Abstract
The incorporation of histone variants into nucleosomes can alter chromatin-based processes. CENP-A is the histone H3 variant found exclusively at centromeres that serves as an epigenetic mark for centromere identity and is required for kinetochore assembly. CENP-A mislocalization to ectopic sites appears to contribute to genomic instability, transcriptional misregulation, and tumorigenesis, so mechanisms exist to ensure its exclusive localization to centromeres. One conserved process is proteolysis, which is mediated by the Psh1 E3 ubiquitin ligase in Saccharomyces cerevisiae (budding yeast). To determine whether there are features of the CENP-A nucleosome that facilitate proteolysis, we performed a genetic screen to identify histone H4 residues that regulate CENP-ACse4 degradation. We found that H4-R36 is a key residue that promotes the interaction between CENP-ACse4 and Psh1. Consistent with this, CENP-ACse4 protein levels are stabilized in H4-R36A mutant cells and CENP-ACse4 is enriched in the euchromatin. We propose that the defects in CENP-ACse4 proteolysis may be related to changes in Psh1 localization, as Psh1 becomes enriched at some 3′ intergenic regions in H4-R36A mutant cells. Together, these data reveal a key residue in histone H4 that is important for efficient CENP-ACse4 degradation, likely by facilitating the interaction between Psh1 and CENP-ACse4.
Keywords: CENP-ACse4, H4, Psh1, proteolysis, centromere, nucleosome
CHROMATIN structure is a barrier to many processes that must gain access to DNA to generate the biomolecules required for cellular viability (Venkatesh and Workman 2015). Chromatin also serves as a platform that promotes the association of supramolecular structures with DNA, such as kinetochores that form on centromeres and capping structures that assemble at telomeres (Verdaasdonk and Bloom 2011; Kupiec 2014). The foundation of chromatin is histones, which associate with and compact DNA. Histones are therefore subject to a variety of regulatory processes, such as post-translational modifications and regulated incorporation or removal at various genomic regions (Rando and Winston 2012). Chromatin is also regulated by the deposition of histone variants, which can alter chromatin structure and accessibility to regulate cellular processes (Li and Fang 2015). One conserved histone is the H3 variant CENP-A, which localizes to centromeres to mediate their epigenetic propagation and to serve as the foundation for kinetochore assembly (Palmer et al. 1987; Van Hooser et al. 2001; Black and Cleveland 2011; Verdaasdonk and Bloom 2011). Consistent with this, CENP-A mislocalization to euchromatin can lead to ectopic kinetochore formation, genome instability, and transcriptional misregulation (Tomonaga et al. 2003; Heun et al. 2006; Amato et al. 2009; Hildebrand and Biggins 2016). It is therefore essential that CENP-A exclusively localizes to centromeres and does not stably incorporate into euchromatin.
One conserved mechanism that prevents ectopic CENP-A localization is ubiquitin-mediated proteolysis (Lomonte et al. 2001; Collins et al. 2004; Moreno-Moreno et al. 2006, 2011; Gross et al. 2012). The most detailed understanding of CENP-A proteolysis is in budding yeast, where the E3 ubiquitin ligase Psh1 mediates CENP-ACse4 degradation to limit its incorporation into euchromatin (Hewawasam et al. 2010; Ranjitkar et al. 2010). Psh1 requires its association with the conserved FACT (Facilitates Chromatin Transcription/Transactions) complex to bind to CENP-ACse4 and mediate ubiquitylation, and a loss of this interaction results in CENP-ACse4 mislocalization and cell death when CENP-ACse4 is overexpressed (Deyter and Biggins 2014). FACT (consisting of the Spt16 and Pob3 proteins) influences many aspects of the dynamic nature of chromatin, and we previously proposed that its role in chromatin disassembly allows Psh1 to access mislocalized CENP-ACse4 for ubiquitylation (Deyter and Biggins 2014). Proline isomerization is also important for CENP-ACse4 degradation because it is impaired in fpr3 and fpr4 proline isomerase mutants (Ohkuni et al. 2014). CENP-ACse4 has several proline residues that may require isomerization to promote an interaction with Psh1 (Ohkuni et al. 2014). Although CENP-A stably associates with histone H4 in CENP-A/H4 dimers and tetramers, the contribution of histone H4 or other features of the centromeric nucleosome to CENP-A proteolysis is unknown (Sekulic et al. 2010; Dechassa et al. 2014).
Here, we report that the histone H4 residue R36 contributes to CENP-ACse4 degradation. CENP-ACse4 overexpression is toxic to H4-R36A mutants, and this lethality is unrelated to centromere dysfunction because kinetochore composition and chromosome segregation are normal in these cells. Instead, overexpressed CENP-ACse4 is stabilized and enriched in the euchromatin in the H4-R36A mutant, indicating a role for histone H4 in preventing CENP-ACse4 mislocalization. Consistent with this, the interaction between Psh1 and CENP-ACse4 is decreased in the H4 mutant. In addition, Psh1 becomes enriched at 3′ ends of genes, similar to the mislocalization pattern of the FACT component Spt16 in H4-R36A mutant cells (Nguyen et al. 2013). The mislocalization of Psh1 to 3′ intergenic regions raises the possibility that it has reduced access to CENP-ACse4 at promoters and intragenic regions, resulting in elevated CENP-ACse4 levels in the euchromatin. Together, our data identify a role for histone H4 in promoting an interaction between Psh1 and CENP-ACse4 to prevent the promiscuous incorporation of CENP-ACse4 into euchromatin.
Materials and Methods
Yeast strain construction and microbial techniques
Microbial techniques and media were performed as described (Deyter and Biggins 2014). For all experiments involving induction of epitope-tagged pGAL-CSE4, budding yeast cells of the indicated strains were grown to log phase (OD 0.55–0.8, Bio-Rad SmartSpec 3000) in lactic acid media at 23° and induced for 2 hr with 2% galactose, unless otherwise indicated. Yeast strains were constructed using standard genetic techniques (Sherman et al. 1974; Rose et al. 1990). Epitope-tagged proteins were constructed using either a PCR integration technique or by the integration of plasmids after restriction digestion (Longtine et al. 1998). Specific plasmids and yeast strains used in this study are described in Supplemental Material, Table S1 and Table S2, respectively. A pGAL-3Flag-CSE4, LEU2 2-μm vector (pSB1730) was constructed by digesting vector YEplac181 with KpnI and SacI (Gietz and Sugino 1988). The vector was ligated with a pGAL-3Flag-CSE4 fragment that was isolated by digesting pSB839 (Ranjitkar et al. 2010) with the same enzymes to create a 2-μm LEU2 plasmid with pGAL-3Flag-CSE4 (pSB1730).
Genetic screen of histone mutant library
A previously constructed budding yeast histone H4 library was used to screen for mutants that are sensitive to CENP-ACse4 overexpression (Dai et al. 2008; Ng et al. 2013). Each H4 amino acid was singly changed to alanine by integrating plasmids from a Histone Mutant Library version 2.1 (HMLv2.1) that had been digested with BciVI to target the HHT2-HHF2 locus in a hht1-hhf1∆ background (Dai et al. 2008; Ng et al. 2013). For convenience in the initial screen, we used a previously constructed library in our lab that also contained an Ipl1-degron protein (Ng et al. 2013). However, the strains were grown under conditions that did not promote Ipl1 degradation. First, the H4 library was transformed in 96-well plate format with a high-copy plasmid expressing Myc-tagged CENP-ACse4 under the galactose promoter [pSB830 (Collins et al. 2004)] and subsequently plated on selective medium containing glucose in 100 × 15-mm plates. After 5 days of growth at 23°, transformants were patched onto selective medium containing glucose and incubated at 23°. Replica plating was performed to analyze the growth of the transformed library on glucose vs. galactose-containing medium. Hits were selected as strains that grew at 23° on glucose but exhibited defective growth on galactose-containing medium. To eliminate histone mutants that are defective in galactose metabolism, positive hits were rescreened to compare their growth on galactose with a control vector [YEplac181 (Gietz and Sugino 1988)] with that of the pGAL-Myc-CSE4 plasmid [pSB830 (Collins et al. 2004)]. Hits were also streaked onto selective medium containing glucose or galactose to confirm the replica plating results. This initial round of screening identified six histone H4 mutants that appeared to have defective growth when CENP-ACse4 was overexpressed: H4-I34A, H4-R36A, H4-K44A, H4-Y51A, H4-T80A, and H4-D85A. Second, the six H4 mutants were obtained from the same initial H4 mutant library and were retransformed with YEplac181 (vector control), pSB830 (Myc-CSE4 plasmid), and pSB1730 (pGAL-3Flag-CSE4, which expresses CENP-ACse4 at higher levels compared to the pGAL-Myc-CSE4 plasmid) to reanalyze their growth on galactose-containing medium. This approach eliminated three of the mutants, leaving H4-R36A, H4-K44A, and H4-Y51A for further testing. These three H4 mutants were then regenerated in an otherwise wild-type (WT) background, where both histone loci were replaced with the mutants in contrast to the parent strains that had a single HHF1-HHT1 locus replaced in the presence of an HHF2-HHT2 deletion. These strains were made for each histone mutant of interest by integrating plasmids from a Histone Mutant Library version 3.0 (HMLv3.0) (a kind gift from Junbiao Dai, Tsinghua University) that were digested with BciVI to target the HHT1-HHF1 locus. The plasmids that were used to make the histone mutants were sequenced to confirm the presence of the desired mutation. They were subsequently crossed to strains with the histone mutant of interest at the HHT2-HHF2 locus to obtain strains with replacements at both histone loci. Plate spot assays (with yeast transformed with YEplac181 or pSB1730) were used to analyze the growth of the mutants on glucose and galactose medium for the final three candidates. The plasmids used to make these strains are listed in Table S1.
