Abstract
Background
After analysis of minor RAS mutations (KRAS exon 3, 4/NRAS) in the FIRE-3 and PRIME studies, an expanded range of RAS mutations were established as a negative predictive marker for the efficacy of anti-EGFR antibody treatment. BRAF and PIK3CA mutations may be candidate biomarkers for anti-EGFR targeted therapies. However, it remains unknown whether RAS/PIK3CA/BRAF tumor mutations can predict the efficacy of bevacizumab in metastatic colorectal cancer. We assessed whether selection according to RAS/PIK3CA/BRAF mutational status could be beneficial for patients treated with bevacizumab as first-line treatment for metastatic colorectal cancer.
Methods
Of the 1001 consecutive colorectal cancer patients examined for RAS, PIK3CA, and BRAF tumor mutations using a multiplex kit (Luminex®), we studied 90 patients who received combination chemotherapy with bevacizumab as first-line treatment for metastatic colorectal cancer. The objective response rate (ORR) and progression-free survival (PFS) were evaluated according to mutational status.
Results
The ORR was higher among patients with wild-type tumors (64.3%) compared to those with tumors that were only wild type with respect to KRAS exon 2 (54.8%), and the differences in ORR between patients with wild-type and mutant-type tumors were greater when considering only KRAS exon 2 mutations (6.8%) rather than RAS/PIK3CA/BRAF mutations (18.4%). There were no statistically significant differences in ORR or PFS between all wild-type tumors and tumors carrying any of the mutations. Multivariate analysis revealed that liver metastasis and RAS and BRAF mutations were independent negative factors for disease progression after first-line treatment with bevacizumab.
Conclusions
Patient selection according to RAS/PIK3CA/BRAF mutations could help select patients who will achieve a better response to bevacizumab treatment. We found no clinical benefit of restricting combination therapy with bevacizumab for metastatic colorectal cancer patients with EGFR-wild type tumors.
Electronic supplementary material
The online version of this article (doi:10.1186/s12885-016-2994-6) contains supplementary material, which is available to authorized users.
Keywords: RAS mutation, PIK3CA mutation, BRAF mutation, Colorectal cancer, bevacizumab
Background
The EGFR signaling pathway has a key role in the proliferation and survival of colorectal cancer cells. Point mutations in exon 2 of the KRAS gene have been shown to be negative predictive markers of the response to anti-EGFR treatment, and consequently anti-EGFR antibodies were not administered to patients with KRAS exon 2 mutant tumors [1]. After a retrospective analysis of minor RAS mutations (e.g. KRAS exon 3 and 4/NRAS) in the FIRE-3 and PRIME studies [2, 3], the so called “all RAS mutation” also came to be regarded as a negative biomarker for anti-EGFR antibody treatment [4]. In addition to RAS, BRAF and PIK3CA mutations are potential biomarkers of response to anti-EGFR targeted therapies [5]. However, it remains unknown whether EGFR pathway mutations affect the efficacy of bevacizumab (Bmab) in metastatic colorectal cancer (mCRC). We evaluated the significance of RAS/PIK3CA/BRAF tumor mutations in patients receiving combination chemotherapy with Bmab as the first-line treatment for mCRC, and we assessed whether these mutations could be used to select patients who would derive the greatest clinical benefit from Bmab.
Methods
Patients
This was a retrospective study conducted at a single Japanese institute and approved by the ethics committee of Cancer Institute Hospital of Japanese Foundation for Cancer Research (No.2009-1048). Of the 1001 consecutive patients with histologically confirmed CRC who were examined for tumor RAS, PIK3CA, and BRAF mutations in our institute between November 2006 and December 2013, 90 patients were administered combination chemotherapy with Bmab as the first-line treatment for mCRC. Patients who received neo-adjuvant chemotherapy (NAC) or adjuvant chemotherapy completed less than 6 months before enrollment to this study were excluded. Patients who had undergone surgery for metastatic sites were included if it had been performed more than 4 weeks earlier. Patients were required to have adequate hematologic, hepatic, cardiac, and renal function. Their medical records were reviewed to obtain data on clinicopathologic variables. All patients provided written informed consent before receiving treatment.
