Abstract
With the rise of both the number and the complexity of traits of interest, control of the false discovery rate (FDR) in genetic association studies has become an increasingly appealing and accepted target for multiple comparison adjustment. While a number of robust FDR-controlling strategies exist, the nature of this error rate is intimately tied to the precise way in which discoveries are counted, and the performance of FDR-controlling procedures is satisfactory only if there is a one-to-one correspondence between what scientists describe as unique discoveries and the number of rejected hypotheses. The presence of linkage disequilibrium between markers in genome-wide association studies (GWAS) often leads researchers to consider the signal associated to multiple neighboring SNPs as indicating the existence of a single genomic locus with possible influence on the phenotype. This a posteriori aggregation of rejected hypotheses results in inflation of the relevant FDR. We propose a novel approach to FDR control that is based on prescreening to identify the level of resolution of distinct hypotheses. We show how FDR-controlling strategies can be adapted to account for this initial selection both with theoretical results and simulations that mimic the dependence structure to be expected in GWAS. We demonstrate that our approach is versatile and useful when the data are analyzed using both tests based on single markers and multiple regression. We provide an R package that allows practitioners to apply our procedure on standard GWAS format data, and illustrate its performance on lipid traits in the North Finland Birth Cohort 66 cohort study.
Keywords: association studies, multiple penalized regression, linkage disequilibrium, FDR
IN the last decade, genome-wide association studies (GWAS) have been the preferential tool to investigate the genetic basis of complex diseases and traits, leading to the identification of an appreciable number of loci (GWAS Catalog; Welter et al. 2014). Soon after the first wave of studies, a pattern emerged: there exists a sizable discrepancy between, on the one hand, the number of loci that are declared significantly associated and the proportion of phenotypic variance they explain (Manolio et al. 2009) and, on the other hand, the amount of information that the entire collection of genotyped single nucleotide polymorphisms (SNPs) appears to contain about the trait (Purcell et al. 2009; Yang et al. 2010). To increase the number of loci discovered (and their explanatory power), substantial efforts have been made to obtain larger sample sizes by genotyping large cohorts (Kvale et al. 2015; UK Biobank, http://www.ukbiobank.ac.uk) and by relying on meta-analysis. However, the gap remains, although not as large as in the original reports. This parallels, in part, the discrepancy between the polygenic model that is used to define complex traits and the simple linear-regression approach to the discovery of associated SNPs which is standard practice, as underscored, for example, in Kang et al. (2010), Stringer et al. (2011), and Sabatti (2013).
Two approaches to bridge the gap emerge quite naturally: (a) an attempt to evaluate the role of genetic variants in the context of multiple linear regression, more closely matching the underlying biology; and (b) relaxing the very stringent significance criteria adopted by GWAS to control the false discovery rate (FDR) (Benjamini and Hochberg 1995) rather than the family-wise error rate (FWER)—a strategy that has been shown to be attractive when prediction is considered as an end goal together with model selection (Abramovich et al. 2006). Both strategies have been pursued, but have encountered a mix of successes and challenges.
The use of multiple linear regression for the analysis of GWAS data has been proposed as early as 2008 (Hoggart et al. 2008; Wu et al. 2009). By examining the distribution of the residuals, it is clear that it provides a more appropriate model for complex traits. However, its use to discover relevant genetic loci has encountered difficulties in terms of computational costs and interpretability of results. On the computational side, progress has been made using approaches based on convex optimization such as the lasso (Zhou et al. 2010), developing accurate methods to screen variables (Fan and Lv 2008; Wu et al. 2010; He and Lin 2011), and relying on variational Bayes (Logsdon et al. 2010; Carbonetto and Stephens 2011). There are, however, remaining challenges. First, the genetics community is, correctly, very sensitive to the need of replicability, and finite-sample guarantees for the selected variants are sought. Unfortunately, this has been difficult to achieve with techniques such as the lasso: Alexander and Lange (2011) attempt to use stability selection; Yi et al. (2015) do a simulation study of a variety of penalized methods, showing that tuning parameters play a crucial role and that standard selection methods for these do not work well; and Frommlet et al. (2012) and Dolejsi et al. (2014) propose some analytical approximation of FDR as an alternative to the lasso. Our recent work (Bogdan et al. 2015) also explores alternative penalty functions that, under some circumstances, guarantee FDR control. Second, multiple linear regression encounters difficulties in dealing with correlated predictors, in that the selection among these is often arbitrary: this is challenging in the context of GWAS, when typically there is a substantial dependence between SNPs in the same genetic region.
The suggestion of controlling FDR rather than FWER in genetic mapping studies that expect to uncover a large number of loci was put forward over a decade ago (Sabatti et al. 2003; Storey and Tibshirani 2003; Benjamini and Yekutieli 2005b) and is accepted in the expression quantitative trait loci (eQTL) community, where FDR is the standard error measure. The existence of strong local dependence between SNPs has also posed challenges for FDR-controlling procedures. While the Benjamini–Hochberg procedure (BH) (Benjamini and Hochberg 1995) might be robust to the correlation between tests that one observes in GWAS, the fact that the same biological association may be reflected in multiple closely located SNPs complicates both the definition and the counting of discoveries, so that it is not immediately evident how FDR should be defined. Prior works (Perone Pacifico et al. 2004; Benjamini and Heller 2007; Siegmund et al. 2011) underscore this problem and suggest solutions for specific settings.
This article proposes a phenotype-aware selective strategy to analyze GWAS data which enables precise FDR control and facilitates the application of multiple regression methodology by reducing the dependency between the SNPs included in final testing. The Methods section starts by briefly recapitulating the characteristics of GWAS, with reference to an appropriate count of discoveries and the identification of a meaningful FDR to control. We introduce our selective strategy and provide some general conditions under which it controls the target FDR. We then describe a specific selection procedure for GWAS analysis and describe how it can be coupled with standard BH for univariate tests, or with SLOPE (Bogdan et al. 2015) to fit multiple regression. In the Results section, we explore the performance of the proposed methodology with simulations and analyze a data set collected in the study of the genetic basis of blood lipids. In both cases, the FDR-controlling procedures we propose allow us to explain a larger portion of the phenotype variability, without a substantial cost in terms of increased false discoveries.
With this article, we are making available an R package geneSLOPE (Brzyski et al. 2016) at the Comprehensive R Archive Network (CRAN). The package can analyze data in the PLINK (Purcell et al. 2007) format.
