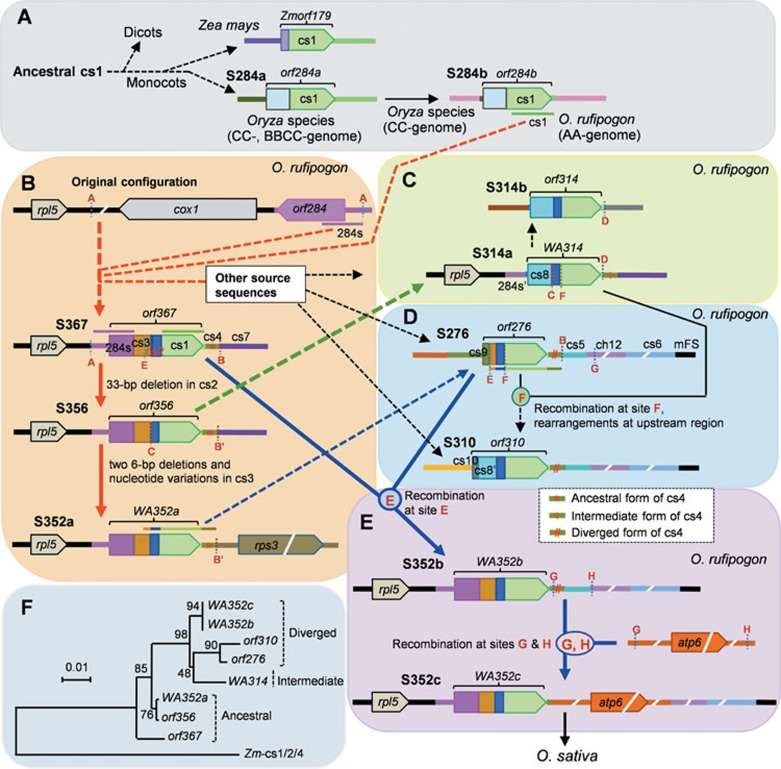
Figure 4.
Tracing the evolutionary history of the CMS-related structures in wild rice. The evolutionary routes of the structures in O. rufipogon were deduced based on their structural compositions and sequence similarities. The red letters A to H (see Supplementary information, Figures S5 and S6) indicate the repeat sites that might mediate the HR or MMEJ recombination events among the source sequences and the donor structures to generate the new structures. The rearrangements without repeat sites are proposed to be based on the cNHEJ mechanism. Arrows with solid lines indicate simple rearrangement by one or two recombination events, while those with dotted lines suggest complex rearrangements. These structures with cs4 were first grouped into ancestral, intermediate, and diverged forms according to the similarities of their cs4 sequences to the referent sequences from other plants (Supplementary information, Figure S5). (A) S284a and S284b are the ancient structures that provided the cs1 source sequence into the other recombinant structures. (B) The rpl5/cox1/orf284 sequence represents the original local structure; at the downstream (site A) of rpl5, the primary recombinant structure S367 was produced by multiple rearrangements. S356 was generated from S367 by 33-bp deletion in the ORF, and S352a was further produced by nucleotide variations in the ORF and a rearrangement with the rps3-containing sequence. This pathway (red arrows) represents the primary evolutionary route for the generation of the protogenes (orf367 and orf356) and the intermediate functional CMS gene (WA352a). (C) S314a was generated likely by translocation of a fragment from S356 and multiple rearrangements, and the WA314 ORF was transferred to another genomic position to produce S314b. (D) S276 arose probably by rearrangements of a sequence from S352a with cs5 (recombination at site B) flanked by ch12/cs6/mFS, and with cs9. S310 was produced from recombination (at site F) between S276 and S314a and other rearrangement events. (E) S352b was generated probably from a recombination between S367 and S276 at site E, then, S352c was produced by integration of the atp6-containing sequence. This indicates the latest evolutionary route (blue arrows) for the origination of the functional CMS genes WA352b and WA352c. The evolutionary routes for the generation of S310 and S352b are supported by the co-presence of the donor structures and the recombined products in the same accessions as shown in Figure 5 and Supplementary information, Figure S7. S352c and some other structures (Table 1) were inherited into O. sativa during domestication, and the materials of O. rufipogon and O. sativa carrying S352c also were used as the cytoplasm donors to breed CMS-WA lines by backcrossing10. (F) A phylogenetic tree based on the cs1, cs2, and cs4 sequences of the recombinant structures in wild rice and those in maize using the neighbor-joining method. Bootstrap values were calculated with 1 000 replications. The sequences are grouped into ancestral, intermediate, and diverged forms.
We traced the possible evolutionary routes connecting these structures based on their similarities in structure and sequence, and assuming that the evolution of the most closely related structures went through the fewest changes. S284a and the maize Zmorf179-containing structure have the same cs1/downstream sequences with a high nucleotide similarity (94%), indicating that they were likely derived from a common ancestral cs1-containing sequence, with sequence divergence of S284a and its homologs since the Oryza species separated from other plants including maize (Figure 4A). S284b could be derived from S284a in the CC-genome species by a rearrangement of the ORF at a different mitochondrial genomic position, and has been inherited by O. rufipogon (with the AA-genome) and O. officinalis (with the CC-genome). Subsequently, the cs1 fragment in S284b, the key source sequence for the CMS function of the CMS genes, became part of the recombinant structures by rearrangements with other donor sequences present in O. rufipogon. The rpl5/cox1/orf284 configuration (Figure 4B) is present in nine of the ten (i.e., except for the CMS-WA line with rpl5/WA352c) sequenced mitochondrial genomes: O. rufipogon (AP012527, AP011076, AP012528) and rice cultivars (DQ167400, JN861111, JF281153, AP011077, DQ167399, and DQ167807)14,15,16,21,26,27,28. Our further PCR analysis showed that the rpl5/cox1/orf284 configuration exists in all or most accessions of the tested five wild rice species with the AA-genome but not in the species with other genome types (Supplementary information, Figure S4). Thus, rpl5/cox1/orf284 could be the original structure in the AA-genome-containing wild rice species. The CMS gene-related structures likely formed at the site downstream of rpl5, by recombination among 284s from orf284 and its promoter region, cs1 from S284b, and other mitochondrial genomic source sequences (Figure 4A and 4B).
