Abstract
Multiresistant Enterococcus faecium is a major cause of hospital acquired infections and outbreaks. Here, we describe the development of multiple-locus variable-number tandem repeat (VNTR) analysis (MLVA) as a novel typing method to assess the genetic relatedness of E. faecium isolates. Six VNTR loci were used to genotype 392 isolates recovered from different animals and human community, hospital survey, and clinical isolates. From 3 to 13 alleles were found per locus, resulting in 127 different MLVA profiles. Clustering of MLVA profiles confirmed the host-specific genogroups found by multilocus sequence typing (MLST) and showed the grouping of clinical and epidemic isolates that belonged to the MLST-C1 cluster in a distinct MLVA-C1 cluster (sensitivity of 97% and specificity of 90%). Furthermore, the discriminatory power of MLVA is comparable to MLST. MLVA profiles appeared to be relatively stable, since isolates from a single outbreak shared the same MLVA profile, which is a prerequisite when MLVA is used to study hospital outbreaks. Our data show that MLVA is a highly reproducible and portable typing method; in contrast to MLST, it is fast, relatively cheap, and easy to perform. Furthermore, it has the abilities of MLST to recognize genetically related and potential epidemic isolates. Submission of MLVA profiles is possible via a Web-based database for international comparison.
During the last decade, vancomycin-resistant Enterococcus faecium (VREF) has emerged as an important cause of nosocomial infections, especially in immunocompromised patients (20). VREF are often resistant to almost all available antibiotics, seriously hampering treatment of infections. Emergence of ampicillin resistance in E. faecium in the United States in the early 1980s preceded the rapid increase of vancomycin resistance (10, 18, 20). Nowadays, VREF is endemic in many hospitals in the United States, and prevalence rates in European hospitals are rising, with VRE rates above 10% in at least six European countries (3, 4, 9, 15, 26, 32; Annual report of the European Antimicrobial Resistance Surveillance System, 2002 [www.earss.rivm.nl]).
Molecular epidemiological studies of both human- and animal-derived E. faecium isolates with amplified fragment length polymorphism (AFLP) and multilocus sequence typing (MLST) revealed the existence of host-specific genogroups (12, 34). Furthermore, a specific genetic lineage (C1), associated with nosocomial outbreaks and infections and clearly distinct from lineages composed of human community- and animal-derived isolates, was identified. This so-called epidemic genetic lineage, C1, was further characterized by ampicillin resistance and the presence of the esp virulence gene (3, 4, 18). The esp gene encodes the enterococcal surface protein (Esp), which was first described for Enterococcus faecalis and is thought to be an adhesin involved in colonization of the urinary tract (24, 27). The esp gene is located on a pathogenicity island in E. faecalis as well as in E. faecium (17, 23).
Important for infection control is to improve recognition and early detection of the potential epidemic isolates as determined by AFLP and MLST. A low-cost typing scheme that is rapid, reproducible, easy to perform, with the portable character of MLST and the ability to recognize the epidemic MLST-C1 genogroup isolates would therefore be a useful tool for outbreak management. AFLP, although rapid, has a poor rate of reproducibility, and interlaboratory data exchange is not possible. In contrast, MLST is highly reproducible and is appropriate for data exchange via the Internet (www.mlst.net). However, this method is labor intensive and therefore time consuming and rather expensive. Multiple-locus variable-number tandem repeat analysis (MLVA) is based on differences in the variable number of tandem repeats (VNTR) on multiple loci on the chromosome of bacteria, which can rapidly be detected by PCRs with specific primers based on the flanking regions of the tandem repeats. MLVA fulfills the previously mentioned criteria. Since MLVA types (MTs) are discriminated by gain and loss of discrete repeats, MLVA also provides an unambiguous assignment and nomenclature of genotypes, making it a portable technique suitable for data exchange.
In this study, a MLVA typing scheme based on six different tandem repeat loci was developed for E. faecium. Here, we show that MLVA typing is as discriminatory as MLST and able to recognize previously identified host-specific genogroups.
MATERIALS AND METHODS
Bacterial isolates.
MLVA was performed with 392 isolates, including isolates from clinical sites like blood, urine, and wounds (126 isolates); hospital surveys (68 isolates); 25 different documented hospital outbreaks (111 isolates); community surveys (17 isolates); and samples from various animals, food, and the environment (70 isolates) (Table 1). Hospital and community survey isolates were derived from fecal samples, and none of these were associated with hospital outbreaks. Hospital outbreak isolates were recovered from clinical sites like blood and urine, as well as from feces. Computer and statistical analyses were performed on all isolates, including one representative isolate from each outbreak (306 isolates).
TABLE 1.
