Abstract
Statistical models in medical and population genetics typically assume that individuals assort randomly in a population. While this simplifies model complexity, it contradicts an increasing body of evidence of nonrandom mating in human populations. Specifically, it has been shown that assortative mating is significantly affected by genomic ancestry. In this work, we examine the effects of ancestry-assortative mating on the linkage disequilibrium between local ancestry tracks of individuals in an admixed population. To accomplish this, we develop an extension to the Wright–Fisher model that allows for ancestry-based assortative mating. We show that ancestry-assortment perturbs the distribution of local ancestry linkage disequilibrium (LAD) and the variance of ancestry in a population as a function of the number of generations since admixture. This assortment effect can induce errors in demographic inference of admixed populations when methods assume random mating. We derive closed form formulae for LAD under an assortative-mating model with and without migration. We observe that LAD depends on the correlation of global ancestry of couples in each generation, the migration rate of each of the ancestral populations, the initial proportions of ancestral populations, and the number of generations since admixture. We also present the first direct evidence of ancestry-assortment in African Americans and examine LAD in simulated and real admixed population data of African Americans. We find that demographic inference under the assumption of random mating significantly underestimates the number of generations since admixture, and that accounting for assortative mating using the patterns of LAD results in estimates that more closely agrees with the historical narrative.
Keywords: admixture, assortative mating, demography, migration, population genetics
ONE of the most common assumptions in human population genetics analyses is that of Hardy–Weinberg Equilibrium (HWE). The HWE assumption in turn enforces a set of additional conditions including the absence of selection, infinite population size, and importantly, random mating. Assortative mating is a common phenomenon (Mathews and Reus 2001; Risch et al. 2009) and many phenotypes including height, education level, and personality traits are correlated between spouses (Merikangas 1982). For Latinos and other admixed populations, the African, Native-American, and European proportions of individual’s genomes can be correlated between spouses. We and others have demonstrated that the genomic ancestry of Latino couples is highly correlated (Risch et al. 2009; Zou et al. 2015), and refer to this as ancestry-assortative mating. Thus, the assumption of random mating and therefore HWE is not satisfied in practice, and the implication of this observation for population and evolutionary genetic studies remains unclear.
The assumption of random mating is used in many types of population and quantitative genetics analyses. Particularly, random mating is assumed both in analysis of population genetics data and when inferring population parameters such as recombination rates, mutation rates, selection, heritability, and others. Moreover, methods for quality control and data cleaning often make the random mating assumption. For example, methods for haplotype phasing typically compute the likelihood of the genotype as the product of the likelihoods of each of the haplotypes, and this derivation is based on the random mating assumption (Marchini et al. 2006). Similarly, such likelihood derivations are also common in methods for the inference of identity-by-descent and inference of ancestry from genomic data (Browning and Browning 2013). Thus far, the sensitivity of these methods to the assumption of assortative mating has not been evaluated. In principle, realistic violations of the random mating assumption may not be detrimental to existing methods; however, this needs to be taken to the test.
In this paper, we explore the robustness of specific genetic features and their inference from genetic data to assortative mating. Because ancestry proportion has been shown to be highly correlated in Latino spouses, we focused our analysis on the behavior of ancestry linkage disequilibrium under assortative mating. We propose a random generative model for population dynamics under assortative mating that is due to population structure. Our model follows the spirit of the Wright–Fisher model, and makes the assumption that the correlation of ancestry proportions between spouses stays fixed across generations. Particularly, when the correlation of ancestry proportions is zero, our model is equivalent to the Wright–Fisher model.
We develop mathematical theory that describes the decay of local ancestry disequilibrium (LAD) as a function of assortative mating strength, migration rate, recombination rate, and the number of generations since admixture began. Thus, one can use these results to infer the demographic history of admixed populations. Several methods for demographic inference in admixed populations exist including ones that use patterns of linkage disequilibrium (LD) decay (Loh et al. 2013), local ancestry track length distribution (Price et al. 2009), and the distribution of identity-by-descent segments (Gravel et al. 2013). However, these methods assume random mating, and under assortative mating LD decay follows a different pattern (Parra et al. 2001). Using simulations, we demonstrate that our mathematical derivation matches empirical LAD decay. Furthermore, we develop the theory with migration rates from the ancestral populations, and we demonstrate that, in the presence of assortative mating, one may erroneously conclude that there has been active migration and vice versa.
