Abstract
Background
Rickettsiae constitute a group of arthropod-borne, Gram-negative, obligate intracellular bacteria that are the causative agents of diseases ranging from mild to life threatening that impact on medical and veterinary health worldwide.
Methods
A total of 6,484 ticks were collected by tick drag from June-October 2013 in the southwestern provinces of the Republic of Korea (ROK) (Jeollanam, n = 3,995; Jeollabuk, n = 680; Chungcheongnam, n = 1,478; and Chungcheongbuk, n = 331). Ticks were sorted into 311 pools according to species, collection site, and stage of development. DNA preparations of tick pools were assayed for rickettsiae by 17 kDa antigen gene and ompA nested PCR (nPCR) assays and the resulting amplicons sequenced to determine the identity and prevalence of spotted fever group rickettsiae (SFGR).
Results
Haemaphysalis longicornis (4,471; 52 adults, 123 nymphs and 4,296 larvae) were the most commonly collected ticks, followed by Haemaphysalis flava (1,582; 28 adults, 263 nymphs and 1,291 larvae), and Ixodes nipponensis (431; 25 adults, 5 nymphs and 401 larvae). The minimum field infection rate/100 ticks (assuming 1 positive tick/pool) was 0.93% for the 17 kDa antigen gene and 0.82% for the ompA nPCR assays. The partial 17 kDa antigen and ompA gene sequences from positive pools of H. longicornis were similar to: Rickettsia sp. HI550 (99.4–100%), Rickettsia sp. FUJ98 (99.3–100%), Rickettsia sp. HIR/D91 (99.3–100%), and R. japonica (99.7%). One sequence of the partial 17 kDa antigen gene for H. flava was similar to Rickettsia sp. 17kd-005 (99.7%), while seven sequences of the 17 kDa antigen gene obtained from I. nipponensis ticks were similar to R. monacensis IrR/Munich (98.7–100%) and Rickettsia sp. IRS3 (98.9%).
Conclusions
SFG rickettsiae were detected in three species of ixodid ticks collected in the southwestern provinces of the ROK during 2013. A number of rickettsiae have been recently reported from ticks in Korea, some of which were identified as medically important. Results from this study and previous reports demonstrate the need to conduct longitudinal investigations to identify tick-borne rickettsiae and better understand their geographical distributions and potential impact on medical and veterinary health, in addition to risk communication and development of rickettsial disease prevention strategies.
Keywords: Rickettsia, Spotted fever group rickettsiae, Ixodid ticks, 17 kDa antigen gene, ompA
Background
Rickettsia species are obligate intracellular bacteria in the order Rickettsiales that infect a variety of vertebrate hosts, including humans via arthropod vectors [1]. The genus Rickettsia has been classified according to morphological, antigenic, and metabolic characteristics, but now with the availability of genetic information, new approaches to phylogenetic inferences have provided new perspectives on rickettsial classification and evolution. Members of the genus Rickettsia are divided into many different phylogenetic groups and this progression will continue with additional phylogeny data. Currently there exists: (i) the spotted fever group Rickettsia (SFGR) (e.g. Rickettsia conorii, R. rickettsii and R. japonica, the causative agents of Mediterranean, Rocky Mountain, and Japanese spotted fever, respectively, that are transmitted by ixodid ticks); (ii) the typhus group (TGR) (e.g. R. typhi, the causative agent of murine typhus transmitted by fleas, and R. prowazekii, the causative agent of epidemic typhus transmitted by the body louse); (iii) the transitional group (TRGR) transmitted by fleas, mites and ticks; (iv) the R. bellii group (ticks); (v) the R. canadensis group; (vi) the Helvetica group; (vii) the Scapularis group; (viii) the Adalia group; and (ix) the Hydra group [1–4].
Ticks, obligate parasites of vertebrates and found in various natural environments throughout the world, are divided into three families: Ixodidae (hard ticks), Argasidae (soft ticks), and Nuttalliellidae (one species in South Africa). Worldwide, ixodid ticks (e.g. Haemaphysalis flava, H. longicornis, Ixodes persulcatus and I. nipponensis in Asia; I. ricinus in Europe; Rhipicephalus sanguineus, Dermacentor andersoni, D. variabilis, Amblyomma americanum and Am. maculatum in America) are the primary vectors/reservoirs of a wide range of rickettsiae of medical and veterinary importance (e.g. R. japonica, R. rickettsii, R. conorii, R. honei, R. sibirica, R. slovaca and R. monacensis) that affect birds, wild and domestic animals, and humans in Japan, Mongolia, South Korea, Russia and China [5–11].
SFGR were first reported in Korea based on serological analysis of acute febrile patients [12, 13]. The first case of Japanese spotted fever and isolation of SFGR from a patient in Korea was reported in 2005 [14]. These serological positive sera were assessed by molecular methods based on sequences of the ompB gene by nested PCR (nPCR) demonstrated similarities to R. conorii, R. akari, R. japonica and R. sibirica.
Haemaphysalis longicornis ticks from Chungju Province were positive for R. japonica using PCR analysis and sequencing of the groEL gene [15]. Moreover, R. japonica and R. monacensis were detected in H. longicornis by nPCR and sequence analysis of the gltA, ompB, and 17 kDa antigen genes [16, 17]. More recently, Rickettsia species have been detected in various arthropods and tick species in Korea that were collected from small mammals, reptiles, and the environment (by tick drag) [10, 18–20].
The purpose of this study was to identify the presence and prevalence of Rickettsia species in ticks collected from the southwestern provinces (Jeollanam, Jeollabuk, Chungcheongbuk and Chungcheongnam) of Korea during 2013 to identify and genetically characterize the rickettsiae based on sequence analysis of the partial 17 kDa antigen and ompA genes.
Methods
Sample collection
A total of 6,484 unengorged ticks (adults, nymphs and larvae) were collected by tick drags when ticks were active during June-October from the southwestern provinces (Jeollanam, Jeollabuk, Chungcheongnam and Chungcheongbuk) of Korea in 2013 as described by Chong et al. [21, 22]. Ticks were identified to species level using morphological keys [23, 24] and placed in 2 ml cryovials according to collection date, species and stage of development (n = 6,484; 311 pools of 1–5 adults, 1–25 nymphs, and 1–69 larvae) (Table 1) [25]. Ticks were washed in 70% ethanol, rinsed twice with sterile PBS, and then homogenized in 600 μl of PBS and stored at -70 °C until used for DNA extraction.
Table 1.
