Kang et al. show that the GCN2–ATF4 pathway induces 4E-BP transcription in response to amino acid deprivation and also during the development of certain Drosophila tissues. 4E-BP has selective effects on translation; therefore, this pathway helps to shift the mRNA expression profiles of cells.
Abstract
Reduced amino acid availability attenuates mRNA translation in cells and helps to extend lifespan in model organisms. The amino acid deprivation–activated kinase GCN2 mediates this response in part by phosphorylating eIF2α. In addition, the cap-dependent translational inhibitor 4E-BP is transcriptionally induced to extend lifespan in Drosophila melanogaster, but through an unclear mechanism. Here, we show that GCN2 and its downstream transcription factor, ATF4, mediate 4E-BP induction, and GCN2 is required for lifespan extension in response to dietary restriction of amino acids. The 4E-BP intron contains ATF4-binding sites that not only respond to stress but also show inherent ATF4 activity during normal development. Analysis of the newly synthesized proteome through metabolic labeling combined with click chemistry shows that certain stress-responsive proteins are resistant to inhibition by 4E-BP, and gcn2 mutant flies have reduced levels of stress-responsive protein synthesis. These results indicate that GCN2 and ATF4 are important regulators of 4E-BP transcription during normal development and aging.
Introduction
It is now established that cells frequently reduce the rate of translational initiation in response to stress as a protective mechanism. One particular form of stress imposed by dietary restriction of amino acids has attracted significant interest in recent decades because of its effect of extending lifespan of a wide-range of organisms, from Saccharomyces cerevisiae to Caenorhabditis elegans to Drosophila melanogaster to mammals (McCay et al., 1935; Klass, 1977; Partridge et al., 1987; Jiang et al., 2000; Kapahi et al., 2004; Kaeberlein et al., 2005).
Inhibition of translational initiation in response to stress mostly occurs through two distinct regulatory mechanisms (Sonenberg and Hinnebusch, 2009). In one type, cells reduce the overall availability of methionine-charged initiator tRNAs to ribosomes. Molecularly, this can be achieved through stress-activated kinases that phosphorylate eIF2α, thereby inhibiting this translational initiation factor’s normal role in helping the 40S ribosome subunit acquire methionyl initiator tRNA (Hinnebusch, 2014). In a second type, another set of stress signals specifically inhibits ribosomes from loading onto the 5′ cap of mRNAs for translational initiation (Hu et al., 1994; Pause et al., 1994).
Stress-activated kinases that phosphorylate eIF2α include GCN2, which is activated by amino acid deprivation (Wek et al., 1989; Dever et al., 1992), and PERK, which responds to ER stress (Harding et al., 1999). Although such conditions reduce the overall rate of translational initiation for most transcripts, a few cellular transcripts have unique 5′ UTRs with regulatory upstream open reading frames that allow them to paradoxically enhance the translation of the main open reading frame under those conditions (Palam et al., 2011; Malzer et al., 2013; Baird et al., 2014; Hinnebusch, 2014). Among the best characterized are the 5′ UTRs of GCN4 of yeast and its metazoan equivalent, ATF4 (Dever et al., 1992; Harding et al., 2000; Kang et al., 2015), which allow these proteins to be specifically synthesized upon eIF2α kinase activation and induce the transcription of several stress-responsive genes, including those involved in amino acid transport and antioxidation. In addition, ATF4 induces the expression of target genes that stimulate the dephosphorylation of eIF2α, thereby restoring the overall translation rate within hours (Harding et al., 2003; Marciniak et al., 2004; Han et al., 2013; Malzer et al., 2013). As several different types of stress-activated eIF2α kinases activate this pathway, this pathway is often referred to as the integrated stress response.
Most eukaryotic mRNAs are translated after eIF-4E recognizes the 5′ cap of mRNAs to load the 40S subunit of the ribosome to mRNAs. A distinct set of stress response mechanisms reduces translation by inhibiting this process. Best characterized is 4E-BP, which directly binds eIF-4E (Hu et al., 1994; Pause et al., 1994). In Drosophila, many cell types do not express 4E-BP under normal healthy conditions, underscoring the importance of regulation at the transcriptional level (Rodriguez et al., 1996). In fact, various types of stress ranging from starvation to pathogen infection trigger the transcriptional induction of 4E-BP, and interestingly, such induction somehow enhances general stress resistance of cells (Bernal and Kimbrell, 2000; Zinke et al., 2002; Teleman et al., 2005; Tettweiler et al., 2005). As an example, it has been shown that mild conditions of amino acid restriction in the diet induce 4E-BP expression, and under certain conditions, such induction mediates lifespan extension in Drosophila (Zid et al., 2009; Partridge et al., 2011). Similarly, artificial activation of 4E-BP enhances the overall stress resistance and extends lifespan in Drosophila (Teleman et al., 2005; Tettweiler et al., 2005; Demontis and Perrimon, 2010).
Although these observations establish 4E-BP’s transcriptional induction as an important effector of dietary restriction and lifespan control, the underlying signaling pathway has remained unclear. Several in vivo Drosophila studies have suggested that the JNK–FOXO pathway is the primary regulator of stress-induced 4E-BP transcription (Jünger et al., 2003; Puig et al., 2003; Wang et al., 2005; Marr et al., 2007). The literature also reports other possible mechanisms, some of which are mutually contradictory. These include a study indicating that PERK directly activates FOXO in Drosophila, and cell culture–based studies in mouse β-islet cells that PERK–ATF4 pathway can induce 4E-BP1 in response to ER stress (Yamaguchi et al., 2008; Zhang et al., 2013). Initial studies in Drosophila suggested FOXO as an inducer of 4E-BP in response to starvation (Teleman et al., 2005; Tettweiler et al., 2005), but more recent studies indicate that FOXO is not required for dietary restriction–induced lifespan extension or 4E-BP induction under these conditions (Giannakou et al., 2008; Min et al., 2008; Zid et al., 2009). Thus, the relevant in vivo pathway that mediates 4E-BP induction upon dietary restriction remains unknown.
