Abstract
The expression of genes encoding extracellular polymer-degrading enzymes and the metabolic pathways required for carbon utilization in fungi are tightly controlled. The control is mediated by transcription factors that are activated by the presence of specific inducers, which are often monomers or monomeric derivatives of the polymers. A D-galacturonic acid-specific transcription factor named GaaR was recently identified and shown to be an activator for the expression of genes involved in galacturonic acid utilization in Botrytis cinerea and Aspergillus niger. Using a forward genetic screen, we isolated A. niger mutants that constitutively express GaaR-controlled genes. Reasoning that mutations in the gaaR gene would lead to a constitutively activated transcription factor, the gaaR gene in 11 of the constitutive mutants was sequenced, but no mutations in gaaR were found. Full genome sequencing of five constitutive mutants revealed allelic mutations in one particular gene encoding a previously uncharacterized protein (NRRL3_08194). The protein encoded by NRRL3_08194 shows homology to the repressor of the quinate utilization pathway identified previously in Neurospora crassa (qa-1S) and Aspergillus nidulans (QutR). Deletion of NRRL3_08194 in combination with RNA-seq analysis showed that the NRRL3_08194 deletion mutant constitutively expresses genes involved in galacturonic acid utilization. Interestingly, NRRL3_08194 is located next to gaaR (NRRL3_08195) in the genome. The homology to the quinate repressor, the chromosomal clustering, and the constitutive phenotype of the isolated mutants suggest that NRRL3_08194 is likely to encode a repressor, which we name GaaX. The GaaR–GaaX module and its chromosomal organization is conserved among ascomycetes filamentous fungi, resembling the quinate utilization activator-repressor module in amino acid sequence and chromosomal organization.
Keywords: gene regulation, galacturonic acid, repressor protein, genomics, transcriptomics, pectin
THE filamentous fungus Aspergillus niger is an important producer of pectin-degrading enzymes that are used in industrial applications including in food and feed processing (Kashyap et al. 2001; Khan et al. 2013). In nature, A. niger is a saprotrophic fungus that feeds on organic matter from decaying plants. The major carbon sources in plant cells are the storage polysaccharides starch, and less frequently inulin, as well as the cell wall polymers cellulose, hemicelluloses, and pectin. Of the different plant polysaccharides, pectin has the most complex structure. Pectin is made up of four substructures, including homogalacturonan, xylogalacturonan, rhamnogalacturonan I, and rhamnogalacturonan II. The abundance of each substructure varies with plant species, but typically homogalacturonan is the most abundant polysaccharide in pectin (65%), followed by rhamnogalacturonan I (25–30%). Xylogalacturonan and rhamnogalacturonan II comprise <10% of the total pectin (Mohnen 2008).
Utilization of plant polysaccharides by fungi, including A. niger, is accomplished by tightly controlled secretion of extracellular enzymes that degrade the polymers into monosaccharides or oligosaccharides that are taken up and catabolised by the fungus. The controlled regulation is not only confined to the expression of genes encoding extracellular proteins. It also includes the controlled expression of genes encoding specific sugar transporters to guarantee efficient uptake of the liberated sugars and the intracellular catabolic pathway enzymes. The precise induction of the network of genes encoding substrate-specific enzymes, transporters, and catabolic pathway enzymes has so far been shown to be mediated via Zn(II)2Cys6 transcription factors. Specific transcription factors in A. niger have been characterized that regulate the utilization of the major polysaccharides. They include AmyR, the regulator for starch utilization (Petersen et al. 1999; Yuan et al. 2008a; vanKuyk et al. 2012); InuR for inulin (Yuan et al. 2008b); ManR, ClrA, and ClrB for cellulose (Raulo et al. 2016); XlnR for xylan (Van Peij et al. 1998; Battaglia et al. 2014); AraR for arabinan (Battaglia et al. 2014); RhaR for rhamnose (Gruben et al. 2014); and GaaR for polygalacturonic acid (PGA) (Alazi et al. 2016). These transcription factors exert coordinated regulation of the target genes by interacting with conserved binding sites that are located upstream of the target genes. Computational analysis has been used to identify the galacturonic acid (GA) responsive element (GARE) of GA-induced genes (Martens-Uzunova and Schaap 2008). The predicted sequence (CCNCCAA) was shown to be required for the induction of GA-responsive genes in A. niger (Niu et al. 2015) and Botrytis cinerea (Zhang et al. 2016). Furthermore, using the yeast one-hybrid method, it was shown in B. cinerea that the GaaR transcription factor interacts specifically with the GARE (Zhang et al. 2016).
Phenotypic characterization of mutants lacking the GA-regulator in both B. cinerea and A. niger has shown that GaaR is required for growth on GA (Alazi et al. 2016; Zhang et al. 2016). Expression analysis in both fungi confirmed that GaaR is required for the induced expression of GA-responsive genes. On complex pectins, growth of B. cinerea and A. niger gaaR deletion mutants was severely reduced and genome-wide expression analysis in A. niger revealed that the residual growth on pectin is likely due to the GaaR-independent expression of pectinases acting on arabinofuranosyl- and galactopyranosyl-containing side chains in rhamnogalacturonan (Alazi et al. 2016).
In addition to the transcription regulation via GaaR, GA-responsive genes are also under carbon catabolite repression (CCR) control (de Vries et al. 1999; de Vries et al. 2002). In filamentous fungi, CreA mediates CCR (Dowzer et al. 1991; Ruijter and Visser 1997). In A. niger, CreA also exerts CCR control on GA-responsive genes (de Vries et al. 1999; Niu et al. 2015). Using an in vivo reporter construct consisting of the promoter of the GA-inducible pgaX gene (PpgaX) and the acetamidase (amdS) gene as a reporter, both the specific induction of pgaX on GA as well as the carbon repression of pgaX via CreA had been demonstrated (Niu et al. 2015). In this study, we have used the PpgaX-amdS reporter strain to isolate mutants displaying constitutive expression of GA-responsive genes. Analysis of the mutants resulted in the identification of a protein that likely acts as a repressor that specifically inhibits GaaR transcription activation activity under noninducing conditions.
Materials and Methods
Strains, media, and growth conditions
All strains in this study are listed in Table 1. Strains were grown in liquid or solidified (1.5% agar) minimal medium (MM) containing 7 mM KCl, 8 mM KH2PO4, 70 mM NaNO3, and 2 mM MgSO4 (pH adjusted to 5.5) as described (Bennett and Lasure 1991). MM was supplemented with 50 mM glucose, 50 mM D-galacturonic acid, 50 mM fructose, or 50 mM sorbitol as carbon source. Complete medium (CM) was also used and consists of MM supplemented with 0.1% casamino acids and 0.5% w/v yeast extract and 50 mM glucose. MM agar plates containing acetamide as sole nitrogen source were made as described previously (Arentshorst et al. 2012).
Table 1. Aspergillus niger strains used in this study.
| Name | Genotype/description | Reference/source |
|---|---|---|
| N402 | cspA1, derivative of N400 | Bos et al. (1988) |
| AB4.1 | pyrG-, derivative of N402 | van Hartingsveldt et al. (1987) |
| MA234.1 | Δku70::DR_amdS_DR in MA169.4 | Alazi et al. (2016) |
| MA70.15 | Δku70::amdS in AB4.1 | Meyer et al. (2007) |
| MA299.2 | Δku70 in MA70.15 | Niu et al. (2015) |
| MA323.1 | Δku70::amdS, ΔnicB-, pyrG- | Niu et al. (2016) |
| JC1.5 | pgaX-amdS in MA299.2, pyrG+ | Niu et al. (2015) |
| JN29.2 | ΔcreA::hygB in JC1.5 | Niu et al. (2015) |
| JN38 | Spontaneous mutation S1 in JN29.2 | This study |
| JN39 | Spontaneous mutation S2 in JN29.2 | This study |
| JN42 | Spontaneous mutation S5 in JN29.2 | This study |
| JN44 | Spontaneous mutation S7 in JN29.2 | This study |
| JN52 | UV1 in JN29.2 | This study |
| JN53 | UV2 in JN29.2 | This study |
| JN54 | UV3 in JN29.2 | This study |
| JN55 | UV4 in JN29.2 | This study |
| JN56 | UV5 in JN29.2 | This study |
| JN57 | UV6 in JN29.2 | This study |
| JN58 | UV7 in JN29.2 | This study |
| JN59 | UV8 in JN29.2 | This study |
| JN60 | UV9 in JN29.2 | This study |
| JN61 | UV10 in JN29.2 | This study |
| JN62 | UV11 in JN29.2 | This study |
| JN63 | UV12 in JN29.2 | This study |
| JN64 | UV13 in JN29.2 | This study |
| JN122.1, JN122.2 and JN122.3 | ΔgaaX::phleo in JN29.2 | This study |
| JN123.1, JN123.2, JN123.3 | ΔgaaX::hygB in JC1.5 | This study |
| JN125.1 | ΔgaaX::nicB in MA323.1 | This study |
| JN126.2, JN126.5, JN126.6 | PgaaX-gaaX::GFP-TgaaX in JN125.1 | This study |
| JN127.1, JN127.2, JN127.3 | PgaaX-GFP::GaaX-TgaaX in JN125.1 | This study |
Isolation of mutants with constitutive expression of genes involved in PGA utilization
A. niger strain JN29.2 (Table 1) was used for the selection of mutants with constitutive expression of genes involved in PGA utilization. Spontaneous mutants were obtained by plating out freshly harvested and myracloth-filtered conidia (1 × 104 conidia per plate) on MM glucose/acetamide plates and incubated at 30° for 5 days. In addition, mutants were obtained after mild UV mutagenesis (80% survival) as described (Damveld et al. 2008). Individual mutants growing on the primary MM-glucose/acetamide selection plates were purified twice on the MM-glucose/acetamide agar plates. In total, 14 spontaneous mutants and 59 UV-mutants were isolated that grew well on MM-glucose/acetamide agar plates and they were potential mutants with constitutive expression of genes involved in PGA utilization. To identify mutants constitutively producing PGA-degrading enzymes, all 73 mutants were grown by inoculating 5 × 107 spores in 50 ml MM-glucose medium for 36 hr at 30° with shaking at 150 rpm. Supernatant of each culture was harvested by filtration. The extracellular culture fluid and the mycelia were stored at −80° for enzymatic assays and RNA extraction, respectively. A total of 10 μl supernatant of each sample was spotted on PGA plates made by dissolving 0.2% PGA (Sigma) in NaAc buffer (pH 4.2) with 1% agarose (Sphaero). The PGA plate assay was modified from the protocol used to detect cellulase activity (Teather and Wood 1982). Plates were incubated at 37° for 17 hr after spotting. PGA was stained by flooding the plates with a filter-sterile 0.05% solution of Congo Red (Sigma) dissolved in Milli-Q water for 15 min. The Congo Red solution was then poured off and the plates were washed with Milli-Q water and further treated by flooding with 1 M NaCl for 15 min. The formation of a clear zone of hydrolysis indicated PGA degradation.
