Abstract
Chronic hepatitis B virus (HBV) infection can cause severe liver disease, including cirrhosis and hepatocellular carcinoma. Lamivudine is a relatively recent alternative to alpha interferon for the treatment of HBV infection, but unfortunately, resistance to lamivudine commonly develops during monotherapy. Lamivudine-resistant HBV mutants display specific mutations in the YMDD (tyrosine, methionine, aspartate, aspartate) motif of the viral polymerase (reverse transcriptase [rt]), which is the catalytic site of the enzyme, i.e., methionine 204 to isoleucine (rtM204I) or valine (rtM204V). The latter mutation is often accompanied by a compensatory leucine-to-methionine change at codon 180 (rtL180M). In the present study, a novel sequencing method, pyrosequencing, was applied to the detection of lamivudine resistance mutations and was compared with direct Sanger sequencing. The new pyrosequencing method had advantages in terms of throughput. Experiments with mixtures of wild-type and resistant viruses indicated that pyrosequencing can detect minor sequence variants in heterogeneous virus populations. The new pyrosequencing method was evaluated with a small number of patient samples, and the results showed that the method could be a useful tool for the detection of lamivudine resistance in the clinical setting.
Chronic hepatitis B virus (HBV) infection can result in severe liver disease, including cirrhosis and hepatocellular carcinoma (10). The magnitude of this problem is very large because approximately 5% of the world's population is chronically infected with HBV. Until relatively recently, alpha interferon was the only drug licensed for use for the treatment of HBV infection, but the efficacy of this treatment has been suboptimal, with a response rate of less than 40% (11). A few years ago the nucleoside analogue lamivudine [(−)2′-deoxy-3′-thiacytidine] was also licensed for use for the treatment of HBV infection. Lamivudine inhibits HBV replication by competing with the natural nucleotide substrate of the HBV polymerase and thereby terminating synthesis of viral DNA (8). Unfortunately, drug-resistant viral strains may emerge after prolonged treatment and may cause treatment failure and rising HBV levels in plasma. The development of lamivudine resistance is especially common among patients who are immunosuppressed, such as transplant recipients and patients coinfected with human immunodeficiency virus (HIV). Lamivudine-resistant HBV mutants are characterized by specific mutations, methionine to isoleucine (rtM204I) or valine (rtM204V), in the YMDD (tyrosine, methionine, aspartate, aspartate) motif of the viral polymerase (reverse transcriptase [rt]), which is the catalytic site of the enzyme (9). The latter mutation is often accompanied by a compensatory leucine-to-methionine change at codon 180 (rtL180 M). Mutants with these mutations are resistant to lamivudine both in vitro and in vivo. HBV resistance to lamivudine is explained by steric hindrance between the oxathiolane ring of lamivudine triphosphate and the β-branched chain of isoleucine and valine (2) in a manner that is similar to the mechanism described for HIV (6).
Lamivudine resistance can be detected in patient samples by PCR followed by conventional automated Sanger sequencing. However, simpler methods for the detection of single-nucleotide polymorphisms (SNPs) could also be used because there are only two positions of interest that are only approximately 50 bases apart. Pyrosequencing is a non-gel-based DNA sequencing technique that is based on the detection of the pyrophosphate (PPi) released during DNA synthesis. In a cascade of enzymatic reactions, visible light is generated at a level that is proportional to the number of incorporated nucleotides (Fig. 1) (5). This method generates 30- to 40-base sequences with each primer, and the throughput is 96 samples in approximately 10 min; i.e., the throughput is much higher than that which can be achieved by conventional Sanger sequencing on gel- or capillary-based automated sequencing machines. Here we describe a new pyrosequencing method for analysis of lamivudine resistance in HBV.
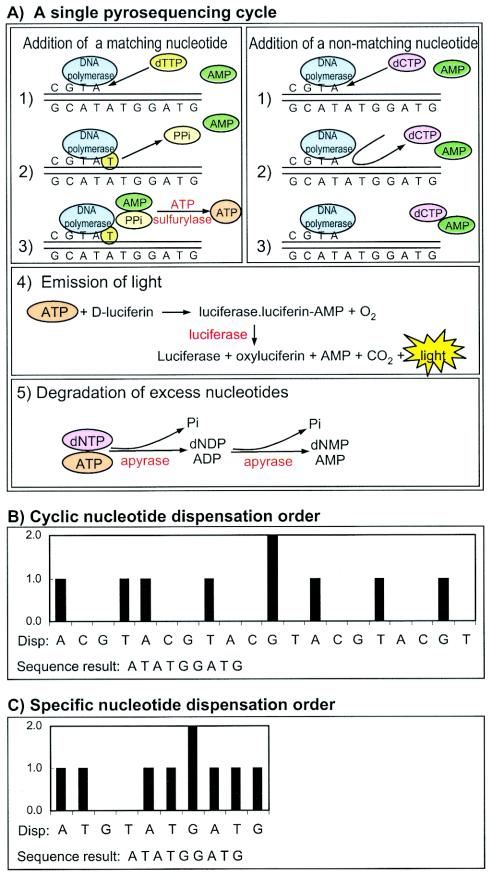
FIG. 1.