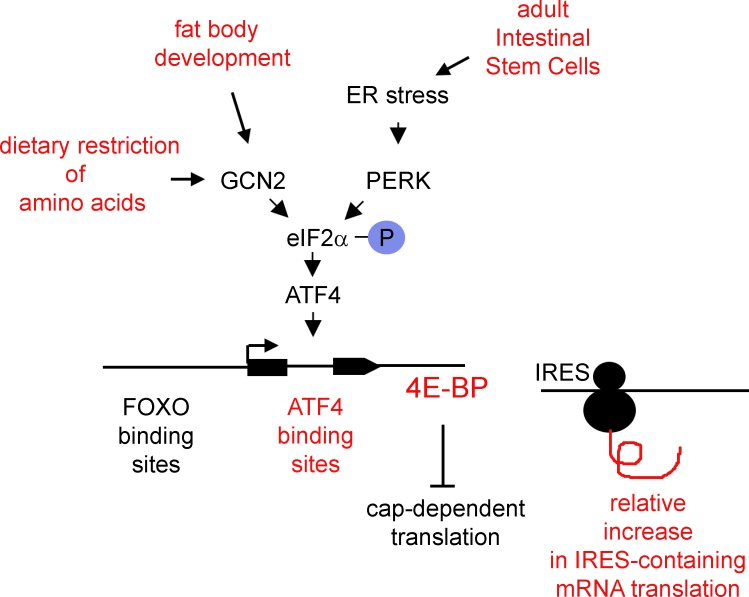
Here, we report that GCN2 and ATF4 mediate 4E-BP induction during normal Drosophila development and upon dietary restriction of amino acids. The 4E-BP intron has ATF4-binding sites that mediate such induction, and a reporter based on this enhancer shows endogenous ATF4 activity in healthy developing tissues, including fat body and adult intestinal stem cells. We find evidence that certain stress-response transcripts have 5′ UTRs that are resistant to 4E-BP inhibition, and such evidence is further supported by directly analyzing the protein synthesis profile using an approach that combines metabolic labeling and azide-alkyne cycloaddition chemistry. In adult flies, reduced availability of yeast extract (YE) prolongs lifespan, and this effect is impaired when gcn2 is lost. These results indicate that the GCN2–eIF2α–ATF4 pathway is an important regulator of 4E-BP transcription in response to dietary restriction and during normal development of certain tissues.
Results
PERK and ATF4, but not FOXO, are required for 4E-BP transcription in response to ER stress
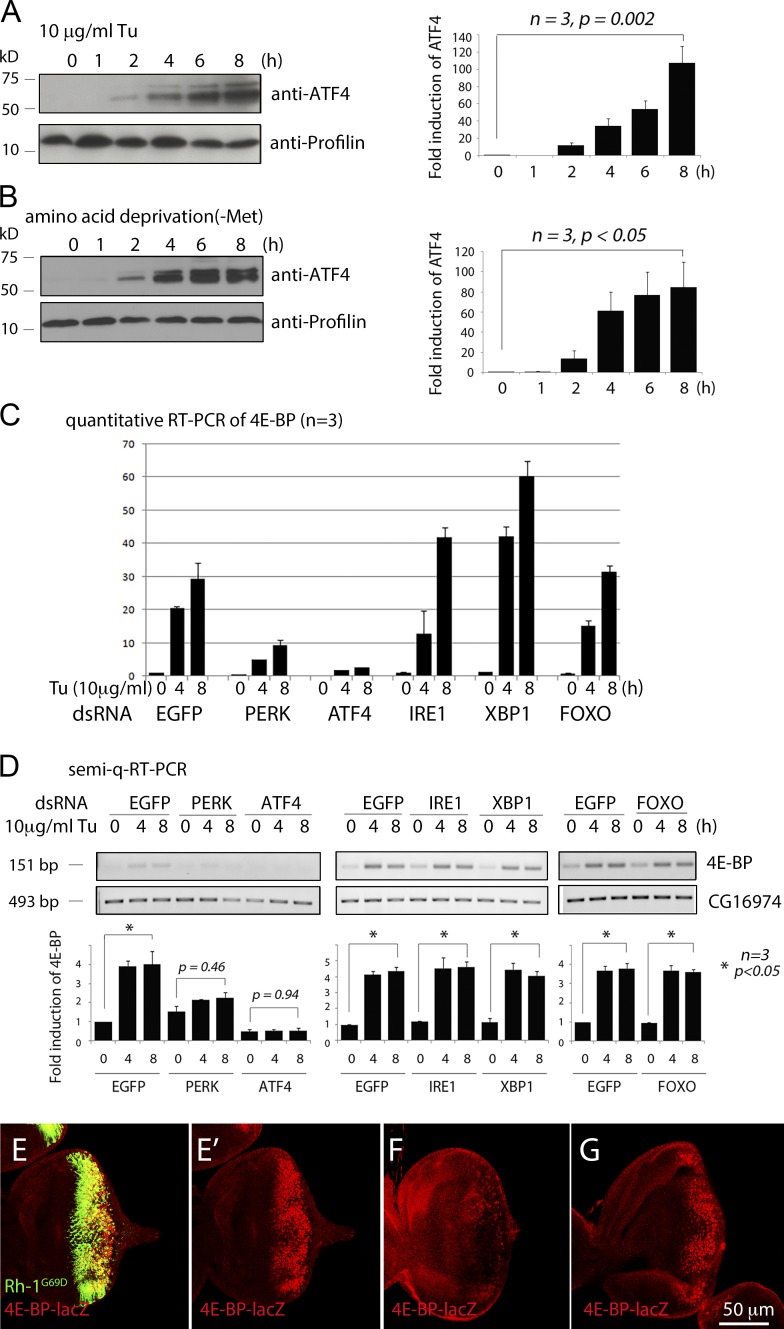
As in mammals, Drosophila ATF4 protein is induced either in response to ER stress, as imposed by tunicamycin (Tu) treatment (Fig. 1 A), or in response to the deprivation of specific amino acids in the culture media (Fig. 1 B). We became interested in the transcriptional regulation of 4E-BP while studying the ER stress response, which activates JNK markers in Drosophila (Kang et al., 2012). Consistent with the idea that 4E-BP is induced by JNK and FOXO, Tu treatment induced 4E-BP within 4 h (Fig. 1, C and D). However, such 4E-BP induction was unaffected by the knockdown of foxo. This led us to explore the alternative possibility that 4E-BP is regulated by ER stress–activated pathways, otherwise referred to as the unfolded protein response (UPR). In fact, we found that the specific knockdown of perk or atf4 in S2 cells blocked Tu-induced 4E-BP transcription. In contrast, 4E-BP induction was not blocked by the knockdown of ire1 or xbp1, which mediates another UPR branch (Fig. 1, C and D).
Figure 1.