Methods
The GWAS design, dependence, and definition of discoveries
The goal of a GWAS study is to identify locations in the genome that harbor variability which influences the phenotype of interest. This is achieved using a sample of n individuals, for whom one acquires trait values and genotypes at a collection of M SNPs that span the genome. Following standard practice, we summarize genotypes by the count of copies of minor alleles that each individual has at each site, resulting in an matrix X, with entries The variant index j is taken to correspond to the order of the position of each SNP in the genome. The true relation between genetic variants and phenotypes can be quite complex. For simplicity, and in agreement with the literature, we assume a linear additive model, which postulates that the phenotype value of subject i depends linearly on her/his allele counts at an unknown set of causal variants. Since there is no guarantee a priori that the variants in are part of the genotype set, we indicate their allele counts with letting
Investigating the relation between y and X is helpful to learn information about the set of causal variants and their effects in two ways: (1) it is possible that some of the causal variants are actually genotyped, so that for some k; and (2) most importantly, the set of M genotyped SNPs contains reasonable proxies for the variants in To satisfy (2), GWAS are designed to capitalize on the local dependence between variable sites in the genome known as linkage disequilibrium (LD), which originates from the modality of transmission of chromosomes from parents to children, with modest recombination. The set of M genotyped SNPs is chosen with some redundancy, so that the correlation between and is expected to be nonzero for k in a certain range: this is to ensure that any untyped casual variant will be appreciably correlated with one (or more) of the typed that are located in the same genomic region. Any discovered association between a SNP and the phenotype y is interpreted as an association between y and some variant in the genomic neighborhood of This design has a number of implications for statistical analysis:
Often, the existence of an association between y and each typed variant is queried via a test statistic which is a function of y and only: these test statistics are “locally” dependent, with consequences for the choice of multiple comparison adjustment, that, for example, might not need to be as stringent as in the case of independence.
When multiple regression models are used to investigate the relation between y and X, one encounters difficulties due to the correlation between regressors—the choice among which is somewhat arbitrary.
The fact that the true causal variants are not necessarily included among the genotyped SNPs makes the definition of a true/false association nontrivial.
We want to underscore the last point. To be concrete, let us assume the role of each variant is examined with the t-statistic for with defined in the univariate regression Even if none of the M genotyped variants are causal, a number of them will have a coefficient in these reduced models; whenever is correlated with one of the variants in should be rejected. Indeed, simulation studies that investigate the power and global error control of different statistical approaches routinely adopt definitions of “true positive” that account for correlation between the known causal variant and the genotyped SNPs (see Yi et al. 2015 for a recent example). At the same time, a rejection of should not be interpreted as evidence of a causal role for in fact, geneticists equate discovery with the identification of a genomic location rather than with the identification of a variant. The rejection of for a number of correlated neighboring SNPs in a GWAS is described in terms of the discovery of one single locus associated with the trait of interest. The number of reported discoveries, then, corresponds to the number of distinct genomic regions (whose variants are uncorrelated) where an association has been established. This discrepancy between the number of rejected hypotheses and the number of discoveries has important implications for FDR-controlling strategies, which have received only a modest attention in the literature. Siegmund et al. (2011) suggest that in situations similar to those of GWAS, neighboring rejections should be grouped and counted as a single rejection and that the global error of interest should be the expected value of the “proportion of clusters that are falsely declared among all declared clusters.” This FDR of clusters—a notion first introduced in Benjamini and Heller (2007)—is not the error rate controlled by the Benjamini–Hochberg procedure on the P-values for the hypotheses. Indeed, because FDR is the expected value of the ratio of the random number of discoveries, its control depends crucially on how one decides to count discoveries. In Peterson et al. (2016) we give another example of how controlling FDR for a collection of hypotheses does not extend to controlling FDR for a smaller group hypotheses logically derived from the initial set. Both in the setting described here and in Peterson et al. (2016), targeting FWER would have resulted in less surprising behavior; assuring that the probability of rejecting at least one null is smaller than a level α and this would also guarantee that the probability of rejecting at least one null cluster of hypotheses is smaller than α. Siegmund et al. (2011) study a setting that is close to our problem and propose a methodology to control their target FDR by relying on a Poisson process distribution for the number of false discoveries. Here we investigate a different approach, one that is more tightly linked to the GWAS design, is adapted to the variable extent of LD across the genome, and capitalizes on results in selective inference (Benjamini and Bogomolov 2014).
Controlling the FDR of interesting discoveries by selecting hypotheses
The approach we study emerged from our interest in using multiple linear regression to analyze the relation between y and X, so it is useful to motivate it in this context. Suppose both and are strongly correlated with the untyped causal variant When univariate regression is used as the analysis strategy, both the test statistics and would have large values, resulting in the discovery of this locus. Instead, the marginal P-values for each of the coefficients of and derived from a multiple linear regression model that includes both variables would be nonsignificant, as and carry roughly the same information and can be substitutes for each other. Model selection strategies would rather arbitrarily result in the inclusion of one or the other regressor, leading to an underestimate of their importance when resampling methods are used to evaluate significance. Using multiple linear regression, one would achieve the best performance if, from the start, only one of and (the most strongly correlated with Zk) is included among the possible regressors. A natural strategy is to prune the set of M-typed SNPs to obtain a subset of m quasi-orthogonal ones and supply these to the model selection procedure of choice. However, this encounters the difficulty that the best proxy for some of the causal variants might have been pruned, resulting in a loss of power. It seems that, ideally, one would select from a group of correlated SNPs the one that has the strongest correlation with the trait to include among the potential regressors. Unfortunately, this initial screening for association would invalidate any guarantees of the model selection strategy, which operates now not on m variables, but on m selected ones. The emerging literature of selective inference, however, suggests that we might be able to appropriately account for this initial selection step, preserving guarantees on error rate control.
Abstracting from the specifics of multiple regression, consider the setting where a collection of M hypotheses with some redundancy is tested to uncover an underlying structure of interest. The hypotheses in can be organized linearly or spatially and are chosen because a priori they provide a convenient and general way of probing the structure; however, it is expected that a large portion of these will be true, and that when one is false, a number of neighboring ones would be also false. In the case of GWAS, these clusters of false hypotheses would correspond to markers correlated with causal mutations. Because of the mismatch between and the underlying structure, the number of scientifically interesting discoveries does not correspond to the number of rejected and strategies that control the FDR defined in terms of these might not lead to satisfactory inference. Specifically, as noted in Siegmund et al. (2011), “a possibly large number of correct rejections at some location can inflate the denominator in the definition of false discovery rate, hence artificially creating a small false discovery rate, and lowering the barrier to possible false detections at distant locations.” This problem was recognized already in Perone Pacifico et al. (2004) and Benjamini and Heller (2007), who introduce the notion of cluster FDR and suggest defining a priori clusters of hypotheses corresponding to signals of interest and applying FDR-controlling strategies to hypotheses relative to these clusters. An implicit example of this approach can be found in the eQTL literature. When investigating the genetic basis of variation in gene expression, the authors in Ardlie et al. (2015) change the unit of inference from SNPs to genes, so as to bypass the redundancy due to many SNPs in the same neighborhood. Here we take a different approach, where “clusters” of hypotheses are defined after looking at the data, and used to select a subset of representative hypotheses. Only this subset is then tested with a procedure that accounts for this initial selection.