Procedure
The treatment regimen was determined by the physician for each patient. The following regimens were employed: modified FOLFOX6 plus Bmab consisted of a fortnightly course of Bmab (5 mg/kg intravenously over 30 to 90 min on day 1), oxaliplatin (85 mg/m2 intravenously over 2 h on day 1) plus l-LV (200 mg/m2 intravenously over 2 h on day 1) and 5-fluorouracil (5-FU) (400 mg/m2 bolus on day 1, followed by infusion of 2400 mg/m2 over 46 h); and CapeOX plus Bmab consisted of oxaliplatin (130 mg/m2 intravenously over 2 h on day 1) plus oral capecitabine (1000 mg/m2 twice daily for 2 weeks in a 3-week cycle). Bmab (7.5 mg/kg) was administered ahead of oxaliplatin intravenously on day 1 every 3 weeks. FOLFIRI plus Bmab consisted of fortnightly courses of Bmab (5 mg/kg intravenously over 30 to 90 min on day 1), irinotecan (150 mg/m2 intravenously over 2 h on day 1) plus l-LV (200 mg/m2 intravenously over 2 h on day 1) and 5-FU (400 mg/m2 bolus on day 1, followed by infusion of 2400 mg/m2 over 46 h).
DNA was extracted from formalin-fixed paraffin-embedded (FFPE) tumor tissue, which was mostly obtained at biopsy. Mutations in KRAS codons 12 and 13 were examined using a kit based on a Luminex assay (MEBGEN KRAS Mutation Detection kit, MBL). A Luminex based kit (GENOSEARCH Mu-PACK, MBL) was also used to detect a total of 36 mutations in KRAS (codons 61 and 146), NRAS (codons 12, 13, and 61), PIK3CA (codons 542, 545, 546, and 1047) and BRAF (codon 600). The concordance of findings based on this newly developed multiplex assay kit with conventional direct sequencing results was confirmed previously [6].
Statistical analysis
The objective response rate (ORR) was evaluated according to the Response Evaluation Criteria in Solid Tumors (RECIST) ver. 1.1. The progression-free survival (PFS) and overall survival (OS) were calculated using the Kaplan-Meier method. PFS was defined as the duration of survival from the start of chemotherapy to the date of recurrence or death from any cause, whichever occurred first. Patients with no recurrence until the cut-off date were regarded as censored on the last date when no recurrence had been proven by imaging. The disease-progression date was retrospectively re-analyzed by the investigator, and was defined as the date on which progression was first detected using a computed tomography (CT) or fluorodeoxyglucose-positron emission tomography (FDG-PET) scan. If treatments were discontinued before or continued after disease-progression due to adverse events or the patient’s request, they were censored at the time of the last radiological examination. OS was defined as survival from the start of chemotherapy to death from any cause. For patients who were lost to follow-up, data were censored on the date when the patient was last known to be alive. The data cut-off date was August 12, 2015. A one-sided Fisher’s exact test was used to assess the statistical significance of the difference between ORRs according to mutational status at a significance level of 2.5%. Both PFS and OS were estimated using the Kaplan-Meier method and compared using the log-rank test at a significance level of 5%. In addition to RAS, PIK3CA, and BRAF tumor mutations, variables with a P value less than 0.05 in a univariate analysis were included in a multivariate Cox regression analysis. All analyses were performed with EZR (Saitama Medical Center, Jichi Medical University, Saitama, Japan), which is a graphical user interface for R software (The R Foundation for Statistical Computing) [7].
Results
Baseline characteristics
The baseline characteristics of the patients are shown in Table 1. Their median age was 63 years (range, 27–79 years). Forty-eight patients (53.3%) were men and 42 patients (46.7%) were women. Almost all of the subjects had a good performance status. Seventy-four patients (82.2%) had colon cancer and 16 (17.8%) had rectal cancer, including right-sided colon cancer (RCC) in 34 patients (37.8%) and left-sided colorectal cancer (LCRC) in 56 patients (62.2%). RCC was defined as a tumor arising from the cecum to the transverse colon, excluding the appendix, while LCRC was defined as a tumor arising from the descending colon to the rectum. Of these 90 patients, the tumors of 43 patients (46.7%) were found to have a mutation in KRAS exon 2. In total 48 patients (53.3%) had a RAS mutation (KRAS/NRAS). Seven patients (8.9%) had a PIK3CA mutation, and another 7 (8.9%) had a BRAF mutation. Thirty-three patients (36.7%) had tumors with no RAS, PIK3CA, or BRAF mutation.
Table 1.