Numbers of pooled ticks collected from the southwestern provinces of Chungcheongnam, Chungcheongbuk, Jeollanam and Jeollabuk in the Republic of Korea
| No. of ticks | No. of pools | |||
|---|---|---|---|---|
| Larvae | Nymphs | Adult males | Adult females | |
| 1–5 | 27 | 50 | 19 | 43 |
| 6–9 | 15 | 10 | 0 | 0 |
| 10–19 | 25 | 6 | 0 | 0 |
| 20–29 | 14 | 7 | 0 | 0 |
| 30–39 | 14 | 0 | 0 | 0 |
| 40–49 | 12 | 0 | 0 | 0 |
| 50–59 | 10 | 0 | 0 | 0 |
| 60–69 | 59 | 0 | 0 | 0 |
| Total | 176 | 73 | 19 | 43 |
DNA extraction
DNA was extracted from 200 μl of tick suspension using the G-spin total DNA extraction kit (iNtRON, Gyeonggi, Korea) according to the manufacturer’s instructions. DNA was eluted into 50 μl TE buffer and stored at -20 °C until PCR amplification.
Nested PCR (nPCR) amplification
Direct amplification by nPCR was performed to identify target genes using the partial 17 kDa and ompA genes for Rickettsia species belonging to the family Rickettsiaceae. The ompA gene encoded for the SFGR-specific 190 kDa outer membrane protein and the partial 17 kDa antigen gene encoded for the Rickettsia genus-specific 17 kDa outer membrane protein.
Rickettsia spp. DNA presence was screened using the 17 kDa antigen gene by nPCR as described previously [26]. Briefly, the PCR was performed in a final reaction volume of 20 μl containing 3 μl DNA, 10 pmol of each primer, and the premix reagent (Maxime PCR PreMix kit/i-starTaqTM GH, iNtRON, Gyeonggi, Korea). nPCR was performed in a final reaction volume of 20 μl containing 3 μl of the initial PCR product, 10 pmol of each primer and the premix reagent. Samples positive for the 17 kDa gene target nPCR (appropriate size band identified following agarose gel electrophoresis) were subsequently assessed for the presence of a fragment of ompA. The premix reagent, reaction volumes, DNA templates and the amount of primers were the same as those used in the 17 kDa reactions (Table 2).
Table 2.
Primer sequences and nested PCR conditions for detection of rickettsial target genes from ticks collected from four southwestern provinces of Chungcheongnam, Chungcheongbuk, Jeollanam, and Jeollabuk in the Republic of Korea
| Target gene | Primer name | Nucleotide sequence (5'–3') | Product size (bp) | PCR profile (°C/s) | Reference | |||
|---|---|---|---|---|---|---|---|---|
| Denaturation | Annealing | Extension | Cycles | |||||
| 17 kDa | Rr17k. 1p | TTTACAAAATTCTAAAAACCAT | 539 | 95/30 | 57/60 | 72/120 | 35 | [23] |
| Rr17k. 539n | TCAATTCACAACTTGCCATT | |||||||
| Rr17k. 90p | GCTCTTGCAACTTCTATGTT | 450 | 95/30 | 57/60 | 72/120 | 35 | ||
| Rr17k. 539n | TCAATTCACAACTTGCCATT | |||||||
| ompA | Rr190k. 71p | TGGCGAATATTTCTCCAAAA | 650 | 95/30 | 42/35 | 60/120 | 35 | [23] |
| Rr190k. 720n | TGCATTTGTATTACCTATTGT | |||||||
| Rr190k. 71p | TGGCGAATATTTCTCCAAAA | 532 | 95/30 | 48/60 | 65/120 | 35 | ||
| Rr190k. 602n | AGTGCAGCATTCGCTCCCCCT | |||||||
Sequencing and phylogenetic analysis
Sequencing of Rickettsia-positive nPCR amplicons was conducted by Macrogen Inc. (Daejeon, Korea). The obtained sequences were compared for similarity to sequences deposited in GenBank using BLAST. Gene sequences, excluding the primer regions, were aligned using the multisequence alignment program in Lasergene version 8 (DNASTAR, USA), and phylogenetic analysis performed using MEGA 6 software.
Phylogenetic trees were constructed in CLUSTAL W of the MegAlign Program (DNASTAR, USA) based on the alignment of rickettsial gene sequences obtained following nPCR using the neighbor-joining method and bootstrap analysis (1,000 reiterations) carried out according to the Kimura 2-parameter method. Pairwise alignments were performed with an open-gap penalty of 10 and a gap extension penalty of 0.5. All positions containing alignment gaps and missing data were eliminated during the pairwise sequence comparison (pairwise deletion).
Results
Collection of ticks
A total of 6,484 ticks belonging to two genera and three species were collected at four southwestern provinces by tick drag (Fig. 1). Haemaphysalis longicornis (4,471; 52 adults, 123 nymphs and 4,296 larvae), was the most commonly collected tick, followed by H. flava (1,582; 28 adults, 263 nymphs and 1,291 larvae), and I. nipponensis (431; 25 adults, 5 nymphs and 401 larvae).
Fig. 1.
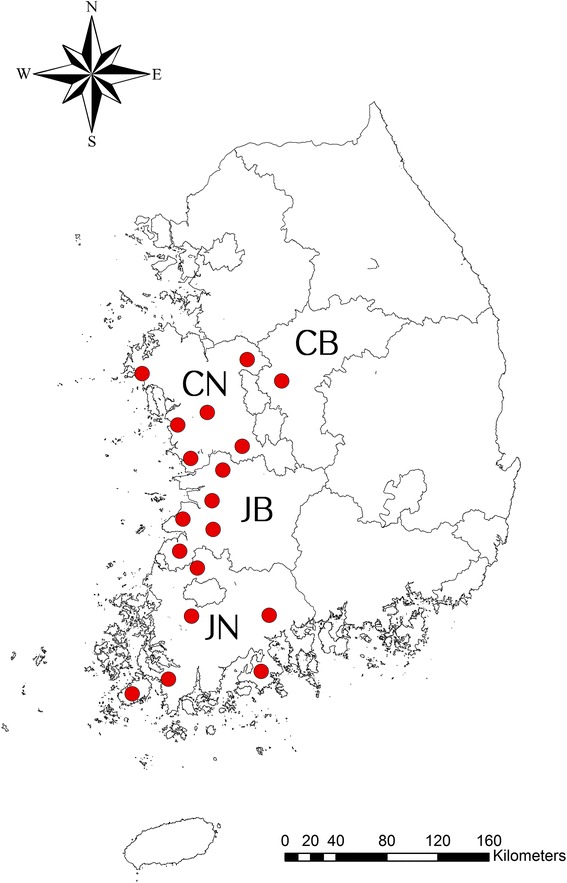
Geographical locations of the tick collection sites in this study. The locations of tick collection sites are marked as red closed circles. This map was created using ArcGIS v.10.3.1 software (Environmental Research System Institute, Redland, CA, USA)]. Abbreviations: CN, Chungcheongnam Province; CB, Chungcheongbuk Province; JN, Jeollanam Province; JB, Jeollabuk Province
Detection and prevalence of rickettsial agents
A total of 60/311 (19.30%) pools from Chungcheongnam (3/1,478; 0.20%), Chungcheongbuk (0/331; 0%), Jeollanam (53/3,995; 1.33%) and Jeollabuk (4/680; 0.59%) provinces, respectively, were positive for Rickettsia spp. using the 17 kDa antigen gene nPCR assay (Table 3). A total of 51/168 (30.36%) and 46/168 (27.38%) of H. longicornis pools were positive for Rickettsia using the 17 kDa antigen and ompA genes, respectively. Only 1/108 (0.93%) and 0/108 (0%) of H. flava were positive for Rickettsia spp. using the partial 17 kDa and ompA genes, respectively, while 7/35 (20.00%) pools of I. nipponensis were positive for R. monacensis.