Formally, let y indicate the data used to test the hypotheses in and let be a selection procedure that, on the basis of the data, identifies a subset of s representative hypotheses. Let be the set of their indexes, so that it is relevant to control the following FDRs:
| (1) |
In other words, the decision of acceptance/rejection is made only for the hypotheses in the selected set and FDRs is a natural notion of global error rate. Naively, to control one might consider applying a BH at level q to the P-values corresponding to the subset of hypotheses However, since these have been chosen by looking at the data—so that, for example, it is acceptable to select the most “promising” hypotheses—the naive approach would not guarantee FDR control. Indeed, consider the case where contains only the hypothesis with the smallest P-value: BH applied to this subset of one hypothesis would compare its P-value with the target rate q, thereby ignoring the original multiplicity. It seems clear that we need to “remember” where the selected hypotheses come from: while we might focus on a subset—to avoid scientific repetition—we need to account for the fact that this subset is selected from an original larger pool, which provided us with a larger freedom margin. A solution that emerges quite naturally consists of using a set of increasing P-value thresholds (just as in BH), but one whose severity is defined in terms of the original large collection of hypotheses: the smallest P-value for should be compared with the second smallest with etc. This can be formally stated by requiring the application of BH to the P-values targeting the more stringent level where the coefficient penalizes for the initial selection. This rule already appears in the literature in slightly different contexts (Benjamini and Yekutieli 2005a; Benjamini and Bogomolov 2014) and it is useful for our problem in that the P-value thresholds are identical to those implied by BH on but the number of hypotheses tested is smaller and the hypotheses are more clearly separated. This prevents the excessive deflation of the BH threshold that results when each true discovery is represented by many rejected hypotheses, and therefore helps to control the number of false discoveries. The following theorem, proven in the Appendix, reassures us that, under some conditions, the rule that we have described not only makes intuitive sense, but indeed guarantees control of
Theorem 1. FDR control for selected hypotheses. Let be a selection procedure, and let be the number of rejections derived by applying BH with target on the selected hypotheses If the P-values satisfy the condition of the positive regression dependence on a subset (PRDS) (Benjamini and Yekutieli 2001) and the selection procedure is such that is nonincreasing in each of the P-values rejecting guarantees control of FDRs.
Two conditions are required for the discussed program to guarantee FDRs control: (1) The P-values have to satisfy the PRDS property; this is a requirement for most of the proofs of FDR control, and—while difficult to verify—it can be loosely interpreted as the requirement of the positive correlation between P-values at linked markers, and it is therefore a reasonable assumption in the GWAS setting (Sabatti et al. 2003). (2) The selection procedure has to be such that if we imagine reducing one of the P-values of the original hypotheses, leaving everything else the same, the final number of rejections does not decrease. This is a property that appears very reasonable and that one would intuitively desire in a testing procedure. An example selection procedure that satisfies the assumptions of the theorem is as follows: the hypotheses are separated in groups a priori and, from each group, selects the hypothesis with the smallest associated P-value. In the next section, we describe a slightly more complicated selection procedure that appears appropriate for the case of GWAS, and where the separation of hypotheses into groups is data driven. While this procedure does not satisfy the assumption that the number of rejections is always a nonincreasing function of the P-values, it does do so for an overwhelming proportion of realistic P-value configurations, and our extensive simulations studies suggest that its use in the context of Theorem 1 still leads to FDRs control.
A GWAS selection procedure: phenotype-aware cluster representatives
In the context of genetic association studies, the selection function defined in Procedure 1 below and illustrated in Figure 1, emerges quite naturally. One starts by evaluating the marginal association of each SNP to the phenotype using the P-value of the t-test for its coefficient in a univariate regression. Then, SNPs with a P-value larger than threshold π are removed from consideration. The collection of remaining SNPs is further pruned to obtain a selected set with low correlation, so that each variant can be equated to a separate discovery. To achieve this, we define clusters of SNPs using their empirical correlation in our sample, starting from the variants with the strongest association to the phenotype, which are selected as cluster representatives.
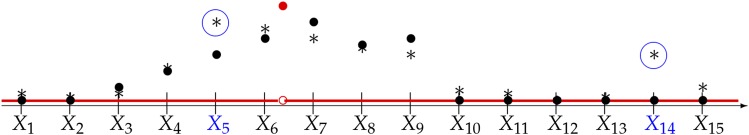
Figure 1.
Phenotype-aware cluster representatives. The x-axis represents the genome, with the locations of genotyped SNPs indicated by tick marks. The true causal effect of each position of the genome is indicated in red; there is only one causal variant in this region, between SNPs and Solid black circles indicate the value of coefficient of in a linear approximation of the conditional expectation Asterisks mark the estimated in the sample. The SNPs and selected as cluster representatives in this schematic diagram, are indicated in blue.
Procedure 1. Selection function to identify cluster representatives.
Input: Screen SNPs:
Calculate the P-value for with defined in the univariate regression as j varies across all SNPs.
Retain in only those SNPs whose P-values are smaller than π.
Cluster SNPs:
Select the SNP j in with the smallest P-value and find all SNPs whose absolute value of the Pearson correlation with this selected SNP are larger than or equal to ρ.
Define this group as a cluster and SNP j as the representative of the cluster. Include SNP j in and remove the entire cluster from
Repeat steps 3–4 until is empty.
Procedure 1 has two parameters: π and ρ, corresponding to the two steps of the selection. The screening in steps 1–2 is similar to that described in Fan and Lv (2008) and Wu et al. (2009) for model selection procedures, where the parameter π controls the stringency of the selection based on univariate association. On the one hand, large values of π result in larger cluster sizes, and hence less precise localization. On the other hand, in the context of multiple regression, it is possible to uncover a role for variants that have weak marginal effects due to masking. To enable this, one must not be too stringent in the initial screening step. In all the simulations and data analyses presented here we have used which seems to be a good compromise. The results in Fan and Lv (2008) and Wu et al. (2009) can provide additional guidance on the choice of π.
Steps 3–5 of Procedure 1 aim to “thin” the set of SNPs on account of the dependency among them. This is related to the selection of tag SNPs (Halperin et al. 2005), for which there is an extensive literature, and is similar to correlation-reduction approaches (Stell and Sabatti 2016). A defining characteristic of Procedure 1, however, is that both the SNP clusters and their representatives are selected with reference to the phenotype of interest. This ensures that the representatives maximize power, and that the location of the true signal is as close as possible to the center of the respective cluster. This also reduces the probability of the selection of more than one SNP per causal variant.
The parameter ρ needs to be chosen to reflect what researchers would consider as independent discoveries. Rather than aggregating discoveries at one locus a posteriori, our procedure simply requires specifying a priori which level of correlation between two SNPs would result in considering the signals at these two SNPs as indistinguishable. Typically, the researcher’s choice would depend on the density of the available markers, sample size, and the expected effect size. We note that as sample size increases, methods based on marginal analysis [with single-marker tests (SMTs)] and multivariate linear regression behave differently. When sample or effect size increases, the signal due to one causal variant is detectable at SNPs with decreasing levels of correlation. Therefore, to avoid excessive true discoveries by SMTs, the researcher might want to choose a correspondingly lower value of ρ. However, with multiple linear regression, an increase in sample size results in an increase of resolution. This means that with increasing probability only the “best” representative of the causal variant will be selected and a meaningful analysis of the data can be carried out with larger values of ρ. We note that this is one advantage of analyzing the data with multiple linear regression rather than relying on marginal tests.
