Abstract
Parainfluenza viruses are a common cause of seasonal respiratory disease, but in high-risk individuals (e.g., young children) these viruses can cause severe clinical manifestations that require hospitalization. Beta-defensins are a subclass of antimicrobial peptides with antiviral activity. Use of adenovirus-mediated beta-defensin gene expression has been proposed as therapy for chronic bacterial infections commonly seen in cystic fibrosis patients; however, its use during parainfluenza virus 3 (PIV3) infection has not been evaluated. The hypothesis in this experiment was that adenovirus expression of human beta-defensin 6 (HBD6) would diminish concurrent PIV3 infection in neonatal lambs. The group infected with adenovirus HBD6 and PIV3 had increased levels of pulmonary neutrophil recruitment compared to those for the group infected with PIV3 or PIV3 and adenovirus, with an increased respiration rate and body temperature late in the course of the PIV3-adenovirus HBD6 infection. Interestingly, the adenovirus-treated groups had higher levels of immunohistochemical staining for PIV3 and syncytial cell formation than the group infected with PIV3, suggesting that treatment with the adenovirus vector, regardless of whether it was carrying a target gene, exacerbated the PIV3 infection. The levels of expression of mRNA for antimicrobial surfactant proteins A and D and sheep beta-defensin 1 were increased by PIV3 and adenovirus treatment, and the increased levels of expression roughly corresponded to the degree of inflammation. While pulmonary administration of a high-dose adenovirus vector has been associated with undesirable inflammation, this is the first study to show that it can exacerbate concurrent viral infection, a concern that needs to be addressed for future studies of adenovirus in the lung. Additionally, this study showed that adenovirus-mediated HBD6 expression increases neutrophil recruitment, a recently described attribute of beta-defensins, with mild accentuation of PIV3 activity and inflammation.
Parainfluenza virus 1 and parainfluenza virus 3 (PIV3) are enveloped, single-stranded, negative-sense RNA viruses of the family Paramyxoviridae and are important, ubiquitous causes of seasonal respiratory disease. Infection in a healthy individual is localized primarily to the upper respiratory tract and is often self-limiting (13, 27). However, human parainfluenza viruses can cause more serious respiratory disease such as severe laryngotracheitis (croupy cough), bronchiolitis, and pneumonia, which may lead to hospitalization and, on rare occasions, death (15). Risk factors for severe manifestations of parainfluenza virus infection include primary lung disease (e.g., cystic fibrosis or asthma), immunocompromise (e.g., transplant recipients), young age, and old age (9, 15, 16). Parainfluenza viruses, along with other viruses, are increasingly recognized in exacerbations of cystic fibrosis lung disease and may predispose individuals to secondary bacterial infections, including infections with Pseudomonas aeruginosa, which is a persistent problem in the lungs of individuals with cystic fibrosis (37). Young children and infants with cystic fibrosis are at increased risk for lower respiratory tract infections caused by these ubiquitous viruses, resulting in increased hospitalization rates and chronic lung damage (16, 36).
Defensins are a conserved class of antimicrobial peptides characterized by a cationic charge, antimicrobial activity, and six to eight highly conserved cysteine residues that form intramolecular disulfide bonds. There are three families of defensins, the alpha-, beta-, and theta-defensins, each of which is characterized by the organization of the intramolecular disulfide bonds (39). Beta-defensins are a class of cationic antimicrobial peptides that are produced primarily in the epithelium at mucosal sites, including the lungs (30). There are more than 30 putative human beta-defensin genes, and many of the products of those genes that have already been characterized have wide spectra of microbicidal activities against bacteria, fungi, and enveloped viruses (1, 31). The antiviral action of beta-defensins is thought to be through disruption of the viral envelope; however, recent work with alpha-defensins suggests that the antiviral activity may also involve intracellular cell signaling for the inhibition of viral replication (6, 30). Inactivation of beta-defensins by the altered local environment (e.g., altered salinity or the presence of airway surface liquid) has been suggested to contribute to the increased susceptibilities of cystic fibrosis patients to pulmonary infection (3, 11).
Perinatal lambs are commonly used as models for pulmonary disease research, including research involving gene therapies (19, 26). Perinatal lambs are susceptible to ovine PIV3 isolates, which have genomic and antigenic properties similar to those of human PIV3 isolates and which cause spontaneous respiratory disease and lesions in sheep (20, 33). Previous work with the human bronchial xenograft model with adenovirus (Ad)-mediated gene therapy and overexpression of a cathelicidin antimicrobial peptide (LL-37 [also known as hCAP-18]) showed success in killing bacteria, suggesting that they may be useful for antimicrobial gene therapy in patients predisposed to PIV3 infections (4); however, evaluation of antimicrobial peptide gene therapy as a way to diminish viral infection has not been applied. Human beta-defensin 6 (HBD6), a recently described beta-defensin selectively expressed in the epididymis, was chosen as the antimicrobial peptide in this model due to its lack of expression in the human lung or in sheep (38). The hypothesis of this study was that HBD6 gene expression in the neonatal lamb lung would diminish concurrent PIV3 infection in neonatal lambs.
MATERIALS AND METHODS
Animals.
All animal studies were approved by the Iowa State University Animal Care and Use Committee. Neonatal lambs (n = 20) were obtained from Iowa State University's Laboratory Animal Resources. At 3 to 5 days after birth (which allowed time for acclimation) the lambs were inoculated intratracheally with two 20-ml volumes at approximately 30-min intervals. The order and composition of the inoculum for each group (n = 4) were as follows: group A, sterile medium and sterile medium (control group); group B, sterile medium and PIV3 (PIV3 group); group C, Ad vector (no gene insertion) and PIV3 (PIV3/Ad group); and group D, Ad HBD6 and PIV3 (PIV3/Ad/HBD6 group). At day 7 of infection the lambs were euthanized by intravenous injection of sodium pentobarbital for collection of tissues. From our preliminary data this time was chosen because it is the time of significant lesion development and active viral clearance. An additional group of lambs were given an intratracheal inoculation (20 ml) of sterile medium (n = 2) or Ad HBD6 (n = 2) containing a lacZ gene insert for cellular localization of transfection. These lambs were euthanized on day 4 of infection, which was near the peak amount for pulmonary Ad vector gene expression (10).