Principle and chemistry of pyrosequencing. Pyrosequencing is based on real-time detection of PPi as it is released when a DNA polymerase incorporates nucleotides into a growing primer-template complex. (A) The five steps of a single pyrosequencing cycle, which takes approximately 60 s to complete and which involves the cooperative actions of four enzymes. The first step is the addition (dispensation) of a new nucleotide triphosphate (panels 1, right and left). If the nucleotide is complementary to the next position in the template (matching), it is incorporated and PPi is released (panel 2, left). If the nucleotide does not match the template, it is not incorporated and no PPi is released (panel 2, right). If PPi has been released, it is used to convert AMP into ATP (panel 3, left). The ATP is used to produce visible light through the action of the firefly enzyme luciferase (panel 4). Thus, a light signal is generated if a complementary nucleotide is added, but no light signal is generated if a noncomplementary nucleotide is added. The cycle is completed when apyrase degrades unused dNTPs and ATP (panel 5). (B) Example of a theoretical pyrogram obtained by using a cyclic nucleotide dispensation order. The intensity of the light generated in each pyrosequencing cycle is visualized as peaks with heights that correspond to the number of incorporated nucleotides. The cyclic nucleotide dispensation order (Disp:) is displayed directly under the pyrogram. The pyrosequencing result is displayed at the bottom of the panel. This type of cyclic nucleotide dispensation order is preferred if the sequence of interest is highlypolymorphic. In our experiments we instead used a specific nucleotide dispensation order (C), because this allows the design of clearly distinguishable patterns for different genotypes (Fig. 2) and also reduces the number of pyrosequencing cycles. (C) An example of a theoretical pyrogram obtained by using a specific nucleotide dispensation order. The specific nucleotide dispensation order (Disp:) is displayed directly under the pyrogram. The pyrosequencing result is displayed at the bottom of the panel. Note that the same sequence result was obtained in panels B and C, even though the number of nucleotide dispensations has been reduced by a factor of 2 (20 dispensations in panel B versus 10 dispensations in panel C).
MATERIALS AND METHODS
Reference material and patient samples.
The initial optimization of the PCR protocol was performed with selected HBV DNA-positive serum samples that had undergone previous routine qualitative or quantitative HBV PCR at Clinical Virology, Huddinge Hospital. The samples were between 1 and 12 months old and had been stored at −20°C. The samples that were used for the optimization of extraction and PCR were subjected to repeated quantification by the Amplicor HBV Monitor assay (Roche Molecular Systems, Roche Diagnostics, Branchburg, N.J.).
For the mixing experiments described below we used plasma samples from one patient who had developed lamivudine resistance following liver transplantation. The sample (sample MUT) was kindly provided by Magnus Lindh at the Department of Clinical Virology, Sahlgrenska University Hospital, Gothenburg, Sweden, and was stored at −20°C after arrival in our laboratory. A sample with wild-type (WT) virus was selected from among those undergoing routine testing at Huddinge University Hospital. The viral loads in these two samples were determined by repeat quantification by the Amplicor HBV Monitor assay. Since the viral loads were very high, both samples were diluted 1:100 in normal plasma before use.
The clinical utility of pyrosequencing was tested with 20 clinical serum samples from patients who had received lamivudine treatment and who showed clinical and laboratory signs of treatment failure, e.g., increasing serum HBV DNA levels. Using these 20 samples, we compared pyrosequencing and Sanger sequencing (see below). The HBV PCR, which was used to prepare templates for both pyrosequencing and Sanger sequencing, was evaluated with 34 additional clinical patient samples that underwent routine testing for lamivudine resistance by Sanger sequencing. The study was approved (approval no. 53/00) by the local research ethics committee at Karolinska Institutet, Huddinge University Hospital.
DNA extraction and nested PCR.