Table 3.
SFGR minimum field infection rates (MFIR) for ticks collected from four southwestern provinces of Chungcheongnam, Chungcheongbuk, Jeollanam and Jeollabuk in the Republic of Korea during 2013 by province
| Province | Species | Stage | No. of ticks (No. of tested pools) | nPCR positive no. of Rickettsia sp. (MFIR, %)a | |
|---|---|---|---|---|---|
| 17 kDa | ompA | ||||
| Chungcheongnam | H. longicornis | Larvab | 533 (22) | 1 (0.19) | 1 (0.19) |
| Nymphc | 16 (9) | 0 | 0 | ||
| Adult (male)d | 1 (1) | 0 | 0 | ||
| Adult (female)d | 22 (10) | 0 | 0 | ||
| H. flava | Larva | 475 (17) | 0 | 0 | |
| Nymph | 30 (8) | 0 | 0 | ||
| Adult (male) | 5 (2) | 0 | 0 | ||
| Adult (female) | 3 (2) | 0 | 0 | ||
| I. nipponensis | Larva | 379 (13) | 0 | 0 | |
| Nymph | 3 (2) | 0 | 0 | ||
| Adult (male) | 5 (3) | 1 (20.00) | 1 (20.00) | ||
| Adult (female) | 6 (2) | 1 (16.66) | 1 (16.66) | ||
| Subtotal | 1,478 (91) | 3 (0.20) | 3 (0.20) | ||
| Chungcheongbuk | H. longicornis | Larva | 147 (6) | 0 | 0 |
| Nymph | 2 (2) | 0 | 0 | ||
| Adult (female) | 3 (2) | 0 | 0 | ||
| H. flava | Larva | 176 (5) | 0 | 0 | |
| Nymph | 1 (1) | 0 | 0 | ||
| I. nipponensis | Larva | 2 (1) | 0 | 0 | |
| Subtotal | 331 (17) | 0 | 0 | ||
| Jeollanam | H. longicornis | Larva | 3,314 (70) | 34 (1.03) | 31 (0.94) |
| Nymph | 103 (18) | 7 (6.79) | 6 (5.82) | ||
| Adult (male) | 6 (4) | 4 (66.66) | 4 (66.66) | ||
| Adult (female) | 20 (11) | 2 (10.00) | 2 (10.00) | ||
| H. flava | Larva | 401 (19) | 1 (0.25) | 0 | |
| Nymph | 105 (18) | 0 | 0 | ||
| Adult (male) | 4 (3) | 0 | 0 | ||
| Adult (female) | 11 (8) | 0 | 0 | ||
| I. nipponensis | Larva | 18 (2) | 1 (5.55) | 0 | |
| Adult (male) | 5 (3) | 2 (40.00) | 2 (40.00) | ||
| Adult (female) | 8 (5) | 2 (25.00) | 2 (25.00) | ||
| Subtotal | 3,995 (161) | 53 (1.33) | 47 (1.18) | ||
| Jeollabuk | H. longicornis | Larva | 302 (11) | 2 (0.66) | 1 (0.33) |
| Nymph | 2 (2) | 1 (50.00) | 1 (50.00) | ||
| H. flava | Larva | 239 (9) | 0 | 0 | |
| Nymph | 127 (11) | 0 | 0 | ||
| Adult (male) | 2 (2) | 0 | 0 | ||
| Adult (female) | 3 (3) | 0 | 0 | ||
| I. nipponensis | Larva | 2 (1) | 1 (50.00) | 1 (50.00) | |
| Nymph | 2 (2) | 0 | 0 | ||
| Adult (male) | 1 (1) | 0 | 0 | ||
| Subtotal | 680 (42) | 4 (0.59) | 3 (0.44) | ||
| Total | 6,484 (311) | 60 (0.93) | 53 (0.82) | ||
aMFIR (minimum field infection rate/100 ticks) = no. of positive pools/no. of examined ticks in pools × 100, by species and stage of development
b1–69 larvae/pool
c1–25 nymphs/pool
d1–5 adults/pool
The overall minimum field infection rates (MFIR) of Rickettsia-positive pools (assuming 1 positive tick/pool) were 0.93% (60/6,484) for the 17 kDa antigen gene and 0.82% (53/6,484) for the ompA gene targets (Table 4) [25]. The overall MFIR for all three species ranged from 0–0.88% for larvae, 0–6.50% for nymphs, and 0–57.1% for adults (Table 4). There were no significant differences (Chi-square test, P = 0.98) observed between the positive rates of the partial 17 kDa and ompA genes.
Table 4.
