Abstract
Numerous studies across a wide range of taxa have demonstrated that immune genes are routinely among the most rapidly evolving genes in the genome. This observation, however, does not address what proportion of immune genes undergo strong selection during adaptation to novel environments. Here, we determine the extent of very recent divergence in genes with immune function across five populations of Drosophila melanogaster and find that immune genes do not show an overall trend of recent rapid adaptation. Our population-based approach uses a set of carefully matched control genes to account for the effects of demography and local recombination rate, allowing us to identify whether specific immune functions are putative targets of strong selection. We find evidence that viral-defense genes are rapidly evolving in Drosophila at multiple timescales. Local adaptation to bacteria and fungi is less extreme and primarily occurs through changes in recognition and effector genes rather than large-scale changes to the regulation of the immune response. Surprisingly, genes in the Toll pathway, which show a high rate of adaptive substitution between the D. melanogaster and D. simulans lineages, show little population differentiation. Quantifying the flies for resistance to a generalist Gram-positive bacterial pathogen, we found that this genetic pattern of low population differentiation was recapitulated at the phenotypic level. In sum, our results highlight the complexity of immune evolution and suggest that Drosophila immune genes do not follow a uniform trajectory of strong directional selection as flies encounter new environments.
Keywords: evolution, local adaptation, population genetics, immunity, Drosophila melanogaster
A current challenge in evolutionary biology is to understand the genetic changes that drive local phenotypic adaptation (Hereford 2009; Stapley et al. 2010; Stephan 2016). In recent years, numerous studies have addressed this challenge by examining genome-wide changes in allele frequencies across clines or between populations (Turner et al. 2008; Stapley et al. 2010; Lamichhaney et al. 2012; Pespeni et al. 2012; Hubner et al. 2013). In limited instances, such genome-wide patterns could even be connected back to specific phenotypic traits or environmental variables, greatly increasing our understanding of the genetic processes underlying phenotypic evolution (Turner et al. 2010; Jones et al. 2012; Jeong and Di Rienzo 2014). While analytic approaches differ, these studies often rely on the detection of outlier loci that display patterns of high population differentiation or other extreme signatures of local selection. Such approaches are particularly adept at detecting single loci that have experienced strong selective sweeps.
Immune genes are often evolutionary outliers in such studies, displaying fast rates of evolution and high population differentiation across multiple taxa including humans (Fumagalli et al. 2011; Daub et al. 2013; Quintana-Murci and Clark 2013), Daphnia (McTaggart et al. 2012), mosquitoes (Waterhouse et al. 2007; Crawford et al. 2010), and bees (Chavez-Galarza et al. 2013; Erler et al. 2014). Across Drosophila species, immune genes are known to evolve faster than the genome average (Sackton et al. 2007), and on shorter timescales, both genome-wide studies and studies of individual genes have highlighted examples of immune genes displaying unusually high population differentiation across Drosophila melanogaster populations (Juneja and Lazzaro 2010; Fabian et al. 2012; Hubner et al. 2013). Similarly, clinical studies have identified numerous immune genes displaying high levels of population differentiation (Kolaczkowski et al. 2011b; Fabian et al. 2012).
These studies highlight the major role pathogens play in shaping the evolution of their hosts. But, while such observations suggest that certain Drosophila immune genes may be subject to unusually strong, spatially variable selection, they provide only a partial picture of how immunity, as an integrated complex phenotypic trait, is evolving. “Immune competence” is in reality a suite of phenotypes that are affected not only by pathogen pressures but also by environmental factors and genotype-by-environment interactions (Lazzaro et al. 2008; McKean et al. 2008; Lazzaro and Little 2009; Howick and Lazzaro 2014). While their resistance to similar types of pathogens may be weakly correlated (Lazzaro et al. 2006), flies show no evidence of cross-resistance to distinct pathogen classes (Kraaijeveld et al. 2012). Similarly, resistance is often decoupled from tolerance, highlighting the numerous physiological processes that influence host fitness (Ayres et al. 2008; Ayres and Schneider 2009). Further complicating the process of adaptation, trade-offs involving immune defense and other key life history traits are documented (Kraaijeveld et al. 2001; McKean et al. 2008; Ye et al. 2009), as are behavioral traits that lie outside the canonical immune system (Kacsoh et al. 2013; Babin et al. 2014). Such factors may lead to a scenario where populations experience broad shifts in immune regulation as they confront different optimal immune strategies in different environments. For such a complex trait, local adaptation may occur through a process of polygenic selection, where multiple variants experience weak selection leading to small allele frequency changes (Pritchard and Di Rienzo 2010; Pritchard et al. 2010; Turchin et al. 2012). Still, the literature contains numerous examples of single alleles that carry strong phenotypic effects against specific pathogens (Magwire et al. 2011; Sironi et al. 2015).
Here, we seek to determine the extent and routes of recent D. melanogaster immune adaptation. As populations adapt to novel environments do they experience large-scale changes across immune genes, or alternatively, is immune adaptation driven by a subset of rapidly evolving genes? Do we find changes across all immune processes or does a limited number of pathogen types drive adaptation? To answer these questions, we undertake a comprehensive analysis of immune gene diversity and divergence across five populations of D. melanogaster. We compare known immune genes with genomic controls matched for size, genome location, and local recombination rate—factors known to affect patterns of polymorphism and the efficacy of selection (Larracuente et al. 2008; Comeron et al. 2012; Castellano et al. 2016). With this approach, we identify not only putative single-gene targets of local selection but also examine whether entire pathways and gene classes show evidence of heightened selection. In addition, we compare patterns of interspecific divergence in Drosophila immunity genes with patterns of population-level differentiation and find both similarities and differences between the results of these two different analyses.
Materials and Methods
Gene and fly line selection
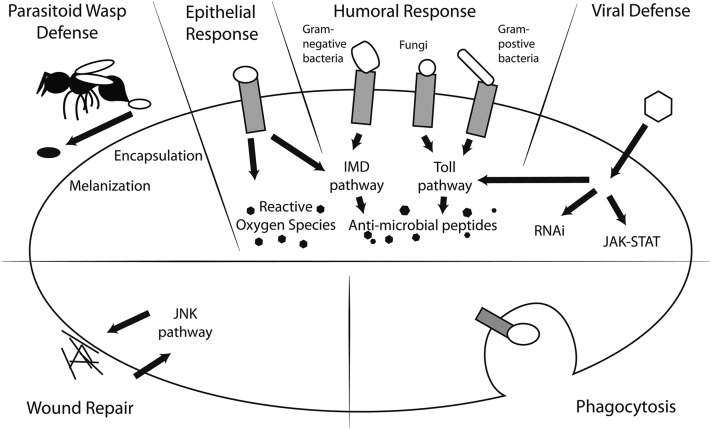
Through literature searches, we assembled a list of 361 genes with well-supported immune function, many of which have appeared in previous large-scale studies (Sackton et al. 2007; Obbard et al. 2009). When possible, we assigned each gene to the immune pathway(s) or process(es) in which it functions (Figure 1 and Table 1). In the humoral response—regulated by the Toll and immune-deficiency (IMD) pathways—pattern recognition receptors bind common bacterial and fungal membrane components, triggering the downstream production of antimicrobial peptides (AMPs) and other microbicidal compounds. The cellular response coordinates the activities of specialized hemocytes such as phagocytes, which engulf foreign particles or necrotic cells, as well as lamellocytes and crystal cells, which participate in defense against parasitic wasps through the encapsulation and melanization of the deposited eggs. Finally, antiviral defense largely operates through RNA interference (RNAi) but also involves the JAK-STAT and Toll pathways. Members of the JNK pathway, together with other genes, contribute to various aspects of immune tolerance and resistance by mediating tissue repair and wound closure. Categories were not mutually exclusive and some were nested within larger umbrella categories (for instance, all Toll and IMD genes were also included in the humoral class). We also assigned a functional class to each gene. These included the broad categories of recognition receptor (identification of pathogens and parasites), signaling molecule (signal transduction), and effector protein (pathogen destruction). When applicable, we used more specific functional assignments (e.g., phagocytosis receptor, AMP). The full list of genes, as well as their pathway and functional class assignments, can be found in Supplemental Material, Table S1.
Figure 1.
Main pathways in the Drosophila immune system.