We applied our analysis to a data set of 1730 African Americans from the Study of African Americans, Asthma, Genes and Environments (SAGE) study (Borrell et al. 2013). The existence of ancestry-assortative mating in African Americans has been previously suggested by indirect examinations of related features including skin color and varying ancestry distributions across geographic regions (Udry et al. 1971; Bryc et al. 2015; Baharian et al. 2016). Here, we present the first direct evidence of ancestry-assortment in African Americans. We used ANCESTOR (Zou et al. 2015) to show that the correlation of African ancestry between the spouses in the last generation is ∼0.32. We then used our analysis to infer the number of generations and migration patterns in the African American population. Under the assumption of no migrations and random mating, an analysis of LAD resulted in an estimate of the number of generations since admixture of three. Adding assortment and migrations, we find that the estimated number of generations since the admixture event is 15. Assuming a generation time of 25 years, this places the initial migrations in the mid-17th century, which is consistent with the history of African Americans (Schroeder et al. 2015).
Methods
The model
We assume the following alternative to Wright–Fisher. Let N be the number of individuals in each population. Each individual has two haplotypes, so the total number of haplotypes is across both populations. Also, we assume the population is a recently admixed population with two ancestral populations (referred to as population 1 and population 2), and let denote the fraction of the genome with population 1 ancestry in individual i.
In the next generation, each individual picks two parents from the current generation, such that the correlation between the ancestry of the two parents is a fixed value P. One way of generating such mating in silica is the following. We randomly pick the set of mothers (with or without replacement) from the original distribution. We then randomly choose the set of fathers (with or without replacements). Now, for each of the parents we give a score where is the global ancestry of the parent, and is drawn from a normal distribution We then sort the mothers and the fathers based on their score and we let the mother with largest score marry the father with the largest score. We then compute the correlation between where are the ancestries of the mother and the father. We search for mate pairs that give us an empirical within 0.01 of P by increasing σ by when the correlation is too large and decreasing σ by when the correlation is too small. Faster algorithms may exist, but this approach works well in practice. We note that our analysis below does not rely on this specific procedure; particularly, the distribution of parents for the new generation can be quite general, and our only assumption is that P is constant across the generations. Note that this assumption may seem restrictive at first, however the case of random mating is far more restrictive, since there one requires that in all generations.
LAD
Denote by the probability of having an allele from ancestry 1 at a given position at generation t. Furthermore, for a pair of positions, let denote the probability of having an allele from ancestry 1 at the two positions. We define a new statistic, termed LAD, denoted by We define We are interested in the expected value of ( at generation t) as a function of the recombination rate r, the number of generations t, and the original LAD
For the following derivations, we will assume that the population and genome size are infinite. We will later show empirically that the infinite population size assumption does not have a substantial effect for realistic values of N. We will first assume that there is no migration and we will relax this assumption in the next section.
Since there is no migration and the population size is infinite, the mean of θ is fixed across the generations (remember that the marginal distribution of the mothers and the fathers is the same and is simply a random draw from the current generation) (Chakraborty and Weiss 1988). We denote and let the ancestry of random individual from generation t where is the onset of admixture. Let be the variance of θ in generation t. Note that the expectations and variances are defined over the set of all individuals in one generation, rather than over multiple realizations of the process. Finally, let be the covariance For we have:
This demonstrates that the variance of genome-wide ancestry is larger when there is assortative mating. Note that previous work has shown that sampling from a finite genome can lead to substantial departures for the distribution of θ across time even under random mating (Gravel 2012). Now, we know
| (1) |
Note that for since there was no assortative mating prior to the admixture event, and therefore for the above calculation gives and To simplify the notation, we change the indices, so that generation corresponds to the time of encounter of the two populations and is the first generation after admixture. Therefore, we have that Equation 1 holds for every
We now find a recursion formula for Let r be the probability for an odd number of recombinations between the two positions in a given meiosis. Hence,
We are now ready to describe our main result:
Lemma 3.1:
Proof. We show this is true by induction. It is easy to verify that since the base case holds. Assume the lemma holds for t and we will prove it for
LAD under migration
We now assume that, in each generation, a fraction of the population is replaced by individuals from the first population (), and a fraction of the population is replaced by individuals from the population We denote by and Since there is migration, the mean global ancestry is changing over time, and we let the average values of θ when an individual is randomly sampled from the population. For simplicity of notation, we denote and we note that is exponentially decreasing. Since we have that and therefore
We now show the following lemma:
Lemma 3.2:
If there is a sequence satisfying the recursion equation where is defined as above, and are abitrary constants, then
where:
Proof. To prove the base of the induction, we need to satisfy which is a simple linear equation. We will show that the induction step adds two more linear equations. Assume the lemma holds for t, and consider
Now, note that Therefore:
Substitution gives the definitions of stated above.