SFGR minimum field infection rates (MFIR) for ticks collected from four southwestern provinces of Chungcheongnam, Chungcheongbuk, Jeollanam and Jeollabuk in the Republic of Korea during 2013 by species of tick using the partial 17 kDa and ompA genes by nPCR
| Collected ticks | Developmental stage | No. of ticks/(no. of tested pools) | nPCR positive no. of Rickettsia sp. (MFIR)a | |
|---|---|---|---|---|
| 17 kDa | ompA | |||
| H. longicornis | Larvab | 4,296 (109) | 38 (0.88) | 33 (0.77) |
| Nymphc | 123 (31) | 8 (6.50) | 7 (5.70) | |
| Adult (male)d | 7 (5) | 4 (57.10) | 4 (57.10) | |
| Adult (female)d | 45 (23) | 2 (4.40) | 2 (4.40) | |
| Subtotal | 4,471 (168) | 52 (1.16) | 46 (1.03) | |
| H. flava | Larva | 1,291 (50) | 0 | 0 |
| Nymph | 263 (38) | 1 (0.38) | 0 | |
| Adult (male) | 11 (7) | 0 | 0 | |
| Adult (female) | 17 (13) | 0 | 0 | |
| Subtotal | 1,582 (108) | 1 (0.06) | 0 | |
| I. nipponensis | Larva | 401 (17) | 1 (0.25) | 1 (0.25) |
| Nymph | 5 (4) | 0 | 0 | |
| Adult (male) | 11 (7) | 3 (27.2) | 3 (27.2) | |
| Adult (female) | 14 (7) | 3 (21.4) | 3 (21.4) | |
| Subtotals | 431 (35) | 7 (1.62) | 7 (1.62) | |
| Totals | 6,484 (311) | 60 (0.93) | 53 (0.82) | |
aMFIR (minimum field infection rate/100 ticks) = No. of positive pools/No. of examined ticks in pools × 100, by species and stage of development
b1–69 larvae/pool
c1–25 nymphs/pool
d1–5 adults/pool
Sequencing and phylogenetic analysis
The partial 17 kDa antigen gene and ompA nPCR amplicons were sequenced and aligned with other rickettsial genes deposited in the GenBank database to identify known sequences with a high degree of similarity using ClustalW [26]. The sequencing electropherograms of all positive pools were confirmed as single peaks, indicating each pool represented a single Rickettsia species.
The amplicon sequences of the partial 17 kDa gene obtained from H. longicornis demonstrated 99.4–100% similarity to previously reported molecular sequences from H. longicornis that phylogenetically clustered with Rickettsia sp. HIR/D91, Rickettsia sp. 71-8, Rickettsia sp. HI550 and Rickettsia sp. LON-2, LON-13. Similarly, ompA sequences of positive pools of H. longicornis demonstrated 99.3–100% similarity to sequences of rickettsiae from H. longicornis previously reported as Rickettsia sp. HIR/D91, Rickettsia sp. LON-2, LON-13, Rickettsia sp. HI550 and Rickettsia sp. FUJ98, that are similar, but distinct from R. japonica. Phylogenetic analysis showed a close relationship between rickettsial isolates from H. longicornis from the southwestern provinces of Korea and rickettsial isolates from H. longicornis from other Asian countries (Figs. 2 and 3).
Fig. 2.
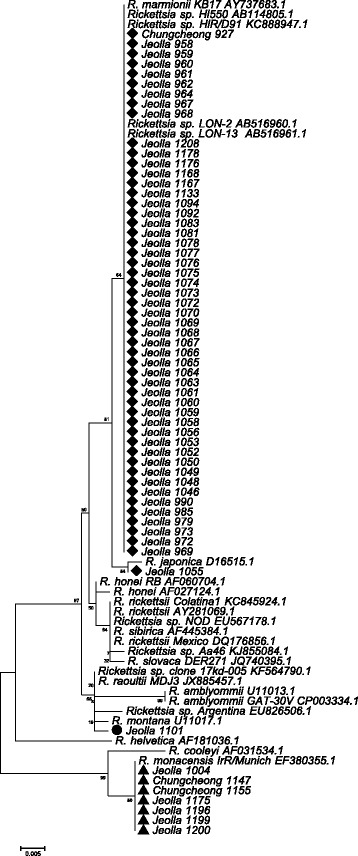
Phylogenetic tree based on 342 bp of the 17 kDa outer membrane protein gene of Rickettsia species
Fig. 3.
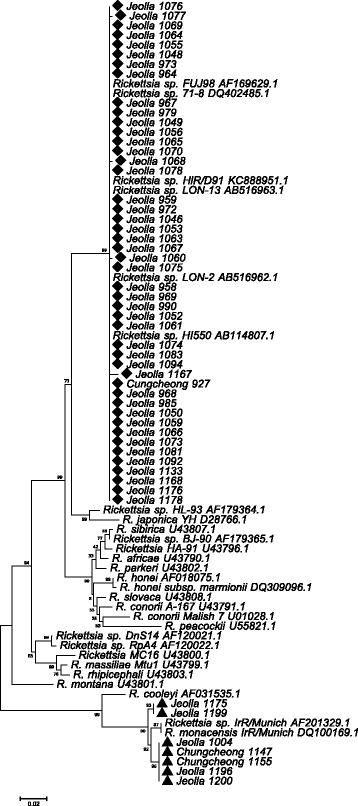
Phylogenetic tree based on 443 bp of ompA gene of Rickettsia species
The amplicon sequence of the partial 17 kDa gene for one positive pool of H. flava demonstrated 99.7% similarity to previously reported Rickettsia sp. 17-kDa-005 that clustered with R. montanensis and R. raoultii (Fig. 2). The amplicon sequences of the partial 17 kDa gene and ompA genes from seven pools of I. nipponensis demonstrated 100% and 98.7–98.9% similarity, respectively, to previously reported R. monacensis IrR/Munich (Figs. 2 and 3).
Discussion
Tick-borne rickettsiae are obligate intracellular bacteria belonging to the genus Rickettsia, many of which are of medical importance [27–29]. Clinically, tick-borne rickettsioses present with mild to life threatening signs and symptoms that include: an eschar (not always indicated) that is present 1–2 days prior to the onset of headache and fever (39.5–40.0 °C), and a characteristic rash 1–2 days after the onset of fever that can last for 2–3 weeks. Tick-borne infections are often reported as non-specific febrile diseases due to the lack of specific clinical signs and symptoms and diagnostic assays effective early in the process of disease [27–29]. In the USA, there were a total of 3,649 cases of rickettsioses reported between 1997–2002 and more than 1,500 cases reported annually since 2005 (www.cdc.gov/rmsf/stats/index.html). This increase in reporting may be due to the fact that rickettsioses are becoming more widely recognized [1, 29]. In addition to disease producing tick-borne rickettsiae, many rickettsiae (e.g. R. bellii, R. canadensis, R. asiatica, R. hoogstraalii, R. montanensis, R. rhipicephali and R. tamurae) have not been identified as pathogens and therefore are often referred to as non-pathogenic or of unknown pathogenicity. To further complicate matters, with the discovery of numerous new Rickettsia spp. in ticks using molecular tools, their role as causative agents of diseases of medical and veterinary importance has not been established, in part owing to lack of diagnostic tools for pathogen detection, rather than for antibodies [29].
With increased interest in tick-borne diseases, surveillance of ticks from reptiles, mammals, birds, and vegetation has led to the identification of known and yet to be described pathogens belonging to genera of Ehrlichia, Anaplasma, Bartonella, Borrelia, Babesia and Rickettsia, in addition to viruses [10, 18–20, 30–36]. Rickettsia akari, a mite-borne pathogen isolated from a rodent, was first reported in Korea in 1957 [37, 38]. Later, acute febrile patients tested positive by serological tests for R. japonica in 2004, 2005 and 2006 [9, 12, 13]. Recently, various gene targets from rickettsial pathogens were identified in various ixodid tick species, including H. longicornis, H. flava, I. nipponensis and I. persulcatus [15–17].