Viruses.
The ovine isolate of PIV3 was grown to 107.8 50% tissue culture infective doses/ml (20 ml per lamb), as described previously (20). The replication-defective human Ad 5 vector had a cytomegalovirus promoter and was commercially acquired with the human HBD6 gene (identification no. 2602699; IMAGE Consortium), the HBD6 gene with a lacZ reporter gene, or no gene insert (ViraQuest, Inc., North Liberty, Iowa). Ad (no gene insert) and Ad/HBD6 had a deletion of the gene for protein E1 (nucleotides 358 to 3328), and the Ad/HBD6/lacZ insert for 5-bromo-4-chloro-3-indolylphosphate (X-Gal) staining had deletions at nucleotides 358 to 3328 and nucleotides 28592 to 30470, with the concurrent loss of protein E1 and E3 functions. Each lamb received approximately 1012 virus particles in 20 ml of sterile saline.
Clinical parameters.
The lambs were assessed daily for different clinical parameters. Body weight, temperature, and respiratory rate were taken at the same time of day, following the morning feeding. The lambs were treated daily with antibiotic (ceftiofur at 10 mg/day subcutaneously) to prevent complications associated with secondary bacterial infections (34).
Tissues.
Lung tissue was consistently collected from the cranial and caudal lobes on the left and right sides. The tissue samples were placed in 10% neutral buffered formalin and processed routinely for hematoxylin-eosin staining or immunohistochemistry. In addition, lung tissue was snap-frozen in liquid nitrogen for RNA isolation and real-time fluorogenic PCR.
RNA isolation.
RNA isolation and cDNA production were performed as described previously (24). Briefly, 0.3 g of tissue and 3 ml of Trizol reagent (Invitrogen) were homogenized for 30 s, and the homogenate was allowed to sit for 5 min at room temperature. Chloroform (0.6 ml) was added, and the mixture was shaken vigorously for 15 s and then allowed to sit for 3 min at room temperature. This mixture was microcentrifuged for 10 min at 4°C, the resulting top aqueous layer was retrieved and mixed with isopropanol (1.5 ml), and the mixture was allowed to sit for 10 min at room temperature. The solution was again microcentrifuged for 10 min at 4°C, the aqueous-isopropanol layer was removed, and the visible pellet was resolubilized in nuclease-free high-pressure liquid chromatography-grade water containing 0.1 mM EDTA. Spectrophotometric measurements (detection at absorbances of 260 and 280 nm with 1:40 sample dilutions) of each RNA sample were then taken to assess the isolates for purity and quantity.
cDNA production.
The RNA samples were treated with a commercially available DNase (RQ1 RNase-free DNase; Promega) to remove potential genomic DNA contamination. Immediately after this, reverse transcription was performed with the reagents available in a commercially available kit (TaqMan reverse transcriptase reagents; PE Applied Biosystems), according to the directions of the manufacturer. Briefly, 2 μg of DNase-treated RNA from each sample and control was added to a reaction mixture which contained final concentrations of 1× TaqMan reverse transcriptase buffer, 5.5 mM MgCl2, 2 mM deoxyribonucleoside triphosphate mixture (500 μM each deoxyribonucleoside triphosphate), 2.5 μM random hexamers, 1.25 U of murine leukemia virus reverse transcriptase per μl, 0.4 to 0.8 U of RNase inhibitor per μl, and high-pressure liquid chromatography-grade water. The following thermocycler conditions were then used: 10 min at 25°C, 30 min at 48°C, and 5 min at 95°C. The resulting cDNA was stored in nuclease-free microcentrifuge tubes at 20°C.
Primer and probe design.
Sequence-specific oligonucleotide primers and a fluorescent probe were designed with software (ABI Prism Primer Express, version 1.5; PE Applied Biosystems), according to the suggestions of the software manufacturer. The similarities of the resultant potential primer and probe sequences to DNA sequences available in databases were determined by use of the search tool BLAST (Basic Local Alignment Search Tool, version 1.4; National Center for Biotechnology Information), and only unique sequences were used for primer and probe design (Table 1). Sequence-specific oligonucleotide primers and a fluorescent probe for detection of cDNA corresponding to the endogenous reference gene, 18S rRNA, to which target cDNA levels were normalized, were purchased commercially (TaqMan rRNA control reagents; PE Biosystems).
TABLE 1.
Primer and probe sequences for expression of mRNAs for SP-A, SP-D, SBD1, and HBD6
| Primer or probe specificity and primer or probe | Sequencea |
|---|---|
| SP-A | |
| Forward | 5′-TGACCCTTATGCTCCTCTGGAT-3′ |
| Reverse | 5′-GGGCTTCCAAGACAAACTTCCT-3′ |
| Probe | 5′-6FAM-TGGCTTCTGGCCTCGAGTGCG-TAMRA-3′ |
| SP-D | |
| Forward | 5′-ACGTTCTGCAGCTGAGAAT-3′ |
| Reverse | 5′-TCGGTCATGCTCAGGAAAGC-3′ |
| Probe | 5′-6FAM-TTGACTCAGCTGGCCACAGCCCAGAACA-TAMRA-3′ |
| SBD1 | |
| Forward | 5′-CCATAGGAATAAAGGCGTCTGTGT-3′ |
| Reverse | 5′-CGCGACAGGTGCCAATCT-3′ |
| Probe | 5′-6FAM-CCGAGCAGGTGCCCTAGACACATGA-TAMRA-3′ |
| HBD6 | |
| Forward | 5′-CCC CAG CCA AGA ATG CAT-3′ |
| Reverse | 5′-TCA TTT TTC CCG CAA TTG TTC-3′ |
| Probe | 5′-6FAM-TTTTGATGAGAAATGCAACAAACTTAAAGGGACA-TAMRA-3′ |
6FAM, 6-carboxyfluorescein (fluorescent reporter dye); TAMRA, 6-carboxytetramethylrhodamine (fluorescent quencher dye).
PCR.
