Abstract
Mycoplasma bovis is an important veterinary pathogen causing pneumonia, arthritis, and mastitis in infected cattle. We investigated the genetic diversity of 53 isolates collected in the United Kingdom between 1996 and 2002 with pulsed-field gel electrophoresis (PFGE), amplified fragment length polymorphism (AFLP), and random amplified polymorphic DNA (RAPD) analysis. In addition, the influence of variable surface protein (Vsp) profiles on the profiles generated with molecular typing techniques was studied. Both AFLP and RAPD separated the isolates into two distinct groups, but PFGE showed less congruence with the other techniques. There was no clear relationship between the geographic origin or year of isolation of the isolates and the profiles produced. No correlation between Vsp profiles and any of the molecular typing techniques was observed. We propose that RAPD and AFLP provide valuable tools for molecular typing of M. bovis.
Mycoplasmas, belonging to the class Mollicutes, are among the smallest free-living microorganisms capable of autoreplication and are highly fastidious, being difficult to culture and slow growing. Many species are important veterinary pathogens, including Mycoplasma mycoides subsp. mycoides small colony, the causative agent of the Office International des Epizooties list A disease contagious bovine pleuropneumonia. In countries that are free of contagious bovine pleuropneumonia, the most important and pathogenic bovine mycoplasma is considered Mycoplasma bovis (15, 16).
M. bovis was first isolated in 1961 in the United States from a cow with severe mastitis (6). Subsequently, infection has been reported throughout the world, including most European countries (16). M. bovis is a primary cause of bovine pneumonia, arthritis, and mastitis and has also been associated with keratoconjunctivitis, otitis, meningitis, infertility, and abortion (16, 18). In the United Kingdom, bovine respiratory disease is thought to affect 1.9 million cattle annually, with M. bovis infection the likely causative agent of at least a quarter to a third of these losses (16). Losses due to mastitis caused by M. bovis may be higher than those due to respiratory disease, with estimates from the United States of up to $108 million per year and infection rates of up to 70% of a herd (22).
As the prevalence of M. bovis varies widely across the world, there are important trade implications and a pressing need to monitor cattle for M. bovis. However, to date, there is no molecular typing scheme in routine use to trace M. bovis outbreaks or screen isolates from imported animals.
Previously it has been shown that M. bovis contains an elaborate genetic system in which numerous genes encoding variable surface lipoproteins (Vsps) undergo spontaneous on-off switching to generate surface antigen variation (2, 20). Site-specific inversions within the vsp locus and intrachromosomal recombination between vsp genes enable Vsp diversification (12, 13, 14). Previous studies have suggested that altered Vsp expression and the associated chromosomal rearrangements may influence molecular typing techniques (5).
This study assessed the suitability of amplified fragment length polymorphism (AFLP), random amplified polymorphic DNA (RAPD), and pulsed-field gel electrophoresis (PFGE) for the typing of M. bovis. The influence of Vsp rearrangements on the profiles generated by these typing techniques is also explored.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The test population in this study included 53 M. bovis field isolates and the type strain PG45. All the field isolates had been isolated from pneumonic lungs submitted to the laboratory for routine diagnostic purpose from cattle herds located in the United Kingdom in the period from 1996 to 2002 (Table 1). The 53 isolates were randomly selected from our culture collection of M. bovis strains, and all originated from different farms. An additional 10 isolates were collected from a single farm in Exeter, Devon, that had an outbreak of M. bovis respiratory disease in July 2003. All isolates were grown in Eaton's medium at 37°C and 5% CO2 for 48 h without aeration (17) and subsequently identified as M. bovis by PCR with primers specific for the uvrC gene of M. bovis as described previously (25).
TABLE 1.
Origins and characteristics of M. bovis isolates
| Strain | Geographic origin | Yr of isolation | RAPD type | AFLP type | PFGE type | Vsp profile |
|---|---|---|---|---|---|---|
| 208B02 | Devon | 2002 | A1 | A4 | A5 | B |
| 162B02 | Cumbria | 2002 | A1 | A5 | Bb4 | BF |
| 217B02 | N. Yorkshire | 2002 | A1 | A8 | A5 | BF |
| 147B02 | Shropshire | 2002 | A1 | A8 | A2 | BF |
| 6B01 | N. Yorkshire | 2001 | A1 | Ba6 | Ba2 | BO |
| 2B01 | Tyne and Wear | 2001 | A2 | A9 | A3 | B |
| 166B01 | Warwickshire | 2001 | A3 | A1 | A5 | BO |
| 125B01. | Lancashire | 2001 | A4 | A3 | nt | BF |
| 124B96 | N. Yorkshire | 1996 | A4 | A9 | A4 | OF |
| 8B01 | N. Yorkshire | 2001 | A4 | Ba7 | Ba1 | BOC |