Baseline patient characteristics (n = 90)
| Characteristic | N (%) |
|---|---|
| Sex | |
| Male | 48 (53.3) |
| Female | 42 (46.7) |
| Median age (range), years | 63 (27–79) |
| ECOG performance status | |
| 0 | 82 (91.1) |
| 1, 2 | 8 (8.9) |
| Site of primary tumor | |
| RCC (cecum to the transverse colon) | 34 (37.8) |
| LCRC (descending to the rectosigmoid colon) | 40 (44.4) |
| Rectum | 16 (17.8) |
| Mode of metastasis | |
| Synchronous | 77 (85.6) |
| Asynchronous | 13 (14.4) |
| Sites of metastasis | |
| Liver | 51 |
| Lung | 38 |
| Distant lymph nodes | 29 |
| Peritoneum | 23 |
| Histology | |
| Differentiated | 83 (92.2) |
| Undifferentiated | 7 (7.8) |
| Number of metastases | |
| 1 | 37 (41.1) |
| ≥ 2 | 53 (58.9) |
| Chemotherapy regimen | |
| FOLFOX4/mFOLFOX6 or XELOX | 86 (95.6) |
| FOLFIRI | 4 (4.4) |
| Prior metastatectomy | |
| Yes | 12 (13.3) |
| No | 78 (86.7) |
| Resection of primary tumor | |
| Yes | 67 (74.4) |
| No | 23 (25.6) |
| Previous oxaliplatin treatment as adjuvant CTx | |
| Yes | 13 (14.4) |
| No | 77 (85.6) |
| KRAS status (codon 12,13) | |
| Wild-type | 47 (52.2) |
| Mutant | 43 (47.8) |
| RAS status (KRAS/NRAS) | |
| Wild-type | 42 (46.7) |
| Mutant | 48 (53.3) |
| PIK3CA status | |
| Wild-type | 82 (91.1) |
| Mutant | 8 (8.9) |
| BRAF status | |
| Wild-type | 82 (91.1) |
| Mutant | 8 (8.9) |
ECOG Eastern Cooperative Oncology Group, RCC right-sided colon cancer, LCRC left-sided colorectal cancer, CTx chemotherapy
Treatment exposure
Almost all patients received an oxaliplatin-containing regimen, which was FOLFOX in 34 cases (37.8%) and XELOX in 52 cases (57.8%). Among them, 13 (14.4%) had been administered oxaliplatin prior to this treatment as an adjuvant therapy. The primary tumor was resected in 67 patients (74.4%) and 13 patients (14.4%) underwent a metastatectomy.
ORR, PFS, and OS
The median follow-up period for all eligible patients was 23.5 (0.8–41.4) months, and 51 patients (56.7%) died by the cut-off date. Seventy-seven of the 90 patients had measurable lesions. The overall ORR was 52.6% whilst the ORR of patients with no detected tumor mutations was 64.3%. The ORRs of patients with a PIK3CA or BRAF tumor mutation were very low (28.6%), and more than 40% of patients with a BRAF tumor mutation had confirmed disease progression at the first evaluation (Fig. 1). Although the ORR varied according to the mutational status of the tumor, these differences were not statistically significant (Table 2). The differences in ORR became gradually greater among patients with wild-type tumors as restricting treatment subjects from only wild type with respect to KRAS exon 2, all RAS wild-type toward all wild-type (RAS/PIK3CA/BRAF). The difference in ORR between the whole population and patients with all wild-type tumors was 11.7% (Fig. 2).
Fig. 1.
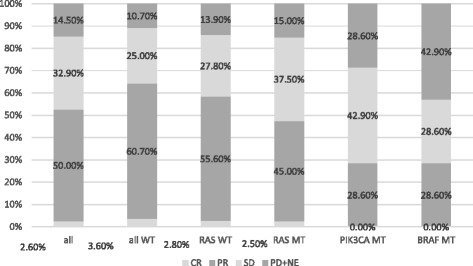
Response according to tumor mutation status
Table 2.
Response according to mutational status
|
KRAS exon 2 wt vs. mt |
RAS
wt vs. mt |
PIK3CA/BRAF
wt vs. mt |
RAS/PIK3CA/BRAF
All wt vs. any mt |
|
|---|---|---|---|---|
| Responder (n) | 23 vs. 17 | 21 vs. 19 | 38 vs. 2 | 18 vs. 22 |
| Non-responder (n) | 19 vs. 18 | 15 vs. 21 | 31 vs. 5 | 10 vs. 26 |
| ORR (%) | 54.8 vs. 48.6 | 58.4 vs. 47.5 | 55.1 vs. 28.6 | 64.3 vs. 45.9 |
| Difference in ORR | 6.8 | 10.9 | 26.5 | 18.4 |
| P value* | 0.651 | 0.368 | 0.246 | 0.155 |
Responder, CR + PR; Non-responder, SD + PD + NE; ORR, overall response rate; wt, wild-type; mt, mutant
*calculated using Fisher’s exact test
Fig. 2.