DNA was extracted from serum samples with the NucliSens isolation kit (Organon Teknika, Boxtel, The Netherlands), according to the recommendations of the manufacturer. Briefly, 500 μl of serum was lysed in 9 ml of lysis buffer, and the solution was mixed. A silica suspension (50 μl) was added, and the samples were incubated for 10 min at room temperature. The silica particles with bound DNA were washed five times: twice with washing buffer, twice with 70% ethanol, and once with acetone. The silica pellet was dried for 10 min at 56°C, and the nucleic acids were eluted in 50 μl of elution buffer and stored at −70°C. The nested PCR was designed to amplify a short fragment (239 bp) of the HBV polymerase gene that includes the resistance mutations at amino acid positions 180 and 204. The primers were designed to anneal to highly conserved regions by using an alignment of 153 HBV polymerase sequences representing all HBV genotypes. The same strategy was used to design primers for Sanger sequencing and pyrosequencing. The reaction mixture for the first PCR contained 2.5 mM MgCl2, 0.4 mM each deoxynucleoside triphosphate (dNTP; Amersham Pharmacia Biotech, Freiburg, Germany), 0.1 μM each primer (Scandinavian Gene Synthesis AB, Köping, Sweden), 0.2 μl of Taq polymerase (5 U/μl; Perkin-Elmer, Roche, Branchburg, N.J.), and 5 μl of DNA template in 1× GeneAmp PCR buffer (Perkin-Elmer, Roche) in a total volume of 50 μl. The outer PCR was performed with primers JA224 (position +457; 5′-AGG TAT GTT GCC CGT TTG TCC TC-3′) and JA227 (position; −982; 5′-AAT TCT TTG ACA TAC TTT CCA ATC AAT-3′); the positions in the HBV ADR sequence are indicated (+, sense; −, antisense). The amplification profile for the first PCR was 5 min at 95°C; 30 cycles of 30 s at 95°C, 30 s at 54°C, and 30 s at 72°C; and 6 min at 72°C. The reaction mixture for the second (nested) PCR contained 2.5 μl of the product from the first PCR and the same reagents used in the first PCR, with the exception that the primers were changed to JA225 (position +600; 5′-AYT GCA CYT GTA TTC CCA TCC CAT-3′) and JA226 (position −817; 5′-biotin-TYA AAT GTA TAC CCA AAG ACA AAA GAA A-3′). The Y in the primer sequences signifies positions that were synthesized with C-T wobbles to accommodate the known sequence variations between published HBV genomes. Primer JA225 was biotinylated for the subsequent pyrosequencing steps (see below). The amplification profile in the second PCR was the same as that in the first PCR. All reactions were carried out in a GeneAmp PCR System 9700 (Perkin-Elmer, Foster City, Calif.). After amplification, the product (5 μl) was detected by ethidium bromide staining on 2% agarose gels, and the remaining volume (45 μl) was used for sequencing.
The sensitivity of the optimized PCR system was investigated by limiting dilution experiments and performing calculations by use of the Poisson distribution formula (1). Serum samples with known HBV DNA levels were diluted in HBV DNA-negative serum. Fivefold dilutions of this material were tested in 5 to 10 replicates. The most diluted material contained less than 1 HBV DNA copy/5 μl, which was the volume used in each PCR. The PCR results followed the Poisson distribution, and both strongly positive and negative results were observed with high dilutions, which indicates that the analytical sensitivity of the PCR was close to 1 HBV DNA copy.
Limiting dilution was also used to amplify single HBV molecules of lamivudine-resistant (MUT) and WT HBV variants for the mixing experiments described below. Serum samples that had been shown to contain primarily lamivudine-resistant or WT virus by direct population sequencing were diluted to contain, on average, less than 1 HBV DNA copy/5 μl. Single HBV molecules were amplified from the diluted material by the nested PCR described above. The PCR products were sequenced on an ABI310 instrument to verify that these “clones” had the expected sequence, i.e., 100% lamivudine resistant and 100% WT, respectively. The PCR products obtained from single HBV molecules (clones) were used in the mixing experiments described below.
Sanger sequencing.
The PCR product (45 μl) was purified with a QIAquick PCR purification kit (Qiagen, Hilden, Germany), according to the recommendations of the manufacturer. The purified amplicons were sequenced with an ABI PRISM BigDye terminator cycle sequencing kit with AmpliTaq DNA polymerase (FS; Perkin-Elmer). The sequencing reaction mixture contained 2 μl of Terminator Ready Reaction Mix, 6 μl of 2.5× Sequencing Buffer, 3 μl of template, 8 μl of deionized water, and 1 μl of either of the two inner PCR primers, primers JA225 and JA226. The cycle sequencing profile was 25 cycles of 96°C for 10 s, 50°C for 5 s, and 60°C for 4 min, followed by incubation at 4°C. The sequencing fragments were purified with 70% ethanol, 95% ethanol, and sodium acetate. Sequencing was performed on an ABI Prism 310 Genetic Analyzer with ABI Prism 310 Collection and Sequencing Analysis software. The sequences generated by the forward and reverse sequencing primers were assembled and analyzed with the software program Sequencher (Gene Codes Corporation, Ann Arbor, Mich.). The resulting complete sequences were translated into amino acid sequences.
Pyrosequencing.