Table 1. Immune gene groupings based on immune process and functional class.
| Immune process | Genes | Basic role(s) |
|---|---|---|
| Cellular | 131 | Hemocyte-mediated responses to all classes of parasites and pathogens |
| Encapsulation | 36 | Initial recognition and coating of parasitoid wasp eggs |
| Phagocytosis | 45 | Cellular uptake of bacteria, fungi, and necrotic cells |
| Epithelial | 30 | Regulation of bacteria and fungi on epithelial surfaces, including the gut, trachea, and reproductive tract |
| Humoral | 141 | Recognition and elimination of bacteria and fungi in the hemolymph through the production of antimicrobial compounds |
| IMD | 57 | Humoral pathway that targets mainly Gram-negative bacteria |
| Toll | 61 | Humoral pathway that targets mainly Gram-positive bacteria and fungi |
| JAK-STAT | 26 | Signaling pathway implicated in viral response, control of hemocyte differentiation, and regulation of humoral response |
| JNK pathway and wound repair | 44 | Epithelial repair and cell growth |
| Melanization and PO production | 32 | Cell-mediated response that responds to wounding and the detection of parasitoid wasp eggs and microbes |
| ROS production | 14 | Production of reactive oxygen species; especially key in epithelial immune regulation |
| Viral defense | 29 | Destruction of viruses through RNAi; elimination of virus-infected cells |
| Functional class | Genes | Basic role(s) |
| Recognition | 58 | Initial detection of invading parasites and pathogens. Includes cell-surface (e.g., phagocytosis) receptors as well as humoral-response pattern recognition proteins (e.g., GNBPs and PGRPs) |
| Signaling | 198 | Signal transduction following pathogen recognition; negative regulation of immune response; cross talk between pathways |
| Effector | 75 | Pathogen destruction or sequestration. Includes AMPs, lysozymes, and ROS |
ROS, reactive oxygen species; GNBPs, Gram-negative bacteria binding proteins; PGRPs, peptidoglycan recognition proteins.
For each immune gene, we identified four control, protein-coding genes that were matched for size, genome location, and local recombination rate. For size and position, we required that control genes had a total length (including introns) within either 1500 bp of, or 0.5–2 times, the total immune gene length, and we preferentially chose genes located within 50 kb of the immune gene. Using the Drosophila melanogaster Recombination Rate Calculator version 2.3 (Fiston-Lavier et al. 2010), we obtained the estimated local recombination rate of each immune gene and required that control gene recombination rates be within 1 cM/Mb of this value. The overall correlation between immune gene recombination rate and the mean control genes’ recombination rates was high, even at low levels of recombination (R2 = 0.9997, Figure S1). We additionally checked the genes using the more fine-scale recombination estimates of Comeron et al. (2012). As their estimates cover a wider range than the Recombination Rate Calculator, the correlation between immune and control genes was weaker (R2 = 0.9443, Figure S2), but we ensured that all control gene recombination rates were still within 2.5 cM/Mb of the immune gene rate. If immune genes were found near the boundaries of known segregating inversions, we ensured that the matched controls were similarly within or outside of the inversion. In the event that more than four control genes fulfilled these requirements for a particular immune gene, we randomly chose four. Because of the restrictions, 15 immune genes had fewer than four controls in the final data set.
We had no way of guaranteeing that the control genes have no immune function. In particular, location matching carries this risk as genes of similar function—including immune function—often cluster in the genome. To minimize this risk, we excluded from the initial control pool 595 genes that had weak evidence of immune involvement (for example, moderate expression changes in large-scale immunity screens or homology with known immune gene families). We performed no further filtering on the control set. For example, we did not exclude genes that are known to be rapidly evolving.
We obtained information on nucleotide polymorphism within these genes for each member of the D. melanogaster Global Diversity Lines. These are a set of 84 inbred lines from five populations (15 from Beijing, China; 19 from Ithaca, NY; 19 from the Netherlands; 18 from Tasmania; and 13 from Zimbabwe) that have been sequenced to an average depth of 12× (Grenier et al. 2015). We masked genomic regions with poor “callability” (as defined in Grenier et al. 2015) to ensure that our analyses excluded genomic regions with large amounts of missing data. Using the Ensembl database Variant Effect Predictor script (Berkeley Drosophila Genome Project 5.25 assembly, release 64), we annotated the putative effect of each SNP within our genes of interest (nonsynonymous coding, synonymous coding, intronic, splice site, 3′-UTR, and 5′-UTR).
Analysis of D. melanogaster-D. simulans divergence
Using FlyBase (version FB2013_06) we determined which of our genes had known one-to-one orthologs in D. simulans. We then downloaded all transcripts for each gene from FlyBase and performed codon-based alignments for all D. melanogaster-D. simulans pairs using PRANK (Loytynoja and Goldman 2005). For each transcript pair, we used custom Perl scripts to count nonsynonymous and fourfold degenerate substitutions using the Nei–Gojobori method (Nei and Gojobori 1986) and to calculate the length of the aligned regions after removing all gaps. For each gene, we then identified the transcript pair with the longest aligned region and the fewest number of nonsynonymous substitutions. We introduced this second, more conservative, criterion to reduce the likelihood of spuriously aligning paralogous transcripts for immune genes that show a high level of alternative splicing. The values and the dN/dS values calculated using the longest transcripts were highly correlated (immune genes, R2 = 0.93; control genes, R2 = 0.97), and our approach did exclude a few incorrect immune gene alignments whose errors were easily detected on visual inspection (Figure S3). We used these transcripts to represent the gene in all downstream analyses.
Incorporating polymorphism data from the ancestral Zimbabwe population, we used DFE-alpha version 2.13 (Eyre-Walker and Keightley 2009) to determine the proportion of adaptive substitutions (α) and the relative rate of adaptive substitutions (ωa) within each pathway or functional class. Both statistics were calculated relative to substitutions at fourfold degenerate sites. In brief, this method uses a maximum likelihood method to infer the distribution of fitness effects of new mutations from the folded site frequency spectrum. We ran DFE-alpha using a two-epoch model, allowing variable mutation-effect sizes and variable shape parameters for the gamma distribution. Gene classes with <10 genes were excluded from the analysis.
Population genetic analyses
Using custom Perl scripts, we calculated derived allele frequency, pairwise nucleotide diversity (π), pairwise FST, and global FST for each SNP. These same statistics, as well as Watterson’s θ, Tajima’s D, KST, Hudson’s nearest neighbor (Snn), and normalized Fay and Wu’s H (Zeng et al. 2006), were calculated at the gene level. Although inbred, all fly lines retained at least some residual heterozygosity. When a fly line was heterozygous for a given site, we randomly sampled a single allele to use in all analyses. In all calculations, we used only biallelic SNPs. For all calculations, we required that there was a base call for at least eight lines in any given population and accounted for missing data by adjusting the sample size at each SNP. We calculated FST at each SNP according to Weir and Cockerham (1984). The numerator and denominator were averaged separately across regions to calculate FST for transcripts. Negative FST estimates were declared to be zero.
Analyzing gene classes using genomic controls
Evolutionary rate at a given gene can be influenced by factors in the local genetic environment, such as recombination rate or selection on nearby alleles. To control for these factors, we leveraged genetic data from our set of matched control genes to more conservatively assess the probability of selection within specific gene classes. We determined whether the genes in a given pathway or functional class deviated, as a set, from control expectations by conducting a paired Wilcoxon signed-rank test that compared the statistic for each immune gene within the class to the mean of its four control genes’ statistics. This test therefore relied only on the relative, not absolute, values of the immune and control statistics and determined whether there was a directional trend in the immune genes relative to the controls. Overall, this made the test less sensitive to genome-wide outliers and more sensitive to local variation. The test was conducted for our major test statistics: π, Fay and Wu’s H, Tajima’s D, and FST. Due to our small sample sizes, we determined significance through 100,000 permutations. For each permutation round, we randomly chose one of the four control genes to serve as the “test” gene and averaged the statistics from the remaining three controls and the immune gene. When genes had no polymorphism in a given population, they were dropped from the permutation round. As a result, the maximum V-statistic varied among permutations and so we used the resulting P-values to construct our null distribution.
We accounted for multiple testing at the level of the statistic (π, Fay and Wu’s H, Tajima’s D, and FST), correcting for multiple testing across multiple gene process groups (n = 13, with RNAi) or gene functional groups (n = 8). To do this, we set a P-value threshold at the 0.19% tails. As many of our groups are nonindependent and their significance values are expected to correlate, we did not attempt to identify P-values that reached study-wide significance. All statistical analyses were carried out in R (R Development Core Team 2011).
Systemic bacterial infections
Using a septic pinprick through the thoracic cuticle, we infected male flies (3–5 days old) with the bacterial pathogen Enterococcus faecalis (Lazzaro et al. 2004). Bacterial cultures had been grown at 37° overnight to an OD600 of 1.0 (±0.06). Flies were reared and maintained on standard glucose-yeast media at 25° with 12 hr light-dark cycles. Infections occurred in the 1–4 hr after the flies’ “dawn.” After 28 (±1) hr, we homogenized pools of three flies and plated the homogenate onto LB agar plates using a Spiral Biotech Autoplate 4000. We counted the number of colonies that grew on each plate with a QCount Colony Counter to infer the number of colony forming units per infected fly. Infections were carried out in a block structure, with each fly line infected on four different days. To assess whether population of origin had a significant effect on bacterial load, we constructed a linear model with infector, experimental day, and experimental block as random effects. The residuals were used to calculate the mean and median for each fly line. We then conducted an ANOVA with both the line means and medians to determine whether there was a population of origin effect. All statistical analysis were carried out in R (R Development Core Team 2011).