The constitutive expression of genes involved in PGA degradation was further determined by Northern blot analysis. Total RNA was isolated from eleven UV-mutants and two spontaneous mutants from frozen mycelia using TRIzol reagent (Invitrogen, Carlsbad, CA). Quantification and purity assessment of total RNA was done by spectrophotometric method (NanoDrop 2000; Thermo Scientific). Total RNA (3.5 μg) was loaded per sample and blotted to a Hybond-N+ nylon membrane (Amersham, GE Healthcare) followed by hybridization with [α-32P]-dCTP–labeled probes (Rediprime II kit; Amersham, GE Healthcare). Probes were PCR-amplified using the N402 genomic DNA and the primer pairs are listed in Supplemental Material, Table S1. Standard molecular techniques were applied as described (Sambrook and Russell 2001).
DNA sequencing and data analysis
Sequencing of the gaaR gene from 11 constitutive mutants was performed by PCR amplification of the gaaR gene, including 137 bp upstream and 152 bp downstream sequences, using genomic DNA of the mutants as template and primers gaaRP7f and gaaRP8r (Table S1). Genomic DNA was isolated as described (Arentshorst et al. 2012). The PCR fragment (2765 bp in size) was sequenced in both directions using gaaR sequencing primers (Table S1). Sequencing was performed by Macrogen Europe (Amsterdam, The Netherlands).
Genomic DNA of three spontaneous mutants and two UV mutants was isolated as described (Arentshorst et al. 2012), and was further purified with DNA Isolation Kit (MO BIO Laboratories) for whole-genome DNA sequencing. The mutant genomes were sequenced at the McGill University Génome Québec Innovation Centre (QC, Canada) using the Illumina HiSeq platform to ∼50-fold coverage. The DNA reads were aligned to the NRRL3 genome with Bowtie2 (Langmead and Salzberg 2012) and sequence differences were detected with Freebayes (Garrison and Marth 2012).
Deletion of gaaX gene
Deletion of the gaaX gene (NRRL3_08194) in the JC1.5, JN29.2, and MA323.1 backgrounds (Table 1) was carried out using the split marker approach (Arentshorst et al. 2015). The 869 bp 5′-flank and 870 bp 3′-flank regions were PCR amplified with the primers listed in Table S1, using N402 genomic DNA as template. These PCR fragments were used in fusion PCRs with hygromycin, phleomycin resistance genes, or the nicB gene (Niu et al. 2016) to generate the split marker fragments. After amplification, the 5′flank-hyg and 3′flank-hyg fragments were transformed to the recipient strain JC1.5, the 5′flank-phleo and 3′flank-phleo fragments were transformed to the recipient strain JN29.2, and the 5′flank-nicB and 3′flank-nicB fragments were transformed to the recipient strain MA323.1. Putative gaaX disruption strains were purified by two consecutive single colony streaks. Genomic DNA was isolated as described (Arentshorst et al. 2012) and Southern blot hybridizations, using PCR-amplified fragments generated with primers listed in Table S1 as probes, were performed to confirm proper deletion and to exclude additional integrations.
Bioreactor cultivation
Controlled bioreactor cultivations for A. niger strains MA234.1 and JN123.1 were performed in 6.6-L BioFlo3000 bioreactors (New Brunswick Scientific) as previously described (Jørgensen et al. 2010). Briefly, autoclaved bioreactor vessels were filled with 5 liter of sterile MM with 0.75% fructose. During cultivation at 30°, the controller was set to maintain pH 3 by addition of titrants (2 M NaOH or 1 M HCl). Sterile air was supplied at a rate of 1 liter/min. Prior to inoculation, 1.5 ml of 10% (w/v) filter-sterilized yeast extract was added to enhance conidial germination. Cultures were inoculated with freshly harvested spores at a concentration of 7.0 × 108 conidia per liter. To reduce the loss of hydrophobic conidia during germination, the stirrer speed was set to 250 rpm and the culture was aerated via the headspace during the first 6 hr after inoculation. Subsequently, the stirrer speed was increased to 750 rpm, 0.5 ml of polypropyleneglycol P2000 was added as an antifoam agent, and air was supplied via the sparger. Cultures broth was harvested at regular intervals from batch cultures and mycelial biomass was retained by vacuum filtration using glass microfiber filters (Whatman). Both biomass and filtrate were quickly frozen in liquid nitrogen and subsequently stored at −80°. Dry biomass concentrations were gravimetrically determined from lyophilized mycelia originating from a known mass of culture broth.
Transcriptome analysis
Mycelia grown in bioreactors to midexponential phase were used to isolate RNA using TRIzol reagent (Invitrogen) and purified with NucleoSpin RNA Clean-up kit (Macherey-Nagel) with DNase treatment. Quantity and quality of the RNA samples were determined with a NanoDrop 2000 spectrophotometer and by RNA gel electrophoresis, respectively. RNA sequencing was conducted by Genome scan (Leiden, the Netherlands). Briefly, messenger RNA was isolated from the total RNA using NEBNext Ultra Directional RNA Library Prep Kit for Illumina, according to the manufacturer’s protocol. After fragmentation of the messenger RNA, complementary DNA was synthesized using random primers; after a second-strand complementary DNA synthesis reaction, fragments were ligated to the sequencing adapters. Clustering and DNA sequencing was performed using the Illumina NextSequation 500 SR75. We refer to A. niger gene IDs based on the most up-to-date and accurate annotation of the A. niger NRRL3 genome (http://genome.fungalgenomics.ca/). The RNA-seq reads were cleaned by correcting sequencing errors with Rcorrector (Song and Florea 2015), trimming sequencing adapters and low quality sequences with Skewer (Jiang et al. 2014), and removing ribosomal RNA with SortMeRNA (Kopylova et al. 2012). The cleaned reads were mapped to NRRL3 transcripts and counted with Salmon (Patro et al. 2016), and the read counts were analyzed for differences in transcript expression between genotypes with DESeq2 (Love et al. 2014).
Construction of strains expressing GaaX-GFP or GFP-GaaX fusion proteins
To construct fusions of GFP to the N-terminus or C-terminus of GaaX, PgaaX_GFP::GaaX_TgaaX and PgaaX_GaaX::GFP_TgaaX constructs were generated using a fusion-PCR approach in which N402 genomic DNA, as well as plasmid PagsA_eGFP_TtrpC (Damveld et al. 2008), were used as template DNA. For constructing the PgaaX_GFP::GaaX_TgaaX construct, the promoter region of gaaX was PCR amplified using primers PgaaX_P7f-NotI and PgaaX_P11r, GFP was PCR amplified from plasmid PagsA_eGFP_TtrpC using primers GFP_P1f and GFP_P3r, gaaX and the terminator region of gaaX was PCR amplified using primers gaaX_P12f and TgaaX_P10r-NotI, and the three fragments were combined together in a two-step fusion PCR. Two amino acids (Gly-Ala) were introduced as spacer between GFP and GaaX. Subsequently, the fusion fragment was cloned into vector pJet1.2 to give plasmid pJN34. For the PgaaX_GaaX::GFP_TgaaX construct, gaaX with the promoter region of gaaX was PCR amplified using primers PgaaX_P7f-NotI and gaaX_P8r, GFP was PCR amplified using primers GFP_P1f and GFP_P2r, the terminator region of gaaX was PCR amplified using primers gaaX_P9f and TgaaX_P10r-NotI, and the three fragments were combined together in a two-step fusion PCR. Again, a Gly-Ala spacer was introduced between GaaX and GFP. Subsequently, the fusion fragment was cloned into vector pJet1.2 to give plasmid pJN35.
Plasmids pJN34 and pJN35 were digested by NotI, and the fragments containing PgaaX_GFP::GaaX_TgaaX and PgaaX_GaaX::GFP_TgaaX were cloned into pMA334 (Arentshorst et al. 2015) to generate pJN36 and pJN37, respectively. The pMA334 plasmid was designed such that the reporter constructs are targeted to the pyrG locus. Plasmids pJN36 and pJN37 were linearized by AscI digestion and purified from gel before transformation to A. niger strain JN125.1. Proper integration of the PgaaX_GFP::GaaX_TgaaX or PgaaX::GaaX_GFP_TgaaX fragments at the pyrG locus was confirmed by Southern blot analysis using PCR-amplified fragments generated with primers listed in Table S1 as probes.
Microscopy
For microscopic analysis, conidia of strains MA323.1, JN125.1, JN126.2, and JN127.3 were inoculated on coverslips in Petri dishes. Liquid MM supplemented with 50 mM GA or 50 mM fructose as the carbon source was used. After incubation at 30° for 16 hr, the coverslips with adherent germlings were mounted upside down on glass slides and observed under a confocal laser scanning microscope (Zeiss Imager; Zeiss, Jena, Germany), equipped with an LSM 5 exciter using 63× objectives. Images were processed by ImageJ with the exact same brightness and contrast adjustments and the median filter (radius 1.0).
Data availability
Strains are listed in Table 1 and are available upon request. Table S2 contains SNPs and indels detected in genomes of mutants. Table S3A contains transcript per million (TPM) values of NRRL3 gene models in wild type and the gaaX mutant, and Table S3B contains their DEseq2 analysis. The DNA reads described in this study are deposited in the Short Read Archive under accession number SRP078415. The RNA reads described in this study are deposited in the Short Read Archive under accession number SRP078485.
Results
Mutants constitutively expressing genes related to galacturonic acid utilization
To identify mutants that constitutively express genes related to PGA degradation and GA utilization in A. niger, we designed a forward screening procedure using a reporter strain containing a PpgaX-amdS reporter construct for positive selection of the desired mutants. We recently showed that the pgaX gene is specifically induced by GA, PGA, and pectin, allowing the reporter PpgaX-amdS strain to grow on acetamide as a nitrogen source when GA, PGA, or pectin is present as a carbon source (Niu et al. 2015). We also showed that deletion of the CCR protein (CreA) did not result in growth of the PpgaX-reporter strain on glucose, indicating that derepression via creA deletion was not sufficient to drive PpgaX-amdS expression to sustain growth. For the mutant screen, we used the PpgaX-amdS reporter strain in the ΔcreA background (JN29.2) to prevent interference with possible CreA pathway-related repression mechanisms. Spores of A. niger strain JN29.2 were UV-mutagenized and surviving spores (80%) were plated on MM-glucose-acetamide plates. After mutagenesis, 59 mutants were isolated based on growth on acetamide. In addition to UV-generated mutants, 14 spontaneous mutants were isolated, resulting in a total of 73 mutants that could grow on glucose/acetamide plates.