PyMOL structures
Preparation of structural figures of the human CENP-A [PDBID 3AN2 (Tachiwana et al. 2011)] and yeast H3 [PDBID 1ID3 (White et al. 2001)] nucleosomes were performed using the PyMOL Molecular Graphics System (Schrodinger).
Protein and immunoprecipitation techniques
Protein extracts to analyze total CENP-ACse4 levels were prepared as described (Minshull et al. 1996; Hildebrand and Biggins 2016). Immunoblots using chemiluminescence were performed as previously described (Minshull et al. 1996; Hildebrand and Biggins 2016). For all immunoblots, the antibody dilutions were as follows: mouse α-Pgk1 monoclonal antibodies (Invitrogen, Carlsbad, CA, catalog # 459250) at a 1:10,000 dilution were used as a loading control. Mouse α-Flag M2 monoclonal antibodies (Sigma [Sigma-Chemical], St. Louis, MO, catalog # F3165) were used at a 1:3000 dilution, mouse α-HA 12CA5 monoclonal antibodies (Roche, catalog # 1-583-816) were used at a 1:10,000 dilution, rabbit α-H2B polyclonal antibodies (Active Motif, catalog # 39237) were used at a 1:3000 dilution, and mouse α-Myc 9E10 monoclonal antibodies were used at a 1:10,000 dilution (Covance, catalog # MMS-150R). Rabbit α-Spc105 antibodies were used at a 1:1000 dilution (Akiyoshi et al. 2010). Rabbit α-Ctf19 antibodies (a generous gift from Arshad Desai) were used at a 1:15,000 dilution. Quantitative immunoblots were carried out according to (Ng et al. 2013) with the modification of using 4% nonfat milk in PBS as the blocking agent for the α-Cse4 immunoblot. Briefly, IRDye anti-mouse and anti-rabbit secondary antibodies from LI-COR were used at a 1:5000 dilution. The immunoblots were imaged on a LI-COR imaging system, and the protein levels were quantified using Image Studio Lite.
Kinetochore purifications were performed from 50 ml cultures as previously described (Akiyoshi et al. 2010). For experiments analyzing ubiquitin conjugates of Cse4, NEM (N-ethylmaleimide) was added to the lysis and wash buffers to a final concentration of 5 mM (Deyter and Biggins 2014). Immunoprecipitates were separated by SDS-PAGE and analyzed by immunoblotting. Co-immunoprecipitation experiments were performed as previously described for Psh1-Myc and Spt16-Flag strains using protein G dynabeads conjugated with α-Myc (A-14, SC-789) and separated on a SDS-PAGE gel (Deyter and Biggins 2014).
Chromatin fractionation
Chromatin fractionation assays were performed as described, followed by analysis on immunoblots (Deyter and Biggins 2014). α-PGK1 was used as a marker and loading control for the soluble fraction, and α-H2B was used as a marker and loading control for the chromatin fraction (Deyter and Biggins 2014).
CENP-ACse4 stability assays
CENP-ACse4 stability assays were performed as in Deyter and Biggins (2014). Briefly, cells were grown in YEP + lactic acid until midlog phase, followed by a 2-hr 3Flag-Cse4 induction with 2% galactose. Glucose was added to a final concentration of 2% to further inhibit 3Flag-Cse4 transcription, and cycloheximide was added to a final concentration of 50 μg/ml. Time point 0 was taken immediately, followed by sample collection at the indicated time points. Extracts were prepared and analyzed as described (Minshull et al. 1996).
Chromosome segregation assay
Analysis of GFP-LacI was performed as described (Straight et al. 1996; Biggins et al. 1999). The lacO array was integrated at the TRP1 locus, ∼12.5 kb from CEN4.
Chromatin immunoprecipitation (ChIP)
ChIP was performed from 50 ml asynchronous yeast cultures with 1% formaldehyde to mediate cross-linking followed by sonication to fragment DNA, as previously described (Ng et al. 2009). α-Flag M2 antibodies conjugated to protein G dynabeads were used for both Flag-Cse4 and Psh1-Flag ChIPs. Quantitative polymerase chain reaction (qPCR) was performed as previously described (Hildebrand and Biggins 2016), on an Applied Biosystems (Foster City, CA) QuantStudio 5 qPCR machine. Oligonucleotide sequences are available in Table S3. Graphpad Prism 7 was used for two-way ANOVA with Tukey’s multiple comparison tests to analyze Psh1 or CENP-ACse4 enrichment, and to calculate Spearman’s correlation coefficient for the Psh1 vs. CENP-ACse4 data in the H4-R36A strain. Normalized ChIP signal for Psh1-Flag and Flag-Cse4 ChIPs was calculated as the mean of the ratio of the % Input for each strain relative to % Input for the WT PSH1-Flag or pGAL-Flag-CSE4 strain, respectively, for three biological replicates.
PSH1 and CSE4 gene expression analysis
Gene expression of PSH1 and CSE4 in H4-R36A compared to WT cells was determined using published microarray data (Jung et al. 2015). The microarray data were accessed using the NCBI GEO accession viewer (GSE29059). A .txt file containing the log2 ratio of each gene in each histone mutant compared to WT was downloaded from this record (GSE29059_log2_mut_wt_ratio.txt.gz). This was imported into Microsoft Excel, and the log2 ratio for the H4-R36A mutant compared to the WT cells was looked up using the Microsoft Excel search function for CSE4 (YKL049C, log2 H4-R36A vs. WT = 0.94562094) and PSH1 (YOL054W, log2 H4-R36A vs. WT = 0.15504214). The paper that published the microarray data used a cutoff of > 1.5-fold up- or downregulation to determine which genes were differentially regulated, and both CSE4 and PSH1 have < 1.5-fold change compared to WT (Jung et al. 2015).
Reagent and data availability
All strains and plasmids are available upon request. Table S1 describes the plasmids used to construct yeast strains, Table S2 lists the genotypes of all yeast strains, and Table S3 reports all oligonucleotide sequences used in this study.
