Abstract
Staphylococcus sciuri is known as an opportunistic pathogen colonizing domesticated animals and has also been associated with wound infections in humans. Particularly over the last decade, oxacillin (methicillin) resistant strains had been emerged, which now increase the medical relevance of this species. This report describes the identification of an oxacillin-resistant S. sciuri isolate from a wound infection of a horse. We determined the absence of coagulase and hyaluronidase activity and analysed the antibiotic resistance profile.
Keywords: Colonization, Horse, Oxacillin resistance, Staphylococcus sciuri
Introduction
Staphylococcus sciuri is a member of the S. sciuri-species group composed of coagulase-negative and novobiocin-resistant bacteria (Nemeghaire et al., 2014a). This group includes S. sciuri (with three subspecies), S. lentus, S. vitulinus, S. fleurettii and S. stepanovicii (Becker et al., 2014a), which are in general considered as commensal animal-associated species (Kloos et al., 1976). S. sciuri possesses a certain pathogenic potential and is able to induce infections in both, animals (Frey et al., 2013; Dos Santos et al., 2015) and humans (Stepanovic et al., 2003). Some isolates of the S. sciuri group are known to carry different homologues of the methicillin resistance genes mecA, B and C and display methicillin/oxacillin resistance (Becker et al., 2014a, b; Harrison et al., 2014).
In the present study, we report the identification of an oxacillin-resistant S. sciuri isolate from a purulent skin lesion of a horse, determined activity of coagulase and hyaluronidase and characterized the antibiotic resistance profile.
Case Details
A “Hannoveraner Hengst” at the age of ten presented a purulent skin lesion on the right forehand pastern. The medical prehistory claimed repeated episodes of purulent skin infections, foremost on the bridge of the nose, which poorly healed untreated within a couple of weeks.
One year later, a closed, swollen abscess-like structure was developed on the right forehand pastern. The abscess erupted within two to three days and presented a bloody skin lesion of ~4 cm in length and ~1 cm in width (Fig. 1).
Fig. 1.
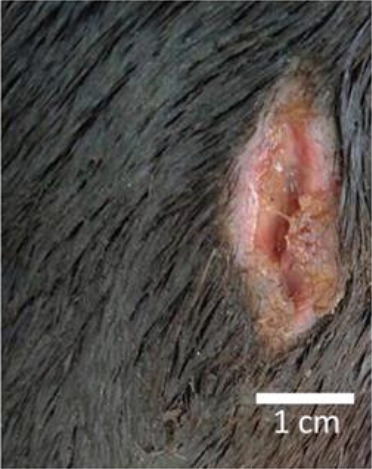
Erupted abscess-like skin lesion of ~4 cm in length and ~1 cm in width on the right forehand pastern of the horse.
The skin lesion became purulent and was treated with non-antibiotic zinc-containing ointment. No systemic clinical signs were detected.
Culture-based analyses/Identification of S. sciuri
A swab specimen was taken from the purulent skin lesion and cultured onto Columbia blood agar plates (Becton Dickenson) containing 5% sheep blood at 37°C and 5% CO2. Morphological analyses revealed growth of uniform, non-hemolytic, white opaque colonies after 24 h at 37 °C (Fig. 2A, Fig. 2B). Light microscopy and Gram-stain indicated a pure culture of Gram positive cocci clustered in grape-like aggregates (Fig. 2C).
Fig. 2.
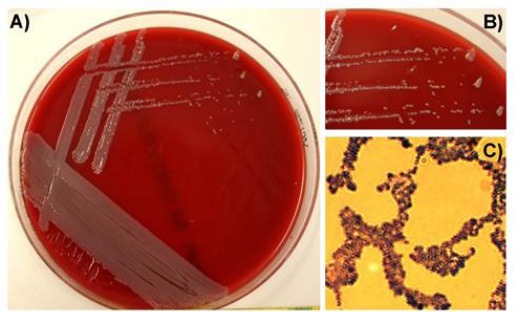
(A): Morphology of Staphylococcus sciuri on Columbia blood agar plates. (B): The zoom in indicates white to light grey staphylococcal colonies without any haemolytic activity. (C): Gram-stain visualized Gram-positive, coccoid bacteria, clustered in grape-like structures.