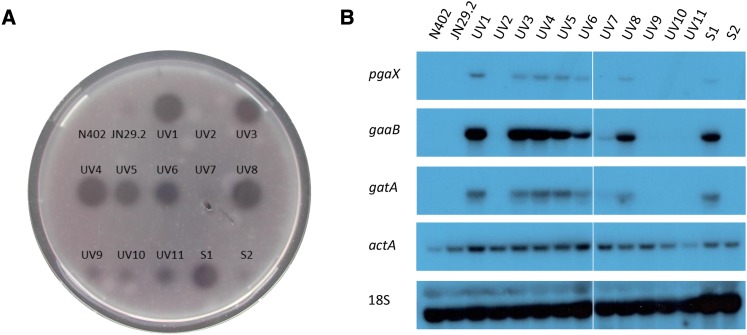
To determine whether mutations were cis- or trans-acting, mutants were cultured in glucose medium for 36 hr and the medium was analyzed for polygalacturonase activity. Initial experiments showed that cultivation of JN29.2 in glucose medium resulted in very low polygalacturonase levels and no halo was formed on Congo Red-stained PGA plates when culture medium was spotted on a PGA plate (Figure 1A). We reasoned that if the mutation is trans-acting, the medium should contain increased levels of both exo- and endo-polygalacturonases, leading to the formation of a halo. On the other hand, if the mutation is cis-acting, thereby only affecting the PpgaX-amdS reporter construct, it would not result in a halo on a PGA plate. Based on this assay, we concluded that the mutations of 65 out of 73 mutants are trans-acting, while the remaining eight mutants carry presumed cis-acting mutations.
Figure 1.
Enzymatic and RNA blot analysis of mutants with constitutive expression of genes involved in polygalacturonic acid utilization. (A) Supernatant (10 µl) from glucose-grown cultures of reference strains N402 and JN29.2, 11 UV mutants, and two spontaneous mutants were spotted on polygalacturonic acid agarose medium to detect polygalacturonase activity. (B) Northern blot analysis of selected GA-responsive genes in the reference strains N402 and JN29.2, 11 UV mutants, and two spontaneous mutants.
To further demonstrate that the presumed trans-acting mutations indeed affected expression of multiple genes related to GA utilization and belonging to the GA-induced genes (Martens-Uzunova and Schaap 2008; Niu et al. 2015; Alazi et al. 2016), the expression of three GA-induced genes (pgaX, gatA, and gaaB) was examined by Northern blot analysis in a subset of mutants after growth on glucose. The pgaX, gatA, and gaaB genes encode an exo-polygalacturonase, a GA-specific transporter, and the L-galactonic acid dehydratase involved in the GA release from PGA, the uptake of GA, and subsequent metabolism of GA, respectively. As shown in Figure 1B, expression of these genes was not detected in the wild type (N402) and the parent JN29.2 (ΔcreA, PpgaX-amdS) strains, whereas these genes were expressed in the constitutive mutants that displayed increased polygalacturonase activity. The presumed cis-acting mutants from the plate assay (UV2, UV7, UV9, UV10, UV11, and S2) did not constitutively express pgaX, gatA, and gaaB, and showed a small halo on PGA plates, indicating the halo assay can be used to discriminate between cis- and trans-acting mutants. To determine whether a cis-acting mutation in the pgaX promoter in front of the amdS gene was responsible for the ability of this class of mutants to grow on acetamide, the pgaX promoter in front of the amdS gene of all eight cis-acting mutants was PCR amplified using pyrG- and amdS-specific primers (Table S1). This analysis revealed no mutations in the pgaX promoter region of any of the eight presumed cis-acting mutants. Hence, the nature of the mutation(s) in these strains that allows growth on acetamide remains unknown. A possibility could be the activation of expression of endogenous amdS genes, as at least four amdS-like genes are present in the genome of A. niger.
Identification of mutations responsible for the constitutive expression of the galacturonic acid utilization genes
A possible explanation for the constitutive expression of GA utilization genes in the mutants is that they carry mutations in the recently identified GaaR transcriptional activator (Alazi et al. 2016). We therefore PCR amplified and sequenced the gaaR locus of eight constitutive mutants obtained after UV mutagenesis (UV1, UV3, UV4, UV5, UV6, UV8, UV12, and UV13) and three spontaneous mutants (S1, S5, and S7). The gaaR coding regions as well as 300 bp flanking regions were sequenced, but no mutation in the gaaR gene in any of these eleven mutants was found (data not shown).
To determine whether the constitutive expression of GA-induced genes in these mutants involves a functioning GaaR transcription factor, we deleted the gaaR gene in seven of the constitutive mutants (UV1, UV3, UV4, UV5, UV6, UV8, and S1) and analyzed constitutive expression using the PpgaX-amdS reporter. All seven mutants were unable to grow on glucose/acetamide plates (data not shown), indicating that the constitutive expression of the GA-induced genes requires a functional gaaR gene.
To identify mutation(s) in the gene(s) responsible for the constitutive phenotype, the genomes of five mutants (UV1, UV8, S1, S5, and S7) and the parental strain JN29.2 were sequenced. Table 2 summarizes the number of SNPs and indels detected in the five mutant strains and Table S2 lists positions and type of all SNPs and indels detected. Spontaneous mutant S7 contains only 11 SNPs or indels, of which 10 are located in intergenic regions and only one SNP mutated a gene (NRRL3_08194). Remarkably, in all four other mutants a mutation was found in the same gene. Two of the mutants carry nonsense mutations (UV1 and S5) and one carries a frame-shift mutation (S3), all leading to premature stop codons and predicted to result in truncated proteins (Table 2). Mutants S7 and UV8 have missense mutations in the C-terminal part of the protein. These results strongly suggest that the constitutive expression of genes encoding pectin-degrading enzymes in the five mutants is caused by a loss of function of the protein encoded by NRRL3_08194.
Table 2. Mutations in the constitutive mutants as compared to the parental strain JN29.2.
| Strain | Total number of SNPs and indels | SNPs or indels in coding region | Mutation in NRRL3_08194 | Position of mutation relative to ATG of NRRL3_08194 | Mutation in codon (bold) | Amino acid change | Predicted protein length (full length protein is 697 amino acids) |
|---|---|---|---|---|---|---|---|
| JN29.2_UV1 | 40 | 19 | G-T | 1372 | GAG-TAG | E to Stop | 457 aa |
| JN29.2_UV8 | 21 | 4 | G-A | 1577 | GGA-GAA | G to E (526) | 697 aa |
| JN29.2_S1 | 68 | 34 | Extra G | 1958 | GTT-GGT | V to G out of frame | 663 aa |
| JN29.2_S5 | 48 | 11 | C-T | 1105 | CAA-TAA | Q to Stop | 368 aa |
| JN29.2_S7 | 11 | 1 | T-C | 2027 | CTG-CCG | L-P (676) | 697 aa |
Deletion of NRRL3_08194 results in the constitutive expression of genes required for PGA breakdown and GA catabolism
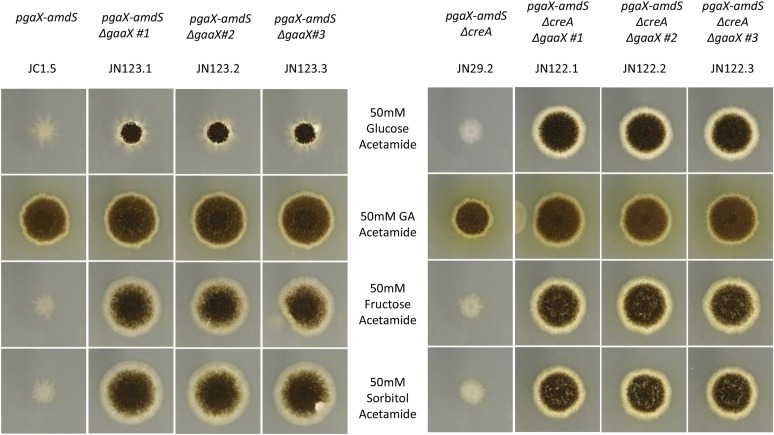
To determine whether the constitutive expression of the target genes of the GaaR transcriptional activator (Alazi et al. 2016) is caused by a loss-of-function mutation in NRRL3_08194, we deleted this gene in the pgaX-amdS reporter strains JC5.1 and JN29.2 (ΔcreA), as well in a parental background without reporter constructs (MA323.1) (Table 1). The deletion mutants were purified and deletion of NRRL3_08194 was confirmed by Southern blot analysis for JC5.1, JN29.2, and MA323.1 (Figure S1 and Figure S2). The verified deletion mutants were tested for growth on acetamide plates containing different carbon sources (Figure 2), as well as for constitutive expression of polygalacturonases (see section GaaX is induced on galacturonic acid and localized in the cytosol). Figure 2 shows that deletion of NRRL3_08194 in the ΔcreA background (JN122) resulted in the ability to grow on glucose/acetamide, fructose/acetamide, and sorbitol/acetamide. The colony size on all three different carbon sources was similar, indicating that the amdS gene was expressed regardless of the carbon source used. However, deletion of NRRL3_08194 in the JC1.5 reporter strain (JN123) resulted in similar growth on fructose/acetamide and sorbitol/acetamide plates as JN122, but reduced growth on glucose. This indicates that glucose-mediated CCR repressed PpgaX-driven amdS expression even in the absence of NRRL3_08194. The ability of the ΔNRRL3_08194 strain to grow on acetamide plates strongly suggests that a loss of function of NRRL3_08194 results in constitutive expression of pgaX and other pectinolytic genes. Furthermore, deletion of gaaX did not result in an altered growth behavior on GA, PGA, apple pectin, glucose, fructose, sorbitol, xylose, and arabinose (data not shown). These results are most easily explained by proposing that NRRL3_08194 encodes a repressor protein, which we name GaaX, that represses the activity of the GaaR transcription factor in the absence of GA. Interestingly, GaaX shows sequence similarity to a previously identified repressor protein, QutR (Grant et al. 1988). Moreover, the transcriptional activator (GaaR, NRRL3_08195) and the repressor (GaaX, NRRL3_08194) are clustered in the genome, similar to the quinic acid utilization transcriptional activator (QutA/Qa-1F) and repressor (QutR/Qa-1S) in Aspergillus nidulans and Neurospora crassa, respectively (Geever et al. 1989; Levesley et al. 1996).
