Targeted therapy using BRAF and MEK inhibitors (BRAFi/MEKi) has transformed melanoma treatment, improving the quality of life and outcomes of patients with metastatic disease. However, responses to BRAFi/MEKi are often incomplete and transient as tumors rapidly become resistant to therapy. Numerous mechanisms of drug resistance identified using in vitro and mouse models have been subsequently validated in patient samples. Known resistance mechanisms mainly involve reactivation of the MAPK pathway, via BRAFV600E amplification or splicing variants, NRAS or MEK1/2 mutations. Activation of alternative survival pathways such as RTK amplification and mutation of nodes within the PI3K/AKT pathway can also occur. Nonetheless, subsets of resistant tumors do not harbor any of the known resistance conferring alterations. Uncovering additional adaptive mechanisms will undoubtedly help develop more effective strategies to prevent and overcome drug resistance.
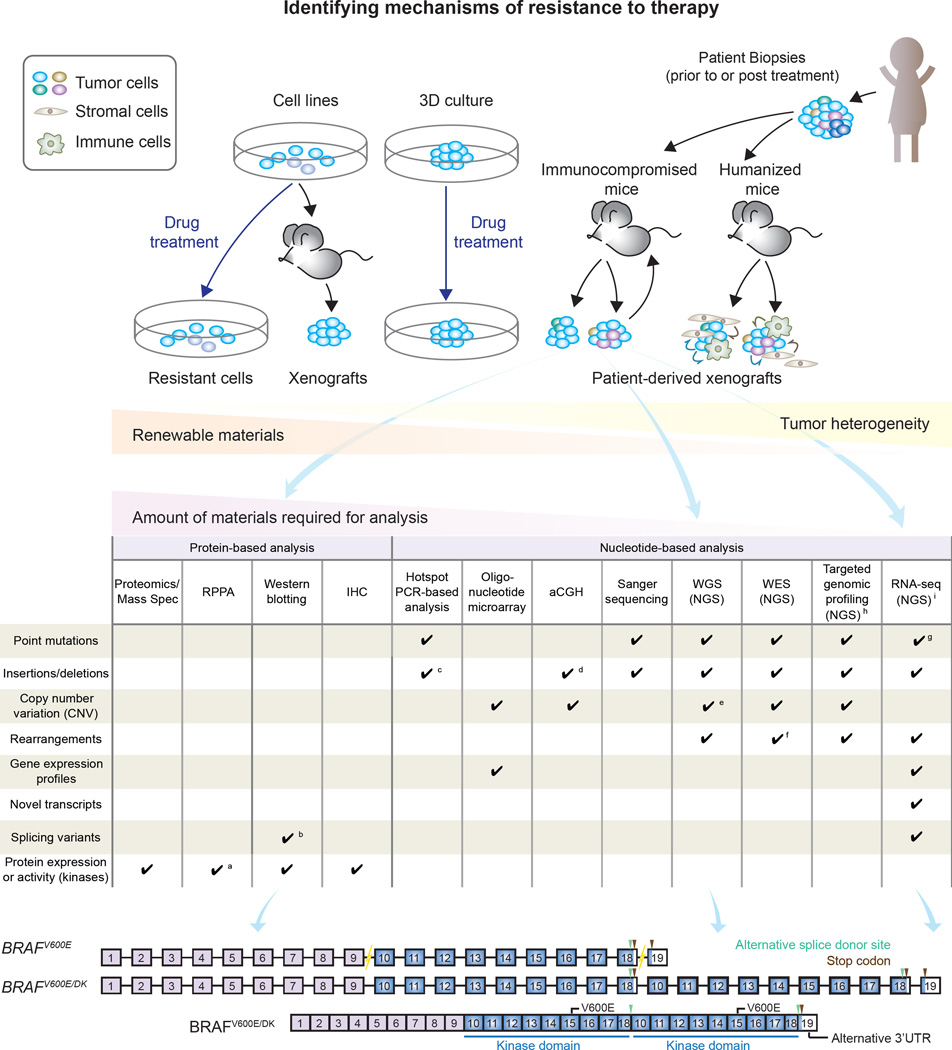
Kemper and colleagues established and thoroughly characterized an extensive collection of patient-derived xenografts (PDX) models derived from metastatic melanoma samples with BRAFV600E mutation, NRASQ61 mutations, or unknown driver mutations (BRAFWT/NRASWT). Moreover, the PDX bank also included paired BRAFV600E tumor samples obtained from the same patients prior to BRAFi treatment or at progression. Analysis of biomarkers, chromosomal aberrations, mutational and RNA expression profiling indicated that melanoma PDXs retain the molecular and biological features of the original patient tumors. Notably, these PDXs preserve the genetic complexity and heterogeneity of melanoma, along with the spectrum of resistance patterns to targeted drugs that were characterized as either “intrinsic” or “acquired”. Importantly, many previously identified mechanisms of drug resistance were recapitulated in this model. This PDX collection, offers a renewable tissue resource, allowing for the discovery of novel resistance mechanisms that may otherwise be missed using limited tumor material (Figure 1). These models can also be used for preclinical testing of personalized therapies and to optimize clinically meaningful drug combinations. Leveraging this PDX collection, the team led by Daniel Peeper identified an unusually larger BRAFV600E protein (~140kDa) exclusively expressed in BRAFi resistant tumors (Figure 1C). This higher molecular weight BRAFV600E turned out to contain a duplicated kinase domain (BRAFV600E/DK), which conferred resistance to BRAFi. Furthermore, the authors tested in the PDX models possible therapies to overcome resistance mediated by BRAFV600E/DK.