Certainly, Procedure 1 is but one possibility for creating clusters. For example, one might want to include information on physical distance in the formation of clusters. In our experiments, however, this has not led to better performance. On the other hand, we note that clusters cannot be defined using information on physical distance alone: it is the correlation r between the SNPs that determines how the signal due to one causal variant leaks across multiple sites. The Result section illustrates some of the properties of the clusters derived from Procedure 1.
We now consider two approaches to the analysis of GWAS data that can be adopted in conjunction with the selection of cluster representatives to control the FDRs.
Univariate testing procedures after selection
By and large, the most common approach to the analysis of GWAS data relies on univariate tests of association between trait and variants. This has advantages in terms of computational costs, handling of missing data, and portability of results across studies. We therefore start by considering how to control relevant FDR in this context.
We consider two different approaches to obtain the P-values for each of the hypotheses: univariate linear regression (which we indicate with SMT) and EMMAX (Kang et al. 2010), a mixed model that allows us to consider polygenic effects. To enable computational scaling, EMMAX only estimates the parameters of the variance component model once rather than for every marker. We use SMTs and EMMAXs to denote the procedures that consist of testing the set of hypotheses corresponding to cluster representatives, using P-values obtained with SMT and EMMAX, respectively, and identifying rejections with the BHs procedure described below.
Procedure 2. Benjamini-Hochberg on selected hypotheses BHs.
Input: M = total number of SNPs (before initial screening), = collection of selected hypotheses (cluster representatives), = desired level for FDRs.
Let be the number of hypotheses in and the vector of their P-values. Apply BH to with target level
GeneSLOPE: FDR control in multiple regression
SLOPE (Bogdan et al. 2015) is a recently introduced extension of the lasso that achieves FDR control on the selection of relevant variables when the design is nearly orthogonal. Specifically, assume the following model:
where X is the design matrix of the dimension is the n-dimensional vector of random errors, and β is the M-dimensional vector of regression coefficients, a significant portion of which is assumed to be zero. For a sequence of nonnegative and nonincreasing numbers the SLOPE estimate of β is the solution to a convex optimization problem
| (2) |
where are sorted absolute values of the coordinates of b.
Defining a discovery as every estimated and a false discovery as the case where but the true Bogdan et al. (2015) show that with a specific sequence of (corresponding to the sequence of decreasing thresholds in BH) the program in (2) controls FDR at a desired level when X is orthogonal. Moreover, the modified sequence λ—described in Procedure 4 in the Appendix—has been shown in simulation studies to achieve FDR control when the regressors are nearly independent and the number of nonzero β’s is not too large.
Note that, as for other shrinkage methods (Tibshirani 1994; Fan and Li 2001), the results of SLOPE depend on the scaling of explanatory variables: the values of the regularizing sequence in Procedure 4 assume that explanatory variables are “standardized” to have zero mean and a unit norm. Moreover, since in most cases the variance of the error term is unknown and needs to be estimated, in Bogdan et al. (2015) an iterative procedure for the joint estimation of σ and the vector of regression coefficients was proposed. This is described in the Appendix as Procedure 5, and closely follows the idea of scaled lasso (Sun and Zhang 2012). All these data preprocessing and analysis steps are implemented in R package SLOPE, available on CRAN.
The fact that SLOPE comes with finite-sample guarantees for the selected parameters makes it an attractive procedure for GWAS analysis. However, the presence of substantial dependence between SNPs (regressors Xj) presents challenges: on the one hand, the FDR-controlling properties have only been confirmed so far when the explanatory variables are quasi-independent; and, on the other hand, the definition of FDR is problematic in a setting where the true causal variants are not measured and X contains a number of correlated proxies. The identification of a subset of variants with Procedure 1 takes care of both aspects: the regressors are not strongly correlated and they represent different locations in the genome, so that we can expect the projection of the true model in the space they span to be sparse and the number of to capture the number of scientifically relevant discoveries. We therefore propose, as a potential analysis pipeline, the application of Procedure 1 followed by Procedure 3, which outlines the application of SLOPE to the selected cluster representatives. Both procedures have been implemented in the R package geneSLOPE, which is available on CRAN and can handle typical GWAS data provided in PLINK format.
Procedure 3. geneSLOPE.
Input: y = vector of trait values, M = total number of SNPs (before initial screening), = selected SNPs (cluster representatives), and = desired level for FDRs.
Initialize
Center y by subtracting its mean, and standardize so that each column has a zero mean and unit norm.
Calculate the sequence λ using Procedure 4 and using M as a total number of regressors, and retain the first elements of it.
Compute the residual sum of squares (RSS) obtained by regressing y onto variables in and set where denotes the cardinality of
Compute the solution for SLOPE as in Equation 2, explaining y as a linear function of with parameters and λ. Set
If stop; if not, set and reiterate steps 3–4.
Data availability
To illustrate the performance of our methods, we used the data from the North Finland Birth Cohort (NFBC66) study (Sabatti et al. 2009), available in the database of Genotypes and Phenotypes (dbGaP) under accession number phs000276.v2.p1 (http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000276.v2.p1).
Results
To test the performance of the proposed algorithms, we relied on simulations and real data analysis. In both cases, genotype data came from the NFBC66 study (Sabatti et al. 2009). The raw genotype matrix contains 364,590 markers for 5402 subjects. We filtered the data in PLINK to exclude copy-number variants and SNPs with Hardy–Weinberg equilibrium P-value <0.0001, minor allele frequency <0.01, or call rate <95%. This resulted in an predictor matrix with and When applying GeneSLOPE, missing genotype data were imputed as the SNP mean.
For simulations, the trait values are generated using the multiple regression model:
| (3) |
where is the standardized matrix of genotypes, is the set of indices corresponding to “causal” mutations, and The number of causal mutations takes the value and in each replicate the k causal features are selected at random from a subset of the M SNPs. For each k, the values of are evenly spaced in the interval with and As a result, the smallest genetic effect is rather weak (heritability in a single quantitative trait loci model h2 = 0.0017), while the strongest effect is relatively large Each scenario is explored with 100 simulations.
In evaluating FDRs and power we adopt the following conventions, which we believe to closely mimic the expectations of researchers in this field: the null hypothesis relative to a SNP/cluster representative is true if the SNP/cluster representative has a correlation <0.3 with any causal variant. Similarly, a causal variant is discovered if at least one of the variants in the rejection set has correlation of at least magnitude 0.3 with it.
In addition to evaluating performance in the context of simulated traits, we apply the proposed procedures to four lipid phenotypes available in NFBC66 (Sabatti et al. 2009): high-density lipoproteins (HDL), low-density lipoproteins (LDL), triglycerides (TG), and total cholesterol (CHOL). We compare the discoveries obtained by the simple and multiple regression approaches on the NFBC data to those reported in the Global Lipids Genetics Consortium (2013), a much more powerful study based on 188,577 subjects.