| 68B98. | Lancashire | 1998 | A5 | A5 | nt | F |
| 86B96 | Lancashire | 1996 | A6 | A4 | Bb7 | BF |
| 144B02 | Dumfries | 2002 | A6 | A5 | A3 | BF |
| 145B02. | Dumfries | 2002 | A6 | A5 | nt | ABF |
| 187B02 | N. Yorkshire | 2002 | A6 | A1 | Ba5 | AB |
| 124B02. | N. Yorkshire | 2002 | A6 | A1 | nt | ABC |
| 29B02 | Nottinghamshire | 2002 | A6 | A1 | A5 | ACOF |
| 163B02 | Shropshire | 2002 | A6 | A2 | A6 | BOF |
| 171B02 | Lancashire | 2002 | A6 | A10 | A3 | B |
| 222B02 | N. Yorkshire | 2002 | A6 | Ba5 | Ba2 | BO |
| 151B02 | Somerset | 2002 | A7 | A4 | A1 | F |
| 233B02. | Warwickshire | 2002 | A8 | A4 | nt | BO |
| 54B98. | N. Yorkshire | 1998 | A9 | A5 | nt | BF |
| 159B00. | Hampshire | 2000 | A10 | A4 | nt | B |
| 17B01. | Cornwall | 2001 | A10 | A1 | nt | BO |
| 189B02. | Cumbria | 2002 | A11 | A7 | nt | B |
| 149B00 | Tyne and Wear | 2000 | A12 | A6 | Bb4 | BF |
| PG45 | USA | 1962 | A13 | A11 | Ba9 | ABCO |
| 139B01. | Tyne and Wear | 2001 | A14 | A1 | nt | AO |
| 158B01 | Shropshire | 2001 | B1 | Bb1 | Bb4 | O |
| 14B01 | Shropshire | 2001 | B1 | Bb6 | Bb6 | BO |
| 235B02 | N. Yorkshire | 2002 | B1 | Bb8 | Bb3 | C |
| 40B01 | N. Yorkshire | 2001 | B2 | Ba2 | Ba1 | O |
| 226B01 | Lancashire | 2001 | B3 | Ba14 | Ba1 | B |
| 76B98 | Cumbria | 1998 | B3 | Bb6 | Bb6 | AB |
| 30B01 | Shropshire | 2001 | B3 | Bb2 | Bb6 | B |
| 148B02 | Cumbria | 2002 | B4 | Ba15 | Bb5 | BO |
| 202B02 | Shropshire | 2002 | B5 | Ba8 | Ba1 | BF |
| 201B02 | Shropshire | 2002 | B6 | Ba3 | Ba10 | B |
| 231B02 | Cumbria | 2002 | B7 | Ba1 | Ba3 | BO |
| 15B01 | Cornwall | 2001 | B8 | A1 | A2 | OF |
| 40B97. | Warwickshire | 1997 | B9 | A4 | nt | B |
| 62B98 | Hampshire | 1998 | B10 | Bb3 | Bb1 | BO |
| 7B99 | Hampshire | 1999 | B10 | Bb5 | Ba8 | BO |
| 164B02. | N. Yorkshire | 2002 | B11 | Ba10 | nt | B |
| 236B02 | N. Yorkshire | 2002 | B12 | Bb8 | Bb3 | BC |
| 21B01 | Borders | 2001 | B13 | Ba4 | Ba1 | BO |
| 232B02 | Shropshire | 2002 | B14 | Bb4 | A1 | BO |
| 152B02 | Cumbria | 2002 | C1 | Ba12 | Ba4 | BO |
| 133B00 | Borders | 2000 | C1 | Bb9 | Ba6 | A |
| 18B01 | N. Yorkshire | 2001 | C2 | Ba9 | Ba7 | BO |
| 150B02 | Cumbria | 2002 | C2 | Ba13 | Ba6 | BO |
| 9B99 | N. Yorkshire | 1999 | C2 | Ba11 | Ba3 | BO |
| 71B01 | Lancashire | 2001 | C2 | Bb7 | Ba8 | B |
Strain not typeable (nt) by PFGE analysis.
DNA extraction.
DNA was extracted from 10-ml aliquots of stationary-phase cells with a phenol-chloroform procedure as described previously (23) and quantified spectrophotometrically.
RAPD analysis.
The single primer Hum4, 5′-ACGGTACACT-3′ (7), was used for the generation of RAPD profiles. Amplification was performed in a 50-μl total reaction volume containing 100 ng of DNA sample, 10 mM Tris-HCl (pH 9.0), 1.5 mM MgCl2, 50 mM KCl, 0.1% Triton X-100, 0.2 mM each deoxynucleoside triphosphate, and 0.5 U of TaqGold (Perkin-Elmer). Cycling conditions included an initial denaturation step at 94°C for 5 min, followed by 40 cycles of 94°C for 15 s, 37°C for 60 s, and 72°C for 90 s (4). The last cycle included a final elongation at 72°C for 7 min. PCR products were resolved by electrophoresis on 10-cm 2% agarose gels at 60 mA for 1.5 h, stained with ethidium bromide, and visualized under UV illumination.
AFLP analysis. (i) DNA restriction and ligation of PCR adapters.
DNA restriction, ligation of AFLP adapters, and amplification of the modified fragments were carried out as described previously (9). Briefly, the template DNA was simultaneously restricted with 5 U of BglII and 5 U of MfeI (New England Biolabs) at 37°C for 2 h in a restriction buffer containing 10 mM Tris acetate (pH 7.5), 10 mM magnesium acetate, 50 mM potassium acetate, 5 mM dithiothreitol, and 50 ng of bovine serum albumin per μl (27). A 5-μl aliquot of the DNA digest was added to 15 μl of a ligation mixture containing 2 pmol of BglII adapter and 20 pmol of MfeI adapter, 1 U of T4 DNA ligase (Amersham Pharmacia Biotech), 2 μl of 10× ligase buffer (supplied with the enzyme), and 8 μl of restriction buffer. Ligation reactions were carried out overnight at room temperature.
(ii) PCR amplification of DNA template and detection of AFLP fragments.
The modified genomic fragments were amplified with a Bgl-2F-0 primer, which was labeled at the 5′ end with 6-carboxyfluorescein (FAM), and an unlabeled MfeI-0 primer, as described previously (9). The PCRs were performed in a 50-μl total volume containing 2 μl (10-fold diluted) of ligation product, 0.2 mM each deoxynucleoside triphosphate, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 2.5 mM MgCl2, 65 ng of each primer, and 1.5 U of Taq polymerase (Gibco-BRL). Cycling conditions included an initial denaturation step of 94°C for 3 min and 25 cycles of denaturation (94°C for 60 s), annealing (56°C for 60 s), and extension (72°C for 90 s). The last cycle also included a final extension step at 72°C for 10 min. The PCR products were detected with an ABI 377 automated DNA sequencer (Perkin-Elmer) as described previously (9).