Results
H4-R36A mutant cells are sensitive to CENP-ACse4 overexpression
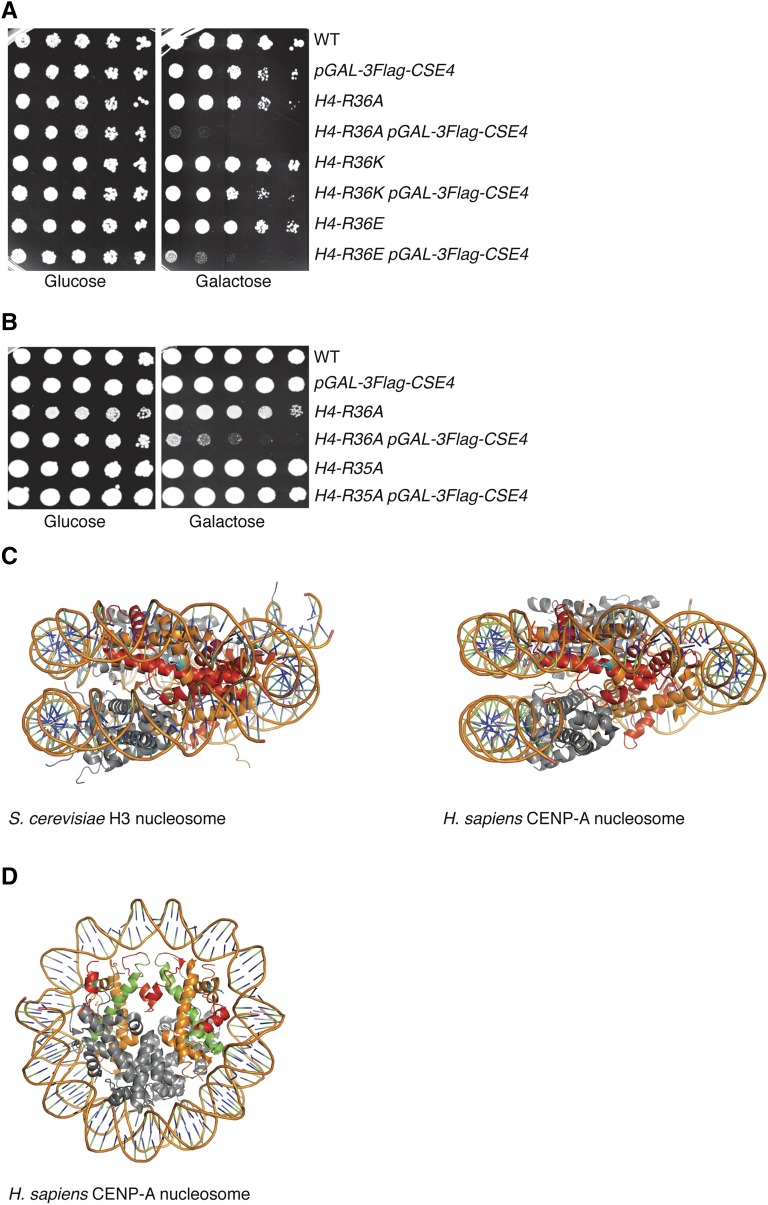
Because CENP-ACse4 proline residues influence its degradation, we hypothesized that additional features of the CENP-ACse4 nucleosome might be important for CENP-ACse4 proteolysis (Ohkuni et al. 2014). CENP-ACse4 binds with high affinity to histone H4 before and after deposition on DNA, so we asked whether histone H4 residues are important for CENP-ACse4 degradation. To do this, we took advantage of the fact that mutants defective in CENP-ACse4 degradation are sensitive to CENP-ACse4 overexpression (Hewawasam et al. 2010; Ranjitkar et al. 2010). We used a previously constructed library of budding yeast histone H4 mutants, where each amino acid was changed to alanine, to screen for mutants that are intolerant to CENP-ACse4 overexpression (Dai et al. 2008; Ng et al. 2013). We transformed the H4 library with a high-copy plasmid expressing N-terminally Myc-tagged CENP-ACse4 under the galactose promoter and analyzed the growth of the cells on glucose- vs. galactose-containing medium. After subsequent rescreening (see Materials and Methods), only one histone H4 mutant, H4-R36A, exhibited significant lethality when CENP-ACse4 was overproduced (Figure 1A and Figure S1A). Since arginine residues are subject to methylation in budding yeast, we created additional amino acid substitutions at H4 position 36 to test whether a post-translational modification might be involved (Rando and Winston 2012). We replaced R36 with a similarly basic residue lysine (R36K) that would inhibit methylation by an arginine methyltransferase. The growth of H4-R36K cells was similar to that of WT cells when CENP-ACse4 was overexpressed, indicating that methylation is not involved. We also replaced H4-R36 with glutamic acid (R36E) to generate a negative charge and found it was nearly as sensitive to increased CENP-ACse4 levels as H4-R36A cells (Figure 1A and Figure S1A). Together, these data indicate that a basic residue at position 36 in histone H4, but not post-translational modification of the amino acid, is required to prevent sensitivity to CENP-ACse4 overexpression. Also, mutation of the neighboring H4-R35 residue did not cause sensitivity to CENP-ACse4 (Figure 1B and Figure S1B), indicating a specific role for H4-R36 in protecting cells from CENP-ACse4-induced lethality. A notable feature of H4-R36 is that it is the only residue in the α-1 helix of H4 whose side chain interacts with nucleosomal DNA near the entry/exit site in canonical H3 nucleosomes and in octameric human CENP-A nucleosomes (Figure 1C) (Luger et al. 1997; Tachiwana et al. 2011). H4-R36 is also located in a region near to the CENP-A targeting domain (CATD) in the human CENP-A nucleosome, which is required for Psh1 recognition of CENP-ACse4 (Figure 1D) (Ranjitkar et al. 2010; Tachiwana et al. 2011). These data suggest that the interaction between H4-R36 and DNA contributes to its function in shielding cells from the potentially lethal consequences of CENP-ACse4 overexpression.
Figure 1.
CENP-ACse4 overexpression is lethal to H4-R36A cells. (A) Fivefold serial dilutions of the indicated strains [WT (SBY13389), pGAL-3Flag-CSE4 (SBY13390), H4-R36A (SBY13391), H4-R36A pGAL-3Flag-CSE4 (SBY13392), H4-R36K (SBY13393), H4-R36K pGAL-3Flag-CSE4 (SBY13394), H4-R36E (SBY13395), and H4-R36E pGAL-3Flag-CSE4 (SBY13396)] were plated on glucose or galactose media at 23°. (B) Fivefold dilutions of the indicated strains [WT (SBY9401), pGAL-3Flag-CSE4 (SBY10025), H4-R36A (SBY9365), H4-R36A pGAL-3Flag-CSE4 (SBY9397), H4-R35A (SBY10510), and H4-R35A pGAL-3Flag-CSE4 (SBY13407)] were plated on glucose or galactose media at 23°. (C) Side views of crystal structures of the S. cerevisiae nucleosome containing histone H3 (left) and the Homo sapiens nucleosome containing CENP-A (right). Histone H4 (orange) residue R36 (cyan) makes side-chain interactions with nucleosomal DNA. Red: histone H3 (left) or CENP-A (right). (D) Top view of the crystal structure of the H. sapiens nucleosome containing CENP-A as in (C), with the CATD indicated (green). WT, wild-type.
Kinetochores are functional in H4-R36A cells overexpressing CENP-ACse4
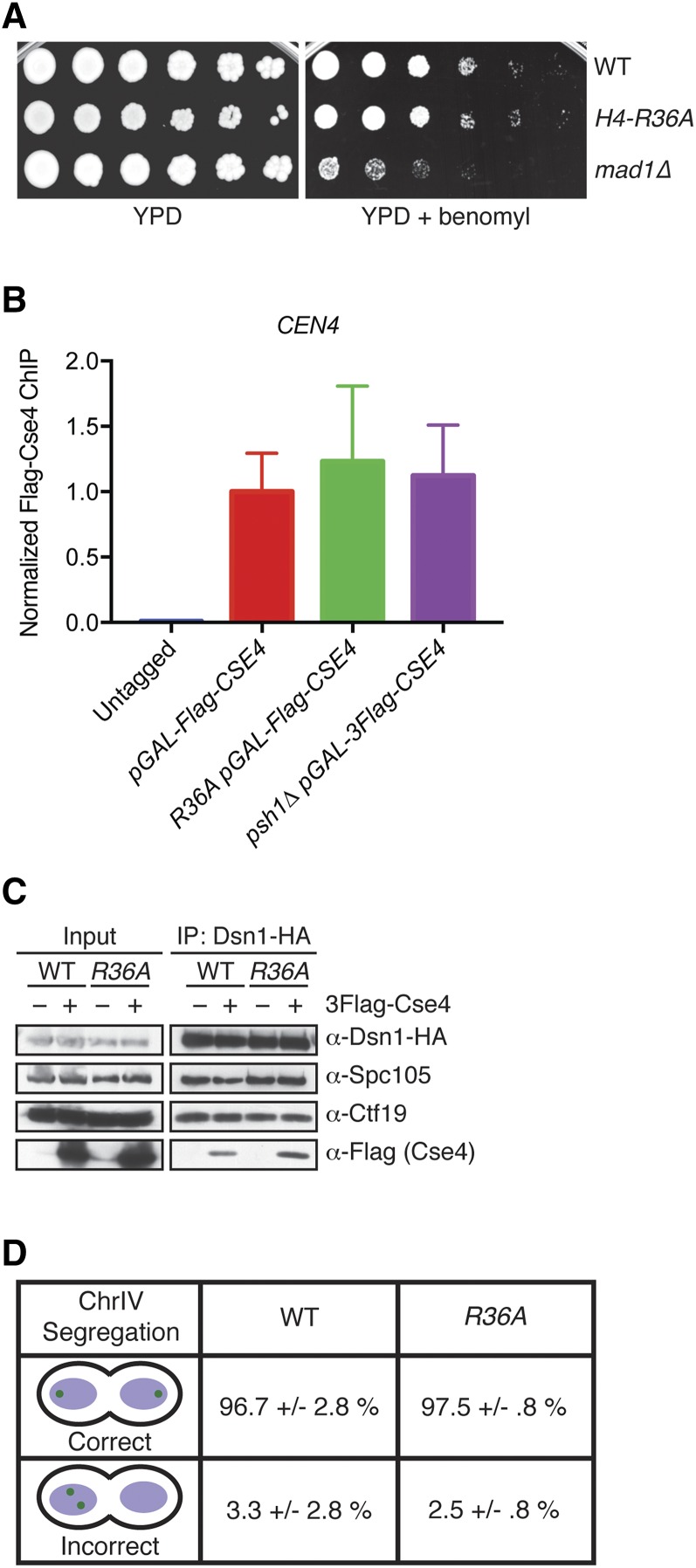
Since CENP-ACse4 is essential for chromosome segregation, we asked if CENP-ACse4 overexpression in H4-R36A cells altered kinetochore composition or function (Stoler et al. 1995). There is a precedent for H4 influencing chromosome segregation, since the hhf1-20 mutation (conferring a H4-T82I/A89V substitution) that weakens the interface between H4 and DNA causes chromosome missegregation, as do other H4 mutations that were more recently identified (Smith et al. 1996; Kawashima et al. 2011; Ng et al. 2013). However, H4-R36A mutants have not been identified in any of these chromosome segregation screens and H4-R36A mutant cells do not show sensitivity to mitotic spindle poisons (Dai et al. 2008). We confirmed that the growth of H4-R36A cells was not affected by benomyl, a microtubule-destabilizing drug that inhibits the growth of kinetochore mutants and spindle assembly checkpoint mutants such as mad1Δ (Figure 2A and Figure S1C). We also tested whether CENP-ACse4 still localizes to the centromere by performing ChIP followed by qPCR and detected similar levels of CENP-ACse4 at CEN4 in WT and H4-R36A cells overexpressing CENP-ACse4 (Figure 2B).