Next, we observe:
By Lemma 3.2, we have for specified in the lemma. Note that, based on the lemma’s proof, Now,
Therefore, noting that we have
Now, recall Therefore, we have the form satisfying Lemma 3.2 with the following values:
Thus, for taken from Lemma 3.2 we have
Plugging in the values of and the fact that we get
| (2) |
Data availability
All genetic data are available via dbGAP with the accession number phs000355.v1.p1 and software is freely available at https://github.com/dpark27/ancassort.
Results
When applied to the genome, we can estimate the value of LAD for known values of r by averaging the observed LAD across the genome. We can now fit the values of and P based on the distribution of the LAD as a function of r in the current generation. Therefore, it is important to understand the dependency of the distribution of LAD for varying values of r as a function of and m. In what follows, we explore the behavior of LAD under different settings.
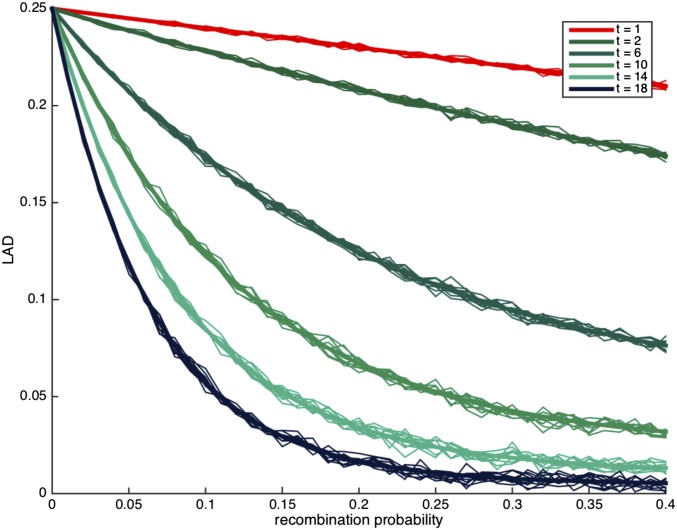
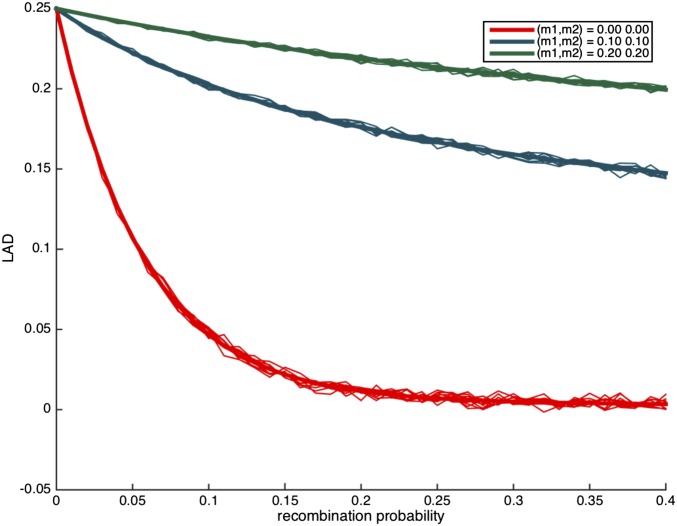
We first consider the case where i.e., there is no migration, and In Figure 1, we observe that there is a clear separation between the different curves for the different numbers of generations since admixture, and it should therefore be easy to estimate the time of admixture event under the assumption of no migration and
Figure 1.
The distribution of local ancestry linkage disequilibrium (LAD) for different values of t with no migration (and ). The thick lines correspond to the expected LAD based on Lemma 3.1, and the thin lines correspond to simulation runs of a single locus in the genome.