Strain identification by sequencing of 16S rRNA was conducted from a single colony as described elsewhere (Weisburg, et al., 1991) using the following oligonucleotides: forward primer: 27f 5’-AGA GTT TGA TCM TGG CTC AG-3’, reverse primer: 1492 r 5’-CGG TTA CCT TGT TAC GAC TT-3’ and was repeated three times using colony material from the same culture plate. After purification of PCR-products using QIA quick PCR Purification Kit (Qiagen) sequencing procedure was performed by GATC-Biotech (Germany).
Blast® search (provided by NCBI, NLM, Bethesda, USA) identified S. sciuri in all three independent probes as primary infectious agent. The listed blast results presented in Table 1 confirm the high sequence identity with up to 98% query coverage to S. sciuri in all of the tested three independent probes. Results further point to the identification of carnicatus or rodentium as respective subspecies but the nominal difference to the results given for the third potential subspecies sciuri were not significant enough to allow a final determination of the subspecies.
Table 1.
16SrRNA-based sequencing results from three independent culture probes as obtained by blast®-sequence alignment (NCBI; NLM).
| sample | hit | S. sciuri | |||
|---|---|---|---|---|---|
| subsp. | carnicatus | rodentium | sciuri | ||
| 1 | 1 | Strain | GTC 1227 | GTC 844 | ATCC 29062 |
| Query cover | 98% | 98% | 98% | ||
| Ident | 85% | 85% | 85% | ||
| Accession | NR 041327.1 | NR 041328.1 | AJ421446.1 | ||
| 2 | Strain | GTC 1227 | GTC 844 | ATCC 29062 | |
| Query cover | 98% | 98% | 40% | ||
| Ident | 85% | 85% | 85% | ||
| Accession | AB233331.1 | AB233332.1 | AY688097.1 | ||
| 3 | Strain | ATCC 700058 | ATCC 70061 | ||
| Query cover | 40% | 40% | |||
| Ident | 82% | 82% | |||
| Accession | AY688095.1 | AY688096.1 | |||
| 2 | 1 | Strain | GTC 1227 | GTC 844 | ATCC 29062 |
| Query cover | 96% | 96% | 95% | ||
| Ident | 86% | 86% | 86% | ||
| Accession | |||||
| 2 | Strain | GTC 1227 | GTC 844 | ATCC 29062 | |
| Query cover | 96% | 96% | 35% | ||
| Ident | 86% | 86% | 82% | ||
| Accession | AB233331.1 | AB233332.1 | AY688097.1 | ||
| 3 | Strain | ATCC 700058 | ATCC 70061 | ||
| Query cover | 35% | 35% | |||
| Ident | 82% | 82% | |||
| Accession | AY688095.1 | AY688096.1 | |||
| 3 | 1 | Strain | GTC 1227 | GTC 844 | ATCC 29062 |
| Query cover | 95% | 95% | 95% | ||
| Ident | 86% | 86% | 86% | ||
| Accession | NR 041327.1 | NR 041328.1 | AJ421446.1 | ||
| 2 | Strain | GTC 1227 | GTC 844 | ATCC 29062 | |
| Query cover | 95% | 95% | 37% | ||
| Ident | 86% | 86% | 82% | ||
| Accession | AB233331.1 | AB233332.1 | AY688097.1 | ||
| 3 | Strain | ATCC 700058 | ATCC 70061 | ||
| Query cover | 37% | 37% | |||
| Ident | 82% | 82% | |||
| Accession | AY688095.1 | AY688096.1 | |||
A bacterial colonization analysis from swabs taken from the horse nostrils did not identified S. sciuri as constant colonizer of the horse, but identified typical members of horse microflora such as Aeromonas viridans and S. vitulinus by Maldi Tof.
Verification of strain-identification and MIC-determination
Species identification of the S. sciuri isolate was confirmed using the standardized API STAPH V5.0 system and revealed an ID value of 88.4%. The specification of the respective isolate was further confirmed using the bioMérieux VITEK®2 system (Germany) according to the manufacturer recommendations and also independently by the “National Reference Center of Staphylococci and Enterococci” of the Robert Koch Institute (RKI) in Wernigerode, Germany.
