Abstract
Long interspersed elements 1 (L1) are active mobile elements that constitute almost 17% of the human genome. They amplify through a “copy-and-paste” mechanism termed retrotransposition, and de novo insertions related to these elements have been reported to cause 0.2% of genetic diseases. Our previous data demonstrated that the endonuclease complex ERCC1-XPF, which cleaves a 3′ DNA flap structure, limits L1 retrotransposition. Although the ERCC1-XPF endonuclease participates in several different DNA repair pathways, such as single-strand annealing, or in telomere maintenance, its recruitment to DNA lesions is best characterized in the nucleotide excision repair (NER) pathway. To determine if the NER pathway prevents the insertion of retroelements in the genome, we monitored the retrotransposition efficiencies of engineered L1 elements in NER-deficient cells and in their complemented versions. Core proteins of the NER pathway, XPD and XPA, and the lesion binding protein, XPC, are involved in limiting L1 retrotransposition. In addition, sequence analysis of recovered de novo L1 inserts and their genomic locations in NER-deficient cells demonstrated the presence of abnormally large duplications at the site of insertion, suggesting that NER proteins may also play a role in the normal L1 insertion process. Here, we propose new functions for the NER pathway in the maintenance of genome integrity: limitation of insertional mutations caused by retrotransposons and the prevention of potentially mutagenic large genomic duplications at the site of retrotransposon insertion events.
Keywords: L1 retrotransposon, nucleotide excision repair, target-primed reverse transcription, DNA damage, genome stability
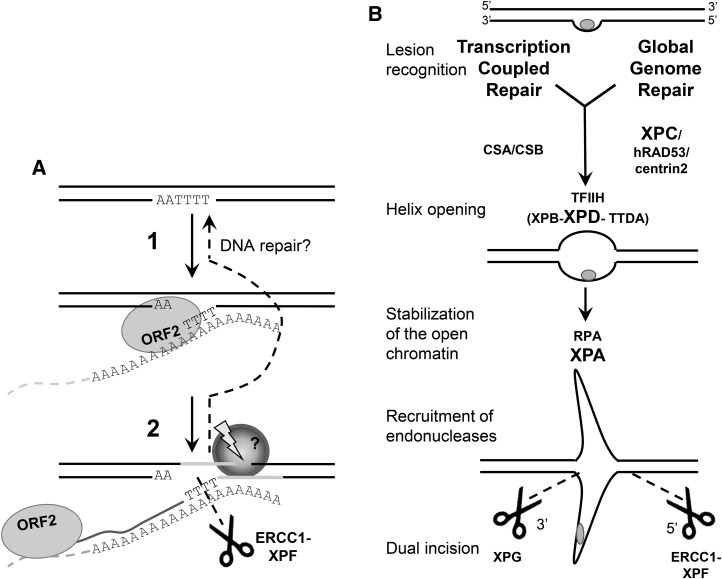
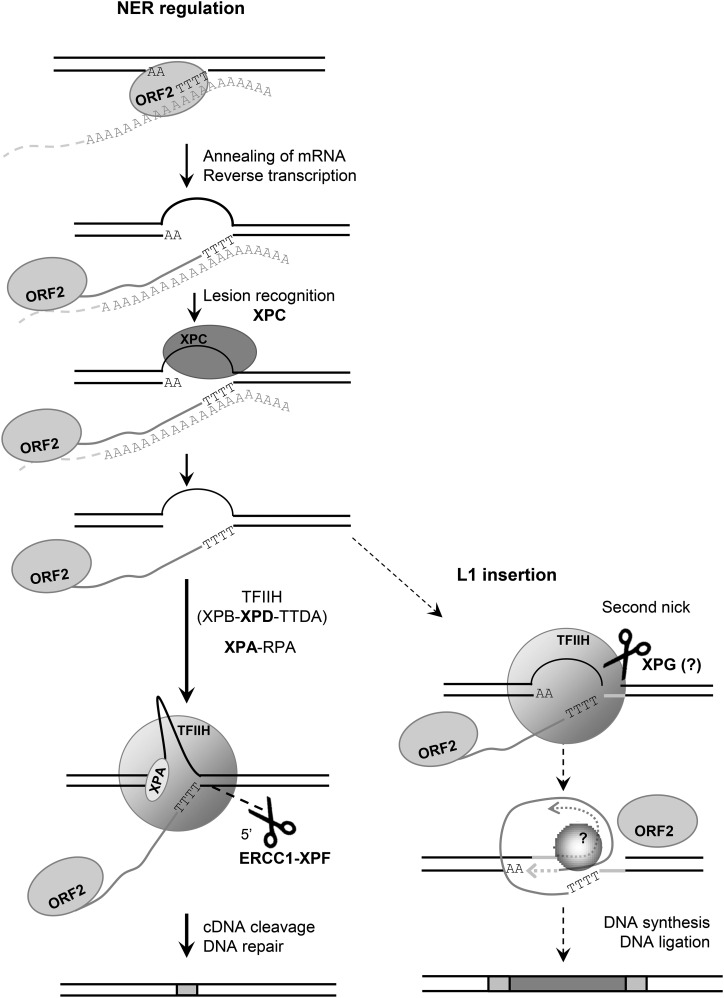
RETROTRANSPOSONS, including Long INterspersed Element 1 (L1), constitute about a third of the human genome (Lander et al. 2001; de Koning et al. 2011). De novo insertions of retrotransposons have been reported as the cause of over 90 genetic diseases, indicating that these elements continue to amplify in the human genome (Ostertag and Kazazian 2001; Xing et al. 2007; Belancio et al. 2008a; Hancks and Kazazian 2012, 2016). Retrotransposons amplify throughout the genome using a “copy-and-paste” mechanism, termed retrotransposition, based on the reverse transcription of an RNA intermediate (Boeke et al. 1985). The L1-encoded proteins, ORF1p and ORF2p, are responsible for the amplification of L1 elements in the genome (Mathias et al. 1991; Feng et al. 1996; Moran et al. 1996). Reverse transcription of L1, a non-LTR (long terminal repeat) retrotransposon, occurs in the nucleus through a proposed process called target-primed reverse transcription (TPRT) (Luan et al. 1993; Feng et al. 1996; Luan and Eickbush 1996), diagrammed in Figure 1A. In the TPRT model, the L1 ORF2p-encoded endonuclease cleaves between the T and the A of a consensus sequence in the DNA (5′-TTTT/AA-3′), freeing a 3′ T-rich DNA end that primes the reverse transcription from the polyA tail of the L1 mRNA. A 3′ flap DNA structure is thought to be generated resulting from the elongation of L1 complementary DNA (cDNA) at the insertion site (Figure 1A) (Feng et al. 1996; Luan and Eickbush 1996; Boeke 1997; Christensen et al. 2006). The factors involved in the completion of the insertion process are unknown but a second nick must be made for second-strand DNA synthesis to occur.
Figure 1.
A similar 3′ flap DNA structure is generated during the L1 insertion process (TPRT) and the NER pathway. (A) Schematic of the first steps of L1-TPRT reaction. (1) ORF2 endonuclease recognizes a consensus sequence 5′-TTTTAA-3′ and cleaves the DNA between the T and A nucleotides resulting in a T-rich free 3′ end. (2) This end is allowed to base-pair with the polyA tail of the L1 mRNA (orange) and prime the reverse transcription of the mRNA. In this model, the proposed structure formed by the elongating cDNA is a 3′ flap intermediate. This 3′ flap intermediate, a known substrate for the structure-specific endonuclease ERCC1-XPF, is proposed to be cleaved, limiting L1 retrotransposition and leading to the restoration of the original DNA sequence. (B) Schematic model of the first steps of the human NER pathway. The NER pathway consists of two subpathways: TCR and GGR. Choice of pathway is determined by DNA lesion recognition. In the TCR subpathway, the base lesion in actively transcribed regions of the genome induces the arrest of transcription elongation by RNAPII. CSA and CSB proteins are recruited to the site of stalled RNAPII and initiate the repair process. In the nontranscribed genomic regions, NER repair occurs through the GGR subpathway. A wide variety of DNA base damage is detected by the DNA lesion-binding protein XPC through the structural distortion of the DNA helix. The next steps of the repair are identical in both subpathways. Once the base lesion is recognized and signaled, the general transcription factor TFIIH, a complex of 10 components including the helicases XPD and XPB, is recruited to the damage site and responsible for DNA unwinding around the lesion. XPA-RPA proteins stabilize the opened chromatin structure and recruit the endonuclease ERCC1-XPF to the damage site. XPG endonuclease is recruited with the complex TFIIH and seems to be required for the DNA unwinding. XPG incises the damaged strand at the 3′ end of the lesion and ERCC1-XPF excises 5′ of the damage. Proteins in bold are the factors evaluated in the present study. cDNA, complementary DNA; GGR, global genome repair; mRNA, messenger RNA; NER, nucleotide excision repair; RNAPII, RNA Polymerase II; TCR, transcription coupled repair; TPRT, target-primed reverse transcription.
We have previously reported that the enzymatic complex ERCC1-XPF, a 3′ flap endonuclease utilized in various DNA repair pathways, limits L1 retrotransposition (Gasior et al. 2008). ERCC1-XPF is a structure-specific endonuclease that nicks double-stranded DNA 5′ of a DNA lesion (de Laat et al. 1998). Therefore, we proposed that ERCC1-XPF cleaves the predicted flap structure formed by the elongating cDNA during L1 insertion (Figure 1A and Gasior et al. 2008). ERCC1-XPF has a global function in cellular DNA damage repair, notably in the removal of the DNA flap structures during single-strand annealing (SSA) repair of DSBs (Sargent et al. 2000; Ahmad et al. 2008; Al-Minawi et al. 2008). ERCC1-XPF is also a component of the telomeric TRF2 complex, involved in the protection of telomeres (Zhu et al. 2003). However, ERCC1-XPF function is best characterized in the cleavage of the damaged DNA strand in the nucleotide excision repair (NER) pathway, an important DNA repair system that removes a wide variety of lesions, including ultraviolet (UV) light-induced cyclobutane pyrimidine dimers (CPDs) and pyrimidine-(6,4)-pyrimidone photoproducts (6–4 PPs), as well as bulky chemical DNA adducts and intrastrand crosslinks (Hoeijmakers 2001; Reardon and Sancar 2005; Sugasawa et al. 2009). Therefore, different pathways could be involved in the recruitment of the ERCC1-XPF complex to limit L1 retrotransposition. Although a DNA flap structure has not been identified as a lesion recognized by the NER pathway, the NER lesion binding protein XPCp can recognize a flap structure in vitro (Sugasawa et al. 2001, 2002). In the present study, we thus investigated the role of the NER pathway in the regulation of L1 retrotransposition.