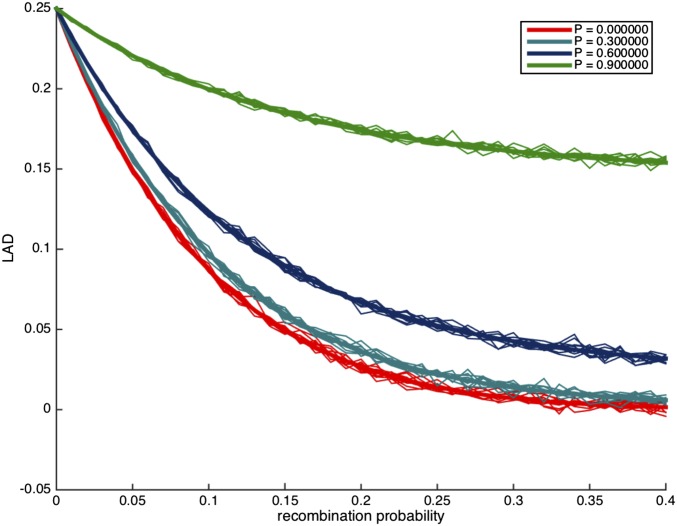
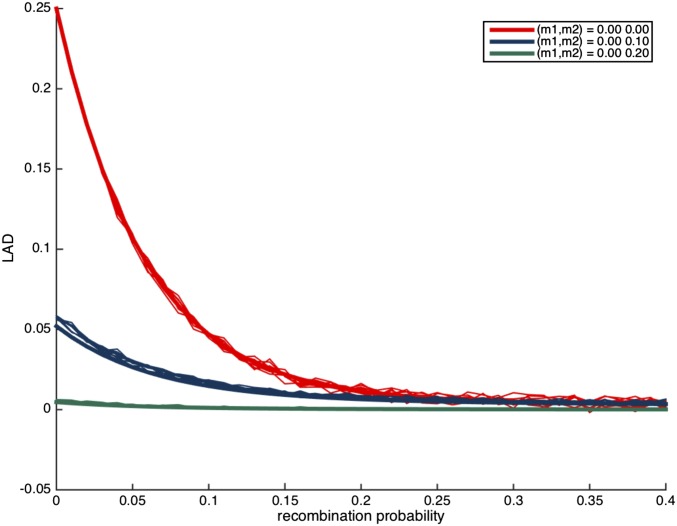
Next, we study the effect of P on the LAD distribution. In Figure 2, we plot the LAD distribution under no migration, after 10 generations of admixture, with varying values of P. Evidently, strong assortative mating with large values of P results in a substantially different levels of LAD. However, we observe that low values of P are harder to distinguish, and therefore we expect that random mating is a robust assumption for any statistic that uses LAD or its derivatives, as long as assortative mating is weak (e.g., ).
Figure 2.
The distribution of local ancestry linkage disequilibrium (LAD) for different values of P with no migration and The thick lines correspond to the expected LAD based on Lemma 3.1, and the thin lines correspond to simulation runs of a single locus in the genome.
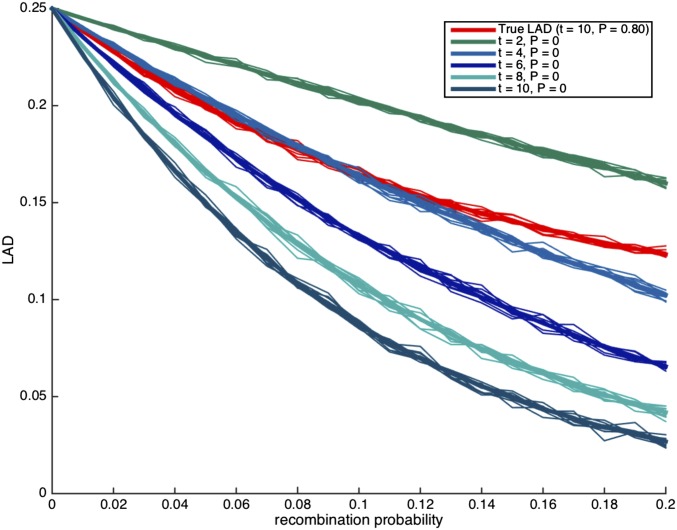
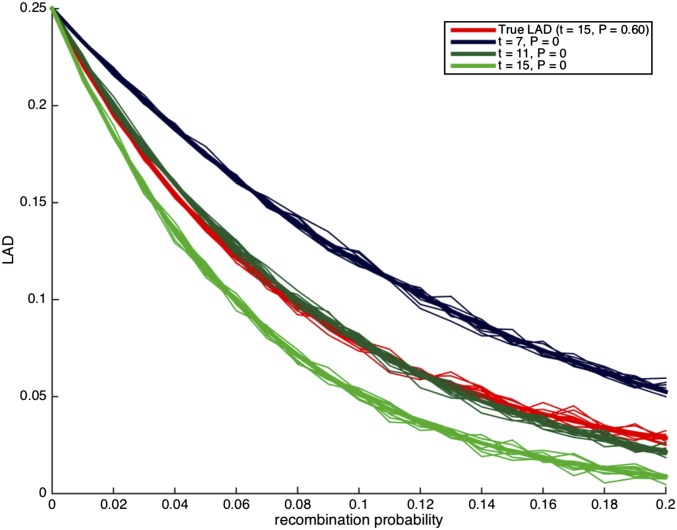
Since typical analysis of genetic data assumes random mating, we attempted to understand the potential risk in making the assumption in the presence of assortative mating. Thus, we consider the case where there is assortative mating, and we try to estimate the time of admixture under the assumption of random mating. For ancient admixture, the difference between the estimates under assortative mating and random mating is not substantial (about 10%, data not shown). For recent admixture (10–20 generations), we observe that there is a considerable difference between the true LAD curve compared to the LAD curve under random mating and, moreover, the true LAD curve is similar to LAD curves that assume random mating but that are substantially more recent. Specifically, in Figure 3, the admixture event occurred 10 generations ago under a strong assortative mating (); however under random mating, the LAD curve that corresponds to is the most similar to the true LAD curve. In Figure 4, the admixture event occurred 15 generations ago under a somewhat weaker assortative mating (), while the estimated number of generations would be 11 under random mating.