Data availability
Table S1 and Table S2 list the names and coordinates of the immune and control genes and transcripts used in the analysis. Sequence data from the Global Diversity Lines are available on the National Center for Biotechnology Information Sequence Read Archive as BioProject PRJNA268111. Custom Perl scripts used for population genetic analysis are available upon request.
Results
Construction and annotation of gene sets
The D. melanogaster immune system encompasses a network of interacting pathways that regulate diverse immune functions (reviewed in Ferrandon et al. 2007). By searching databases and the D. melanogaster literature, we assembled a list of 361 genes with well-supported immune roles and categorized each gene according to 12 immune pathways or processes and three functional classes, as outlined in Table 1. Categories were not mutually exclusive and some genes were assigned to multiple categories. While our list is expanded and updated, it maintains substantial overlap with previous large-scale studies of Drosophila immune genes (Sackton et al. 2007; Obbard et al. 2009).
We acquired SNP calls for each gene from 84 inbred D. melanogaster lines from five populations (Zimbabwe; the Netherlands; Beijing, China; Ithaca, NY; Tasmania, Australia; Grenier et al. 2015). We discarded three genes that had <25% sequence coverage within these lines, leaving us with a final study set of 358 immune genes. For each gene, we chose four control genes that were matched for size, location, and recombination rate. In the case of 15 immune genes, we were unable to identify four adequate control genes, leaving us with a final set of 1402 control genes. Sequence data for each control gene was similarly acquired for each of the 84 fly lines. In total, we identified 129,083 SNPs, 21,519 of which were in coding regions (6193 were nonsynonymous while 15,326 were synonymous; Figure S4).
Patterns of D. melanogaster-D. simulans divergence differ among gene classes
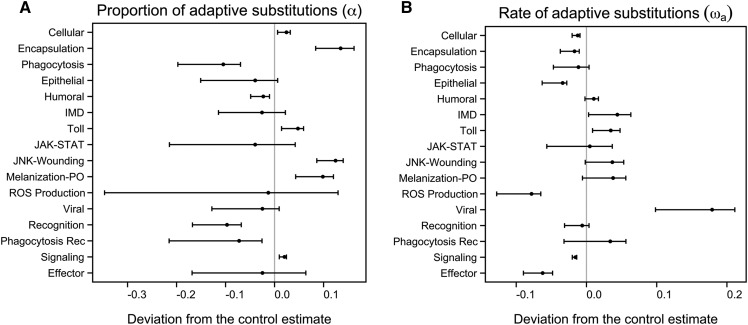
Within our full set of genes, there were 1208 control genes and 280 immune genes with one-to-one orthologs in D. simulans. For each of these genes, we calculated nucleotide divergence at nonsynonymous and fourfold degenerate sites. Combining these values with polymorphism data from the ancestral Zimbabwe population, we then estimated the proportion of adaptive substitutions (α) and the relative rate of adaptive substitutions (ωa) for each gene class (Eyre-Walker and Keightley 2009). The absolute values we obtained for α and ωa are given in Table S3. We acknowledge that the values we obtained for these statistics are relatively high compared to some other studies (but see Schneider et al. 2011). Our aim, however, is not to present an absolute measure of adaptation, but rather compare the relative α and ωa values of our various groups.
Consistent with previous cross-species comparisons (Sackton et al. 2007; Obbard et al. 2009), we found that there is apparent heterogeneity in patterns of adaptive substitution across immune gene classes (Figure 2). The Toll pathway, cellular defense genes, encapsulation genes, wound-repair genes, and genes involved in melanization and phenoloxidase (PO) production showed a significantly elevated α relative to the control estimate; consistent with a role of natural selection driving a high proportion of adaptive, amino acid-changing substitutions. Contrary to this, genes involved in humoral defense and phagocytosis had low proportions of adaptive substitutions relative to the control set (Figure 2A). The estimated α for viral response genes was not elevated, a result that may appear surprising given that viral response genes, and in particular genes involved in RNAi, are among the fastest evolving genes across the Drosophila phylogeny (Obbard et al. 2006, 2009; Kolaczkowski et al. 2011a). These genes, however, did have a markedly higher rate of adaptive substitution (ωa) relative to control estimates (Figure 2B). When considered together, these values of α and ωa suggest that viral response genes have fixed adaptive mutations at a high rate (high ωa), but during this process have also accrued a substantial number of substitutions that do not carry a fitness advantage (nonsignificant α). In addition, ωa was significantly elevated in IMD and Toll genes. Consistent with previous observations (Obbard et al. 2009), the effector class of AMPs showed little evidence of adaptive evolution at the nucleotide level (Table S3).
Figure 2.
Adaptive divergence of Drosophila immune gene groups. (A) Proportion of adaptive substitutions (α) and (B) relative rate of adaptive substitutions (ωa) calculated from D. melanogaster-D. simulans alignments using the DFE-alpha method (Eyre-Walker and Keightley 2009). Shown are the mean estimates with 95% C.I. calculated with jackknife resampling at the gene level. The vertical gray lines denote the 95% C.I. as calculated with a set of 1208 control genes. Values are presented as deviations from the control gene estimates. Rec, recognition; ROS, reactive oxygen species.
General patterns of polymorphism and FST are comparable for immune and control genes
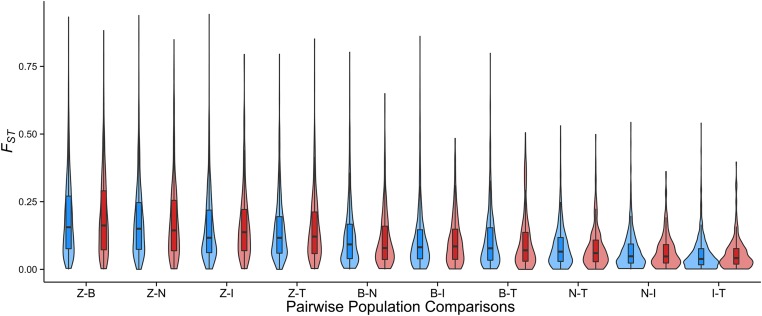
Using the polymorphism data from our 84 inbred D. melanogaster lines, we measured nucleotide diversity (π and θ), Tajima’s D, Fay and Wu’s H (Zeng et al. 2006), and Weir and Cockerham’s unbiased estimator of FST (Weir and Cockerham 1984) for each gene. The general population genetic patterns of both control and immune genes conformed to genome-wide observations made within these lines (Grenier et al. 2015). For both sets of genes, nucleotide diversity and FST between population pairs reflected known demographic patterns in D. melanogaster, which arose in sub-Saharan Africa, spread into Europe and Asia 10,000 years ago, and finally reached Australia and North America from Europe in the last 200 years (Figure 3; David and Capy 1988; Thornton and Andolfatto 2006; Keller 2007; Laurent et al. 2011). In the non-African populations, genes on the X chromosome harbored less nucleotide diversity at coding sites than genes on the four major autosomes (both π and θw, Mann–Whitney U-test, P < 0.001 for all control and immune comparisons). This is the expected pattern of polymorphism recovery following a founding event. For the Zimbabwe flies, there was no significant difference between nucleotide diversity on the X and autosomes.
Figure 3.
Patterns of pairwise FST for control (blue) and immune (red) genes. Levels of FST reflect D. melanogaster’s spread out of its ancestral range in sub-Saharan Africa (represented by the Zimbabwe population). Flies likely spread to Europe and Asia 10,000 years ago, only reaching Tasmania and North America through colonization by European populations within the last 200 years. Population samples are from Zimbabwe (Z); Beijing (B); the Netherlands (N); Ithaca, New York (I); and Tasmania (T).
We next compared large-scale polymorphism patterns within immune and control genes. Taken as a whole, immune genes did not display elevated population differentiation in any of the pairwise or global FST comparisons at nonsynonymous sites (Figure 3, Wilcoxon signed-rank test, P > 0.05). When comparing chromosomes individually, we did find differences between the groups, but there was no consistent trend and none of the P-values survived multiple-test correction.
Only two immune processes show evidence of augmented population differentiation
Since general patterns of evolution are comparable across all immune and control genes, we next investigated whether adaptation was elevated in certain immune processes or functions. To do so, we divided genes into two types of groups: (1) immune process or pathway, and (2) functional class (Table 1). Classifying genes by immune process allowed us to determine whether certain pathogen classes exert unusually strong, spatially variable selection on immune genes in flies. Since the innate immune response is somewhat compartmentalized, this would lead to signatures of selection being enriched within specific gene classes (Figure 1). For instance, observing high signatures of selection within the IMD pathway would suggest that Gram-negative bacteria display global variation in abundance or virulence, leading to differential selection pressures across populations. In addition, we analyzed genes grouped by functional class, a description of the gene’s position or function within a pathway. Past work has found that species-level rates of evolution differ among Drosophila genes based on their functional classification as recognition receptors, signaling molecules, or effector proteins (Sackton et al. 2007). This method of grouping allowed us to determine whether genetic adaptation was concentrated in particular locations throughout immune pathways.