Figure 2.
The overproduction of CENP-ACse4 in the H4-R36A mutant does not perturb chromosome segregation. (A) WT (SBY9401), H4-R36A (SBY9979), and mad1∆ (SBY3256) cells were serially diluted fivefold and plated on YPD (left) or YPD + benomyl (right) medium and incubated at 23°. (B) 3Flag-Cse4 ChIP in WT (SBY16469), H4-R36A (SBY13812), and psh1∆ (SBY11189) cells with pGAL-3Flag-CSE4 overexpression. An untagged strain (SBY3) was used as a negative control. qPCR for the CEN4 locus (using primers SB3061 + SB3062) was performed. Normalized Flag-Cse4 ChIP is the ratio of the % Input in each strain relative to the % Input in the pGAL-3Flag-CSE4 strain. Mean Flag-Cse4 ChIP enrichment ± 1 SE of the mean (SEM) for three biological replicates is shown. (C) Anti-HA-conjugated beads were used to immunoprecipitate kinetochores via Dsn1-3HA from WT (SBY10450) or H4-R36A (SBY10453) cells containing pGAL-3Flag-CSE4 in the presence (+) or absence (−) of galactose. The levels of Spc105 and Ctf19 were analyzed by immunoblotting with antibodies raised against the respective proteins. Immunoblotting with α-HA antibodies revealed the levels of Dsn1-3HA in the immunoprecipitates while α-Flag antibodies detected the incorporation of overexpressed 3Flag-Cse4. (D) Asynchronous cultures of WT (SBY10494) or H4-R36A (SBY10391) cells with mad1∆ and expressing pGAL-3Flag-CSE4 were grown in galactose for 4 hr and chromosome IV segregation was monitored in anaphase cells (defined by cells with DNA masses at opposite poles). At least 100 cells were counted for each strain from two independent replicates. Percent correct or incorrect segregation ± 1 SD is shown. ChIP, chromatin immunoprecipitation; Chr, chromosome; WT, wild-type.
We next analyzed the integrity of kinetochores by purification of the Dsn1 kinetochore protein, a method we previously developed to isolate native kinetochore particles (Akiyoshi et al. 2010). We immunoprecipitated Dsn1-3HA from WT and H4-R36A cells in the presence or absence of CENP-ACse4 overexpression (Figure 2C). The levels of copurifying Ctf19 and Spc105, representative components of the inner and outer kinetochore, respectively, were similar to those of kinetochores isolated from WT and H4-R36A cells (Figure 2C). These data suggest that the overall kinetochore composition is not affected. To assess kinetochore function, we monitored chromosome segregation in H4-R36A cells that also carried a mad1∆ to inactivate the spindle assembly checkpoint and ensure similar cell cycle progression for all strains (Hardwick and Murray 1995). Cells containing a fluorescently marked chromosome IV were analyzed for segregation to opposite poles at anaphase (Straight et al. 1996). There was no significant difference in chromosome segregation between cells overexpressing CENP-ACse4 in the presence or absence of the H4-R36A mutation (Figure 2D), indicating that the H4-R36 mutation does not significantly alter kinetochore composition or function.
Cells with reduced H3 levels are not sensitive to CENP-ACse4 overexpression
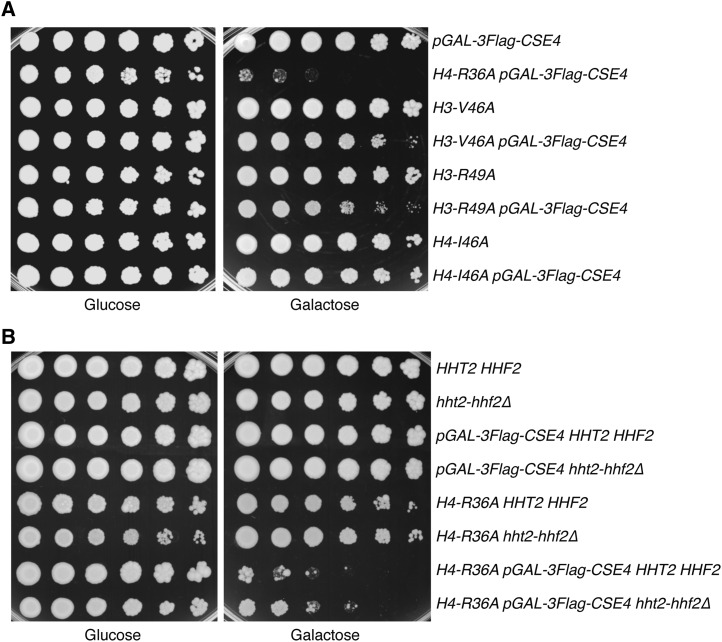
The cellular levels of canonical H3 and CENP-ACse4 are tightly regulated, and defects in H3 chromatin assembly can lead to CENP-ACse4 incorporation (Sharp et al. 2002; Collins et al. 2004; Singh et al. 2009; Hewawasam et al. 2010; Ranjitkar et al. 2010; da Rosa et al. 2011; Eriksson et al. 2012; Kurat et al. 2014). Interestingly, H4-R36A is one of several histone mutants that have decreased H3 occupancy at highly transcribed genes (Hainer and Martens 2011). Therefore, we hypothesized that the sensitivity of H4-R36A cells to CENP-ACse4 levels may be due to “gap-filling” in which defects in H3 chromatin occupancy provide nucleosome-depleted genomic regions that promote CENP-ACse4 incorporation. If this were true, other histone mutants that are defective in H3 occupancy should also be sensitive to increased CENP-ACse4 levels. To test this, we analyzed the growth of three other histone mutants in this class (H3-V46A, H3-R49A, and H4-I46A) when CENP-ACse4 was overexpressed (Figure 3A and Figure S2A) (Hainer and Martens 2011). H3-V46A and H3-R49A cells exhibited a slight sensitivity to increased CENP-ACse4 levels, while H4-I46A cell growth was unaffected by CENP-ACse4 overproduction. Notably, the H4-I46A mutant has a reduction in H3 chromatin assembly that is comparable to R36A cells, strongly suggesting that the lethality of CENP-ACse4 overexpression in H4-R36A cells is not related to gap-filling. As an additional test of this model, we directly reduced H3 levels. H3 and H4 are transcribed from two loci, HHT1-HHF1 and HHT2-HHF2. The major locus is HHT2-HHF2, and deleting this locus decreases the soluble pools of histones H3 and H4 (Liang et al. 2012). Therefore, we analyzed the growth of hht2-hhf2Δ cells when CENP-ACse4 was overexpressed (Figure 3B and Figure S2B) and found no growth defect, indicating that reduced H3 levels do not sensitize cells to CENP-ACse4 incorporation. Additionally, H4-R36A hht2-hhf2Δ cells did not show a more severe phenotype with CENP-ACse4 overexpression compared to H4-R36A cells. Altogether, these data suggest that altered H3 levels likely play a minimal role in the sensitivity of H4-R36A cells to increased CENP-ACse4 protein.
Figure 3.
The sensitivity of H4-R36A cells to CENP-ACse4 overexpression is not due to H3 depletion. (A) Fivefold serial dilutions of the indicated strains [pGAL-3Flag-CSE4 (SBY10025), H4-R36A pGAL-3Flag-CSE4 (SBY9397), H3-V46A (SBY10392), H3-V46A pGAL-3Flag-CSE4 (SBY10511), H3-R49A (SBY10393), H3-R39A pGAL-3Flag-CSE4 (SBY10512), H4-I46A (SBY10394), and H4-I46A pGAL-3Flag-CSE4 (SBY10513)] were plated on glucose or galactose media at 23°. (B) Fivefold dilutions of the indicated strains [HHT2-HHF2 (SBY9401), hht2-hhf2∆ (SBY10730), pGAL-3Flag-CSE4 HHT2-HHF2 (SBY10025), pGAL-3Flag-CSE4 hht2-hhf2∆ (SBY10734), H4-R36A HHT2-HHF2 (SBY9365), H4-R36A hht2-hhf2∆ (SBY10778), H4-R36A pGAL-3Flag-CSE4 HHT2-HHF2 (SBY9397), and H4-R36A pGAL-3Flag-CSE4 hht2-hhf2∆ (SBY10782)] were plated on glucose or galactose media at 23°.