Haemaphysalis longicornis, H. flava and I. nipponensis are commonly collected throughout Korea, while H. phasiana, A. testudinarium, I. pomerantzevi, I. persulcatus and I. ovatus have a limited geographical/habitat distribution and are collected much less frequently [22, 38, 39]. Tick-borne disease surveillance usually includes the detection of pools of ticks, as it is costly and untimely to assay for multiple agents within individual ticks [25]. However, it is important to assay ticks from specific habitats (e.g. forests and grasses/herbaceous vegetation) and hosts over their geographical range to determine the potential association with man and domestic animals, as well as the distribution of associated pathogens [8, 10, 16, 17, 19, 20, 31, 39].
The conserved 17 kDa antigen gene was used in this study to screen for the presence of rickettsiae in tick pools. Subsequently, the 17 kDa Rickettsia-positive pools (n = 60) were assessed for the presence of ompA (also by nPCR). All but seven of the 60 Rickettsia-positive pools were positive for the more variable ompA gene. A previous report also showed that ompA genes were not detected in several different rickettsial genotypes [26].
Results of the partial 17 kDa antigen and ompA gene sequences obtained from H. longicornis pools showed that the rickettsial agents detected were closely related to Rickettsia sp. HIR/D91, 71-8 identified in Korea, Rickettsia sp. HI550 and LON-2, LON-13 identified in Japan, and Rickettsia sp. FuJ98 identified in China. Only one Rickettsia-positive H. flava pool sequenced demonstrated a high similarity to Rickettsia sp. 17-kDa-005 identified in China, while all seven Rickettsia-positive pools of I. nipponensis were similar to R. monacensis and Rickettsia sp. IrR/Munich identified in Europe. Rickettsia monacensis, a known human pathogen, was first isolated from I. ricinus collected from an English garden in Germany in 1998 [1]. While R. monacensis was generally observed only in I. ricinus mainly from southern and eastern Europe [40], it has been detected in I. nipponensis collected from rodents captured in Korea (Jeollanam Province in 2006 and Gyeonggi and Gangwon provinces in 2008) [10, 17].
Haemaphysalis longicornis is commonly collected from grasses and herbaceous vegetation, while H. flava is more commonly associated with forest habitats and I. nipponensis is collected similarly from both habitats throughout the ROK [22]. Haemaphysalis longicornis is commonly found in grassy areas that expose civilians and military populations to tick bites and associated pathogens, which not only include Rickettsia spp., but other bacteria and viruses of medical and veterinary importance [32–35, 41]. Rickettsia monacensis has been detected in I. nipponensis, which are more frequently reported in tick bites [42–45]. While H. longicornis, the primary vector of the severe fever with thrombocytopenia syndrome (SFTS) virus, has not been frequently reported to bite humans, with 36, 51, and 78 cases of SFTS infections among civilians in the ROK reported from 2013–2015, respectively, indicates that most bites go unreported with the potential for the transmission of rickettsiae to both civilian and military populations.
Additional analysis of gene sequences of Rickettsia spp. will allow for their specific identification and the development of species-specific PCR assays and determination of their medical and veterinary importance. SFGR infections are not reportable events in the ROK and some cases are likely included as scrub typhus since the symptoms, including fever, eschar and rash, are similar. Analysis of eschar tissue by PCR would allow for the detection and identification of Rickettsia spp. and scrub typhus strains among patients with similar disease presentation [46]. The identification of the SFGR diseases and scrub typhus is essential to determine tick- and mite-borne disease risks and develop appropriate disease prevention strategies. Additional investigations to determine the identification of rickettsiae associated with each of the tick species using single tick analysis and the geographical/habitat distribution of each of the tick species and associated pathogens are needed to identify disease risks to both civilian and military populations in the ROK.
Conclusion
Rickettsial pathogens pose a potential health threat to military and civilian communities in the ROK. Ixodes nipponensis has been shown to be infected with R. monacensis, a human pathogen, and H. longicornis and H. flava have been shown to be infected with SFGR of unknown pathogenicity. More intense and longitudinal surveillance of ticks and their hosts in the ROK is needed to determine their geographical and habitat distributions, and the geographical distribution and prevalence of their associated pathogens. The characterization of SFGR is essential to identify agents of tick-borne human diseases and their relative pathogenicity. Lastly, the detection and identification data of the rickettsiae reported herein will provide for the development of species-specific diagnostic assays that are essential for rapid detection of SFGR in the vectors, vertebrate hosts and patients.
Acknowledgements
Not applicable.
Funding
This research was supported by the Center for Disease Control & Prevention, the Ministry of Health & Welfare (grant no. 4800-4838-303), the Armed Forces Health Surveillance Branch, Global Emerging Infections Surveillance and Response Systems (AFHSB-GEIS) work unit A1402 (ALR), and the Medical Department Activity-Korea, 65th Medical Brigade. The opinions expressed herein are those of the authors and are not to be construed as official or reflecting the views of the U.S. Departments of the Army, Navy or Defense.