PFGE analysis.
Aliquots (10 ml) of stationary-phase M. bovis cultures (.600 of approximately 0.3) were used for PFGE analysis. Cells were harvested by centrifugation (3,500 × . for 20 min at 4°C), washed three times with Tris-EDTA buffer, and resuspended in 300 μl of cold Tris-EDTA buffer. Agarose plugs were made from a 1:1 mixture of 2% low-melting-point agarose (Bio-Rad) and the cell suspension. The plugs were incubated in lysis buffer (10 mM Tris-HCl, 1 mM EDTA, 1% lauroyl sarcosine, 1 mg of proteinase K per ml) for 48 h at 56°C. The plugs were washed four times with Tris-EDTA buffer for 30 min at 4°C. Slices (2 mm) were cut aseptically from the plugs and equilibrated in restriction buffer (Promega) for 1 h. Subsequently, restriction digestion was performed with 30 U of SmaI (Promega) for 16 h according to the manufacturer's instructions. The fragments were resolved on 1% pulsed-field-certified agarose (Bio-Rad) gels with a CHEF-DRIII system (Bio-Rad) at 6 V/cm, with a running time of 20 h at 14°C; included angle of 120°; initial pulse time of 4 s; and final pulse time of 40s. Gels were stained with ethidium bromide (0.5 μg/ml) for 15 min, destained in distilled water for 1 h, and photographed under UV light. A lambda ladder PFGE marker (Sigma) was used for fragment size determination.
Determination of Vsp expression profiles.
The expression of Vsp-related antigens was examined with monoclonal antibodies 4D7, 2A8, 1E5, and 9F1 (kindly provided by Konrad Sachse, Jena, Germany) and Western blotting, as described by Beier et al. (2). The reactivities of the monoclonal antibodies were defined as follows: 1E5, Vsps A, B, and C; 4D7, VspA, VspB, and VspC (distinct from 1E5); 9F1, VspF; and 2A8, Vsps C and O.
Data analysis.
Vsp expression profiles were analyzed by visual inspection of immunoblots. DNA fingerprint patterns obtained with AFLP, RAPD, and PFGE were analyzed with BioNumerics software (Applied Maths). Similarity between fingerprints was calculated with the coefficient of Jaccard (8). Dendrograms were constructed by the unweighted pair group method with arithmetic means (24).
RESULTS
RAPD analysis.
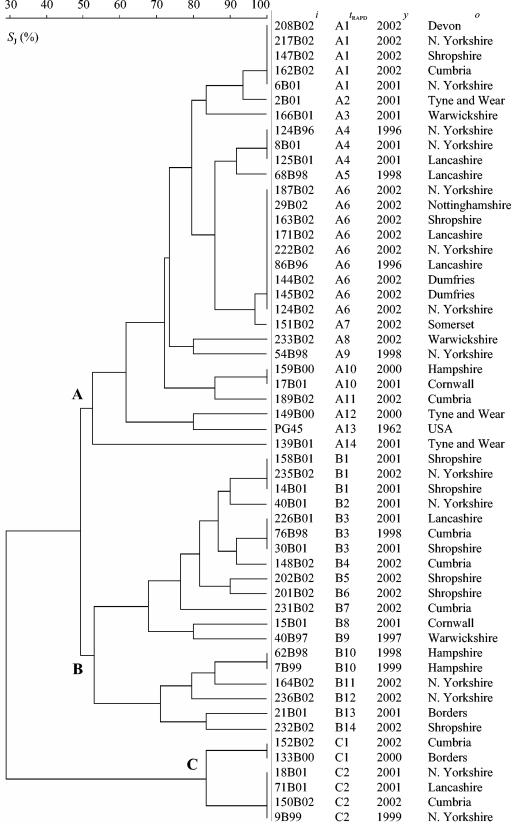
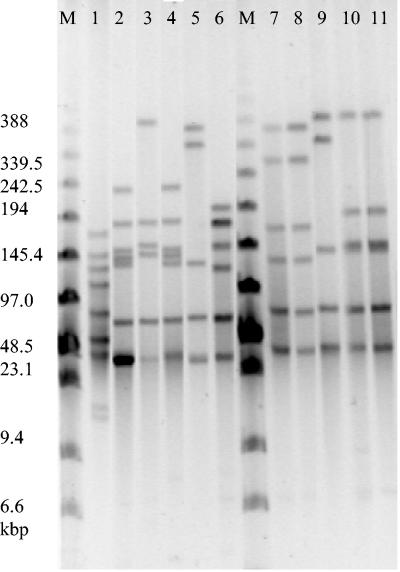
RAPD analysis with the single primer Hum4 produced between two and nine bands ranging from approximately 200 to 1,600 bp. In total, 30 different RAPD profiles were obtained from 54 M. bovis strains analyzed. The isolates could be divided into two major groups (A and B), with approximately 49% similarity between the groups, and a third, smaller group (C), which showed about 28% similarity to the two major groups (Fig. 1). Isolates were unevenly distributed among the groups, with 54% of the isolates in group A, 35% of the isolates in group B, and 11% of the isolates in group C. All group B and C isolates had characteristic bands of approximately 1,400 or 2,000 bp that were not found in group A isolates (Fig. 2). Group C isolates were unusual compared with all the other isolates tested as they showed a very simple fingerprint pattern consisting of two or three fragments. The type strain PG45 clustered within group A and was most closely related to the field isolate 149B00, with about 73% similarity.