The principle of pyrosequencing is illustrated in Fig. 1. Two primers were designed to anneal adjacent to the two resistance sites of interest, i.e., positions 180 and 204. The primer sequences and their positions in the HBV ADR sequence are provided (+, sense; −, antisense). Primer JA228 (position +645; 5′-AGT GGG CCT CAG TCC GTT TC-3′) was used for analysis of position 204, and JA229 (position −718; 5′-CAC TGT TTG GCT TTC AGT T-3′) was used for analysis of position 180. Serum samples were prepared for pyrosequencing by sample extraction and nested PCR as described above. The inner PCR primer JA226 was biotinylated to allow immobilization of the PCR product on streptavidin-coated beads and preparation of single-stranded DNA for pyrosequencing. Sample preparation was done in the 96-well format with the Magnatrix 1200 sample preparation robot (Magnetic BioSolutions AB, Stockholm, Sweden), according to the instructions of the manufacturer, by using paramagnetic beads (Dynabeads M280; Dynal, Oslo, Norway). Primer annealing was done at 50°C for 10 min. The single-stranded products with the annealed primer were moved from the robot and immediately used for pyrosequencing. Pyrosequencing was performed at 25°C in a total volume of 50 μl in the automated 96-well pyrosequencer, according to the instructions of the manufacturer, with the PSQ SNP 96 reagent kit (Pyrosequencing AB, Uppsala, Sweden). We used a specific rather than a cyclic nucleotide dispensation strategy (order of nucleotide additions) because it allowed us to design clearly distinguishable patterns for different genotypes (see results and Fig. 1B and C). Thus, the nucleotide dispensation order was designed on the basis of our prior knowledge about the WT and MUT HBV sequence variants. An additional advantage of the specific nucleotide dispensation strategy is that it reduces the number of pyrosequencing cycles and thereby increases the read length and sequence quality. It was possible to use a specific nucleotide dispensation strategy because lamivudine resistance involves only a few sequence variants. If the sequence of interest had been highly polymorphic, we would have been forced to choose the less attractive cyclic nucleotide dispensation strategy.
Mixing experiments.
Mixtures of WT and resistant virus were created to evaluate the abilities of the Sanger and pyrosequencing methods to accurately detect and quantify minor sequence variants. PCR products from single HBV molecules (clones) representing WT and lamivudine-resistant (MUT) variants were prepared by limiting dilution as described above. The two viruses were both of genotype A, and apart from the two resistance mutations at positions 180 and 204, the two viruses had identical sequences in the region analyzed. The PCR products were quantified with a DNA 7500 Labchip kit (Agilent Technologies), according to the recommendations of the manufacturer. Briefly, a gel-dye mixture was prepared from a gel-matrix mixture and a gel-dye mixture. The gel-dye mixture was then added to the DNA chip, together with a marker mixture, a DNA 7500 ladder for measurement of size, and the samples. The chip was inserted in a 2100 Bioanalyzer (Agilent Technologies). The DNA content in each sample was analyzed three times, and the mean of the three runs was calculated. The quantified PCR products were diluted to the same concentration, and mixtures ranging from 100% WT-0% MUT to 0% WT-100% MUT were prepared at increments of 10%. The WT-MUT mixtures were analyzed by traditional Sanger sequencing and pyrosequencing.
RESULTS
Optimization of PCR and pyrosequencing.
The PCR system was optimized by using selected serum samples from HBV-infected patients. The final optimized PCR protocol for amplification of the two positions in the HBV rt that are involved in development of resistance to lamivudine (positions 180 and 204) is described in Materials and Methods. The sensitivity of the optimized PCR was investigated by limiting dilution experiments and calculations performed by using the Poisson distribution formula. The results indicated that the analytical sensitivity of the PCR system was close to 1 HBV DNA copy per PCR (data not shown). Thus, it should be possible to perform the resistance test with samples with as little as 20 to 50 HBV DNA copies/ml if 500 μl of serum is used for sample preparation. This includes all samples for which testing for lamivudine resistance may be clinically relevant because patients infected with lamivudine-resistant HBV generally have much higher virus titers.
The principle of pyrosequencing is illustrated in Fig. 1. For pyrosequencing, two primers were designed for sequence analysis for mutations at positions 180 and 204. The primers were designed to anneal closely upstream of the positions of interest since pyrosequencing has a read length of approximately 40 bp. Special attention was paid to the specificity of primer annealing because pyrosequencing is performed at room temperature, which increases the risk of mispriming, which can generate a background sequence. We decided to add the nucleotides in a specific order rather than a cyclic order (Fig. 1B and C). This was possible because the sequence of the HBV genome is relatively conserved in the areas that surround the mutations of interest and because we aimed at sequencing only two specific codons in the HBV genome (positions 180 and 204). This allowed us to design distinct theoretical pyrogram patterns for resistant and WT virus sequences. Thus, for each primer we calculated the theoretical pyrogram patterns for each possible sequence variant that might be encountered during pyrosequencing (Fig. 2). Another reason for choosing a specific rather than a cyclic nucleotide dispensation order was that the quality of the pyrosequencing peaks decreases relatively rapidly and the read length is therefore limited to approximately 40 nucleotides. A specific nucleotide dispensation order reduces the number of pyrosequencing cycles required and thereby increases the pyrosequencing quality (Fig. 1B and C). Figure 2 shows that the actual pyrosequencing results displayed relatively high, distinguishable peaks that agreed well with the expected pyrogram patterns for both the WT and MUT viruses at positions 180 and 204. As expected, the signal-to-noise ratio decreased toward the end of the sequence, but the pyrogram patterns for the mutant and WT viruses at the two sites were clearly distinguishable.