Availability of data and materials
The datasets supporting the conclusions of this article are included with NCBI accession numbers: Chungcheong 927 (KX418671, KX418672), Jeolla 958 (KX418673), Jeolla 959 (KX418675, KX418676), Jeolla 960 (KX418677), Jeolla 961 (KX418678), Jeolla 962 (KX418740), Jeolla 964 (KX418765, KX418766), Jeolla 967 (KX418741, KX418742), Jeolla 968 (KX418767, KX418768), Jeolla 969 (KX418743, KX418744), Jeolla 972 (KX418754, KX418755), Jeolla 973 (KX418745, KX418746), Jeolla 979 (KX418756, KX418757), Jeolla 985 (KX418758, KX418759), Jeolla 990 (KX418760, KX418761), Jeolla 1004 (KX418663, KX418664), Jeolla 1046 (KX418747, KX418748), Jeolla 1048 (KX418679, KX418680), Jeolla 1049 (KX418681, KX418682), Jeolla 1050 (KX418683, KX418684), Jeolla 1052 (KX418685, KX418686), Jeolla 1053 (KX418687, KX418688), Jeolla 1055 (KX418689, KX418690), Jeolla 1056 (KX418691, KX418692), Jeolla 1058 (KX418749), Jeolla 1059 (KX418750, KX418751), Jeolla 1060 (KX418693, KX418694), Jeolla 1061 (KX418695, KX418696), Jeolla 1063 (KX418697, KX418698), Jeolla 1064 (KX418699, KX418700), Jeolla 1065 (KX418762, KX418763), Jeolla 1066 (KX418701, KX418702), Jeolla 1067 (KX418703, KX418704), Jeolla 1068 (KX418705, KX418706), Jeolla 1069 (KX418707, KX418708), Jeolla 1070 (KX418709, KX418710), Jeolla 1072 (KX423490), Jeolla 1073 (KX418711, KX418712), Jeolla 1074 (KX418713, KX418714), Jeolla 1075 (KX418715, KX418716), Jeolla 1076 (KX418717, KX418718), Jeolla 1077 (KX418719, KX418720), Jeolla 1078 (KX418721, KX418722), Jeolla 1081 (KX418723, KX418724), Jeolla 1083 (KX418725, KX418726), Jeolla 1092 (KX418727, KX418728), Jeolla 1094 (KX418729, KX418730), Jeolla 1101 (KX418764), Jeolla 1133 (KX418731, KX418732), Chungcheong 1147 (KX418657, KX418658), Chungcheong 1155 (KX418665, KX418666), Jeolla 1167 (KX418752, KX418753), Jeolla 1168 (KX418733, KX418734), Jeolla 1175 (KX418659, KX418660), Jeolla 1176 (KX418735, KX418736), Jeolla 1178 (KX418737, KX418738), Jeolla 1196 (KX418667, KX418668), Jeolla 1199 (KX418661, KX418662), Jeolla 1200 (KX418669, KX418670), Jeolla 1208 (KX418739).
Authors’ contributions
Conceived the study, drafted the manuscript, and performed the experiments: YTN, YSL, HKL and SYK. Collected and identified ticks: TAK, HCK, and STC. Reviewed the manuscript: TN, HCK, TAK, ALR, JJ and HKL. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Abbreviations
- 17 kDa
Rickettsia-specific outer membrane antigen gene
- CB
Chungcheongbuk Province
- CN
Chungcheongnam Province
- JB
Jeollabuk Province
- JN
Jeollanam Province
- MFIR
minimum field infection rate per 100 ticks
- nPCR
nested PCR
- ompA
spotted fever group-specific 190 kDa outer membrane protein A gene
- ROK
Republic of Korea
Contributor Information
Yoontae Noh, Email: gionyt@korea.kr.
Yeong Seon Lee, Email: yslee07@korea.kr.
Heung-Chul Kim, Email: hungchol.kim2.ln@mail.mil.
Sung-Tae Chong, Email: sungtae.chong.ln@mail.mil.
Terry A. Klein, Email: terry.a.klein2.civ@mail.mil
Ju Jiang, Email: ju.jiang2.ctr@mail.mil.
Allen L. Richards, Email: allen.l.richards.civ@mail.mil
Hae Kyeong Lee, Email: hkglee@korea.kr.
Su Yeon Kim, Email: tenksy@korea.kr.
References
- 1.Parola P, Paddock CD, Raoult D. Tick-borne rickettsioses around the world: emerging diseases challenging old concepts. Clin Microbiol Rev. 2005;18:719–56. doi: 10.1128/CMR.18.4.719-756.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gillespie JJ, Joardar V, Williams KP, Driscoll T, Hostetler JB, Nordberg E, et al. Rickettsia genome overrun by mobile genetic elements provides insight into the acquisition of genes characteristic of an obligate intracellular lifestyle. J Bacteriol. 2012;194:376–394. doi: 10.1128/JB.06244-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Merhej V, Angelakis E, Socolovschi C, Raoult D. Genotyping, evolution and epidemiological findings of Rickettsia species. Infect Genet Evol. 2014;25:122–37. doi: 10.1016/j.meegid.2014.03.014. [DOI] [PubMed] [Google Scholar]
- 4.Murray GGR, Weinert LA, Rhule EL, Welch JJ. The Phylogeny of Rickettsia using different evolutionary signatures: how tree-like is bacterial evolution? Syst Biol. 2016;65:265–279. doi: 10.1093/sysbio/syv084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sekeyova Z, Fournier PE, Rehacek J, Raoult D. Characterization of a new spotted fever group Rickettsia detected in Ixodes ricinus (Acari: Ixodidae) collected in Slovakia. J Med Entomol. 2000;37:707–13. doi: 10.1093/jmedent/37.5.707. [DOI] [PubMed] [Google Scholar]
- 6.Tian ZC, Liu GY, Shen H, Xie JR, Luo J, Tian MY. First report on the occurrence of Rickettsia slovaca and Rickettsia raoultii in Dermacentor silvarum in China. Parasit Vectors. 2012;5:19. doi: 10.1186/1756-3305-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Speck S, Derschum H, Damdindorj T, Dashdavaa O, Jiang J, Kaysser P, et al. Rickettsia raoultii, the predominant Rickettsia found in Mongolian Dermacentor nuttalli. Ticks Tick Borne Dis. 2012;3:227–31. doi: 10.1016/j.ttbdis.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 8.Cheng C, Fu W, Ju W, Yang L, Xu N, Wang YM, et al. Diversity of spotted fever group Rickettsia infection in hard ticks from Suifenhe, Chinese-Russian border. Ticks Tick Borne Dis. 2016;7:715–9. [DOI] [PubMed]
- 9.Chung MH, Lee SH, Kim MJ, Lee JH, Kim ES, Kim MK, et al. Japanese spotted fever, South Korea. Emerg Infect Dis. 2006;12:1122–4. doi: 10.3201/eid1207.051372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shin SH, Seo HJ, Choi YJ, Choi MK, Kim HC, Klein TA, et al. Detection of Rickettsia monacensis from Ixodes nipponensis collected from rodents in Gyeonggi and Gangwon Provinces, Republic of Korea. Exp Appl Acarol. 2013;61:337–47. doi: 10.1007/s10493-013-9699-1. [DOI] [PubMed] [Google Scholar]