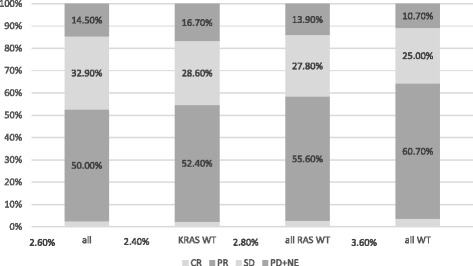
Responses among patients with wild-type tumor KRAS exon 2, RAS, and RAS/PIK3CA/BRAF
The overall median OS and PFS were 27.7 and 13.3 months, respectively. The ORR of patients with KRAS exon 2 wild-type and mutant-type tumors differed by 6.2%, and these populations had nearly identical Kaplan-Meier curves for PFS (Fig. 3). The difference between ORRs (18.4%) was larger when comparing patients with wild-type tumors to those with a tumor carrying any mutation. There was no statistically significant difference in PFS between these groups, although there was a slightly larger difference in Kaplan-Meier curves (Fig. 3).
Fig. 3.
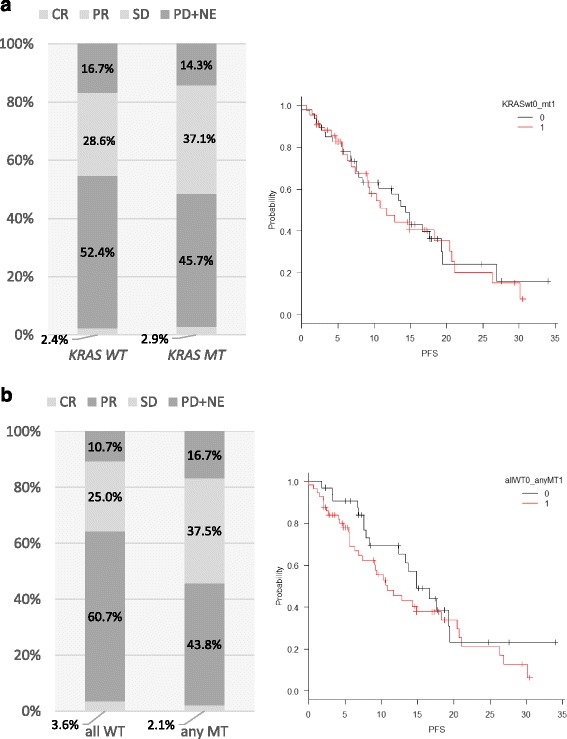
Relationship between overall response rate (ORR) and progression free survival (PFS) in patients with wild-type or mutant (a) KRAS exon 2 and (b) RAS/PIK3CA/BRAF tumors
Univariate analysis revealed that an elevated serum C-reactive protein (CRP) level (>0.05 mg/dl), an unresectable primary tumor, and liver metastases were associated with a significantly shorter PFS (Table 3). Multivariate analysis that included RAS, PIK3CA, and BRAF tumor mutations and baseline prognostic variables revealed that liver metastasis, unresectable primary tumor, RAS and BRAF tumor mutations had independent prognostic value for early progression (Table 3).
Table 3.
Univariate and multivariate analysis of pr4gression-free survival (PFS) (n = 90)
| Variable | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | |
| Sex | ||||
| Male vs. female | 1.0 vs. 0.72 (0.42–1.24) | 0.236 | ||
| Age | ||||
| < 63 y.o vs. ≥63 y.o | 1.0 vs. 0.82 (0.48–1.40) | 0.472 | ||
| ECOG PS | ||||
| 0 vs. 1,2 | 1.0 vs. 0.66 (0.21–2.14) | 0.490 | ||
| Site of primary tumor | ||||
| RCC vs. LCRC | 1.0 vs. 0.88 (0.51–1.52) | 0.643 | ||
| Differentiated-type | ||||
| yes vs. no | 1.0 vs. 1.78 (0.75–4.22) | 0.188 | ||
| Synchronous mets | ||||
| yes vs. no | 1.0 vs. 0.61 (0.29–1.30) | 0.201 | ||
| Sites of metastasis | ||||
| Non-liver vs. liver | 1.0 vs. 1.84 (1.06–3.20) | 0.031 | 1.0 vs. 3.26 (1.57–6.77) | 0.002 |
| Non-lung vs. lung | 1.0 vs. 1.17 (0.68–2.00) | 0.567 | ||
| Non-LN vs. LN | 1.0 vs. 0.67 (0.37–1.20) | 0.177 | ||
| Non-P vs. P | 1.0 vs. 0.95 (0.51–1.78) | 0.878 | ||
| Number of metastases | ||||
| 1 vs. ≥2 | 1.0 vs. 0.87 (0.51–1.50) | 0.628 | ||
| Primary resection | ||||
| yes vs. no | 1.0 vs. 2.00 (1.09–3.66) | 0.024 | 1.0 vs. 2.13 (1.05–4.29) | 0.035 |
| Prior L-OHP | ||||
| yes vs. no | 1.0 vs. 0.75 (0.35–1.60) | 0.457 | ||
| ALP (/) | ||||
| ≤ ULN vs. >ULN | 1.0 vs. 1.07 (0.56–2.04) | 0.836 | ||
| LDH (/IU) | ||||
| ≤ ULN vs. >ULN | 1.0 vs. 1.27 (0.74–2.21) | 0.387 | ||
| CRP (mg/dl) | ||||
| ≤ ULN vs. >ULN | 1.0 vs. 2.35 (1.32–4.17) | <0.001 | 1.0 vs. 1.57 (0.79–3.10) | 0.196 |
| CEA | ||||
| ≤ ULN vs. >ULN | 1.0 vs. 0.90 (0.49–1.65) | 0.735 | ||
| CA19-9 | ||||
| ≤ ULN vs. >ULN | 1.0 vs. 1.63 (0.95–2.80) | 0.076 | ||
| RAS status | ||||
| Wild-type vs. mutant | 1.0 vs. 1.36 (0.79–2.32) | 0.264 | 1.0 vs. 2.01 (1.07–3.76) | 0.030 |
| PIK3CA status | ||||
| Wild-type vs. mutant | 1.0 vs. 1.00 (0.36–2.77) | 0.993 | 1.0 vs. 0.66 (0.21–2.03) | 0.466 |
| BRAF status | ||||
| Wild-type vs. mutant | 1.0 vs. 1.60 (0.68–3.75) | 0.285 | 1.0 vs. 3.87 (1.38–10.9) | 0.010 |
CI confidence interval, HR hazard ratio, RCC right-sided colon cancer, LCRC left-sided colorectal cancer, mets metastasis, LN lymph node, P peritoneum, ULN upper limit of normal, NA not assessable. All data in italics are with p-value <0.05
Discussion