FIG. 1.
Genetic relationships between M. bovis strains based on comparison of RAPD electrophoretic patterns obtained with the primer Hum4. The dendrogram was produced with the unweighted pair group method with arithmetic means (UPGMA) method with Jaccard similarity coefficient (.J) matrix. ., strain designation; .RAPD, arbitrarily assigned RAPD type; ., year of isolation; ., origin of strain.
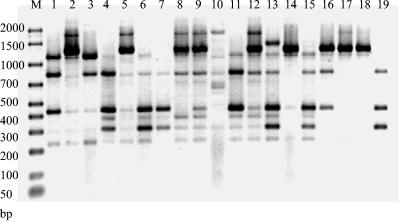
FIG. 2.
Representative profiles produced by RAPD with the primer Hum4. Lane M, Bio-Rad markers; lane 1, 222B02 (profile A6); lane 2, 235B02 (profile B1); lane 3, 6B01 (profile A1); lane 4, 151B02 (profile A7); lane 5, 226B01 (profile B3); lane 6; 86B96 (profile A6); lane 7, 2B01 (profile A2); lane 8, 14B01 (profile B1); lane 9, 76B98 (profile B3); lane 10, 164B02 (profile B11); lane 11, 8B01 (profile A4); lane 12, 30B01 (profile B3); lane 13, 68B98 (profile A5); lane 14, 236B02 (profile B12); lane 15, 145B02 (profile A6); lane 16, 21B02 (profile B13); lane 17, 18B01 (profile C2); lane 18, 71B01 (profile C2); lane 19, 139B01 (profile A14).
The technique proved to be reproducible, with the same banding pattern obtained for replicate isolates even if DNA extractions were done on separate occasions (results not shown). The only variation between replicate samples was in terms of the intensity of bands rather than the presence or absence of bands.
There was no obvious relationship between geographical and/or temporal origin of strains and RAPD profiles. Isolates with close geographical origins often had highly variable fingerprint patterns. Conversely, in many cases, isolates from different origins had the same fingerprint type. For example, RAPD type A1 included isolates from widely varied areas of the United Kingdom such as Devon, Shropshire, and north Yorkshire, which are approximately 515 km apart. Similarly, the most common profile (profile A6, which was found in nine isolates) was found among isolates from different origins, such as Dumfries, Lancashire, and Nottinghamshire, and over a relatively long period of time, from November 1996 to September 2002. There were differences in the overall level of heterogeneity of the groups. Group B showed the most variability, as 74% of the isolates had unique profiles, whereas only 33 and 48% of group A and C profiles were unique, respectively.
The stability of isolate profiles within an individual herd was demonstrated, as all 10 isolates from the M. bovis outbreak in Exeter, Devon, produced identical RAPD profiles (results not shown).
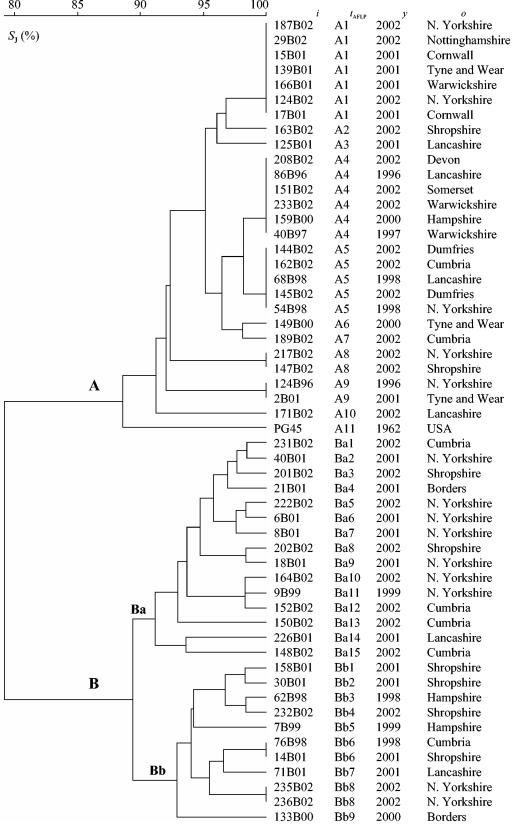
AFLP analysis.
In total, 54 isolates were typed with AFLP, and 35 different profiles were obtained. By comparison of AFLP fragments in the size range 50 to 400 bp, the M. bovis isolates could be separated into two distinct groups (A and B), with about 79% similarity between the groups (Fig. 3). Several bands were detected that were not shared between members of the two groups: fragments of 199, 269, and 337 bp were detected only among the members of group B, while fragments of 223, 230, 248, and 387 bp were present only among the members of group A. In addition, all of the members of group A had a fragment of either 254 or 255 bp that was not detected among any of the group B strains.
FIG. 3.
Genetic relationship between M. bovis strains based on comparison of AFLP profiles (50- to 400-bp fragment size range) produced by amplification of BglII and MfeI DNA templates with nonselective primers. The dendrogram was produced with the UPGMA method with Jaccard similarity coefficient (.J) matrix. ., strain designation; .AFLP, arbitrarily assigned AFLP type; ., year of isolation; ., origin of strain.
Distinct differences in heterogeneity were seen between group A and group B. Group B exhibited the greatest genetic diversity, with 93% of isolates having unique profiles, compared with 39% of group A isolates. Group B could also be further divided into two subgroups (Ba and Bb). Three fragments (157, 267, and 287 bp) were present exclusively among the strains of subgroup Ba. Interestingly, a single fragment (137 bp) which was present in all subgroup Ba strains and all group A strains but none of the group Bb strains was detected.