We compared each immune gene class to its set of genomic control genes that were carefully matched for size, position, and local recombination rate. This approach partially controlled for potential evolutionary confounders that can influence the signatures of evolution observed across genomes. We compared each immune group to the mean of its relevant control group with a paired Wilcoxon signed-rank test and determined the significance of these comparisons through permutations. All statistics that fell within the 2.5 and 0.19% (equivalent to a significance level of 0.05 after Bonferroni correction) tails of the permuted null distributions are listed in Table 2 and Table S6.
Table 2. Statistics showing different patterns in immune vs. control gene classes.
| Gene class | Population(s) | Statistic | Direction of immune vs. control statistica |
|---|---|---|---|
| Cellular | Neth-Ith | FST | Greater |
| Phagocytosis | Global | FST | Greater |
| Ith-Tas | FST | Greater | |
| Neth-Ith | FST | Greater | |
| Neth-Tas | FST | Greater | |
| Epithelial | Netherlands | H | Less |
| IMD | Zimbabwe | π | Greater |
| Toll | Zim-Neth | FST | Less |
| JAK-STAT | Tasmania | D | Less |
| Viral defense | Zim-Ith | FST | Greater |
| Zim-Tas | FST | Greater | |
| RNAi | Beijing | H | Less |
| Zimbabwe | H | Less | |
| Neth-Tas | FST | Greater | |
| Zim-Ith | FST | Greater | |
| Recognition | Beijing | π | Greater |
| Ithaca | π | Greater | |
| Netherlands | π | Greater | |
| Tasmania | π | Greater | |
| Zimbabwe | π | Greater | |
| Ith-Tas | FST | Greater | |
| Neth-Ith | FST | Greater | |
| Neth-Tas | FST | Greater | |
| Phagocytosis recognition receptors | Beijing | π | Greater |
| Ithaca | π | Greater | |
| Ith-Tas | FST | Greater | |
| Neth-Ith | FST | Greater | |
| Neth-Tas | FST | Greater | |
| Zim-Beij | FST | Greater | |
| Zim-Tas | FST | Greater | |
| Humoral recognition receptors | Beijing | π | Greater |
| Ithaca | π | Greater | |
| Netherlands | π | Greater | |
| Tasmania | π | Greater | |
| Zimbabwe | π | Greater | |
| Netherlands | D | Greater | |
| Beijing | H | Less | |
| Ithaca | H | Less | |
| Tasmania | H | Less | |
| Effectors | Tasmania | π | Greater |
| AMPs | Tasmania | π | Greater |
Neth, the Netherlands; Ith, Ithaca, NY; Tas, Tasmania; Zim, Zimbabwe; Beij, Beijing.
All statistics were corrected for multiple testing and were significant at an α of 0.05, as determined by permutation.
Viral defense and phagocytosis are the only immune processes showing signs of rapid population differentiation:
Consistent with past studies, we found evidence of directional selection and high population differentiation within viral defense genes—and in particular RNAi genes. At a significance level corrected for multiple testing, two population pairs showed high FST for viral defense (Zimbabwe-Ithaca and Zimbabwe-Tasmania) and RNAi (the Netherlands-Tasmania and Zimbabwe-Ithaca). For both gene groups, the tests for heightened FST between Zimbabwe and all derived populations fell in the 2.5% tail. In addition, values of Fay and Wu’s H were significantly lower than the background for RNAi genes in Beijing and Zimbabwe, suggestive of higher levels of directional selection on derived alleles in these genes.
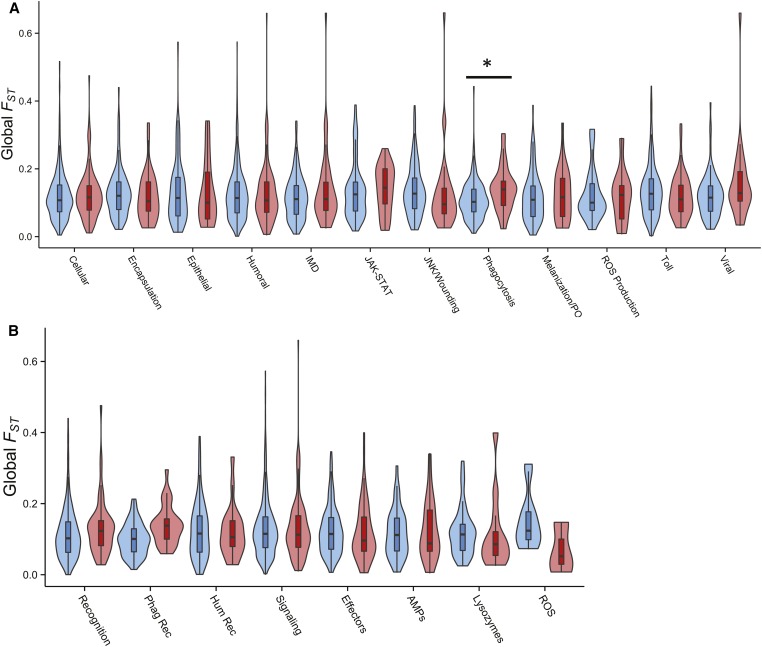
Genes involved in phagocytosis also showed evidence of unusually high population differentiation. In fact, the signal we observed for phagocytosis genes was even stronger than that seen for viral defense: global FST (Figure 4 and Figure S7) as well as three pairwise FST comparisons were significantly elevated compared to controls. As discussed below, however, this pattern was driven by recognition receptor genes. When we limited the analysis to phagocytosis genes without receptor function, we detected no deviations from background patterns. The larger class of cellular response genes (of which phagocytosis genes are a subset) also showed significantly higher FST between a single population pair: the Netherlands and Ithaca.
Figure 4.
Levels of global FST within gene classes at nonsynonymous sites. Immune genes (red) were categorized according to (A) immune process or (B) functional class, and then matched with four control genes (blue) based on chromosomal location, gene length, and local recombination rate. When compared with a Wilcoxon signed-rank test, only phagocytosis genes displayed a significant difference between the two groups after multiple-testing correction (α ≤ 0.05). ROS, reactive oxygen species.
Rapid population differentiation is not a universal mark of immune gene classes:
The only additional statistically significant FST comparison was lower—not higher—than the control expectation. This was the pairwise FST between the Netherlands and Zimbabwe for Toll genes. While this population pair was the only one to survive multiple-testing correction, it was not an anomaly among Toll gene comparisons. Tests for low FST fell within the 2.5% tail for four additional population pairs (Beijing-Tasmania, the Netherlands-Tasmania, Zimbabwe-Ithaca, and Zimbabwe-Tasmania).
Signatures of adaptation differ across functional classes:
Analyzing genes based on their functional class showed that recognition receptors are highly diverse and differentiated across populations. Recognition molecules had elevated nucleotide diversity relative to controls in all five populations, and higher than background FST in three pairwise comparisons. When tested individually, both subclasses of recognition receptors (phagocytosis and humoral) also showed strong evidence of high polymorphism and population divergence. Effector proteins only showed significantly elevated polymorphism in a single population (Tasmania) despite having a mean level of polymorphism that is higher than that of recognition genes. This somewhat counterintuitive observation is a result of our genomic control approach, which accounted for recombination rate, gene location, and gene size. In other words, effector molecules display patterns of polymorphism and divergence that are largely consistent with other small genes (mean transcript length = 792 nt), whereas recognition genes are unusual among the transcripts within their longer length class (mean transcript length = 1465 nt). In contrast to recognition and effector molecules, which function at the ends of their pathways, internal signaling molecules trended toward lower nucleotide diversity in all populations, although none of these tests were significant after multiple-testing correction.
It is worth noting that other genetic processes may contribute to the adaptation of effector molecules and recognition receptors. Both classes encompass large gene family groups, and effector gene families, in particular, are known to expand and contract at an unusually rapid pace (Sackton et al. 2007). Our focus on single nucleotide variants does not reflect this source of genetic novelty, and we note that these fly lines do contain two segregating duplications in the drosomycin family, a group of antifungal AMPs (Cardoso-Moreira et al. 2016).
Populations display similar levels of resistance to a generalist bacterial pathogen
In our gene class analysis, we found a trend toward lower-than-expected population differentiation in the canonical genes of the Toll pathway, which is the primary response pathway to fungal and Gram-negative bacterial infections. We were next interested in testing how the flies compared phenotypically. To do so, we infected the 84 lines with the generalist Gram-positive bacterial pathogen, E. faecalis. We chose to use a generalist pathogen to detect general, large-scale changes in immune response and regulation. This type of phenotypic difference might result from geographic variation in environmental factors like temperature (Lazzaro et al. 2008), rather than adaptations to unique population-specific pathogens. Questions of how populations are adapting to Drosophila-specific and geographically variable pathogens will need to be addressed once we gain a better understanding of the identity and geographic distribution of Drosophila pathogens.