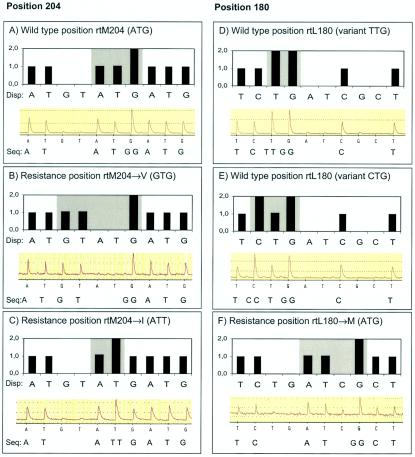
FIG. 2.
Theoretical pyrogram patterns (top of each panel) and examples of raw data from pyrosequencing (bottom of each panel). (A) WT pattern (ATG; methionine) at position rt204; (B) valine mutation pattern at position rt204 (GTG; rtM204→V); (C) isoleucine mutation pattern at position rt204 (ATT; rtM204→I); this mutation is sometimes seen during the transition from the WT sequence to a sequence with a valine mutation; (D) WT pattern (TTG; leucine) at position rt180; (E) WT pattern (CTG; leucine) at position rt180; the patterns in both panels D and E are considered WT and encode a leucine; (F) methionine mutation pattern at position rt180 (ATG; rtL180→M); this mutation is often seen in combination with the more important mutation at position rt204. The letters under the black bars show the dispensation (Disp:) order. The actual sequence detected by pyrosequencing is indicated below the panels after “Seq:”; note that when the pyrogram has a relative peak high of 2.0, the sequence is interpreted as two consecutive identical nucleotides (e.g., the double G in panel B). The light gray area shows the pyrogram for the codon of interest. Note that the nucleotide dispensation order has been designed so that the pyrogram patterns for WT and resistant variants differ at several positions, even though the sequences differ by only a single point mutation.
Detection of mixtures of resistant and WT virus.
To test the abilities of pyrosequencing and Sanger sequencing to detect and quantify minor sequence variants, we prepared artificial mixtures of lamivudine-resistant (MUT) and WT viruses that contained 100 to 0% resistant virus in increments of 10%. Figure 3A shows the theoretical pyrogram patterns for mixtures of different proportions. These patterns were used as templates for interpretation of the results from the mixing experiments. Figure 3B shows representative results for the detection of mutations at position rt180 by pyrosequencing and Sanger sequencing. The preparation and analysis of the mixtures were repeated three times, with similar results each time. Both sequencing methods could detect minor variants in the mixtures made from WT and MUT viruses. The results obtained with 100% WT virus and 100% mutant virus were correct by both methods. In the mixture with 50% WT virus and 50% MUT virus, the pyrogram indicated the presence of both WT and MUT viruses in similar amounts. In contrast, by Sanger sequencing the MUT sequence appeared to dominate and only traces of WT sequence could be detected. With the mixture with 80% WT virus and 20% MUT virus, both pyrosequencing and Sanger sequencing detected the presence of both virus variants, but the pyrosequencing results agreed better with the expected ratio between the WT and MUT sequences. With the mixture with 20% WT virus and 80% MUT virus, Sanger sequencing did not detect WT virus, but pyrosequencing still detected the WT virus. The artificial mixtures were also used to evaluate the abilities of the methods to detect sequence polymorphisms at position rt204. For this position, the results of pyrosequencing also agreed somewhat better than those of Sanger sequencing with the expected ratio of the WT virus/MUT virus sequences (data not shown).
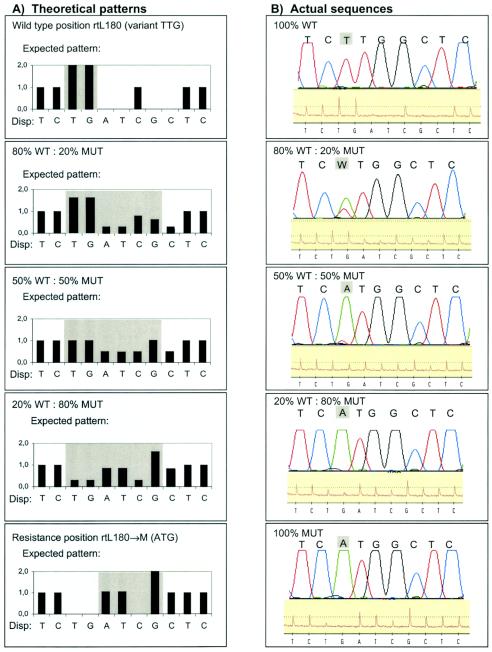
FIG. 3.