Furthermore, automated antimicrobial susceptibility testing was performed at the “National Reference Center for Staphylococci and Enterococci” of the Robert Koch Institute in Wernigerode, Germany via microbouillon-dilution, including the following antibiotics: ß-lactams (benzylpenicillin, oxacillin), macrolides (erythromcycin), lincosamides (clindamycin), oxazolidinone (linezolid), fucidanes (fusidic acid), aminoglycosides (gentamycin), ansamycins (rifampicin), tetracycline (oxytetracyclin), glycopeptides (vancomycin, teicoplanin), gylcylcyclins (tigecyclin), fluoroquinolons (ciprofloxacin, moxifloxacin), cyclic lipopeptides (daptomycin), the epoxid fosfomycin and the folate synthesis inhibitor cotrimoxazol. MIC value determination was evaluated according to the EUCAST standards for human medicine and revealed sensitivity to most of the tested antibiotics (Table 2).
Table 2.
Analyses of MIC of different antibiotics by the “National Reference Center of Staphylococci and Enterococci” of the Robert Koch Institute in Wernigerode, Germany, evaluated by EUCAST-based interpretation.
| Antibiotics | MIC (µg/ml) | interpretation |
|---|---|---|
| Benzylpenicillin | 0.125 | S |
| Oxacillin | 1.0 | R |
| Fosfomycin | 8.0 | R |
| Gentamycin | 0.5 | S |
| Linezolide | 1.0 | S |
| Erythromycin | 0.5 | S |
| Clindamycin | 0.25 | S |
| Oxytetracyclin | 0.5 | S |
| Tigecyclin | 0.125 | S |
| Vancomycin | 1.0 | S |
| Teicoplanin | 4.0 | R |
| Ciprofloxacin | 0.5 | S |
| Moxifloxacin | 0.25 | S |
| Daptomycin | 1.0 | S |
| Co-trimoxazole | 0.5 | S |
| Rifampicin | 0.063 | S |
| Fusidic acid | 8.0 | R |
Interestingly, the antibiotic resistance profile covering 17 antibiotics indicated a resistance against fosfomycin, fusidic acid and teicoplanin (Table 2).
Teicoplanin resistance was additionally tested by plating a higher inoculum. In sum, the results point to a heterogenic teicoplanin resistant strain. Moreover, according to the EUCAST standards, the S. sciuri strain is resistant against oxacillin. This is remarkable since detection of the common resistance genes mecA and mecC via specific PCR by Reference Center of the RKI (Wernigerode, Germany) was negative.
Additionally, the particular S. sciuri was tested negative for coagulase and hyaluronidase activity, respectively. The activities of both virulence factors was analysed by tube test with human and rabbit plasma and by decapsulation test with S. equi as described by Essers and Radebold (1980).
Discussion
S. sciuri is mostly recovered from skin and mucous membrane of animals and has long been considered as a non-pathogenic commensal bacterium (Adegoke, 1986). During the last decade of years, it has been associated with several cases of bovine mastitis (Lüthje and Schwarz, 2006; Nam et al., 2010; Frey et al., 2013), as well as from goats suffering from peste des petites ruminants (PPR) (Ugochukwu and Agwu, 1991), from cases of canine dermatitis (Hauschild and Wójcik, 2007; Hauschild et al., 2010), and from several outbreaks of fatal exudative epidermitis in piglets (Chen et al., 2007; Nemeghaire et al., 2014c).
The recurrent manifestation of skin lesions monitored in the present case, initially suggested a permanent colonization of the horse with S. sciuri. In contrast to several reports pointing to nasal colonization of horses with S. sciuri, so far no data on permanent skin colonization has been reported for horses (Bagcigil et al., 2007; Aslantas et al., 2012; Karakulska et al., 2012). The lack of S. sciuri in cultures of nasal swabs in this case may point to the occurrence of a single colonization event or may suggest repeated episodes of temporary colonization.
Interestingly, a transmission in between healthy domestic animals colonized with S. sciuri was repeatedly observed (Moodley and Guardabassi, 2009; Aslantas et al., 2012). This transmission may be promoted by insects serving as transmission vectors. In this respect, a report also suggested that the possible source of S. sciuri colonization in surgical wounds may be flies perching on open wounds (Kolawole and Shittu, 1997). Thus, it is assumed that frequent contact with healthy domestic and farm animals may also contribute to an at least temporary colonization of the skin, and subsequently the wounds, by S. sciuri (Kloos et al., 1976; Nemeghaire et al., 2014b).