Figure 2.
Regulation of the pgaX expression is controlled by GaaX and by CreA-mediated glucose repression. Growth of pgaX-amdS reporter strains in gaaX deletion and their parental strains was examined on various carbon sources. Parental strains and corresponding deletion strains were grown on MM-acetamide supplemented with 50 mM glucose, galacturonic acid (GA), fructose, or sorbitol. All strains carry the PpgaX-amdS reporter construct. Strain JC1.5 is the parent of three independent transformants (JN123.1, JN123.2, and JN123.3) that contain a deletion in the gaaX gene. Strain JN29.2 carries the ΔcreA marker and is the parent of three independent transformants (JN122.1, JN122.2, and JN122.3) that contain a deletion in the gaaX gene.
The GaaR–GaaX target gene regulon
We posit that the regulation of GA-responsive genes is likely to be negatively controlled by the repressor protein GaaX. A possible mode of action is that the repressor GaaX inhibits the activity of the transcriptional activator GaaR in the absence of an inducer. This would imply that deletion of the repressor or activation of the transcription factor by growth on GA would result in activation of the same set of genes. To show that the loss of function of the repressor activates the GA regulon and to identify the genes repressed by GaaX under noninducing conditions, RNA-seq profiles of the ΔgaaX and its parental strain (MA234.1) were compared after growth on fructose, a nonrepressing carbon source. Controlled cultivations in bioreactors showed that the growth rates [μmax parental strain 0.214 ± 0.007 g dry wt/kg/hr (n = 3); μmax ΔgaaX 0.223 ± 0.004 g dry wt/kg/hr (n = 2)] as well as biomass yields [Ymax parental strain 4.15 ± 0.13 g dry wt/kg (n = 3); Ymax ΔgaaX 4.29 ± 0.19 g dry wt/kg (n = 2)] of the two strains were highly comparable, indicating the gaaX deletion did not result in major physiological changes affecting the growth and biomass yield.
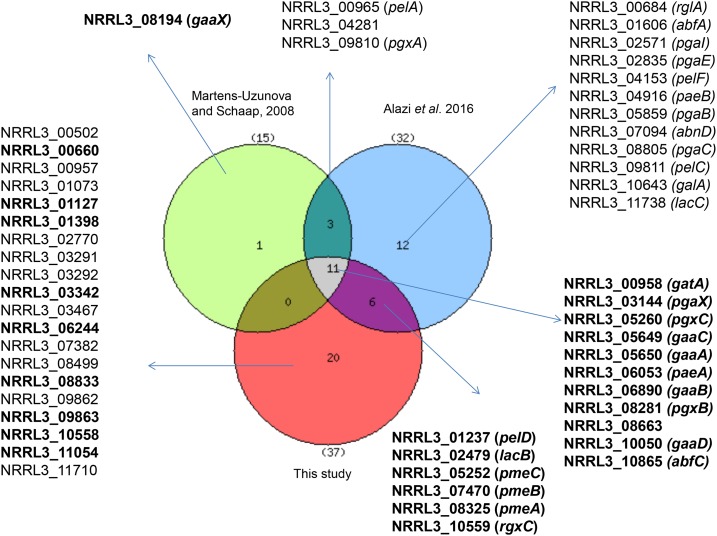
To identify differentially expressed genes in the ΔgaaX strain as compared to its parental strain, RNA-seq was performed on RNA isolated from exponentially growing cells at the time point at which about 75–80% of the maximum biomass yield was reached. RNA-seq reads were mapped to the NRRL3/N400 genome as this is the parent of the laboratory strain N402 and derivatives used in this study. TPM values were calculated using Salmon (Patro et al. 2016) (Table S3). Analysis of differential gene expression, based on a stringent false discovery rate (FDR) <0.001 and a fold change (FC) >4.0, identified 37 upregulated genes (Table 3). Gene Ontology (GO) enrichment analysis using FetGOat (Nitsche et al. 2012) and manual inspection of the genes upregulated in the ΔgaaX mutant indicated that genes involved in pectin catabolism were highly enriched. Of the 37 genes, 16 are predicted to encode extracellular enzymes acting on the GA backbone of pectin or acting on pectin side chains (Table 3). Nine genes in the group of 37 upregulated genes in the ΔgaaX strain are predicted to encode intracellular proteins. Four of these nine genes (gaaA–gaaD) are required for the conversion of GA into pyruvate and glycerol (Martens-Uzunova and Schaap 2008). The exact role of the other five genes and their possible role in GA catabolism is currently unknown. The group of 37 upregulated genes also includes seven genes predicted to encode sugar transporter proteins. Of these seven transporter-encoding genes, only GatA has been studied in detail and shown to be able to transport GA (Sloothaak et al. 2014). Apart from the genes encoding extracellular enzymes (16), transporters (seven), and enzymes possibly involved in GA catabolism (nine), the remaining five genes in this group encode proteins with unknown functions or with similarities to known proteins that, for now, cannot be directly linked to GA metabolism. The deletion of gaaX has the most profound effect on the transcript levels of the genes encoding the first three steps of the GA utilization pathway (gaaA, gab, and gaaC) and on the expression of gatA. Deletion of gaaX resulted in a 1.24-fold (P = 0.000035) increase in gaaR gene activity. Since the upregulation of gaaR in the ΔgaaX mutant is modest, it is likely that the repressing activity of GaaX is mediated at the protein level (e.g., by interacting with GaaR) rather than by transcriptional control of gaaR. Seventeen of the 37 genes upregulated in the ΔgaaX mutant were previously identified as part of the GA regulon (Martens-Uzunova and Schaap 2008; Alazi et al. 2016) (Figure 3 and Table 3). Sixteen of the 17 genes found in common with previous studies are predicted or demonstrated to encode extracellular pectin-degrading enzymes. These results indicate that loss of function of gaaX affects the expression of the GA regulon. The other 20 genes were identified as significantly upregulated in the gaaX mutant, but these were not identified previously as being part of the GA regulon (Figure 3 and Table 3). A re-examination of the expression of these 20 genes in the RNA-seq data published earlier (Alazi et al. 2016) indicated that 10 of the genes (indicated in Table 3 by the asterisk, Table S4) were also GA-induced or GaaR-dependent for induction in this previous study. On the other hand, 15 genes identified to be GA-induced in a GaaR-dependent manner in the previous study (Alazi et al. 2016) were not significantly upregulated in the gaaX deletion strain (Figure 3). These results therefore suggest that full induction of GA-inducible genes requires more than the loss of GaaX activity, and that an additional induction mechanism plays a role.
Table 3. Comparative RNA-seq analysis between wild-type and ΔgaaX strains: genes (37) upregulated in ΔgaaX strain.
| Gene ID (NRRL3) | Gene ID (CBS513.88) | Gene name | Description | Average expression value wta | Average expression value ΔgaaXa | FC ΔgaaX vs. wtb | False discovery rateb | Predicted localization | GARE elementc |
|---|---|---|---|---|---|---|---|---|---|
| NRRL3_05649 | An02g07720d,e | gaaC | L-threo-3-deoxy-hexylosonate aldolase | 12.54 | 2283.77 | 169.39 | 0.00E+00 | Intracellular | (-) −292, −606 |
| NRRL3_09863f | An11g03500 | α-Hydroxy acid dehydrogenase | 0.82 | 165.84 | 160.07 | 0.00E+00 | Intracellular | (+)−543, (−)-182 | |
| NRRL3_00958 | An14g04280d,e | gatA | MFS-type sugar/inositol transporter | 3.47 | 524.95 | 140.38 | 0.00E+00 | Membrane | (+)−360 |
| NRRL3_06890 | An16g05390d,e | gaaB | L-galactonic acid dehydratase | 47.77 | 6256.70 | 121.49 | 0.00E+00 | Intracellular | (+)−326 |
| NRRL3_03291 | An12g05600 | Heterokaryon incompatibility protein | 0.00 | 6.78 | 82.98 | 6.48E-78 | Intracellular | (+)−737, −325 | |
| NRRL3_05650 | An02g07710d,e | gaaA | D-galacturonic acid reductase | 19.92 | 1515.44 | 71.84 | 0.00E+00 | Intracellular | (+)−414, −100 |
| NRRL3_03144 | An12g07500d,e | pgaX | Exo-polygalacturonase | 1.36 | 51.63 | 32.66 | 2.60E-272 | Extracellular | (+)−388 |
| NRRL3_05252 | An02g12505e | pmeC | Pectin methylesterase | 1.06 | 31.32 | 25.03 | 7.10E-189 | Extracellular | (+)−275, −246, −35 |
| NRRL3_06244f | An02g00140 | Glycoside hydrolase family 43 protein | 0.81 | 22.19 | 23.46 | 8.13E-193 | Extracellular | (+)−96, (−)−712 | |
| NRRL3_08281 | An03g06740d,e | pgxB | Exo-polygalacturonase Pgx28B | 0.00 | 2.10 | 22.95 | 3.59E-43 | Extracellular | (−)−298, −823 |
| NRRL3_10559 | An18g04810e | rgxC | Glycoside hydrolase family 28 protein | 0.08 | 3.11 | 17.32 | 1.33E-45 | Extracellular | (+)−852, (−)−250 |
| NRRL3_05260 | An02g12450d,e | pgxC | Exo-polygalacturonase Pgx28C | 0.95 | 16.77 | 15.22 | 1.83E-144 | Extracellular | (+)−268, (−)−642 |
| NRRL3_08663 | An03g01620d,e | MFS-type sugar/inositol transporter | 0.27 | 5.62 | 14.28 | 8.28E-46 | Membrane | (+)−673 | |
| NRRL3_03342f | An12g04990 | Short-chain dehydrogenase/reductase | 0.81 | 14.46 | 13.51 | 2.10E-62 | Intracellular | None | |
| NRRL3_10865 | An08g01710d,e | abfC | α-N-arabinofuranosidase | 0.80 | 11.21 | 11.92 | 1.19E-84 | Extracellular | None |
| NRRL3_09862 | An11g03510 | Hypothetical protein | 0.00 | 0.93 | 11.05 | 4.32E-18 | Unknown | (−)−517, −829, −843 | |
| NRRL3_10050 | An11g01120d,e | gaaD/larA | NADPH-dependent erythrose reductase Err1 | 256.41 | 2732.37 | 10.16 | 0.00E+00 | Intracellular | (−)−538, −583, −801 −813 |
| NRRL3_00957 | An14g04260 | B3/B4 domain-containing protein | 1.53 | 18.56 | 10.05 | 2.46E−64 | Unknown | None | |
| NRRL3_00502 | An09g06200 | Hypothetical protein | 0.93 | 12.38 | 9.36 | 1.08E−32 | Unknown | (−)−189 | |
| NRRL3_10558f | An18g04800 | α-L-rhamnosidase | 0.35 | 3.85 | 9.09 | 2.19E−61 | Extracellular | (+)−365 | |