In clinical practice, it is often not as easy to conduct CT scans at regular intervals as it is in clinical trials. We considered that the ORR would be a relatively rigid parameter for evaluating the efficacy of first-line treatment in clinical practice. In this study, treatment was discontinued due to not only disease progression, but also adverse events or patient refusal, and 21 patients discontinued treatment before disease progression was confirmed by imaging. Time to treatment failure (TTF) is sometimes chosen as an endpoint instead of PFS in clinical trials. However, we considered that PFS would be a more suitable endpoint to evaluate the biological activity of the tumor and drug resistance compared to TTF. OS was the most rigid endpoint but would be determined by not only the first-line treatment but also by second-line and subsequent treatments. The difference in OS may be a result of anti-EGFR therapy after the first-line treatment in patients who have KRAS or RAS wild-type tumors. We therefore assessed the relationship between clinicopathologic factors including RAS, PIK3CA, and BRAF tumor mutation status and PFS.
In this study, we found that patients with a RAS, PIK3CA, or BRAF tumor mutation had a lower ORR than patients with tumors that did not carry these mutations, although this difference was not statistically significant. Patient selection according to tumor mutations in the EGFR pathway might improve the overall response to combination therapy with Bmab as a first-line treatment for mCRC. However, these differences in ORRs could not translate into an improved PFS. Multivariate analysis revealed a negative predictive value of RAS and BRAF tumor mutations with respect to first-line Bmab treatment. In this study, all patients were treated with Bmab, and hence, we could not clarify whether these gene mutations had predictive or prognostic value.
Data from preclinical research has indicated that changes in the EGFR signaling pathway might be related to the efficacy of anti-VEGF therapy [8]. Post-analysis of the AVF2107g trial revealed that adding Bmab to cytotoxic chemotherapy was beneficial regardless of KRAS exon 2 mutation status [9]. KRAS exon 2 mutations are not regarded as predictive markers of Bmab treatment. However, we found that the ORR, PFS, and OS differed between patients with KRAS wild-type and mutant tumors (ORR, 60.0% vs. 41.2%; PFS, 13.5 months vs. 9.3 months; OS, 27.7 months vs. 19.9 months). These findings are mostly similar to those of other trials comparing clinical outcomes between patients with KRAS exon 2 wild-type and mutant tumors [10–12]. Although statistically significant differences in OS and PFS between patients with KRAS exon 2 wild-type and mutant-type tumors were only shown in the MACRO trial [12], there were numerical differences shown all other trials. KRAS exon 2 wild-type tumors may predict a favorable prognosis. In a retrospective analysis of the PEAK, FIRE-3, and CALGB/SWOG80405 trials, this trend was also apparent when including subjects with any RAS tumor mutation, rather than just those with KRAS exon 2 tumor mutations [13–15]. A recent retrospective analysis of data from the TRIBE trial suggested that tumor mutations in both BRAF and RAS genes predicted a poor outcome for patients undergoing first-line treatment with Bmab plus FOLFIRI or FOLFOXIRI, although RAS mutations had less impact than BRAF mutations [16]. Larger patient numbers would be needed to translate the difference in ORR between patients with wild-type and mutant tumors into an improved clinical outcome, which may be why only the MACRO trial, with the largest number of patients, revealed statistically significant differences in outcome between patients with KRAS wild-type and mutant-type tumors.