Figure 1. Models and strategies to identify mechanisms of drug resistance in melanoma.
(A) Strategies used to model drug resistance in melanoma include chronically exposing cells in vitro or in vivo to drugs, or propagating tumors derived from patients who progress on therapy in immunocompromised mice (patient-derived xenografts/PDXs). Additional models that have not been extensively exploited thus far include 3D cultures and humanized mice. PDXs models closely resemble the original patient samples and preserve tumor heterogeneity. This model provides a renewable source of tumor material for identification of resistance mechanisms by analyses requiring large amounts of sample. (B) Various molecular and biochemical approaches can be used to investigate mechanisms mediating drug resistance. (C) By integrating whole genome sequencing, Western blotting and RNA sequencing, Kemper et al. identified a novel BRAFV600E mutant (BRAFV600E/DK). BRAFV600E/DK contains duplicated kinase domains (DK), and confers resistance to first generation BRAF inhibitors in melanoma.
a. Limited to antibodies on the arrays. b. Detecting those having discernible molecular weight/motility in electrophoresis. c. Limited to small alterations within targeted areas. d. Detecting alterations larger than 100 kb. e. Precisely delineating the breakpoints. f. Limited to those within or near exons. g. Restricted to targeted regions. h. Limited to expressing (coding and non-coding) regions. i. Challenging in low-expressing genes, affected by RNA editing.
Abbreviations: Mass Spec: mass spectrometry. RPPA: reverse phase protein array. IHC: immunohistochemistry. aCGH: array comparative genomic hybridization. WGS: Whole genome sequencing. NGS: next generation sequencing. WES: Whole exome sequencing. RNA-seq: RNA sequencing.
Gene duplication is a fairly common genomic mechanism underlying molecular adaptation to highly selective pressure and has been associated with resistance to antibiotics and cancer therapy. BRAF kinase domain duplication (KDD) has been previously reported in gliomas (Rodriguez et al., 2012). More recently, analysis of 50,000 samples spanning multiple tumor types found that BRAF-KDD comprises 0.5% of BRAF alterations (Klempner et al., 2016). Kemper’s is the first report of a structural alteration on BRAFV600E protein identified in BRAFi-resistant melanomas. The tandem kinase domain duplication (KDD) results from breaks in introns of the BRAFT1799A gene between exons 9–10 and exons 18–19. Using an alternative splice donor site in exon 18, the mRNA splices out the conventional stop codon in exon 18, leading to duplication of exons 10–18 and an alternative 3’UTR from exon 19 (Figure 1C). The authors further demonstrated that BRAFV600E/DK was sensitive to a pan-RAF inhibitor (LY3009120) that targets both RAF monomers and dimers thereby inhibiting downstream MAPK signaling. This pan-RAF inhibitor is currently being evaluated in phase I clinical trials and could provide a therapeutic option for patients who carry this resistance mechanism. This novel mechanism of drug resistance (BRAFV600E/DK) is estimated to occur in 10% of melanomas that progress on BRAFi. An application of these findings would be adding this molecular trait into a future tumor profiling platform aimed at matching driver molecular alterations with the most suitable treatment options.
The molecular basis whereby this mutation causes resistance is yet to be investigated. KDD mutants in other kinases have been identified, including EGFR, FGFR1 and BRAF and could offer some clues for how BRAFV600E/DK confers resistance. For example, EGFR with a duplicated kinase domain can undergo intra-molecular dimerization becoming constitutively active (Gallant et al., 2015). Since BRAFV600E/DK is resistant to BRAF inhibitors that target BRAF monomers (and fail to occupy both ATP binding sites of RAF dimers), but is sensitive to a pan-RAF inhibitor that targets both ATP binding sites of RAF dimers, it is tempting to speculate that the tandem kinase domain duplication may create a situation similar to RAF dimerization. Resolving the mode of action of BRAFV600E/DK could help develop strategies to inhibit this mutant form of BRAF, as well as provide insights for other KDD mutants, as this is not exclusive to BRAF. Intragenic partial duplication is likely a common mechanism leading cancer cells to acquire new protein isoforms, as tumor cells are exposed to diverse stressors which can trigger genomic instability and alternative RNA splicing. Occasionally, alternative splicing leads to a different 3’UTR (as in the case of BRAFV600E/DK), which may affect regulation of mRNA expression, fostering evolution of drug resistance tumors. However, whether the alternative 3’UTR in the BRAF gene is utilized under normal physiological settings remains to be determined.