Figure 3.
Demonstrating the effect of a random mating assumption when truly All curves correspond to scenarios with no migrations. The thick lines correspond to the expected local ancestry linkage disequilibrium (LAD) based on Lemma 3.1, and the thin lines correspond to simulation runs of a single locus in the genome.
Figure 4.
Demonstrating the effect of a random mating assumption when truly All curves correspond to scenarios with no migrations. The thick lines correspond to the expected local ancestry linkage disequilibrium (LAD) based on Lemma 3.1, and the thin lines correspond to simulation runs of a single locus in the genome.
Next, we explore the effect of migration on the LAD function. We consider both the case where the two populations migrate at the same rate () as shown in Figure 5, as well as the case in which as shown in Figure 6. Evidently, the theoretical calculations capture the empirical well in the sense that they allow for a clear distinction between different migration rates.
Figure 5.
The distribution of local ancestry linkage disequilibrium (LAD) for different values of with equal migration rates from both populations. The thick lines correspond to the expected LAD based on Equation 2, and the thin lines correspond to simulation runs of a single locus in the genome.
Figure 6.
The distribution of local ancestry linkage disequilibrium (LAD) for different values of with no migration from population 1. The thick lines correspond to the expected LAD based on Equation 2, and the thin lines correspond to simulation runs of a single locus in the genome.
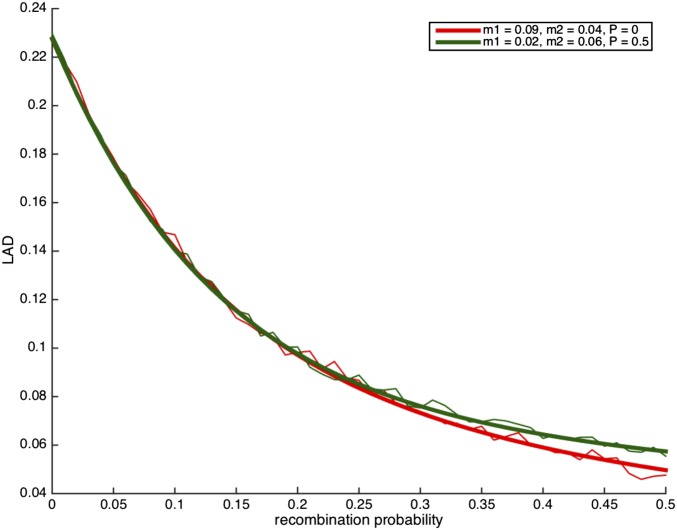
We note that migration and assortative mating can result in similar LAD decay. We estimated the LAD curve using the formula of Lemma 3.1 under random mating with migration, as well as under assortative mating with different values of migration. Since the parameter space () is large, there are triplets of values with very similar LAD curves, thus in practice the model parameters will not necessarily be identifiable. In Figure 7 we present an example where identifiability requires the comparison of LAD decay over dozens of megabases.
Figure 7.
The expected local ancestry linkage disequilibrium (LAD) decay under two conditions, one with assortative mating and another with random mating. In the presence of migration, the two curves almost overlap, and distinguishing between the two cases will be challenging in practice, particularly if LAD is measured only up to a few dozen centimorgans.
Results on real data
To examine the properties of our model in real data, we used genetic data from 1730 African American individuals from the SAGE study. The individuals in the SAGE data were genotyped at 800,000 SNPs on the Affymetrix Axiom Genome-Wide LAT 1 Array, and genotype calling and quality control (QC) were performed as previously described (Torgerson et al. 2012).
To compute LAD, we first called local ancestry using the LAMP-LD software package (Pasaniuc et al. 2013) and genome-wide ancestry was inferred from the mean value of local ancestry for each individual. We measured the LAD decay in 164 10-Mb overlapping windows with a 1 Mb overlap. We calculated the mean LAD decay across all windows as well as the squared distance of each window to the mean. Regions that are under selection or in which the estimates of recombination rates are inaccurate will result in a different LAD decay. Therefore, we performed additional QC by removing windows with a LAD decay > 2 SD from the mean. We repeated this process until convergence, leaving 96 windows.