Rather than recognizing specific base modifications, NER senses structural DNA distortion and nonhydrogen-bonded bases, caused by DNA lesions (Sugasawa et al. 2009; Clement et al. 2010). In humans, defects in NER are associated with several autosomal recessive disorders, such as xeroderma pigmentosum (XP) and Cockayne syndrome (Lehmann et al. 2011; Laugel 2013). Patients suffering from these diseases exhibit an extreme sensitivity to sun exposure, neurological disorders, and a predisposition to cancers. Complementation studies have revealed nine factors (XPA through XPG, CSA, and CSB) involved in NER activity (for review, see Cleaver et al. 2009; Nouspikel 2009). NER consists of two subpathways that have two distinct mechanisms of lesion recognition but share a common central repair pathway: global genome repair (GGR) and transcription-coupled repair (TCR) (Figure 1B) (Vermeulen and Fousteri 2013; Puumalainen et al. 2016). TCR only repairs DNA lesions in the transcribed strand of active genes (Mellon et al. 1987). In TCR, the CSA and CSB proteins are recruited to the site of a stalled RNA Polymerase II (RNAPII) to initiate the repair (Troelstra et al. 1992; Henning et al. 1995; Groisman et al. 2003; van den Boom et al. 2004). GGR is the most active branch that detects base lesions in the remainder of the genome through the XPC complex, consisting of the XPC protein, as well as hRAD53 and centrin2 proteins (Sugasawa et al. 1998; Riedl et al. 2003). After the recognition step, both pathways converge to use a common set of proteins involved in the removal of the damaged strand (Schärer 2011). Two structure-specific endonucleases, the ERCC1-XPF complex and XPG, excise the damaged strand on the 5′ and 3′ sides of the lesion, respectively, followed by DNA replication and ligation to fill the gap (O’Donovan et al. 1994; Sijbers et al. 1996).
Our data show that multiple proteins of the GGR pathway are required to limit the mobility of the non-LTR retrotransposon L1. Additionally, new L1 inserts, recovered in NER-deficient cells, show abnormally large duplications, suggesting a potential role of NER proteins in the L1 insertion process. Therefore, we hypothesize that the GGR pathway can recognize the elongating L1 cDNA in a TPRT model of retrotransposition and excise it, inhibiting the damage caused by de novo L1 inserts.
Materials and Methods
Cell lines and culture conditions
HeLa cells (American Type Culture Collection, Rockville, MD: CCL2) were grown in Eagle’s Minimal Essential Medium (EMEM) supplemented with 10% Fetal Bovine Serum, 0.1 mM nonessential amino acids (Life Technologies), and 1 mM sodium pyruvate (Life Technologies) at 37° in a 5% carbon dioxide environment. The following cell lines were obtained from the Coriell Cells Repository: XPC-SV40-transformed fibroblast (GM15983), XPD-SV40-transformed fibroblast (GM08207), the stably complemented version of XPD-cell line (XPD+) (GM15877) and the isogenic pair of XPA-SV40-transformed fibroblasts (GM04312), and the stably complemented version (XPA+) (GM15876). XPC-, XPD-, and XPA- cell lines were grown in EMEM supplemented with 10% Fetal Bovine Serum and 0.1 mM nonessential amino acids (Life Technologies) at 37° in a 5% carbon dioxide environment. The XPD+ cell line was grown in the same conditions with the presence of 500 μg/ml G418 (Life Technologies). The XPA+ cell line was grown in Dulbecco’s Modified Eagle Medium supplemented with 10% Fetal Bovine Serum (Life Technologies).
Plasmids
TAM102/L1.3 contains the cytomegalovirus (CMV) promoter upstream of the L1.3 element, deleted for L1 5′UTR, and the mblastI indicator cassette cloned in pCEP4 (Morrish et al. 2002).
TAMD702A/L1.3 derives from TAM102/L1.3 and contains the reverse transcriptase-deficient mutant of the L1.3 element and the mblastI indicator cassette cloned in pCEP4 (Morrish et al. 2002).
TAMH230A/L1.3 derives from TAM102/L1.3 and contains the endonuclease-deficient mutant of the L1.3 element and the mblastI indicator cassette cloned in pCEP4 (Morrish et al. 2002).
The synL1_neo vector used for the recovery of de novo L1 inserts was previously described (Gasior et al. 2007).
The pBS-L1PA1CH_blast rescue vector is a pBS-L1PA1CH plasmid (Wagstaff et al. 2011) tagged with a Blast rescue cassette for the recovery of de novo L1 inserts. (Addgene 69611)
pCMV6-XPC (ORIGENE) consists of XPC cDNA driven by the CMV promoter.
pCMV6-XPCΔ, control vector, was constructed by deleting 342 bp, between BglII (nt 753) and BamHI (nt 1095) sites, in the XPC cDNA to create a defective deletion product.
pIRES2-EGFP (Clontech) contains a neomycin resistance gene expressed from a SV40 promoter that allows for the selection of transfected cells, which is used as a toxicity control. The vector also contains a multi-cloning site downstream of a CMV promoter and upstream of an IRES and eGFP marker.
pCMV-Bsd (Life Technologies) contains a blasticidin resistance gene.
Both plasmids above were used in parallel in the retrotransposition assays as a combined control for transfection and growth variations between the cell lines used.
All plasmid DNA was purified by alkaline lysis and twice purified by cesium chloride buoyant density centrifugation. DNA quality was also evaluated by the visual assessment of ethidium bromide-stained agarose gel electrophoresed aliquots.
Retrotransposition assays
For cells with lower transfection efficiencies such as XPA-, XPA+, XPD-, XPD+, and XPC- fibroblasts, L1 retrotransposition assays were performed as described in Moran et al. (1996) using the L1 episomal vectors. Briefly, 5 × 106 cells were seeded in T75 flasks. Cells were transfected the next day at about 90% confluence using Lipofectamine 2000 (Life Technologies) following the manufacturer’s protocol. For L1 retrotransposition assays, cells were transfected with 1 or 3 μg of L1.3 or L1.3-RT (-) construct tagged with the mblast retrotransposition cassette (TAM102/L1.3 or TAMD702A/L1.3) in T75 flasks. Next, 3 µg of XPC expression vector were used to transiently complement XPC-deficient cells. Cells were selected for the presence of the L1 plasmid in selective medium containing 200 µg/ml hygromycin (Life Technologies) for 5 days. The selection was removed and cells were grown for 7 days in growth medium. After the growing period, hygroR cells were trypsinized and reseeded in six-well plates in triplicate at serial dilutions, from 106 to 103 cells, and selected for transposition events in medium containing 10 µg/ml blasticidin (InvivoGen). After 15 days, cells were fixed and stained with crystal violet solution (0.2% crystal violet in 5% acetic acid and 2.5% isopropanol). The number of blastR or neoR colonies was counted in each well and the relative number of colonies per 106 transfected cells was determined.
L1 toxicity and colony formation assay
L1 toxicity and colony formation assays were performed using the L1 episomal vectors. Briefly, 5 × 106 cells were seeded in T75 flasks. Cells were transfected the next day at about 90% confluence using Lipofectamine 2000 (Life Technologies) following the manufacturer’s protocol. Cells were transfected with 3 μg of L1.3, L1.3-RT (-), or L1.3-EN (-) construct tagged with the mblast retrotransposition cassette (TAM102/L1.3, TAMD702A/L1.3, or TAMH230A/L1.3). To transiently complement XPC-deficient cells, 3 µg of XPC expression vector were used. Cells were selected for the presence of the L1 plasmid in selective medium containing 200 µg/ml hygromycin (Life Technologies) for 14 days. The cells were then fixed and stained with crystal violet solution (0.2% crystal violet in 5% acetic acid and 2.5% isopropanol). The number of hygroR colonies were counted in each flask.
Quantitative RT-PCR (RT-qPCR)
First, 5 × 106 cells were seeded in T75 flasks. Cells were transfected the next day at about 90% confluence using Lipofectamine 2000 (Life Technologies) following the manufacturer’s protocol. Cells were transfected with 3 μg of L1.3 construct tagged with the mblast retrotransposition cassette (TAM102/L1.3). To allow for the expression of the vector, total RNA was extracted the next day from each flask using TRIzol Reagent (Life Technologies). We then carried out chloroform extraction and isopropanol precipitation. RNA was suspended in 80 µl DEPC-treated water. The cDNA was synthesized using the Reverse Transcription System (Promega, Madison, WI), following the manufacturer’s protocol. Briefly, 1 µg total RNA was denatured at 75° for 5 min. Reverse transcription reaction was primed with Oligo(dT)15 primers and incubated at 42° for 1 hr in a thermocycler (Bio-Rad, Hercules, CA: C1000 Touch). The enzyme was then heat-inactivated at 99° for 5 min. To avoid contamination with L1 DNA from the genome or the expression vector, and to make sure that only the L1 transcript was quantified, we analyzed the spliced mblast reporter cassette in L1 RNA using the TAQMAN qPCR approach. The PCR reaction (SsoFast Probes Supermix, Bio-Rad) was performed in a qPCR thermocycler (Bio-Rad, CFX96) using 1 µl of cDNA and the blasticidin primers at 250 mM final concentration each: Primer F (5′-GCAGATCGAGAAGCACCTGT-3′), Primer R (5′-TGGTGTCAATGTATATCATTTTACTGG-3′) and the 5′-FAM-labeled, ZEN internal quencher, Iowa-Black Fluorescent Quencher (IDT) probe at 150 mM final concentration (5′−/56-FAM/AGGTTGCCA/ZEN/GCTGCCGCA/3IABkFQ/-3′) as follows: initial denaturation at 95°, 45 cycles of 95° for 10 sec, 60° for 20 sec. The probe was designed to span the splice junction of the mature RNA so that it would not anneal to the unspliced DNA. The fluorescence was read during the annealing/elongation step (60°; 20 sec). For normalization, predeveloped TaqMan Assay Reagents for Human β-actin (cat# 4326315E, Life Technologies) were used. Reactions were done in triplicate. The values of the Cq for each PCR reaction were determined using Bio-Rad CFX Manager software (Bio-Rad).