Isolates used in this study
| Sourcea | No. of isolates | MLVA genogroup | Reference or Source |
|---|---|---|---|
| Community surveillance | 17 | 12A, 3B, 2C | 30, 31, 33-35 |
| Clinical isolates | 126 | 3A, 17B, 18C, 84C1, 4R | 1, 4-6, 14, 18, 22, 32-35 |
| Hospital surveillance | 68 | 17A, 7B, 17C, 24C1, 3R | 4, 14, 19, 26, 32-35 |
| Hospital outbreak Australia-1 | 2 | C1 | 1, 33 |
| Hospital outbreak Australia-2 | 2 | C1 | W. Grubbb |
| Hospital outbreak DK-1 | 1 | C1 | 25 |
| Hospital outbreak DK-2 | 1 | C1 | 25 |
| Hospital outbreak DK-3 | 1 | C1 | 25 |
| Hospital outbreak GR-1 | 2 | C1 | 21 |
| Hospital outbreak GR-2 | 3 | C1 | 21 |
| Hospital outbreak GR-3 | 2 | C1 | 21 |
| Hospital outbreak NL-1-1 | 32 | C1 | 32 |
| Hospital outbreak NL-1-2 | 7 | C1 | 32 |
| Hospital outbreak NL-2-1 | 18 | C1 | 19 |
| Hospital outbreak NL-2-2 | 4 | C1 | 19 |
| Hospital outbreak NL-3-1 | 4 | R | 26 |
| Hospital outbreak NO-1 | 1 | C1 | 15 |
| Hospital outbreak TZA-1 | 5 | C1 | B. Blombergb |
| Hospital outbreak UK-1 | 4 | C1 | 14 |
| Hospital outbreak US-1 | 10 | C1 | 5 |
| Hospital outbreak US-2-1 | 1 | C1 | 2 |
| Hospital outbreak US-2-2 | 1 | C1 | 2 |
| Hospital outbreak US-2-3 | 1 | C1 | 2 |
| Hospital outbreak US-2-4 | 1 | C1 | 2 |
| Hospital outbreak US-2-5 | 1 | C1 | 2 |
| Hospital outbreak US-2-6 | 1 | C1 | 2 |
| Hospital outbreak US-2-7 | 1 | C1 | 2 |
| Hospital outbreak US-3 | 5 | C1 | S. Slaughterb |
| Poultry | 13 | B | 14, 31 |
| Pig | 20 | 16A, 3B, 1C | 8, 14, 30 |
| Domestic pet | 6 | 3B, 3C | 29 |
| Calf | 19 | B | 34 |
| Miscellaneous | 12 | 1A, 2B, 4C, 3C1, 2R | 8, 34 |
| Total | 392 |
Abbreviations: DK, Denmark; GR, Greece; NL, The Netherlands; NO, Norway; TZA, Tanzania; UK, United Kingdom; US, United States.
Personal communication.
Tandem repeat search.
A search for tandem repeats in the unfinished genome sequence of E. faecium published on the Internet site of the DOE Joint Genome Institute (http://www.jgi.doe.gov/JGI_microbial/html/index.html) was performed using the program repeat finder (http://tandem.bu.edu). From the list of tandem repeats, a selection of 10 different loci was made. This selection was based on the following criteria: (i) minimum repeat size of 20 bp, allowing differentiation of the polymorphic VNTR loci by size on agarose gels, (ii) conservation between the tandem repeats (>90%), and (iii) presence in noncoding regions. Initially the 10 VNTR loci were tested on a set of 72 isolates from different origins designated VNTR-1 to VNTR-10. Eventually, six VNTR loci were used and their characteristics are listed in Table 2.
TABLE 2.
VNTR characteristics and specific primers used in MLVA
| Locus name | Repeat length (bp) | Range of repeats | No. of alleles | % Conservation | Primer sequence | PCR program temperature | Estimated size range (bp) | % Agarose gel |
|---|---|---|---|---|---|---|---|---|
| VNTR-1 | 123 | 0-8 | 8 | 95 | F: CTGTGATTTGGAGTTAGATGG | 30 cycles, 52°C | 250-1,012 | 2 |
| R: CATTGTCCAGTAGAATTAGATTTG | ||||||||
| VNTR-2 | 279 | 1-14 | 13 | 96 | F: GATGCTTATTTCCACTGCTTGTTG | TD, 70-60°C | 724-4,351 | 1 |
| R: GTTTTACCCTCTCTTTTAAGGTCAATG | ||||||||
| VNTR-7 | 121 | 1-7 | 7 | 98 | F: CTATCAGTTTCAGCTATTCCATC | TD, 65-55°C | 416-1,021 | 2 |
| R: CTGGTACGAATCAAATCAAGTG | ||||||||
| VNTR-8 | 121 | 1-7 | 7 | 96 | F: GGGGAGTGGCAAAAAATAGTGTG | TD, 70-60°C | 237-963 | 2 |
| R: CAGATCATCAACTATCAACCGCTG | ||||||||
| VNTR-9 | 121 | 1-3 | 3 | 93 | F: CTGCATCTAATAACAAGGACCCATG | TD, 70-60°C | 205-447 | 2 |
| R: ACATTCCGATTAACGCGAAATAAG | ||||||||
| VNTR-10 | 121 | 0-3 | 4 | 96 | F: CCTACAGAAAATCCAGACGG | TD, 65-55°C | 174-474 | 2 |
| R: TTTTTTCCATCCTCT TGAATTG |
DNA preparation and VNTR PCR.
Bacterial isolates were grown overnight on Columbia blood agar plates. Three colonies of bacterial cells were suspended in 20 μl of lysis buffer (0.25% sodium dodecyl sulfate, 0.05 N NaOH) and incubated at 95°C for 5 min. The cell lysate was spin by short centrifugation and diluted with 180 μl of buffer (10 mM Tris-HCl [pH 8.5]). After the lysate was thoroughly mixed, another centrifugation for 5 min at 16,000 × g was performed to remove cell debris. Supernatants were frozen at −20°C until further use.