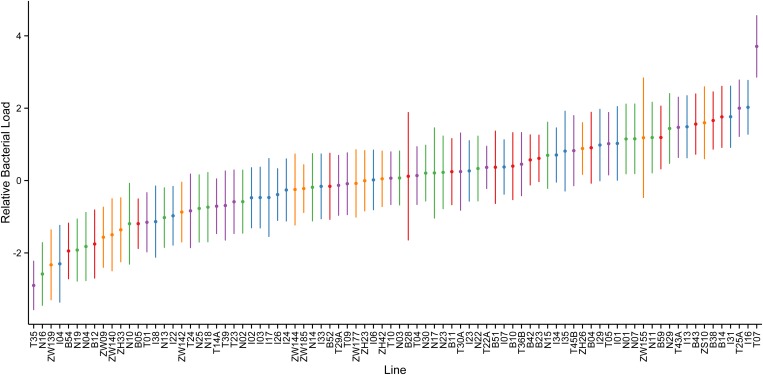
Individually, the 84 lines showed significant variation in bacterial load 28 hr after infection (Figure 5; ANOVA, d.f. = 83, F = 1.7, P = 6.9 × 10−5). This result shows that flies harbor extensive variation in immune resistance. On a population level, however, we detected no significant difference in bacterial load (ANOVA, d.f. = 4, F = 1.11, P = 0.36). This relative phenotypic homogeneity across populations contrasts with the population-level variation observed in a number of other phenotypes assayed in these same lines, including lipid content (Scheitz et al. 2013), metabolic regulation (Greenberg et al. 2011), and transposable element defense (Shiqi Luo, Andrew G. Clark and Jian Lu, unpublished data). Bacterial load is, of course, only one metric of immune competence (Ayres and Schneider 2009; Howick and Lazzaro 2014), and we only measured responses under a single environmental condition (Lazzaro et al. 2008). Nevertheless—and especially when coupled with our genetic analysis—these results suggest that global fly populations have not undergone widespread changes in their magnitude of response to generalist bacterial infection.
Figure 5.
Relative bacterial load in flies 28 hr after E. faecalis systemic infection. Flies were infected with the bacterial pathogen via septic pinprick and total bacterial load was measured after 28 hr. The values plotted are the mean residuals (± SE) from a model that accounted for sources of experimental error. Higher values correspond to a higher bacterial load and lower immune resistance. The color represents population of origin: Zimbabwe (orange); Beijing, China (red); the Netherlands (green); Ithaca, New York (blue); Tasmania (purple).
Immune genes with high population differentiation
Both our genetic and phenotypic analyses suggest that D. melanogaster populations do not vary widely in basic immune processes outside of viral defense and phagocytosis. Still, the populations may have experienced more limited—perhaps pathogen-specific—adaptations in a small number of genes. We therefore identified the individual genes showing signatures most consistent with local adaptation. Because we were primarily interested in identifying changes that are most likely to carry phenotypic effects, we continued to focus on statistics calculated at nonsynonymous sites (Table S5). It is important to note, therefore, that a number of genes’ statistics were calculated with data from a very small number of SNPs. For comparison, statistics calculated across the entire coding region are available in Table S4.
Table 3 lists the 20 immune genes that showed the highest degree of global population differentiation at nonsynonymous sites as measured by FST. Demonstrating that these patterns of population differentiation are robust to different sampling schemes, population structure within several of these genes has been previously discussed in the literature. These include Tak1 (Yukilevich et al. 2010), lectin-24A (Keebaugh and Schlenke 2012), and CHKov1 (Aminetzach et al. 2005; Magwire et al. 2011). Naturally segregating haplotypes of CHKov1 (Magwire et al. 2011) and DptA (Unckless et al. 2016) are even known to possess differential efficacy against certain viruses and bacterial strains, respectively. To our knowledge, natural variation in the remaining genes in Table 3 has not been discussed in the literature, and so these genes may represent novel candidate loci of local adaptation.
Table 3. Immune genes showing the highest global FST at nonsynonymous sites.
| Gene | Chr | S | Nucleotide diversity (π) (×1000) | Fay and Wu’s H | Population differentiation (FST) | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Zim | Neth | Beij | Ith | Tas | Zim | Neth | Beij | Ith | Tas | Zim-Beij | Zim-Neth | Zim-Ith | Zim-Tas | Beij-Neth | Beij-Ith | Beij-Tas | Neth-Tas | Neth-Ith | Ith-Tas | Global | |||
| Tak1 | X | 7 | 0.278 | 0.204 | 0.085 | 0.254 | 0.142 | −1.804 | 0.477 | 0.284 | −4.974 | −5.894 | 0.885 | 0.851 | 0.797 | 0.854 | 0.052 | 0.062 | 0.028 | 0.051 | 0.074 | 0.000 | 0.662 |
| lectin-24A | 2L | 24 | 4.904 | 2.644 | 6.891 | 2.364 | 4.061 | −4.875 | −1.292 | 0.860 | −2.839 | −1.049 | 0.626 | 0.732 | 0.750 | 0.692 | 0.243 | 0.218 | 0.092 | 0.065 | 0.022 | 0.053 | 0.476 |
| LysP | 3L | 4 | 2.740 | 0.321 | 0.814 | 0 | 0.339 | 0.821 | −4.216 | 0.402 | NA | −4.162 | 0.470 | 0.510 | 0.580 | 0.499 | 0.036 | 0.043 | 0.035 | 0.000 | 0.024 | 0.028 | 0.401 |
| DptA | 2R | 5 | 4.330 | 2.595 | 0.000 | 3.996 | 5.455 | −0.215 | −3.465 | NA | −1.703 | −0.988 | 0.776 | 0.609 | 0.473 | 0.270 | 0.056 | 0.172 | 0.311 | 0.137 | 0.047 | 0.048 | 0.342 |
| Ect4 | 3L | 23 | 0.875 | 0.333 | 0.598 | 0.403 | 0.556 | 1.538 | −0.572 | −3.446 | −3.677 | −3.655 | 0.494 | 0.610 | 0.590 | 0.518 | 0.055 | 0.107 | 0.032 | 0.035 | 0.033 | 0.029 | 0.342 |
| slpr | X | 15 | 1.136 | 0.270 | 0.456 | 0.107 | 0.203 | 0.294 | 0.646 | 0.426 | 0.537 | 0.610 | 0.202 | 0.394 | 0.439 | 0.413 | 0.418 | 0.479 | 0.432 | 0.012 | 0.038 | 0.000 | 0.340 |
| Eb1 | 2R | 2 | 0.662 | 0.000 | 0.355 | 0.810 | 0.459 | 0.578 | NA | 0.480 | −2.043 | −2.644 | 0.592 | 0.726 | 0.429 | 0.525 | 0.122 | 0.015 | 0.000 | 0.069 | 0.164 | 0.006 | 0.337 |
| hep | X | 19 | 0.731 | 0.167 | 0.690 | 0.716 | 0.750 | 0.911 | −1.777 | −1.500 | −2.023 | −1.252 | 0.462 | 0.732 | 0.527 | 0.452 | 0.265 | 0.048 | 0.003 | 0.221 | 0.087 | 0.037 | 0.336 |
| Tl | 3R | 36 | 2.940 | 1.137 | 1.067 | 0.803 | 1.154 | 0.040 | 1.191 | 1.095 | −1.434 | 1.597 | 0.393 | 0.401 | 0.528 | 0.390 | 0.023 | 0.403 | 0.075 | 0.053 | 0.346 | 0.325 | 0.335 |
| PGRP-LE | X | 9 | 0.540 | 1.384 | 0.307 | 1.637 | 1.895 | 0.595 | −0.309 | 0.480 | 0.679 | 0.552 | 0.762 | 0.450 | 0.254 | 0.322 | 0.652 | 0.411 | 0.394 | 0.076 | 0.109 | 0.008 | 0.332 |
| Dsp1 | X | 1 | 0 | 0 | 0.520 | 0 | 0 | NA | NA | 0.546 | NA | NA | 0.292 | NA | NA | NA | 0.326 | 0.342 | 0.334 | NA | NA | NA | 0.317 |
| dnr1 | 2R | 5 | 0.261 | 0.434 | 0.113 | 0.541 | 0.272 | 0.451 | −1.145 | 0.284 | 0.647 | 0.477 | 0.039 | 0.583 | 0.126 | 0.052 | 0.588 | 0.076 | 0.000 | 0.499 | 0.287 | 0.028 | 0.308 |
| Vps33B | 3R | 33 | 3.326 | 0.613 | 3.352 | 1.383 | 1.791 | 0.883 | −2.409 | −0.549 | −4.373 | −4.415 | 0.265 | 0.628 | 0.541 | 0.446 | 0.209 | 0.143 | 0.061 | 0.082 | 0.021 | 0.033 | 0.304 |
| Adgf-A | 3L | 14 | 0.606 | 1.002 | 1.168 | 1.086 | 1.410 | −3.002 | −0.831 | −0.351 | −0.077 | −1.049 | 0.388 | 0.630 | 0.543 | 0.153 | 0.285 | 0.132 | 0.073 | 0.343 | 0.066 | 0.212 | 0.301 |
| modSP | 3R | 8 | 0.589 | 0.291 | 0.285 | 0.196 | 0.292 | −0.537 | 0.607 | 0.502 | 0.537 | 0.611 | 0.502 | 0.516 | 0.433 | 0.300 | 0.060 | 0.082 | 0.158 | 0.176 | 0.106 | 0.005 | 0.299 |
| crq | 2L | 4 | 0.135 | 0.092 | 0.117 | 0.450 | 0 | 0.318 | 0.234 | 0.284 | 0.331 | NA | 0.037 | 0.037 | 0.323 | 0.058 | 0.031 | 0.337 | 0.043 | 0.024 | 0.363 | 0.398 | 0.297 |
| Atf-2 | 2R | 5 | 1.534 | 0.535 | 0.941 | 0.741 | 1.035 | −0.436 | 0.595 | −1.134 | 0.810 | 0.828 | 0.236 | 0.475 | 0.395 | 0.255 | 0.474 | 0.357 | 0.361 | 0.100 | 0.000 | 0.071 | 0.292 |
| Eph | 4 | 5 | 0.202 | 0.152 | 0.053 | 0.221 | 0.203 | −0.693 | 0.546 | 0.284 | 0.575 | 0.704 | 0.556 | 0.481 | 0.469 | 0.449 | 0.082 | 0.209 | 0.098 | 0.005 | 0.155 | 0.045 | 0.284 |