Recovery of de novo L1 inserts
De novo L1 insert recovery was performed as previously described with slight modifications, as noted below (Morrish et al. 2002; El-Sawy et al. 2005). Briefly, 3 × 106 XPA-, XPD-, XPC-, or XPD+ cells were transfected with 3 µg synL1_neo rescue vector (Gasior et al. 2007) or pBS-L1PA1CH_blast rescue vector using Lipofectamine 2000 reagent (Life Technologies). Cells were selected with 200 µg/ml hygromycin for 5 days, and then put under 400 µg/ml of G418 or 10 µg/ml of Blasticidin for 14 days to allow for colony formation. NeoR and blastR cells were harvested by trypsinization and genomic DNA was extracted using a QIAGEN DNeasy Blood and Tissue kit (QIAGEN, Valencia, CA). Genomic DNA was digested with 100 U HindIII (NEB) overnight at 37°. The following day, digested genomic DNA was self-ligated using 1200 U T4 DNA ligase (NEB) in a volume of 1 ml overnight at room temperature. DNA was purified and concentrated using centrifugal filters (Amicon Ultra, 0.5 ml, 50K, Millipore). Purified DNA was transformed by electroporation into competent DH5α Escherichia coli (Life Technologies). Individual kanamycin- or blasticidin-resistant colonies were grown and plasmid DNA was harvested using a SV Wizard miniprep kit (Promega). The 5′ end of the de novo L1 insert was sequenced using primers specific to the L1 rescue plasmid and primer walking until the 5′ end of the insert was recovered as described in Morrish et al. (2002). Because sequencing through a long adenosine tract at the 3′ end of the L1 inserts is not effective, the 3′ flanking genomic region was sequenced by ligation-mediated PCR based on Yuanxin et al. (2003) and Streva et al. (2015). Briefly, a pool of five to six L1 rescue vectors was digested with StuI (NEB) to relax supercoils, and then sheared by sonication using a Bioruptor (Diagenode, high, 30 sec on, 90 sec off, for 12 min). Sheared plasmid DNA was primer extended using an oligo specific to the 3′ end of the L1 rescue plasmid (3′_rescue_1: 5′-ATATATGAGTAACCTGAGGC-3′ or 3′_rescue_1_secondpA: 5′-GTGGGCATTCTGTCTTGTTC-3′). Duplexed T-linkers were ligated using 10 U T4 DNA ligase and PCR was performed using the primers: linker-specific (5′-ACACTCTTTCCCTACACGACGCTCTTCCGATCT-3′) and 3′_rescue_1 (or 3′_rescue_1_secondpA) primers as follows: initial denaturation at 94°, 20 cycles of 94° for 30 sec, 60° for 1 min, 72° for 1 min, and a final extension for 10 min at 72°. PCR reactions were run on a 1% agarose gel and a light smear between 400 and 700 nt was gel extracted with the Qiaquick gel extraction kit (QIAGEN). Gel-extracted DNA (1 μl) was subject to an additional 15 cycles of PCR amplification as described above using linker-specific and nested 3′ rescue vector primers (3′_rescue_2: 5′-TGAGTAACCTGAGGCTATGCTG-3′ or 3′_rescue_2_secondpA: 5′-TTCTGTCTTGTTCCGGTTCTTAAT-3′) primers. The nested PCR product was run on a 1% agarose gel and the resulting smear was gel extracted and cloned into TOPO-TA (Life Technologies). Cloned PCR products were Sanger sequenced using M13 forward and reverse primers to determine 3′ end junctions. Samples were sent for sequencing to Elim Biopharmaceuticals (Hayward, CA). Lasergene 10 SeqBuilder software was utilized for sequence analysis. Flanking regions were mapped on human reference genome hg19 (build 37) using the Blat tool (https://genome.ucsc.edu/cgi-bin/hgBlat). We verified the presence of the 5′ and 3′ flanking sequences within a L1 rescue plasmid with a combination of PCR between the two regions (Supplemental Material, Table S3 for primers and Figure S8B for results). Sequences from rescues are included in File S1.
Immunoblot analysis
To evaluate expression of NER proteins in the cells, XPA-, XPA+, XPC-, XPD-, and XPD+ cells were haverested in 300 µl of lysis buffer (50 mM Tris, pH 7.2; 150 mM NaCl; 0.5% Triton X-100; 10 mM EDTA, and 0.5% SDS). After 10 min of sonication (Bioruptor, Diagenode, manufacturer’s recommended settings), lysates were clarified by centrifugation for 15 min at 4° at 13,000 rpm and the protein concentration was determined by Bradford assay (Bio-Rad). 40 µg of proteins were loaded on 4–12% bis-tris polyacrylamide or 3–8% tris-acetate polyacrylamide gels (Life Technologies). Proteins were transferred to a nitrocellulose membrane using an iBlot gel transfer system from Life Technologies (manufacturer’s settings). The membrane was blocked for 1 hr at room temperature in PBS (pH 7.4), 0.1% Tween 20 (Sigma [Sigma Chemical], St. Louis, MO), 5% skimmed milk powder (OXOID), and then incubated overnight at 4° with an anti-XPC monoclonal antibody (D-10, sc-74410, Santa Cruz Biotechnology), anti-XPD monoclonal antibody (ab54676, Abcam), or anti-XPA polyclonal antibody (H-273, sc-853, Santa Cruz Biotechnology) diluted at 1:1000 in PBS, 0.1% Tween 20, and 3% nonfat dry milk. Membrane was then incubated for 1 hr at room temperature with the secondary donkey anti-rabbit HRP-conjugated antibody (sc-2317, Santa Cruz Biotechnology) diluted at 1:100,000 in PBS, 0.1% Tween 20, 3% nonfat milk. Signals were detected using Super Signal West Femto Chemiluminescent Substrate (Pierce Chemical, Rockford, IL).
To determine XPC protein expression, 24 hr after transfection with 3 μg of XPC expression vector or control vector, XPC-deficient cells and untransfected HeLa cells were harvested in 300 µl of lysis buffer (50 mM Tris, pH 7.2; 150 mM NaCl; 0.5% Triton X-100; 10 mM EDTA; and 0.5% SDS). After 10 min of sonication (Bioruptor, Diagenode, manufacturer’s recommended settings), lysates were clarified by centrifugation for 15 min at 4° at 13,000 rpm and the protein concentration was determined by Bradford assay (Bio-Rad). 20 µg of proteins for transfected cells and 40 µg of proteins for HeLa cells were loaded on a 3–8% tris-acetate polyacrylamide gel (Life Technologies). Proteins were transferred to a nitrocellulose membrane using an iBlot gel transfer system from Life Technologies (manufacturer’s settings). The membrane was blocked for 1 hr at room temperature in PBS (pH 7.4), 0.1% Tween 20 (Sigma Chemical), 5% skimmed milk powder (OXOID), and then incubated overnight at 4° with an anti-XPC polyclonal antibody (H-300, sc-30156, Santa Cruz Biotechnology) diluted at 1:1000 in PBS, 0.1% Tween 20, and 3% nonfat dry milk. The membrane was then incubated for 1 hr at room temperature with the secondary donkey anti-rabbit HRP-conjugated antibody (sc-2317, Santa Cruz Biotechnology) diluted at 1:100,000 in PBS, 0.1% Tween 20, and 3% nonfat milk. Signals were detected using Super Signal West Femto Chemiluminescent Substrate (Pierce Chemical).
UV sensitivity assay
The protocol was adapted from Emmert et al. (2000). Briefly, 5 × 105 cells were seeded in 6-cm plates and grown in growth medium for 24 hr. The growth medium was removed and the cells were irradiated in the presence of 1 ml of 1 × PBS with a bactericidal UVC lamp (254 nm, 1.57 J/m2/sec) at 0, 3, 6, 9, and 12 J/m2 UVC dose. The PBS was removed and replaced with growth medium. After 4 days, cells were counted with a hemocytometer to determine cell survival. Cell survival was calculated as the percent of live cells in the irradiated sample relative to the untreated sample. To determine the efficiency of the XPC transient complementation, 3 × 106 XPC-deficient cells were transfected with 3 μg of XPC expression vector or the control vector in T75 flasks using Lipofectamine 2000 (Life Technologies) and were reseeded in 6-cm plates the next day in the conditions described above. The UV assay was then performed as mentioned above.
Cell cycle analysis
Cells from confluent T75 plates were harvested and fixed for over 24 hr with ice-cold 70% ethanol at −20°. After fixation, cells were washed once with PBS and incubated in PI/Triton X-100 staining solution (20 μg/ml propidium iodide, 0.1% (v/v) Triton X-100, and 0.2 mg/ml RNAse A in PBS) for at least 2 hr. Flow cytometry of the stained cells was carried out on a Becton Dickinson LSRII using DiVA Software (Louisiana Cancer Research Consortium FACS Core) and 50,000 events were collected. The data were analyzed using Modfit Software (Verity Software House).
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
Two essential proteins of the NER pathway, XPA and XPD, limit L1 retrotransposition
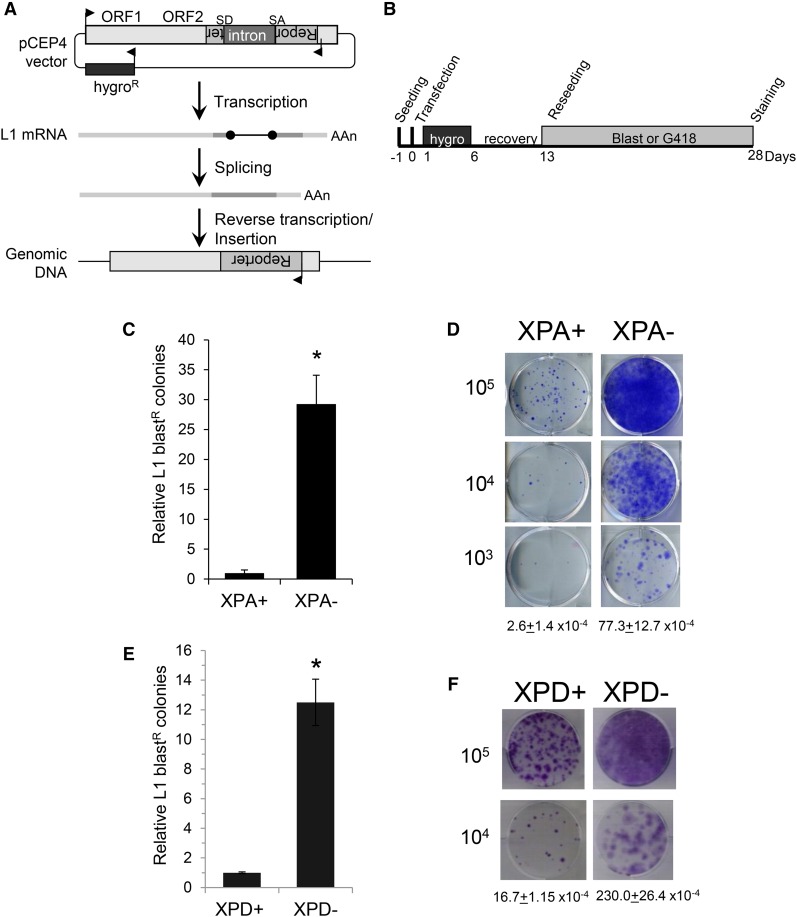
To determine whether the NER pathway controls L1 retrotransposition and is responsible for the role of the ERCC1-XPF complex in preventing L1 mobility, we investigated the effect of the deficiency of XPA and XPD, two crucial factors of the NER pathway, on L1 retrotransposition. XPA recruits ERCC1-XPF to the site of damage by directly binding the complex (Figure 1B) (Volker et al. 2001; Saijo et al. 2004). The helicase activity of the XPD protein forms part of the core structure of the transcriptional/DNA repair complex TFIIH, which opens the chromatin around the lesion during the NER process (Figure 1B and Coin et al. 1998). We performed L1 retrotransposition assays with marked L1 elements (Moran et al. 1996) in XPA- and XPD-deficient cell lines, and their stably complemented partners, XPA+ and XPD+ cells. We transfected the cells with the episomal pCEP4 vector carrying a L1.3 element (TAM102/L1.3) tagged at its 3′ end with the blasticidin (mblast) retrotransposition cassette (Morrish et al. 2002). The principle of the assay (Figures 2, A and B) is based on the introduction of an intron into the reporter gene in a manner that the reporter gene becomes functional after transcription of the L1 element, splicing of L1 mRNA, and target-primed reverse transcription. Thus, the formation of blasticidin-resistant colonies, due to the expression of the reporter gene, indicates that retrotransposition had occurred. The results of the L1 retrotransposition assays in XPA-, XPA+, XPD-, and XPD+ cells showed that stable complementation of NER-deficiency greatly decreased the number of blastR colonies (Figure 2, C–F). These results are not due to differences in L1 expression in the cells, because the quantification of L1 mRNA by RT-qPCR showed equal amounts of RNA in all the cells, whether deficient or proficient for the NER pathway (Figure S1). In addition, immunoblot analysis verified that XPA and XPD protein expression in the complemented XPA- and XPD-deficient cell lines was restored (Figure S2A). A UV sensitivity assay (Levy et al. 1995) validated that the repair of UV-caused damage is not efficient in XPA- and XPD-deficient cells (low cell survival at high UV dose exposure), and becomes efficient again in their complemented versions (Figure S2, B and C). The increase in L1 retrotransposition in NER-deficient cells does not seem to be explained by the growth rate of the different cell lines, because the cell cycle analysis by FACS revealed a slightly longer cycle for NER-deficient cells, as a few more cells accumulated in G2 phase (Figure S3). A longer cell cycle does not seem to be a significant factor of a retrotransposition rate increase in our studies, because previous publications have shown that, although transiting the cell cycle does not seem to be required for retrotransposition (Kubo et al. 2006), any arrest in the cell cycle seemd to lead to a decrease in L1 retrotransposition (Kubo et al. 2006; Shi et al. 2007; Xie et al. 2013).