Simulation study
Cluster characteristics:
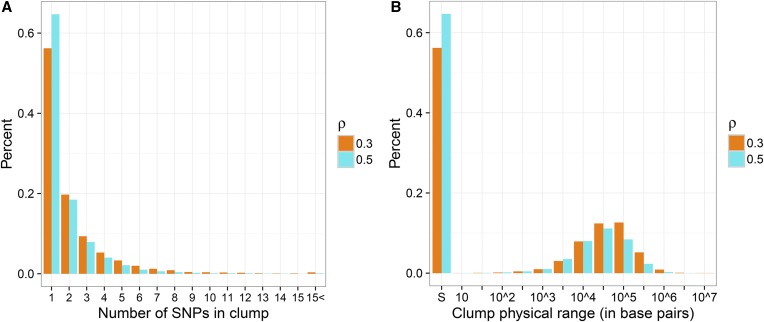
We begin by exploring the distribution of the size of clusters created according to Procedure 1 for two values of Figure 2A illustrates the size of clusters when the trait was generated according to the model in Equation 3 with and genotypes from the NFBC data set. Most of the clusters are rather small and do not include more than five SNPs, and in fact >50% of them is comprised of one SNP only. Figure 2B reports the maximal distance in base pairs between the elements of one cluster: apart from the spike at zero (corresponding to clusters with one SNP only), the median distance spanned by clusters is for respectively. Of course, differences in the genotype density would result in differences in the cluster sizes obtained.
Figure 2.
(A) Histograms of the number of SNPs included in each cluster when Procedure 1 is applied to P-values with and or (B) Histogram of the maximal distance between SNPs in the same cluster. The symbol “S” on the x-axis corresponds to clusters that contain only one SNP.
Error control with EMMAX and SMT:
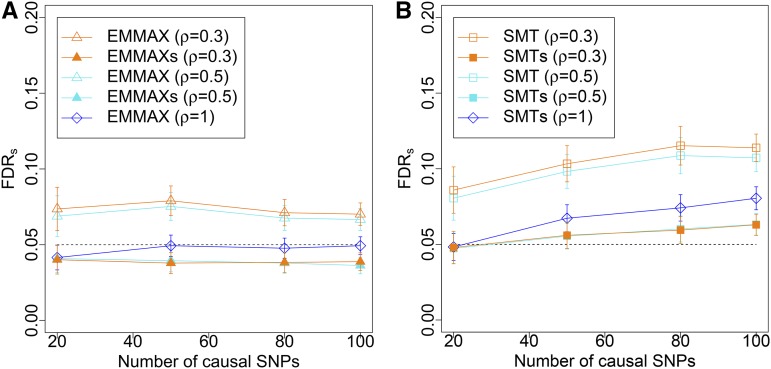
Figure 3 illustrates the results of simulations exploring the FDRs control properties of BH applied to the complete set of M P-values obtained from EMMAX or SMT (i.e., with no prescreening or clustering of the hypotheses) and the corresponding two-step approaches we recommend (EMMAXs and SMTs), where cluster representatives are first chosen using Procedure 1 and then discoveries are identified with Procedure 2. The FDRs for the traditional version of EMMAX and SMT is calculated by mimicking what researchers typically do in practice to interpret GWAS results. Specifically, the SNPs for which the null hypotheses are rejected using BH are supplied to Procedure 1 to identify clusters. The realized FDRs is defined as the average across 100 iterations of the fraction of falsely selected clusters over all clusters obtained.
Figure 3.
FDRs for the described procedures: in (A) we report results relative to EMMAX and in (B) relative to SMT. The dashed black line represents the target FDRs level of 0.05. Note that EMMAXs with (i.e., with no clustering) coincides with EMMAX, and that the FDRs for this specific case corresponds to the regular FDR. Shapes indicate the procedures: empty triangles for the application of BH to the collection of P-values from EMMAX for all hypotheses followed by clustering of the discoveries; filled triangles for the selective procedure EMMAXs; empty squares for the application of BH to the collection of P-values from SMTs for all hypotheses followed by clustering of the discoveries; filled squares for the selective procedure SMTs; and empty diamonds for the application of BH to the full collection of P-values with no clustering. Colors indicate the parameters for clustering: orange for turquoise for and blue for
Figure 3 illustrates that, in agreement with Theorem 1, EMMAXs controls FDRs at all levels of ρ and for any number of causal SNPs. In contrast, BH applied to the full set of P-values obtained from EMMAX with post hoc clustering of the discoveries results in a somewhat elevated FDRs due to the deflation of the BH threshold. Moreover, EMMAXs offers better control of FDRs than SMTs, particularly as the number of causal SNPs increases. This makes sense given that EMMAX better accounts for the polygenic effects than the SMT.
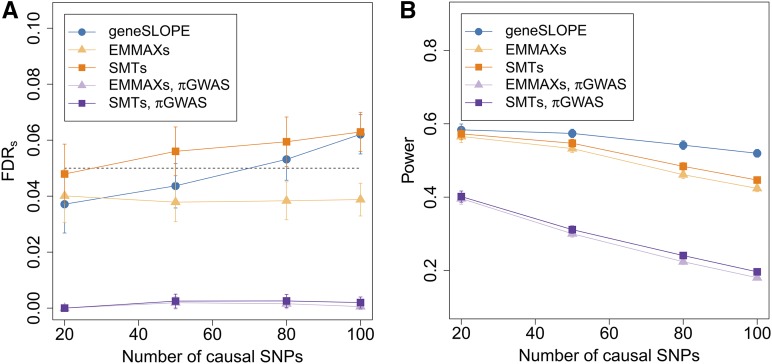
GeneSLOPE error control and power:
Figure 4 illustrates the performance of geneSLOPE in terms of FDRs and power in the context of the performance of EMMAXs and SMTs for the same setting and range of k. For all procedures, power decreases as k increases, with a slower decay for geneSLOPE. Note that the average power of geneSLOPE is systematically larger than the power of SMTs, with the difference increasing with k, while the FDRs of geneSLOPE is always smaller than that of SMTs. Figure 4 also demonstrates how using the standard genome-wide significance threshold setting results in a very substantial loss of power as compared to procedures controlling FDR.
Figure 4.
(A) FDRs and (B) power for geneSLOPE. Clustering is done with and the target FDRs level 0.05 (marked with a dashed line). Values for geneSLOPE are in blue. For comparison, we reproduce from Figure 3 the curves indicating the performance of EMMAXs and SMTs for the same setting (marked in shades of orange). We also include the values of (A) FDRs and (B) power when EMMAXs and SMTs are carried out using cluster representatives selected with the standard GWAS genome-wide significance threshold (marked in shades of purple). Shapes indicate the procedures: Filled circles for geneSLOPE, filled triangles for EMMAXs, and filled squares for SMTs.
Real data analysis
To analyze the lipid phenotypes, we adopted the protocol described in Sabatti et al. (2009): subjects that had not fasted or were being treated for diabetes (n = 487) were excluded, leaving a set of subjects for further analysis. All phenotypes were adjusted for sex, pregnancy, oral contraceptive use, and population structure as captured by the first five genotype principal components (computed using Eigensoft, Price et al. 2006); the residuals were used as the trait values in the subsequent association analysis.