The 96-microwell plates were designed to enable testing of two replicates of both the target gene and the endogenous 18S rRNA of a 1:5 dilution of cDNA from all lambs, a negative control (diethyl pyrocarbonate-treated water), and five progressive 1:5 dilutions of cDNA from a control sheep for generation of the standard curve. On each plate, the wells with the target gene and the endogenous reference rRNA were run simultaneously by PCR with the tissues represented on that plate. The plate was run in a sequence detection system (GeneAmp 5700, version 1.3; PE Applied Biosystems) under conditions identical to those used in the optimization and validation tests. The resultant data gathered by the detection system were exported from the machine to a floppy disk, and all final data processing was performed by using a departmentally designed Excel software file to compare the target to the reference cDNA signals in order to create graphs of the relative levels of mRNA expression.
PIV3 immunohistochemistry.
Formalin-fixed, paraffin-embedded sections were deparaffinized in xylene, followed by passage through a series of alcohol baths; and pronase E (0.1 g of protease XIV and 0.1 g of CaCl2 per 100 ml of Tris-buffered saline [pH 7.4]) was applied for antigen retrieval. After the sections were washed (with phosphate-buffered saline), the background blocker (20% normal swine serum), followed by primary antibody (goat polyclonal anti-bovine PIV3, 1:1,000 [Veterinary Medical Research & Development, Inc.]), was applied to the tissues. These sections were washed, hydrogen peroxide (3% in methanol) was applied to block endogenous peroxidases, and the sections were washed again. Secondary antibody (1:400 biotinylated rabbit anti-goat antibody) was applied, followed by washing in supersensitive-horseradish peroxidase and chromogen (Vector Red; Vector). The slides were counterstained with hematoxylin, dehydrated in a series of alcohol baths and then with xylene, and placed under a coverslip, to which Permount was applied.
Morphometry.
Measurement of microscopic morphological changes, including cellular staining, rate of mitosis, neutrophil density, and syncytial cell formation, were assessed by a pathologist blinded from the study using a grid (18). Cranial and caudal lung lobes (bilaterally) were used for the study. For each slide, the pathologist randomly chose 10 sites under low power, and then the selected morphological alteration was counted under high power. This was done for each segment of lung, and the results were totaled and averaged for the grid area (13,000 or 52,000 μm2).
Ad distribution.
OCT-embedded frozen sections were cut to a thickness of 6 μm and placed in a bath of 4% formaldehyde for 5 min, and the slides were then washed twice in Hanks balanced salt solution (HBSS). X-Gal staining solution (20 mg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside per ml of N′,N′-dimethyl formamide) was added (20 μl/ml) to a fresh solution of 5 mM potassium ferricyanide-5 mM potassium ferrocyanide-2 mM magnesium chloride in HBSS for the final X-Gal working solution. The slides were then exposed to the final X-Gal working solution for 10 h at 37°C. The slides were washed in HBSS counterstained with nuclear fast red and placed under a coverslip. Sections were then examined for cells that stained blue, which is indicative of lacZ expression (23).
Statistics.
For assessment of clinical and fluorogenic real-time PCR data, significance was determined to be a P value of <0.05. Nonparametric Wilcoxon tests were used to assess samples of unequal sizes for statistical differences. t tests that assumed unequal variances were used to assess the means for groups of equal sizes for differences.
RESULTS
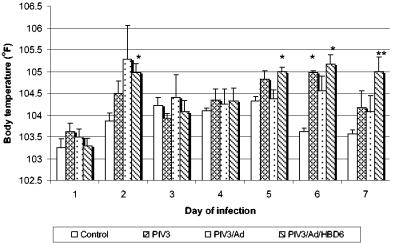
The daily body temperatures showed early and late increases through the course of infection. On day 2, the body temperatures of all animals in the PIV3-infected groups were generally increased, with the animals in the PIV3/Ad/HBD6 group having a significantly higher temperature than those in the control group (P < 0.01) (Fig. 1). Starting on day 5 the animals in the PIV3/Ad/HBD6 group had significantly increased body temperatures compared with those of the animals in the control group, and these continued throughout the duration of the experiment (P < 0.05). The animals in the PIV3 and PIV3/Ad groups had moderately increased body temperatures that were not statistically significant different except for those for the animals in the PIV3 group on day 6 (P < 0.01).
FIG. 1.
Body temperatures for the treatment groups. The animals in the PIV3 group had significantly increased body temperatures compared with those of the animals in the control group on day 6, while the animals in the PIV3/Ad/HBD6 group had increased body temperatures compared with those of the animals in the control group on days 2, 5, 6, and 7. *, P < 0.01 compared with the results for the controls; **, P < 0.05 compared with the results for the controls.
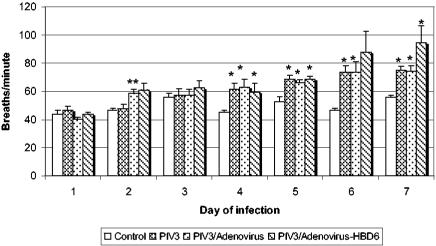
The respiration rate on day 2 was significantly higher in the animals in the PIV3/Ad group (P < 0.05) and moderately elevated in the animals in the PIV3/Ad/HBD6 group (P = 0.08) compared to that in the animals in the PIV3 group (Fig. 2). By day 4 the respiration rates in the animals in the PIV3-infected groups were generally elevated (P < 0.05) compared with those in the animals in the control group, except for an increase that lacked statistical significance for the PIV3/Ad/HBD6 group on day 6. Interestingly, on days 6 and 7 the animals in the PIV3/Ad/HBD6 group had a slightly higher respiration rate than those in the PIV3 or PIV3/Ad group.
FIG. 2.
Respiration rates for treatment groups. On day 2 the animals in the PIV3/Ad group and the PIV3/Ad/HBD6 group (P = 0.08) had increased respiration rates compared to those for the animals in PIV3 group. Starting on day 4 the animals in the PIV3-infected groups had increased respiration rates compared to those of the control animals, with that for the animals in the PIV3/Ad/HBD6 group being slightly higher on days 6 and 7. *, P < 0.05 compared with the results for the controls; **, P < 0.05 compared with the results for the PIV3 group.