PERK and ATF4, but not FOXO, mediate 4E-BP transcription in response to ER stress. (A) ATF4 protein Western blot from S2 cells challenged with the ER stress–causing chemical tunicamycin (Tu). A graph showing the quantified band intensities is shown on right. (B) ATF4 protein levels of S2 cells cultured in media lacking methionine. A graph of the quantified bands is shown on the right. (C) Quantitative RT-PCR for 4E-BP in S2 cells challenged with Tu. The values were normalized to Rp49 levels. Cells were pretreated with double-stranded RNA (dsRNA) targeting EGFP (negative control), PERK, ATF4, IRE1, XBP1, or FOXO. The numbers above the gels indicate hours after Tu treatment. (D) Semiquantitative RT-PCR (semi-q-RT-PCR) against 4E-BP under conditions equivalent to C. CG16974 was performed as a loading control (bottom). PCR band intensities were quantified and are shown in the graphs below the gel. (E–G) Eye imaginal discs expressing Rh-1G69D through the GMR-Gal4 driver. (E) In atf4+ discs, Rh-1G69D expressing cells (green) also express 4E-BP-lacZ (red). (E′) Anti–β-galactosidase single-channel image of E. atf4crc1/R6 −/− eye imaginal disc (F) and foxoΔ94 −/− disc (G) in otherwise identical genetic backgrounds with E. A scale bar for E–G is shown at the bottom of G. Error bars show standard error (SE). P-values were derived from Student’s t tests.
We validated these results in vivo by using a mutant rhodopsin 1 (Rh-1) allele, Rh-1G69D, whose product fails to fold properly in the ER and impose stress (Ryoo et al., 2007). Expression of Rh-1G69D through the eye-specific GMR-Gal4 driver strongly induced the 4E-BP-lacZ reporter, an enhancer trap line (Fig. 1 E), but such induction was abolished in the atf4 hypomorphic allele crc1 over the null allele R6 (atf4crc1/R6) background (Fig. 1 F). Consistent with the S2 cell culture results, Rh-1G69D expression induced 4E-BP-lacZ even in the foxoΔ 94 −/− background (Fig. 1 G). These experiments indicate that the PERK–ATF4 branch of the UPR induces 4E-BP transcription in response to stress.
ATF4 is required for 4E-BP transcriptional induction in response to amino acid deficiency
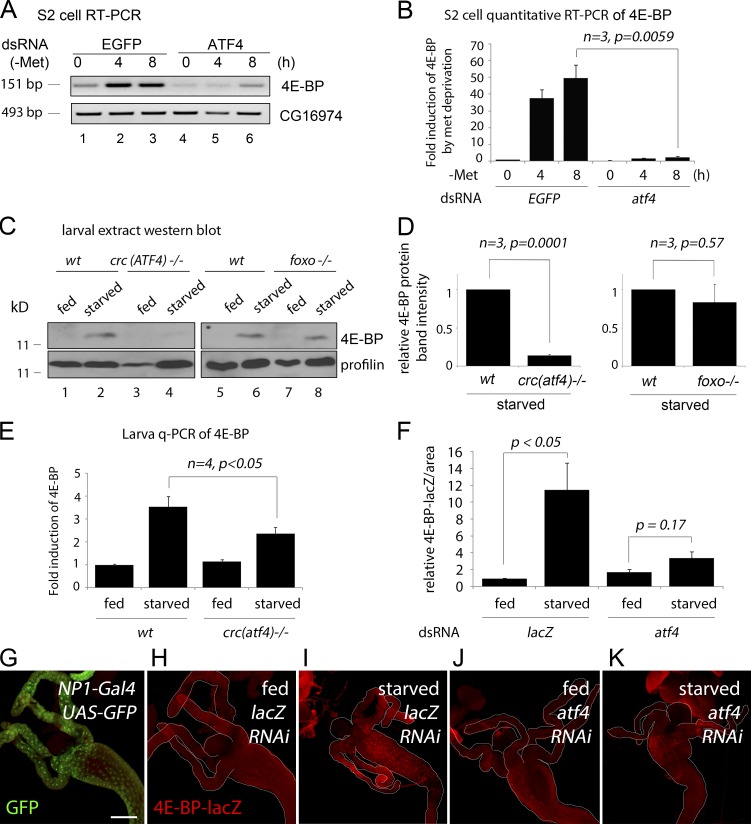
4E-BP transcription in Drosophila is strongly induced by amino acid deficiency in the diet, and most studies have focused on the role of FOXO, with conflicting results (Teleman et al., 2005; Tettweiler et al., 2005; Zid et al., 2009). Because ATF4 is a common mediator of the integrated stress response, we considered ATF4 as an alternative candidate mediator of 4E-BP induction in response to amino acid deprivation. 4E-BP transcripts were strongly induced in S2 cells when cultured in a methionine-deficient media, and such induction was abolished when atf4 was knocked down (Fig. 2, A and B). As reported previously (Zinke et al., 2002; Teleman et al., 2005; Tettweiler et al., 2005), second-instar larvae had very low basal levels of 4E-BP expression, but larvae reared in amino acid–deficient food (5% sucrose in PBS) for 18 h had elevated expression of 4E-BP protein, as detected through Western blot (Fig. 2 C). We did not see an obvious reduction of 4E-BP levels in foxoΔ94 −/− larvae, but such induction was strongly impaired in the atf4crc1 /R6 background (Fig. 2, C and D). Immunolabeling experiments with 4E-BP-lacZ indicated that this reporter is most prominently induced in the anterior intestine upon as short as 4 h of amino acid deprivation (Fig. 2, F, H, and I). When the intestine-specific NP1-Gal4 driver (Fig. 2 G) was used to knock down atf4, starvation-induced 4E-BP-lacZ expression was impaired (Fig. 2, J and K). Such effect was quantified through the measurement of pixel intensities from representative images (Fig. 2 F). Together, these results indicate that ATF4 is a major mediator of 4E-BP induction in response to amino acid deprivation in Drosophila larvae.
Figure 2.
ATF4 is required for 4E-BP transcription in response to amino acid deficiency. (A) Semiquantitative RT-PCR of 4E-BP in S2 cells cultured in methionine deficient media (upper gel). Cells were pretreated with either mock dsRNA (lanes 1–3) or dsRNA that targets ATF4 (lanes 4–6). CG16974 RT-PCR is shown as a control (bottom gel). (B) Quantitative RT-PCR of 4E-BP transcripts. Conditions are identical to those in A, except for the use of Rp49 transcript levels as a normalization control. (C) Anti–4E-BP Western blot on second-instar larval extracts. Wild type (wt) and atf4crc1/R6 −/− larvae were reared with either standard cornmeal food (marked “fed”) or in 5% sucrose dissolved in PBS (marked “starved”) for 18 h. The top panel shows the anti–4E-BP blot, and the bottom panel shows antiprofilin as a control. Similar analysis with foxoΔ 94 −/− larvae is shown in lanes 5–8. (D) The normalized band intensities of gels in C were quantified and shown as a graph. (E) Quantitative RT-PCR of 4E-BP (normalized to Rp49 levels) from whole larval extracts, reared under conditions identical to those in C and D. Error bars show SE. P values are from t tests. (F–K) 4E-BP-lacZ expression in second-instar larval intestine reared in standard food (H and J) or with only PBS for 4 h (I and K). NP1-Gal4 intestine driver was used to knock down a control RNAi line targeting lacZ (H and I) or atf4 (J and K). (G) NP1-Gal4 active tissue, as shown by the uas-GFP expression pattern (green). (F) The 4E-BP-lacZ pixel intensities from representative microscope images were quantified. Error bars indicate SE. P values are based on t tests. Bar, 200 µm.