| NRRL3_11710 | An06g00620 | MFS-type sugar/inositol transporter | 2.86 | 25.29 | 8.05 | 1.39E−118 | Membrane | (+)−487, (−)−368 | |
| NRRL3_06053 | An02g02540d,e | paeA | Carbohydrate esterase family 16 protein | 2.06 | 17.62 | 7.76 | 5.39E−107 | Extracellular | (+)−1238 |
| NRRL3_01073 | An14g05840 | O-methyltransferase, COMT-type | 0.54 | 5.57 | 7.16 | 1.06E−21 | Intracellular | (+)−300 | |
| NRRL3_07382 | An16g00540 | α-L-fucosidase | 0.04 | 0.67 | 7.07 | 5.92E−14 | Extracellular | (−)−606 | |
| NRRL3_00660f | An14g00860 | Carboxylesterase | 0.21 | 1.87 | 6.98 | 1.47E−31 | Extracellular | None | |
| NRRL3_08499 | An03g03960 | Uncharacterized protein | 0.48 | 4.58 | 6.71 | 3.84E−21 | Unknown | None | |
| NRRL3_07470 | An04g09690e | pmeB | Pectin methylesterase | 0.75 | 5.72 | 6.36 | 3.59E−43 | Extracellular | (+)−389 |
| NRRL3_02479 | An01g10350e | lacB | Exo-β-1,4-galactanase | 3.48 | 22.29 | 6.03 | 1.37E−179 | Extracellular | None |
| NRRL3_03292 | An12g05590 | Carboxylesterase | 0.18 | 1.86 | 5.26 | 1.75E−09 | Extracellular | (−)−1000 | |
| NRRL3_01237 | An19g00270e | pelD | Pectin lyase | 0.17 | 1.47 | 5.25 | 6.14E−14 | Extracellular | (−)−409, −465 |
| NRRL3_03467 | An12g03550 | MFS-type transporter | 1.13 | 6.73 | 5.05 | 4.03E−28 | Membrane | None | |
| NRRL3_01127f | An14g06500 | DHAK | Dihydroxyacetone kinase | 18.90 | 100.64 | 4.98 | 1.61E−102 | Intracellular | (+)−69 |
| NRRL3_11054f | An08g04040 | MFS-type sugar/inositol transporter | 6.62 | 31.89 | 4.57 | 2.57E−126 | Membrane | (+)−778, −726, (−)−646 | |
| NRRL3_08833f | N.a. | Hypothetical protein | 0.31 | 2.02 | 4.52 | 1.58E−12 | Unknown | None | |
| NRRL3_01398 | An13g02090 f | MFS-type transporter | 2.81 | 13.10 | 4.16 | 5.62E−30 | Membrane | (−)−270 | |
| NRRL3_08325 | An03g06310e | pmeA | Pectin methylesterase Pme8A | 0.04 | 0.56 | 4.10 | 1.49E−07 | Extracellular | (+)−983, (−)−308 |
| NRRL3_02770 | An01g13880 | MFS-type transporter | 0.87 | 4.07 | 4.06 | 1.57E−20 | Membrane | (+)−578 |
Gray boxes indicate genes proposed to be part of the GaaR/GaaX regulon.
Expression values (TPM) are averages of biological triplicates for the wild type (wt) and duplicates for the ΔgaaX strain.
Fold changes and FDR-values are calculated with DESeq2, and genes with fold changes ≥4 and FDR-values ≤0.001 are listed.
GA-responsive element (GARE) position (CCNCCAA) is given with respect to the translational start site.
Genes identified as the GA-regulon by Martens-Uzunova and Schaap (2008).
Genes identified as the GA regulon by Alazi et al. (2016).
Gene not identified as components of the GA regulons in the studies by Martens-Uzunova and Schaap (2008) and Alazi et al. (2016), but are likely to be under the regulation of GaaR (see text for details).
Figure 3.
Venn diagram showing the overlaps between upregulated genes in the wt_fructose vs. wt_GA study (Martens-Uzunova and Schaap 2008), the upregulated genes between ΔgaaR-GA vs. wt-GA (Alazi et al. 2016), and the upregulated genes in wt_fructose vs. ΔgaaX-fructose (this study) to identify the GA regulon. The 27 genes defining the GaaR–GaaX core regulon are indicated in bold.
An additional GA-induced gene identified in the study of Martens-Uzunova and Schaap (2008) but missing in the GaaR study is gaaX itself. Expression of gaaX was not examined in the Alazi et al. (2016) study as its function was not yet directly linked to GA utilization. However, re-evaluation of the dataset revealed that the induced expression of gaaX on GA is dependent on GaaR (FC of wild type vs. ΔgaaR: 18.7; P = 0.003; Table S4). Combining the expression data of the ΔgaaX mutant (this study), the ΔgaaR mutant (Alazi et al. 2016) and the genes induced on GA (Martens-Uzunova and Schaap 2008), we propose a panregulon of 53 GaaR–GaaX controlled genes and a core GaaR–GaaX regulon of at least 27 genes (Figure 3, Table 3, and Table S4). These 27 genes include 11 genes present in the intersection of all three data sets, six genes present in the intersection of the ΔgaaX data and the ΔgaaR data (Alazi et al. 2016), nine genes identified by examining the gaaX dataset with supporting evidence from previous studies, and gaaX (Figure 3). Of these 27 genes, all except NRRL3_00660 (carboxyesterase), NRRL3_10865 (α-N-arabinofuranosidase), NRRL3_03342 (short-chain dehydrogenase/reductase), NRRL3_08833 (hypothetical protein), and NRRL3_02479 (β-galactosidase), have at least one predicted GARE motif in the upstream regions of the coding region (Table 3). It is interesting to note that among the genes listed in Figure 3 and Table 3 that are upregulated in the ΔgaaX, some of them (NRRL3_00957 and NRRL3_00958; NRRL3_09862 and NRRL3_09863; NRRL3_03291 and NRRL3_03292) are clustered. Except for NRRL3_00958, which encodes a GA-specific transporter (Sloothaak et al. 2014), the possible role of these genes in pectin degradation is currently unknown.
GaaX is induced on galacturonic acid and localized in the cytosol
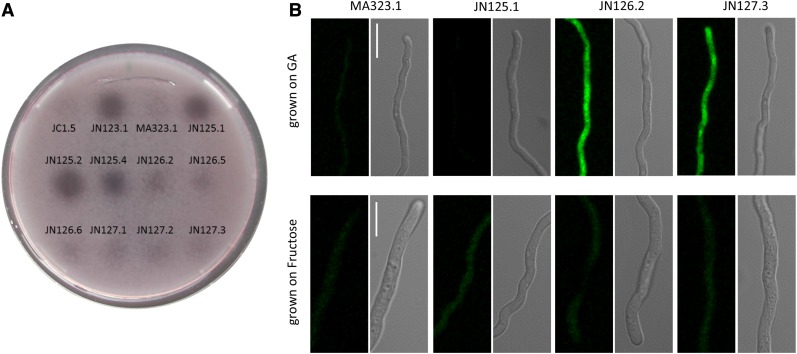
GaaX was previously identified as a GA-induced gene with unknown function (Martens-Uzunova and Schaap 2008). To monitor the induction of GaaX and to localize the GaaX protein in the cell, GaaX was fused to GFP at either the N- or C-terminal part of GaaX and expressed from the endogenous GaaX promoter. Fusion constructs were targeted to the pyrG locus of A. niger in a strain lacking endogenous gaaX (JN125.1) to be able to test complementation of the GFP-GaaX and GaaX-GFP fusion proteins (Figure S3). As shown in Figure 4A, JN125.1 (ΔgaaX::nicB) constitutively expressed pectinases indicated by the halo on PGA plates, while both the C-terminally tagged as well as the N-terminally tagged versions of GaaX (JN126.2 and JN127.3, respectively) complemented the constitutive expression phenotype, indicating that both fusion proteins are functional. Confocal fluorescent microscopy was performed on GFP-tagged strains to localize GaaX (Figure 4B). Spores were germinated either on GA or on fructose (a nonrepressing carbon source) and a fluorescent signal was only detectable in the GFP-labeled strains after growth on GA. This observation confirms the results from the expression data that indicate that GaaX is lowly expressed under noninducing conditions and is induced on GA. The expression of GaaX is low on fructose and no GFP signal above the background level was detected on fructose. Based on the fluorescent pictures, GaaX is likely to be localized in the cytosol.
Figure 4.
(A) Complementation analysis of GaaX-GFP fusions. Polygalacturonase activities of gaaX deletion strains, gaaX-GFP and GFP-gaaX complementation strains, and their parental strains were detected by spotting 50 µl supernatant from fructose-grown cultures on polygalacturonic acid agarose. (B) Subcellular localization of GaaX-GFP and GFP-GaaX in A. niger germlings. Strains were grown on coverslips in Petri dishes with minimal medium (pH 5.8) supplemented with either galacturonic acid or fructose as carbon source. Bar, 10 µm.
Discussion
The forward genetic screen with a positive selection strategy for the isolation of A. niger mutants with constitutive expression of genes involved in PGA degradation resulted in the identification of a repressor protein (NRRL3_08194), which we named GaaX. Both the genome sequencing of five independently obtained mutants, as well as the analysis of a targeted deletion mutant (ΔgaaX), showed that the loss of function of gaaX leads to constitutive expression of genes previously identified as GA-induced genes (Martens-Uzunova and Schaap 2008) and genes encoding pectinolytic enzymes that are activated via the transcription factor GaaR (Alazi et al. 2016). Deletion of gaaX did not result in a growth alteration on any carbon source tested (Figure 2 and data not shown). Transcriptome analysis (Table S3) strongly suggests that deletion of gaaX only affects the expression of genes related to the degradation and metabolism of (poly)galacturonic acid. Genes encoding enzymes involved in the hydrolysis of nonpectin polysaccharides are not differentially regulated in ΔgaaX. In addition, GO enrichment analysis of ΔgaaX transcriptome shows a strong correlation only between the activity of GaaX and the expression of GA-induced genes. In agreement with these observations, the phenotype of the gaaR deletion mutant was specific for (poly)galacturonic acid, with no growth defect observed on other substrates tested (glucuronic acid, rhamnose, xylose, and arabinose) (Alazi et al. 2016). Taken together, these findings indicate that GaaR and GaaX are specifically involved in the regulation of pectin catabolism.