A total of 2.5 μl of lysate was used in the PCR. Chromosomal DNA was extracted from isolates that did not yield a PCR product with the QiaAmp Blood kit (QIAGEN, Inc., Valencia, Calif.) according to the manufacturer's instructions for gram-positive bacteria, with some minor changes in the lyses of the bacteria. From an overnight culture, 1.5 ml was spin for 2 min, suspended in 200 μl of 10 mM Tris-1 mM EDTA (pH 8.0) and 10 μl of a 50-mg/ml solution of egg white lysozyme (Roche), and incubated at 37°C for 15 min. The bacteria were lysed by the addition of 30 μl of 10% sodium dodecyl sulfate and 20 μl of a 20-mg/ml proteinase K (Merck) solution and subsequently incubated at 65°C for 1 h. Subsequently, the protocol according to the manufacturer's instructions was used. The PCR conditions were not the same for all of the amplification reaction mixtures (Table 2). In all cases, initial denaturation was at 95°C for 15 min, and a final extension step consisted of 5 min at 72°C. For VNTR-1, 35 cycles, each consisting of 30 s at 94°C, 30 s at 52°C, and 30 s at 72°C, were performed. For VNTR-2, VNTR-8, and VNTR-9, a touchdown (TD) PCR was used that included 10 cycles, each consisting of 30 s at 94°C, 30 s at 70°C down to 60°C, and 30 s at 72°C. The annealing temperature during the first cycle was 70°C and decreased 1°C at each cycle during the next nine cycles. During the remaining 25 cycles, an annealing temperature of 60°C was used. For VNTR-7 and VNTR-10, the initial annealing temperature was 65°C and was decreased to 55°C. Reactions were performed in 25-μl volumes with HotStar Taq polymerase and HotStar master mix buffers from QIAGEN. PCR fragments were separated on 1 or 2% agarose gels with a 50-bp, 100-bp, or 1-kb ladder as a size marker (Invitrogen) (Table 2).
Computer analysis of MLVA data.
An MLVA profile was created from the number of repeats for each of the VNTR loci. For each MLVA profile, an MT was assigned. Clustering of the MLVA profiles was performed with BioNumerics software (version 3.5; Applied Maths) by the unweighted pair group method using arithmetic averages (UPGMA) with the categorical coefficient of similarity and with the eBURST algorithm described by Feil et al. (7), initially developed for MLST but also suitable for MLVA. This algorithm is implemented as a Java applet at http://eburst.mlst.net.
Statistics.
To compare the discriminatory ability of MLVA, MLST, and AFLP, Simpson's index of diversity (D) and 95% confidence intervals (CI) were calculated for 78 isolates typed by MLST, MLVA, and AFLP according to the formulas described by Grundmann et al. and Hunter et al. (11, 13). To determine whether MLVA was able to identify E. faecium genotypes belonging to the epidemic MLST-C1 genogroup, the sensitivity and specificity of MLVA were calculated with a set of 291 isolates. In addition, positive and negative predictive values (PPVs and NPVs) of different MLVA profile combinations were calculated to determine to what extent isolates belonging to the MLST-C1 genogroup were identified.
Isolate characterization using MLVA via the Internet.
Comparable to the MLST Internet site (http://www.mlst.net), an Internet site has been developed for the E. faecium MLVA scheme (http://www.mlva.umcutrecht.nl). Through this site, submission of MLVA profiles and assignment of MLVA types are possible. Furthermore, a database with MLVA profiles and strain information can be queried.
RESULTS
Characteristics of VNTR loci.