Figure 2.
XPD and XPA proteins limit L1 retrotransposition. (A) Schematic of the L1-retrotransposition assay. The L1 vector is a pCEP4 episomal vector carrying a hygromycin resistance (hygroR) gene, for the selection of transfected cells, and a full length L1 element tagged at the 3′ end with a retrotransposition cassette. The retrotransposition cassette consists of a reporter gene such as blasticidin resistance (blastR) or a neomycin resistance (neoR) gene interrupted with an intron. The reporter gene is in the reverse orientation in comparison to the L1 element and its transcription is driven by its own promoter, whereas the intron is in the sense orientation (direction of transcription is indicated by arrows). The splice donor and acceptor of the intron are indicated as SD and SA. The resistance gene becomes functional after L1 transcription, splicing, and TPRT of a new L1 copy into the genome. Therefore, only when retrotransposition of the cassette occurs can cells grow under selection conditions (blastR or neoR colonies). (B) Schematic of the protocol of the L1-retrotransposition assay used with the hygroR pCEP4 episomal vectors. The day after transfection, cells containing plasmid are selected with hygromycin for 3 days. Then, selection medium is removed and replaced by growth medium for recovery. Ten days post-transfection, cells were reseeded in six-well plates at serial dilution, from 106 to 103 cells, in triplicate and grown under the appropriate selection. Two weeks later, cells were fixed and stained in crystal violet solution and the number of blastR or neoR colonies in each well is counted. (C) Relative retrotransposition rates of a blastR-tagged L1 reporter element (TAM102/L1.3) were determined in an XPA-deficient cell line (XPA-) and a commercially available stably complemented XPA-deficient cell line (XPA+). The results were normalized relative to XPA+, which was arbitrarily set to 1.0. L1 retrotransposition assay was performed three times independently. Bars represent the average and SEM from the three independent experiments. Statistical significance is indicated by * P-value = 1 × 10−15 (two-tailed, two-sample t-test). (D) Representative example of blastR colony formation resulting from L1 retrotransposition assay performed in XPA+ and XPA- cells. The L1 retrotransposition rate for each cell line is indicated below. (E) Relative number of blastR colonies resulting from L1 retrotransposition assay performed in XPD+ and XPD- cell lines. The results were normalized relative to the XPD+, which was arbitrarily set to 1.0. L1 retrotransposition assay was performed three times independently. Bars represent the average with SEM from the three independent experiments. Statistical significance is indicated by * P-value = 6.5 × 10−6 (two-tailed, two-sample t-test). (F) Representative example of blastR colony formation resulting from L1 retrotransposition assay performed in XPD+ and XPD- cells. The L1 retrotransposition rate for each cell line is indicated below.
As a control, we tested for the possibility that NER-deficiency would promote the retrotransposition of L1 elements defective in the endonuclease (EN) as has been seen in other DNA repair defects (Morrish et al. 2002). No blastR colonies were observed performing retrotransposition assays in NER-deficient cells, using L1 constructs individually defective in either endonuclease or reverse transcriptase [TAMH230A102/L1.3, EN(-) or TAMD702A/L1.3, RT(-)] (Figure S4). In parallel, to rule out any bias engendered by various transfection efficiency, growth rates, and L1-caused toxicity, we verified that L1 expression did not alter colony formation in a manner that could cause retrotransposition rate differences in any of the cells used in the experiment (Figure S5).
The GGR DNA lesion binding protein, XPC, also contributes to the limitation of L1 retrotransposition
NER can recognize and repair a wide variety of lesions that are not related in their chemical structures, such as bulky chemical DNA adducts and UV-induced lesions (Gillet and Schärer 2006). Some reports suggest that it is the helical distortion that attracts the NER repair proteins (Clement et al. 2010; Naegeli and Sugasawa 2011). Whether the DNA lesion occurs in a transcriptionally active or inactive region in the genome will influence the NER sensor used for the repair. Because L1 insertion occurs throughout the genome (Deininger and Batzer 2002; Morrish et al. 2002) and not just in transcribing genes, we evaluated the GGR DNA lesion-binding protein XPC for its ability to limit L1 retrotransposition. We monitored L1 retrotransposition events in XPC-deficient cells and in transiently complemented (XPC+) versions of the same cells.
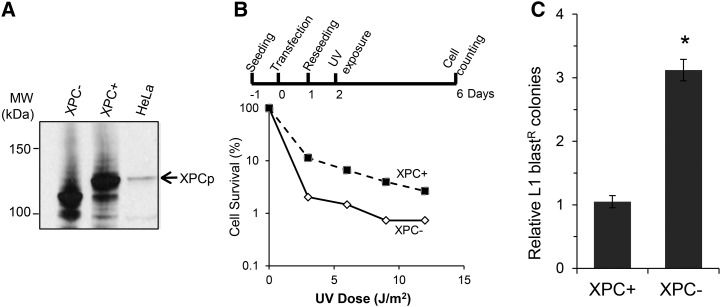
Monitoring XPC protein (XPCp) expression level and repair efficiency of the UV-induced DNA damage, we determined that the transient transfection of XPC-deficient cells resulted in a partial complementation of the deficiency (Figure 3, A and B). In fact, the cell survival rate was under 10% for moderate UV dose (6 J/m2), while the cell survival rates of stably complemented XPA+ and XPD+ cell lines and HeLa cells were about 50% for the same UV dose (Figure S2, B and C). However the cell survival of transiently complemented XPC- cells (XPC+) using a vector expressing XPC cDNA (pCMV6-XPC), which expresses a large amount of XPCp (Figure 3A), was significantly higher than the cell survival of the XPC-deficient cells (XPC-) transfected with the vector control (pCMV6-XPCΔ) (P < 0.0001, Chi-square test for trend) (Figure 3B). Showing the same trend as in XPA- and XPD-deficient cells, L1 retrotransposition increased threefold in the XPC-deficient cells (XPC-) in comparison to the partially complemented cells (XPC+) (Figure 3C). These results indicate that the lesion binding protein in the GGR pathway, XPCp, is likely involved in the regulation of L1 retrotransposition.
Figure 3.
XPC limits L1 retrotransposition. (A) Expression of XPC protein (XPCp) in HeLa cells and in XPC-deficient cells, transfected with either wild-type XPC (pCMV6-XPC) (XPC+) or mutant XPC (pCMV6-XPCΔ) (XPC-) expression vector was analyzed by immunoblotting 24 hr post-transfection. The extra bands observed in XPC+ and XPC- lanes can be the truncated form of XPC protein, present in the XPC-deficient cell line and caused by two deletions in the XPC gene sequence, generating a frameshift and a truncated protein (Li et al. 1993). The truncated protein might be stabilized by the overexpression of the wild-type or mutant XPC protein. (B) Determination of the nucleotide excision repair competency of the transient complementation of the XPC-deficient cells with a wild-type XPC (pCMV6-XPC)- or mutant XPC (pCMV6-XPCΔ)-expressing vector by using a UV sensitivity assay. Top panel shows the schematic of the UV sensitivity assay (details in Materials and Methods). XPC- cells and its transiently complemented version, XPC+ cells, were exposed to 0, 3, 6, 9, or 12 J/m2 UVC dose. Cell survival was determined 4 days post UV exposure. Data represent the logarithm of the percentage of cell survival plotted against the UV dose. The assay was performed at least three times independently for each condition, a representative is shown. (C) Relative number of blastR colonies resulting from the L1 retrotransposition assay performed in the XPC-deficient cell line and its transiently complemented version. XPC- cells were cotransfected with XPC cDNA (XPC+) or control (XPC-) vectors and the tagged L1 vector (TAM102/L1.3). The L1 retrotransposition assay was performed as described for Figure 2. The results were normalized to the XPC+ result, which was arbitrarily set to 1.0. Data correspond to the relative number of blastR colonies determined in each condition. L1 retrotransposition assays have been performed independently at least three times. Data represent the average with SEM. Statistical significance is indicated by *P-value: 0.0001 (two-tailed, two-sample t-test). MW, molecular weight.
The deficiency in NER proteins does not have an impact on the size of de novo L1 inserts, but de novo L1 inserts present abnormally large target site duplications in NER-deficient cell lines
Because complementation of NER-deficiency leads to a decrease in the apparent retrotransposition rate, suggesting that NER proteins inhibit retrotransposition events, we wished to investigate whether mutations in NER proteins impact the characteristics of de novo L1 inserts. We extracted the genomic DNA of XPC-, XPA-, and XPD-deficient cells and in the stably complemented partner, XPD+, expressing the marker of retrotransposition. We then sequenced de novo inserts and the flanking regions to evaluate the features of the inserts, such as the length of the insert, the length of the polyA tail, the size of the target site duplication (TSD), and the sequence of the L1 endonuclease cleavage site. The principle of the method is presented in Figure S6A. The previously published method (Morrish et al. 2002; Yuanxin et al. 2003; El-Sawy et al. 2005; Gasior et al. 2007) was slightly modified to facilitate analysis of the 3′ flanking sequences of the L1 elements recovered as plasmids (details in Materials and Methods). Because the stably complemented cell lines were resistant to neomycin, we could not use the previously described synL1_neo vector (El-Sawy et al. 2005). As a result, we designed a new rescue vector, pBS-L1PA1CH_blast, using a blasticidin retrotransposition cassette (Figure S6B). Except for the insert size that depends on the length of the reporter cassette, the characteristics of de novo L1 inserts obtained with the new L1PA1CH_blast rescue vector in HeLa cells were similar to those previously published in (Gilbert et al. 2002, 2005; Symer et al. 2002) (Table S1 and Table S2).