To deduce the evolutionary relationships among these recombinant structures after S284a and S284b formed, we first inferred the ancestral and diverged types of these structures and then compared their structural and sequence similarities. As the cs4 sequences present in all these structures have a number of nucleotide variations, we compared these cs4 sequences with the homologous sequences from maize, wheat, rye and Aegilops speltoides as the outgroup references (Supplementary information, Figure S5). The cs4 sequences of S367, S356, and S352a are the most similar to these outgroup reference sequences; those of S276, S310, S352b, and S352c have more diverged nucleotide sequences, while that of S314a has both the conserved sites and some of the diverged sites (Figure 4B-4E and Supplementary information, Figure S5). Thus, S367, S356, and S352a probably are the ancestral forms, S314a is an intermediate form, and S276, S310, S352b, and S352c might be the most recently derived. Furthermore, at three varied sites (in cs1), the nucleotides of S367 are the same as those in the outgroup (Supplementary information, Figure S5) but different from the other structures, indicating that S367 is the closest to S284b.
S356 has the highest sequence similarity to S367; thus it was likely derived from S367 by deletion of the 33-bp in cs2 of orf367 (Figure 4B and Supplementary information, Figure S4). S352a could be derived from S356 by the two 6-bp deletions and nucleotide variation in cs3, and by recombination of the downstream region at the site B' with an rps3-containing sequence (Figure 4B and Supplementary information, Figures S4 and S5). This S284b-rpl5/S367-S356-S352a history is proposed as the primary evolutionary route for producing the functional CMS gene WA352a. Since S314a contains the rpl5/284s' and the cs2/cs1/cs4 sequences, just as in S356, it could be derived from S356 by integration of the cs8 sequence (by a recombination event at site C) to replace cs3 and a part of 284s; this forms a branch for generating WA314 (Figure 4C), another functional CMS gene (Figure 2). Further, the WA314 ORF region moved to another mitochondrial genomic position, by a MMEJ recombination at site D and a cNHEJ recombination at the 5′ of the ORF, to produce S314b, whose upstream and downstream sequences differ from those of S314a (Figure 4C).
Furthermore, a sequence containing the cs3′/cs2/cs1/cs4 segments derived from WA352a could rearrange by recombination at site B with cs5 (flanked by the ch12/cs6/mFS sequences) to produce S276 (Figure 4D). A recombination between S314a and S276 at site F, plus cNHEJ-based rearrangements in the upstream region, could produce S310 (Figure 4D and Supplementary information, Figure S5). As S352b possesses the S276 sequences (from the site E to mFS), and it co-exists with S367 and S276 in some wild rice accessions (see below), we proposed that S352b could be generated from recombination between S367 and S276 at site E (Figure 4E and Supplementary information, Figure S5). S352c differs from S352b by having an atp6-containing sequence replacing cs5; thus S352c must have arisen later, by transferring the atp6-containing sequence from its original location into S352b via recombination at the sites G and H (Figure 4B, 4D, 4E and Supplementary information, Figure S6). Therefore, this S367/S276-S352b-S352c path forms a branch route for generating WA352b and WA352c. In summary, the functional CMS genes and CMS-like ORFs originated from complex but related evolutionary routes. A phylogenetic tree based on the cs1, cs2, and cs4 sequences supports the inferred evolutionary relationships among these structures (Figure 4F). Some of these structures, including S352c, were inherited by O. sativa cultivars (Table 1) during the domestication of O. sativa from O. rufipogon populations, as well as into CMS-WA lines by the hybrid breeding programs.
Dramatic copy-number variation of the recombinant structures
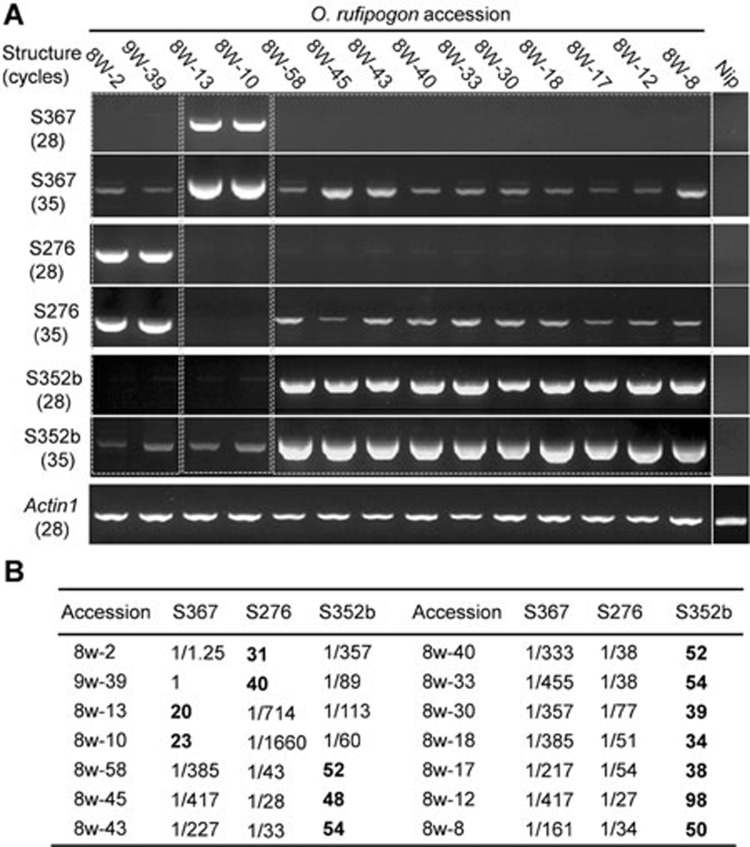
Since mitochondrial genomes are largely matrilineally inherited, the donor structures and the recombinant new structures will be present in the same mitochondrial genome. We selected some O. rufipogon accessions that contain S367, S276, and S352b (Table 1), and performed semi quantitative PCR to test these structures. The results confirmed that S367, S276, or S352b (or at least two of them) co-exist in all these accessions (Figure 5A), supporting the idea that S352b was derived from recombination between S367 and S276 (Figure 4B, 4D and 4E). Furthermore, the copy numbers of these structures varied dramatically in the same or different accessions. For example, the donor structures S367 or S276 were predominant (at high copy numbers) in some accessions, but in other accessions the product structure S352b has become predominant (34-98 copies/cell), whereas S367 and S276 were present as substoichiometric DNAs (sublimons, ca. 1 copy per 161-455 cells and per 27-77 cells, respectively) (Figure 5A and 5B). Similar co-existence and copy-number variation for the S314a/S276/S310 route also were observed in some O. rufipogon accessions (Supplementary information, Figure S7), supporting the inference that S310 was derived from recombination between S314a and S276. These results suggest that the donor structures and the generated new recombinant structures have undergone SSS during the evolution, from low to high copy-number variation for the new structures, and from high to low copy-number variation for the donor structures. The above results strongly support the likelihood that these CMS-related recombinant structures were generated through integration of multiple conserved mitochondrial sequences into a specific region followed by complex evolutionary routes based on the dynamic recombination, sequence variation and SSS.