The 4E-BP intron contains a regulatory element that is controlled by ATF4
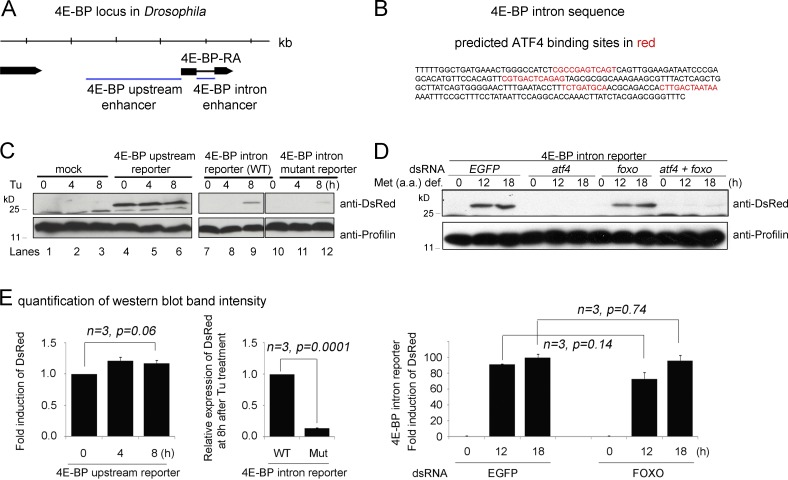
To examine whether ATF4 regulates 4E-BP directly in Drosophila, we used the r-VISTA genome browser (http://genome.lbl.gov) and found four predicted ATF4 binding sites that are clustered in the first intron (Fig. 3, A and B). Consistently, we found that the intron sequence drove the expression of the dsRed reporter when S2 cells were subjected to Tu treatment or when cultured in methionine-deficient media (Fig. 3, C and D). The upstream intergenic regions, which harbor FOXO-binding sites (Puig et al., 2003), did not respond to Tu treatment (Fig. 3 C, lanes 4–6). We mutated the putative Drosophila ATF4-binding sites within the intron reporter and found that the degree of reporter induction by stress was significantly reduced (Fig. 3, C and E). Furthermore, knockdown of atf4, but not foxo, blocked the intron reporter induction (Fig. 3, D and E), supporting the idea that ATF4 regulates 4E-BP induction through the intron enhancer.
Figure 3.
Functional ATF4-binding sites are present in the 4E-BP intron. (A) A diagram of the 4E-BP (thor) genomic locus. Putative regulatory sequences (marked as blue lines) were subcloned upstream of dsRed to generate the upstream enhancer and the intron enhancer reporters. (B) The sequence of the intron enhancer. Predicted ATF4 binding sites are marked in red. (C) Upstream and intron (wild type [WT] and mutant for ATF4-binding sites) enhancer reporter expression in response to tunicamycin (Tu) treatment as shown by Western blot. The numbers above indicate hours of Tu treatment. The upstream enhancer-dsRed expression is shown in lanes 4–6, whereas the intron enhancer-dsRed is in lanes 7–9. The intron enhancer-dsRed with putative ATF4-binding sites mutated is shown in lanes 10–12. Antiprofilin blots are shown as controls (bottom). (D) 4E-BP intron reporter expression detected through Western blot after being cultured in a methionine-deficient media. The indicated genes (EGFP, atf4, and foxo) were knocked down before the analysis. The antiprofilin blot is shown as a control for those lanes (bottom). (E) Graphs showing quantified and normalized Western blot bands from gels in C and D. Error bars show SE. A t test was used to derive p-values.
The ATF4–4E-BP pathway is inherently active in healthy tissues
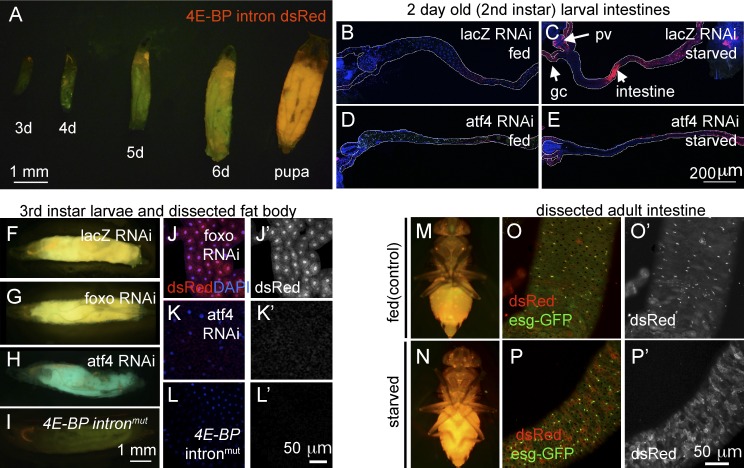
Although ATF4 is mostly studied as a pathway that responds to exogenously imposed stress, Drosophila ATF4 is a developmentally essential gene (Hadorn and Gloor, 1943; Fristrom, 1965), indicating that this pathway must be also active in normally developing tissues. To visualize those cells, we generated a Drosophila transgenic line harboring the 4E-BP intron driving a dsRed reporter (see Materials and methods). Reflecting the endogenous expression pattern of 4E-BP transcripts, the intron reporter activity remained low and limited to the salivary glands up until the second instar larval stage (Fig. 4 A). However, rearing second-instar larvae in food without protein (5% sucrose) for 6 h led to the induction of 4E-BP intron dsRed expression in parts of the intestine, gastric caeca, and proventriculus (Fig. 4, B and C). Such induction was impaired when atf4 was knocked down using the intestine specific NP1-Gal4, further supporting the role of ATF4 in mediating 4E-BP induction through the intron element (Fig. 4 E).
Figure 4.