Despite the rare occurrence of S. sciuri in humans (Marsou et al., 1999; Couto et al., 2000; Nagase et al., 2002), some reports furthermore point to the role of S. sciuri as opportunistic pathogens isolated from various clinical specimen and causing serious infections in humans such as endocarditis, peritonitis, septic shock, and wound infections (Hedin and Widerstrom, 1998; Wallet et al., 2000; Horii et al., 2001; Stepanovic et al., 2002, 2003;). Moreover, despite the lack of data regarding S. sciuri colonization of the handler, a recurrent transmission from the handler to the horse cannot be excluded. The isolated S. sciuri strain was tested negative for coagulase and hyaluronidase activity and the antibiotic profiling confirmed sensitivity against most of the tested antibiotics, which suggested a low general pathogenicity of this strain. Nevertheless, the S. sciuri strain revealed resistance against fosfomycin, fusidic acid and teicoplanin (Table 2). According to the information provided by the Reference Center of the RKI in Wernigerode, approximately 80% of the tested S. sciuri strains reveal resistance against fusidic acid.
Interestingly, based on the EUCAST definition, the present strain is also resistant against Oxacillin. The S. sciuri species cluster group is represented by three S. siuri subspecies and also contains the species S. vitulinus. This cluster group carries different mecA homologues and has been proposed as origin and reservoir of the S. aureus mecA gene (Becker et al., 2014a, b; Nemeghaire et al., 2014a). In the genome of the present S. sciuri isolate, neither a mecA nor a mecC gene mediating methicillin/oxacillin resistance could be amplified by specific PCR at the National Reference Center at the RKI in Wernigerode. Thus, it has been reported that phenotypic methicillin (and other β-lactam) -resistance in Staphylococcaceae members is conferred not only by mecA, but also by different mecA allotypes and also by homologous genes such as mecB and mecC (Becker et al., 2014a, b). Moreover, a hybrid SCCmec consisting of a mecA-encoding SCCmec type VII element and a separate mecC region in terms of a ΨSCCmec element was published for S. sciuri (Harrison et al., 2014; Becker et al., 2014a). These reports might suggest the presence of a further mec-homolog or a mec gene hybrid within the genome of the isolated S. sciuri strain, which could not be amplified by the mecA and mecC-specific oligonucleotides.
Nevertheless, based on genomic and plasmid encoded genes, multiresistant S. sciuri isolates carrying resistance genes against all major classes of antibiotics have already been reported (Li et al., 2016) and support the potential to temporarily serve as a “bacterial shuttle” e.g. by transmitting genetic information between other bacterial species of the horse´s skin microbiome.
In sum, these results suggest the identification of a coagulase-negative Staphylococcus exhibiting moderate virulence.
Conflict of interest
The Authors declare that there is no conflict of interest.
References
- Adegoke GO. Characteristics of staphylococci isolated from man, poultry and some other animals. J. Appl. Bacteriol. 1986;60:97–102. doi: 10.1111/j.1365-2672.1986.tb03365.x. [DOI] [PubMed] [Google Scholar]
- Aslantas O, Turkyilmaz S, Yilmaz MA, Erdem Z, Demir C. Isolation and molecular characterization of methicillin-resistant staphylococci from horses, personnel and environmental sites at an equine hospital in Turkey. J. Vet. Med. Sci. 2012;74:1583–1588. doi: 10.1292/jvms.12-0124. [DOI] [PubMed] [Google Scholar]