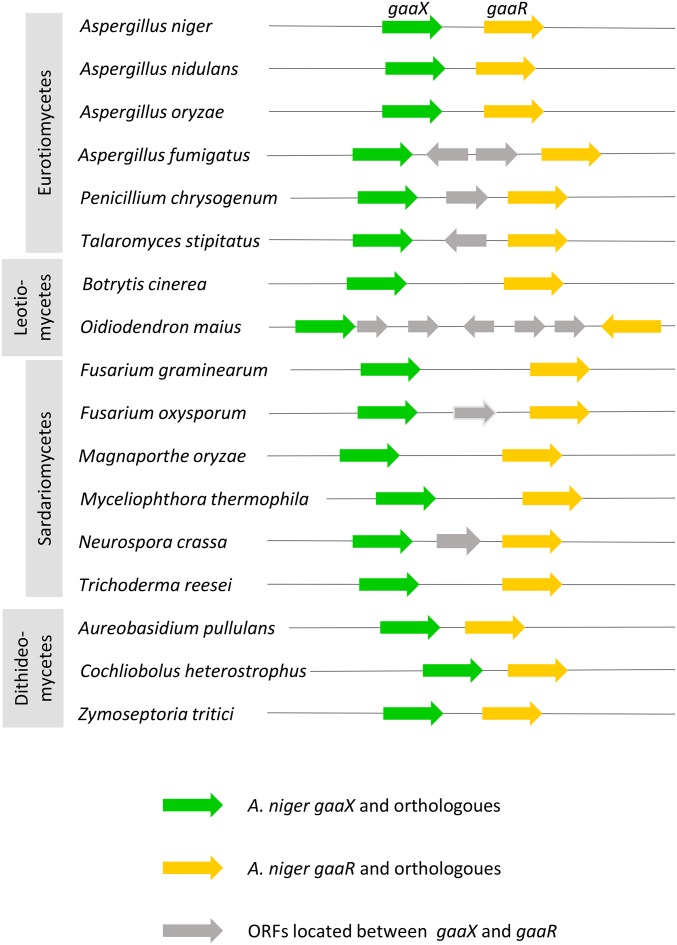
Interestingly, the gaaX gene is located next to the recently identified GA-specific transcriptional activator gaaR (NRRL3_08195). The GaaR transcriptional activator is conserved in 19 out of the 20 Aspergillus species for which genomic sequences are available via Aspergillus Genome Database (AspGD), and only absent in Aspergillus glaucus (Alazi et al. 2016), which corresponds with the inability of A. glaucus to grow on GA (http://www.fung-growth.org/). In all 19 Aspergillus species containing GaaR, a GaaX ortholog could be identified adjacent to GaaR. Only in Aspergillus fumigatus (Figure 5) and Aspergillus wentii (data not shown) were ORFs predicted to be present in between gaaX and gaaR. The ORFs between gaaX and gaaR in A. fumigatus are Afu4g06430 and Afu4g06450. Afu4g06430 is predicted to encode a 128-aa long protein that has no ortholog in other aspergilli. According to available expression data (Lind et al. 2015), this gene is not expressed, and it is questionable whether this predicted gene actually encodes a protein. Afu4g06450 is predicted to encode a Tan1-related transposase of the DDE family. This type of transposase is found in both A. nidulans and A. niger, as well as in many other organisms. This transposase is also lowly expressed in A. fumigatus (Lind et al. 2015).
Figure 5.
Schematic overview of the conservation of the gaaX-gaaR gene pair in 17 Pezizomycotina species. GaaX orthologs (green), GaaR orthologs (yellow), and ORFs between gaaX and gaaR (gray) are indicated. Arrow heads indicate the direction of transcription.
Like gaaR, gaaX is also missing in A. glaucus. BLASTP and synteny analysis between A. niger and A. glaucus revealed that the GaaR/GaaX encoding genes have been excised, as surrounding genes are conserved. Despite the loss of GaaX and GaaR, A. glaucus still possesses the GA-specific catabolic genes gaaA (Aspgl1_0124049), gaaB (Aspgl1_0091535), and gaaC (Aspgl1_0065497).
The GaaR transcriptional activator has previously been reported to be conserved in other Ascomycetes belonging to the Pezizomycotina subdivision, including members of the Eurotiomycetes (Penicillium, Talaromyces spp), Leotiomycetes (Botrytis, Oidiodendron), Sordariomycetes (Neurospora, Myceliophthora, Magnaporthe, Trichoderma, and Fusarium spp.), and Dothideomycetes [Zymoseptoria (Mycosphaerella), Aureobasidium, and Cochliobolus spp.] (Zhang et al. 2016). Synteny analysis of 17 species belonging to four classes of Pezizomycetes (Eurotiomycetes, Leotiomycetes, Sordariomycetes, and Dothideomycetes) revealed a strong conservation of the genomic clustering of gaaR and gaaX orthologs (Figure 5 and Table S5). For most fungal species analyzed, gaaR and gaaX are next to each other on the chromosome or close to each other and separated by one to five genes (Figure 5). The head to tail orientation of gaaR–gaaX driving expression of gaaR and gaaX from different promoters is conserved in all species except in Oidiodendron maius. Like GaaR, GaaX was found only in the Pezizomycotina and not in ascomycete yeasts, zygomycetes, or basidiomycetes.
The strategy to identify the responsible mutation by sequencing five independently obtained mutants has been successful and efficient. Clearly, sequencing only a limited number of mutants leads only to successful identification when the mutants isolated in the screen all belong to a single complementation group. If more complementation groups are involved, more mutants would need to be sequenced. It is interesting to note that in addition to mutations in gaaX which were present in all five mutants, we noticed that two mutants (S1 and UV1) also contained allelic mutations in NRRL3_06175 (Table S2). The protein encoded by this gene is predicted to encode a cocaine esterase and belongs to a protein subfamily of hydrolases that included cocaine esterase (CocE), several glutaryl-7-ACA acylases, and the putative diester hydrolase NonD of Streptomyces griseus. This family shows extensive, low-level similarity to a family of Xaa-Pro dipeptidyl-peptidases. Whether this gene also contributes to the constitutive expression of GA-dependent genes remains to be determined, but this is unlikely as mutants without mutations in this gene display essentially the same constitutive phenotype.
Previous studies have identified genes specifically induced by GA (Martens-Uzunova and Schaap 2008) and pectinolytic genes that were dependent on the GaaR transcriptional activator for induction by GA (Alazi et al. 2016). Eleven of the 15 GA-induced genes identified by Martens-Uzunova and Schaap were upregulated in the gaaX mutant (Figure 3 and Table 3). The three genes that are considered GA-inducible but not detected as differentially expressed in the gaaX mutant are predicted to encode a transporter (NRRL3_04281), an exo-polygalacturonase (NRRL3_09810, pgxA), and a pectin lyase (NRRL3_00965, pelA). These three genes were not classified as differentially expressed according to the stringent statistical settings in our current study. The fourth gene induced on GA in the study of Martens-Uzunova and Schaap (2008), but missing in our study, is gaaX itself.
In our recent study on the GaaR transcriptional activator, we identified 32 pectinolytic genes whose expression on GA was dependent on GaaR (Alazi et al. 2016). These genes overlap largely with the previously identified GA-responsive genes (Martens-Uzunova and Schaap 2008) (Figure 3 and Table 3), but also include 18 new potential GaaR target genes. Six of these genes [including NRRL3_02479 (lacB), NRRL3_05252 (pmeC), NRRL3_08325 (pmeA), NRRL3_07470 (pmeB), NRRL3_10559 (rgxC), and NRRL3_01237 (pelD)] were also found to be significantly upregulated in ΔgaaX (Figure 3 and Table 3) and are therefore considered to be part of the core GA regulon. The remaining 12 genes identified as being GaaR dependent for induction on GA (Alazi et al. 2016) were not identified as differentially expressed based on the stringent settings in this study. Whether these genes are indeed directly controlled by GaaR and GaaX, and therefore part of the core GA regulon, awaits further study.
The GaaX protein is predicted to be 697 aa long and displays significant similarity to the last three domains in the C-terminal half of the AROM protein. AROM is a large (1586 aa in A. niger) pentafunctional protein composed of five domains and the individual domains are involved in five different enzymatic steps representing the prechorismate shikimate pathway, which is required for aromatic amino acid biosynthesis (Duncan et al. 1987; Hawkins and Smith 1991). The last three domains of the AROM protein encode the shikimate kinase, 3-dehydroquinate dehydratase, and shikimate dehydrogenase and are homologous to the respective bacterial enzymes (aroL, aroD, and aroE) (Lamb et al. 1996). The AROM protein is present in fungi, including yeasts, and Euglena. The evolutionary origin of AROM is likely to be bacterial and it has been suggested that the AROM protein is the result of gene fusion events (Richards et al. 2006). Sequence alignment and BLASTP searches showed that the GaaX protein has significant sequence homology with the last three domains of the AROM protein. The observation of a transcriptional activator (GaaR) located next to a possible repressor protein (GaaX) that displays significant homology to AROM is analogous to the clustered transcriptional activator/repressor module regulating quinic acid utilization (Geever et al. 1989; Lamb et al. 1990). Like GaaX, the quinate repressor protein shows significant sequence similarities with the last three C-terminal domains of AROM (Lamb et al. 1996).
The regulation of metabolic enzymes required for quinic acid utilization has been a classical example of gene regulation both in N. crassa and A. nidulans (Geever et al. 1989; Leversley et al. 1996). In A. nidulans and N. crassa, the transcriptional activator and repressor are located in a gene cluster which consists of the activator and repressor and other genes involved in quinic acid catabolism and transport (Geever et al. 1989; Lamb et al. 1990). A. niger also has a quinic acid gene cluster that includes, besides the qutA gene (NRRL3_11038) and qutR gene (NRRL3_11039), a catabolic 3-dehydroquinase (NRRL3_11037) and an Major facilitator superfamily (MFS) transporter possibly involved in quinate uptake (NRRL3_11036). In contrast to the quinic acid gene cluster in which the regulatory genes (activator and repressor) are clustered with structural genes, no structural genes involved in GA utilization were clustered with GaaR and GaaX. Deletion of the qutA transcription factor (NRRL3_11038) in A. niger results in a quinate nonutilizing mutant (M. Arentshorst and A. F. J. Ram, unpublished results). Both in A. nidulans and N. crassa, the regulation of genes involved in quinic acid metabolism has been studied in detail and is characterized by the presence of a transcriptional activator (named QutA in A. nidulans, and qa-1F in N. crassa) located next to a repressor protein (QutR in A. nidulans, and qa-1S in N. crassa). Loss of function of quinic acid repressor qutR or qa-1S in A. nidulans and N. crassa, respectively, leads to constitutive expression of quinic acid utilization genes (Giles et al. 1985; Lamb et al. 1996), very similar to the effect observed for the loss of function of GaaX, resulting in constitutive expression of GA utilization genes. Based on the phenotype of the gaaX mutant and the analogy to the organization of the quinic acid utilization gene cluster, our current working hypothesis is that gaaX encodes a repressor protein which is required to keep the transcriptional activator GaaR in an inactive form in the absence of the inducer molecule.