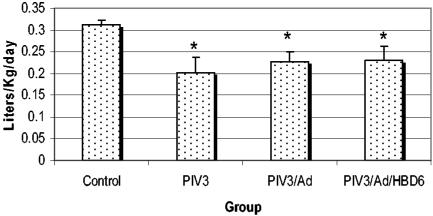
Daily milk consumption (in liters per kilogram of body weight) was evaluated to determine if there were any clinical differences in appetite. For the duration of the experiment, the animals in the PIV3, PIV3/Ad, and PIV3/Ad/HBD6 groups had decreased (P < 0.05) average milk consumption compared to that for the animals in the control group, with no significant difference among the PIV3-treated groups (Fig. 3).
FIG. 3.
Daily milk consumption for the treatment groups. All PIV3-infected groups had similar decreases in milk consumption. *, P < 0.05 compared with the results for the controls.
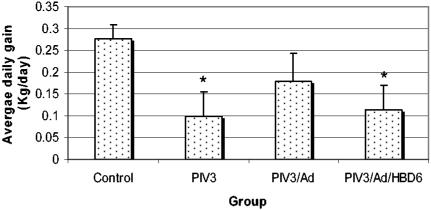
The growth of the lambs (average daily gain, in kilograms per day) during the experiment corresponded to the milk consumption, with significantly decreased growth of the animals in the PIV3 (P < 0.05) and PIV3/Ad/HBD6 (P < 0.05) groups (Fig. 4). The animals in the PIV3/Ad group showed a similar decrease in growth, but the decrease was not statistically significant.
FIG. 4.
Average daily weight gain for the treatment groups. The animals in the PIV3 and PIV3/Ad/HBD6 groups had significant decreases in growth rates, while the animals in the PIV3/Ad group had a slightly decreased growth rate compared to that of the control animals. *, P < 0.05 compared with the results for the controls.
Grossly, the control lambs had no visible lesions, while the lambs in the PIV3, PIV3/Ad, and PIV3/Ad/HBD6 groups had a cranioventral to hilar distribution of multifocal plumb-red consolidation. There was a slightly wider lesion distribution in the lambs in the PIV3/Ad and PIV3/Ad/HBD6 groups compared with that in the lambs in the PIV3 group. Microscopically, the PIV3-induced lesions consisted of a multifocal thickening of the alveolar septa by macrophages and a small number of lymphocytes, neutrophils, and plasma cells. The bronchiolar lumens contained small aggregates of neutrophils and sloughed cellular debris. Within these areas, the bronchiolar epithelium was thickened due to hyperplasia, with folds of piled cells having pale cytoplasms, increased mitotic figures, and infrequent syncytial cells. The animals in the PIV/3/Ad/HBD6 group appeared to have relatively higher numbers of neutrophils within the lungs compared to the numbers in the lungs of the animals in the group inoculated with PIV3 alone.
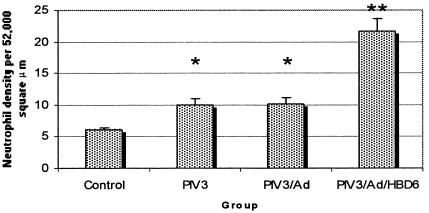
To evaluate neutrophil recruitment to the lungs following treatment, pulmonary neutrophil density was determined by morphometry. The lungs of each of the PIV3-infected groups had significantly increased (P < 0.05) neutrophilic infiltrates compared with the lungs of the controls (Fig. 5). In addition, the PIV3/Ad/HBD6 group had a higher neutrophil count than either the PIV3 or the PIV3/Ad group (P < 0.05).
FIG. 5.
Pulmonary neutrophil density in the treatment groups. All PIV3-infected groups had higher neutrophil densities than the control group. The PIV3/Ad/HBD6 group had increased neutrophil density compared with those of the PIV3 and PIV3/Ad groups. *, P < 0.05 compared with the results for the controls; **, P < 0.05 compared with the results for the PIV3 or PIV3/Ad group.
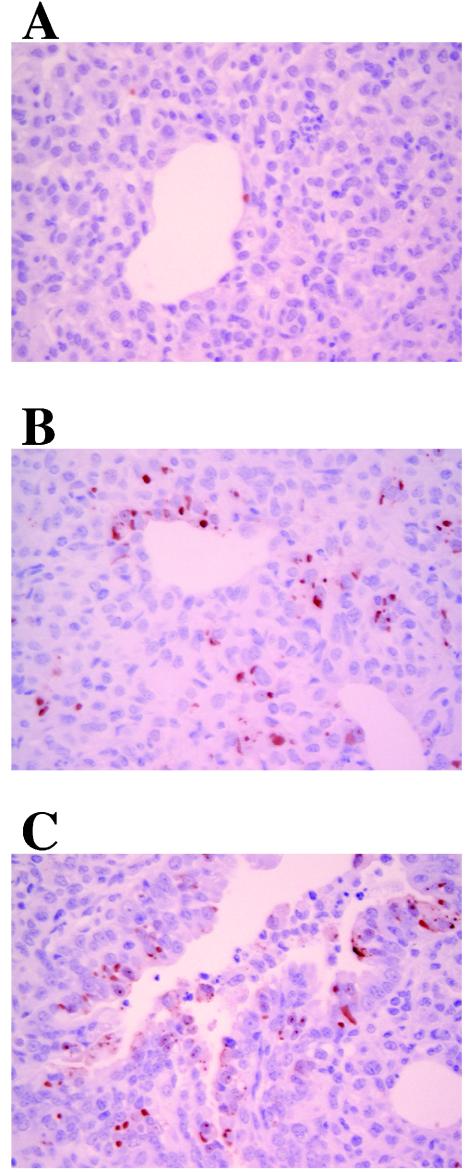
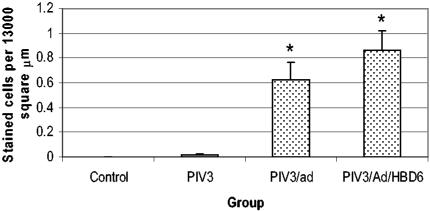
PIV3 immunohistochemistry produced no staining in the control lungs, while cellular staining was present in the lungs of all lambs in the PIV3-infected groups. This was characterized by small granular red staining within the cytoplasm (Fig. 6). Affected cells most often were epithelial cells of the distal bronchi to terminal bronchioles. Morphometric quantification of the cellular PIV3 immunostaining showed increased numbers of immunoreactive cells in the PIV3/Ad (P < 0.05) and PIV3/Ad/HBD6 (P < 0.05) groups compared to the number in the PIV3 group (Fig. 7).