As with RAPD, there was no geographical or temporal relationship between isolate origins and the profiles produced. For example, group A4 isolates were collected between 1996 and 2002 from diverse regions of the United Kingdom, such as Devon and Lancashire, which lie 435 km apart.
There was a high degree of congruence between AFLP and RAPD, with 90% of the isolates in RAPD group A also in AFLP group A and 80% of the isolates in RAPD group B also in AFLP group B. As was found by RAPD analysis, the type strain PG45 clustered within group A.
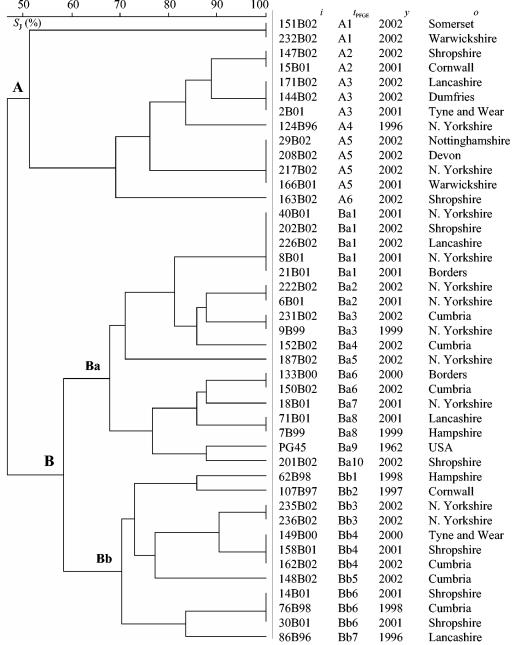
PFGE analysis.
PFGE analysis with SmaI produced profiles for 42 field isolates and the type strain PG45 (Fig. 4). The profiles consisted of between five and nine bands, with a range of approximately 2 to 150 kb (Fig. 5). PFGE analysis could separate the isolates into two main groups (A and B), with approximately 47% similarity between the groups (Fig. 4). Group B demonstrated greater heterogeneity than group A and could be further subdivided into two groups (Ba and Bb), with about 57% similarity between the groups. Overall, 23 different profiles were produced with PFGE.
FIG. 4.
Genetic relationship between M. bovis strains based on comparison of PFGE profiles obtained with the SmaI restriction enzyme. The dendrogram was produced with the UPGMA method with Jaccard similarity coefficient (.J) matrix. ., strain designation; .PFGE, arbitrarily assigned PFGE type; ., year of isolation; ., origin of strain.
FIG. 5.
Representative PFGE fingerprint patterns of M. bovis strains produced with the SmaI restriction enzyme. Lane M, pulsed-field markers (Sigma); lane 1, 201B02 (profile Ba10), lane 2, 222B02 (profile Ba2); lane 3, 124B96 (profile A4); lane 4, 6B01 (profile Ba2); lane 5, 171B02 (profile A2); lane 6, 152B02 (profile Ba4); lane 7, 29B02 (profile A5); lane 8, 217B02 (profile A5); lane 9, 147B02 (profile A2); lane 10, 151B02 (profile A1); lane 11, 233B02 (profile A1).
In some cases there was concordance between the groupings obtained with PFGE and AFLP/RAPD; for example, 76B98 and 30B01 had identical profiles by PFGE and RAPD and were placed in the same subgroup, Bb, with AFLP. Similarly, isolates 235B02 and 236B02 gave indistinguishable profiles by both PFGE and AFLP and were placed in the same cluster with RAPD. Overall there was a high degree of congruence between group A isolates, as 77% of PFGE group A isolates fell in RAPD group A and 85% fell into AFLP group A. However, PFGE group B isolates showed less concordance with the other typing techniques, as 38 and 66% of PFGE group Ba isolates fell into RAPD and AFLP group B, respectively, and 66% of group Bb isolates fell into AFLP and RAPD group B. In contrast with AFLP and RAPD, the type strain PG45 fell into group Ba and was most closely related to 201B02.
As with AFLP and RAPD, there seemed to be no clear relationship between the geographical origin of the isolates and the profile produced. The stability of isolate profiles within an individual herd was demonstrated, as all 10 isolates from the M. bovis outbreak in Exeter, Devon, produced identical PFGE profiles (results not shown).
A number of the isolates (11 of 54) were untypeable by PFGE. These isolates produced smeared fingerprints, probably due to DNase activity. Attempts to remove DNase activity and thus improve the fingerprints by the addition of thiourea to the running buffer and formalin treatment of the cells was unsuccessful.
Determination of Vsp profiles.
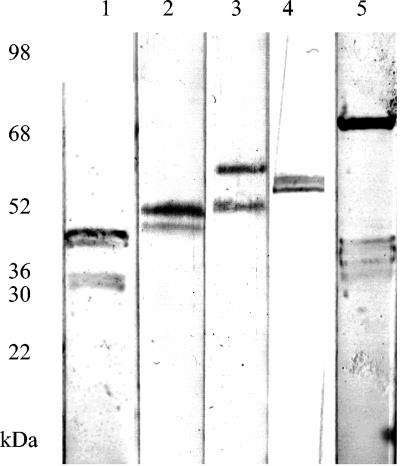
In total, 54 isolates were assayed for the presence of Vsps by Western blotting with Vsp-specific monoclonal antibodies (Fig. 6). The most common Vsps were VspB (46 kDa), O (40 kDa) and F (55 kDa), which were found in 60, 45, and 26% of isolates, respectively. Less common Vsps were VspA (63 kDa) and VspC (75 kDa), which were found in 12 and 6% of isolates, respectively. In total, 16 different combinations of Vsp were found. The most frequently found Vsp profiles were BO (16 isolates), B (11 isolates), and BF (9 isolates). Other Vsp profiles were O (two isolates), AB (two isolates), OF (two isolates), and F (two isolates); all other Vsp combinations were found only in single isolates (Table 1). The type strain showed the greatest diversity of Vsp expression with Vsps A, B, C, and O detected.