As noted earlier, gaaX (NRRL3_08194) was identified as an upregulated gene when an A. niger culture pregrown for 18 hr with 2% fructose was transferred to a medium containing 1% GA as the sole carbon source (Martens-Uzunova and Schaap 2008). The expression of a functional GFP-tagged version of GaaX confirmed the induced expression and showed cytosolic localization of GaaX in the presence of GA (Figure 4). In the promoter region of gaaX, a GA-responsive element (GARE) was found, suggesting that activation of the transcription factor results in increased levels of repressor protein. Although this might seem contradictory at the first glance, it could be an elegant mechanism to ensure that the expression of GA-induced genes is tightly controlled and quickly responds to the presence or absence of GA. The induction of the expression of the repressor is partially analogous the activation/repression system of the qa cluster in N. crassa. In N. crassa it has been shown that both the activator (qa-1F) and the repressor (qa-1S) are transcriptionally induced in the presence of quinic acid (Patel et al. 1981; Giles et al. 1991). In the GA regulation system of A. niger, only the repressor protein is induced and not the activator. It should be noted that in almost all of 17 species analyzed, the gaaX and gaaR genes do not share the same promoter region (head to tail orientation; Figure 5), while the qa-1S and qa-1F genes of N. crassa share the same promoter region, which might function as a bidirectional promoter.
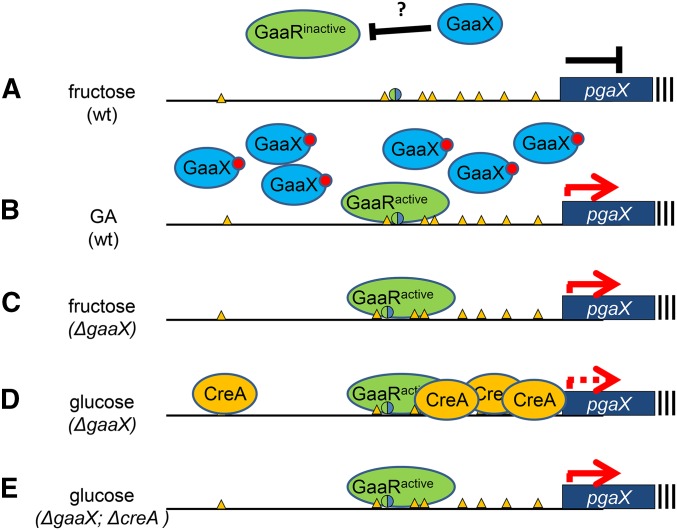
As a working model (Figure 6), we postulate that in the presence of GA, the inducer molecule, which could be GA or a derivative of GA, binds in the cytosol to repressor protein GaaX. Binding of the inducer to the GaaX repressor is posited to result in the activation of the transcription factor GaaR. Active GaaR is expected to induce the expression of GA-responsive genes involved in GA release, uptake and metabolism, but also induces the expression of repressor protein. As long as the inducer is present in sufficient amounts, the GaaX repressor is predicted to be inactive as a repressor and thereby the GaaR transcription factor remains active. When the concentration of inducer decreases, it is reasonable to suggest that repressor proteins lacking bound inducer could inactivate the GaaR transcriptional activator, thereby restraining the expression of GA-responsive genes. Thus, high expression of the repressor could serve as a sensitive system to ensure that, when intracellular GA levels decrease, the cell can tightly turn off expression of GA-responsive genes. Moreover, this mechanism also ensures the rapid response to the presence of GA as it does not require de novo synthesis of GaaR. Induction simply requires the binding of inducer to the repressor and subsequent activation of GaaR via post-translational mechanisms, as the expression of GaaR is not dramatically induced by GA (Alazi et al. 2016) or in the gaaX mutant (this study). The expression of GA-induced genes is also controlled via CreA mediated CCR (de Vries et al. 2002; Niu et al. 2015). The analysis of the PpgaX-amdS reporter strain (Figure 2) suggests that the expression of pgaX is carbon catabolite repressed even in the ΔgaaX strain. This suggests that CreA directly represses pgaX expression via CreA binding sites in the pgaX promoter, independent of GaaX repression (Figure 6).
Figure 6.
Model for the regulation of GA-induced gene expression in A. niger. (A) GA-induced gene expression, with pgaX as an example, is controlled via interaction of the transcriptional activator (GaaR) and transcriptional repressor (GaaX) in combination with CreA-mediated carbon catabolite repression. (A) In the presence of fructose (a nonrepressible, noninducing carbon source) pgaX expression is prevented because GaaX inhibits GaaR activation. The question mark indicates that the mechanism by which GaaX controls GaaR activity is unknown. (B) In the presence of GA, GA itself or a derivative of GA is predicted to bind to GaaX. The binding of the inducer to GaaX is expected to activate GaaR. GaaX is induced and remains cytosolic but the presence of the inducer keeps GaaX inactive. (C) In the ΔgaaX strain, GaaR is no longer kept inactive by GaaX and therefore is constitutively active, resulting in constitutive expression of pgaX. (D) In the ΔgaaX strain, the presence of glucose leads to CreA-mediated repression leading to reduced expression of pgaX and possibly other pectinolytic genes. (E) Deletion of both gaaX and creA results in constitutive expression of pgaX even in the presence of glucose. The yellow triangles represent putative CreA binding sites. The green/blue circle represents a putative GaaR-binding site. The red circle represents the postulated inducing sugar.
The proposed model for the mechanism by which GaaR and GaaX regulate gene expression resembles in some aspects the Gal3/Gal4/Gal80 module of Saccharomyces cerevisiae, but shows at least two important differences. Whereas the Gal4 regulatory system consists of three proteins (Gal4 as the transcriptional activator, Gal80 as the repressor, and Gal3 as possible galactose sensor), we have identified two genes/proteins involved in GA regulation and no evidence for a third member. Also in the regulation of quinate metabolism, no third regulatory gene has been identified even though saturating mutant screens have been performed. These observations do not exclude the possibility that a third factor is involved in the GA or quinic acid regulation, but it is unlikely with the available evidence. Whereas the sensor (Gal3)/repressor (Gal80) function is mediated via two different proteins in the Gal regulatory system in S. cerevisiae, in the GA and quinic acid regulatory systems, the sensor/repressor function might well be performed by a single protein, GaaX and QutR, respectively. Another important difference is that GaaX and QutR do not show homology to Gal80 or Gal3, nor do Gal80 or Gal3 display homology to AROM. Based on these observations, we suggest that the GAL repressor module has evolved independently from that of GaaX/QutR.
In addition to GaaX (NRRL3_08194) and QutR (NRRL3_11039), we identified two additional paralogues in the A. niger genome (NRRL3_08276 and NRRL3_07605). All four paralogues showed significant homology to the A. niger AROM protein, as well as limited homology towards each other. Both NRRL3_08276 and NRRL3_07605 are also located next to predicted Zn(II)2Cys6 domain transcription factors, NRRL3_08275 and NRRL3_07604, respectively. Whereas the function of the GaaR/GaaX and QutA/QutR modules are related to GA and quinic acid metabolism, respectively, the function of the two other pairs that are present in A. niger remains to be elucidated. The sequence similarity of NRRL3_08276 and NRRL3_07605 to QutR and GaaX and their genome clustering with predicted transcription factors suggest that the proposed activator/repressor modules observed for GaaR–GaaX and QutA-QutR is an evolutionarily conserved mechanism to control gene expression in filamentous ascomycete fungi. The number of similar activator/repressor modules varies among Pezizomycotina species (Figure S4 and Figure S5). Most Pezizomycotina species contain the galacturonic acid and quinic acid related transcriptional activator/repressor modules. It is interesting to note that some fungi, e.g., Talaromyces stipitatus and B. cinerea, seem to have lost the quinic acid specific repressor, which suggests they might have lost the capacity to utilize quinic acid. The GaaR/GaaX and QutA/QutR activator/repressor modules and their variants are specific for Pezizomycotina and missing in ascomycete yeasts, zycomycetes, and basidiomycetes.
Acknowledgments
We thank Cees van den Hondel for helpful discussions and Tim Knetsch and Jos Reijngoud for assistance with bioreactor cultivations. J.N. is supported through a grant from the China Scholarship Council. E.A. is supported by a grant from BE-Basic within flagship 10 Advanced Microbial Biofuel and chemicals production (AMBIC). This work was in part supported by Genome Canada and Génome Québec.
Footnotes
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.194050/-/DC1.