| CHKov1 | 3R | 15 | 5.429 | 1.783 | 3.623 | 2.430 | 4.302 | 0.281 | −8.288 | −5.975 | −7.584 | −3.864 | 0.402 | 0.571 | 0.530 | 0.356 | 0.038 | 0.037 | 0.000 | 0.066 | 0.000 | 0.048 | 0.273 |
| DptB | 2R | 8 | 6.550 | 2.893 | 0.488 | 4.407 | 4.041 | 1.153 | −2.478 | 0.284 | −0.959 | −0.673 | 0.638 | 0.453 | 0.306 | 0.280 | 0.191 | 0.206 | 0.305 | 0.043 | 0.053 | 0.007 | 0.272 |
Values that fall in the extreme 5% of the population’s distribution are marked with italic font. Chr, chromosome; S, number of segregating sites; Zim, Zimbabwe; Neth, the Netherlands; Beij, Beijing; Ith, Ithaca, NY; Tas, Tasmania.
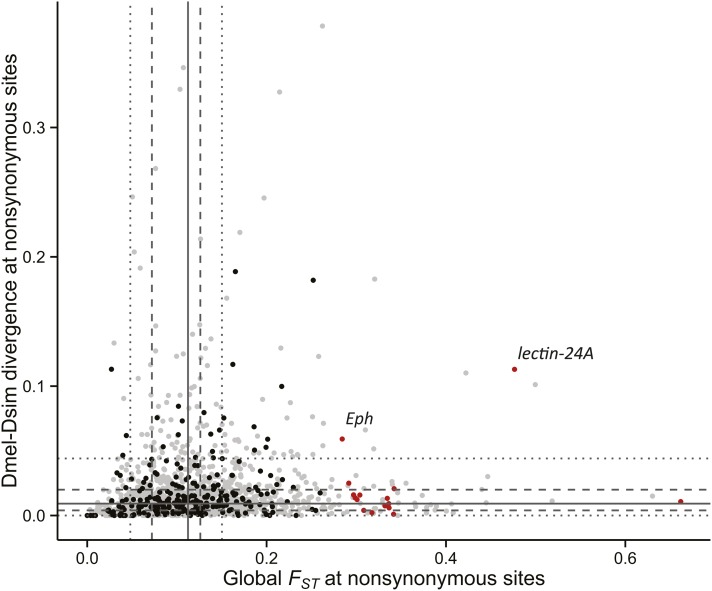
Previously, Maruki et al. (2012) demonstrated that loci experiencing higher purifying selection show lower levels of population differentiation. This can be a confounding factor in outlier approaches such as ours, biasing results toward genes experiencing relaxed selective constraint. We, too, find a positive correlation within our immune genes between global FST at nonsynonymous sites and nonsynonymous divergence between D. melanogaster and D. simulans (Spearman’s ρ = 0.236, P = 0.0001; Figure 6). The pattern in two genes—Eph and lectin-24A—is consistent with a model of relaxed constraint as they are outliers for both global nonsynonymous FST and Dn. Importantly, however, these are the only two high FST genes that are outliers for both statistics, suggesting that alleles within the other genes may not experience relaxed constraint.
Figure 6.
Gene divergence and global population differentiation at nonsynonymous sites. Across 264 immune genes (black), D. melanogaster-D. simulans nucleotide divergence and global FST correlate (Spearman’s ρ = 0.236, P = 0.0001). The solid, dashed, and dotted lines mark the median, the lower and upper quartiles, and the quartiles ±1.5 interquartile range, respectively, as calculated with the control genes (gray). The genes identified in Table 3 are marked in red.
For 10 of the outlier genes in Table 3, the high global FST is largely driven by differences between the ancestral Zimbabwe population and the other four derived populations. This pattern is consistent with D. melanogaster’s known demographic history, and so might be attributed to neutral forces. To further assess these genes’ validity as selection candidates, we therefore included other statistics like nucleotide diversity and Fay and Wu’s H, which specifically consider changes in the derived allele frequency (Table 3). Interestingly, two of the outlier genes (lectin-24A and Adgf-A) have low values of H that fall within the bottom 5% of African haplotypes. This suggests that the differences in population allele frequencies are partially attributed to recent changes in Africa and not solely the result of an out-of-Africa bottleneck.
There are also instances where a single derived population largely drives the global signature of population differentiation. For instance, the patterns of pairwise FST, reduced π, and low H suggest that LysP and Vps33B may have experienced directional selection in the Netherlands before the recent colonization of North America and Australia.
Discussion
The results in this study demonstrate that rapid adaptation is the exception, not the rule, among immune genes in D. melanogaster. As has been previously demonstrated (Obbard et al. 2006, 2009), genes involved in viral defense—and in particular the RNAi portion of this process—display signs of elevated selection. Our analysis of other pathways, however, shows that the extent of this adaptive signature is unusual, not only in a genome-wide context but also in comparison to other immune processes. Indeed, several immune processes even showed unexpectedly low levels of population differentiation that were nominally significant when compared to the genetic background. In addition, instead of occurring through small changes in numerous genes throughout a pathway—as in the polygenic selection model—most local immune adaptation appears to progress through allele frequency changes in a few genes at the periphery that are likely to interact directly with pathogens. This suggests that abiotic factors in the flies’ environment may not be major drivers of local immune adaptation at the amino acid level.
Across all populations, recognition genes showed high amino acid diversity and signs of heightened population differentiation. While we have not shown that this variation drives changes in pathogen recognition, previous lines of evidence suggest that the variation in recognition receptors does carry phenotypic effects. First, this observation is in line with theoretical predictions that recognition molecules serve as the main locus of host-pathogen coevolution (Nuismer and Dybdahl 2016) and with observations in mice that upstream initiation genes are more likely to undergo positive selection (Casals et al. 2011; Webb et al. 2015). Second, past quantitative genetic studies in Drosophila have suggested that natural variation in immune resistance is largely driven by polymorphisms in recognition and, to a lesser extent, signaling genes (Lazzaro et al. 2006; Sackton et al. 2010). This implies that D. melanogaster recognition genes carry a substantial level of functional standing variation. Selection could act on this standing variation in a population-specific manner, leading to differentiation across populations. Alternatively, recognition genes, like effector genes (Unckless and Lazzaro 2016), may experience balancing selection, and the resulting asynchronous fluctuations across populations could heighten the population differentiation measured in our samples.
Balancing selection and temporal fluctuations in allele frequencies might also play a role in one of the more intriguing results from this analysis: the discordance between the species-level and population-level analyses of adaptation. Phagocytosis genes and recognition receptors showed no evidence of elevated rates of adaptation in the divergence data, but showed heightened levels of population differentiation. These gene classes maintain a large number of polymorphic sites (Figure S5 and Figure S6), a common hallmark of balancing selection, which would drive down values of α and ωa. Conversely, we found that both the Toll and IMD pathways showed elevated rates of adaptive substitution, but Toll genes trended toward lower levels of population differentiation. The full reason for this incongruity across different timescales remains an open question (Messer et al. 2016), but one likely factor is that our analyses are static snapshots of dynamic processes. Our population samples captured a single time point that provides no information on the known temporal fluctuations that occur in wild populations (Bergland et al. 2014). Similarly, species divergence data focus on fixed differences, an endpoint that is largely blind to the trajectory alleles took on their way to fixation. Antiviral genes are perhaps the only class where directional selection is sufficiently strong to create robust signatures of positive selection in the genome on both timescales. Further research on the interactions between the Drosophila host and its pathogens, the variability in pathogen distributions, and the temporal dynamics of immune processes will greatly extend our understanding of these evolutionary patterns.