- Bagcigil FA, Moodley A, Baptiste KE, Jensen VF, Guardabassi L. Occurrence, species distribution, antimicrobial resistance and clonality of methicillin-and erythromycin-resistant staphylococci in the nasal cavity of domestic animals. Vet. Microbiol. 2007;121(3-4):307–315. doi: 10.1016/j.vetmic.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Becker K, Ballhausen B, Köck R, Kriegeskorte A. Methicillin resistance in Staphylococcus isolates: the “mec alphabet” with specific consideration of mecC, a mec homolog associated with zoonotic S. aureus lineages. Int. J. Med. Microbiol. 2014a;304(7):794–804. doi: 10.1016/j.ijmm.2014.06.007. [DOI] [PubMed] [Google Scholar]
- Becker K, Heilmann C, Peters G. Coagulase-negative staphylococci. Clin. Microbiol. Rev. 2014b;27(4):870–926. doi: 10.1128/CMR.00109-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Wang Y, Chen F, Yang H, Gan M, Zheng SJ. A highly pathogenic strain of Staphylococcus sciuri caused fatal exudative epidermitis in piglets. PLoS One. 2007;2:e147. doi: 10.1371/journal.pone.0000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couto I, Santos Sanches I, Sá-Leão E, de Lencastre H. Molecular characterization of Staphylococcus sciuri strains isolated from humans. J. Clin. Microbiol. 2000;38:1136–1143. doi: 10.1128/jcm.38.3.1136-1143.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos F, Mendoca LC, Reis DR, Guimaraes AS, Lange CC, Ribeiro JB, Machado MA, Brito MA. Presence of a mecA-positive multidrug-resistant Staphylococcus epidermidis in bovine milk samples in Brazil. J. Dairy Sci. 2015;99(2):1374–1382. doi: 10.3168/jds.2015-9931. [DOI] [PubMed] [Google Scholar]
- Essers L, Radebold K. Rapid and Reliable Identification of Staphylococcus aureus by a Latex Agglutination Test. J. Clin. Microbiol. 1980;12:641–643. doi: 10.1128/jcm.12.5.641-643.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey Y, Rodriguez JP, Thomann A, Schwendener S, Perreten V. Genetic characterization of antimicrobial resistance in coagulase-negative staphylococci from bovine mastitis milk. J. Dairy Sci. 2013;96:2247–2257. doi: 10.3168/jds.2012-6091. [DOI] [PubMed] [Google Scholar]
- Harrison EM, Paterson GK, Holden MT, Ba X, Rolo J, Morgan FJ, Pichon B, Kearns A, Zadoks RN, Peacock SJ, Parkhill J, Holmes MA. A novel hybrid SCC mec-mecC region in Staphylococcus sciuri. J. Antimicrob. Chemother. 2014;69:911–918. doi: 10.1093/jac/dkt452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauschild T, Slizewski P, Masiewicz P. Species distribution of staphylococci from small wild mammals. Syst. Appl. Microbiol. 2010;33:457–460. doi: 10.1016/j.syapm.2010.08.007. [DOI] [PubMed] [Google Scholar]
- Hauschild T, Wojcik A. Species distribution and properties of staphylococci from canine dermatitis. Res. Vet. Sci. 2007;82:1–6. doi: 10.1016/j.rvsc.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Hedin G, Widerstrom M. Endocarditis due to Staphylococcus sciuri. Eur. J. Clin. Microbiol. Infect. Dis. 1998;17:673–675. doi: 10.1007/BF01708356. [DOI] [PubMed] [Google Scholar]
- Horii T, Suzuki Y, Kimura T, Kanno T, Maekawa M. Intravenous catheter-related septic shock caused by Staphylococcus sciuri and Escherichia vulneris. Scand. J. Infect. Dis. 2001;33(12):930–932. doi: 10.1080/00365540110076750. [DOI] [PubMed] [Google Scholar]
- Karakulska J, Fijałkowski K, Nawrotek P, Pobucewicz A, Poszumski F, Czernomysy-Furowicz D. Identification and methicillin resistance of coagulase-negative staphylococci isolated from nasal cavity of healthy horses. J. Microbiol. 2012;50(3):444–451. doi: 10.1007/s12275-012-1550-6. [DOI] [PubMed] [Google Scholar]
- Kloos WE, Zimmerman RJ, Smith RF. Preliminary studies on the characterization and distribution of Staphylococcus and Micrococcus species on animal skin. Appl. Environ. Microbiol. 1976;31:53–59. doi: 10.1128/aem.31.1.53-59.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolawole DO, Shittu AO. Unusual recovery of animal staphylococci from septic wounds of hospital patients in Ile-Ife, Nigeria. Lett. Appl. Microbiol. 1997;24:87–90. doi: 10.1046/j.1472-765x.1997.00337.x. [DOI] [PubMed] [Google Scholar]