We recovered 29 de novo L1 inserts in XPD-deficient cells (XPD-), 9 in XPA-deficient cells (XPA-), and 26 in XPC-deficient cells (XPC-) using the synL1_neo vector and 17 in stably complemented XPD-deficient cells (XPD+) using the L1PA1CH_blast rescue vector (Table 1, Table 2, Table 3, and Table 4). We particularly focused on recovering L1 inserts in the XPD-deficient cell lines to allow a statistically significant analysis. The recovered inserts were dispersed randomly throughout the genome with no noticeable preference for a specific site or orientation on chromosomes. No full-length de novo L1 insertions were recovered in NER-deficient cells or in the stably complemented cell line XPD+. The inserts were all 5′ truncated and the size of the inserts varied from 1.8 to 2.8 kb in the complemented cells and from 2.3 to 4.3 kb in NER-deficient cells (the minimal recoverable size of the insert is 1.8 kb using L1PA1CH_blast rescue vector for the assay or 2.3 kb using synL1_neo vector as the presence of the selection marker, blasticidin, or neomycin, respectively, and the bacterial origin of replication at the 3′ end of the L1 construct are required for recovery). In comparison with data obtained in HeLa cells, we detected no significant difference in the median length of L1 inserts in NER-deficient and proficient cells lines (P-value Mann–Whitney U-test > 0.05, Figure S7, A and B). Therefore, NER proteins do not seem to influence the size of the de novo L1 inserts.
Table 1. Characteristics of de novo L1 inserts recovered in XPD-deficient cells.
| Clone # | Insert Size | Chromosome | Orientation | Position (5′–3′ End) | TSD Length | TSD Verification | pA Length | Cleavage Site |
|---|---|---|---|---|---|---|---|---|
| DM.1 | 2663 | Chr5 | + | 137,495,110–137,495,108 | 2 | S | 41 nt | TCTT/a |
| DM.2 | 2883 | Chr18 | — | 57,193,322–57,193,327 | 5 | S | 55 nt | TTTA/a |
| DM.3 | 3147 | Chr12 | + | 112,775,935–112,775,928 | 7 | S | 45 nt | CTTT/a |
| DM.4 | 2780 | Chr7 | — | 61,968,906–61,968,919 | 13 | S | 81 nt | TTTC/a |
| DM.5 | 2827 | Chr12 | — | 98,026,422–98,026,438 | 16 | S | 30 nt | TTTT/a |
| DM.6 | 3929 | Chr8 | + | 128,728,905–128,728,851 | 54 | S | 58 nt | TCTT/a |
| DM.7 | 3989 | Chr17 | — | 13,596,243–13,596,460 | 217 | S | 83 nt | TTTT/a |
| DM.8 | 2804 | Chr17 | — | 50,628,899–50,629,306 | 407 | S/P | — | CTAG/a |
| DM.9 | 3626 | Chr1 | — | 121,484,567–121,484,978 | 411 | S | 73 nt | TTCT/g |
| DM.10 | 3686 | Chr3 | — | 174,149,054–174,149,550 | 496 | S/P | 84 nt | TCTT/a |
| DM.11 | 2550 | Chr7 | + | 144,760,424–144,759,911 | 513 | S/P | 34 nt | TTTT/g |
| DM.12 | 3008 | Chr3 | + | 29,453,614–29,453,069 | 545 | S/P | 25 nt | TTTT/a |
| DM.13 | 3167 | Chr11 | + | 105,190,706–105,190,152 | 554 | S/P | 46 nt | ACTT/g |
| DM.14 | 2559 | ChrX | — | 10,708,604–10,709,170 | 571 | S/P | 77 nt | TTCT/a |
| DM.15 | 3719 | Chr15 | + | 30,302,496–30,301,796 | 700 | S/P | — | CCCA/g |
| DM.16 | 3103 | Chr8 | — | 58,341,572–58,342,341 | 769 | S/P | 97 nt | TTTC/a |
| DM.17 | 3411 | Chr14 | — | 77,523,239–77,524,010 | 771 | S/P | 69 nt | TTTT/a |
| DM.18 | 3610 | Chr9 | + | 5,534,460–5,533,597 | 863 | 37 nt | TTTT/a | |
| DM.19 | 2928 | ChrX | + | 33,775,626–33,774,740 | 886 | S/P | 33 nt | TTTA/g |
| DM.20 | 2582 | Chr7 | — | 130,375,857–130,376,914 | 1057 | S/P | — | TGGT/c |
| DM.21 | 2380 | Chr9 | + | 40,823,585–40,822,515 | 1070 | S/P | 56 nt | CTTT/a |
| DM.22 | 2894 | ChrX | + | 122,778,988–122,777,801 | 1187 | 39 nt | TTTC/a | |
| DM.23 | 2536 | Chr5 | + | 64,034,619–64,033,086 | 1533 | P | 19 nt | TTTT/c |
| DM.24 | 3917 | Chr11 | + | 14,727,287–14,725,642 | 1645 | 57 nt | TTTC/a | |
| DM.25 | 3857 | Chr1 | — | 120,251,234–120,249,499 | 1735 | S/P | 65 nt | GTTT/g |
| DM.26 | 3991 | Chr3 | + | 29,455,524–29,453,069 | 2455 | P | 22 nt | TTTT/a |
| DM.27 | 3940 | Chr15 | — | 28,869,801–28,873,325 | 3361 | P | 87 nt | TATT/a |
| DM.28 | 3996 | Chr13 | — | 44,857,642–44,861,667 | 4025 | P | 57 nt | TTTG/a |
| DM.29 | 2655 | Chr3 | + | 29,458,420–29,453,069 | 5896 | 30 nt | TTTT/a |
TSD, target site duplication; Chr, chromosome; S, sequence; P, PCR.
Table 2. Characteristics of de novo L1 inserts recovered in (XPD+) complemented XPD-deficient cells.
| Clone # | Insert Size | Chromosome | Orientation | Position (5′–3′ End) | TSD Length | Deletion Length | pA Length | Cleavage Site |
|---|---|---|---|---|---|---|---|---|
| DP.1 | 1956 | Chr3 | + | 166,704,010–166,704,008 | 3 | 31 nt | TTTT/a | |
| DP.2 | 1989 | Chr8 | + | 121,836,950–121,836,947 | 4 | 31 nt | TCTT/a | |
| DP.3 | 2318 | Chr20 | + | 52,652,085–52,652,079 | 7 | 38 nt | TTTA/a | |
| DP.4 | 2270 | Chr3 | — | 156,453,738–156,453,742 | 4 | 28 nt | CTTT/a | |
| DP.5 | 2269 | Chr7 | + | 90,201,740–90,201,726 | 15 | 45 nt | TTTC/a | |
| DP.6 | 2038 | Chr16 | — | 53,268,517–53,268,532 | 16 | 33 nt | TTTT/a | |
| DP.7 | 1866 | Chr8 | + | 30,763,655–30,763,645 | 11 | 46 nt | TTAT/a | |
| DP.8 | 2013 | Chr2 | — | 24,215,408–24,216,927 | 1520 | 35 nt | TTTT/a | |
| DP.9 | 1977 | Chr17 | + | 35,458,775–35,457,164 | 1612 | 40 nt | TCTT/a | |
| DP.10 | 2704 | Chr12 | — | 18,901,143–18,901,142 | — | — | 52 nt | TCTT/a |
| DP.11 | 1866 | Chr15 | — | 87,439,350–87,439,347 | 2 | 52 nt | TTTA/a | |
| DP.12 | 2791 | Chr6 | + | 99,948,429–99,948,435 | 5 | 19 nt | TTTT/a | |
| DP.13 | 1900 | Chr14 | — | 51,873,941–51,873,933 | 7 | 25 nt | TTTG/a | |
| DP.14 | 2062 | Chr1 | — | 94,735,344–94,735,334 | 9 | 34 nt | TCTT/a | |
| DP.15 | 2380 | Chr4 | — | 30,539,318–30,539,303 | 14 | 25 nt | TTTT/a | |
| DP.16 | 2002 | Chr2 | — | 59,684,354–59,684,338 | 15 | 110 nt | TTCT/a | |
| DP.17 | 2741 | Chr6 | — | 248,880–248,410 | 469 | 55 nt | TTTT/a |
TSD, target site duplication; Chr, chromosome.
Table 3. Characteristics of de novo L1 inserts recovered in XPC-deficient cells.
| Clone # | Insert Size | Chromosome | Orientation | Position (5′–3′ End) | TSD Length | Deletion Length | pA Length | Cleavage Site |
|---|---|---|---|---|---|---|---|---|
| CM.1 | 2491 | Chr15 | — | 52,326,493–52,326,495 | 2 | 25 nt | TTTT/a | |
| CM.2 | 2736 | Chr4 | + | 58,109,300–58,109,280 | 20 | 29 nt | TTTA/a | |
| CM.3 | 2421 | Chr4 | — | 81,232,879–81,233,154 | 275 | — | ATTT/a | |
| CM.4 | 3167 | Chr5 | + | 41,311,414–41,311,137 | 277 | 60 nt | TCTT/g | |
| CM.5 | 2886 | Chr15 | + | 33,641,486–33,640,983 | 503 | 15 nt | TTTT/a | |
| CM.6 | 2661 | Chr8 | + | 23,490,649–23,489,121 | 516 | 18 nt | TGAA/a | |
| CM.7 | 2946 | Chr2 | + | 182,583,931–182,583,379 | 552 | — | AATA/a | |
| CM.8 | 2886 | Chr6 | — | 85,235,896–85,236,510 | 614 | — | AATT/t | |
| CM.9 | 2567 | Chr5 | — | 65,162,148–65,162,763 | 615 | — | AATG/a | |
| CM.10 | 2609 | Chr16 | — | 9,557,849–9,558,491 | 642 | 10 nt | TTTT/a | |
| CM.11 | 3386 | Chr8 | + | 26,232,474–26,231,790 | 684 | 25 nt | TCTT/a | |
| CM.12 | 2677 | Chr9 | + | 66,811,576–66,810,877 | 699 | 15 nt | TTTT/g | |
| CM.13 | 3293 | Chr17 | + | 49,728,154–49,727,312 | 842 | 6 nt | GATT/g | |
| CM.14 | 3112 | Chr11 | — | 114,392,300–114,393,187 | 887 | 23 nt | TCTT/a | |
| CM.15 | 2894 | Chr3 | + | 34,830,281–34,829,315 | 966 | — | AGTT/c | |
| CM.16 | 2604 | Chr1 | — | 211,947,488–211,948,623 | 1135 | — | CTGC/a | |
| CM.17 | 2684 | Chr8 | + | 127,633,155–127,631,920 | 1235 | 84 nt | TTCT/a | |
| CM.18 | 3044 | Chr3 | + | 5,007,476–5,005,443 | 2033 | — | ATTT/t | |
| CM.19 | 3096 | Chr17 | + | 48,833,262–48,831,219 | 2043 | 21 nt | TTTC/a | |
| CM.20 | 2718 | Chr20 | — | 12,381,555–12,384,581 | 3026 | — | ACCT/g | |
| CM.21 | 3035 | Chr18 | + | 20,321,307–20,317,622 | 3685 | — | TAAG/c | |
| CM.22 | 2890 | Chr2 | — | 137,563,056–137,567,601 | 4545 | 31 nt | TTTT/a | |
| CM.23 | 4006 | Chr19 | — | 27,732,520–27,739,285 | 6765 | 38 nt | TTTT/g | |
| CM.24 | 2700 | Chr18 | — | 22,016,206–22,016,201 | 5 | 21 nt | CTTT/a | |
| CM.25 | 4306 | Chr14 | — | 67,575,620–67,575,614 | 6 | 20 nt | TTAA/g | |
| CM.26 | 4192 | Chr9 | + | 115,010,933–115,010,942 | 9 | 13 nt | TTTT/a |
TSD, target site duplication; Chr, chromosome.