Mutations in the BRAF gene have been shown to be markers of a poor prognosis following mCRC treatment [17, 18] and have a stronger prognostic value than RAS mutations [19]. However, BRAF mutant cases were very rare, only 8 in this cohort. It is therefore very difficult to obtain an adequate number of these cases to show statistically significant differences.
Taking into account these previous data, RAS and BRAF mutations may be associated with the inferior efficacy of Bmab treatment. However, due to the relatively small effect of RAS mutations and the rarity of BRAF mutations, we were unable to show statistically significant differences in the ORR and PFS of patients undergoing first-line treatment with Bmab for mCRC.
The ORR and PFS in patients with any of the examined mutations were 45.9% and 10.8 months, respectively, in this study. These were comparable with those of patients with KRAS exon 2 mutant tumors treated using FOLFOX4 alone in the OPUS study (ORR, 52.0%; PFS, 8.6 months) [20]. However, in the cetuximab arm of the OPUS and CRYSTAL trials, the ORR and PFS of patients with KRAS mutant tumors were much worse (ORR, 26.0 and 31.3% respectively; PFS, 5.5 and 7.4 months, respectively) than patients with no tumor mutations in our study [20, 21]. Our study did not show that mCRC patients with a tumor mutation in RAS, PIK3CA, or BRAF had a poorer response to combination chemotherapy with Bmab compared with patients who had none of these mutations. As such, there are insufficient data to justify the exclusion of patients with a RAS, PIK3CA, or BRAF tumor mutation from Bmab treatment regimens.
Our study has several limitations. Firstly, this was retrospective cohort study conducted at a single institute. Secondly, there was selection bias in the treatment of patients with KRAS wild-type tumors, especially those who had metastases only in the liver. In our institute, patients with KRAS wild-type tumors and liver only metastases were administered anti-EGFR therapy as an initial standard treatment in order to achieve conversion to metastatectomy. Patients with KRAS wild-type tumors and liver metastases in this cohort had relatively unfavorable factors, such as a high tumor burden or multiple organ metastases, and patients with relatively favorable factors were usually excluded. This may explain why liver metastasis was an independent predictor of a poor prognosis in this cohort. Patients with RAS wild-type tumors in this cohort might have had a poor prognosis, compared to those included in other studies, and this selection bias might have affected the outcome of patients with tumors that do not carry mutations in the genes studied here. Thirdly, due to the rare incidence of PIK3CA and BRAF mutation in CRC, we could evaluate only small number patients with these mutations.
Conclusion
There were no statistically significant differences in ORR and PFS according to mutations in EGFR pathway genes in patients receiving cytotoxic chemotherapy with Bmab as the first-line treatment for mCRC. RAS/PIK3CA/BRAF mutations could help identify tumors that will respond to both anti-EGFR antibodies and Bmab. However, this study did not find a clinical benefit for restricting Bmab treatment to mCRC patients with tumors that have wild-type EGFR pathway genes.
Acknowledgments
The authors would like to thank the staff who managed patients at the ambulatory treatment center and on the ward. We also thank Editage (www.editage.jp) for English language editing.
Fundings
There is no funding to be declared for publication in this article.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its Additional file 1.
Authors’ contributions
IN analyzed the clinical data and wrote the original manuscript. IN, ES, TM, TW, MO, TI, MO, DT, MS, KC, NM and KY were all involved in the administration of chemotherapy. All authors contributed to editing the manuscript and approved the final version.
Competing interests
Eiji Shinozaki: Honoria from Merck Serono Co.,Ltd, Takeda Co.,Ltd, Brystol Myers Squibb Japan Co.,Ltd, Taiho Co.,Ltd, Chugai Co., Ltd, Ono Co,.Ltd
The remaining authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The present study was conducted according to the principles of the declaration of Helsinki and all participating patients provided written informed consent. This study was approved by the ethics committee of Cancer Institute Hospital of Japanese Foundation for Cancer Research (No.2009-1048).