- 11.Uchida T, Uchiyama T, Kumano K, Walker DH. Rickettsia japonica sp. nov., the etiological agent of spotted fever group rickettsiosis in Japan. Int J Systematic Bacteriol. 1992;42:303–5. doi: 10.1099/00207713-42-2-303. [DOI] [PubMed] [Google Scholar]
- 12.Jang WJ, Kim JH, Choi YJ, Jung KD, Kim YG, Lee SH, et al. First serologic evidence of human spotted fever group rickettsiosis in Korea. J Clin Microbiol. 2004;42:2310–3. doi: 10.1128/JCM.42.5.2310-2313.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jang WJ, Choi YJ, Kim JH, Jung KD, Ryu JS, Lee SH, et al. Seroepidemiology of spotted fever group and typhus group rickettsioses in humans, South Korea. Microbiol Immunol. 2005;49:17–24. doi: 10.1111/j.1348-0421.2005.tb03635.x. [DOI] [PubMed] [Google Scholar]
- 14.Choi YJ, Lee SH, Park KH, Koh YS, Lee KH, Baik HS, et al. Evaluation of PCR-based assay for diagnosis of spotted fever group rickettsiosis in human serum samples. Clin Diagn Lab Immunol. 2005;12:759–63. doi: 10.1128/CDLI.12.6.759-763.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JHAS, Park HS, Jeong EJ, Choi HG, Jang WJ, Kang SS, et al. Prevalence of spotted fever group Rickettsia from Haemaphysalis ticks in Chungju province. J Bacteriol Virol. 2005;35:203–7. [Google Scholar]
- 16.Moon BCJJ, Choi YJ, Kim JE, Seo HJ, Shin EH, Song BGLH, et al. Detection and identification of the spotted fever group rickettsial agents from Haemaphysalis ticks in Jeju Island, Korea. J Bacteriol Virol. 2009;39:317–27. doi: 10.4167/jbv.2009.39.4.317. [DOI] [Google Scholar]
- 17.Lee KM, Choi YJ, Shin SH, Choi MK, Song HJ, Kim HC, et al. Spotted fever group Rickettsia closely related to Rickettsia monacensis isolated from ticks in South Jeolla province, Korea. Microbiol Immunol. 2013;57:487–95. [DOI] [PubMed]
- 18.Ko S, Kim HC, Yang YC, Chong ST, Richards AL, Sames WJ, et al. Detection of Rickettsia felis and Rickettsia typhi and seasonal prevalence of fleas collected from small mammals at Gyeonggi Province in the Republic of Korea. Vector Borne Zoonotic Dis. 2011;11:1243–51. doi: 10.1089/vbz.2010.0261. [DOI] [PubMed] [Google Scholar]
- 19.Chae JS, Yu DH, Shringi S, Klein TA, Kim HC, Chong ST, et al. Microbial pathogens in ticks, rodents and a shrew in northern Gyeonggi-do near the DMZ, Korea. J Vet Sci. 2008;9:285–93. doi: 10.4142/jvs.2008.9.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim CM, Yi YH, Yu DH, Lee MJ, Cho MR, Desai AR, et al. Tick-borne rickettsial pathogens in ticks and small mammals in Korea. Appl Environ Microbiol. 2006;72:5766–76. doi: 10.1128/AEM.00431-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chong ST, Kim HC, Lee IY, Kollars TM, Jr, Sancho AR, Sames WJ, et al. Comparison of dragging and sweeping methods for collecting ticks and determining their seasonal distributions for various habitats, Gyeonggi Province, Republic of Korea. J Med Entomol. 2013;50:611–8. doi: 10.1603/ME12032. [DOI] [PubMed] [Google Scholar]
- 22.Chong ST, Kim HC, Lee IY, Kollars TM, Jr, Sancho AR, Sames WJ, et al. Seasonal distribution of ticks in four habitats near the demilitarized zone, Gyeonggi-do (Province), Republic of Korea. Korean J Parasitol. 2013;51:319–25. doi: 10.3347/kjp.2013.51.3.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamaguti N, Tipton VJ, Keegan HL, Toshioka S. Ticks of Japan, Korea, and the Ryukyu Islands. Brigham Young Univ Sci Bull, Biol Ser. 1971;15:1–226. doi: 10.5962/bhl.part.25691. [DOI] [Google Scholar]
- 24.Robbins RG, Keirans JE. Systematics and ecology of the subgenus Ixodiopsis (Acari: Ixodidae: Ixodes) Lanham: Thomas Say Foundation Monograph XIV. Entomological Society of America; 1992. p. 159. [Google Scholar]
- 25.Gu W, Lampman R, Novak RJ. Assessment of arbovirus vector infection rates using variable size pooling. Med Vet Entomol. 2004;18:200–4. doi: 10.1111/j.0269-283X.2004.00482.x. [DOI] [PubMed] [Google Scholar]
- 26.Ishikura M, Ando S, Shinagawa Y, Matsuura K, Hasegawa S, Nakayama T, et al. Phylogenetic analysis of spotted fever group rickettsiae based on gltA, 17-kDa, and rOmpA genes amplified by nested PCR from ticks in Japan. Microbiol Immunol. 2003;47:823–32. doi: 10.1111/j.1348-0421.2003.tb03448.x. [DOI] [PubMed] [Google Scholar]
- 27.Raoult D, Roux V. Rickettsioses as paradigms of new or emerging infectious diseases. Clin Microbiol Rev. 1997;10:694–719. doi: 10.1128/cmr.10.4.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parola P, Davoust B, Raoult D. Tick- and flea-borne rickettsial emerging zoonoses. Vet Res. 2005;36:469–92. doi: 10.1051/vetres:2005004. [DOI] [PubMed] [Google Scholar]
- 29.Walker DH. Rickettsiae and rickettsial infections: the current state of knowledge. Clin Infect Dis. 2007;45:S39–44. doi: 10.1086/518145. [DOI] [PubMed] [Google Scholar]
- 30.Tateno M, Sunahara A, Nakanishi N, Izawa M, Matsuo T, Setoguchi A, et al. Molecular survey of arthropod-borne pathogens in ticks obtained from Japanese wildcats. Ticks Tick Borne Dis. 2015;6:281–9. doi: 10.1016/j.ttbdis.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 31.Kang SW, Doan HT, Choe SE, Noh JH, Yoo MS, Reddy KE, et al. Molecular investigation of tick-borne pathogens in ticks from grazing cattle in Korea. Parasitol Int. 2013;62:276–82. doi: 10.1016/j.parint.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 32.Kim KH, Yi J, Oh WS, Kim NH, Choi SJ, Choe PG, et al. Human granulocytic anaplasmosis, South Korea, 2013. Emerg Infect Dis. 2014;20:1708–11. doi: 10.3201/eid2010.131680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moon S, Gwack J, Hwang KJ, Kwon D, Kim S, Noh Y, et al. Autochthonous lyme borreliosis in humans and ticks in Korea. Osong Public Health Res Perspect. 2013;4:52–6. doi: 10.1016/j.phrp.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yun SM, Song BG, Choi W, Park WI, Kim SY, Roh JY, et al. Prevalence of tick-borne encephalitis virus in ixodid ticks collected from the Republic of Korea during 2011–2012. Osong Public Health Res Perspect. 2012;3:213–21. [DOI] [PMC free article] [PubMed]