Figure 5.
Co-existence of related recombinant structures and their substoichiometric shifting. (A) Semi-quantitative PCR using structure-specific primers with 28 and 35 cycles was performed to detect the structures and estimate their abundance in leaves of the selected O. rufipogon accessions. The products that could be detected with strong signals by 28 cycles are present in high copy numbers, while those could be detected only by 35 cycles are at low copy numbers. The nuclear gene OsActin1 was used as a control. (B) Estimation of copy numbers per cell of S367, S276, and S352b in leaves by quantitative PCR. Actin1 (two allelic copies per cell) was used as the internal control for normalization.
Functionalization of the CMS genes by sequence variation
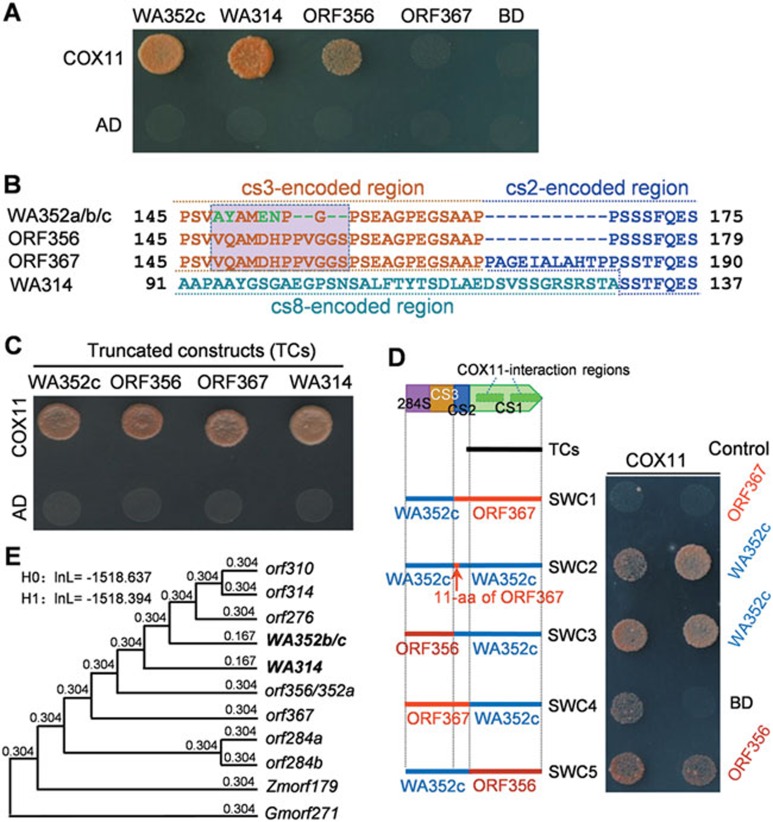
To know the reason that orf367 and orf356 had no CMS function (Figure 2), we investigated the biochemical function of the proteins they encode. WA352a/b/c, orf367, orf356, and WA314 encode putative transmembrane proteins (Supplementary information, Figure S8). Despite the constitutive expression of WA352c mRNA, the WA352c protein accumulates specifically in the tapetum regulated by unknown mechanism, where it interacts with COX11 to impair COX11's function in hydrogen peroxide degradation, thus triggering specifically premature programmed cell death of the tapetum and leading to pollen abortion10. Therefore, we carried out yeast two-hybrid (Y2H) assays to test whether ORF367, ORF356, and WA314 can also interact with COX11. The results indicated that WA314, like WA352c, could interact with COX11. However, ORF356 showed a weaker interaction with COX11 and ORF367 showed no interaction (Figure 6A), suggesting that their non-CMS property is probably due to their poor interaction with COX11.
Figure 6.
Functionalization and natural selection of the CMS-related genes. (A) Yeast two-hybrid assays of the interaction of WA352c, ORF356, ORF367, and WA314 (in pGBKT7 vector, BD) with COX11 (in pGADT7, AD). The yeast cells were grown on SD/-Leu/-Trp/-His/-Ade medium. (B) The aa changes (in green) and the 11-aa deletion in WA352a, WA352b, and WA352c, and the replacement of the cs3-encoded sequence in WA314. These variations release the interaction potential with COX11. (C) Yeast two-hybrid assays of the COX11-interaction of four truncated constructs (169 aa in length) as shown in (D). (D) Test of the swapped constructs (SWC1-SWC5) and the controls for the interaction with COX11. (E) Estimation of non-synonymous to synonymous substitution rate ratios (dN/dS) of the cs1 sequences in the ORFs using the one-ratio model (for the null hypothesis H0) and the branch model (for the alternative hypothesis H1) of the PAML/CODEML program. H0 assumes that all the cs1 branches in the phylogenetic tree have the same evolutionary rate; H1 assumes that the branch cs1 sequences of the functional WA352b/c and WA314 have a different evolutionary rate from the others. No significant difference (P > 0.05) of the log likelihoods (InL) was detected between H0 and H1. The cs1 sequences of Zmorf179 and Gmorf271 are from maize and soybean, respectively.