4E-BP intron dsRed reporter marks cells with active ATF4 signaling. Shown are 4E-BP intron dsRed reporters in larvae (A–L) and adult flies (M–P). Unless specified otherwise, 4E-BP intron dsRed is marked in red and GFP in green. (A) The reporter expression during larval development (numbers indicate days after egg laying). (B–E) Second-instar larval intestines. Anterior is to the left, and posterior is to right. The outlines of the intestines are marked in white. Before dissection, the larvae were either well fed (B and D) or deprived of amino acids for 6 h (C and E). These intestines were expressing RNAi lines against control lacZ (B and C) or atf4 (D and E). dsRed reporter induction in response to amino acid deprivation (C) can be seen in the proventriculus (pv), gastric caeca (gc), and the intestine (arrows), which is suppressed when atf4 is knocked down (E). (F–I) Third-instar larvae show inherent activation of ATF4 as shown by 4E-BP intron reporter expression. The indicated RNAi lines were driven together with uas-GFP using the fat body–specific cg-Gal4 driver. (J–L) Dissected larval fat bodies containing the 4E-BP intron dsRed reporter. RNAi-mediated knockdown of foxo (J and J′) had no effect, whereas atf4 knockdown blocked the reporter expression (K and K′). (L and L′) The expression of the 4E-BP intronmut reporter, with the putative ATF4-binding sites mutated. (J′, K′, and L′) dsRed-only channels of J, K, and L. Adult flies reared under well-fed conditions (M and O) or those starved overnight (N and P). (M and N) Low-resolution images under dissecting microscopes. (O and P) Dissected intestinal epithelium under confocal microscopy. The posterior intestine, where esg-GFP–positive ISCs are found, was focused. (O′ and P′) dsRed-only channels of O and P. Note that in the epithelium of well-fed flies, the most prominent dsRed signals colocalize with the small nuclear cells that are positive for esg-GFP. In contrast, starved epithelium shows reporter activity spreading to esg-GFP–negative cells with large nuclei.
By the early third-instar larval stage, the most prominent 4E-BP intron reporter signal emerged in the larval fat body, and by the pupal stage, the reporter was ubiquitously expressed (Fig. 4, A and F). We used fat body–specific cg-Gal4 lines to knock down candidate genes and confirmed their knockdown efficiency through quantitative RT-PCR (Fig. S1). Under these conditions, knockdown of atf4 by driving RNAi (Vienna Drosophila Resource Center ID 109014) abolished 4E-BP intron reporter expression in third-instar larvae (Fig. 4, H and K). On the other hand, knockdown of foxo had no detectable effect on the reporter expression as expected by the specificity of the reporter (Fig. 4, G and J). Mutation in the putative ATF4-binding sites of the 4E-BP intron reporter also abolished dsRed expression in the fat body (Fig. 4, I and L). We also performed converse experiments in which we overexpressed candidate genes in early third-instar larval fat body. atf4 overexpression significantly enhanced the 4E-BP intron dsRed signal, but foxo overexpression did not show such effects (Fig. S2).
In the third-instar larval stage, 4E-BP transcripts are detected in selected tissues that include the fat body, testis, gut, and ring glands (Rodriguez et al., 1996). 4E-BP-lacZ is an enhancer trap line whose reporter expression largely mirrors the mRNA distribution pattern (Rodriguez et al., 1996). The 4E-BP intron dsRed reporter is active in a subset of tissues that express 4E-BP-lacZ, which includes the larval fat body, salivary glands, and the gastric caeca attached to the intestine (Fig. S3, A, B, E, and H). The 4E-BP upstream dsRed reporter is also expressed in normally developing larvae, at lower levels and complementary to the intron dsRed reporter (Fig. S3, A, D, F, and G). The 4E-BP upstream dsRed reporter expression was not abolished in the foxoΔ94 −/− background, indicating that there are FOXO independent-regulatory inputs on the upstream element (Fig. S4). On the other hand, the strict dependence of the 4E-BP intron dsRed reporter on ATF4 indicates that ATF4 signaling is active in certain tissues undergoing normal development.
ATF4–4E-BP signaling in the adult intestinal epithelium
4E-BP intron dsRed is also expressed in adult flies (Fig. 4, M–P), and starvation enhanced the reporter signal, which was noticeable even under dissection microscopes (Fig. 4 N). In the intestinal epithelium of well-fed flies, the 4E-BP intron reporter signal was most prominent in intestinal stem cells (ISCs) and their immediate daughter cells, enteroblasts, which are present frequently in the posterior part of the intestine and are marked by escargot-GFP (esg-GFP; Fig. 4 O). Detection of basal ATF4–4E-BP activity is consistent with our previous observation that loss of perk specifically in ISCs reduced their proliferation (Wang et al., 2015). When adult flies were starved for 16 h, 4E-BP intron dsRed expression spread beyond the esg-GFP–positive ISCs/enteroblasts and into cells with larger nuclei that are indicative of enterocytes (Fig. 4 P).
GCN2 promotes 4E-BP induction under conditions of dietary restriction
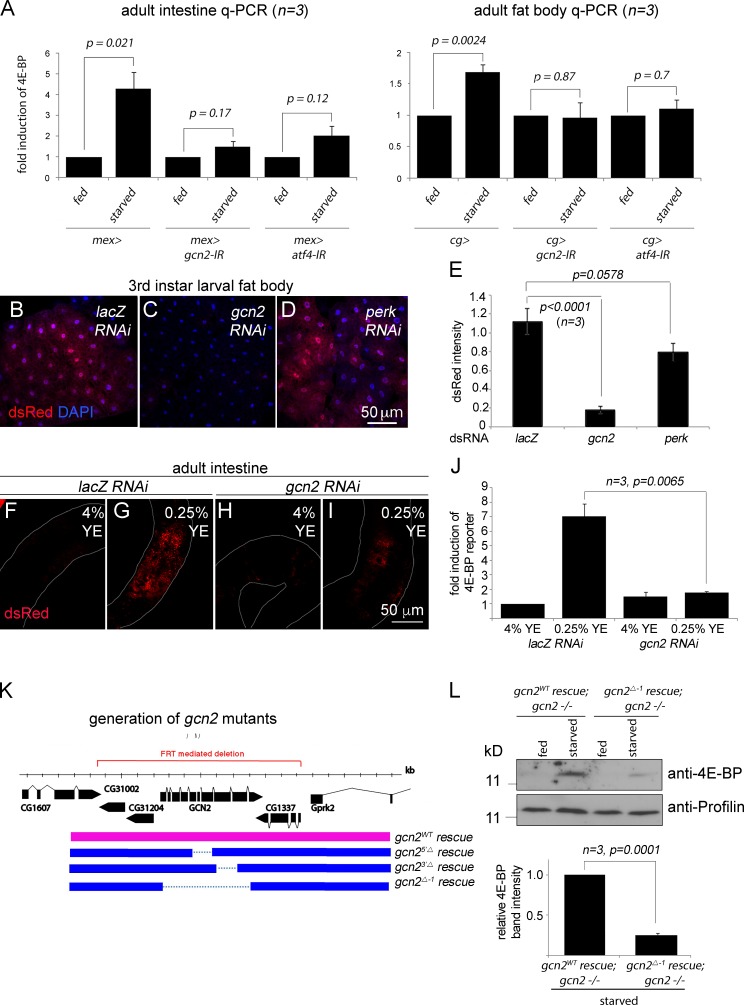
We turned our attention to GCN2, an ATF4-upstream kinase whose role in the amino acid deprivation response is conserved in Drosophila (Malzer et al., 2010; Chakrabarti et al., 2012; Bjordal et al., 2014; Fig. 5 A). Starvation of adult flies triggers an increase in 4E-BP transcripts, as detected by quantitative RT-PCR from dissected intestines and fat bodies (Fig. 5 A). Knockdown of gcn2 or atf4 using the intestine-specific mex-Gal4 driver or the fat body–specific cg-Gal4 driver suppressed such 4E-BP transcript induction in the respective tissues (Fig. 5 A). In the developing third-instar larva, gcn2 knockdown abolished the inherent 4E-BP intron dsRed expression, whereas neither a control lacZ RNAi nor perk RNAi showed any effect (Fig. 5, B–E). In adults, 4E-BP intron dsRed levels were low in flies fed with food that contains 4% YE (Fig. 5 F), but such dsRed signals were induced when the flies were reared in 0.25% YE food (Fig. 5 G). We conclude that gcn2 mediates this effect, as gcn2 knockdown with the mex-Gal4 driver impaired such reporter induction (Fig. 5, I and J).