CENP-ACse4 is mislocalized in H4-R36A cells
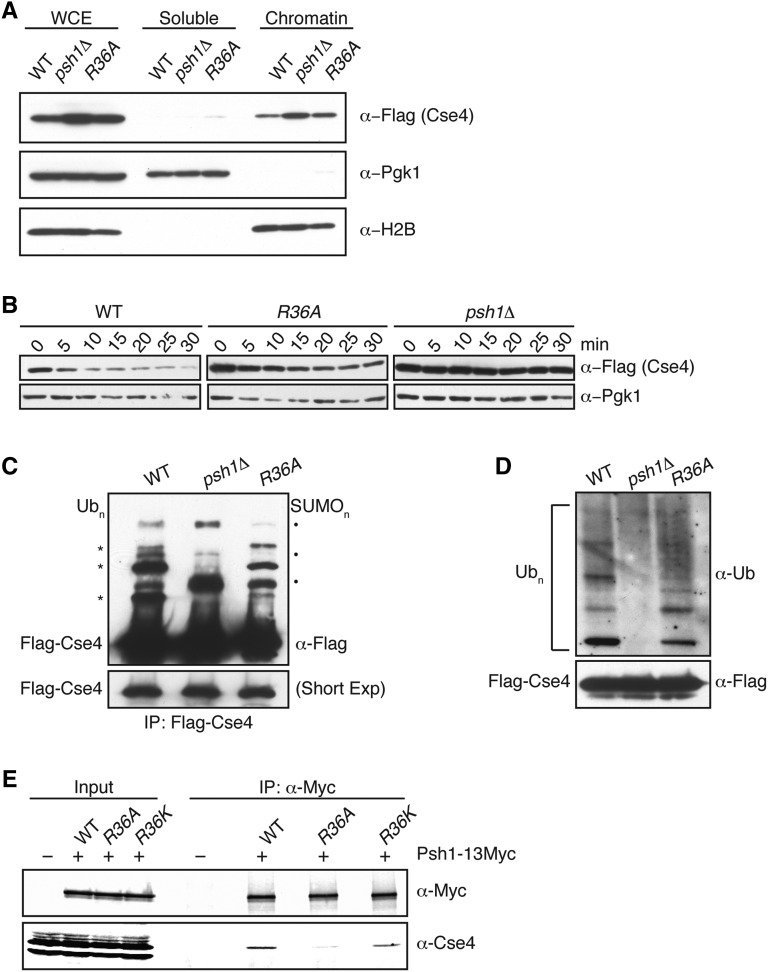
Because the sensitivity of cells to CENP-ACse4 overexpression correlates with the level of CENP-ACse4 misincorporation into euchromatin, we asked whether CENP-ACse4 localizes to euchromatin in H4-R36A cells (Hewawasam et al. 2010; Ranjitkar et al. 2010; Hildebrand and Biggins 2016). To test this, we compared the levels of overexpressed CENP-ACse4 in soluble and chromatin fractions from WT, H4-R36A, and psh1Δ cells. As we previously showed, the levels of CENP-ACse4 are higher in psh1∆ than WT since CENP-ACse4 is stabilized (Ranjitkar et al. 2010) (Figure 4A). Similarly, CENP-ACse4 levels are also higher in H4-R36A cells compared to WT cells, and it is enriched in the chromatin fraction of H4-R36A cells, although to a lesser extent than psh1Δ cells (Figure 4A and Figure S3).
Figure 4.
H4-R36A cells have defects in CENP-ACse4 degradation. (A) WCE from WT (SBY10025), psh1∆ (SBY10590), and H4-R36A (SBY9397) cells expressing pGAL-3Flag-CSE4 were fractionated into soluble and chromatin fractions. 3Flag-Cse4 levels were monitored in each fraction with α-Flag antibodies. Pgk1 and H2B are markers of the soluble and chromatin fractions, respectively. Note that a longer exposure of the blot for CENP-ACse4 is included in Figure S3, which shows CENP-ACse4 in the soluble fraction. (B) WT (SBY10025), H4-R36A (SBY9397), and psh1∆ (SBY10590) cells expressing pGAL-3Flag-CSE4 were grown in galactose, protein synthesis was inhibited by cycloheximide addition at time zero, and lysates were monitored for 3Flag-Cse4 levels at the indicated time points with α-Flag antibodies. Pgk1 served as a loading control. (C) WT (SBY10025), psh1∆ (SBY10590), and H4-R36A (SBY9397) cells expressing pGAL-3Flag-CSE4 were grown in galactose and 3Flag-Cse4 was immunoprecipitated with α-Flag antibodies. Unmodified 3Flag-Cse4 and its higher-mobility species (Ubn) were detected on immunoblots by probing with α-Flag antibodies, these bands are marked on the left by asterisks. Note that SUMO conjugates appear on Cse4 in the absence of ubiquitylation (SUMOn), these bands are marked on the right by solid dots (Ohkuni et al. 2016). The lower panel shows a shorter exposure of the immunoblot to display the levels of unmodified 3Flag-Cse4 in the immunoprecipitates and serves as a loading control. (D) An immunoblot probed with α-ubiquitin antibodies of the α-Flag immunoprecipitates isolated from the strains denoted in (C) reveals the ubiquitin conjugates of 3Flag-Cse4. The lower panel shows the levels of unmodified 3Flag-Cse4 by immunoblotting with α-Flag antibodies. (E) Psh1-13Myc was immunoprecipitated from WT (SBY15335), H4-R36A (SBY15624), and H4-R36K (SBY16468) cells, and immunoblots were probed with α-Myc and α-Cse4 antibodies. Cells expressing untagged Psh1 (SBY3) served as a control. The Cse4/Psh1 ratio ± 1 SEM was calculated vs. WT for H4-R36A (0.5 ± 0.03) and H4-R36K (3.0 ± 0.6) from three biological replicates. IP, immunoprecipitation; WCE, whole cell extract; WT, wild-type.
The interaction between Psh1 and CENP-ACse4 is altered in the H4-R36A mutant
The increased CENP-ACse4 protein levels in H4-R36A cells suggested a defect in CENP-ACse4 proteolysis because there are no changes in PSH1 or CSE4 transcription in these cells [see Materials and Methods and Jung et al. (2015)]. To determine if CENP-ACse4 degradation is impaired in H4-R36A cells, we monitored CENP-ACse4 protein levels after transient overexpression followed by translational repression with cycloheximide. CENP-ACse4 was partially stabilized in H4-R36A cells compared to WT cells (Figure 4B). To determine whether this is a result of a reduction in ubiquitin-mediated proteolysis, we analyzed the level of CENP-ACse4 ubiquitin conjugates in WT, psh1Δ, and H4-R36A cells. As expected, CENP-ACse4 isolated from WT cells displayed several higher mobility species, most of which correspond to ubiquitin conjugates of CENP-ACse4 (Figure 4, C and D). Consistent with previous work, ubiquitin conjugates of CENP-ACse4 are decreased in psh1Δ cells (Hewawasam et al. 2010; Ranjitkar et al. 2010). The upper CENP-ACse4 forms that are present in the psh1Δ cells correspond to SUMO conjugates that become detectable when ubiquitylation is reduced (Ranjitkar et al. 2010; Ohkuni et al. 2016) (Figure 4C and data not shown). CENP-ACse4 isolated from H4-R36A cells showed lower levels of most ubiquitin conjugates and increased sumoylation compared to WT cells, consistent with its partial stabilization. Together, these results show that H4-R36A cells are partially defective in CENP-ACse4 degradation and accumulate CENP-ACse4 in euchromatin.
The increased stability of CENP-ACse4 in H4-R36A cells suggested that the interaction between CENP-ACse4 and Psh1 might be altered in the mutant. Indeed, the association between Psh1 and CENP-ACse4 is reduced when Psh1 is immunoprecipitated from H4-R36A cells compared to WT cells (Figure 4E). The CENP-ACse4/Psh1 ratio ± 1 SE of the mean (SEM) vs. WT for H4-R36A cells was 0.5 ± 0.03. Consistent with our data suggesting that residue 36 of H4 needs to be basic to protect cells from lethality when CENP-ACse4 is overproduced (Figure 1A), the Psh1-CENP-ACse4 interaction is retained in the H4-R36K mutant that restores the basic charge (Figure 4E). The CENP-ACse4/Psh1 ratio ± 1 SEM vs. WT for H4-R36K cells was 3.0 ± 0.6. Taken together, these results suggest that the defects in CENP-ACse4 proteolysis are likely due to a decreased interaction between Psh1 and CENP-ACse4 in H4-R36A.
Psh1 is enriched at the 3′ ends of genes in H4-R36A cells
The altered Psh1-CENP-ACse4 interaction could be due to a change in CENP-ACse4 nucleosome structure that directly affects the binding of CENP-ACse4 to Psh1, and/or it could be due to changes in the localization of Psh1 such that it is no longer available to bind to CENP-ACse4 nucleosomes. Psh1 is present in both the soluble and chromatin fractions of cells; however, its euchromatic localization pattern has not been analyzed (Hewawasam et al. 2010; Ranjitkar et al. 2010). In addition, Psh1 is known to associate with FACT (Ranjitkar et al. 2010; Deyter and Biggins 2014), although their level of colocalization throughout the genome is unknown. The FACT complex exhibits an altered localization pattern in H4-R36A mutants and becomes enriched at the 3′ untranslated region (UTR) of highly transcribed genes (Nguyen et al. 2013). This phenotype is also shared with other histone mutants including H4-R31E and H3-L61W (Duina et al. 2007; Nguyen et al. 2013). These residues map to a distinct area of the nucleosome, and it was proposed that this nucleosomal region releases FACT from the end of transcribed units, although the exact mechanism or signal for the dissociation remains unclear (Duina et al. 2007; Nguyen et al. 2013).