Looking to studies performed in other organisms, we see that our results are not without precedent. In a laboratory-based study with the insect Tribolium castaneum, Berenos et al. (2011) demonstrated that evolution in the presence of parasites counteracted the effects of drift, leading to higher levels of allelic diversity within populations and lower levels of differentiation between populations. Although more diverged and in possession of an adaptive immune response, humans also show patterns in their innate immune genes that are similar to what we find here: while numerous single genes show signs of directional selection, innate immune genes have globally experienced stronger purifying selection than nonimmune genes (Mukherjee et al. 2009; Deschamps et al. 2016). Together, these results highlight that immune evolution is more complex than the outlier scenarios that garner the greatest attention in the literature. To fully understand immune adaptation, it will therefore be key to not only search out the points of rapid divergence, but also consider what drives—or allows for—the maintenance of low genetic variability across diverse environments. This more inclusive view of immune evolution will no doubt yield increased insight into the process of adaptation in this key ecological trait.
Acknowledgments
We thank Brian Lazzaro, Tim Connallon, Charles Aquadro, Todd Schlenke, Richard Harrison, and members of the Clark and Aquadro laboratory groups for helpful discussions, insights, and comments on this work and on an earlier version of this manuscript. This work was supported by the National Institutes of Health (grant number R01 AI-064950 to A.G.C. and Brian Lazzaro) and by a National Science Foundation graduate research fellowship to A.M.E.
Footnotes
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.195016/-/DC1.
Communicating editor: M. W. Hahn
Literature Cited
- Aminetzach Y. T., Macpherson J. M., Petrov D. A., 2005. Pesticide resistance via transposition-mediated adaptive gene truncation in Drosophila. Science 309: 764–767. [DOI] [PubMed] [Google Scholar]
- Ayres J. S., Schneider D. S., 2009. The role of anorexia in resistance and tolerance to infections in Drosophila. PLoS Biol. 7: e1000150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayres J. S., Freitag N., Schneider D. S., 2008. Identification of Drosophila mutants altering defense of and endurance to Listeria monocytogenes infection. Genetics 178: 1807–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babin A., Kolly S., Schneider F., Dolivo V., Zini M., et al. , 2014. Fruit flies learn to avoid odours associated with virulent infection. Biol. Lett. 10: 20140048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berenos C., Wegner K. M., Schmid-Hempel P., 2011. Antagonistic coevolution with parasites maintains host genetic diversity: an experimental test. Proc. Biol. Sci. 278: 218–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergland A. O., Behrman E. L., O’Brien K. R., Schmidt P. S., Petrov D. A., 2014. Genomic evidence of rapid and stable adaptive oscillations over seasonal time scales in Drosophila. PLoS Genet. 10: e1004775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso-Moreira M., Arguello J. R., Gottipati S., Harshman L. G., Grenier J. K., et al. , 2016. Evidence for the fixation of gene duplications by positive selection in Drosophila. Genome Research 26: 787–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casals F., Sikora M., Laayouni H., Montanucci L., Muntasell A., et al. , 2011. Genetic adaptation of the antibacterial human innate immunity network. BMC Evol. Biol. 11: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano D., Coronado-Zamora M., Campos J. L., Barbadilla A., Eyre-Walker A., 2016. Adaptive evolution is substantially impeded by Hill-Robertson interference in Drosophila. Mol. Biol. Evol. 33: 442–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez-Galarza J., Henriques D., Johnston J. S., Azevedo J. C., Patton J. C., et al. , 2013. Signatures of selection in the Iberian honey bee (Apis mellifera iberiensis) revealed by a genome scan analysis of single nucleotide polymorphisms. Mol. Ecol. 22: 5890–5907. [DOI] [PubMed] [Google Scholar]
- Comeron J. M., Ratnappan R., Bailin S., 2012. The many landscapes of recombination in Drosophila melanogaster. PLoS Genet. 8: e1002905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford J. E., Guelbeogo W. M., Sanou A., Traore A., Vernick K. D., et al. , 2010. De novo transcriptome sequencing in Anopheles funestus using Illumina RNA-seq technology. PLoS One 5: e14202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daub J. T., Hofer T., Cutivet E., Dupanloup I., Quintana-Murci L., et al. , 2013. Evidence for polygenic adaptation to pathogens in the human genome. Mol. Biol. Evol. 30: 1544–1558. [DOI] [PubMed] [Google Scholar]
- David J. R., Capy P., 1988. Genetic-variation of Drosophila melanogaster natural-populations. Trends Genet. 4: 106–111. [DOI] [PubMed] [Google Scholar]
- Deschamps M., Laval G., Fagny M., Itan Y., Abel L., et al. , 2016. Genomic signatures of selective pressures and introgression from archaic hominins at human innate immunity genes. Am. J. Hum. Genet. 98: 5–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erler S., Lhomme P., Rasmont P., Lattorff H. M., 2014. Rapid evolution of antimicrobial peptide genes in an insect host-social parasite system. Infect. Genet. Evol. 23: 129–137. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A., Keightley P. D., 2009. Estimating the rate of adaptive molecular evolution in the presence of slightly deleterious mutations and population size change. Mol. Biol. Evol. 26: 2097–2108. [DOI] [PubMed] [Google Scholar]
- Fabian D. K., Kapun M., Nolte V., Kofler R., Schmidt P. S., et al. , 2012. Genome-wide patterns of latitudinal differentiation among populations of Drosophila melanogaster from North America. Mol. Ecol. 21: 4748–4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandon D., Imler J. L., Hetru C., Hoffmann J. A., 2007. The Drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nat. Rev. Immunol. 7: 862–874. [DOI] [PubMed] [Google Scholar]
- Fiston-Lavier A. S., Singh N. D., Lipatov M., Petrov D. A., 2010. Drosophila melanogaster recombination rate calculator. Gene 463: 18–20. [DOI] [PubMed] [Google Scholar]
- Fumagalli M., Sironi M., Pozzoli U., Ferrer-Admetlla A., Pattini L., et al. , 2011. Signatures of environmental genetic adaptation pinpoint pathogens as the main selective pressure through human evolution. PLoS Genet. 7: e1002355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg A. J., Hackett S. R., Harshman L. G., Clark A. G., 2011. Environmental and genetic perturbations reveal different networks of metabolic regulation. Mol. Syst. Biol. 7: 563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenier J. K., Arguello J. R., Moreira M. C., Gottipati S., Mohammed J., et al. , 2015. Global diversity lines—a five-continent reference panel of sequenced Drosophila melanogaster strains. G3 (Bethesda) 5: 593–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hereford J., 2009. A quantitative survey of local adaptation and fitness trade-offs. Am. Nat. 173: 579–588. [DOI] [PubMed] [Google Scholar]
- Howick V. M., Lazzaro B. P., 2014. Genotype and diet shape resistance and tolerance across distinct phases of bacterial infection. BMC Evol. Biol. 14: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner S., Rashkovetsky E., Kim Y. B., Oh J. H., Michalak K., et al. , 2013. Genome differentiation of Drosophila melanogaster from a microclimate contrast in Evolution Canyon, Israel. Proc. Natl. Acad. Sci. USA 110: 21059–21064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong C., Di Rienzo A., 2014. Adaptations to local environments in modern human populations. Curr. Opin. Genet. Dev. 29: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones F. C., Grabherr M. G., Chan Y. F., Russell P., Mauceli E., et al. , 2012. The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484: 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juneja P., Lazzaro B. P., 2010. Haplotype structure and expression divergence at the Drosophila cellular immune gene eater. Mol. Biol. Evol. 27: 2284–2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacsoh B. Z., Lynch Z. R., Mortimer N. T., Schlenke T. A., 2013. Fruit flies medicate offspring after seeing parasites. Science 339: 947–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keebaugh E. S., Schlenke T. A., 2012. Adaptive evolution of a novel Drosophila lectin induced by parasitic wasp attack. Mol. Biol. Evol. 29: 565–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A., 2007. Drosophila melanogaster’s history as a human commensal. Curr. Biol. 17: R77–R81. [DOI] [PubMed] [Google Scholar]
- Kolaczkowski B., Hupalo D. N., Kern A. D., 2011a Recurrent adaptation in RNA interference genes across the Drosophila phylogeny. Mol. Biol. Evol. 28: 1033–1042. [DOI] [PubMed] [Google Scholar]