FIG. 6.
Immunohistochemistry for PIV3 in the PIV3 group (A), PIV3/Ad group (B), and PIV3/Ad/HBD6 group (C). The PIV3/Ad and animals in the PIV3/Ad/HBD6 group had increased cellular staining compared with that for the animals in the PIV3 group. No staining was detected for the animals in the control group (not shown). Magnifications, ×400.
FIG. 7.
Cellular PIV3 antigen staining in the treatment groups. The PIV3/Ad and PIV3/Ad/HBD6 groups each had significant increases in PIV3 antigen staining compared with that for the PIV3 group. The control group had no staining. *, P < 0.05 compared with the results for the PIV3 group.
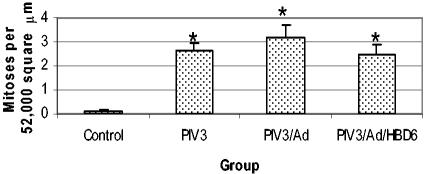
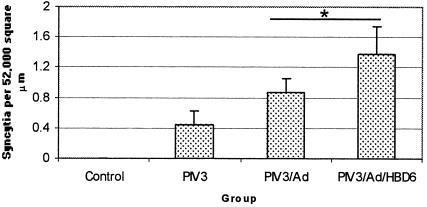
The mitotic rate was significantly increased in all of the PIV3-infected groups compared with that in the control group (Fig. 8). No significant changes in the PIV3-infected groups were evident. PIV3-induced syncytial cell formation was not present in the control group but was present in all of the PIV3-infected groups. The combined Ad gene therapy groups had higher syncytial cell densities than the PIV3-infected groups (P < 0.05) (Fig. 9).
FIG. 8.
Mitotic rate in the treatment groups. The PIV3, PIV3/Ad, and PIV3/Ad/HBD6 groups had similar significant increases in the mitotic rate compared with that for the control group, with no significant differences among the PIV3-infected groups. *, P < 0.05 compared with the results for the control group.
FIG. 9.
Syncytial cell formation in the treatment groups. The PIV3/Ad and PIV3/Ad/HBD6 groups combined had significantly higher syncytial cell formation than the PIV3 group. *, P < 0.05 compared with the results for the PIV3 group.
The Ad vector was localized by X-Gal staining to a small number (<5%) of individual epithelial cells of the bronchi and terminal bronchioles (data not shown). This staining was not seen in samples from two control lambs inoculated with sterile medium.
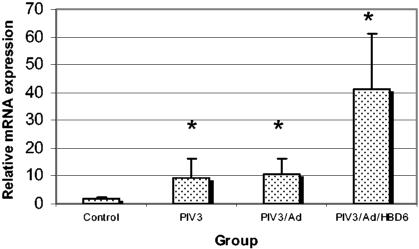
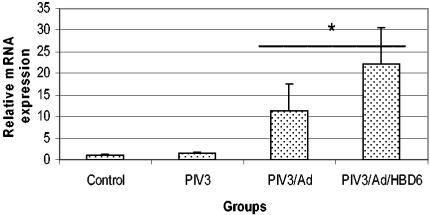
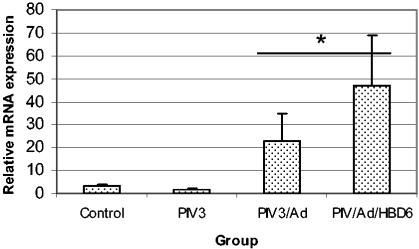
Real-time fluorogenic PCR was used to assess the lambs for transfection of HBD6 and for alterations in host SBD1, SP-A, and SP-D mRNA. No amplification of HBD6 was detected in the control groups (inoculated with PIV3/Ad) by the fluorogenic PCR, while all four of the HBD6-inoculated lambs had late-cycle amplification, indicating weak HBD6 expression (data not shown). Expression of SP-D in each PIV3-infected group was significantly elevated compared with that in the control group, and the levels of expression tended to increase from the PIV3/Ad group to the PIV3/Ad/HBD6 group (Fig. 10). Similar trends in SP-A and SBD1 mRNA expression were present, with statistically significant differences seen for the levels of expression in the PIV3/Ad and PIV3/Ad/HBD6 groups combined compared with that in the PIV3 group for both SP-A and SBD1 (P < 0.05) (Fig. 11 and 12).
FIG. 10.
SP-D mRNA expression in the treatment groups. The levels of expression were increased in the PIV3, PIV3/Ad, and PIV3/Ad/HBD6 groups compared to those in the control group, with a moderate increase in the PIV3/Ad/HBD6 group compared to that for the PIV3/Ad group *, P < 0.05 compared with the results for the control group.
FIG. 11.
SP-A mRNA expression in the treatment groups. The levels of SP-A mRNA expression in the PIV3/Ad and PIV3/Ad/HBD6 groups combined was significantly increased compared to that in the PIV3 group. *, P < 0.05 compared with the results for the PIV3 group.
FIG. 12.
SBD1 mRNA expression in the treatment groups. The levels of SBD1 mRNA expression in the PIV3/Ad and PIV3/Ad/HBD6 groups combined were significantly increased compared to that in the PIV3 group. *, P < 0.05 compared with the results for the PIV3 group.