- Li D, Wang Y, Schwarz S, Cai J, Fan R, Li J, Feßler AT, Zhang R, Wu C, Shen J. Co-location of the oxazolidinone resistance genes optrA and cfr on a multiresistance plasmid from Staphylococcus sciuri. J. Antimicrob. Chemother. 2016;71(6):1474–1478. doi: 10.1093/jac/dkw040. [DOI] [PubMed] [Google Scholar]
- Lüthje P, Schwarz S. Antimicrobial resistance of coagulase-negative staphylococci from bovine subclinical mastitis with particular reference to macrolide-lincosamide resistance phenotypes and genotypes. J. Antimicrob. Chemother. 2006;57:966–969. doi: 10.1093/jac/dkl061. [DOI] [PubMed] [Google Scholar]
- Marsou R, Bes M, Boudouma M, Brun Y, Meugnier H, Freney J, Vandenesch F, Etienne J. Distribution of Staphylococcus sciuri subspecies among human clinical specimens, and profile of antibiotic resistance. Res. Microbiol. 1999;150:531–541. doi: 10.1016/s0923-2508(99)00104-7. [DOI] [PubMed] [Google Scholar]
- Moodley A, Guardabassi L. Clonal spread of methicillin-resistant coagulase-negative staphylococci among horses, personnel and environmental sites at equine facilities. Vet. Microbiol. 2009;137:397–401. doi: 10.1016/j.vetmic.2009.01.034. [DOI] [PubMed] [Google Scholar]
- Nagase N, Sasaki A, Yamashita K, Shimizu A, Wakita Y, Kitai S, Kawano J. Isolation and species distribution of staphylococci from animal and human skin. J. Vet. Med. Sci. 2002;64:245–250. doi: 10.1292/jvms.64.245. [DOI] [PubMed] [Google Scholar]
- Nam HM, Lim SK, Kim JM, Kang HM, Moon JS, Jang GC, Wee SH, Joo YS, Jung SC. Antimicrobial susceptibility of coagulase-negative staphylococci isolated from bovine mastitis between 2003 and 2008 in Korea. J. Microbiol. Biotechnol. 2010;20:1446–1449. doi: 10.4014/jmb.1005.05034. [DOI] [PubMed] [Google Scholar]
- Nemeghaire S, Argudin MA, Fessler AT, Hauschild T, Schwarz S, Butaye P. The ecological importance of the Staphylococcus sciuri species group as a reservoir for resistance and virulence genes. Vet. Microbiol. 2014a;171:342–356. doi: 10.1016/j.vetmic.2014.02.005. [DOI] [PubMed] [Google Scholar]
- Nemeghaire S, Argudin MA, Haesebrouck F, Butaye P. Molecular epidemiology of methicillin-resistant Staphylococcus sciuri in healthy chickens. Vet. Microbiol. 2014b;171:357–363. doi: 10.1016/j.vetmic.2014.01.041. [DOI] [PubMed] [Google Scholar]
- Nemeghaire S, Vanderhaeghen W, Argudin MA, Haesebrouck F, Butaye P. Characterization of methicillin-resistant Staphylococcus sciuri isolates from industrially raised pigs, cattle and broiler chickens. J. Antimicrob. Chemother. 2014c;69:2928–2934. doi: 10.1093/jac/dku268. [DOI] [PubMed] [Google Scholar]
- Stepanovic S, Dakic I, Djukic S, Lozuk B, Svabic-Vlahovic M. Surgical wound infection associated with Staphylococcus sciuri Scand. J. Infect. Dis. 2002;34:685–686. doi: 10.1080/00365540110076949a. [DOI] [PubMed] [Google Scholar]
- Stepanovic S, Jezek P, Vukovic D, Dakic I, Petras P. Isolation of members of the Staphylococcus sciuri group from urine and their relationship to urinary tract infections. J. Clin. Microbiol. 2003;41:5262–5264. doi: 10.1128/JCM.41.11.5262-5264.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugochukwu EI, Agwu CO. Aerobic bacteria from nasal discharge of goats suffering from clinical PPR: isolation and identification. Microbios. 1991;65(263):81–85. [PubMed] [Google Scholar]
- Wallet F, Stuit L, Boulanger E, Roussel-Delvallez M, Dequiedt P, Courcol R. J. Peritonitis due to Staphylococcus sciuri in a patient on continuous ambulatory peritoneal dialysis. Scand. J. Infect. Dis. 2000;32:697–698. doi: 10.1080/003655400459667. [DOI] [PubMed] [Google Scholar]
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]