Detection of mixtures of WT and lamivudine-resistant (MUT) virus at position rtL180. (A) Theoretical pyrogram patterns for 100% WT (variant TTG), 100% MUT, and WT-MUT mixtures at position rtL180. Note that the 50% WT-50% MUT mixture is expected to create a pyrogram pattern that at each position represents an average for the WT and MUT patterns. The light gray area shows the critical positions for evaluation of the pryosequencing and the Sanger sequencing results. (B) Results from actual sequence analysis of mixtures of WT and MUT HBV variants by Sanger sequencing (top of each panel) and pyrosequencing (bottom of each panel). The panels show 100, 80, 50, 20, and 0% WT. Mixtures were also made to contain 90, 70, 60, 40, 30, and 10% WT (data not shown). The “W” in the second panel of Fig. 3B is a International Union of Pure and Applied Chemistry code which signifies that the ABI310 software identified a mixture of the A and the T nucleotides at this position.
Note that small background peaks were observed at certain positions by both Sanger sequencing and pyrosequencing (Fig. 3B). By Sanger sequencing, a background G nucleotide was observed together with the C nucleotide at position 7 in all five panels in Fig. 3B. By pyrosequencing, background peaks were observed at positions 3 (T) and 7 (C) and were best visualized in the bottom panel of Fig. 3B (100% MUT). By pyrosequencing the distinction between background peaks and real polymorphisms was aided by the fact that several peaks in the pyrogram were used to evaluate a single polymorphic position. Thus, position 4 in the pyrogram for 100% MUT has no signal, which indicates that the peaks at positions 3 and 7 represent background noise.
Analysis of clinical patient samples.
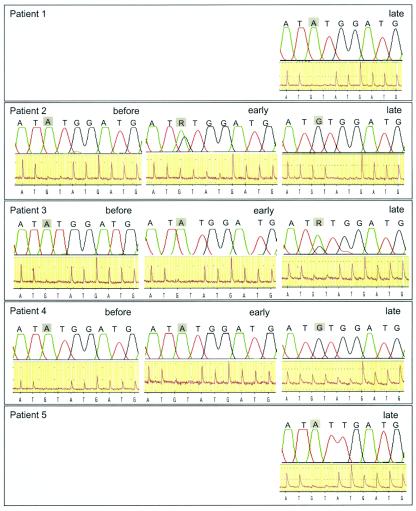
The clinical utility of pyrosequencing was evaluated with serum samples from 20 HBV-infected patients that had been routinely analyzed for the presence of lamivudine-resistant virus at Clinical Virology, Huddinge Hospital. For most patients the resistance test was carried out to investigate lamivudine treatment failures, as evidenced by increasing HBV levels in serum. In addition, the HBV PCR was tested with routine samples from an additional 34 patients. All samples with detectable HBV (>400 HBV DNA copies/ml by the Amplicor HBV Monitor assay) were successfully analyzed. The pyrosequencing and Sanger sequencing results for five representative patients are shown in Fig. 4. For three patients whose viruses displayed lamivudine resistance, we also analyzed a sample obtained before treatment and a sample obtained earlier during treatment. The results of Sanger sequencing and pyrosequencing agreed for patient 1, with both methods detecting a WT pattern. The early treatment sample from patient 2 had a mixture of WT and mutant (rtM204→V) viruses. Sanger sequencing indicated that approximately 30% of the virus population was resistant, while pyrosequencing indicated that the proportion of resistant virus was smaller (approximately 10%). The late treatment sample from patient 2 showed mutant virus by both sequencing techniques. The last sample from patient 3 tested had a mixture of WT and resistant (rtM204→V) viruses. By pyrosequencing approximately 15% of the virus population appeared to be lamivudine resistant, whereas by Sanger sequencing a larger proportion (approximately 30%) appeared to be resistant. The two earlier samples from patient 3 had the WT sequence by both sequencing methods. The last sample from patient 4 showed a mixture of WT and resistant (rtM204→V) viruses by pyrosequencing but only resistant virus by Sanger sequencing. The two earlier samples showed WT virus by both methods. The virus from patient 5 displayed an rtM204→I mutation by both methods.
FIG. 4.
Comparison of Sanger sequencing and pyrosequencing for detection of mutations at position rtM204 in clinical serum samples from five patients who failed lamivudine treatment. For patients 2 to 4, the results of analysis of three samples obtained before the start of treatment and early and late during treatment are shown. For patients 1 and 5, only the results from analysis of the last available on-treatment samples are shown. “R” is an International Union of Pure and Applied Chemistry code which signifies that the ABI310 software identified a mixture of the A and the G nucleotides at this position.
DISCUSSION
In this study we have designed a pyrosequencing method for the detection of lamivudine resistance in clinical samples from HBV-infected individuals. The method was shown to be at least as sensitive as traditional Sanger sequencing for the detection of minority virus populations and had advantages in terms of throughput.