Because Psh1 association with FACT is important for CENP-ACse4 ubiquitylation and degradation (Deyter and Biggins 2014), we analyzed Psh1 localization in WT vs. H4-R36A cells at the 5′, 3′, and coding regions of two highly transcribed genes, ADH1 and PMA1. It was previously found that the Spt16 component of FACT is mislocalized to the 3′ UTRs of these genes in H4-R36A or H3-L61W mutant cells (Duina et al. 2007; Nguyen et al. 2013). We confirmed the enrichment of Spt16 to the 3′ UTRs of the ADH1 and PMA1 genes in the H4-R36A mutants in our strain background using ChIP-PCR (data not shown). Strikingly, ChIP-qPCR revealed that Psh1 also shows a strong enrichment at the 3′ UTRs of these genes in H4-R36A cells compared to WT, while the levels at the promoter and gene regions were similar to WT cells (Figure 5, A–D). The H4-R36K mutant, which rescues the Psh1-CENP-ACse4 interaction, also restores the WT Psh1 localization pattern (Figure 5, C and D). Together, these data suggest that altered Psh1 localization could contribute to the CENP-ACse4 stability phenotype in H4-R36A mutant cells.
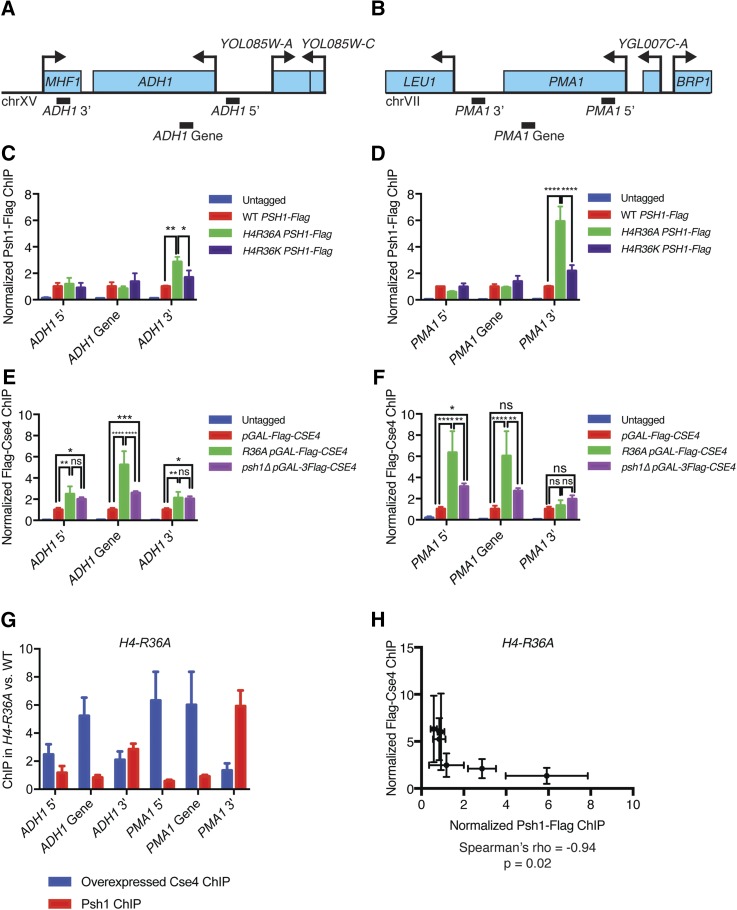
Figure 5.
Psh1 and CENP-ACse4 are mislocalized in the H4-R36A mutant cells. (A) Diagram of the ADH1 genomic locus showing regions amplified by 5′, 3′, and intragenic primer sets (not to scale). (B) Diagram of the PMA1 genomic locus showing regions amplified by the 5′, 3′, and intragenic primer sets (not to scale). (C) Psh1-3Flag ChIP from WT (SBY13723), H4-R36A (SBY13725), and H4-R36K (SBY16594) cells. Cells expressing untagged Psh1 (SBY3) were used as controls. qPCR was performed for the primer sets shown in (A), ADH1 5′ (SB5059 and SB5060), ADH1 gene (SB4470 and SB4471), and ADH1 3′ (SB4472 and SB4473). Normalized Psh1-Flag ChIP is the ratio of the % Input in each strain relative to the % Input in the WT PSH1-Flag strain. Mean Psh1-Flag ChIP enrichment ± 1 SEM from three biological replicates is shown. Results from two-way ANOVA with Tukey’s multiple comparisons test are shown. * P < 0.05, ** P ≤ 0.01, *** P ≤ 0.001, **** P ≤ 0.0001. (D) Psh1-Flag ChIP as in (C), with PCR primers as shown in (B) for PMA1 5′ (SB5029 and SB5030), PMA1 gene (SB4076 and SB4077), and PMA1 3′ (SB5027 and SB5028). (E) ChIP of overexpressed Flag-Cse4 from WT (SBY16469), H4-R36A (SBY13812), and psh1∆ (SBY11189) strains. Cells expressing untagged CENP-ACse4 were used as a control (SBY3). PCR primers at the ADH1 locus were used as in (C). qPCR was performed, normalized Flag-Cse4 ChIP is the ratio of the % Input in each strain relative to the % Input in the pGAL-3Flag-CSE4 strain. Mean Flag-Cse4 ChIP enrichment ± 1 SEM from three biological replicates is shown. (F) Flag-Cse4 ChIP as in (E) for PMA1 primers as in (D). (G) Bar plot of the average CENP-ACse4 (blue) or Psh1 (red) ChIP signal at each locus for the H4-R36A strains ± 1 SEM. (H) Scatter plot of Psh1 ChIP signal vs. CENP-ACse4 ChIP signal at each locus as in (G) (average ± 1 SD) in the H4-R36A strains. Spearman’s correlation coefficient was calculated from these data (ρ = −0.94, P = 0.02). ChIP, chromatin immunoprecipitation; chr, chromosome; qPCR, quantitative PCR; WT, wild-type; ns, not significant.
CENP-ACse4 mislocalization is negatively correlated with Psh1 enrichment in H4-R36A cells
We previously discovered that overexpressed CENP-ACse4 is mislocalized to nucleosomes in both tandem and divergent intergenic regions in the absence of Psh1 (Hildebrand and Biggins 2016). To test whether this is also true in the H4-R36A mutant cells, we performed ChIP-qPCR on overexpressed CENP-ACse4 from WT, H4-R36A, and psh1∆ cells, and analyzed CENP-ACse4 enrichment at the ADH1 and PMA1 promoters, genes, and 3′ regions compared to the WT pGAL-3Flag-CSE4 strain. For both genes, overexpressed CENP-ACse4 is enriched in the promoter and coding regions in H4-R36A cells compared to WT and psh1∆ mutant cells (Figure 5, E and F). In contrast, the level of CENP-ACse4 mislocalization at the 3′ ends of these genes is either much lower (at the ADH1 3′ end) or not enriched at all (at the PMA1 3′ end). Strikingly, correlation analysis of Psh1 and CENP-ACse4 localization in the H4-R36A strain reveals a significant negative correlation (Spearman’s ρ = −0.9429, P = 0.0167) (Figure 5, G and H). This finding is consistent with the possibility that the local enrichment of Psh1 mediates the ubiquitylation and degradation of CENP-ACse4 at specific regions.
Discussion
In this study, we performed the first systematic screen to identify mutations in H4 that affect the regulation of the histone variant CENP-ACse4. The H4 mutant R36A exhibited the strongest sensitivity to CENP-ACse4 overexpression of those analyzed. Our genetic and biochemical analyses support a model whereby a positive charge at this residue is required for CENP-ACse4 regulation by ensuring that the Psh1 ubiquitin ligase interacts with CENP-ACse4.
The H4-R36A cells overexpressing CENP-ACse4 are most likely inviable due to mislocalization of the centromeric histone variant to the euchromatin. We previously determined that CENP-ACse4 overexpression is lethal in cells lacking Psh1 due to ectopic localization of the centromeric histone variant, and here we found that Psh1 exhibits reduced binding to its substrate in the H4-R36A mutant cells (Ranjitkar et al. 2010; Deyter and Biggins 2014). Consistent with this, there was an increase in CENP-ACse4 stability and a reduction in its ubiquitylated forms in the H4-R36A mutant cells. Since H4-R36A cells have transcriptional defects as well as decreased nucleosome occupancy at transcribed genes, it is possible that increased transcriptional stress in the presence of mislocalized CENP-ACse4 contributes to cell death (Dai et al. 2008; Hainer and Martens 2011; Nguyen et al. 2013; Jung et al. 2015; Hildebrand and Biggins 2016). However, the lethality induced by CENP-ACse4 overexpression is rescued in the H4-R36K mutant, which restores the interaction between Psh1 and CENP-ACse4 but not all of the other H4-R36A phenotypes (Dai et al. 2008). Together, these data strongly suggest that the reduced interaction between Psh1 and CENP-ACse4 is related to the cell death caused by CENP-ACse4 overexpression.