FIG. 6.
Detection of Vsps in M. bovis by immunoblotting. Lane 1, 40B01 (Vsp O); lane 2, 164B02 (Vsp B); lane 3, 187B02 (Vsps B and A); lane 4, 68B98 (Vsp F); lane 5, 235B02 (Vsp C).
There appeared to be no clear relationship between Vsp expression and the profiles obtained by any of the molecular typing methods used in this study. Many isolates that were identical or highly similar by molecular typing gave widely variable Vsp profiles; for example, 76B98 and 30B01 were identical by both RAPD and PFGE but possessed different Vsp profiles. In addition, isolates 217B02 and 147B02 were identical by AFLP and RAPD and had the same Vsp profile but exhibited different PFGE profiles.
Nucleotide sequence analysis of the Vsps detected in this study was undertaken (NCBI accession numbers AF396970, AF162138, AF396971, AF162149, and AF162140), and it was found that only Vsps A and C possessed a SmaI restriction site (Webcutter version 2.0).
DISCUSSION
M. bovis infection represents a major disease burden for cattle producers worldwide, emphasizing the need for a reliable method of molecular typing for outbreak investigation and epidemiological surveillance. This study represents the first attempt to assess the genetic variability of M. bovis isolates in the United Kingdom.
Following molecular typing with RAPD and AFLP, a distinct pattern of genetic variability emerged, with United Kingdom isolates divided into two markedly different groups (A and B) with about 79% similarity by AFLP and 49% similarity by RAPD. A small number of isolates were placed in RAPD group C, but all of these isolates possessed group B-specific bands and all fell into AFLP group B, indicating that these isolates are probably more accurately described as members of group B. With RAPD, it was found that all group B (and C) isolates had a common band that was absent in all group A isolates, while AFLP analysis revealed a total of nine fragments that were not shared between the two main groups. Such a distinct pattern of chromosomal variances clearly indicates that the two major clusters represent two divergent lines of descent that arose during the evolutionary development of the species. However, in light of the fact that all the strains analyzed in this study except the type strain PG45 derived from a relatively narrow geographical area (the United Kingdom) and were isolated in a relatively short period of time (6 years), it is not possible to draw a general conclusion about the population structure of M. bovis. It would therefore be interesting to examine genomic polymorphism between M. bovis strains that were isolated in a broader time frame and at other geographic localities in order to see whether the species consists of more than two clonal lineages, as detected in this study.
In terms of the overall diversity of M. bovis in the United Kingdom, AFLP and RAPD analysis showed that group B isolates were relatively more heterogeneous than isolates of the A group, which may indicate slightly different rates of genetic drift between the two putative clonal lines. On the basis of the AFLP results, group B isolates could be further divided into two subgroups, Ba and Bb. As group Bb isolates lacked a band that was present in group A and group Ba isolates, it can be speculated that the mutational event responsible for the disappearance of the fragment from Bb probably occurred after the divergence of groups A and B.
Previous studies with RAPD analysis on M. bovis in Europe and the United States have indicated some degree of intraspecies heterogeneity and concluded that isolates were likely to have derived from more than one source. However, these studies only analyzed a limited number of strains and are not directly comparable to the present study (3, 7). On the other hand, in a directly comparable AFLP study, Danish M. bovis strains were found to be relatively homogeneous and to cluster closely with the type strain PG45, which has a distinct geographical and temporal origin (10). When compared with the findings of this study, it seems probable that the Danish strains are of the same lineage as the group A isolates, as they also cluster closely with the type strain. This notion is supported by the fact that the Danish isolates had cluster bands in common with United Kingdom group A isolates (B. Kokotovic, unpublished data).
It is possible to speculate that the two distinct groups identified by RAPD and AFLP in this study represent two clonal lines of descent. It is hypothesized that group A may have entered the United Kingdom from Europe around the time of the creation of the European Union single market in 1993, when large numbers of cattle came into the United Kingdom from continental Europe. Interestingly, at this time M. bovis was isolated for the first time in Ireland, which had previously been free of the disease (16). Group B could have a different and possibly older ancestral origin and have derived from the first strains of M. bovis isolated in the United Kingdom in 1975 (26). Clearly, further work on a molecular typing scheme incorporating strains from both the United States and continental Europe will be necessary to clarify the origin of the United Kingdom isolates.
In this study, a lack of correlation between geographical origin, time of isolation, and genomic profiles obtained for the United Kingdom strains was noted, and in some instances, strains from farms over 400 km apart had identical profiles. These findings are similar to those in a study of European M. bovis strains with restriction enzyme analysis, which indicated that there was no correlation between the geographical origin, genetic heterogeneity, or isolation date (21). It is likely that the findings of the present study reflect the extensive cattle movement that occurred within the United Kingdom and that the cattle had diverse origins and in most cases did not originate on the farm where they were sampled. This further underlines the need for a molecular typing scheme, as animal movements mean that outbreaks are likely to disseminate rapidly throughout the United Kingdom.
The genetic stability of M. bovis isolates from an outbreak on a single farm was also examined, and all isolates were found to be identical, indicating homogeneity of profiles within this farm. However, this is an area which requires much further research as it seems likely that the stability of profiles within a given herd will be influenced by the management of that herd, that is, whether the herd is closed (with all cattle being brought in at the same time) or open (with cattle brought in from different sources over a period of time). In the latter case, it seems probable that M. bovis outbreaks could involve multiple strains in the event that two or more cattle enter the herd carrying M. bovis infection from different origins. The Exeter herd used in this study, although not entirely closed, had only obtained cattle from a single source within the 6 months preceding sampling for this study.