WA352c contains two COX11-interaction regions at aa 218-292 (interaction region 1) and aa 294-352 (interaction region 2) encoded by the cs1 sequence; rice transformation with truncated WA352c constructs that expressed proteins with and without the COX11-interaction domains verified that the COX11-interaction is required for induction of CMS10. However, the cs1-encoded sequences show no common aa difference between the non-CMS group (ORF367 and ORF356) and the functional group (WA352a/b/c and WA314) (Supplementary information, Figure S8). Instead, the ORF356/ORF367 group and WA352a/b/c group have eight common aa variations (including residue substitutions and deletions) in the aa 148-159 region encoded by cs3, and ORF367 has an additional 11-aa encoded by cs2 (Figure 6B). Indeed, Y2H assays of the truncated constructs encoded by the cs1 sequences of WA352c, WA314, orf367, and orf356 showed that they all interacted with COX11 (Figure 6C), indicating that ORF356 and ORF367 also have the potential for the COX11-interaction. Thus, we propose that ORF367 and ORF356 (with the additional 11-aa segment and/or the distinct sequence in the aa 148-159 region) can adopt special conformations that prevent the interaction of the cs1-encoded domains with COX11, whereas the variations in WA352a/b/c (the 11-aa deletion and the aa changes in the aa 148-159 region) and in WA314 (replacement of the 11-aa region and the whole cs3-encoded sequence with cs8) might cause conformational changes that allow the COX11-interaction.
To test this hypothesis, we prepared five swapped constructs (SWC1-SWC5) for Y2H analysis (Figure 6D). The results showed that the expressed products of the constructs with the 11-aa segment of ORF367 (SWC1, SWC2, and SWC4) in combination with CS1 of ORF367 or WA352c showed greatly reduced COX11-interaction strength to different extents (Figure 6D). By contrast, the combination of CS2/CS3 of WA352c with CS1 of ORF356 (SWC5) showed a slightly stronger interaction than ORF356. The reduction of the interaction strength due to presence of the 11-aa segment of ORF367 (SWC2) was more severe than that due to the ancestral-type aa 148-159 region of ORF356 (SWC3). This analysis confirmed that the 11-aa segment in ORF367 and the ancestral-type 148-159 aa sequence in ORF367 and ORF356 could interrupt the interaction with COX11. On the other hand, SWC2 (containing the 11-aa segment and WA352c-CS1) could interact weakly with COX11, but SWC1 (containing the 11-aa segment and ORF367-CS1) showed no interaction (Figure 6D), suggesting that the three aa substitutions (A225S, H233Y, and I263M) in the interaction region 1 of WA352c (Supplementary information, Figure S8) may enhance its interaction strength. This view implies that orf367 and orf356 are self-inhibited sequences that can be viewed as protogenes for CMS; the sequence changes, first in cs2 (the 33-bp deletion in generating orf356) then in cs3 (the SNPs and two 6-bp deletions in generating WA352a), and the replacement of the whole cs3/36-bp (cs2) segment in generating WA314 (Figure 4B, 4C and Supplementary information, Figure S5), are key processes for the functionalization of these CMS genes. In summary, the cs1 sequences provide the potential in these ORFs for the interaction with COX11, but the ancestral and the derived cs2/cs3 sequences act as “off” and “on” switches to the COX11-interaction, respectively, for the functionality of the CMS genes.
Purifying selection on the CMS genes
Generally, coding sequences of plant mitochondrial essential genes have relatively low nucleotide mutation rates in comparison with nuclear genes29. However, the cs1 sequences in these ORFs exhibit relatively high levels of nucleotide variation (Supplementary information, Figure S5). Our Y2H assays showed that all the cs1 sequences of these ORFs, including those from orf314, maize Zmorf179 and soybean Gmorf271, could interact with COX11, albeit with different interaction strengths (Supplementary information, Figure S9). This suggests that the cs1-contaning ORFs may have potential biological function.
Protein-coding genes generally have non-synonymous substitution rates (dN) lower than synonymous substitution rates (dS), giving the dN/dS ratios less than 1 and suggesting a purifying selection on the genes30. To learn whether these cs1-containing ORFs have undergone natural selection, we estimated the dN/dS ratios of these cs1 sequences using the one-ratio model (for the null hypothesis H0) and the branch model (for the alterative hypothesis H1) of the CODEML program in the PAML package31, assuming WA352b/c and WA314 (the later generated functional CMS genes) as the branches different from the other ORFs in H1. We then tested the maximum likelihood of H0 and H1 by likelihood rate test. The dN/dS ratios of all these branches were 0.304 or less (Figure 6E). The log likelihoods of the two hypotheses did not significantly differ, meaning that the branching cs1 sequences of WA352b/c and WA314 might not have experienced different evolutionary rates. This analysis suggests that the diverged cs1 sequences in these ORFs have been subjected to purifying selection, which might functionally constrain the protein sequences by retaining their potential to interact with COX11 and/or their unknown biological function(s).
Discussion
New gene origination plays a crucial role in the origin and evolution of new phenotypes and ultimately, biological diversity. Although being of widely interest, new gene origination is often difficult to study, especially for those very young genes not yet fixed in the species, as it is hard to identify the new genes, their pre-structures, and intermediate products from the existing populations32. The recent vast expansion of genome sequencing data, however, may greatly facilitate the finding of new genes and their source sequences. It has been proposed that genes younger than 10-30 million years have not experienced much sequence evolution and are thus a valid system to investigate the early evolution of young genes and to understand their properties32.
In this study, we provide the first identification of a relatively detailed evolutionary network by which new mitochondrial genes originated, depicting the dynamic, multiple steps, and different levels of variation in the structure, sequence, copy number and function for their microevolutionary process. We identified 10 recombinant structures in the wild rice O. rufipogon, and another one S284a in several other Oryza species, showing that CMS and CMS-like genes are widespread in the wild rice.