An MLVA scheme for the molecular typing of E. faecium was developed. All 10 VNTR loci were initially tested with a set of 72 isolates of different origins. PCRs were performed on crude bacterial lysate with the exception of the VNTR-2 PCR, which was also performed on QiaAmp-purified DNA. None of the 10 VNTR PCRs yielded PCR products when E. faecalis DNA was used as a template. Four of the 10 VNTR loci were unsuitable for typing purposes, due to insufficient polymorphism (one locus) and absence in a number of isolates (three loci). The remaining six VNTR loci were used to type the entire strain set. None of the six VNTR loci were found within open reading frames, and they were present on different contigs of the preliminary genome sequence, indicating that the six VNTR loci are probably scattered over the chromosome. The repeat size of the six VNTR loci ranged from 121 to 279 bp, and the number of alleles ranged from 3 for VNTR-9 up to 13 for VNTR-2 (Table 2). The percentage of sequence conservation of the repeats was above 90% for each VNTR locus.
Genogrouping of MLVA profiles.
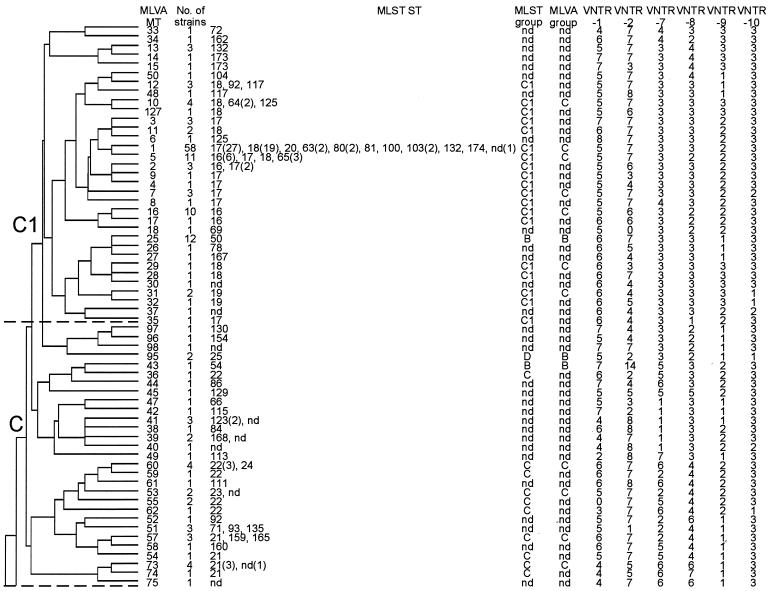
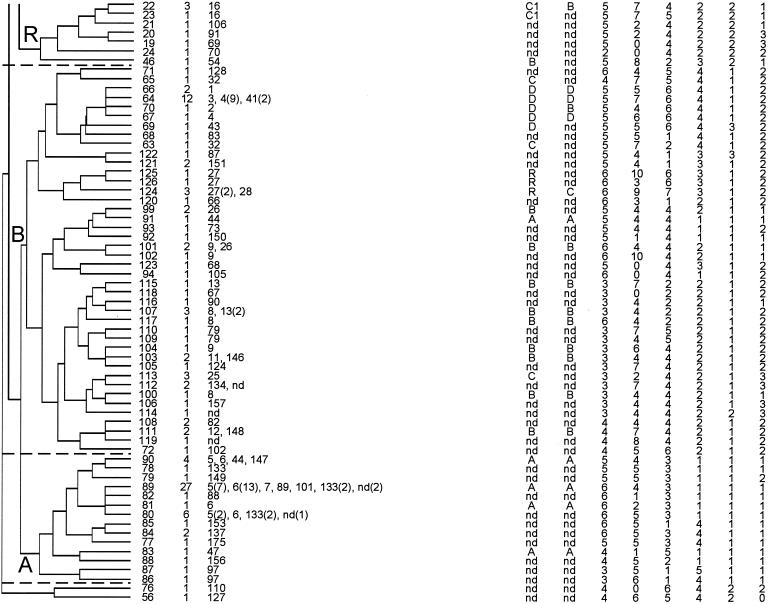
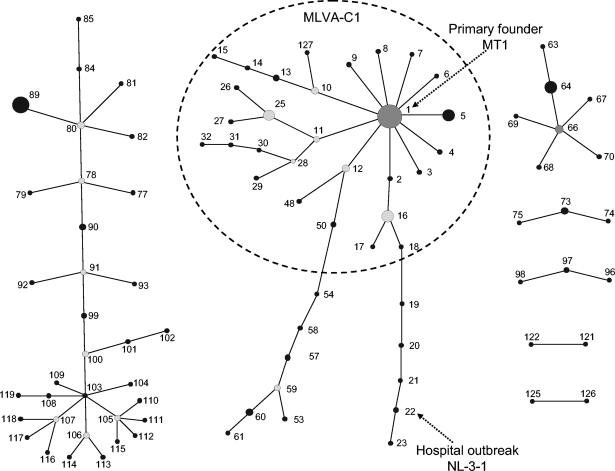
MLVA typing of 392 isolates resulted in 127 different MTs. An UPGMA clustering of the MLVA profiles revealed the existence of five MLVA genogroups (A, B, C, C1, and R) (Fig. 1). Isolates within each group shared repeat numbers in at least two of the six loci (>33%). Naming of the MLVA genogroups was based on MLST and AFLP classifications (12, 34). The majority of isolates (74%) clustered similarly when typed by either MLVA, MLST, or AFLP (Fig. 1). With MLVA, the majority of community survey isolates (12 of 17) and pigs (16 of 20) clustered in genogroup A. Isolates from poultry (13 of 13) and calf (19 of 19) clustered in genogroup B, clinical infection isolates (102 of 126) clustered in genogroups C and C1, and all but 1 hospital outbreak isolate (24 of 25) clustered in genogroup C1. The hospital survey isolates were proportionally represented among genogroups A (17 isolates), B (7 isolates), C (17 isolates), and C1 (24 of 68 isolates). Finally, a small number of isolates of miscellaneous origin clustered in genogroup R (Table 1). The UPGMA clustering of MLVA profiles confirmed clustering of isolates that belonged to the MLST-C1 group (12 isolates) into one MLVA-C1 group with the exception of a single hospital outbreak clone, NL-3-1 (MT-22) (Table 1 and Fig. 1). In addition, three outbreaks—GR-3 (MT-5), NL-2-2 (MT-25), and US-1 (MT-1)—that did not cluster in the epidemic C1 genogroup by MLST (data not shown) grouped within the MLVA-C1 cluster. UPGMA clustering was used to assess the genetic relationship of MLVA profiles and to define and compare genogroups by MLST and AFLP. UPGMA was not suitable for obtaining insight in the evolutionary descent of E. faecium MTs. For this purpose, the eBURST algorithm was used. eBURST was originally developed for the analysis of large MLST data sets to reveal biologically meaningful clusters of sequence types and patterns of evolutionary descent from predicted founder types (7). As both MLST and MLVA profiles are based on a combination of numbers, eBURST should also be suitable for cluster analysis of MLVA profiles (Fig. 2). In Fig. 2, the dotted circle surrounds all MLVA profiles belonging to the MLVA-C1 genogroup based on UPGMA clustering except for MT-33, MT-34, MT-35, and MT-37, which are double-locus variants and therefore not connected to the MLVA-C1 complex. The eBURST clustering suggested that MT-1 is the primary founder of the other MLVA-C1 types. In the MT-1 group, 12 of the 25 documented hospital outbreak isolates, 36 clinical infection isolates, and 12 hospital survey isolates were found. These 60 isolates originated from geographically widespread regions (the United States, Europe, Israel, Tanzania, and Brazil).
FIG. 1.
UPGMA clustering of the 127 different MLVA profiles with a categorical similarity coefficient. For each MT, the number of isolates, MLST sequence types, MLST and AFLP genogroups, and MLVA profile are depicted. C1, epidemic isolates (24 of 25 isolates); C and C1, clinical infection (102 of 126 isolates); B, calf (19 of 19 isolates) and poultry (13 of 13 isolates); A, community survey (12 of 17 isolates) and pigs (16 of 20 isolates). A (17 isolates), B (7 isolates), C (17 isolates), and C1 (24 of 68 isolates), hospital survey; R, miscellaneous origin.