Figure 5.
GCN2 is required for 4E-BP induction. (A) Quantitative RT-PCR (q-PCR) of 4E-BP from dissected adult intestines (left) and fat bodies (right). Mex-Gal4 was used to knock down the indicated genes in the intestine, whereas cg-Gal4 was used to knock down those genes in the fat body. Error bars show SE. The P values are based on t tests. (B–D) 4E-BP intron dsRed containing larval fat bodies with the indicated genes knocked down through RNAi. (E) Quantification of 4E-BP intron dsRed intensity from images in B–D. dsRed pixel intensity was normalized to the signal from uas-GFP that was also expressed with cg-Gal4 driver. (F–I) 4E-BP intron dsRed signals from adult intestines. The samples (F and H) were from flies reared with 4% YE in food and (G and I) from those reared with 0.25% YE food for 2 d. Control lacZ knockdown flies (F and G) show enhanced dsRed signal in response to 0.25% YE diet, but such induction is suppressed under the condition of gcn2 RNAi (H and I). Quantification of the dsRed signal is shown in J. (K) A schematic diagram of the gcn2 locus and the design of the deletion and rescue lines. The red line indicates the locus deleted through FRT-mediated recombination. In this background, genomic DNA containing either the wild-type gcn2 locus (pink bar) or equivalent DNA with the gcn2 coding sequences deleted (blue bars) were reintroduced. (L) Anti–4E-BP Western blot (top gel) in the larvae of the indicated genotypes that were fed food with either standard cornmeal food (marked fed) or in 5% sucrose dissolved in PBS (marked starved) for 18 h. Bottom gel shows antiprofilin blots as loading controls. The normalized anti–4E-BP band intensities are quantified in the graph below the gels.
To further investigate the role of gcn2 in vivo, we generated loss-of-function alleles. Specifically, we deleted a 12-kb region that includes the gcn2 locus on the third chromosome (henceforth referred to as gcn2FRT 12kb), and in this background, a bacterial artificial chromosome clone containing either the wild-type gcn2 (referred to as gcn2wt rescue) or equivalent clones with deletions in the gcn2 coding sequence (gcn25′Δ rescue, gcn23′Δ rescue, and gcn2Δ-1 rescue) were reintroduced to a targeted locus on the second chromosome through the phiC31 integrase system (Fig. 5 K; see also Materials and methods). The gcn2 mutants generated through this strategy (genotype: gcn2mutant rescue; gcn2FRT 12kb −/−) were homozygous viable and fertile under standard conditions of fly husbandry. When reared in food with low yeast content (0.25% YE), the wild-type rescued adults had elevated levels of 4E-BP protein as detected through Western blot (Fig. 5 L, lane 2), but the gcn2 mutants (genotype gcn2Δ-1 rescue; gcn2FRT 12kb −/−) had comparatively reduced 4E-BP levels (Fig. 5 L). Although the gcn2 mutants were viable, they had smaller ovaries as compared with the wild-type control when reared in low-YE food (Fig. S5), consistent with a recent study based on gcn2 RNAi (Armstrong et al., 2014).
Loss of gcn2 impairs lifespan extension in response to dietary restriction
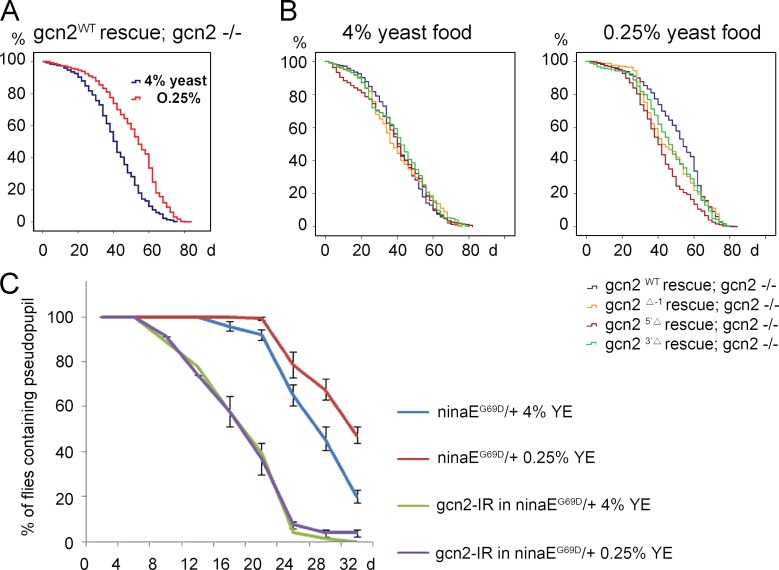
Wild-type flies show extended lifespan when reared with limited YE content in food, and under certain conditions, such effects are lost in the 4E-BP mutant flies (Tettweiler et al., 2005; Zid et al., 2009; Partridge et al., 2011). This prompted us to test if the newly generated gcn2 mutant flies have altered lifespan extension under similar conditions. Two different types of food were used: high protein content (4% YE) and low protein content (0.25% YE). As expected, gcn2 mutant flies that were rescued with wild-type gcn2 transgene (genotype: gcn2WT rescue; gcn2FRT 12kb −/−) significantly extended median lifespan by 12 d when reared in food with reduced YE content (survival curve shown in Fig. 6 A; P < 0.0001). The wild-type and mutant gcn2 rescued flies had indistinguishable lifespan when reared in food with high yeast content. In contrast, the extent of lifespan extension in response to food with lower yeast content was reduced or mostly abolished in the three independent gcn2 mutant flies that were examined, as compared with wild-type rescued flies (Fig. 6 B). Median lifespan changes and the statistical analyses of these results are shown in Table S1.
Figure 6.