We measured the assortative mating over the last generation by applying the method ANCESTOR (Zou et al. 2015) to the data. ANCESTOR takes as input local and global ancestry and determines the ancestral proportions of the mother and the father of each individual. The Pearson correlation coefficient between the parental ancestries was estimated across all individuals. This establishes that there was strong spousal ancestry correlation in African Americans in the last generation. If this ancestry-based assortative mating exists in previous generations, our theory shows that LAD decay will be affected. Under the assumption that this correlation was stable throughout history, one can use this estimate to constrain the potential demographic histories of African Americans inferred via LAD.
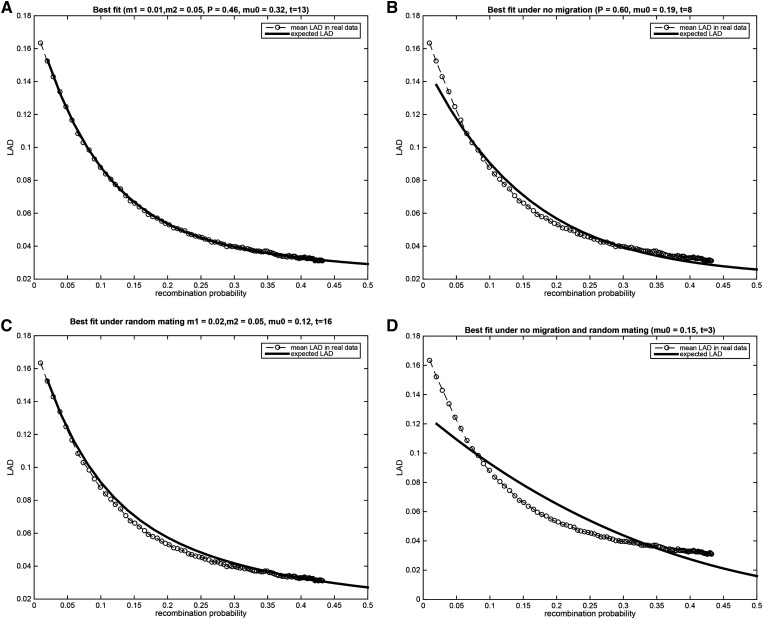
We fitted the migration and assortative mating parameters using a grid search over the entire range of parameters. The best fit resulted in an estimate of generations, with migration rates and assortative mating (Figure 8A). Next, we made the assumption of no migration by searching the grid but with the constraint but we allowed for assortative mating. In this case, the number of generations was dramatically shortened to eight generations, and the assortative mating value increased dramatically to (Figure 8B). Similarly, we search the grid with the constraint to study the case of random mating with migration. In this case the number of generations was 16, and the migration values slightly increased to (Figure 8C). Finally, under random mating and no migration the estimated number of generation is which is clearly a vast underestimate of the true number based on the known history of African Americans (Figure 8D). Notably, there is no good fit under random mating and no migration, and the best fit is obtained in the presence of both migration and assortative mating.
Figure 8.
Each of the plots shows the best fit of the parameters to the mean local ancestry linkage disequilibrium (LAD) in the Study of African Americans, Asthma, Genes and Environments (SAGE) data set. (A) The parameters were searched over the entire grid, resulting in the best fit with estimated number of generations 13, migration rates and correlation (B) The best fit under the assumption of no migration. The number of generations was estimated to be eight, and (C) The best fit under the assumption of random mating with migration. The number of generations is estimated as 16. (D) The best fit under the assumption of random mating and no migration – the number of generations is estimated as 3.
Clearly, the LAD decay is only one summary statistic that depends on the parameters and other statistics may give somewhat different results. For example, it may be possible to examine the distribution of IBD (Gravel et al. 2013), local ancestry (Price et al. 2009), and LD (Loh et al. 2013) under an assortative mating model. Moreover, the LAD decay is not identifiable since different sets of parameters often lead to similar LAD decay. In particular, in the case of the African Americans in SAGE, the best fit was followed by a few different sets of parameters. Under the assumption that is fixed across the generations, the best fit was with generations, and the migration rates were Due to the computational complexity of the grid search used to estimate model parameters, it was not feasible to estimate confidence intervals. However, as was the case in simulations, migration rates and generation times could be altered to accommodate the removal of assortative mating from the model.
Discussion
We presented an adaption of the Wright–Fisher model that incorporates ancestry-assortative mating in admixed populations. We demonstrated that, under this model, the LAD between markers is a function of their recombination rate, the ancestral population migration rates, and the strength of ancestry-based assortment. Assortative mating is likely impacting other estimates of population and medical genetic parameters, both within admixed and continental populations including identity-by-descent distributions, estimates of heritability, joint site frequency spectra, runs of homozygosity, and the distribution of local ancestry track lengths.