FIG. 2.
eBURST clustering of the 127 different MLVA profiles. In the eBURST algorithm, each MT is represented as a node (solid black dot). For clarity, only clusters or clonal complexes of related MTs are depicted. Dark-gray dot, primary founder. Light-gray dots, subgroups or secondary founders. The clinical relevant genogroup MLVA-C1 based on the UPGMA clustering is surrounded with a dotted circle.
Comparing the level of discrimination of MLVA, MLST, and AFLP.
The discriminatory ability of MLVA, MLST, and AFLP was determined and compared by calculating the genetic diversity (D) with 95% CIs of 78 isolates typed by all three methods. MLVA showed the same level of discrimination as MLST and AFLP (Table 3). When the D values of different combinations of VNTR loci and the complete MLVA scheme of 291 isolates were compared to MLST, the genetic diversity of MLST and the complete MLVA were comparable. The D value of MLVA based on combinations of a limited number of VNTR loci was lower (Table 3). One exception was the combination of VNTR loci 1, 7, 8, and 10, in which the 95% CI of the genetic diversity of this profile just overlapped with that of MLST. Using MLST as a reference, sensitivities of individual VNTR loci and profiles of different VNTR loci combinations to identify isolates that belong to the MLST-C1 cluster ranged from 76% for typing based on VNTR-8 to 97% for MLVA of the complete set of loci. Remarkably, 97% of the MLST-C1 isolates appeared to have three repeats for VNTR-7. The specificity for VNTR-7 was the lowest 65%, but increased to 90% for MLVA on the complete set of loci and for MLVA with combinations of VNTR loci 7, 8, and 9 or VNTR-7, VNTR loci 8, 9, and 10 (Tables 3 and 4). The PPVs and NPVs to identify the MLST-C1 cluster were calculated for the complete MLVA profile and for a combination of either VNTR loci 7, 8, 9, and 10 or VNTR loci 7, 8, and 10. The PPV was 87% and the NPV was 97% for all three VNTR locus combinations analyzed (Table 4).
TABLE 3.
Comparison of Simpson's index of diversity (D) and 95% CI
| Typing method | Isolates typed by:
|
|||
|---|---|---|---|---|
| AFLP, MLST, and MLVAa
|
MLST and MLVAb
|
|||
| D | CI | D | CI | |
| AFLP | 0.94 | 0.90-0.97 | ||
| MLST | 0.94 | 0.91-0.97 | 0.96 | 0.95-0.97 |
| MLVA | 0.93 | 0.90-0.97 | 0.95 | 0.92-0.97 |
| VNTR loci 1, 7, 8, 9, 10 | 0.93 | 0.91-0.95 | ||
| VNTR loci 1, 7, 8, 10 | 0.91 | 0.89-0.93 | ||
| VNTR loci 2, 7, 8, 10 | 0.90 | 0.87-0.93 | ||
| VNTR loci 7, 8, 9, 10 | 0.91 | 0.89-0.93 | ||
| VNTR loci 7, 8, 10 | 0.86 | 0.83-0.89 | ||
| VNTR loci 1, 8, 10 | 0.90 | 0.88-0.92 | ||
| VNTR loci 1, 8, 9 | 0.90 | 0.88-0.92 | ||
| VNTR loci 2, 7, 10 | 0.86 | 0.82-0.90 | ||
A total of 78 isolates were tested.
A total of 291 isolates were tested.
TABLE 4.
MLVA of 291 isolates to identify isolates belonging to the MLST-C1 group
| Procedure (no. of repeats) | Result
|
Profiles indicative of MLST-C1c
|
||||||
|---|---|---|---|---|---|---|---|---|
| Sensitivity (%) | Specificity (%) | PPVa | NPVb | VNTR-7 | VNTR-8 | VNTR-9 | VNTR-10 | |
| Complete MLVA | 97 | 90 | 87 | 98 | ||||
| MLVA with VNTR loci 7, 8, 9, 10 | 97 | 90 | 87 | 98 | 3 | 3 | 2 | 3 |
| 3 | 2 | 2 | 3 | |||||
| 3 | 3 | 2 | 2 | |||||
| 4 | 3 | 2 | 3 | |||||
| 3 | 3 | 3 | 3 | |||||
| 3 | 3 | 1 | 3 | |||||
| 3 | 4 | 3 | 3 | |||||
| 3 | 3 | 3 | 1 | |||||
| 4 | 3 | 3 | 3 | |||||
| 4 | 2 | 3 | 3 | |||||
| 3 | 1 | 2 | 3 | |||||
| 3 | 4 | 1 | 3 | |||||
| MLVA with VNTR loci 7, 8, 10 | 97 | 90 | 87 | 98 | 3 | 3 | 3 | |
| 3 | 2 | 3 | ||||||
| 3 | 3 | 2 | ||||||
| 4 | 3 | 3 | ||||||
| 3 | 4 | 3 | ||||||
| 4 | 2 | 3 | ||||||
| 3 | 3 | 1 | ||||||
| VNTR-1 (5) | 82 | 66 | ||||||
| VNTR-2 (6, 7) | 92 | 68 | ||||||
| VNTR-7 (3) | 97 | 65 | ||||||
| VNTR-8 (3) | 76 | 82 | ||||||
| VNTR-9 (2) | 79 | 83 | ||||||
| VNTR-10 (3) | 93 | 66 | ||||||
PPVs are the number of MLST-C1 and MLVA-C1 isolates divided by the number of MLST-C1 and MLVA-C1 plus MLST-nonC1 and MLVA-C1 isolates.
NPVs are the number of MLST-nonC1 and MLVA-nonC1 negative isolates divided by the number of MLST-nonC1 and MLVA-nonC1 plus MLST-C1 and MLVA-nonC1 isolates.