We compare the results of geneSLOPE, EMMAXs, and classically applied EMMAX. GeneSLOPE (Procedure 1 followed by Procedure 3) was applied using ρ = 0.3 or 0.5, and q = 0.05 or 0.1 (for a total of four versions) to a centered and normalized version of the genotype matrix where each column has mean 0 and norm 1. EMMAXs (Procedure 1 followed by Procedure 2) was applied with ρ = 0.3 or 0.5, and q = 0.05 or 0.1. To mimic the standard GWAS analysis, we ran EMMAX identifying as significant those SNPs with P-value to obtain comparable numbers of discovered SNPs we applied Procedure 1 to cluster the results.
We compare the discoveries of these three methods on the NFBC data to those reported in Global Lipids Genetics Consortium (2013), a much more powerful study based on 188,577 subjects. We compute the realized selected false discovery proportion FDPs for each method assuming that SNPs within 1 Mb of a discovery (defined as P < 5 × 10−8) in the comparison study are true positives (even if, of course, the biological truth for the given study population is not known, and the association statistics in Global Lipids Genetics Consortium 2013 are based on univariate tests and may therefore not fully capture the genetic underpinnings of these complex traits). We also seek to understand what proportion of the trait heritability is captured by the selected SNPs. To this end, we estimate the proportion of phenotypic variance explained by the set of genome-wide autosomal SNPs using Genome-wide Complex Trait Analysis (GCTA) (Yang et al. 2011), and compare this to the adjusted obtained from a multiple regression model including the selected cluster representatives as predictors.
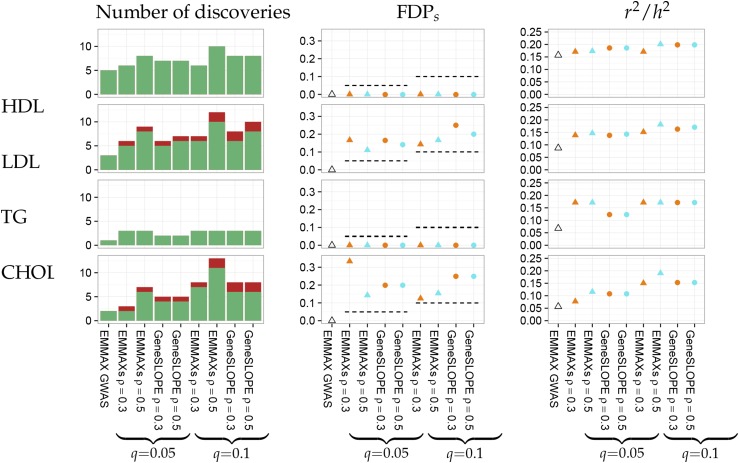
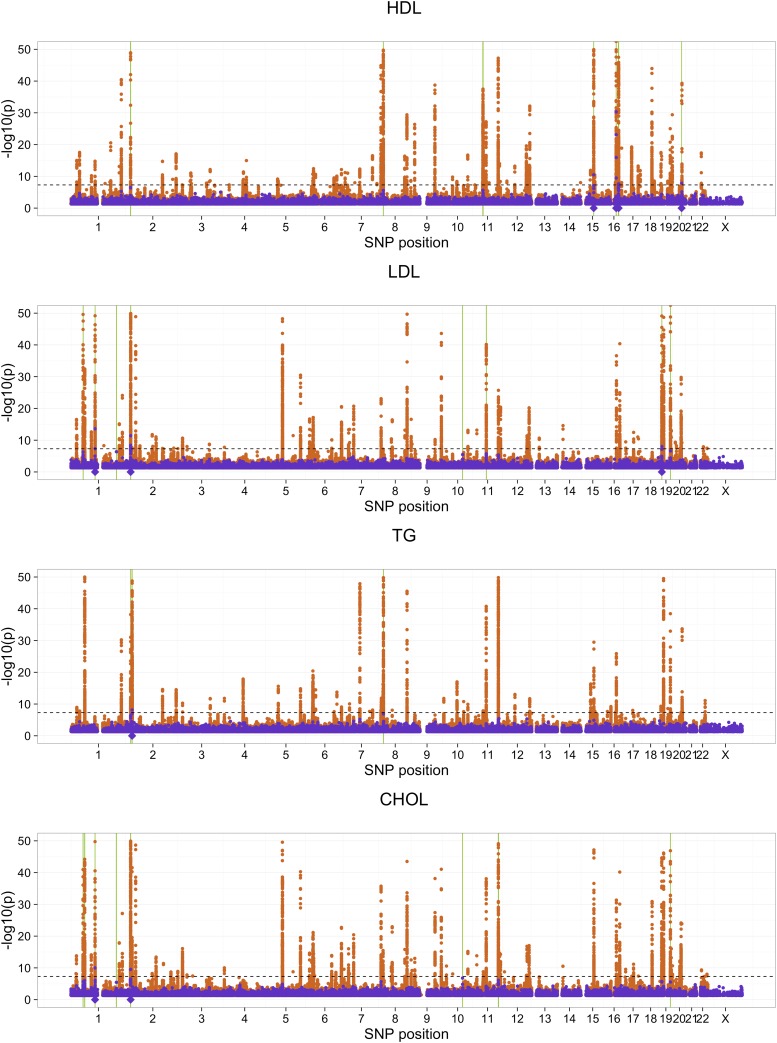
The estimated proportion of phenotypic variance explained by genome-wide SNPs is 0.34, 0.32, 0.10, and 0.29 for HDL, LDL, TG, and CHOL, respectively. A comparison of the number of discoveries (i.e., the number of selected cluster representatives), number of true discoveries, FDPs, and across methods is given in Figure 5. As an illustrative example, geneSLOPE selections with π = 0.05, q = 0.1, and ρ = 0.5 are shown in Figure 6 along with P-values obtained using EMMAX and those obtained in the more highly powered comparison study (Global Lipids Genetics Consortium 2013).
Figure 5.
Study of four lipid traits. Comparison of results for HDL, LDL, TG, and CHOL. “Number of discoveries” corresponds to the number of selected cluster representatives under each method; true and false discoveries are marked in green and red, respectively. FDPs is the realized selected false discovery proportion, and is the adjusted obtained when using the set of selected cluster representatives as predictors in a multiple regression model divided by the proportion of phenotype variance explained by genome-wide SNPs obtained using GCTA.
Figure 6.
Localization of lipid signals. GeneSLOPE selections using ρ = 0.5, and target FDRs 0.1 are marked using solid green bars for cluster representatives and semitransparent bars for the remaining members of the cluster. P-values from EMMAX (purple) and the Global Lipids Genetics Consortium comparison study (orange) are plotted on the −log10 scale. The horizontal dashed line marks a significance cut off of 5 × 10−8, and the purple diamonds below the x-axis represents selected cluster representatives under EMMAX using and a P-value threshold of 5 × 10−8.