The reduced Psh1-CENP-ACse4 interaction in the H4-R36A mutant cells could be due to a change in the nucleosome or (CENP-ACse4-H4)2 tetramer structure that directly alters the binding site of Psh1 for CENP-ACse4. The CATD of CENP-ACse4 is required for Psh1 binding and H4-R36 is not predicted to directly contact the CATD in an octameric nucleosome (Ranjitkar et al. 2010; Tachiwana et al. 2011). However, H4-R36 contacts the DNA near the DNA entry/exit site on the nucleosome and this region is close to the CATD (White et al. 2001; Tachiwana et al. 2011). Therefore, this residue may directly contribute to Psh1 recognition of the nucleosome and help distinguish nucleosome-bound vs. soluble CENP-ACse4.
The decreased binding between Psh1 and CENP-ACse4 may also be due to an indirect mechanism related to altered localization of the ubiquitin ligase. We found that Psh1 becomes enriched at 3′ UTRs in H4-R36A cells at two loci. If this is a widespread phenomenon, it could explain the decreased Psh1-CENP-ACse4 interaction, since Psh1 may be sequestered at 3′ UTRs. Consistent with this, there appears to be a negative correlation between the enrichment of Psh1 and CENP-ACse4 in H4-R36A cells at these loci, raising the possibility that there is localized degradation of CENP-ACse4 at the 3′ sites where Psh1 accumulates. In the future, ChIP-seq assays to determine the genome-wide mislocalization pattern of CENP-ACse4 and Psh1 in H4-R36A cells may further elucidate the underlying mechanism.
It is unclear what mechanism underlies the Psh1 3′ UTR enrichment. This mislocalization pattern is similar to what has been seen for the FACT complex component Spt16 in H4-R36A cells (Nguyen et al. 2013). Since Psh1 binds to Spt16 and this interaction is essential for controlling CENP-ACse4 localization in vivo, one possibility is that the mislocalization of FACT in H4-R36A cells leads to Psh1 enrichment at 3′ UTRs (Ranjitkar et al. 2010; Deyter and Biggins 2014). However, there are intragenic Spt16 suppressors that relieve its accumulation at the 3′ UTR in the H4-R36A mutant cells, but only some of them suppress the lethality of CENP-ACse4 overexpression (data not shown) (Duina et al. 2007; Nguyen et al. 2013). Therefore, the underlying mechanism appears to be more complicated than just FACT mislocalization leading to the accumulation of Psh1 at 3′ UTR regions. Consistent with this, while both Psh1 and Spt16 are enriched at 3′ UTRs in H4-R36A cells, only Spt16 shows depletion at promoters and intragenic regions (Nguyen et al. 2013).
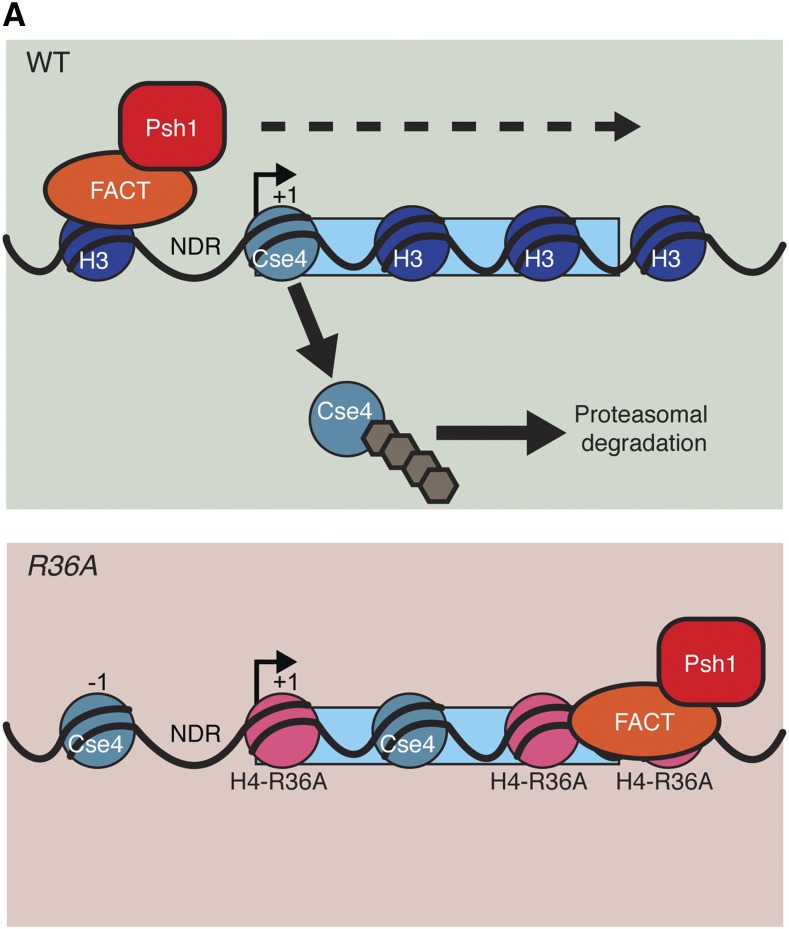
While the precise reason for the Psh1 mislocalization to 3′ ends of genes is not clear, it is correlated with altered CENP-ACse4 localization. Therefore, we propose that Psh1 needs to dynamically associate with chromatin to regulate CENP-ACse4 (Figure 6). This is consistent with our previous conclusion that Psh1 needs to be traveling with FACT and elongating RNA polymerases to recognize CENP-ACse4 that is incorporated into nucleosomes (Deyter and Biggins 2014). By disrupting chromatin, FACT would allow Psh1 to access the CATD, which is normally buried in the core of the nucleosome. The position of H4-R36 near the nucleosome entry/exit site may facilitate the ability of FACT to disrupt nucleosome structure. When this residue is disrupted, it may alter Psh1 and FACT localization in a manner that impairs CENP-ACse4 proteolysis, possibly by preventing the dynamic movement of FACT and Psh1 (Figure 6).
Figure 6.
Model for control of CENP-ACse4 localization via a dynamic Psh1-chromatin interaction. In WT cells containing canonical nucleosomes (dark blue), Psh1 (red) and FACT (orange) interact dynamically with genes during transcription (dynamic interaction is indicated by dashed arrow). This allows Psh1 to recognize mislocalized CENP-ACse4 (teal) at 5′, intragenic, and 3′ regions and mediate its ubiquitylation (brown), leading to its proteasomal degradation. Psh1-mediated recognition and degradation enables the cells to control CENP-ACse4 localization and to live when CENP-ACse4 is overexpressed (green shaded box). When H4-R36 is mutated to alanine (pink), FACT and Psh1 lose their dynamic interaction with transcribed genes and are sequestered at 3′ UTRs, compromising the Psh1/CENP-ACse4 interaction. CENP-ACse4 that incorporates at 5′ or intragenic regions is not adequately ubiquitylated and is retained at ectopic sites, leading to cell death (red shaded box). WT, wild-type.
In summary, our work has identified another facet of the regulation of CENP-ACse4 protein levels and localization. Histone H4 residue (R36) facilitates the Psh1-CENP-ACse4 physical interaction, likely by allowing Psh1 and FACT to travel dynamically through genes during transcription to facilitate CENP-ACse4 removal and degradation. Because it is essential to maintain the exclusive localization of the centromeric histone to prevent genomic instability and transcriptional misregulation (Hewawasam et al. 2010; Ranjitkar et al. 2010; Hildebrand and Biggins 2016), it will be important to further elucidate the precise nature of the interaction between Psh1 and the centromeric nucleosome, as well as the dynamics of Psh1 localization to chromatin in the future.
Acknowledgments
We thank Junbiao Dai for providing the plasmids required to generate the histone mutant strains, and Arshad Desai for sharing α-Ctf19 antibodies. We thank members of the Tsukiyama and Biggins laboratories for helpful discussions and advice and T. Tsukiyama and M. Basrai for critical reading of the manuscript. Research was funded by a National Institutes of Health grant Regulation of Centromeric Chromatin (R01 GM-078069) to S.B., an American Cancer Society fellowship to G.M.R.D., a National Institutes of Health Chromosome Metabolism and Cancer Training grant (T32CA009657), and a National Science Foundation Graduate Research Fellowship Program to E.M.H. S.B. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Communicating editor: J. Surtees
Supplemental material is available online at http://www.genetics.org/cgi/content/full/genetics.116.194027/DC1.