Based on structure/sequence comparison and functional analysis, we deduced that WA352c and the related CMS genes in O. rufipogon have most likely evolved through the following steps: (1) The rpl5/cox1/orf284 configuration, which widely exists in the AA-genome-containing wild and cultivated rice species, likely represents the original locus for the new gene formation. Multiple rearrangements among the promoter-containing sequence (from the orf284 region) and other mitochondrial genomic source sequences (including the key sequence cs1 from S284b), involving HR, MMEJ, and cNHEJ, occurred at the specific site downstream of rpl5, generating the transcription-competent pre-structure. Homologous recombination between certain sites in the plant mitochondrial genomes may be reversible and reproducible under the control of some nuclear genes (MSH1 and homologs of RecA and MutS)33,34, however, the MMEJ- and cNHEJ-based recombination events at the specific sites could be rare, suggesting that these recombinant structures were likely unique evolutionary products. (2) Formation of the CMS-like protogenes (orf367 and orf356) that are expressed but functionally self-inhibitory; (3) Generation of intermediate, functional CMS genes (WA352a/b, WA314) by gradual variations in the protogene sequence and the structures, and acquisition of the biochemical function through emancipating their COX11-interaction potential, coupled with SSS and natural (purifying) selection; (4) Formation of WA352c via further subtle sequence variation, e.g., insertion of the atp6-containing sequence into the downstream of WA352c, of which a part sequence serves as a new signal for transcription termination10. The functionalization of these CMS genes allows integration of the physiological consequence into the specific cellular process, e.g., triggering the premature tapetal programmed cell death10. These dynamic genomic and functional variations reflect the characteristics of the plant mitochondrial genomes, although the mechanisms for the mitochondrial genomic recombination, DSB repair, SSS, and heteroplasmic sorting behaviors in the plant mitochondrial genomes are still largely unknown.
Recent studies found that in the human mitochondrial genome, besides the known genes, there are dozens of new genes for short peptides, and some have roles in regulating metabolism as well as either promoting or quelling apoptosis, suggesting that mitochondrial genomes of eukaryotes contain far more functional genes than we previously appreciated35,36. In plants, the large mitochondrial genomes contain more genes and putative ORFs; many of them are of unknown function. For example, in the mitochondrial genomes of O. rufipogon and O. sativa, besides the known functional genes, there are 32 and 21 putative genes (ORFs) of unknown function, respectively21,27. This study demonstrates that during the new gene origination process in the mitochondrial genomes, other non-CMS genes, together with the CMS genes, could also be generated. We found that orf314, orf276, and orf310 likely arose from the CMS genes (WA314 and WA352a) by rearrangements (Figure 4C and 4D) that might change the expression pattern of the genes (Figure 3); this might cause gene neo-functionalization assuming that they have certain biological functions. As orf314 and orf310 are widespread in O. rufipogon and cultivated rice (Table 1), presumably they may confer some kind of beneficial effects to the plants, which have yet to be discovered.
New nuclear genes may evolve through transitory protogenes32,37. In this study we show the first case of protogenes identified in eukaryotic cytoplasmic genomes, and suggest that the generation of protogenes, as well as SSS, had served as a buffer, thus could also be a critical step for the establishment of CMS genes in natural populations. Our results also suggest that the previously recognized general features of new nuclear gene origination, including (1) recombination of existing genes and/or previously non-coding sequences, leading to a hybrid gene structure, (2) sequence evolution driven by natural selection, and (3) acquisition of new functions32,38,39, may also hold true for new mitochondrial gene formation. However, the microevolutionary process of the new mitochondrial genes we demonstrate here are much complicated than that of new nuclear genes reported so far. Notably, unlike most of the reported CMS genes that involve sequences of mitochondrial essential genes5, we show here that WA352c and the related CMS genes could have arisen from conserved mitochondrial genomic sequences of unknown function, but not necessarily involving those essential genes (such as for ATPase, cytochrome c oxidase, and ribosomal proteins), although WA352c is partially co-transcribed with rpl510.
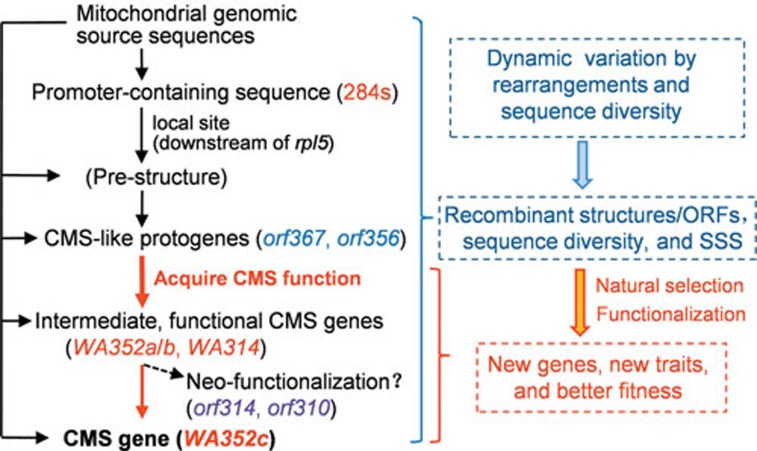
Based on these investigations, we propose a multi-step model for the formation and evolution of new mitochondrial genes, via a “multi-recombination/protogene formation/functionalization” mechanism (Figure 7). This model has some features distinct from the current models for the origination of new nuclear genes32,37,40,41,42,43,44, including the “RNA-first” and “ORF-first” routes for generation of de novo genes44. Also notably, the CMS gene origination processes from the protogene orf367 to the functional genes WA352a/b/c and WA314 were completed within a relatively short timeframe in O. rufipogon (less than two million years19). This process appears to be much faster than the origination of most nuclear new genes, such as the new Drosophila gene Umbrea whose origination has spent 15 million years45, thus reflecting the remarkably dynamic properties of plant mitochondrial genomes.
Figure 7.
A model of multi-step origination of new CMS genes in plant mitochondrial genomes by a “multi-recombination/protogene formation/functionalization” mechanism. The dynamic, multiple steps of rearrangements among the promoter-active sequence and other mitochondrial genomic source sequences at the specific site (downstream of rpl5 in this case) generated the transcription-competent pre-structures and function-inhibited protogenes, then new functional genes and the consequent new traits and better fitness evolved by acquiring the biological functions via further sequence variations, rearrangements, substoichiometric shifting (SSS), and natural selection.
According to Darwin's “principle of descent with modification”, it is anticipated that adaptive modifications within populations of a species over time, a process now referred as microevolution, would be a process of continuous and gradual changes46. Natural selection and genetic drift could operate on both facets of new gene evolution: the fixation of new gene loci and their acquisition of beneficial functions32. We provide evidence that purifying selection has acted upon the cs1-coding sequences of the CMS genes for the functional constraint. It is likely that the other ORFs might have unknown functions that have attributed to the purifying selection to their cs1 sequences during the evolution.