Pyrosequencing and Sanger sequencing differ in several aspects. The most important difference is that pyrosequencing is faster than Sanger sequencing, even when the latter is done on an automated sequencing machine. An ABI310 machine has a throughput of approximately 1 sample per h, whereas the newer ABI3100 machine can analyze 16 samples per h. By pyrosequencing it is possible to sequence 96 samples in approximately 10 min. Another advantage with pyrosequencing compared with Sanger sequencing is that preparation of the PCR product before sequencing is easier and quicker. Cycle sequencing by the Sanger method involves purification of the PCR product, sequencing, and precipitation of DNA before the sample is loaded in the sequencing machine. By pyrosequencing all of these steps are replaced by a fully automated sample preparation step; thus, the time needed for presequencing preparation is approximately one-fifth of the time needed for that for conventional sequencing. Finally, pyrosequencing has the advantage that the nucleotide dispensation order can be designed so that sequence variants that differ by only a single nucleotide generate distinct pyrograms that differ at several peaks rather than at a single peak (Fig. 2). In addition, the use of a specific rather than a cyclic nucleotide dispensation strategy also reduces the number of pyrosequencing cycles and thereby increases sequence quality and the read length. In our experiments it was possible to use a specific nucleotide dispensation strategy because lamivudine resistance involves only a few sequence variants. If the sequence of interest had been highly polymorphic, we would have been forced to choose the less attractive cyclic nucleotide dispensation strategy.
One present limitation of pyrosequencing is that relatively few laboratories have access to pyrosequencing equipment, while many laboratories have automated Sanger sequencing machines. A limitation inherent to the pyrosequencing technique is that the read length is short compared to that of Sanger sequencing: approximately 40 bp versus more than 500 bp. However, this is not a problem during analysis of point mutations, such as the lamivudine resistance mutations in HBV described here. Thus, pyrosequencing is well suited for large-scale screening for known point mutations but not for scanning of larger regions in a viral genome.
Pyrosequencing, like other PCR and sequencing techniques, requires careful primer and probe design to avoid problems due to primer-template mismatches. In this study we designed PCR, pyrosequencing, and Sanger sequencing primers that anneal to regions of the HBV genome that are highly conserved among HBV genotypes A to G. The HBV sequences in all 54 serum samples with detectable HBV levels that we have analyzed thus far have successfully been amplified by PCR. These samples included HBV isolates of genotypes A, B, C, and D (data not shown). This indicates that false-negative results due to primer-template mismatches should be rare, even though more extensive evaluations with samples representing all genotypes are required to formally prove that this is the case.
Today pyrosequencing is mainly used for the detection of SNPs in the human genome. In most cases SNP analysis can have three outcomes: homozygous nonmutant, heterozygous, and homozygous mutant. Analysis of viral genomes is more complicated because polymorphic nucleotide positions can contain mixtures of anything from 0 to 100% of the variant sequence. Sanger-based cycle sequencing is known to generate uneven peak heights, and therefore, quantification of mixed nucleotide positions may be difficult. This has been shown for HIV type 1 resistance testing (7). Pyrosequencing has a theoretical advantage in such an analysis because the nucleotide dispensation order can be designed so that several peaks are used to evaluate a single polymorphic nucleotide position. This should allow identification of polymorphic positions that is more reliable than that which can be obtained by traditional sequencing, in which quantification is based on only one peak or nucleotide position in a chromatogram. Background noise or peaks occur by both pyrosequencing and Sanger sequencing, but they are usually easier to distinguish from true sequence polymorphisms by pyrosequencing because more than one peak is evaluated. The background in pyrosequencing can be due to a low signal level, which can be caused, for example, by a suboptimal PCR and thereby low levels of pyrosequencing template. By pyrosequencing a cutoff level for minimal peak height is objectively calculated to avoid the interpretation of baseline flutter as a minor peak. Background can also appear at specific nucleotide positions as a result of sequence-dependent nucleotide misincorporations or small impurities in the dNTP reagent mixture. This type of background is often reproducible within runs and is directly analogous to similar problems that occur during Sanger sequencing. Because it is reproducible, it usually can be distinguished from true sequence polymorphisms by the inclusion of proper controls. However, interpretation of pyrosequencing results requires some training, just like the interpretation of results of Sanger sequencing. We are working on developing automated methods for pattern recognition that could facilitate a more rapid and reliable identification of polymorphic positions. In our experiments with artificial mixtures of WT and MUT variants, the results of pyrosequencing agreed somewhat better with the expected results than the results of Sanger sequencing did. More experiments with other HBV genotypes and clinical specimens are needed to definitively establish if one method is superior to the other, but our data and the theoretical considerations described above suggest that pyrosequencing is at least as efficient as traditional sequencing for the detection of minor sequence variants. Our conclusion that minor viral variants can be detected by pyrosequencing is supported by a previous study in which pyrosequencing was used to detect resistance to antiretroviral drugs in patients infected with HIV type 1 (4). However, it should be stressed that it remains to be shown if it is clinically relevant to be able to accurately quantify the proportion of resistant virus to WT virus in HBV-infected patients. The mixing experiments were done with clones generated by limiting dilution rather than conventional molecular cloning. Even though these two procedures differ, we feel confident that our results and conclusions would have been very similar if traditional cloning had been used, because both Sanger and pyrosequencing indicated that the clones were pure (within the limits of detection of the two sequencing methods).