Literature Cited
- Akiyoshi B., Sarangapani K. K., Powers A. F., Nelson C. R., Reichow S. L., et al. , 2010. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature 468: 576–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato A., Schillaci T., Lentini L., Di Leonardo A., 2009. CENP-A overexpression promotes genome instability in pRb-depleted human cells. Mol. Cancer 8: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S., Severin F. F., Bhalla N., Sassoon I., Hyman A. A., et al. , 1999. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 13: 532–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black B. E., Cleveland D. W., 2011. Epigenetic centromere propagation and the nature of CENP-A nucleosomes. Cell 144: 471–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K. A., Furuyama S., Biggins S., 2004. Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr. Biol. 14: 1968–1972. [DOI] [PubMed] [Google Scholar]
- da Rosa J. L., Holik J., Green E. M., Rando O. J., Kaufman P. D., 2011. Overlapping regulation of CenH3 localization and histone H3 turnover by CAF-1 and HIR proteins in Saccharomyces cerevisiae. Genetics 187: 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J., Hyland E. M., Yuan D. S., Huang H., Bader J. S., et al. , 2008. Probing nucleosome function: a highly versatile library of synthetic histone H3 and H4 mutants. Cell 134: 1066–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechassa M. L., Wyns K., Luger K., 2014. Scm3 deposits a (Cse4–H4)2 tetramer onto DNA through a Cse4–H4 dimer intermediate. Nucleic Acids Res. 42: 5532–5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyter G. M. R., Biggins S., 2014. The FACT complex interacts with the E3 ubiquitin ligase Psh1 to prevent ectopic localization of CENP-A. Genes Dev. 28: 1815–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duina A. A., Rufiange A., Bracey J., Hall J., Nourani A., et al. , 2007. Evidence that the localization of the elongation factor Spt16 across transcribed genes is dependent upon histone H3 integrity in Saccharomyces cerevisiae. Genetics 177: 101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson P. R., Ganguli D., Nagarajavel V., Clark D. J., 2012. Regulation of histone gene expression in budding yeast. Genetics 191: 7–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R. D., Sugino A., 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74: 527–534. [DOI] [PubMed] [Google Scholar]
- Gross S., Catez F., Masumoto H., Lomonte P., 2012. Centromere architecture breakdown induced by the viral E3 ubiquitin ligase ICP0 protein of herpes simplex virus type 1. PLoS One 7: e44227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainer S. J., Martens J. A., 2011. Identification of histone mutants that are defective for transcription-coupled nucleosome occupancy. Mol. Cell. Biol. 31: 3557–3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick K., Murray A. W., 1995. Mad1p, a phosphoprotein component of the spindle assembly checkpoint in budding yeast. J. Cell Biol. 131: 709–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heun P., Erhardt S., Blower M. D., Weiss S., Skora A. D., et al. , 2006. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev. Cell 10: 303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewawasam G., Shivaraju M., Mattingly M., Venkatesh S., Martin-Brown S., et al. , 2010. Psh1 is an E3 ubiquitin ligase that targets the centromeric histone variant Cse4. Mol. Cell 40: 444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand E. M., Biggins S., 2016. Regulation of budding yeast CENP-A levels prevents misincorporation at promoter nucleosomes and transcriptional defects. PLoS Genet. 12: e1005930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung I., Seo J., Lee H. S., Stanton L. W., Kim D., et al. , 2015. Global mapping of the regulatory interactions of histone residues. FEBS Lett. 589: 4061–4070. [DOI] [PubMed] [Google Scholar]
- Kawashima S., Nakabayashi Y., Matsubara K., Sano N., Enomoto T., et al. , 2011. Global analysis of core histones reveals nucleosomal surfaces required for chromosome bi-orientation. EMBO J. 30: 3353–3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupiec M., 2014. Biology of telomeres: lessons from budding yeast. FEMS Microbiol. Rev. 38: 144–171. [DOI] [PubMed] [Google Scholar]
- Kurat C. F., Recht J., Radovani E., Durbic T., Andrews B., et al. , 2014. Regulation of histone gene transcription in yeast. Cell. Mol. Life Sci. 71: 599–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Fang Y., 2015. Histone variants: the artists of eukaryotic chromatin. Sci. China Life Sci. 58: 232–239. [DOI] [PubMed] [Google Scholar]
- Liang D., Burkhart S. L., Singh R. K., Kabbaj M. H., Gunjan A., 2012. Histone dosage regulates DNA damage sensitivity in a checkpoint-independent manner by the homologous recombination pathway. Nucleic Acids Res. 40: 9604–9620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomonte P., Sullivan K. F., Everett R. D., 2001. Degradation of nucleosome-associated centromeric histone H3-like protein CENP-A induced by herpes simplex virus type 1 protein ICP0. J. Biol. Chem. 276: 5829–5835. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., III, Demarini D. J., Shah N. G., Wach A., et al. , 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Luger K., Mader A. W., Richmond R. K., Sargent D. F., Richmond T. J., 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389: 251–260. [DOI] [PubMed] [Google Scholar]
- Minshull J., Straight A., Rudner A., Dernburg A., Belmont A., et al. , 1996. Protein phosphatase 2A regulates MPF activity and sister chromatid cohesion in budding yeast. Curr. Biol. 6: 1609–1620. [DOI] [PubMed] [Google Scholar]
- Moreno-Moreno O., Torras-Llort M., Azorin F., 2006. Proteolysis restricts localization of CID, the centromere-specific histone H3 variant of Drosophila, to centromeres. Nucleic Acids Res. 34: 6247–6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Moreno O., Medina-Giró S., Torras-Llort M., Azorín F., 2011. The F box protein partner of paired regulates stability of Drosophila centromeric histone H3, CenH3CID. Curr. Biol. 21: 1488–1493. [DOI] [PubMed] [Google Scholar]
- Ng T. M., Waples W. G., Lavoie B. D., Biggins S., 2009. Pericentromeric sister chromatid cohesion promotes kinetochore biorientation. Mol. Biol. Cell 20: 3818–3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng T. M., Lenstra T. L., Duggan N., Jiang S., Ceto S., et al. , 2013. Kinetochore function and chromosome segregation rely on critical residues in histones H3 and H4 in budding yeast. Genetics 195: 795–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H. T., Wharton W., II, Harper J. A., Dornhoffer J. R., Duina A. A., 2013. A nucleosomal region important for ensuring proper interactions between the transcription elongation factor Spt16 and transcribed genes in Saccharomyces cerevisiae. G3 (Bethesda) 3: 929–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkuni K., Abdulle R., Kitagawa K., 2014. Degradation of centromeric histone H3 variant Cse4 requires the Fpr3 peptidyl-prolyl cis-trans isomerase. Genetics 196: 1041–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkuni K., Takahashi Y., Fulp A., Lawrimore J., Au W. C., et al. , 2016. SUMO-Targeted Ubiquitin Ligase (STUbL) Slx5 regulates proteolysis of centromeric histone H3 variant Cse4 and prevents its mislocalization to euchromatin. Mol. Biol. Cell 27: 1500–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer D. K., O’Day K., Wener M. H., Andrews B. S., Margolis R. L., 1987. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J. Cell Biol. 104: 805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando O. J., Winston F., 2012. Chromatin and transcription in yeast. Genetics 190: 351–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjitkar P., Press M. O., Yi X., Baker R., MacCoss M. J., et al. , 2010. An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol. Cell 40: 455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. D., Winston F., Heiter P., 1990. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sekulic N., Bassett E. A., Rogers D. J., Black B. E., 2010. The structure of (CENP-A-H4)2 reveals physical features that mark centromeres. Nature 467: 347–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp J. A., Franco A. A., Osley M. A., Kaufman P. D., 2002. Chromatin assembly factor I and Hir proteins contribute to building functional kinetochores in S. cerevisiae. Genes Dev. 16: 85–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Fink G., Lawrence C., 1974. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Singh R. K., Kabbaj M.-H. M., Paik J., Gunjan A., 2009. Histone levels are regulated by phosphorylation and ubiquitylation-dependent proteolysis. Nat. Cell Biol. 11: 925–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. M., Yang P., Santisteban M. S., Boone P. W., Goldstein A. T., et al. , 1996. A novel histone H4 mutant defective in nuclear division and mitotic chromosome transmission. Mol. Cell. Biol. 16: 1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler S., Keith K. C., Curnick K. E., Fitzgerald-Hayes M., 1995. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 9: 573–586. [DOI] [PubMed] [Google Scholar]
- Straight A. F., Belmont A. S., Robinett C. C., Murray A. W., 1996. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr. Biol. 6: 1599–1608. [DOI] [PubMed] [Google Scholar]
- Tachiwana H., Kagawa W., Shiga T., Osakabe A., Miya Y., et al. , 2011. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature 476: 232–235. [DOI] [PubMed] [Google Scholar]
- Tomonaga T., Matsushita K., Yamaguchi S., Oohashi T., Shimada H., et al. , 2003. Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res. 63: 3511–3516. [PubMed] [Google Scholar]
- Van Hooser A. A., Ouspenski I. I., Gregson H. C., Starr D. A., Yen T. J., et al. , 2001. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J. Cell Sci. 114: 3529–3542. [DOI] [PubMed] [Google Scholar]
- Venkatesh S., Workman J. L., 2015. Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell Biol. 16: 178–189. [DOI] [PubMed] [Google Scholar]
- Verdaasdonk J. S., Bloom K., 2011. Centromeres: unique chromatin structures that drive chromosome segregation. Nature Publishing Group 12: 320–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White C. L., Suto R. K., Luger K., 2001. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 20: 5207–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]