Abbreviations
- 5-FU
5-fluorouracil
- Bmab
bevacizumab
- CT
Computed tomography
- EGFR
Epidermal growth factor receptor
- FFPE
Formalin-fixed paraffin-embedded
- LCRC
Left-sided colon
- mCRC
Metastatic colorectal cancer
- NAC
Neo-adjuvant chemotherapy
- ORR
Objective response rate
- OS
Overall survival
- PET
Positron emission tomography
- PFS
Progression free survival
- RCC
Right-sided colon
- RECIST
Response Evaluation Criteria in Solid Tumors
- TTF
Time to treatment failure
Additional file
Row data about clinicopathological features and clinical outcomes with omitting personally identifiable information of patients. (XLSX 27 kb)
Contributor Information
Izuma Nakayama, Email: izuma.nakayama@jfcr.or.jp.
Eiji Shinozaki, Phone: +81-3-3520-0111, Email: eiji.shinozaki@jfcr.or.jp.
Tomohiro Matsushima, Email: tomohiro.matsushima@jfcr.or.jp.
Takeru Wakatsuki, Email: takeru.wakatsuki@jfcr.or.jp.
Mariko Ogura, Email: mariko.ogura@jfcr.or.jp.
Takashi Ichimura, Email: takashi.ichimura@jfcr.or.jp.
Masato Ozaka, Email: masato.ozaka@jfcr.or.jp.
Daisuke Takahari, Email: daisuke.takahari@jfcr.or.jp.
Mitsukuni Suenaga, Email: mitsukuni.suenaga@jfcr.or.jp.
Keisho Chin, Email: kchin@jfcr.or.jp.
Nobuyuki Mizunuma, Email: mizunuma@jfcr.or.jp.
Kensei Yamaguchi, Email: kensei.yamaguchi@jfcr.or.jp.
References
- 1.Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, et al. K-ras Mutations and Benefit from Cetuximab in Advanced Colorectal Cancer. N Engl J Med. 2013;369:1023–1034. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 2.Heinemann V, von Weikersthal LF, Decker T, Kiani A, Vehling-Kaiser U, Al-Batran SE, et al. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab as first-line treatment for patients with metastatic colorectal cancer (FIRE-3): a randomised, open-label, phase 3 trial. Lancet Oncol. 2014;15:1065–1075. doi: 10.1016/S1470-2045(14)70330-4. [DOI] [PubMed] [Google Scholar]
- 3.Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369:1023–1034. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 4.Sorich MJ, Wiese MD, Rowland A, Kichenadasse G, McKinnon RA, Karapetis CS, et al. Extended RAS mutations and anti-EGFR monoclonal antibody survival benefit in metastatic colorectal cancer: a meta-analysis of randomized, controlled trials. Ann Oncol. 2015;26:13–21. doi: 10.1093/annonc/mdu378. [DOI] [PubMed] [Google Scholar]
- 5.De Roock W, De Vriendt V, Normanno N, Ciardiello F, Tejpar S. KRAS, BRAF, PIK3CA, and PTEN mutations: implications for targeted therapies in metastatic colorectal cancer. Lancet Oncol. 2011;12:594–603. doi: 10.1016/S1470-2045(10)70209-6. [DOI] [PubMed] [Google Scholar]
- 6.Bando H, Yoshino T, Shinozaki E, Nishina T, Yamazaki K, Yamaguchi K, et al. Simultaneous identification of 36 mutations in KRAS codons 61 and 146, BRAF, NRAS, and PIK3CA in a single reaction by multiplex assay kit. BMC Cancer. 2013;13:405. doi: 10.1186/1471-2407-13-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanda Y. Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transplant. 2013;48:452–458. doi: 10.1038/bmt.2012.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ince WL, Jubb AM, Holden SN, Holmgren EB, Tobin P, Sridhar M, et al. Association of k-ras, b-raf, and p53 Status With the Treatment Effect of bevacizumab. J Natl Cancer Inst. 2005;97:981–989. doi: 10.1093/jnci/dji174. [DOI] [PubMed] [Google Scholar]
- 9.Hurwitz HI, Yi J, Ince W, Novotny WF, Rosen O. The clinical benefit of bevacizumab in metastatic colorectal cancer is independent of K-ras mutation status: analysis of a phase III study of bevacizumab with chemotherapy in previously untreated metastatic colorectal cancer. Oncologist. 2009;14:22–28. doi: 10.1634/theoncologist.2008-0213. [DOI] [PubMed] [Google Scholar]
- 10.Hecht JR, Mitchell E, Chidiac T, Scroggin C, Hagenstad C, Spigel D, et al. A randomized phase IIIB trial of chemotherapy, bevacizumab, and panitumumab compared with chemotherapy and bevacizumab alone for metastatic colorectal cancer. J Clin Oncol. 2009;27:672–680. doi: 10.1200/JCO.2008.19.8135. [DOI] [PubMed] [Google Scholar]
- 11.Price TJ, Hardingham JE, Lee CK, Weickhardt A, Townsend AR, Wrin JW, et al. Impact of KRAS and BRAF Gene Mutation Status on Outcomes From the Phase III AGITG MAX Trial of Capecitabine Alone or in Combination With Bevacizumab and Mitomycin in Advanced Colorectal Cancer. J Clin Oncol. 2011;29:2675–2682. doi: 10.1200/JCO.2010.34.5520. [DOI] [PubMed] [Google Scholar]
- 12.Díaz-Rubio E, Gómez-España A, Massutí B, Sastre J, Reboredo M, Manzano JL, et al. Role of Kras status in patients with metastatic colorectal cancer receiving first-line chemotherapy plus bevacizumab: a TTD group cooperative study. PLoS One. 2012;7:e47345. doi: 10.1371/journal.pone.0047345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karthaus M. Updated Overall Survival Analysis of Novel Predictive KRAS /NRAS Mutations beyond KRAS exon 2 in PEAK: A 1st-line Phase 2 Study of FOLFOX6 plus Panitumumab or Bevacizumab in Metastatic Colorectal Cancer. Poster session presented at: The European Cancer Congress 2013. 38th annual conference of the European Society for Medical Oncology; 2013 27 Sep-01 Oct: Amsterdam, Netherlands.