DISCUSSION
The hypothesis of this study was that beta-defensin (HBD6) expression during concurrent PIV3 infection would diminish disease progression. Clinical parameters were monitored during the course of infection, and day 7 of infection was chosen for necropsy, as the lesions were usually marked and active viral clearance was seen in the lambs (7, 20). All PIV3-infected groups had clinical evidence of infection (e.g., elevated temperature and tachypnea and depressed milk consumption and growth), gross and microscopic lesions consistent with PIV3 infection, and immunohistochemical staining in PIV3-infected animals only. Ad transfection was confirmed by X-Gal staining of the Ad/HBD6/lacZ-infected airway epithelium on day 4 and by HBD6 mRNA expression on day 7 of infection.
The animals in the Ad-treated groups (the PIV3/Ad and PIV3/Ad/HBD6 groups) had increased temperatures and respiration rates on day 2 of infection. This was consistent with previous work in which BALB/c mice showed increased levels of the proinflammatory cytokines tumor necrosis factor-alpha (TNF-α) and interleukin-6 (IL-6) within even hours of intrapulmonary inoculation of human Ad 5 (40). TNF-α and IL-6 are endogenous pyrogens that account for the increased body temperatures, and TNF-α mediates bronchial and vascular smooth muscle contractility, which, along with pyrexia, could have increased the respiratory rate (2, 35). Other clinical alterations compared to the clinical conditions of the controls were generally attributable to PIV3 infection. Of particular interest was the fact that the animals in the Ad-treated groups had increased staining of PIV3 and syncytial cell formation compared with that in the animals in the PIV3 groups not treated with Ad. Antigen staining occurs at sites of PIV3 protein synthesis, and syncytial cell formation is mediated by the fusion protein and is regulated by the hemagglutinin-neuraminidase protein; both indicators are by-products of PIV3 replication (28).
The increased immunostaining and syncytial cell formation were both indicative of enhanced PIV3 activity (21, 28). This novel finding suggests that pulmonary Ad-mediated gene therapy can exacerbate concurrent PIV3 infection. Proteins of Ads, including proteins E1A, E1B, and E3, have been shown to have immunomodulating functions, most of which are immunosuppressing (17, 29). The vector used in this study had an E1 deletion, but the E3 gene was intact. E3 gene products inhibit peptide processing for major histocompatibility complex (MHC) I presentation; activation of nuclear factor-kappa beta (NF-κβ); and apoptosis through the fatty acid synthase, TNF-related apoptosis-inducing ligand, and TNF-α pathways (5, 8, 17, 29, 32). Processing for MHC I presentation and apoptosis of virus-infected cells are important mechanisms for viral clearance, and the synthesis of antiviral type I interferon along with other inflammatory mediators are mediated in part through NF-κβ activation. Early protein E3 inhibition of antiviral pathways could have allowed more enhanced PIV3 replication and infection. While the immunosuppressive effects of the Ad E3 gene may participate in enhancing PIV3 infection, the low level of cellular distribution of Ad suggests that this may not be a significant component. Another mechanism for enhanced PIV3 infection may be the general inflammatory and cellular changes caused by the addition of the Ad vector. In previous work, pretreatment with 4-ipomeanol similarly enhanced PIV3 activity in a calf model (21). The compound 4-ipomeanol is biotransformed in Clara cells into a free-radical metabolite which causes necrotizing bronchiolitis and interstitial pneumonia. The investigators that used the PIV3-infected calf model hypothesized that 4-impomeanol may (i) suppress pulmonary innate defenses, (ii) increase the number of susceptible cells, or (iii) increase the levels of factors (e.g., proteases) that enhance viral replication. Early immunosuppression by Ad proteins and/or altered cellular susceptibility to PIV3 infection and replication may contribute to this increased PIV3 activity in Ad-treated lambs. Further work needs to be done to clarify and define the mechanism.
We further evaluated the levels of mRNA for innate antimicrobial peptides and proteins to determine if Ad gene therapy altered mRNA expression. SP-D levels were increased in each PIV3-infected group compared with those in the control group (P < 0.05), with moderate increases in the PIV3/Ad/HBD6 group compared with those in the PIV3/Ad and PIV3 groups. Similar trends were seen for SP-A and SBD1 mRNA, with significant increases in the PIV3/Ad and PIV3/Ad/HBD6 groups combined compared with those in the PIV3 group (P < 0.05). Surfactant protein and beta-defensin mRNA expression is partially regulated by the status of pulmonary inflammation and lung injury (22, 30). The increases seen here were likely a coordinated response to the exacerbated inflammation and injury associated with inoculation of the Ad vector and exacerbated PIV3 infection.
The effect of HBD6 expression in vivo on PIV3 infection was unanticipated, as it tended to be proinflammatory and aggravated the overall clinical situation. Microscopically, neutrophil infiltration was increased in the PIV3/Ad/HBD6 group, and this was confirmed by morphometry, in which the neutrophil density was increased (P < 0.05) compared with those in either the PIV3 or the PIV3/Ad group. The clinical scores for body temperature and respiration rates were increased for the PIV3/Ad/HBD6 group compared with those for the control group and with slight increases compared with those for the PIV3/Ad group. While beta-defensins are increasingly known for their antimicrobial properties, they have additional functions, such as leukocyte chemotaxis and immunomodulation (30). Recently, HBD2 was shown to function in vitro as a chemoattractant for TNF-α-primed neutrophils, and the HBD2 ligand interaction with CC-chemokine-receptor-6 (CCR6) was considered the mechanism for chemotaxis (25). TNF-α expression is an early consequence of pulmonary Ad-mediated gene therapy and may prime the neutrophils for CCR6-mediated chemotaxis (40). HBD6 expression in these lambs both stimulated increased neutrophilic infiltrates and accentuated the clinical parameters of PIV3 disease without decreasing the activity of PIV3; rather, HBD6 expression accentuated the activity of PIV3. Although HBD6 expression is the likely source for these events, a specific interaction between the Ad vector and HBD6 expression cannot be ruled out. While the efficacy of HBD6 against pathogenic organisms has not been established, this study did show for the first time the in vivo recruitment of neutrophils by beta-defensin expression and that HBD6 expression during gene therapy results in biological activity in the neonatal lamb lung.