It has been proposed that CMS may confer female advantage47,48. However, for successful reproduction of the progeny from outcrossing, male fertility of CMS-carrying plants has to be rescued by the nuclear restorer genes. Thus, CMS genes and the corresponding nuclear restorer genes most likely have undergone co-evolution through some kind of cytoplasmic-nuclear interaction and communication5,47,48. How the cytoplasmic-nuclear genomes coordinate their adaptive evolution remains an interesting and challenging question5,47,48. The series of CMS genes identified here may provide ideal materials for studying the molecular mechanism of the interaction between the CMS and restorer genes. Furthermore, the new findings of this study will open the door to study the origin and functionalization of new mitochondrial genes in other important crops, such as maize, Brassica, wheat and sugar beet, in which multiple types of CMS cytoplasm have been identified. New mitochondrial genes may provide unique sources of genetic variation that have phenotypic consequences. These consequences can benefit natural adaptation and prove useful for crop breeding10,42,49,50,51. In addition, the new CMS genes identified in this study could greatly expand the germplasm for hybrid rice breeding.
Materials and Methods
Plant materials
A total of 808 accessions or cultivars of O. sativa, O. rufipogon, and 13 other Oryza species were selected from the “Ding's Rice Collection” preserved in South China Agricultural University for detection of the WA352c-related recombinant structures. The CMS-WA maintainer line ZS97B was used for transformation and backcrosses and Arabidopsis (ecotype Columbia) was used for transformation. All plants, including transgenic plants and backcross lines, were planted in experimental fields or in a phytotron (for growth of seedlings in some cultivars) at South China Agricultural University in Guangzhou.
PCR and hiTAIL-PCR
The sequences of S352a/b, S367, S356, S314a, S310, S276, S284a, and S284b were isolated by long PCR or hiTAIL-PCR13 with specific primers (Supplementary information, Tables S1 and S2). The amplified fragments were recovered, cloned and sequenced. The sequences were analyzed by BLAST (http://www.ncbi.nlm.nih.gov/BLAST/).
Preparation of binary constructs and plant transformation
The sequences of WA314, orf356, and orf367 were amplified by PCR with specific primers (Supplementary information, Table S3) and used to replace the orf79 sequence of the binary construct P35S::MTS:orf79, where the MTS sequence (1-105 bp) was derived from Rf1b of the rice CMS-BT system20. The binary constructs were transferred into the rice maintainer line ZS97B and Arabidopsis (ecotype Columbia) by Agrobacterium-mediated transformation52,53. The in vitro germination of Arabidopsis pollen was carried out as described54.
Quantitative RT-PCR and quantitative PCR
Structure-specific primers (Supplementary information, Tables S4 and S5) were used for semi-qPCR and qPCR to determine the relative copy number of the structures. Total RNA was extracted from rice tissues using TRIzol reagent (Invitrogen). About 1.0 μg total RNA treated with DNase (Promega) was used for first-strand cDNA synthesis using M-MLV Reverse Transcriptase (Promega) with specific primers (Supplementary information, Table S6). Expression assays were performed by qRT-PCR. The relative expression levels (i.e., ratio of target gene/atp6) were normalized with the atp6 expression as the internal standard using the ΔCT method (2−ΔCT), where ΔCT = CT (target) − CT (atp6) and shown as mean ± SD (3 biological replicates).
To analyze the copy-number variation of the structures S367, S276, and S352b, and semi qPCR and qPCR analyses of total genomic DNA of leaf tissue of the selected O. rufipogon accessions were performed using specific primers (Supplementary information, Table S4A and S4B). The copy numbers per cell were estimated using the single-copy Actin 1 gene (two allelic copies per cell) as the internal standard using the ΔCT method (2−ΔCT × 2), where ΔCT = CT (target) − CT (Actin 1).
Y2H assay
DNA fragments of WA352c, WA314, orf356, and orf367 of various lengths and the swapped constructs were amplified by PCR and Ω-PCR55 using primers shown in Supplementary information, Table 7, and cloned into the bait vector pGBKT7 (ClonTech). The prey construct of COX11 was described in a previous study10. The bait and prey constructs were co-transformed into yeast strain AH109 and the transformed cells were plated on SD/-Leu/-Trp medium and incubated at 30 °C for 2 days, then diluted and spotted onto SD/-Leu/-Trp/-His/-Ade medium and SD/-Leu/-Trp medium (as loading control), and cultured at 30 °C for 3–3.5 days.
Evolutionary analysis
The phylogenetic trees based on the cs1, cs2, and cs4 sequences of the recombinant structures (Figure 4F) were constructed using the neighbor-joining method with the MEGA software (v4.3)56. To investigate whether the mitochondria cs1 sequences in wild rice have experienced selective pressure, PAML (v4.5)31 was employed to estimate their evolutionary pressure. The alignment of the cs1 sequences (Supplementary information, Figure S5) and the phylogenetic tree construction (Figure 6E) were performed with MEGA (v4.3) based on the maximum likelihood approach. This alignment and the phylogenetic tree were used to analyze the evolutionary pressure. Subsequently, two statistical hypotheses were formulated, the null hypothesis (H0, based on the one-ratio model) assuming that all of the branches on the phylogenetic tree have the same evolutionary rate, and the alternative hypothesis (H1, based on the branch model) supposing that the branch cs1 sequences of the functional WA352b/c and WA314 have different rates of evolution from the others. The program CODEML of the PAML package was used to estimate dN (number of non-synonymous substitutions per non-synonymous site) and dS (number of synonymous substitutions per synonymous site) of each hypothesis and the dN/dS ratios. A likelihood rate test was used to analyze whether the difference is significant by calculating the maximum likelihood of these two hypotheses.
Data access
The sequences of the recombinant structures have been submitted to GenBank under accession numbers KX255850, KX255851, KX255852, KX255853, KX255854, KX255855, KX255856, KX255857, KX255858, KX255859, and KX255860.
Author Contributions
HT performed most of the experiments; XZhe, CL, YC, LC, XZha, XX, HZ, and JZ performed some of the experiments. JG, Y-GL, and HT analyzed data. Y-GL and JG conceived and supervised the project, and wrote the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
Acknowledgments
We thank X Liu for providing some of the wild and cultivated rice materials preserved in South China Agricultural University, and H Yu for assistance in the sequence analysis with the CODEML program. We also thank H Wang, D Charlesworth, W Wang, C-I Wu, X-L He, H Ma, Q Zhang, Y Ouyang, and K Tsunewaki for comments on this study and/or the manuscript. This work was supported by grants from the National Nature Science Foundation of China (31230052), the Ministry of Science and Technology of China (2013CBA01401 and 2013CB126904), the Nature Science Foundation of Guangdong Province, China (2014A030310399), and the Postdoctoral Science Foundation of China (2015M570717).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Primers used for detection of the recombinantstructures (Figure 1, Figure S4).