Values are the number of repeats.
Stability of tandem repeats in outbreaks and in vitro.
To analyze the stability of MLVA profiles, MLVA profiles of isolates recovered during two related outbreaks in the years 2000 to 2003 were determined. The collection comprised 32 isolates collected during hospital outbreak NL-1-1 and 18 isolates collected during hospital outbreak NL-2-1. With the exception of two isolates, all isolates from both outbreaks were shown to have MT-16. One patient carried the outbreak strain for more than 2 years. The MLVA profiles of the isolates recovered from this patient remained unchanged (data not shown). These data suggest that MLVA profiles are stable among strains that are recovered during a hospital outbreak. Two patients from outbreak NL-2-1 acquired colonization with a VREF belonging to MT-17, which is a single-locus variant of MT-16, in which VNTR-1 changed from five to six repeats. The stability of the MLVA profiles in vitro was determined by repeated subculturing of various isolates with a known MLVA profile; no change in MLVA profiles was observed in any case (data not shown).
DISCUSSION
The availability of a fast, reproducible, cheap, and highly discriminatory bacterial typing method is essential for hospital epidemiology. MLVA is a typing technique that combines these characteristics and has been used to type and study the transmission of various bacterial species (16, 28). In this study, we have developed an MLVA typing scheme for E. faecium and compared its discriminatory ability to that of AFLP and MLST. The data presented here show that MLVA is extremely useful for studying the genetic relatedness of E. faecium isolates. Furthermore, MLVA can be used to study the local and global epidemiology of E. faecium.
The comparison of Simpson's index of diversity revealed that MLVA achieved the same degree of discrimination as MLST or AFLP. Moreover, UPGMA cluster analysis of MLVA profiles showed a degree of clustering of isolates in genogroups previously found by AFLP and MLST (12, 34). One specific MLVA genogroup designated MLVA-C1 harbored the majority of clinical isolates and all but one of the hospital outbreak-associated isolates. This genogroup was highly comparable to the MLST-C1 genogroup, which contained epidemic and clinical isolates (12). One hospital outbreak did not cluster in both the MLVA-C1 and the AFLP-C genogroup. Isolates from this outbreak clustered together with the other outbreak-associated isolates only by MLST. In contrast, three other hospital outbreak isolates with three different MLST sequence types that clustered outside the MLST-C1 group did group within MLVA-C1. eBURST confirmed the grouping of clinical relevant isolates in the MLVA-C1 cluster and revealed that MT-1 was the primary founder of this cluster. The existence of a cluster of clinical and epidemic E. faecium isolates was also reported previously (12, 18, 34).
We determined the sensitivity, specificity, PPV, and NPV of MLVA based on combinations of various VNTR loci to identify isolates that belong to the MLST-C1 cluster. It appeared that a single VNTR locus, VNTR-7, yielded the same sensitivity (97%) in identifying genotypes belonging to MLST-C1 as the complete MLVA profile, yet specificity and PPV were rather low (65%). Addition of the VNTR loci 8, 9, and 10 increased specificity to 90% and PPV to 87%, comparable to the complete MLVA profile. Consequently, for rapid screening to identify epidemic isolates, a multistage approach can be used, starting with a single PCR to determine the number of repeats in VNTR-7. Subsequent PCRs with VNTR loci 8, 9, and 10 can confirm the potential epidemic nature of the isolates, when the single PCR revealed three repeats for VNTR-7. However, for library typing and study of the epidemiology of E. faecium, it is recommended that MTs be assigned that based on all six VNTR loci, since the index of diversity decreased strongly when loci were excluded from the full MLVA profile.
MLVA of multiple isolates from two hospital outbreaks (NL-1-1 and NL-2-1) (19, 32) were used to analyze the stability of the tandem repeats and thus the MLVA profiles; such analysis is a prerequisite for identifying and studying hospital outbreaks. The first isolates from these outbreaks date from 2000 and were found in patients from the nephrology departments of both hospitals. Most of the patients were hemodialysis patients who came to the hospital on a regular basis and were often treated with antibiotics, including vancomycin. From the 50 isolates collected during a 3-years period, only 2 isolates isolated from two different patients were found to have a divergent MLVA profile. However, this was only due to the addition of a single repeat in one of the six VNTR loci. This means that MLVA profiles generally remain unchanged during hospital outbreaks, demonstrating that MLVA can be used to study local outbreaks.
The MLVA method described in this study is much faster and cheaper than MLST, because it is a PCR-based method that utilizes simple agarose gels for analysis. AFLP is also considered a fast typing method, but the reproducibility and portability of AFLP are problematic. The fact that this MLVA uses agarose gels to detect and size amplicons also makes it an attractive typing method for use in local laboratories to determine patient-to-patient transmission and to identify potential epidemic isolates. MLVA profiles can easily be stored in a database, facilitating the exchange of MLVA typing data through a Web-based database. Therefore, an Internet site (http://www.mlva.umcutrecht.nl) has been developed for the submission of MLVA profiles to assign the MTs. Furthermore, a strain database is available on the Web site, which contains MTs as well as strain characteristics like MLST sequence type, isolate source, and country of isolate origin.
In the E. faecium MLVA scheme, tandem repeat loci were chosen that, according to the partially annotated E. faecium genome sequence (http://genome.ornl.gov/microbial/efae/), were not located within known open reading frames. Therefore, we assume that changes in repeat numbers are not the result of selective pressure, which means that MLVA data can also be used to study the phylogeny of E. faecium. However, since the E. faecium genome sequence is not yet annotated completely, we cannot exclude that some of the loci are located within coding or regulatory regions.