GCN2 and ATF4 contribute to lifespan extension and delay in photoreceptor degeneration upon dietary restriction. (A and B) Kaplan–Meier plot survival curve of male adult flies reared in food containing 4% or 0.25% YE. The genotypes and food conditions are marked on each graph. (A) The gcn2WT rescue; gcn2FRT 12kb −/− flies showed consistent extension of lifespan after dietary restriction of YE (P < 0.0001 based on Cox proportional hazard analysis). (B) Lifespan comparison between the wild-type gcn2 control and the three gcn2 mutant lines. When reared in 4% yeast food, no significant difference was observed. Specifically, the P values between the gcn2WT and the gcn2 mutants alleles Δ-1, 5′Δ, and 3′Δ are P = 0.582, P = 0.375, and P = 0.027, respectively. However, in 0.25% yeast food, the gcn2 mutants have shorter lifespan than the wild-type control. The P values under these conditions between the wild-type and Δ-1, 5′Δ, and 3′Δ alleles are P = 0.012, P < 0.0001, and P = 0.001, respectively. (C) Quantification of age-related retinal degeneration in ninaEG69D/+ flies. Rh1-GFP signal from the pseudopupils of live flies were assessed. RNAi lines targeting either gcn2 or control white were expressed in the photoreceptors through the combined action of Rh1-Gal4 and GMR-Gal4 drivers. For each genotype, the graph shows the percentage of flies with intact pseudopupils (mean of at least five independent trials). Dietary restriction delays the course of retinal degeneration of ninaEG69D/+ flies (six independent trials, total number of flies analyzed = 258, P < 0.0001). In contrast, the beneficial effect on retinal degeneration by dietary restriction disappeared after gcn2 knockdown (five independent trials, total number of flies analyzed = 221, P = 0.16). Error bars show SE.
In addition to the lifespan analysis, we examined whether reduced protein in the diet can delay age-related diseases and whether any such effect requires gcn2. As a tool, we used a Drosophila model for autosomal-dominant retinitis pigmentosa, in which a mutant allele of the Rh-1 gene, ninaEG69D, causes age-related retinal degeneration (Colley et al., 1995; Kurada and O’Tousa, 1995). An Rh-1>GFP reporter–based fluorescent pseudopupil assay was used to follow retinal degeneration (see Materials and methods). ninaEG69D/+ flies had their pseudopupils disappear beginning at 16 d, in a progressive manner, with only 20% of these flies showing intact pseudopupils at 32 d after eclosion (Fig. 6 C, blue line). Those ninaEG69D/+ flies fed with reduced YE content in the food showed a delayed course of retinal degeneration, with only approximately half of the examined flies with pseudopupil loss at day 32 (Fig. 6 C, red line; 47.6 ± 7.37%, P < 0.0001). On the other hand, ninaEG69D/+ flies with gcn2 knocked down in their photoreceptors no longer showed a positive effect of reduced yeast content in the diet on the course of retinal degeneration (Fig. 6 C; 0% at 4% YE [green line] vs. 4.2 ± 3.17% at 0.25% YE [purple line] in gcn2-IR flies at 32 d, P = 0.16). We speculate that activation of GCN2 by nutrient deprivation provide additional resistance to the ER stress caused by mutant Rh-1 expression, as part of a preconditioning process or an additive effect of ATF4-mediated stress response.
Bicistronic assays suggest the presence of transcripts that evade suppression by 4E-BP
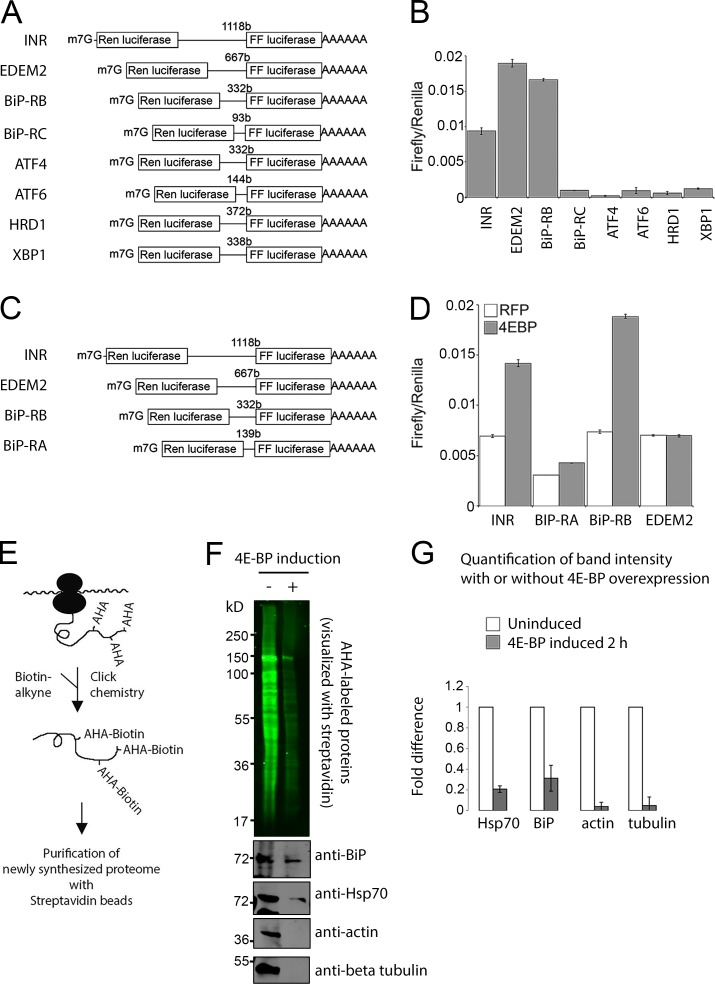
The presence of two translational inhibition steps in a pathway, triggered by eIF2α phosphorylation and 4E-BP induction, was intriguing as independent studies have shown that the overall attenuation of translation in the integrated stress response, as measured by 35S-methionine incorporation, can be fully attributed to eIF2α phosphorylation (Harding et al., 1999; Marciniak et al., 2004; Han et al., 2013). In contrast, a recent study found that a subset of transcripts remains inhibited in their translation even after eIF2α activity is restored, suggestive of a second node or translational inhibition with more selective effects (Preston and Hendershot, 2013). Thus, we explored the possibility that 4E-BP exerts a more selective effect on translation. We specifically considered the possibility that 4E-BP cannot inhibit transcripts with internal ribosome entry sites (IRESs; Marr et al., 2007; Zid et al., 2009; Olson et al., 2013). To test for the presence of IRESs in the stress-response transcripts, we first used the widely used “bicistronic assay,” where the first cistron (renilla luciferase) is translated through a cap-dependent mechanism and the second (firefly luciferase) requires IRES activity (Fig. 7, A and C). The 5′ UTRs of several ER stress–responsive genes were analyzed, and among these, those of EDEM2 and the RB splice isoform of BiP (annotated in FlyBase as hsp70-3 or hsc3) scored positive in this assay (Fig. 7, B and D). The firefly values were comparable to a positive control, InR, which had been previously shown to contain IRESs (Marr et al., 2007; Olson et al., 2013). In particular, we found that the RB isoform of BiP maintained firefly luciferase activity even when the constitutively active 4E-BP LLAA variant, which binds eIF-4E more efficiently and cannot be phosphorylated by TOR (Miron et al., 2003), was overexpressed to inhibit cap-dependent translation (Fig. 7 D).