Communicating editor: M. Freitag
Literature Cited
- Alazi E., Niu J., Kowalczyk J. E., Peng M., Aguilar Pontes M. V., et al. , 2016. The transcriptional activator GaaR of Aspergillus niger is required for release and utilization of D-galacturonic acid from pectin. FEBS Lett. 590: 1804–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arentshorst M., Ram A. F., Meyer V., 2012. Using non-homologous end-joining-deficient strains for functional gene analyses in filamentous fungi. Methods Mol. Biol. 835: 133–150. [DOI] [PubMed] [Google Scholar]
- Arentshorst M., Niu J., Ram A. F., 2015. Efficient Generation of Aspergillus niger knock out Strains by combining NHEJ mutants and a split marker approach, pp. 263–272 in Genetic Transformation Systems in Fungi, Vol. 1, edited by van den Berg M. A., Maruthachalam K. Springer International Publishing, New York. [Google Scholar]
- Battaglia E., Zhou M., de Vries R. P., 2014. The transcriptional activators AraR and XlnR from Aspergillus niger regulate expression of pentose catabolic and pentose phosphate pathway genes. Res. Microbiol. 165: 531–540. [DOI] [PubMed] [Google Scholar]
- Bennett J. W., Lasure L., 1991. Growth media, pp. 441–458 in More Gene Manipulations in Fungi, edited by Bennett J. W., Lasure L. L. Academic Press, San Diego. [Google Scholar]
- Bos C. J., Debets A. J., Swart K., Huybers A., Kobus G., et al. , 1988. Genetic analysis and the construction of master strains for assignment of genes to six linkage groups in Aspergillus niger. Curr. Genet. 14: 437–443. [DOI] [PubMed] [Google Scholar]
- Damveld R. A., Franken A., Arentshorst M., Punt P. J., Klis F. M., et al. , 2008. A novel screening method for cell wall mutants in Aspergillus niger identifies UDP-galactopyranose mutase as an important protein in fungal cell wall biosynthesis. Genetics 178: 873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries R. P., Visser J., de Graaff L. H., 1999. CreA modulates the XlnR-induced expression on xylose of Aspergillus niger genes involved in xylan degradation. Res. Microbiol. 150: 281–285. [DOI] [PubMed] [Google Scholar]
- de Vries R. P., Jansen J., Aguilar G., Parenicova L., Joosten V., et al. , 2002. Expression profiling of pectinolytic genes from Aspergillus niger. FEBS Lett. 530: 41–47. [DOI] [PubMed] [Google Scholar]
- Dowzer C. E., Kelly J. M., 1991. Analysis of the creA gene, a regulator of carbon catabolite repression in Aspergillus nidulans. Mol. Cell. Biol. 11: 5701–5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan K., Edwards R. M., Coggins J. R., 1987. The pentafunctional arom enzyme of Saccharomyces cerevisiae is a mosaic of monofunctional domains. Biochem. J. 246: 375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison, E., and G. Marth, 2012 Haplotype-based variant detection from short-read sequencing. ArXiv 1207.3907 [q-bio.GN].
- Geever R. F., Huiet L., Baum J. A., Tyler B. M., Patel V. B., et al. , 1989. DNA sequence, organization and regulation of the qa gene cluster of Neurospora crassa. J. Mol. Biol. 207: 15–34. [DOI] [PubMed] [Google Scholar]
- Giles N. H., Case M. E., Baum J., Geever R., Huiet L., et al. , 1985. Gene organization and regulation in the qa (quinic acid) gene cluster of Neurospora crassa. Microbiol. Rev. 49: 338–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles N. H., Geever R. F., Asch D. K., Avalos J., Case M. E., 1991. The Wilhelmine E. Key 1989 invitational lecture. Organization and regulation of the qa (quinic acid) genes in Neurospora crassa and other fungi. J. Hered. 82: 1–7. [DOI] [PubMed] [Google Scholar]
- Grant S., Roberts C.F., Lamb H., Stout M., Hawkins A.R. 1988. Genetic regulation of the quinic acid utilization (QUT) gene cluster in Aspergillus nidulans. J. Gen. Microbiol. 134: 347–358. [DOI] [PubMed] [Google Scholar]
- Gruben B. S., Zhou M., Wiebenga A., Ballering J., Overkamp K. M., et al. , 2014. Aspergillus niger RhaR, a regulator involved in L-rhamnose release and catabolism. Appl. Microbiol. Biotechnol. 98: 5531–5540. [DOI] [PubMed] [Google Scholar]
- Hawkins A. R., Smith M., 1991. Domain structure and interaction within the pentafunctional arom polypeptide. Eur. J. Biochem. 196: 717–724. [DOI] [PubMed] [Google Scholar]
- Jiang H., Lei R., Ding S., Zhu S., 2014. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics 15: 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen T. R., Nitsche B. M., Lamers G. E., Arentshorst M., van den Hondel C. A., et al. , 2010. Transcriptomic insights into the physiology of Aspergillus niger approaching a specific growth rate of zero. Appl. Environ. Microbiol. 76: 5344–5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap D. R., Vohra P. K., Chopra S., Tewari R., 2001. Applications of pectinases in the commercial sector: a review. Bioresour. Technol. 77: 215–227. [DOI] [PubMed] [Google Scholar]
- Khan M., Nakkeeran E., Umesh-Kumar S., 2013. Potential application of pectinase in developing functional foods. Annu. Rev. Food Sci. Technol. 4: 21–34. [DOI] [PubMed] [Google Scholar]
- Kopylova E., Noé L., Touzet H., 2012. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28: 3211–3217. [DOI] [PubMed] [Google Scholar]
- Lamb H. K., Hawkins A. R., Smith M., Harvey I. J., Brown J., et al. , 1990. Spatial and biological characterisation of the complete quinic acid utilisation gene cluster in Aspergillus nidulans. Mol. Gen. Genet. 223: 17–23. [DOI] [PubMed] [Google Scholar]
- Lamb H. K., Moore J. D., Lakey J. H., Levett L. J., Wheeler K. A., et al. , 1996. Comparative analysis of the QUTR transcription repressor protein and the three C-terminal domains of the pentafunctional AROM enzyme. Biochem. J. 313(Pt 3): 941–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S., 2012. Fast gapped-read alignment with Bowtie2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesley I., Newton G. H., Lamb H. K., van Schothorst E., Dalgleish R. W., et al. , 1996. Domain structure and function within the QUTA protein of Aspergillus nidulans: implications for the control of transcription. Microbiology 142(Pt 1): 87–98. [DOI] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S., 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens-Uzunova E. S., Schaap P. J., 2008. An evolutionary conserved D-galacturonic acid metabolic pathway operates across filamentous fungi capable of pectin degradation. Fungal Genet. Biol. 45: 1449–1457. [DOI] [PubMed] [Google Scholar]
- Meyer V., Arentshorst M., El-Ghezal A., Drews A. C., Kooistra R., et al. , 2007. Highly efficient gene targeting in the Aspergillus niger kusA mutant. J. Biotechnol. 10: 770–775. [DOI] [PubMed] [Google Scholar]
- Mohnen D., 2008. Pectin structure and biosynthesis. Curr. Opin. Plant Biol. 11: 266–277. [DOI] [PubMed] [Google Scholar]
- Nitsche B. M., Jørgensen T. R., Akeroyd M., Meyer V., Ram A. F., 2012. The carbon starvation response of Aspergillus niger during submerged cultivation: insights from the transcriptome and secretome. BMC Genomics 13: 380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu J., Homan T. G., Arentshorst M., de Vries R. P., Visser J., et al. , 2015. The interaction of induction and repression mechanisms in the regulation of galacturonic acid-induced genes in Aspergillus niger. Fungal Genet. Biol. 82: 32–42. [DOI] [PubMed] [Google Scholar]
- Niu J., Arentshorst M., Seelinger F., Ram A. F., Ouedraogo J. P., 2016. A set of isogenic auxotrophic strains for constructing multiple gene deletion mutants and parasexual crossings in Aspergillus niger. Arch. Microbiol. 198: 861–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel V. B., Schweizer M., Dykstra C. C., Kushner S. R., Giles N. H., 1981. Genetic organization and transcriptional regulation in the qa gene cluster of Neurospora crassa. Proc. Natl. Acad. Sci. USA 78: 5783–5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R., Duggal G., Love M. I., Irizarry R. A., Kingsford C., 2016. Salmon provides accurate, fast, and bias-aware transcript expression estimates using dual-phase inference. bioRxiv 021592. [Google Scholar]
- Petersen K. L., Lehmbeck J., Christensen T., 1999. A new transcriptional activator for amylase genes in Aspergillus. Mol. Gen. Genet. 262: 668–676. [DOI] [PubMed] [Google Scholar]
- Raulo R., Kokolski M., Archer D. B., 2016. The roles of the zinc finger transcription factors XlnR, ClrA and ClrB in the breakdown of lignocellulose by Aspergillus niger. AMB Express 6: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards T. A., Dacks J. B., Jenkinson J. M., Thornton C. R., Talbot N. J., 2006. Evolution of filamentous plant pathogens: gene exchange across eukaryotic kingdoms. Curr. Biol. 16: 1857–1864. [DOI] [PubMed] [Google Scholar]
- Ruijter G. J. G., Visser J., 1997. Carbon repression in Aspergilli. FEMS Microbiol. Lett. 151: 103–114. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D. W., 2001. Molecular Cloning: A Laboratory Manual, 3rd Ed Cold Spring Harbor Press, New York. [Google Scholar]
- Sloothaak J., Schilders M., Schaap P. J., de Graaff L. H., 2014. Overexpression of the Aspergillus niger GatA transporter leads to preferential use of D-galacturonic acid over D-xylose. AMB Express 4: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Florea L., 2015. Rcorrector: efficient and accurate error correction for Illumina RNA-seq reads. Gigascience 4: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teather R. M., Wood P. J., 1982. Use of Congo red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl. Environ. Microbiol. 43: 777–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hartingsveldt W., Mattern I. E., van Zeijl C. M., Pouwels P. H., van den Hondel C. A., 1987. Development of a homologous transformation system for Aspergillus niger based on the pyrG gene. Mol. Gen. Genet. 206: 71–75. [DOI] [PubMed] [Google Scholar]
- van Peij N. N., Visser J., de Graaff L. H., 1998. Isolation and analysis of xlnR, encoding a transcriptional activator co-ordinating xylanolytic expression in Aspergillus niger. Mol. Microbiol. 27: 131–142. [DOI] [PubMed] [Google Scholar]
- vanKuyk P. A., Benen J. A., Wösten H. A., Visser J., de Vries R. P., 2012. A broader role for AmyR in Aspergillus niger: regulation of the utilisation of D-glucose or D-galactose containing oligo- and polysaccharides. Appl. Microbiol. Biotechnol. 93: 285–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X. L., van der Kaaij R. M., van den Hondel C. A., Punt P. J., van der Maarel M. J., et al. , 2008a Aspergillus niger genome-wide analysis reveals a large number of novel alpha-glucan acting enzymes with unexpected expression profiles. Mol. Genet. Genomics 279: 545–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X. L., Roubos J. A., van den Hondel C. A., Ram A. F., 2008b Identification of InuR, a new Zn(II)2Cys6 transcriptional activator involved in the regulation of inulinolytic genes in Aspergillus niger. Mol. Genet. Genomics 279: 11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Lubbers R. J., Simon A., Stassen J. H., Vargas Ribera P. R., et al. , 2016. A novel Zn2Cys6 transcription factor BcGaaR regulates D-galacturonic acid utilization in Botrytis cinerea. Mol. Microbiol. 100: 247–262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains are listed in Table 1 and are available upon request. Table S2 contains SNPs and indels detected in genomes of mutants. Table S3A contains transcript per million (TPM) values of NRRL3 gene models in wild type and the gaaX mutant, and Table S3B contains their DEseq2 analysis. The DNA reads described in this study are deposited in the Short Read Archive under accession number SRP078415. The RNA reads described in this study are deposited in the Short Read Archive under accession number SRP078485.