Other simple methods for the detection of drug-resistant mutations in HBV have been described, e.g., the INNO-LiPA HBV DR line probe assay (LiPA). This method is based on reverse hybridization of amplified HBV DNA fragments with specific nucleotide probes immobilized on nitrocellulose strips. It has been shown that this method can detect resistance in some patient samples earlier than Sanger sequencing can (3) and can also detect mixtures of WT and resistant virus before viral breakthrough. We plan to compare our pyrosequencing method with LiPA.
In conclusion, we have developed a new assay for the identification of lamivudine resistance in clinical samples from HBV-infected patients. We show that the method has a high throughput and can detect minor sequence variants. Our pyrosequencing method should be useful for the analysis of clinical specimens for the presence of virus variants with lamivudine resistance.
Acknowledgments
This study was supported by grants from the Swedish Medical Research Council. Genotyping was performed at the Wallenberg Consortium North (WCN)-supported KTH SNP genotyping facility.
We thank Peter Simmonds for sharing an alignment of complete HBV genomes and Ola Weiland and Magnus Lindh for valuable discussions and the provision of reference serum samples.
REFERENCES
- 1.Brinchmann, J. E., J. Albert, and F. Vartdal. 1991. Few infected CD4+ T cells but a high proportion of replication-competent provirus copies in asymptomatic human immunodeficiency virus type 1 infection. J. Virol. 65:2019-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Das, K., X. Xiong, H. Yang, C.E. Westland, C. S. Gibbs, S. G. Sarafianos, and E. Arnold. 2001. Molecular modeling and biochemical characterization reveal the mechanism of hepatitis B virus polymerase resistance to lamivudine (3TC) and emtricitabine (FTC). J. Virol. 75:4771-4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lok, A. S. F., F. Zoulim, S. Locarnini, A. Mangia, G. Niro, H. Decraemer, G. Maertens, F. Hulstaert, K. de Vreese, and E. Sablon. 2002. Monitoring drug resistance in chronic hepatitis B virus (HBV)-infected patients during lamivudine therapy: evaluation of performance of INNO-LiPA HBV DR assay. J. Clin. Microbiol. 40:3729-3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O'Meara, D., K. Wilbe, T. Leitner, B. Hejdeman, J. Albert, and J. Lundeberg. 2001. Monitoring resistance to human immunodeficiency virus type 1 protease inhibitors by pyrosequencing. J. Clin. Microbiol. 39:464-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ronaghi, M., M. Uhlén, and P. Nyrén. 1998. A sequencing method based on real-time pyrophosphate. Science 281:363-365. [DOI] [PubMed] [Google Scholar]
- 6.Sarafianos, S. G., K. Das, A. D. Clark, Jr., J. Ding, P. L. Boyer, S. H. Hughes, and E. Arnold. 1999. Lamivudine (3TC) resistance in HIV-1 reverse transcriptase involves steric hindrance with beta-branched amino acids. Proc. Natl. Acad. Sci. USA 96:10027-10032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schuurman, R., D. Brambilla, T. de Groot, D. Huang, S. Land, J. Bremer, I. Benderes, and C. A. B. Boucher. 2002. Underestimation of HIV type 1 drug resistance mutations: results from the ENVA-2 genotyping proficiency program. AIDS Res. Hum. Retrovir. 18:243-248. [DOI] [PubMed] [Google Scholar]
- 8.Severini, A., X. Y. Liu, J. S. Wilson, and D. L. J. Tyrrell. 1995. Mechanism of inhibition of duck hepatitis B virus polymerase by (−)-β-l-2′,3′-dideoxy-3′-thiacytidine. Antimicrob. Agents Chemother 39:1430-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stuyver, L. J., S. A. Locarnini, A. S. Lok, D. D. Richman, W. F. Carman, J. L. Dienstag, and R. F. Schinazi. 2001. Nomenclature for antiviral-resistant human hepatitis B virus mutations in the polymerase region. Hepatology 33:751-757. [DOI] [PubMed] [Google Scholar]
- 10.Tiollais, P., and M. Buendia. 1991. Hepatitis B virus. Sci. Am. 246:116-123. [DOI] [PubMed] [Google Scholar]
- 11.Wong, D. K., A. M. Cheung, K. O'Rourke, C. D. Naylor, A. S. Detsky, and J. Heathcote. 1993. Effects of alpha-interferon treatment in patients with hepatitis B e antigen positive chronic hepatitis B: a meta-analysis. Ann. Intern. Med 119:312-323. [DOI] [PubMed] [Google Scholar]