Previous studies with RAPD also found that the typing profiles of M. bovis corresponded well with husbandry conditions, with a single profile found in closed herds with one source of infection but outbreaks in open herds producing multiple profiles indicating several sources of infection (3). Although this study has not looked specifically at the stability of profiles over time, it does seem likely that profiles obtained are relatively stable, as indistinguishable profiles (albeit in different geographical areas) were found over a 6 year period in this study. Stability of isolate profiles was also demonstrated in a comparable study where identical AFLP profiles were found over a 10 year period in the same area (10).
All of the isolates used in this study were obtained from pneumonic cattle, as M. bovis less frequently causes mastitis and arthritis in the United Kingdom. In contrast, in the United States M. bovis is more commonly associated with mastitis (22). It would be informative to compare isolates associated with different disease types and also determine whether groups A and B found in this study are associated with various levels of severity of disease.
This study has compared and evaluated different molecular epidemiological typing techniques for M. bovis. Previous studies of RAPD with the Rep1R-I/Rep2-I primer combination found RAPD to be a reliable and rapid method of typing M. bovis strains (3). RAPD requires the least specialist equipment of all of the typing methods studied but the differing intensity of banding patterns could be difficult to interpret without an automated gel analysis package, such as BioNumerics. Also RAPD requires very careful sample preparation to ensure reproducibility.
AFLP has many advantages as a typing technique: it is highly discriminatory, robust and reproducible, and suitable for high- throughput analysis and automation, which means that databases can be created for long-term data storage and interlaboratory comparison. Previous studies have also demonstrated the utility of AFLP for typing M. bovis and found it to have a discriminatory power at least comparable to that of PFGE (10).
In this study, PFGE showed the lowest level of congruence with the other typing techniques, was the most time-consuming and had the additional disadvantage that many isolates could not be typed as a result of DNase activity. In contrast, earlier studies of AFLP and PFGE have indicated concordance between techniques in terms of the overall level of genetic heterogeneity but these findings are not comparable to the present study as the relationship between individual isolates was not examined with PFGE (10, 11). The limitations of PFGE for M. bovis typing have also been highlighted previously and it has been suggested that the chromosomal rearrangements responsible for altered Vsp expression may influence PFGE profiling, particularly if SmaI is used, as the vsp locus may contain a SmaI restriction site (5). However, in this study the presence of different Vsp combinations was not found to influence PFGE profiles or, incidentally, AFLP or RAPD profiles obtained. Many isolates had identical or highly similar molecular typing profiles but widely different Vsp profiles. These results are not surprising, as sequence analysis of the Vsp operons indicates that there is only a restriction site for SmaI in Vsps A and C, and these Vsps are found very infrequently in the United Kingdom (see below). The lack of correlation between Vsp profiles and RAPD profiles has already been reported for M. bovis (4) but the relationship between PFGE and Vsp profiles has not previously been studied.
Interestingly, this study found that a high proportion of isolates had Vsps B, O, or F, but Vsps A and C were found infrequently. In contrast, previous studies have reported that VspA was ubiquitous in M. bovis isolates obtained from French cattle (19). However, it is important to note that this study only looked at the expression of Vsp proteins by Western blotting and therefore may only indicate phenotypic variation and that genetic analysis of the vsp locus (by Southern blotting) would be necessary to confirm the Vsps present in each isolate.
In conclusion, United Kingdom isolates can be separated into two distinct groups by AFLP and RAPD analysis. These techniques may provide a valuable tool for epidemiological typing of M. bovis isolates.
Acknowledgments
We thank the Department for the Environment Food and Rural Affairs for continued funding.
We thank Colin Churchward for technical assistance.
REFERENCES
- 1.Behrens, A., M. Heller, H. Hirchhoff, D. Yogev, and R. Rosengarten. 1994. A family of phase- and size-variant membrane surface lipoprotein antigens (Vsps) of Mycoplasma bovis. Infect. Immun. 62:5075-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beier, T., H. Hotzel, I. Lysnyansky, C. Grajetzki, M. Heller, B. Rabeling, D. Yogev, and K. Sachse. 1998. Intraspecies polymorphism of vsp genes and expression profiles of variable surface protein antigens (Vsps) in field isolates of Mycoplasma bovis. Vet. Microbiol. 63:189-203. [DOI] [PubMed] [Google Scholar]
- 3.Butler, J. A., C. C. Pinnow, J. U. Thomson, S. Levisohn, and R. F. Rosenbusch. 2001. Use of arbitrarily primed polymerase chain reaction to investigate Mycoplasma bovis outbreaks. Vet. Microbiol. 78:175-181. [DOI] [PubMed] [Google Scholar]