- 35.Christova I, Van De Pol J, Yazar S, Velo E, Schouls L. Identification of Borrelia burgdorferi sensu lato, Anaplasma and Ehrlichia species, and spotted fever group rickettsiae in ticks from Southeastern Europe. Eur J Clin Microbiol Infect Dis. 2003;22:535–42. doi: 10.1007/s10096-003-0988-1. [DOI] [PubMed] [Google Scholar]
- 36.Choi YJ, Lee EM, Park JM, Lee KM, Han SH, Kim JK, et al. Molecular detection of various rickettsiae in mites (Acari: Trombiculidae) in southern Jeolla Province Korea. Microbiol Immunol. 2007;51:307–12. doi: 10.1111/j.1348-0421.2007.tb03912.x. [DOI] [PubMed] [Google Scholar]
- 37.Jackson EB, Danauskas JX, Coale MC, Smadel JE. Recovery of Rickettsia akari from the Korean vole Microtus fortis pelliceus. Am J Hyg. 1957;66:301–8. doi: 10.1093/oxfordjournals.aje.a119903. [DOI] [PubMed] [Google Scholar]
- 38.Kim HC, Kim JH, Jo YS, Chong ST, Sames WJ, Klein TA, et al. Records of Ixodes pomeranzevi Serdyukova, 1941 (Acari: Ixodidae) from small mammals in northern Gyeonggi and Gangwon provinces, Republic of Korea. Sys Applied Acarol. 2009;14:129–35. [Google Scholar]
- 39.Sames WJ, Kim HC, Chong ST, Lee IY, Apanaskevich DA, Robbins RG, et al. Haemaphysalis (Ornithophysalis) phasiana (Acari: Ixodidae) in the Republic of Korea: two province records and habitat descriptions. Sys Applied Acarol. 2008;13:43–50. [Google Scholar]
- 40.Katargina O, Geller J, Ivanova A, Varv K, Tefanova V, Vene S, et al. Detection and identification of Rickettsia species in Ixodes tick populations from Estonia. Ticks Tick Borne Dis. 2015;6:689–94. doi: 10.1016/j.ttbdis.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 41.Suh JH, Kim HC, Yun SM, Lim JW, Kim JH, Chong ST, et al. Detection of SFTS virus in Ixodes nipponensis and Amblyomma testudinarium (Ixodida: Ixodidae) collected from reptiles in the Republic of Korea. J Med Entomol. 2016;53:584–90. doi: 10.1093/jme/tjw007. [DOI] [PubMed] [Google Scholar]
- 42.Cho BK, Nam HW, Cho SY, Lee WK. A case of tick bite by a spontaneously retreated Ixodes nipponensis. Korean J Parasitol. 1995;33:239–42. doi: 10.3347/kjp.1995.33.3.239. [DOI] [PubMed] [Google Scholar]
- 43.Ko JH, Cho DY, Chung BS, Kim SI. Two human cases of tick bite caused by Ixodes nipponensis. Korean J Parasitol. 2002;40:199–203. doi: 10.3347/kjp.2002.40.4.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jeon WS, Kim HS, Lee JD, Cho SH. Tick bite. Ann Dermatol. 2014;26:127–8. doi: 10.5021/ad.2014.26.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee SH, Chai JY, Kho WG, Hong SJ, Chung YD. A human case of tick bite by Ixodes nipponensis on the scalp. Kisaengchunghak Chapchi. 1989;27:67–9. doi: 10.3347/kjp.1989.27.1.67. [DOI] [PubMed] [Google Scholar]
- 46.Luce-Fedrow A, Mullins K, Kostick AP, St John HK, Jing J, Richards AL. Strategies for detecting rickettsiae and diagnosing rickettsial diseases. Future Microbiol. 2015;10:537–64. doi: 10.2217/fmb.14.141. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the conclusions of this article are included with NCBI accession numbers: Chungcheong 927 (KX418671, KX418672), Jeolla 958 (KX418673), Jeolla 959 (KX418675, KX418676), Jeolla 960 (KX418677), Jeolla 961 (KX418678), Jeolla 962 (KX418740), Jeolla 964 (KX418765, KX418766), Jeolla 967 (KX418741, KX418742), Jeolla 968 (KX418767, KX418768), Jeolla 969 (KX418743, KX418744), Jeolla 972 (KX418754, KX418755), Jeolla 973 (KX418745, KX418746), Jeolla 979 (KX418756, KX418757), Jeolla 985 (KX418758, KX418759), Jeolla 990 (KX418760, KX418761), Jeolla 1004 (KX418663, KX418664), Jeolla 1046 (KX418747, KX418748), Jeolla 1048 (KX418679, KX418680), Jeolla 1049 (KX418681, KX418682), Jeolla 1050 (KX418683, KX418684), Jeolla 1052 (KX418685, KX418686), Jeolla 1053 (KX418687, KX418688), Jeolla 1055 (KX418689, KX418690), Jeolla 1056 (KX418691, KX418692), Jeolla 1058 (KX418749), Jeolla 1059 (KX418750, KX418751), Jeolla 1060 (KX418693, KX418694), Jeolla 1061 (KX418695, KX418696), Jeolla 1063 (KX418697, KX418698), Jeolla 1064 (KX418699, KX418700), Jeolla 1065 (KX418762, KX418763), Jeolla 1066 (KX418701, KX418702), Jeolla 1067 (KX418703, KX418704), Jeolla 1068 (KX418705, KX418706), Jeolla 1069 (KX418707, KX418708), Jeolla 1070 (KX418709, KX418710), Jeolla 1072 (KX423490), Jeolla 1073 (KX418711, KX418712), Jeolla 1074 (KX418713, KX418714), Jeolla 1075 (KX418715, KX418716), Jeolla 1076 (KX418717, KX418718), Jeolla 1077 (KX418719, KX418720), Jeolla 1078 (KX418721, KX418722), Jeolla 1081 (KX418723, KX418724), Jeolla 1083 (KX418725, KX418726), Jeolla 1092 (KX418727, KX418728), Jeolla 1094 (KX418729, KX418730), Jeolla 1101 (KX418764), Jeolla 1133 (KX418731, KX418732), Chungcheong 1147 (KX418657, KX418658), Chungcheong 1155 (KX418665, KX418666), Jeolla 1167 (KX418752, KX418753), Jeolla 1168 (KX418733, KX418734), Jeolla 1175 (KX418659, KX418660), Jeolla 1176 (KX418735, KX418736), Jeolla 1178 (KX418737, KX418738), Jeolla 1196 (KX418667, KX418668), Jeolla 1199 (KX418661, KX418662), Jeolla 1200 (KX418669, KX418670), Jeolla 1208 (KX418739).