In conclusion, we developed a fast, reproducible, cheap, and portable typing method that can be used as a tool to study the epidemiology of E. faecium in general and to rapidly detect potential epidemic isolates. We suggest using MLVA as an initial method to screen and type E. faecium in hospital laboratories. Subsequently, MLVA profiles could be added to a Web-based database for international comparison, and representative isolates could be subjected to MLST to gain insight into the global epidemiology of particular MLVA profiles.
Acknowledgments
We thank Jan van Embden (National Institute for Public Health and the Environment) for a critical review of the manuscript, Bob van der Putten for the development of a web-based database on the Internet, and Erik Drenth for hosting the database.
REFERENCES
- 1.Bell, J. M., J. C. Paton, and J. Turnidge. 1998. Emergence of vancomycin-resistant enterococci in Australia: phenotypic and genotypic characteristics of isolates. J. Clin. Microbiol. 36:2187-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonten, M. J., M. K. Hayden, C. Nathan, T. W. Rice, and R. A. Weinstein. 1998. Stability of vancomycin-resistant enterococcal genotypes isolated from long-term-colonized patients. J. Infect. Dis. 177:378-382. [DOI] [PubMed] [Google Scholar]
- 3.Bonten, M. J., R. Willems, and R. A. Weinstein. 2001. Vancomycin-resistant enterococci: why are they here, and where do they come from? Lancet Infect. Dis. 1:314-325. [DOI] [PubMed] [Google Scholar]
- 4.Coque, T. M., R. Willems, R. Canton, R. del Campo, and F. Baquero. 2002. High occurrence of esp among ampicillin-resistant and vancomycin-susceptible Enterococcus faecium clones from hospitalized patients. J. Antimicrob. Chemother. 50:1035-1038. [DOI] [PubMed] [Google Scholar]
- 5.Dunne, W. M., Jr., and W. Wang. 1997. Clonal dissemination and colony morphotype variation of vancomycin-resistant Enterococcus faecium isolates in metropolitan Detroit, Michigan. J. Clin. Microbiol. 35:388-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Endtz, H. P., N. van den Braak, A. van Belkum, J. A. Kluytmans, J. G. Koeleman, L. Spanjaard, A. Voss, A. J. Weersink, C. M. Vandenbroucke-Grauls, A. G. Buiting, A. van Duin, and H. A. Verbrugh. 1997. Fecal carriage of vancomycin-resistant enterococci in hospitalized patients and those living in the community in The Netherlands. J. Clin. Microbiol. 35:3026-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franz, C. M., A. B. Muscholl-Silberhorn, N. M. Yousif, M. Vancanneyt, J. Swings, and W. H. Holzapfel. 2001. Incidence of virulence factors and antibiotic resistance among enterococci isolated from food. Appl. Environ. Microbiol. 67:4385-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goossens, H., D. Jabes, R. Rossi, C. Lammens, G. Privitera, and P. Courvalin. 2003. European survey of vancomycin-resistant Enterococci in at-risk hospital wards and in vitro susceptibility testing of ramoplanin against these isolates. J. Antimicrob. Chemother. 51:(Suppl. S3)iii5-iii12. [DOI] [PubMed] [Google Scholar]
- 10.Grayson, M. L., G. M. Eliopoulos, C. B. Wennersten, K. L. Ruoff, P. C. De Girolami, M. J. Ferraro, and R. C. Moellering, Jr. 1991. Increasing resistance to β-lactam antibiotics among clinical isolates of Enterococcus faecium: a 22-year review at one institution. Antimicrob. Agents Chemother. 35:2180-2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grundmann, H., S. Hori, and G. Tanner. 2001. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J. Clin. Microbiol. 39:4190-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Homan, W. L., D. Tribe, S. Poznanski, M. Li, G. Hogg, E. Spalburg, J. D. Van Embden, and R. J. Willems. 2002. Multilocus sequence typing scheme for Enterococcus faecium. J. Clin. Microbiol. 40:1963-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jordens, J. Z., J. Bates, and D. T. Griffiths. 1994. Faecal carriage and nosocomial spread of vancomycin-resistant Enterococcus faecium. J. Antimicrob. Chemother. 34:515-528. [DOI] [PubMed] [Google Scholar]
- 15.Jureen, R., J. Top, S. C. Mohn, S. Harthug, N. Langeland, and R. J. Willems. 2003. Molecular characterization of ampicillin-resistant Enterococcus faecium isolates from hospitalized patients in Norway. J. Clin. Microbiol. 41:2330-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keim, P., L. B. Price, A. M. Klevytska, K. L. Smith, J. M. Schupp, R. Okinaka, P. J. Jackson, and M. E. Hugh-Jones. 2000. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J. Bacteriol. 182:2928-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leavis, H., J. Top, N. Shankar, K. Borgen, M. Bonten, J. van Embden, and R. J. Willems. 2004. A novel putative enterococcal pathogenicity island linked to the esp virulence gene of Enterococcus faecium and associated with epidemicity. J. Bacteriol. 186:672-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leavis, H. L., R. J. Willems, J. Top, E. Spalburg, E. M. Mascini, A. C. Fluit, A. Hoepelman, A. J. de Neeling, and M. J. Bonten. 2003. Epidemic and nonepidemic multidrug-resistant Enterococcus faecium. Emerg. Infect. Dis. 9:1108-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mascini, E. M., A. C. Gigengack-Baars, R. J. Hene, T. E. Kamp-Hopmans, A. J. Weersink, and M. J. Bonten. 2000. Epidemiologic increase of various genotypes of vancomycin-resistant Enterococcus faecium in a university hospital. Ned. Tijdschr. Geneeskd. 144:2572-2576. (In Dutch.) [PubMed] [Google Scholar]