Table 4. Characteristics of de novo L1 inserts recovered in XPA-deficient cells.
| Clone # | Insert Size | Chromosome | Orientation | Position (5′–3′ End) | TSD Length | Deletion Length | pA Length | Cleavage Site |
|---|---|---|---|---|---|---|---|---|
| AM.1 | 2490 | Chr2 | — | 36,487,725–36,487,738 | 13 | 35 nt | TTTC/a | |
| AM.2 | 4230 | Chr3 | — | 47,259,213–47,259,253 | 40 | 79 nt | TTTC/a | |
| AM.3 | 2678 | Chr11 | + | 85,736,502–85,736,455 | 47 | 42 nt | TCTT/a | |
| AM.4 | 3409 | Chr15 | — | 28,728,325–28,728,567 | 242 | 25 nt | TTTC/a | |
| AM.5 | 2265 | Chr9 | — | 135,357,719–135,359,341 | 1622 | 5 nt | TTTT/g | |
| AM.6 | 2712 | Chr2 | + | 89,248,581–89,246,458 | 2123 | — | ACTG/a | |
| AM.7 | 2756 | Chr12 | — | 19,866,410–19,866,397 | 13 | 9 nt | TTTT/a | |
| AM.8 | 2306 | Chr3 | + | 36,006,942–36,010,967 | 4025 | 50 nt | TGTT/a |
TSD, target site duplication; Chr, chromosome.
The majority of de novo L1 inserts occurred at typical L1 endonuclease cleavage sites (Table 1, Table 2, Table 3, and Table 4 and Morrish et al. 2002). However, 1 out of the 9 (11%) recovered inserts (clone #AM7, Table 4) in XPA-deficient cells, 9 out of 26 (35%) in XPC- cells (clones #CM3, #CM7, #CM8, #CM9, #CM15, #CM16, #CM18, #CM20, and #CM21, Table 3), and 3 out of 28 (11%) in XPD- cells (clones #DM8, #DM15, and #DM20, Table 1) occurred at an atypical cleavage site and also did not contain the polyA tail at the 3′ end of the L1 sequence, a hallmark of an L1 insertion event. Inserts similar to these have been previously described as the result of endonuclease-independent insertion events in DNA repair-deficient cells (Morrish et al. 2002). However, in the present study, the frequency of these events was significantly higher in XPC-deficient cells than in the other NER-deficient cells and HeLa cells (Gilbert et al. 2005), and we did not observe any retrotransposition events in NER-deficient cells transfected with a L1 construct defective in the endonuclease activity of ORF2p (Figure S4). Therefore, endonuclease-independent insertion of L1 in NER-deficient cells does not seem to explain these events, and these atypical inserts may be the results of recombination events between L1 and genomic sequences occurring at the insertion site during the insertion process.
Genomic deletions at the L1 insertion site were observed in 3 out of 26 (12%) in XPC-deficient cells (clones #CM24 to #CM26, Table 3) and 2 out of 9 (22%) in XPA-deficient cells (clones #AM8 and #AM9, Table 4). These observations are similar in frequency to small deletions found from an analysis of polymorphic genomic L1 inserts (Konkel et al. 2007), although our deletions were slightly longer in general. A significantly higher rate of deletions was observed in the complemented XPD+ cells (8 out of 17, 47%). For nearly half of these events, deletions were generated by homologous recombination between the sequences of the L1 element in the genome at the insertion site and the elongating de novo L1 (see TPRT process, Figure 1A). This type of event has been previously reported and described in HeLa cells, although not at this high rate (Gilbert et al. 2005).
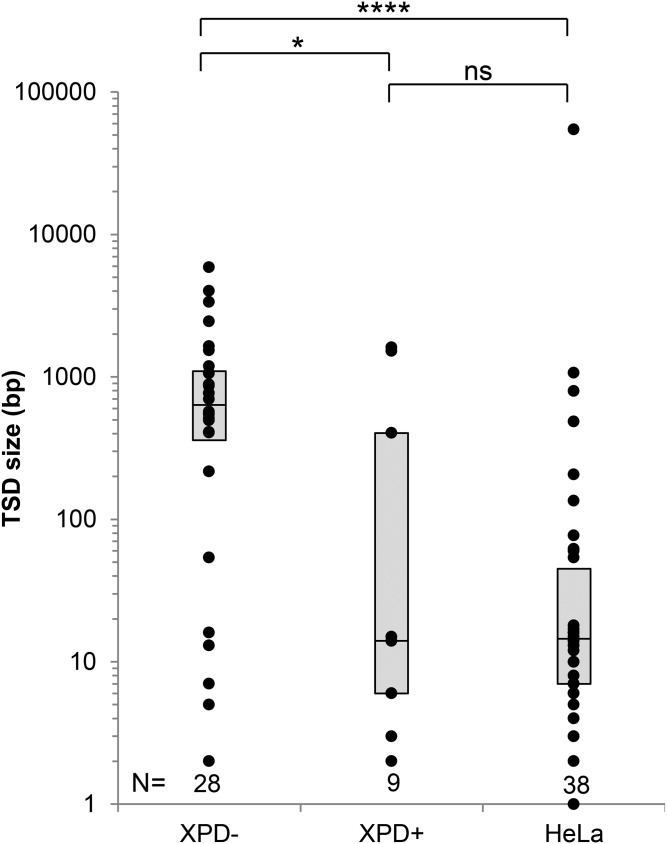
One area in which NER-deficient L1 inserts differed from those in NER-proficient cells is the size of the target size duplication. In NER-deficient cells, TSDs ranged from 5 bp to 5.3 kb in size with a median length of 1495 bp, while in XPD+ cells, TSDs ranged from 2 bp to 1.6 kb in length with a median of 14 bp (Table 1, Table 2, Table 3, and Table 4). TSD length in NER-deficient cells was significantly larger than those observed in either cells stably complemented to correct the NER deficiency or in NER-proficient HeLa cells (P-value Mann–Whitney U-test < 0.05, Figure 4). Previous studies have also reported that the typical size of a TSD from L1 inserts recovered from normal culture cells and from an L1 insert naturally occurring in the genome is usually 5–30 bp (Morrish et al. 2002; Symer et al. 2002; Gilbert et al. 2005). Events generating large TSDs (> 100 bp) have been observed in HeLa cells, but the frequency of these events never exceeded 10% of the total recovered events (Table S1 and Table S2) (Gilbert et al. 2002, 2005; Symer et al. 2002). In the NER-deficient cell lines tested in the present study, 4 out of 7 (57%) de novo inserts on XPA- cells, 21 out of 23 (91.3%) in XPC- cells, and 22 out of 28 (78.6%) in XPD- cells presented TSDs over 100 bp in length (Figure 4 and Figure S8) and of these, 18 (38%) had TSDs of over 1 kb in length. Only three out of nine (33%) events presented TSDs over 100 bp in the stably complemented cells (XPD+), a difference that is statistically significant (P-value Fisher’s exact test < 0.05). It seems possible that the slightly elevated levels of longer TSDs in the complemented cells relative to previous studies in HeLa, for instance, may be due to the complementation not necessarily bringing the NER pathway to maximum efficiency. Therefore, our findings seem to imply that the expression of NER proteins is required to prevent large genomic rearrangements at the L1 insertion site.
Figure 4.
Recovered de novo L1 inserts in NER-deficient cells have large TSDs. Dot plot representation of the size of TSDs of de novo L1 inserts recovered from XPD-deficient cells (XPD-), stably complemented XPD-deficient cells (XPD+) and HeLa cells. The boxes represent the interquartile range (the range between the first and third quartiles) of the distribution of TSD size for each cell line. The line in the middle of the box represents the median. The count (n) of recovered inserts for each cell line is indicated above the name of the cell line. Statistical significance is indicated by *P-value = 0.032, ****P-value = 3.13 × 10−7 (P-value = 0.38) (Mann–Whitney U test). NER, nucleotide excision repair; TSD, target site duplication.
Discussion
L1 elements have a tremendous capacity to cause damage to the human genome, potentially causing disease, cell death, and aging (Belgnaoui et al. 2006; Gasior et al. 2006; Wallace et al. 2010; Belancio et al. 2014). Because L1 retrotransposition is a threat to genome integrity, it is not surprising that numerous cellular factors control the mobility of these elements at different levels. For example, L1 expression is maintained at a low level by methylation (Alves et al. 1996), premature polyadenylation, and alternative RNA splicing (Belancio et al. 2008b). In addition, at a post-transcriptional level, members of the human cytidine deaminase APOBEC3 family, involved in the response against viral infection, inhibit the activity of L1 retrotransposons (Bogerd et al. 2006; Kinomoto et al. 2007; Lovsin and Peterlin 2009; Richardson et al. 2014).
Our data demonstrate that the entire global NER pathway represents an additional mechanism regulating L1 activity. The ERCC1-XPF complex, previously identified as an L1 regulator, is involved in many DNA repair pathways and is more essential for life than the other NER components. All three NER proteins tested in this study (XPAp, XPDp, and XPCp, Figure 2 and Figure 3) show a similar ability to suppress L1 retrotransposition as previously reported for the ERCC1-XPF complex (Gasior et al. 2008). This suppression seems likely to be the result of an altered ability to complete the retrotransposition process successfully, rather than an ability to make longer inserts in NER deficiency, given that we detected no significant differences in insert length (Figure S7).
It seems likely that the NER pathway is primarily responsible for the inhibition of L1 retrotransposition by ERCC1-XPF, as the magnitude of the effect of XPA, XPD, and ERCC1 deficiencies on L1 retrotransposition were very similar. However, this does not rule out the possibility that some of the other sensors and pathways that recruit ERCC1-XPF to DNA lesions may also be involved, to a lesser extent, in the control of L1 insertion. In particular, TCR, the other NER subpathway, may also contribute to the sensing of L1 insertion events. However, because TCR only protects the portion of the genome that is being actively transcribed in that specific cell type (Mellon et al. 1987), the TCR subpathway could only have a more limited effect on L1 insertion rates. Because the TCR pathway is strand-specific, its influence would primarily be limited to influencing the orientation preference of the insertion within genes rather than on the global rate of retrotransposition events.
Previously, the Garfinkel laboratory reported that Ssl2 and Rad3, two DNA helicases of the TFIIH complex and homologs of human XPB and XPD proteins, respectively, can inhibit Ty1 retrotransposition in Saccharomyces cerevisiae (Lee et al. 1998). Ssl2 and Rad3 helicases are proposed to induce the degradation of Ty1 cDNA in the cytoplasm by destabilizing its structure. However, Ty1 and L1 elements have very different integration mechanisms. The former is a virus-like retrotransposon, similar to retroviruses, and its reverse transcription occurs in a pseudoviral particle in the cytoplasm. The cDNA is imported into the nucleus and is integrated in the genome. In contrast, L1 mRNA is reverse transcribed inside the nucleus (TPRT), directly at the site of the insertion of the new copy. Therefore, it is not expected that these two kinds of elements would induce a similar cellular response. Intriguingly, in both cases, it seems likely that the NER components are involved in limiting the insertion by limiting the synthesis/stability of the cDNA intermediate.