The application on real data illustrates how FDRs controlling procedures are more powerful than the standard practice of identifying significant SNPs using a P-value threshold of Both EMMAXs and geneSLOPE attain realized selected false discovery proportions that are consistent with the nominal targeted FDRs. There does not appear to be a power advantage of multiple linear regression (geneSLOPE) over univariate tests (EMMAXs) in this example. This is consistent with the results in our simulations, which indicate that multivariate analysis is really more powerful when there are many (detectable) signals contributing to the phenotype. While it is by now established that 100s of different loci contribute to lipid levels, the signal strength in our data set (which has a modest sample size) is such that only a handful can be identified. In this regime, we find no evidence of an increased power for the multiple linear model. However, this data analysis also shows the potential advantage on the multivariate methods with respect to signal resolution. Changing the value of ρ from 0.3 to 0.5 had a negligible influence on the number of discoveries made by geneSLOPE but substantially increased the number of discoveries by EMMAXs. This suggests that some of the clusters corresponding to were split into smaller clusters for and that in the case of SMT, the resolution of is not sufficient to prevent representing one biological discovery by two or more clusters. This observation goes along with the simulation results reported in the previous section.
Discussion
Following up on an initial suggestion by Siegmund et al. (2011) and reflecting on the elements of the standard practice, we argue that discoveries in a GWAS study should not be counted in terms of the number of SNPs for which the hypothesis of no association is rejected, but in terms of the number of clusters of such SNPs. We propose a strategy to control the FDR of these discoveries that consists in identifying groups of hypotheses on the basis of the observed data, selecting a representative for each group, and applying a modified FDR-controlling procedure to the P-values for the selected hypotheses. We present two articulations of this strategy: in one case we rely on marginal tests of association and modify the target rate of BH on the selected hypotheses, and in the other case we build on our previous work on SLOPE to fit a multiple linear regression model. We show with simulations and real data analysis that the suggested approaches appear to control FDRs and allow an increase in power with respect to the standard analysis methods for GWAS.
The idea of identifying groups of hypotheses and somehow transferring the burden of FDR control from the single hypothesis level to a group one is not new (Perone Pacifico et al. 2004; Benjamini and Heller 2007). In particular, two recent contributions to the literature can be considered parallel to our suggestions. In the context of tests for marginal association, Foygel Barber and Ramdas (2015) propose a methodology to control FDR both at the level of single hypotheses and groups. In the context of multiple regression, Brzyski et al. (2016) extend SLOPE to control the FDR for the discoveries of groups of predictors. Both these contributions, however, are substantially different from ours in that they require a definition of groups prior to observation of the data. Instead, our clusters are adaptive to the signal, and are identified starting from the data. This assures that the group of hypotheses are centered around the locations with strongest signal.
Defining cluster representatives that are input into a multiple regression framework allows us to think more carefully about what FDR means in the context of a regression model that does not include among the regressor the true causal variants; where one is substantially looking for relevant proxies. In their recent work, Foygel Barber and Candès (2016) take a different approach, deciding to focus on the directional FDR. The knock-off filter provides an attractive methodology to analyze GWAS data. However, it still requires an initial selection step: top performance can be achieved only when the selected features are optimally capturing the signal present in a given data set. We believe that the cluster-representatives approach has a substantial edge at this level over, for example, running lasso with only a modest penalization parameter.
Here, we consider a fairly simple strategy to construct clusters of SNPs, exploring two possible levels of resolution corresponding to and In reality, depending on sample size and genotype density, each data set might have a different achievable level of resolution. The study of how this can be adaptively learned is deferred to future work.
It should be noted that while we conduct formal testing only on the selected set of cluster representatives, when the null hypothesis of no association is rejected for a selected SNP, the entire cluster is implicated. In other words, in follow-up studies, the entire region spanned by the cluster should be considered associated with the trait in question. This is entirely similar to what is standard practice after localizing association to a region: all variants in LD with the signal are implicated, and to sort through them, multiple regression models are employed (Hormozdiari et al. 2014).
It is common practice in GWAS studies to rely on the imputation of untyped SNPs to augment the power to detect association. In this context, a cluster should be formed using both the typed and imputed SNPs, so that representatives with maximal power might be selected. The adjusted thresholds for significance (or the penalization coefficients in the case of SLOPE), however, should be determined on the basis of the number of typed SNPs only; since this defines the degrees of freedom of the problem.
Finally, we would like to underscore how, even if we have focused on the case of GWAS here, adopting a selective approach might have wide range applications whenever there is not an exact correspondence between the hypotheses conveniently tested and the granularity of the scientific discoveries. Further studies of the emerging literature on selective inference should lead to better understanding of the theoretical properties of the method we propose, as well as to the identification of other possible strategies.
Acknowledgments
D.B. would like to thank Jerzy Ombach for significant help with the process of obtaining access to the data. We are grateful to dbGap for accession to the Stamped North Finland Birth Cohort data set. This research is supported by the European Union’s Seventh Framework Programme for research, technological development, and demonstration under grant agreement number 602552, cofinanced by the Polish Ministry of Science and Higher Education under grant agreement 2932/7.PR/2013/2 and by National Institutes of Health grants HG-006695, MH-101782, and MH-108467.
Appendix: Proof of Theorem 1
Throughout, R is the number of rejections the two-step procedure commits. Note that the critical values for the BH procedure after selection are of the form
By definition, letting of cardinality be the set of nulls,
Setting we have that for all
| (4) |
The calculation is now as in Benjamini and Yekutieli (2001). The key observation is that since is an increasing set, we have
So consider the first two terms of the sum (4):
Continuing in this fashion, we have that
Hence, for each
Remark: Under independence, we know that if we select everything, i.e., almost surely, then Here, when we select less while retaining the monotonicity assumption, it is possible to have an FDR less than
Procedure 4. Sequence of penalties λ for SLOPE.
Input:
set for
- define
find the largest index, such that
- put
Procedure 5. Selecting λ when σ is unknown.
Input: y, X, and basic sequence λ
initialize:
And repeat
compute RSS obtained by regressing y onto variables in S
set where |S| is the number of elements in S
compute the solution to SLOPE with parameter sequence
set (i.e., is the set of regressors selected by SLOPE in step 5).