While the focus of this work is the definition and presentation of the ancestry-assortative model and its properties, we also estimated the parameters of the model in a real African American data set. Our estimate of 15 generations since admixture in African Americans is larger than previous estimates (Price et al. 2009; Bryc et al. 2015; Baharian et al. 2016), and is consistent with admixture beginning with the slave trade in the mid-17th century and a 25-year generation time. This suggests that taking assortative mating into account may, in some cases, be critical to obtain the correct demographic history or other population parameters.
Previous work has also leveraged LD properties of admixed genomes to infer aspects of demographic history (Moorjani et al. 2011; Loh et al. 2013). These Alder and Roloff statistics use a similar idea to the LAD statistic, but rely on linkage disequilibrium between genotypes as opposed to local ancestry. However, they assume random mating, which likely results in an underestimate of the number of generations in the presence of assortative mating. In future work, it will be interesting to examine the Alder/Roloff statistics in the presence of assortative mating.
The approach we presented for estimating the number of generations since admixture using LAD has its limitations. First, this approach involves a very inefficient grid search, resulting in an inability to provide errors around estimates via bootstrap. Second, in some cases, both migration and assortative mating can give rise to similar LAD distributions, and therefore in those cases one can mistakenly believe that the migration is higher and assortative mating is lower or vice versa. However, the latter raises an interesting question; in previous attempts to learn the demographic histories of humans and other species, is it the case that the migration coefficients were inflated, or that the number of generations since admixture were deflated, due to assortative mating?
Going forward, it will be interesting to determine if assortative mating has biased other recent estimates of demographic events, such as the introgression of Neanderthals (Sankararaman et al. 2014) or the domestication of dogs and pigs (Freedman et al. 2014; Frantz et al. 2015). We will also explore extensions to multi-way admixed populations and the use of MCMC to provide confidence intervals for parameter estimates. In addition to altering the distribution of LAD, we have shown that assortative mating increases the variance of global ancestry. Under certain polygenic models this will induce a concomitant increase in phenotypic variance, which may have implications for selection and evolution.
Our method makes several strong assumptions, which are likely incorrect, such as constant ancestry-assortment strength and migration rates. However, these are a relaxation of previous methods, since, for example under the standard Wright–Fisher model, both random mating and no migration are assumed, and thus both migration rates and ancestry-assortative strengths are fixed across the generations in this case (fixed with value 0). While assortative mating has been well-studied, to the best of our knowledge this is the first attempt to include ancestry-assortment in the estimation of demographic histories. We also reported, for the first time, the strength of ancestry-assortment in African Americans in the previous generation. In future work, we intend to examine the effect of ancestry-assortment on other genetic features as well as the resulting impact in population and medical genetics.
Acknowledgments
The authors acknowledge the patients, families, recruiters, health care providers, and community clinics for their participation. In particular, the authors thank Sandra Salazar for her support as the Study of African Americans, Asthma, Genes and Environments (SAGE) II study coordinator. This work was supported in part by the Sandler Foundation, the American Asthma Foundation, the Robert Wood Johnson Foundation (RWJF) Amos Medical Faculty Development Program, Harry Wm. and Diana V. Hind Distinguished Professor in Pharmaceutical Sciences II, and the National Institutes of Health (NIH) (ES015794, R01Hl128439, and MD006902). N.Z. was supported by an NIH career development award from the National Heart, Lung, and Blood Institute (NHLBI) (K25HL121295) and NIH grant (U01HG009080). E.H. was supported by the Israel Science Foundation (grant 1425/13), United States–Israel Binational Science Foundation (grant 2012304), German–Israeli Foundation (grant 1094-33.2/2010), and by the National Science Foundation (grant III-1217615). The SAGE study was supported by the Sandler Family, the American Asthma Foundation, NIH/National Institute on Minority Health and Health Disparities (NIHMD) grants 1P60 MD006902, 1R01MD010443, and U54MD009523, NIH/NHLBI grant 1R01HL117004-01, NIH/National Institute of Environmental Health Sciences grant R21ES24844-01, and the Tobacco-Related Disease Research Program 24RT-0025.