- 4.Byrne, W., M. McCormack, H. Hotzel, and K. Sachse. 2000. Variable surface protein expression profiles and PCR fingerprints of clinical isolates of Mycoplasma bovis from Irish cattle, p. 14-17. In J. B. Poveda, A. Fernandez, K.-E. Johansson, and J. Frey (ed.), Mycoplasmas of ruminants: pathogenicity, diagnostics, epidemiology and molecular genetics, vol. 5. European Commission, Brussels, Belgium. [Google Scholar]
- 5.Citti, C., A. Lischewski, K. Siebert-Gulle, and R. Rosengarten. 2000. Limitation of pulsed field gel electrophoresis analysis for the typing of Mycoplasma bovis, p. 46-49. In J. B. Poveda, A. Fernandez, K.-E. Johansson, and J. Frey (ed.), Mycoplasmas of ruminants: pathogenicity, diagnostics, epidemiology and molecular genetics, vol. 5. European Commission, Brussels, Belgium. [Google Scholar]
- 6.Hale, H. H., C. F. Helmboldt, W. N. Plastridge, and E. F. Stula. 1962. Bovine mastitis caused by Mycoplasma species. Cornell Vet. 52:582-591. [PubMed] [Google Scholar]
- 7.Hotzel, H., B. Schneider, and K. Sachse. 1998. Investigation of Mycoplasma bovis field isolates using PCR fingerprinting, p. 17-19. In G. Leori, F. Santini, E. Scanziani, and J. Frey (ed.), Mycoplasmas of ruminants: pathogenicity, diagnostics, epidemiology and molecular genetics, vol. 2. European Commission, Brussels, Belgium. [Google Scholar]
- 8.Jaccard, P. 1908. Nouvelles recherches sur la distribution florale. Bull. Soc. Vaudoise Sci. Nat. 44:223-270. [Google Scholar]
- 9.Kokotovic, B., N. F. Friis, J. S. Jensen, and P. Ahrens. 1999. Amplified-fragment length polymorphism fingerprinting of Mycoplasma species. J. Clin. Microbiol. 37:3300-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kusiluka, L. J. M., B. Kokotovic, B. Ojeniyi, N. F. Friis, and P. Ahrens. 2000. Genetic variations among Mycoplasma bovis strains isolated from Danish cattle. FEMS Microbiol. Lett. 192:113-118. [DOI] [PubMed] [Google Scholar]
- 11.Kusiluka, L. J. M., B. Ojeniyi, and N. F. Friis. 2000. Increasing prevalence of Mycoplasma bovis in Danish cattle. Acta Vet. Scand. 41:139-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lysnyansky, I., Y. Ron, K. Sachse, and D. Yogev. 2001. Intrachromosomal recombination within the vsp locus of Mycoplasma bovis generates a chimeric variable surface lipoprotein antigen. Infect. Immun. 69:3703-3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lysnyansky, I., Y. Ron, and D. Yogev. 2001. Juxtaposition of an active promoter to vsp genes via site-specific DNA inversions generates antigenic variation in Mycoplasma bovis. J. Bacteriol. 183:5698-5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lysnyansky, I., R. Rosengarten, and D. Yogev. 1996. Phenotypic switching of variable surface lipoprotein in Mycoplasma bovis involves high-frequency chromosomal rearrangements. J. Bacteriol. 178:5395-5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicholas, R. A. J. 1998. The veterinary significance of mycoplasmas. Methods Mol. Biol. 104:17-24. [DOI] [PubMed] [Google Scholar]
- 16.Nicholas, R. A. J., and R. D. Ayling. 2003. Mycoplasma bovis: disease, diagnosis and control. Res. Vet. Sci. 74:105-112. [DOI] [PubMed] [Google Scholar]
- 17.Nicholas, R. A. J., and S. E. Baker. 1998. Recovery of mycoplasmas from animals. Methods Mol. Biol. 104:37-43. [DOI] [PubMed] [Google Scholar]
- 18.Pfützner, H., and K. Sachse. 1996. Mycoplasma bovis as an agent of mastitis, pneumonia, arthritis and genital disorders. Rev. Sci. Tech. Off. Int. Epiz. 15:1477-1494. [DOI] [PubMed] [Google Scholar]
- 19.Poumarat, F., D. Le Grand, M. Solsona, R. Rosengarten, and C. Citti. 1999. Vsp antigens and vsp-related DNA sequences in field isolates of Mycoplasma bovis. FEMS Microbiol. Lett. 173:103-110. [DOI] [PubMed] [Google Scholar]
- 20.Poumarat, F., M. Solsona, and M. Boldini. 1994. Genomic, protein and antigenic variability of Mycoplasma bovis. Vet. Microbiol. 40:305-321. [DOI] [PubMed] [Google Scholar]
- 21.Rosengarten, R., A. Behrens, A. Stetefield, M. Heller, M. Ahrens, K. Sachse, D. Yogev, and H. Kirchhoff. 1994. Antigen heterogeneity among isolates of Mycoplasma bovis is generated by high frequency variation of diverse membrane surface proteins. Infect. Immun. 62:5066-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosengarten, R., and C. Citti. 1999. The role of ruminant mycoplasma in systemic infection, p. 14-17. In L. Stipkovits, R. Rosengarten, and J. Frey (ed.), Mycoplasmas of ruminants: pathogenicity, diagnostics, epidemiology and molecular genetics, vol. 3. European Commission, Brussels, Belgium. [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., vol. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Sneath, P. H. A., and R. R. Sokal. 1973. Numerical taxonomy. The principle and practice of numerical classification. W. H. Freeman & Co., San Francisco, Calif.
- 25.Subramaniam, S., D. Bergonier, F. Poumarat, S. Capaul, Y. Schlatter, J. Nicolet, and J. Frey. 1998. Species identification of Mycoplasma bovis and Mycoplasma agalactiae based on the uvrC genes by PCR. Mol. Cell. Probes 12:161-169. [DOI] [PubMed] [Google Scholar]
- 26.Thomas, L. H., C. J. Howard, and R. N. Gourlay. 1975. Isolation of Mycoplasma agalactiae var. bovis from a calf pneumonia outbreak in the south of England. Vet. Rec. 97:55-56. [DOI] [PubMed] [Google Scholar]
- 27.Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. van de Lee, M. Hornes, A. Frijters, J. Pot, J. Peleman, M. Kupier, and M. Zabeau. 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23:4407-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]