Ad-mediated gene therapy in the lung has been hindered by the inflammatory response that it generates. In this study a high concentration of Ad (23, 26) was used for transfection, which accentuated the activity of PIV3. Recent technological advances, such as pretreatment of airways with sodium caprate or complexing of Ad with DEAE dextran, have been used to provide a means of reducing the Ad titer required and thus minimizing the adverse effects (12). Another limitation of this study is the small number of lambs used in each group, a common problem associated with work with large animals, which sometimes makes the finding of statistical significance challenging due to interanimal variations (24). However, studies with large animals (as in this study) can provide novel insights into disease mechanisms not readily observed in rodent or other laboratory animal models (14). For the neonatal lamb study we used an ovine isolate of PIV3 which is comparable to human PIV3 in terms of the disease and the lesions that it causes (7, 15, 20). This is the first study to show that high-dose Ad gene therapy may exacerbate a concurrent PIV3 infection, a concern that needs to be further addressed for future Ad gene therapy. Additionally, this is the first study to show that in vivo HBD6 expression has biological activity, specifically, neutrophil recruitment and mild aggravation of concurrent PIV3 infection.
Acknowledgments
This project was funded through USDA/CRIS award 2003-35204-13492.
We thank Vanessa Preast, Erin Costello, and Rachel Dersheid for technical assistance and Ronald E. Haskell (ViraQuest, Inc.) for supplying the Ad vectors.
REFERENCES
- 1.Aarbiou, J., K. F. Rabe, and P. S. Hiemstra. 2002. Role of defensins in inflammatory lung disease. Ann. Med. 34:96-101. [DOI] [PubMed] [Google Scholar]
- 2.Amrani, Y., H. Chen, and R. A. Panettieri, Jr. 2000. Activation of tumor necrosis factor receptor 1 in airway smooth muscle: a potential pathway that modulates bronchial hyper-responsiveness in asthma? Respir. Res. 1:49-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bals, R., D. J. Weiner, R. L. Meegalla, F. Accurso, and J. M. Wilson. 2001. Salt-independent abnormality of antimicrobial activity in cystic fibrosis airway surface fluid. Am. J. Respir. Cell Mol. Biol. 25:21-25. [DOI] [PubMed] [Google Scholar]
- 4.Bals, R., D. J. Weiner, R. L. Meegalla, and J. M. Wilson. 1999. Transfer of a cathelicidin peptide antibiotic gene restores bacterial killing in a cystic fibrosis xenograft model. J. Clin. Investig. 103:1113-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bennett, E. M., J. R. Bennink, J. W. Yewdell, and F. M. Brodsky. 1999. Cutting edge: adenovirus E19 has two mechanisms for affecting class I MHC expression. J. Immunol. 162:5049-5052. [PubMed] [Google Scholar]
- 6.Chang, T. L., F. Francois, A. Mosoian, and M. E. Klotman. 2003. CAF-mediated human immunodeficiency virus (HIV) type 1 transcriptional inhibition is distinct from alpha-defensin-1 HIV inhibition. J. Virol. 77:6777-6784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cutlip, R. C., and H. D. Lehmkuhl. 1982. Experimentally induced parainfluenza type 3 virus infection in young lambs: pathologic response. Am. J. Vet. Res. 43:2101-2107. [PubMed] [Google Scholar]
- 8.Feuerbach, D., S. Etteldorf, C. Ebenau-Jehle, J. P. Abastado, D. Madden, and H. G. Burgert. 1994. Identification of amino acids within the MHC molecule important for the interaction with the adenovirus protein E3/19K. J. Immunol. 153:1626-1636. [PubMed] [Google Scholar]
- 9.Garbino, J., M. W. Gerbase, W. Wunderli, L. Kolarova, L. P. Nicod, T. Rochat, and L. Kaiser. 2004. Respiratory viruses and severe lower respiratory tract complications in hospitalized patients. Chest 125:1033-1039. [DOI] [PubMed] [Google Scholar]
- 10.Gill, D. R., L. A. Davies, I. A. Pringle, and S. C. Hyde. 2004. The development of gene therapy for diseases of the lung. Cell. Mol. Life Sci. 61:355-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldman, M. J., G. M. Anderson, E. D. Stolzenberg, U. P. Kari, M. Zasloff, and J. M. Wilson. 1997. Human beta-defensin-1 is a salt-sensitive antibiotic in lung that is inactivated in cystic fibrosis. Cell 88:553-560. [DOI] [PubMed] [Google Scholar]
- 12.Gregory, L. G., R. P. Harbottle, L. Lawrence, H. J. Knapton, M. Themis, and C. Coutelle. 2003. Enhancement of adenovirus-mediated gene transfer to the airways by DEAE dextran and sodium caprate in vivo. Mol. Ther. 7:19-26. [DOI] [PubMed] [Google Scholar]
- 13.Hall, C. B. 2001. Respiratory syncytial virus and parainfluenza virus. N. Engl. J. Med. 344:1917-1928. [DOI] [PubMed] [Google Scholar]
- 14.Hein, W. R., and P. J. Griebel. 2003. A road less travelled: large animal models in immunological research. Nat. Rev. Immunol. 3:79-84. [DOI] [PubMed] [Google Scholar]
- 15.Henrickson, K. J. 2003. Parainfluenza viruses. Clin. Microbiol. Rev. 16:242-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hiatt, P. W., S. C. Grace, C. A. Kozinetz, S. H. Raboudi, D. G. Treece, L. H. Taber, and P. A. Piedra. 1999. Effects of viral lower respiratory tract infection on lung function in infants with cystic fibrosis. Pediatrics 103:619-626. [DOI] [PubMed] [Google Scholar]
- 17.Horwitz, M. S. 2004. Function of adenovirus E3 proteins and their interactions with immunoregulatory cell proteins. J. Gene Med. 6:S172-S183. [DOI] [PubMed] [Google Scholar]
- 18.Keeney, S. E., M. J. Mathews, A. K. Haque, and F. C. Schmalstieg. 1995. Comparison of pulmonary neutrophils in the adult and neonatal rat after hyperoxia. Pediatr. Res. 38:857-863. [DOI] [PubMed] [Google Scholar]