- 20.Murray, B. E. 2000. Vancomycin-resistant enterococcal infections. N. Engl. J. Med. 342:710-721. [DOI] [PubMed] [Google Scholar]
- 21.Routsi, C., E. Platsouka, R. J. Willems, M. J. Bonten, O. Paniara, G. Saroglou, and C. Roussos. 2003. Detection of enterococcal surface protein gene (esp) and amplified fragment length polymorphism typing of glycopeptide-resistant Enterococcus faecium during its emergence in a Greek intensive care unit. J. Clin. Microbiol. 41:5742-5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schouten, M. A., R. J. Willems, W. A. Kraak, J. Top, J. A. Hoogkamp-Korstanje, and A. Voss. 2001. Molecular analysis of Tn1546-like elements in vancomycin-resistant enterococci isolated from patients in Europe shows geographic transposon type clustering. Antimicrob. Agents Chemother. 45:986-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shankar, N., A. S. Baghdayan, and M. S. Gilmore. 2002. Modulation of virulence within a pathogenicity island in vancomycin-resistant Enterococcus faecalis. Nature 417:746-750. [DOI] [PubMed] [Google Scholar]
- 24.Shankar, N., C. V. Lockatell, A. S. Baghdayan, C. Drachenberg, M. S. Gilmore, and D. E. Johnson. 2001. Role of Enterococcus faecalis surface protein Esp in the pathogenesis of ascending urinary tract infection. Infect. Immun. 69:4366-4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simonsen, G. S., L. Smabrekke, D. L. Monnet, T. L. Sorensen, J. K. Moller, K. G. Kristinsson, A. Lagerqvist-Widh, E. Torell, A. Digranes, S. Harthug, and A. Sundsfjord. 2003. Prevalence of resistance to ampicillin, gentamicin and vancomycin in Enterococcus faecalis and Enterococcus faecium isolates from clinical specimens and use of antimicrobials in five Nordic hospitals. J. Antimicrob. Chemother. 51:323-331. [DOI] [PubMed] [Google Scholar]
- 26.Timmers, G. J., W. C. van der Zwet, I. M. Simoons-Smit, P. H. Savelkoul, H. H. Meester, C. M. Vandenbroucke-Grauls, and P. C. Huijgens. 2002. Outbreak of vancomycin-resistant Enterococcus faecium in a haematology unit: risk factor assessment and successful control of the epidemic. Br. J. Haematol. 116:826-833. [DOI] [PubMed] [Google Scholar]
- 27.Toledo-Arana, A., J. Valle, C. Solano, M. J. Arrizubieta, C. Cucarella, M. Lamata, B. Amorena, J. Leiva, J. R. Penades, and I. Lasa. 2001. The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Appl. Environ. Microbiol. 67:4538-4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Belkum, A., S. Scherer, L. van Alphen, and H. Verbrugh. 1998. Short-sequence DNA repeats in prokaryotic genomes. Microbiol. Mol. Biol. Rev. 62:275-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Belkun, A., N. van den Braak, R. Thomassen, H. Verbrugh, and H. Endtz. 1996. Vancomycin-resistant enterococci in cats and dogs. Lancet 348:1038-1039. [DOI] [PubMed] [Google Scholar]
- 30.van den Bogaard, A. E., P. Mertens, N. H. London, and E. E. Stobberingh. 1997. High prevalence of colonization with vancomycin- and pristinamycin-resistant enterococci in healthy humans and pigs in The Netherlands: is the addition of antibiotics to animal feeds to blame? J. Antimicrob. Chemother. 40:454-456. [DOI] [PubMed] [Google Scholar]
- 31.van den Bogaard, A. E., R. Willems, N. London, J. Top, and E. E. Stobberingh. 2002. Antibiotic resistance of faecal enterococci in poultry, poultry farmers and poultry slaughterers. J. Antimicrob. Chemother. 49:497-505. [DOI] [PubMed] [Google Scholar]
- 32.van der Steen, L. F., M. J. Bonten, E. van Kregten, J. J. Harssema-Poot, R. Willems, and C. A. Gaillard. 2000. Vancomycin-resistant Enterococcus faecium outbreak in a nephrology ward. Ned. Tijdschr. Geneeskd. 144:2568-2572. (In Dutch.) [PubMed] [Google Scholar]
- 33.Willems, R. J., W. Homan, J. Top, M. Santen-Verheuvel, D. Tribe, X. Manzioros, C. Gaillard, C. M. Vandenbroucke-Grauls, E. M. Mascini, E. van Kregten, J. D. Van Embden, and M. J. Bonten. 2001. Variant esp gene as a marker of a distinct genetic lineage of vancomycin-resistant Enterococcus faecium spreading in hospitals. Lancet 357:853-855. [DOI] [PubMed] [Google Scholar]
- 34.Willems, R. J., J. Top, N. van den Braak, A. van Belkum, H. Endtz, D. Mevius, E. Stobberingh, A. van Den Bogaard, and J. D. van Embden. 2000. Host specificity of vancomycin-resistant Enterococcus faecium. J. Infect. Dis. 182:816-823. [DOI] [PubMed] [Google Scholar]
- 35.Willems, R. J. L., J. Top, N. van den Braak, A. van Belkum, D. J. Mevius, G. Hendriks, M. van Santen-Verheuvel, and J. D. van Embden. 1999. Molecular diversity and evolutionary relationships of Tn1546-like elements in enterococci from humans and animals. Antimicrob. Agents Chemother. 43:483-491. [DOI] [PMC free article] [PubMed] [Google Scholar]