Specific primers used for hiTAIL-PCR (Figure 1).
Primers used for preparation of the plant transformation vector constructs (Figure 2A).
Primers used for quantitative PCR (Figure 5B).
Primers used for qRT-PCR (Figure 3).
Primers used for construction of yeast two-hybrid vectors (Figure 6A, C, D and Figure S7).
Male sterility and fertility and the co-segregation with the transgenes in the rice and Arabidopsis transgenic plants.
Fertility phenotypes of Arabidopsis T1 plants with MTS-WA314.
Male phenotype of the backcrossed rice lines.
Detection of therpl5/cox1/orf284 configuration in Oryza species.
Comparison among the conserved sequences of the recombinant structures and those from other plant species as references.
The repeat sites for recombination events among the donor (source) sequences to generate the new structures.
Coexistence of S314a, S276 and S310 and their substoichiometric shifting.
Alignment of the protein sequences of the CMS genes and CMS-related ORFs.
Yeast two-hybrid assay of the COX11-interaction of the cs1-encoded polypeptides of the CMS genes and the related ORFs.
References
- Dyall SD, Brown MT, Johnson PJ. Ancient invasions: from endosymbionts to organelles. Science 2004; 304:253–257. [DOI] [PubMed] [Google Scholar]
- Lynch M, Koskella B, Schaack S. Mutation pressure and the evolution of organelle genomic architecture. Science 2006; 311:1727–1730. [DOI] [PubMed] [Google Scholar]
- Woloszynska M. Heteroplasmy and stoichiometric complexity of plant mitochondrial genomes-though this be madness, yet there's method in't. J Exp Bot 2010; 61:657–671. [DOI] [PubMed] [Google Scholar]
- Arrieta-Montiel MP, Mackenzie SA. Plant mitochondrial genomes and recombination. Plant Mitochon 2011; 1:65–82. [Google Scholar]
- Chen L, Liu YG. Male sterility and fertility restoration in crops. Annu Rev Plant Biol 2014; 65:579–606. [DOI] [PubMed] [Google Scholar]
- Laser KD, Lersten NR. Anatomy and cytology of microsporogenesis in cytoplasmic male angiosperms. Bot Rev 1972; 38:425–454. [Google Scholar]
- Huang X, Kurata N, Wei X, et al. A map of rice genome variation reveals the origin of cultivated rice. Nature 2012; 490:497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Yuan L. Hybrid Rice Breeding in China. Innovative Approaches to Rice Breeding 1980; Los Banos, Philippines, 35–51.
- Cheng SH, Zhuang JY, Fan YY, Du JH, Cao LY. Progress in research and development on hybrid rice: a super-domesticate in China. Ann Bot 2007; 100:959–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D, Xu H, Liu Z, et al. A detrimental mitochondrial-nuclear interaction causes cytoplasmic male sterility in rice. Nat Genet 2013; 45:573–577. [DOI] [PubMed] [Google Scholar]
- Tang H, Luo D, Zhou D, et al. The rice restorer Rf4 for wild–abortive cytoplasmic male sterility encodes a mitochondrial-ocalized PPR protein that functions in reduction of WA352 transcripts. Mol Plant 2014; 7:1497–1500. [DOI] [PubMed] [Google Scholar]
- Khush GS. Origin, dispersal, cultivation and variation of rice. Plant Mol Biol 1997; 35:25–34. [PubMed] [Google Scholar]
- Liu YG, Chen Y. High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques 2007; 43:649–656. [DOI] [PubMed] [Google Scholar]
- Okazaki M, Kazama T, Murata H, Motomura K, Toriyam K. Whole mitochondrial genome sequencing and transcriptional analysis to uncover an RT102-type cytoplasmic male sterility-associated candidate gene derived from Oryza rufipogon. Plant Cell Physiol 2013; 54:1560–1568. [DOI] [PubMed] [Google Scholar]
- Igarashi K, Kazama T, Motomura K, Toriyama K. Whole genomic sequencing of RT98 mitochondria derived from Oryza rufipogon and northern blot analysis to uncover a cytoplasmic male sterility-associated gene. Plant Cell Physiol 2013; 54:237–243. [DOI] [PubMed] [Google Scholar]
- Fujii S, Kazama T, Yamada M, Toriyama K. Discovery of global genomic re-organization based on comparison of two newly sequenced rice mitochondrial genomes with cytoplasmic male sterility-related genes. BMC Genomics 2010; 11:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazama T, Itabashi E, Fujii S, Nakamura T, Toriyama K. Mitochondrial ORF79 levels determine pollen abortion in cytoplasmic male sterile rice. Plant J 2016; 85:707–716. [DOI] [PubMed] [Google Scholar]
- Zou XH, Zhang FM, Zhang JG, et al. Analysis of 142 genes resolves the rapid diversification of the rice genus. Genome Biol 2008; 9:R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Ge S. Phylogenetic relationships among A-genome species of the genus Oryza revealed by intron sequences of four nuclear genes. New Phytol 2005; 167:249–265. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zou Y, Li X, et al. Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. Plant Cell 2006; 18:676–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notsu Y, Masood S, Nishikawa T, et al. The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol Genet Genomics 2002; 268:434–445. [DOI] [PubMed] [Google Scholar]
- Zhang QY, Liu YG. Rice mitochondrial genes are transcribed by multiple promoters that are highly diverged. J Integr Plant Biol 2006; 48:1473–1477. [Google Scholar]
- Paull TT, Gellert M. A mechanistic basis for Mre11-directed DNA joining at microhomologies. Proc Natl Acad Sci USA 2000; 97:6409–6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata M, Sasaki MS, Sonoda E, et al. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J 1998; 17:5497–5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert I, Vu GTH. Genome stability and evolution: attempting a holistic view. Trends Plant Sci 2016; 21:749–757. [DOI] [PubMed] [Google Scholar]
- Tian X, Zheng J, Hu S, Yu J. The rice mitochondrial genomes and their variations. Plant Physiol 2006; 140:401–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentolila S, Stefanov S. A reevaluation of rice mitochondrial evolution based on the complete sequence of male-fertile and male-sterile mitochondrial genomes. Plant Physiol 2012; 158:996–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Hu S, Zhang G, et al. The organelle genomes of Hassawi rice (Oryza sativa L.) and its hybrid in Saudi Arabia: genome variation, rearrangement, and origins. PLoS One 2012; 7:e42041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T, Mikami T. Organization and variation of angiosperm mitochondrial genome. Physiol Plant 2007; 129:6–13. [Google Scholar]
- Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol 1998; 15:568–573. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 2007; 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Long M, VanKuren NW, Chen S, Vibranovski MD. New gene evolution: little did we know. Annu Rev Genet 2013; 47:307–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shedge V, Arrieta-Montiel M, Christensen AC, Mackenzie SA. Plant mitochondrial recombination surveillance requires unusual RecA and MutS homologs. Plant Cell 2007; 19:1251–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrieta-Montiel MP, Shedge V, Davila J, Christensen AC, Mackenzie SA. Diversity of the Arabidopsis mitochondrial genome occurs via nuclear-controlled recombination activity. Genetics 2009; 183:1261–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Zeng J, Drew BG, et al. The mitochondrial-derived peptide MOTS-c promotes metabolic homeostasis and reduces obesity and insulin resistance. Cell Metab 2015; 21:443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb LJ, Lee C, Xiao J, et al. Naturally occurring mitochondrial-derived peptides are age-dependent regulators of apoptosis, insulin sensitivity, and inflammatory markers. Aging 2016; 8:796–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvunis AR, Rolland T, Wapinski I, et al. Proto-genes and de novo gene birth. Nature 2012; 487:370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Zhang J, Alvarez C, Llopart A, Long M. The origin of the jingwei gene and the complex modular structure of its parental gene, Yellow Emperor, in Drosophila melanogaster. Mol Biol Evol 2000; 17:1294–1301. [DOI] [PubMed] [Google Scholar]
- Zhang J, Dean AM, Brunet F, Long M. Evolving protein functional diversity in new genes of Drosophila. Proc Natl Acad Sci USA 2004; 101:16246–16250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A. Darwinian alchemy: human genes from noncoding DNA. Genome Res 2009; 19:1693–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Dong Y, Jiang Y, Jiang H, Cai J, Wang W. A de novo originated gene depresses budding yeast mating pathway and is repressed by the protein encoded by its antisense strand. Cell Res 2010; 20:408–420. [DOI] [PubMed] [Google Scholar]
- Ding Y, Zhou Q, Wang W. Origins of new genes and evolution of their novel functions. Annu Rev Ecol Evol Syst 2012; 43:345–363. [Google Scholar]
- Neme R, Tautz D. Evolution: dynamics of de novo gene emergence. Curr Biol 2014; 24:R238–R240. [DOI] [PubMed] [Google Scholar]
- McLysaght A, Guerzoni D. New genes from non-coding sequence: the role of de novo protein-coding genes in eukaryotic evolutionary innovation. Phil Trans R Soc B 2015; 370:20140332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross BD, Rosin L, Thomae AW, et al. Stepwise evolution of essential centromere function in a Drosophila neogene. Science 2013; 340:1211–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznick DN, Ricklefs RE. Darwin's bridge between microevolution and macroevolution. Nature 2009; 457:837–842. [DOI] [PubMed] [Google Scholar]
- Caruso CM, Case AL, Bailey MF. The evolutionary ecology of cytonuclear interactions in angiosperms. Trends Plant Sci 2012; 17:638–643. [DOI] [PubMed] [Google Scholar]
- Greiner S, Bock R. Tuning a menage a trois: co-evolution and co-adaptation of nuclear and organellar genomes in plants. Bioessays 2013; 35:354–365. [DOI] [PubMed] [Google Scholar]
- Shedge V, Davila J, Arrieta-Montiel MP, Mohammed S, Mackenzie SA. Extensive rearrangement of the Arabidopsis mitochondrial genome elicits cellular conditions for thermotolerance. Plant Physiol 2010; 152:1960–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YZ, Arrieta-Montiel MP, Virdi KS, et al. MutS HOMOLOG1 is a nucleoid protein that alters mitochondrial and plastid properties and plant response to high light. Plant Cell 2011; 23:3428–3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virdi KS, Laurie JD, Xu YZ, et al. Arabidopsis MSH1 mutation alters the epigenome and produces heritable changes in plant growth. Nat Commun 2015; 6:6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 1994; 6:271–282. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 1998; 16:735–743. [DOI] [PubMed] [Google Scholar]
- Fan LM, Wang YF, Wang H, Wu WH. In vitro Arabidopsis pollen germination and characterization of the inward potassium currents in Arabidopsis pollen grain protoplasts. J Exp Bot 2001; 52:1603–1614. [PubMed] [Google Scholar]
- Chen L, Wang F, Wang X, Liu Y-G. Robust one-tube Ω-PCR strategy accelerates precise sequence modification of plasmids for functional genomics. Plant Cell Physiol 2013; 54:634–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 2007; 24:1596–1599. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used for detection of the recombinantstructures (Figure 1, Figure S4).
Specific primers used for hiTAIL-PCR (Figure 1).
Primers used for preparation of the plant transformation vector constructs (Figure 2A).
Primers used for quantitative PCR (Figure 5B).
Primers used for qRT-PCR (Figure 3).
Primers used for construction of yeast two-hybrid vectors (Figure 6A, C, D and Figure S7).
Male sterility and fertility and the co-segregation with the transgenes in the rice and Arabidopsis transgenic plants.
Fertility phenotypes of Arabidopsis T1 plants with MTS-WA314.
Male phenotype of the backcrossed rice lines.
Detection of therpl5/cox1/orf284 configuration in Oryza species.
Comparison among the conserved sequences of the recombinant structures and those from other plant species as references.
The repeat sites for recombination events among the donor (source) sequences to generate the new structures.
Coexistence of S314a, S276 and S310 and their substoichiometric shifting.
Alignment of the protein sequences of the CMS genes and CMS-related ORFs.
Yeast two-hybrid assay of the COX11-interaction of the cs1-encoded polypeptides of the CMS genes and the related ORFs.