until
Footnotes
Communicating editor: E. Eskin
Literature Cited
- Abramovich F., Benjamini Y., Donoho D. L., Johnstone I. M., 2006. Adapting to unknown sparsity by controlling the false discovery rate. Ann. Stat. 34: 584–653. [Google Scholar]
- Alexander D. H., Lange K., 2011. Stability selection for genome-wide association. Genet. Epidemiol. 35: 722–728. [DOI] [PubMed] [Google Scholar]
- Ardlie K. G., Deluca D. S., Segre A. V., Sullivan T. J., Young T. R., et al. , 2015. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348: 648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Bogomolov M., 2014. Selective inference on multiple families of hypotheses. J. R. Stat. Soc. Series B Stat. Methodol. 76: 297–318. [Google Scholar]
- Benjamini Y., Heller R., 2007. False discovery rates for spatial signals. J. Am. Stat. Assoc. 102: 1272–1281. [Google Scholar]
- Benjamini Y., Hochberg Y., 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300. [Google Scholar]
- Benjamini Y., Yekutieli D., 2001. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 29: 1165–1188. [Google Scholar]
- Benjamini Y., Yekutieli D., 2005a False discovery rate-adjusted multiple confidence intervals for selected parameters. J. Am. Stat. Assoc. 100: 71–93. [Google Scholar]
- Benjamini Y., Yekutieli D., 2005b Quantitative trait loci analysis using the false discovery rate. Genetics 171: 783–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdan M., van den Berg E., Sabatti C., Su W., Candès E., 2015. SLOPE—adaptive variable selection via convex optimization. Ann. Appl. Stat. 9: 1103–1140, 26709357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzyski, D., A. Gossmann, W. Su, and M. Bogdan, 2016 Group SLOPE - adaptive selection of groups of predictors. ArXiv: 1610.04960. [DOI] [PMC free article] [PubMed]
- Brzyski D., Peterson C., Sobczyk P., Candès E. J., Bogdan M., et al. , 2016. geneSLOPE: genome-wide association study with SLOPE. R package. Available at: https://CRAN.R-project.org/package=geneSLOPE. [Google Scholar]
- Carbonetto P., Stephens M., 2011. Scalable variational inference for Bayesian variable selection, and its accuracy in genetic association studies. Bayesian Anal. 6: 1–42.22247752 [Google Scholar]
- Dolejsi E., Bodenstorfer B., Frommlet F., 2014. Analyzing genome-wide association studies with an FDR controlling modification of the Bayesian information criterion. PLoS One 9: e103322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J., Li R., 2001. Variable selection via nonconcave penalized likelihood and its oracle properties. J. Am. Stat. Assoc. 96: 1348–1360. [Google Scholar]
- Fan J., Lv J., 2008. Sure independence screening for ultrahigh dimensional feature space. J. R. Stat. Soc. B 70: 849–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foygel Barber, R., and E. J. Candès, 2016 A knockoff filter for high-dimensional selective inference. ArXiv: 1602.03574.
- Foygel Barber, R., and A. Ramdas, 2015 The p-filter: multi-layer FDR control for grouped hypotheses. ArXiv: 1512.03397.
- Frommlet F., Ruhaltinger F., Twaróg P., Bogdan M., 2012. Modified versions of Bayesian Information Criterion for genome-wide association studies. Comput. Stat. Data Anal. 56: 1038–1051. [Google Scholar]
- Global Lipids Genetics Consortium , 2013. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 45: 1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halperin E., Kimmel G., Shamir R., 2005. Tag SNP selection in genotype data for maximizing SNP prediction accuracy. Bioinformatics 21: i195–i203. [DOI] [PubMed] [Google Scholar]
- He Q., Lin D. Y., 2011. A variable selection method for genome-wide association studies. Bioinformatics 27: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoggart C. J., Whittaker J. C., Iorio M. D., Balding D. J., 2008. Simultaneous analysis of all SNPs in genome-wide and re-sequencing association studies. PLoS Genet. 4: e1000130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormozdiari F., Kostem E., Kang E. Y., Pasaniuc B., Eskin E., 2014. Identifying causal variants at loci with multiple signals of association. Genetics 198: 497–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H. M., Sul J. H., Service S. K., Zaitlen N. A., Kong S., et al. , 2010. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 42: 348–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvale M. N., Hesselson S., Hoffmann T. J., Cao Y., Chan D., et al. , 2015. Genotyping informatics and quality control for 100,000 subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) cohort. Genetics 200: 1051–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon B. A., Hoffman G. E., Mezey J. G., 2010. A variational Bayes algorithm for fast and accurate multiple locus genome-wide association analysis. BMC Bioinformatics 11: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio T. A., Collins F. S., Cox N. J., Goldstein D. B., Hindorff L. A., et al. , 2009. Finding the missing heritability of complex diseases. Nature 461: 747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perone Pacifico M., Genovese C., Verdinelli I., Wasserman L., 2004. False discovery control for random fields. J. Am. Stat. Assoc. 99: 1002–1014. [Google Scholar]
- Peterson C. B., Bogomolov M., Benjamini Y., Sabatti C., 2016. Many phenotypes without many false discoveries: error controlling strategies for multitrait association studies. Genet. Epidemiol. 40: 45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A. L., Patterson N. J., Plenge R. M., Weinblatt M. E., Shadick N. A., et al. , 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38: 904–909. [DOI] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A., et al. , 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S. M., Wray N. R., Stone J. L., Visscher P. M., O’Donovan M. C., et al. , 2009. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460: 748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatti C., 2013. Multivariate linear models for GWAS, pp. 188–208 in Advances in Statistical Bioinformatics. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- Sabatti C., Service S., Freimer N., 2003. False discovery rate in linkage and association genome screens for complex disorders. Genetics 164: 829–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatti C., Service S. K., Hartikainen A. L., Pouta A., Ripatti S., et al. , 2009. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat. Genet. 41: 35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegmund D. O., Yakir B., Zhang N., 2011. The false discovery rate for scan statistics. Biometrika 98: 979–985. [Google Scholar]
- Stell L., Sabatti C., 2016. Genetic variant selection: learning across traits and sites. Genetics 202: 439–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey J. D., Tibshirani R., 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100: 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer S., Wray N. R., Kahn R. S., Derks E. M., 2011. Underestimated effect sizes in GWAS: fundamental limitations of single SNP analysis for dichotomous phenotypes. PLoS One 6: e27964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T., Zhang C., 2012. Scaled sparse linear regression. Biometrika 99: 879–898. [Google Scholar]
- Tibshirani R., 1994. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. B 58: 267–288. [Google Scholar]
- Welter, D., J. MacArthur, J. Morales, T. Burdett, P. Hall et al., 2014 The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Research 42:AD1001–D1006. Available at: http://www.ebi.ac.uk/gwas/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Devlin B., Ringquist S., Trucco M., Roeder K., 2010. Screen and clean: a tool for identifying interactions in genome-wide association studies. Genet. Epidemiol. 34: 275–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. T., Chen Y. F., Hastie T., Sobel E., Lange K., 2009. Genome-wide association analysis by lasso penalized logistic regression. Bioinformatics 25: 714–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Benyamin B., McEvoy B. P., Gordon S., Henders A. K., et al. , 2010. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42: 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Lee S. H., Goddard M. E., Visscher P. M., 2011. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88: 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H., Breheny P., Imam N., Liu Y., Hoeschele I., 2015. Penalized multimarker vs. single-marker regression methods for genome-wide association studies of quantitative traits. Genetics 199: 205–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Sehl M. E., Sinsheimer J. S., Lange K., 2010. Association screening of common and rare genetic variants by penalized regression. Bioinformatics 26: 2375–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
To illustrate the performance of our methods, we used the data from the North Finland Birth Cohort (NFBC66) study (Sabatti et al. 2009), available in the database of Genotypes and Phenotypes (dbGaP) under accession number phs000276.v2.p1 (http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000276.v2.p1).