In addition to the role of the NER factors in limiting the insertion of a new copy of L1, our L1 insert recovery data suggest that the NER pathway may also be involved in the less well-defined last step of the L1 TPRT process, as a high number of extremely large TSDs was observed in NER-deficient cell lines. These results suggest that the NER factors present at the site of L1 insertion may influence the formation or selection of the second nick necessary for the completion of the TPRT process at a location close to the cleavage site of the L1 endonuclease (Figure 5). Although speculative, in NER-deficient cells, the location of the second nick is highly variable and might depend on a more random event that can occur at a location very far away from the primary cleavage site, thereby generating extremely large TSDs. The distant location of the second nick can be the result of the direct influence of the chromosomal region on the L1 insertion process, such as exposed DNA ends generated during replication or DNA repair processes, serving as primers for the synthesis of the second L1 cDNA strand. An alternative possibility is that the predicted cDNA flap intermediate may persist in ERCC1-XPF-deficient cells, giving the insertion process a longer time to reach a naturally occurring nick at a more distant location through the strand displacement activities of helicases recruited to the site of L1 insertion by DNA repair machinery.
Figure 5.
Model of limitation of damage caused by L1 TPRT via the GGR-NER pathway. The 3′ flap DNA structure is generated by L1 elongating cDNA during TPRT process. The XPC complex recognizes the structure and recruits the other proteins of the NER pathway to the L1 insertion site. ERCC1-XPF endonuclease cleaves the elongating L1 cDNA, inhibiting the insertion of a new L1 element in the genome and leading to the restoration of the original DNA sequence. To a lesser extent, the NER pathway seems to be involved in generating a second nick during the L1 insertion process, in close proximity to the first cleavage generated by the L1 endonuclease. Therefore, in WT cells, the de novo insert is flanked by small (< 100 bp) TSDs, whereas in NER-deficient cells, a more distal unrelated nick may be used to complete the retroelement insertion resulting in large TSDs. cDNA, complementary DNA; GGR, global genome repair; mRNA, messenger RNA; NER, nucleotide excision repair; RNAPII, RNA Polymerase II; TCR, transcription coupled repair; TPRT, target-primed reverse transcription; TSD, target site duplication.
In addition to the long TSDs in these L1 insertions, the only other unusual feature is an unusually high number of events that do not look like they went through the normal TPRT process on the polyA tail. Although most of the inserts have the typically long A tails expected of de novo L1 insertions, these “tailless” inserts seem to indicate some other undefined aberrant processes during the insertion.
The extremely large TSDs found in NER-deficient L1 insertions fall within the size range of genomic copy number variations (Eichler 2001). This large size makes them potentially more deleterious as insertions. However, it also has the consequence of duplicating regions that may have functional significance, such as exons. Thus, they represent a unique source of this type of genomic duplication. However, it is also worth considering that large segments of homology near one another also have a propensity to recombine with one another (Stankiewicz and Lupski 2002). This would be similar to the formation of solo LTRs following retroviral insertion (Belshaw et al. 2007). Thus, this type of event might be expected to have a shortened life span in the genome and some L1 insertions may be eliminated by recombination between their long TSDs. Our data suggest that individuals with defects in NER, such as XP patients, are likely to be subject to higher levels of DNA damage caused by L1 elements. Other studies reported that NER capacity varies between individuals (Spitz et al. 2001; Tomescu et al. 2001; Hou et al. 2002) in response to different stimuli such as circadian rhythm (Gaddameedhi et al. 2011), or are specifically repressed in some cancers (Xu et al. 2011). Under those conditions and in combination with a deregulation of L1 expression, NER deficiency may increase the potential impact of L1 retrotransposition on the human genome, favoring somatic genetic instability and the generation of age-related diseases, such as cancer.
Acknowledgments
This research was supported by the National Institutes of Health (NIH) grants P20 P20GM103518/P20RR020152 (P.L.D., V.P.B., and A.M.R.-E.), R01GM079709A (A.M.R.-E.), and R01GM45668, an Experimental Program to Stimulate Competitive Research/Board of Reagents Support Fund grant (P.L.D.), and NIH/National Institute on Aging grant 5K01AG030074 (V.P.B.). V.P.B. is also supported by The Ellison Medical Foundation New Scholar in Aging award (AG-NS-0447-08) and the Kay Yow foundation. This paper’s contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Center for Research Resources or NIH.
Author contributions: G.S. was involved in the design and, with support from R.S.D. and M.N., performed most of the experimental assays and data collection. M.N. performed the XPA experiments and complementation experiments. V.A.S. performed experiments to process de novo L1 insertion events and performed their analysis with G.S. R.S.D., and V.P.B. performed immunoblot analysis. G.S. contributed to the data analysis and editing of the manuscript. P.L.D. and A.M.R.-E. performed the overall conception and study design, some data collection, analysis, interpretation, and data writing of the paper and made the decision to submit it for publication.
Footnotes
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.188680/-/DC1.
Communicating editor: L. B. Jorde
Literature Cited
- Ahmad A., Robinson A. R., Duensing A., van Drunen E., Beverloo H. B., et al. , 2008. ERCC1-XPF endonuclease facilitates DNA double-strand break repair. Mol. Cell. Biol. 28: 5082–5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Minawi A. Z., Saleh-Gohari N., Helleday T., 2008. The ERCC1/XPF endonuclease is required for efficient single-strand annealing and gene conversion in mammalian cells. Nucleic Acids Res. 36: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves G., Tatro A., Fanning T., 1996. Differential methylation of human LINE-1 retrotransposons in malignant cells. Gene 176: 39–44. [DOI] [PubMed] [Google Scholar]
- Belancio V. P., Hedges D. J., Deininger P., 2008a Mammalian non-LTR retrotransposons: for better or worse, in sickness and in health. Genome Res. 18: 343–358. [DOI] [PubMed] [Google Scholar]
- Belancio V. P., Roy-Engel A. M., Deininger P., 2008b The impact of multiple splice sites in human L1 elements. Gene 411: 38–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belancio V. P., Blask D. E., Deininger P., Hill S. M., Jazwinski S. M., 2014. The aging clock and circadian control of metabolism and genome stability. Front. Genet. 5: 455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgnaoui S. M., Gosden R. G., Semmes O. J., Haoudi A., 2006. Human LINE-1 retrotransposon induces DNA damage and apoptosis in cancer cells. Cancer Cell Int. 6: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belshaw R., Watson J., Katzourakis A., Howe A., Woolven-Allen J., et al. , 2007. Rate of recombinational deletion among human endogenous retroviruses. J. Virol. 81: 9437–9442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., 1997. LINEs and Alus–the polyA connection. Nat. Genet. 16: 6–7. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., Garfinkel D. J., Styles C. A., Fink G. R., 1985. Ty elements transpose through an RNA intermediate. Cell 40: 491–500. [DOI] [PubMed] [Google Scholar]
- Bogerd H. P., Wiegand H. L., Doehle B. P., Lueders K. K., Cullen B. R., 2006. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res. 34: 89–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen S. M., Ye J., Eickbush T. H., 2006. RNA from the 5′ end of the R2 retrotransposon controls R2 protein binding to and cleavage of its DNA target site. Proc. Natl. Acad. Sci. USA 103: 17602–17607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaver J. E., Lam E. T., Revet I., 2009. Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity. Nat. Rev. Genet. 10: 756–768. [DOI] [PubMed] [Google Scholar]
- Clement F. C., Camenisch U., Fei J., Kaczmarek N., Mathieu N., et al. , 2010. Dynamic two-stage mechanism of versatile DNA damage recognition by xeroderma pigmentosum group C protein. Mutat. Res. 685: 21–28. [DOI] [PubMed] [Google Scholar]
- Coin F., Marinoni J. C., Rodolfo C., Fribourg S., Pedrini A. M., et al. , 1998. Mutations in the XPD helicase gene result in XP and TTD phenotypes, preventing interaction between XPD and the p44 subunit of TFIIH. Nat. Genet. 20: 184–188. [DOI] [PubMed] [Google Scholar]
- de Koning A. P., Gu W., Castoe T. A., Batzer M. A., Pollock D. D., 2011. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 7: e1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat W. L., Appeldoorn E., Jaspers N. G., Hoeijmakers J. H., 1998. DNA structural elements required for ERCC1-XPF endonuclease activity. J. Biol. Chem. 273: 7835–7842. [DOI] [PubMed] [Google Scholar]
- Deininger P. L., Batzer M. A., 2002. Mammalian retroelements. Genome Res. 12: 1455–1465. [DOI] [PubMed] [Google Scholar]
- Eichler E. E., 2001. Recent duplication, domain accretion and the dynamic mutation of the human genome. Trends Genet. 17: 661–669. [DOI] [PubMed] [Google Scholar]
- El-Sawy M., Kale S. P., Dugan C., Nguyen T. Q., Belancio V., et al. , 2005. Nickel stimulates L1 retrotransposition by a post-transcriptional mechanism. J. Mol. Biol. 354: 246–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmert S., Kobayashi N., Khan S. G., Kraemer K. H., 2000. The xeroderma pigmentosum group C gene leads to selective repair of cyclobutane pyrimidine dimers rather than 6–4 photoproducts. Proc. Natl. Acad. Sci. USA 97: 2151–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q., Moran J. V., Kazazian H. H., Jr, Boeke J. D., 1996. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 87: 905–916. [DOI] [PubMed] [Google Scholar]
- Gaddameedhi S., Selby C. P., Kaufmann W. K., Smart R. C., Sancar A., 2011. Control of skin cancer by the circadian rhythm. Proc. Natl. Acad. Sci. USA 108: 18790–18795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasior S. L., Wakeman T. P., Xu B., Deininger P. L., 2006. The human LINE-1 retrotransposon creates DNA double-strand breaks. J. Mol. Biol. 357: 1383–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasior S. L., Preston G., Hedges D. J., Gilbert N., Moran J. V., et al. , 2007. Characterization of pre-insertion loci of de novo L1 insertions. Gene 390: 190–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasior S. L., Roy-Engel A. M., Deininger P. L., 2008. ERCC1/XPF limits L1 retrotransposition. DNA Repair (Amst.) 7: 983–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert N., Lutz-Prigge S., Moran J. V., 2002. Genomic deletions created upon LINE-1 retrotransposition. Cell 110: 315–325. [DOI] [PubMed] [Google Scholar]
- Gilbert N., Lutz S., Morrish T. A., Moran J. V., 2005. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol. Cell. Biol. 25: 7780–7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillet L. C., Schärer O. D., 2006. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem. Rev. 106: 253–276. [DOI] [PubMed] [Google Scholar]