- Kolaczkowski B., Kern A. D., Holloway A. K., Begun D. J., 2011b Genomic differentiation between temperate and tropical Australian populations of Drosophila melanogaster. Genetics 187: 245–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraaijeveld A. R., Limentani E. C., Godfray H. C., 2001. Basis of the trade-off between parasitoid resistance and larval competitive ability in Drosophila melanogaster. Proc. Biol. Sci. 268: 259–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraaijeveld A. R., Layen S. J., Futerman P. H., Godfray H. C., 2012. Lack of phenotypic and evolutionary cross-resistance against parasitoids and pathogens in Drosophila melanogaster. PLoS One 7: e53002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhaney S., Martinez Barrio A., Rafati N., Sundstrom G., Rubin C. J., et al. , 2012. Population-scale sequencing reveals genetic differentiation due to local adaptation in Atlantic herring. Proc. Natl. Acad. Sci. USA 109: 19345–19350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larracuente A. M., Sackton T. B., Greenberg A. J., Wong A., Singh N. D., et al. , 2008. Evolution of protein-coding genes in Drosophila. Trends Genet. 24: 114–123. [DOI] [PubMed] [Google Scholar]
- Laurent S. J., Werzner A., Excoffier L., Stephan W., 2011. Approximate Bayesian analysis of Drosophila melanogaster polymorphism data reveals a recent colonization of Southeast Asia. Mol. Biol. Evol. 28: 2041–2051. [DOI] [PubMed] [Google Scholar]
- Lazzaro B. P., Little T. J., 2009. Immunity in a variable world. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364: 15–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro B. P., Sceurman B. K., Clark A. G., 2004. Genetic basis of natural variation in D. melanogaster antibacterial immunity. Science 303: 1873–1876. [DOI] [PubMed] [Google Scholar]
- Lazzaro B. P., Sackton T. B., Clark A. G., 2006. Genetic variation in Drosophila melanogaster resistance to infection: a comparison across bacteria. Genetics 174: 1539–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro B. P., Flores H. A., Lorigan J. G., Yourth C. P., 2008. Genotype-by-environment interactions and adaptation to local temperature affect immunity and fecundity in Drosophila melanogaster. PLoS Pathog. 4: e1000025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loytynoja A., Goldman N., 2005. An algorithm for progressive multiple alignment of sequences with insertions. Proc. Natl. Acad. Sci. USA 102: 10557–10562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magwire M. M., Bayer F., Webster C. L., Cao C., Jiggins F. M., 2011. Successive increases in the resistance of Drosophila to viral infection through a transposon insertion followed by a duplication. PLoS Genet. 7: e1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruki T., Kumar S., Kim Y., 2012. Purifying selection modulates the estimates of population differentiation and confounds genome-wide comparisons across single-nucleotide polymorphisms. Mol. Biol. Evol. 29: 3617–3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKean K. A., Yourth C. P., Lazzaro B. P., Clark A. G., 2008. The evolutionary costs of immunological maintenance and deployment. BMC Evol. Biol. 8: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McTaggart S. J., Obbard D. J., Conlon C., Little T. J., 2012. Immune genes undergo more adaptive evolution than non-immune system genes in Daphnia pulex. BMC Evol. Biol. 12: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer P. W., Ellner S. P., Hairston N. G., Jr, 2016. Can population genetics adapt to rapid evolution? Trends Genet. 32: 408–418. [DOI] [PubMed] [Google Scholar]
- Mukherjee S., Sarkar-Roy N., Wagener D. K., Majumder P. P., 2009. Signatures of natural selection are not uniform across genes of innate immune system, but purifying selection is the dominant signature. Proc. Natl. Acad. Sci. USA 106: 7073–7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Gojobori T., 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3: 418–426. [DOI] [PubMed] [Google Scholar]
- Nuismer S. L., Dybdahl M. F., 2016. Quantifying the coevolutionary potential of multistep immune defenses. Evolution 70: 282–295. [DOI] [PubMed] [Google Scholar]
- Obbard D. J., Jiggins F. M., Halligan D. L., Little T. J., 2006. Natural selection drives extremely rapid evolution in antiviral RNAi genes. Curr. Biol. 16: 580–585. [DOI] [PubMed] [Google Scholar]
- Obbard D. J., Welch J. J., Kim K. W., Jiggins F. M., 2009. Quantifying adaptive evolution in the Drosophila immune system. PLoS Genet. 5: e1000698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pespeni M. H., Garfield D. A., Manier M. K., Palumbi S. R., 2012. Genome-wide polymorphisms show unexpected targets of natural selection. Proc. Biol. Sci. 279: 1412–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Di Rienzo A., 2010. Adaptation - not by sweeps alone. Nat. Rev. Genet. 11: 665–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Pickrell J. K., Coop G., 2010. The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr. Biol. 20: R208–R215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana-Murci L., Clark A. G., 2013. Population genetic tools for dissecting innate immunity in humans. Nat. Rev. Immunol. 13: 280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team , 2011. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Sackton T. B., Lazzaro B. P., Schlenke T. A., Evans J. D., Hultmark D., et al. , 2007. Dynamic evolution of the innate immune system in Drosophila. Nat. Genet. 39: 1461–1468. [DOI] [PubMed] [Google Scholar]
- Sackton T. B., Lazzaro B. P., Clark A. G., 2010. Genotype and gene expression associations with immune function in Drosophila. PLoS Genet. 6: e1000797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheitz C. J., Guo Y., Early A. M., Harshman L. G., Clark A. G., 2013. Heritability and inter-population differences in lipid profiles of Drosophila melanogaster. PLoS One 8: e72726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A., Charlesworth B., Eyre-Walker A., Keightley P. D., 2011. A method for inferring the rate of occurrence and fitness effects of advantageous mutations. Genetics 189: 1427–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironi M., Cagliani R., Forni D., Clerici M., 2015. Evolutionary insights into host-pathogen interactions from mammalian sequence data. Nat. Rev. Genet. 16: 224–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapley J., Reger J., Feulner P. G., Smadja C., Galindo J., et al. , 2010. Adaptation genomics: the next generation. Trends Ecol. Evol. 25: 705–712. [DOI] [PubMed] [Google Scholar]
- Stephan W., 2016. Signatures of positive selection: from selective sweeps at individual loci to subtle allele frequency changes in polygenic adaptation. Mol. Ecol. 25: 79–88. [DOI] [PubMed] [Google Scholar]
- Thornton K., Andolfatto P., 2006. Approximate Bayesian inference reveals evidence for a recent, severe bottleneck in a Netherlands population of Drosophila melanogaster. Genetics 172: 1607–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchin M. C., Chiang C. W., Palmer C. D., Sankararaman S., Reich D., et al. , 2012. Evidence of widespread selection on standing variation in Europe at height-associated SNPs. Nat. Genet. 44: 1015–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner T. L., Levine M. T., Eckert M. L., Begun D. J., 2008. Genomic analysis of adaptive differentiation in Drosophila melanogaster. Genetics 179: 455–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner T. L., Bourne E. C., Von Wettberg E. J., Hu T. T., Nuzhdin S. V., 2010. Population resequencing reveals local adaptation of Arabidopsis lyrata to serpentine soils. Nat. Genet. 42: 260–263. [DOI] [PubMed] [Google Scholar]
- Unckless R. L., Lazzaro B. P., 2016. The potential for adaptive maintenance of diversity in insect antimicrobial peptides. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371: 20150291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unckless R. L., Howick V. M., Lazzaro B. P., 2016. Convergent balancing selection on an antimicrobial peptide in Drosophila. Curr. Biol. 26: 257–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse R. M., Kriventseva E. V., Meister S., Xi Z., Alvarez K. S., et al. , 2007. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science 316: 1738–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb A. E., Gerek Z. N., Morgan C. C., Walsh T. A., Loscher C. E., et al. , 2015. Adaptive evolution as a predictor of species-specific innate immune response. Mol. Biol. Evol. 32: 1717–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir B. S., Cockerham C. C., 1984. Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370. [DOI] [PubMed] [Google Scholar]
- Ye K., Schulz M. H., Long Q., Apweiler R., Ning Z., 2009. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25: 2865–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yukilevich R., Turner T. L., Aoki F., Nuzhdin S. V., True J. R., 2010. Patterns and processes of genome-wide divergence between North American and African Drosophila melanogaster. Genetics 186: 219–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng K., Fu Y. X., Shi S., Wu C. I., 2006. Statistical tests for detecting positive selection by utilizing high-frequency variants. Genetics 174: 1431–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Table S1 and Table S2 list the names and coordinates of the immune and control genes and transcripts used in the analysis. Sequence data from the Global Diversity Lines are available on the National Center for Biotechnology Information Sequence Read Archive as BioProject PRJNA268111. Custom Perl scripts used for population genetic analysis are available upon request.