- 19.Kramer, B. W., A. H. Jobe, C. J. Bachurski, and M. Ikegami. 2001. Surfactant protein A recruits neutrophils into the lungs of ventilated preterm lambs. Am. J. Respir. Crit. Care Med. 163:158-165. [DOI] [PubMed] [Google Scholar]
- 20.Lehmkuhl, H. D., and R. C. Cutlip. 1983. Experimental parainfluenza type 3 infection in young lambs: clinical, microbiological, and serological response. Vet. Microbiol. 8:437-442. [DOI] [PubMed] [Google Scholar]
- 21.Li, X., and W. L. Castleman. 1991. Effects of 4-ipomeanol on bovine parainfluenza type 3 virus-induced pneumonia in calves. Vet. Pathol. 28:428-437. [DOI] [PubMed] [Google Scholar]
- 22.McCormack, F. X., and J. A. Whitsett. 2002. The pulmonary collectins, SP-A and SP- D, orchestrate innate immunity in the lung. J. Clin. Investig. 109:707-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCray, P. B., Jr., K. Armstrong, J. Zabner, D. W. Miller, G. A. Koretzky, L. Couture, J. E. Robillard, A. E. Smith, and M. J. Welsh. 1995. Adenoviral-mediated gene transfer to fetal pulmonary epithelia in vitro and in vivo. J. Clin. Investig. 95:2620-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyerholz, D. K., J. M. Gallup, B. M. Grubor, R. B. Evans, B. F. Tack, P. B. McCray, Jr., and M. R. Ackermann. 2004. Developmental expression and distribution of sheep beta-defensin-2. Dev. Comp. Immunol. 28:171-178. [DOI] [PubMed] [Google Scholar]
- 25.Niyonsaba, F., H. Ogawa, and I. Nagaoka. 2004. Human beta-defensin-2 functions as a chemotactic agent for tumour necrosis factor-alpha-treated human neutrophils. Immunology 111:273-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peebles, D., L. G. Gregory, A. David, M. Themis, S. N. Waddington, H. J. Knapton, M. Miah, T. Cook, L. Lawrence, M. Nivsarkar, C. Rodeck, and C. Coutelle. 2004. Widespread and efficient marker gene expression in the airway epithelia of fetal sheep after minimally invasive tracheal application of recombinant adenovirus in utero. Gene Ther. 11:70-78. [DOI] [PubMed] [Google Scholar]
- 27.Piedra, P. A., J. A. Englund, and W. P. Glezen. 2002. Respiratory syncytial virus and parainfluenza viruses, p. 763-790. In D. D. Richman, R. J. Whitley, and F. G. Hayden (ed.), Clinical virology, 2nd ed. ASM Press, Washington, D.C.
- 28.Porotto, M., M. Murrell, O. Greengard, and A. Moscona. 2003. Triggering of human parainfluenza virus 3 fusion protein (F) by the hemagglutinin-neuraminidase (HN) protein: an HN mutation diminishes the rate of F activation and fusion. J. Virol. 77:3647-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaack, J., M. L. Bennett, J. D. Colbert, A. V. Torres, G. H. Clayton, D. Ornelles, and J. Moorhead. 2004. E1A and E1B proteins inhibit inflammation induced by adenovirus. Proc. Natl. Acad. Sci. USA 101:3124-3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schutte, B. C., and P. B. McCray, Jr. 2002. β-Defensins in lung host defense. Annu. Rev. Physiol. 64:709-748. [DOI] [PubMed] [Google Scholar]
- 31.Schutte, B. C., J. P. Mitros, J. A. Bartlett, J. D. Walters, H. P. Jia, M. J. Welsh, T. L. Casavant, and P. B. McCray, Jr. 2002. Discovery of five conserved beta-defensin gene clusters using a computational search strategy. Proc. Natl. Acad. Sci. USA 99:2129-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shisler, J., C. Yang, B. Walter, C. F. Ware, and L. R. Gooding. 1997. The adenovirus E3-10.4K/14.5K complex mediates loss of cell surface Fas (CD95) and resistance to Fas-induced apoptosis. J. Virol. 71:8299-8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suzu, S., Y. Sakai, T. Shioda, and H. Shibuta. 1987. Nucleotide sequence of the bovine parainfluenza 3 virus genome: the genes of the F and HN glycoproteins. Nucleic Acids Res. 15:2945-2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Viuff, B., K. Tjornehoj, L. E. Larsen, C. M. Rontved, A. Uttenthal, L. Ronsholt, and S. Alexandersen. 2002. Replication and clearance of respiratory syncytial virus: apoptosis is an important pathway of virus clearance after experimental infection with bovine respiratory syncytial virus. Am. J. Pathol. 161:2195-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wagner, E. M. 2000. TNF-alpha induced bronchial vasoconstriction. Am. J. Physiol. Heart Circ. Physiol. 279:H946-H951. [DOI] [PubMed] [Google Scholar]
- 36.Wat, D. 2003. Impact of respiratory viral infections on cystic fibrosis. Postgrad. Med. J. 79:201-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wat, D., and I. Doull. 2003. Respiratory virus infections in cystic fibrosis. Paediatr. Respir. Rev. 4:172-177. [DOI] [PubMed] [Google Scholar]
- 38.Yamaguchi, Y., T. Nagase, R. Makita, S. Fukuhara, T. Tomita, T. Tominaga, H. Kurihara, and Y. Ouchi. 2002. Identification of multiple novel epididymis-specific beta-defensin isoforms in humans and mice. J. Immunol. 169:2516-2523. [DOI] [PubMed] [Google Scholar]
- 39.Yang, D., O. Chertov, and J. J. Oppenheim. 2001. The role of mammalian antimicrobial peptides and proteins in awakening of innate host defenses and adaptive immunity. Cell. Mol. Life Sci. 58:978-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zsengeller, Z., K. Otake, S. A. Hossain, P. Y. Berclaz, and B. C. Trapnell. 2000. Internalization of adenovirus by alveolar macrophages initiates early proinflammatory signaling during acute respiratory tract infection. J. Virol. 74:9655-9667. [DOI] [PMC free article] [PubMed] [Google Scholar]