- Groisman R., Polanowska J., Kuraoka I., Sawada J., Saijo M., et al. , 2003. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 113: 357–367. [DOI] [PubMed] [Google Scholar]
- Hancks D. C., Kazazian H. H., 2012. Active human retrotransposons: variation and disease. Curr. Opin. Genet. Dev. 22: 191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancks D. C., Kazazian H. H., 2016. Roles for retrotransposon insertions in human disease. Mob. DNA 7: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henning K. A., Li L., Iyer N., McDaniel L. D., Reagan M. S., et al. , 1995. The Cockayne syndrome group A gene encodes a WD repeat protein that interacts with CSB protein and a subunit of RNA polymerase II TFIIH. Cell 82: 555–564. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers J. H., 2001. Genome maintenance mechanisms for preventing cancer. Nature 411: 366–374. [DOI] [PubMed] [Google Scholar]
- Hou S. M., Falt S., Angelini S., Yang K., Nyberg F., et al. , 2002. The XPD variant alleles are associated with increased aromatic DNA adduct level and lung cancer risk. Carcinogenesis 23: 599–603. [DOI] [PubMed] [Google Scholar]
- Kinomoto M., Kanno T., Shimura M., Ishizaka Y., Kojima A., et al. , 2007. All APOBEC3 family proteins differentially inhibit LINE-1 retrotransposition. Nucleic Acids Res. 35: 2955–2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkel M. K., Wang J., Liang P., Batzer M. A., 2007. Identification and characterization of novel polymorphic LINE-1 insertions through comparison of two human genome sequence assemblies. Gene 390: 28–38. [DOI] [PubMed] [Google Scholar]
- Kubo S., Seleme M. C., Soifer H. S., Perez J. L., Moran J. V., et al. , 2006. L1 retrotransposition in nondividing and primary human somatic cells. Proc. Natl. Acad. Sci. USA 103: 8036–8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E. S., Linton L. M., Birren B., Nusbaum C., Zody M. C., et al. , 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Laugel V., 2013. Cockayne syndrome: the expanding clinical and mutational spectrum. Mech. Ageing Dev. 134: 161–170. [DOI] [PubMed] [Google Scholar]
- Lee B. S., Lichtenstein C. P., Faiola B., Rinckel L. A., Wysock W., et al. , 1998. Posttranslational inhibition of Ty1 retrotransposition by nucleotide excision repair/transcription factor TFIIH subunits Ssl2p and Rad3p. Genetics 148: 1743–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann A. R., McGibbon D., Stefanini M., 2011. Xeroderma pigmentosum. Orphanet J. Rare Dis. 6: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D. D., Saijo M., Tanaka K., Kraemer K. H., 1995. Expression of a transfected DNA repair gene (XPA) in xeroderma pigmentosum group A cells restores normal DNA repair and mutagenesis of UV-treated plasmids. Carcinogenesis 16: 1557–1563. [DOI] [PubMed] [Google Scholar]
- Li L., Bales E. S., Peterson C. A., Legerski R. J., 1993. Characterization of molecular defects in xeroderma pigmentosum group C. Nat. Genet. 5: 413–417. [DOI] [PubMed] [Google Scholar]
- Lovsin N., Peterlin B. M., 2009. APOBEC3 proteins inhibit LINE-1 retrotransposition in the absence of ORF1p binding. Ann. N. Y. Acad. Sci. 1178: 268–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan D. D., Eickbush T. H., 1996. Downstream 28S gene sequences on the RNA template affect the choice of primer and the accuracy of initiation by the R2 reverse transcriptase. Mol. Cell. Biol. 16: 4726–4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan D. D., Korman M. H., Jakubczak J. L., Eickbush T. H., 1993. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72: 595–605. [DOI] [PubMed] [Google Scholar]
- Mathias S. L., Scott A. F., Kazazian H. H., Jr, Boeke J. D., Gabriel A., 1991. Reverse transcriptase encoded by a human transposable element. Science 254: 1808–1810. [DOI] [PubMed] [Google Scholar]
- Mellon I., Spivak G., Hanawalt P. C., 1987. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell 51: 241–249. [DOI] [PubMed] [Google Scholar]
- Moran J. V., Holmes S. E., Naas T. P., DeBerardinis R. J., Boeke J. D., et al. , 1996. High frequency retrotransposition in cultured mammalian cells. Cell 87: 917–927. [DOI] [PubMed] [Google Scholar]
- Morrish T. A., Gilbert N., Myers J. S., Vincent B. J., Stamato T. D., et al. , 2002. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat. Genet. 31: 159–165. [DOI] [PubMed] [Google Scholar]
- Naegeli H., Sugasawa K., 2011. The xeroderma pigmentosum pathway: decision tree analysis of DNA quality. DNA Repair (Amst.) 10: 673–683. [DOI] [PubMed] [Google Scholar]
- Nouspikel T., 2009. DNA repair in mammalian cells: so DNA repair really is that important? Cell. Mol. Life Sci. 66: 965–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donovan A., Davies A. A., Moggs J. G., West S. C., Wood R. D., 1994. XPG endonuclease makes the 3′ incision in human DNA nucleotide excision repair. Nature 371: 432–435. [DOI] [PubMed] [Google Scholar]
- Ostertag E. M., Kazazian H. H., Jr, 2001. Biology of mammalian L1 retrotransposons. Annu. Rev. Genet. 35: 501–538. [DOI] [PubMed] [Google Scholar]
- Puumalainen M. R., Rüthemann P., Min J. H., Naegeli H., 2016. Xeroderma pigmentosum group C sensor: unprecedented recognition strategy and tight spatiotemporal regulation. Cell. Mol. Life Sci. 73: 547–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon J. T., Sancar A., 2005. Nucleotide excision repair. Prog. Nucleic Acid Res. Mol. Biol. 79: 183–235. [DOI] [PubMed] [Google Scholar]
- Richardson S. R., Narvaiza I., Planegger R. A., Weitzman M. D., Moran J. V., 2014. APOBEC3A deaminates transiently exposed single-strand DNA during LINE-1 retrotransposition. eLife 3: e02008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl T., Hanaoka F., Egly J. M., 2003. The comings and goings of nucleotide excision repair factors on damaged DNA. EMBO J. 22: 5293–5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo M., Matsuda T., Kuraoka I., Tanaka K., 2004. Inhibition of nucleotide excision repair by anti-XPA monoclonal antibodies which interfere with binding to RPA, ERCC1, and TFIIH. Biochem. Biophys. Res. Commun. 321: 815–822. [DOI] [PubMed] [Google Scholar]
- Sargent R. G., Meservy J. L., Perkins B. D., Kilburn A. E., Intody Z., et al. , 2000. Role of the nucleotide excision repair gene ERCC1 in formation of recombination-dependent rearrangements in mammalian cells. Nucleic Acids Res. 28: 3771–3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schärer O. D., 2011. Multistep damage recognition, pathway coordination and connections to transcription, damage signaling, chromatin structure, cancer and aging: current perspectives on the nucleotide excision repair pathway. DNA Repair (Amst.) 10: 667. [DOI] [PubMed] [Google Scholar]
- Shi X., Seluanov A., Gorbunova V., 2007. Cell divisions are required for L1 retrotransposition. Mol. Cell. Biol. 27: 1264–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijbers A. M., van der Spek P. J., Odijk H., van den Berg J., van Duin M., et al. , 1996. Mutational analysis of the human nucleotide excision repair gene ERCC1. Nucleic Acids Res. 24: 3370–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz M. R., Wu X., Wang Y., Wang L. E., Shete S., et al. , 2001. Modulation of nucleotide excision repair capacity by XPD polymorphisms in lung cancer patients. Cancer Res. 61: 1354–1357. [PubMed] [Google Scholar]
- Stankiewicz P., Lupski J. R., 2002. Genome architecture, rearrangements and genomic disorders. Trends Genet. 18: 74–82. [DOI] [PubMed] [Google Scholar]
- Streva V. A., Jordan V. E., Linker S., Hedges D. J., Batzer M. A., et al. , 2015. Sequencing, identification and mapping of primed L1 elements (SIMPLE) reveals significant variation in full length L1 elements between individuals. BMC Genomics 16: 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugasawa K., Ng J. M., Masutani C., Iwai S., van der Spek P. J., et al. , 1998. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol. Cell 2: 223–232. [DOI] [PubMed] [Google Scholar]
- Sugasawa K., Okamoto T., Shimizu Y., Masutani C., Iwai S., et al. , 2001. A multistep damage recognition mechanism for global genomic nucleotide excision repair. Genes Dev. 15: 507–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugasawa K., Shimizu Y., Iwai S., Hanaoka F., 2002. A molecular mechanism for DNA damage recognition by the xeroderma pigmentosum group C protein complex. DNA Repair (Amst.) 1: 95–107. [DOI] [PubMed] [Google Scholar]
- Sugasawa K., Akagi J., Nishi R., Iwai S., Hanaoka F., 2009. Two-step recognition of DNA damage for mammalian nucleotide excision repair: directional binding of the XPC complex and DNA strand scanning. Mol. Cell 36: 642–653. [DOI] [PubMed] [Google Scholar]
- Symer D. E., Connelly C., Szak S. T., Caputo E. M., Cost G. J., et al. , 2002. Human l1 retrotransposition is associated with genetic instability in vivo. Cell 110: 327–338. [DOI] [PubMed] [Google Scholar]
- Tomescu D., Kavanagh G., Ha T., Campbell H., Melton D. W., 2001. Nucleotide excision repair gene XPD polymorphisms and genetic predisposition to melanoma. Carcinogenesis 22: 403–408. [DOI] [PubMed] [Google Scholar]
- Troelstra C., van Gool A., de Wit J., Vermeulen W., Bootsma D., et al. , 1992. ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne’s syndrome and preferential repair of active genes. Cell 71: 939–953. [DOI] [PubMed] [Google Scholar]
- van den Boom V., Citterio E., Hoogstraten D., Zotter A., Egly J. M., et al. , 2004. DNA damage stabilizes interaction of CSB with the transcription elongation machinery. J. Cell Biol. 166: 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen W., Fousteri M., 2013. Mammalian transcription-coupled excision repair. Cold Spring Harb. Perspect. Biol. 5: a012625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volker M., Mone M. J., Karmakar P., van Hoffen A., Schul W., et al. , 2001. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell 8: 213–224. [DOI] [PubMed] [Google Scholar]
- Wagstaff B. J., Barnerssoi M., Roy-Engel A. M., 2011. Evolutionary conservation of the functional modularity of primate and murine LINE-1 elements. PLoS One 6: e19672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace N. A., Belancio V. P., Faber Z., Deininger P., 2010. Feedback inhibition of L1 and alu retrotransposition through altered double strand break repair kinetics. Mob. DNA 1: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Mates L., Ivics Z., Izsvák Z., Martin S. L., et al. , 2013. Cell division promotes efficient retrotransposition in a stable L1 reporter cell line. Mob. DNA 4: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J., Witherspoon D. J., Ray D. A., Batzer M. A., Jorde L. B., 2007. Mobile DNA elements in primate and human evolution. Am. J. Phys. Anthropol. 134 (Suppl. 45): 2–19. [DOI] [PubMed] [Google Scholar]
- Xu X. S., Wang L., Abrams J., Wang G., 2011. Histone deacetylases (HDACs) in XPC gene silencing and bladder cancer. J. Hematol. Oncol. 4: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuanxin Y., Chengcai A., Li L., Jiayu G., Guihong T., et al. , 2003. T-linker-specific ligation PCR (T-linker PCR): an advanced PCR technique for chromosome walking or for isolation of tagged DNA ends. Nucleic Acids Res. 31: e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X. D., Niedernhofer L., Kuster B., Mann M., Hoeijmakers J. H., et al. , 2003. ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol. Cell 12: 1489–1498. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.