Footnotes
Communicating editor: R. Nielsen
Literature Cited
- Baharian S., Barakatt M., Gignoux C. R., Shringarpure S., Errington J., et al. , 2016. The great migration and African-American genomic diversity. PLoS Genet. 12(5): e1006059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrell L. N., Nguyen E. A., Roth L. A., Oh S. S., Tcheurekdjian H., et al. , 2013. Childhood obesity and asthma control in the gala ii and sage ii studies. Am. J. Respir. Crit. Care Med. 187(7): 697–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning S. R., Browning B. L., 2013. Identity-by-descent-based heritability analysis in the northern Finland birth cohort. Hum. Genet. 132(2): 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryc K., Durand E. Y., Macpherson J. M., Reich D., Mountain J. L., 2015. The genetic ancestry of African Americans, Latinos, and European Americans across the United States. Am. J. Hum. Genet. 96(1): 37–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty R., Weiss K. M., 1988. Admixture as a tool for finding linked genes and detecting that difference from allelic association between loci. Proc. Natl. Acad. Sci. USA 85(23): 9119–9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz L. A., Schraiber J. G., Madsen O., Megens H. J., Cagan A., et al. , 2015. Evidence of long-term gene flow and selection during domestication from analyses of Eurasian wild and domestic pig genomes. Nat. Genet. 47(10): 1141–1148. [DOI] [PubMed] [Google Scholar]
- Freedman A. H., Gronau I., Schweizer R. M., Ortega-Del Vecchyo D., Han E., et al. , 2014. Genome sequencing highlights the dynamic early history of dogs. PLoS Genet. 10(1): e1004016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S., 2012. Population genetics models of local ancestry. Genetics 191(2): 607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S., Zakharia F., Moreno-Estrada A., Byrnes J. K., Muzzio M., et al. , 2013. Reconstructing native American migrations from whole-genome and whole-exome data. PLoS Genet. 9(12): e1004023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh P. R., Lipson M., Patterson N., Moorjani P., Pickrell J. K., et al. , 2013. Inferring admixture histories of human populations using linkage disequilibrium. Genetics 193(4): 1233–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J., Cutler D., Patterson N., Stephens M., Eskin E., Halperin, et al. , 2006. A comparison of phasing algorithms for trios and unrelated individuals. Am. J. Hum. Genet. 78(3): 437–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews C. A., Reus V. I., 2001. Assortative mating in the affective disorders: a systematic review and meta-analysis. Compr. Psychiatry 42(4): 257–262. [DOI] [PubMed] [Google Scholar]
- Merikangas K. R., 1982. Assortative mating for psychiatric disorders and psychological traits. Arch. Gen. Psychiatry 39(10): 1173–1180. [DOI] [PubMed] [Google Scholar]
- Moorjani P., Patterson N., Hirschhorn J. N., Keinan A., Hao L., et al. , 2011. The history of African gene flow into Southern Europeans, Levantines, and Jews. PLoS Genet. 7(4): e1001373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra E. J., Kittles R. A., Argyropoulos G., Pfaff C. L., Hiester K., et al. , 2001. Ancestral proportions and admixture dynamics in geographically defined African Americans living in South Carolina. Am. J. Phys. Anthropol. 114(1): 18–29. [DOI] [PubMed] [Google Scholar]
- Pasaniuc B., Sankararaman S., Torgerson D. G., Gignoux C., Zaitlen N., et al. , 2013. Analysis of Latino populations from gala and mec studies reveals genomic loci with biased local ancestry estimation. Bioinformatics 29(11): 1407–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A. L., Tandon A., Patterson N., Barnes K. C., Rafaels N., et al. , 2009. Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genet. 5(6): e1000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N., Choudhry S., Via M., Basu A., Sebro R., et al. , 2009. Ancestry-related assortative mating in Latino populations. Genome Biol. 10(11): R132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankararaman S., Mallick S., Dannemann M., Prufer K., Kelso J., et al. , 2014. The genomic landscape of Neanderthal ancestry in present-day humans. Nature 507(7492): 354–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder H., Avila-Arcos M. C., Malaspinas A. S., Poznik G. D., Sandoval-Velasco M., et al. , 2015. Genome-wide ancestry of 17th-century enslaved Africans from the Caribbean. Proc. Natl. Acad. Sci. U S A 112(12): 3669–3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torgerson D. G., Capurso D., Ampleford E. J., Li X., Moore W. C., et al. , 2012. Genome-wide ancestry association testing identifies a common European variant on 6q14.1 as a risk factor for asthma in African American subjects. J. Allergy Clin. Immunol. 130(3): 622–629.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udry J. R., Bauman K. E., Chase C., 1971. Skin color, status, and mate selection. Am. J. Sociol. 76(4): 722. [Google Scholar]
- Zou J. Y., Halperin E., Burchard E., Sankararaman S., 2015. Inferring parental genomic ancestries using pooled semi-Markov processes. Bioinformatics 31(12): i190–i196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All genetic data are available via dbGAP with the accession number phs000355.v1.p1 and software is freely available at https://github.com/dpark27/ancassort.