Why hasn’t this novel BRAFV600E/DK been previously found, despite rigorous efforts to uncover mechanisms driving BRAFi-resistance in melanoma? One possible reason is the inability of PCR-based hotspot analysis and standard exome sequencing to detect this type of aberration (Figure 1B). With more whole-genome sequencing analysis to come, more mechanisms of this type are likely to be uncovered. Additionally, the availability and characterization of PDX models will likely expand the spectrum of resistance mechanisms. Compared to in vitro culture and cell line-derived xenograft models, PDXs provide a more powerful and physiologically relevant system that more faithfully resembles and preserves the original patient’s tumor architecture and heterogeneity (Figure 1A).
Furthermore, the readiness of almost unlimited PDX-derived material, offers a unique opportunity for using experimental methods (e.g. reverse phase protein arrays and mass spectrometry) that are much more restricted with patient specimens due to the paucity of material (Figure 1). Particularly, PDX platforms could be used to identify proteomic alterations that contribute to drug resistance and hence expand the spectrum of tumor cell autonomous resistance mechanisms. Nevertheless, PDX models still have a number of limitations. In addition to being both costly and time consuming, discovery and analysis of non-cell autonomous mechanisms is much more limited. This is partly due to the replacement of human stroma by mouse elements and the lack of immune cell components in the PDX model; a platform with those components reconstituted such as humanized mice is required and eagerly awaited(Figure 1A).
In addition to BRAFV600E, this well-characterized PDX collection also includes NRASQ61 and BRAFWT/NRASWT tumors. Along with short-term cultures and similar genetically and clinically-annotated models (Krepler et al., 2016; Kwong et al., 2015), this platform offers an invaluable resource to study melanoma biology, identify tumor vulnerabilities or novel drug resistance mechanisms, as well as to screen and optimize drug combinations. Of course, we need to think about models that will enable us to study determinants of sensitivity and resistance to immunotherapies and the next generation of therapies that will likely involve combinations of targeted agents with immune checkpoint inhibitors. Resources like the one established by Kemper et al. will enable the melanoma research community to continue moving forward in its quest for improving the treatment options available for melanoma patients.
Acknowledgments
Work in our laboratory is supported by NIH/NCI (grants K01 CA175269, PO1 CA114046, P30 CA010815), American Cancer Society, V Foundation for Cancer Research, Melanoma Research Alliance, Melanoma Research Foundation and Martha W. Rogers Trust.
References
- Gallant JN, Sheehan JH, Shaver TM, Bailey M, Lipson D, Chandramohan R, Red Brewer M, York SJ, Kris MG, Pietenpol JA, et al. EGFR Kinase Domain Duplication (EGFR-KDD) Is a Novel Oncogenic Driver in Lung Cancer That Is Clinically Responsive to Afatinib. Cancer Discov. 2015;5:1155–1163. doi: 10.1158/2159-8290.CD-15-0654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klempner SJ, Bordoni R, Gowen K, Kaplan H, Stephens PJ, Ou SH, Ali SM. Identification of BRAF Kinase Domain Duplications Across Multiple Tumor Types and Response to RAF Inhibitor Therapy. JAMA Oncol. 2016;2:272–274. doi: 10.1001/jamaoncol.2015.4437. [DOI] [PubMed] [Google Scholar]
- Krepler C, Xiao M, Sproesser K, Brafford PA, Shannan B, Beqiri M, Liu Q, Xu W, Garman B, Nathanson KL, et al. Personalized Preclinical Trials in BRAF Inhibitor-Resistant Patient-Derived Xenograft Models Identify Second-Line Combination Therapies. Clin Cancer Res. 2016;22:1592–1602. doi: 10.1158/1078-0432.CCR-15-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong LN, Boland GM, Frederick DT, Helms TL, Akid AT, Miller JP, Jiang S, Cooper ZA, Song X, Seth S, et al. Co-clinical assessment identifies patterns of BRAF inhibitor resistance in melanoma. J Clin Invest. 2015;125:1459–1470. doi: 10.1172/JCI78954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez FJ, Ligon AH, Horkayne-Szakaly I, Rushing EJ, Ligon KL, Vena N, Garcia DI, Cameron JD, Eberhart CG. BRAF duplications and MAPK pathway activation are frequent in gliomas of the optic nerve proper. J Neuropathol Exp Neurol. 2012;71:789–794. doi: 10.1097/NEN.0b013e3182656ef8. [DOI] [PMC free article] [PubMed] [Google Scholar]