- 14.Heinemann V, et al. Analysis of KRAS /NRAS and BRAF Mutations in FIRE-3: A Randomized Phase III Study of FOLFIRI plus Cetuximab or Bevacizumab as First-line Treatment for Wild-type KRAS (exon 2) Metastatic Colorectal Cancer Patients. Poster session presented at: The European Cancer Congress 2013. 38th annual conference of the European Society for Medical Oncology; 2013 27 Sep - 01 Oct: Amsterdam, Netherlands.
- 15.Lenz HJ, et al. CALGB/SWOG 80405: Phase III Trial of Irinotecan/5-FU/Leucovorin (FOLFIRI) or Oxaliplatin/5-FU/Leucovorin (mFOLFOX6) with Bevacizumab (BV) or Cetuximab (CET) for Patients (Pts) with Untreated Metastatic Adenocarcinoma of the Colon or Rectum (MCRC): Expanded RAS analyses. Poster session presented at: The European Cancer Congress 2014. 39th annual conference of the European Society for Medical Oncology; 2014 26 Sep-30 Sep: Madrid, Spain.
- 16.Cremolini C, Loupakis F, Antoniotti C, Lupi C, Sensi E, Lonardi S, et al. FOLFOXIRI plus bevacizumab versus FOLFIRI plus bevacizumab as first-line treatment of patients with metastatic colorectal cancer: updated overall survival and molecular subgroup analyses of the open-label, phase 3 TRIBE study. Lancet Oncol. 2015;16:1306–1315. doi: 10.1016/S1470-2045(15)00122-9. [DOI] [PubMed] [Google Scholar]
- 17.Brookmeyer C, Van Cutsem E, Rougier P, Ciardiello F, Heeger S, Schlichting M, et al. Addition of cetuximab to chemotherapy as first-line treatment for KRAS wild-type metastatic colorectal cancer: pooled analysis of the CRYSTAL and OPUS randomised clinical trials. Eur J Cancer. 2012;48:1466–1475. doi: 10.1016/j.ejca.2012.02.057. [DOI] [PubMed] [Google Scholar]
- 18.Tie J, Gibbs P, Lipton L, Christie M, Jorissen RN, Burgess AW, et al. Optimizing targeted therapeutic development: analysis of a colorectal cancer patient population with the BRAF (V600E) mutation. Int J Cancer. 2011;128:2075–2084. doi: 10.1002/ijc.25555. [DOI] [PubMed] [Google Scholar]
- 19.Richman SD, Seymour MT, Chambers P, Elliott F, Daly CL, Meade AM, et al. KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: results from the MRC FOCUS trial. J Clin Oncol. 2009;27:5931–5937. doi: 10.1200/JCO.2009.22.4295. [DOI] [PubMed] [Google Scholar]
- 20.Bokemeyer C, Bondarenko I, Hartmann JT, de Braud F, Schuch G, Zubel A, et al. Efficacy according to biomarker status of cetuximab plus FOLFOX-4 as first-line treatment for metastatic colorectal cancer: the OPUS study. Ann Oncol. 2011;22:1535–1546. doi: 10.1093/annonc/mdq632. [DOI] [PubMed] [Google Scholar]
- 21.Van Cutsem E, Köhne CH, Láng I, Folprecht G, Nowacki MP, Cascinu S, et al. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol. 2011;29:2011–2019. doi: 10.1200/JCO.2010.33.5091. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its Additional file 1.