Figure 7.
BiP 5′ UTR contains an IRES and evades suppression by 4E-BP. (A–D) Bicistronic assays show evidence of IRESs. (A and C) Schematic diagrams of the bicistronic assay reporters used in B and D, respectively. The first cistron (renilla luciferase) is expressed through cap-dependent translation, but the second cistron (firefly luciferase) requires an IRES element upstream to be translated. 5′ UTRs of the indicated genes were inserted in between the two cistrons. (B) The firefly/renilla luciferase ratio of the bicistronic assay constructs is shown in A. (D) The firefly/renilla luciferase ratio of the bicistronic assay constructs is shown in C. INR (insulin receptor) 5′ UTR was used as a positive control. White bars show the values in cells expressing a control RFP gene, whereas the gray bars show values from cells overexpressing the constitutively active 4E-BP. (E–G) BONCAT analysis shows 4E-BP–resistant new protein synthesis. (E) The schematic diagram shows AHA incorporation into newly synthesized peptides. The reactive azide group of AHA can be covalently conjugated to biotin-PEG4-alkyne using Click chemistry (azide-alkyne cycloaddition). (F) Visualization of the newly synthesized proteome with streptavidin-IRDye800 (top gel, green). S2 stable cell lines were used to induce 4E-BPLLAA for 2 h and were metabolically labeled with AHA for 1 h before being “clicked” to biotin. The AHA-biotin–labeled proteome was further purified with streptavidin beads and probed with anti-BiP, anti-Hsp70, antiactin, and anti–β-tubulin antibodies (bottom). (G) Quantification of band intensities from three independent experiments. Error bars represent SD of mean.
Direct visualization of new protein synthesis through BONCAT identifies stress responsive genes that resist suppression by 4E-BP
To directly visualize new protein synthesis and validate the aforementioned results, we metabolically labeled newly synthesized proteins through bio-orthogonal noncanonical amino acid tagging (BONCAT). This method utilizes a methionine analogue, azidohomoalanine (AHA), which has a chemically reactive azide group that can be covalently conjugated to biotin-alkyne through the azide-alkyne cycloaddition click chemistry reaction (Dieterich et al., 2006). This allows the AHA-containing peptides to be conjugated to biotin and visualized with a fluorophore-conjugated streptavidin or purified with streptavidin beads (Fig. 7 E). We overexpressed 4E-BPLLAA and found that overall AHA labeling was reduced significantly (Fig. 7 F, top). To assess the effect on specific proteins, we affinity purified all AHA-labeled peptides with streptavidin agarose beads and probed the bound fraction with specific antibodies. We found that the synthesis of actin and β-tubulin was almost completely suppressed by 4E-BPLLAA, indicating that these transcripts strictly rely on eIF-4E–dependent translation (Fig. 7 F). However, transcripts that are implicated to contain IRESs, such as BiP (Fig. 7 D) and Hsp70 (Hernández et al., 2004), continued to be translated in the presence of active 4E-BP (Fig. 7, F and G). 4E-BP expression reduced BiP synthesis moderately, but not completely, and we speculate that this is because other splice isoforms of BiP do not contain IRES.
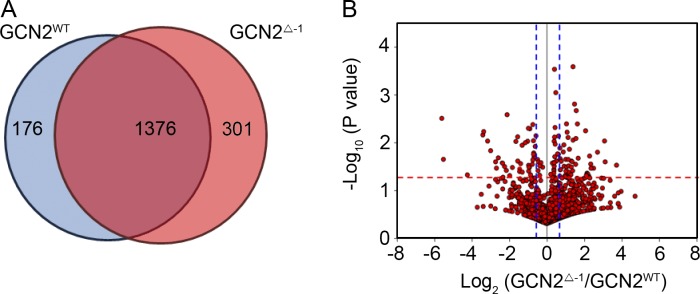
To further characterize the change in protein synthesis by GCN2 under dietary restriction, we analyzed the AHA-labeled proteome of wild-type and gcn2 mutants (gcn2Δ-1 rescue; gcn2FRT 12kb −/−) flies (see Materials and methods). When compared with wild-type flies (gcn2WT rescue; gcn2FRT 12kb −/−), there was a shift in the proteome in gcn2 mutant flies (gcn2Δ-1 rescue; gcn2FRT 12kb −/−; Fig. 8 and Table S2). Gene ontology (GO) term analysis indicated that 22% of the proteins that significantly decreased their synthesis in the gcn2 mutants were involved in oxidation reduction, 9% were involved in the generation of metabolic precursors, and 6% were involved in the electron transport chain (Table S3). These results are reminiscent of the translational profiling studies with 4E-BP mutants, which revealed that mitochondrial proteins were prominently affected in those mutants (Zid et al., 2009). Among the individual proteins that had reduced levels was BiP (heat shock 70-kD protein cognate 3, isoform E, gcn2 mutant/wild type ratio of 0.9, P = 0.038 in Table S2), which scored positive in the IRES assay and was resistant to 4E-BP overexpression (Fig. 7, D, F, and G). In contrast, levels of actin 5C and β-tubulin, which were sensitive to 4E-BP overexpression, did not change significantly (P = 0.262 and P = 0.350, respectively). These results indicate that GCN2 has a selective effect on gene expression and helps to extend lifespan upon dietary restriction.
Figure 8.
Proteomic analysis of the newly synthesized proteins in gcn2WT and gcn2 mutant flies. (A) Venn diagram for protein number identified from gcn2WT rescue; gcn2FRT 12kb −/− (blue) and gcn2Δ-1 rescue; gcn2FRT 12kb −/− flies (red). (B) Volcano plot of 1,376 proteins that were identified in both groups, with the log2 value of ratio (gcn2Δ-1/gcn2WT) on the x axis and log10 value of P values on the y axis. The red dot line indicates the P value of 0.05, and the blue dotted lines indicate the ratio of 1.5 (right line) and 0.67 (left), respectively. A full list of the peptides and GO term analyses can be found in Tables S2 and S3.
Discussion
Previous studies had established the importance of 4E-BP transcription by FOXO in several distinct biological contexts, including the regulation of cell number, metabolism, response to oxidative stress, and cardiac function (Puig et al., 2003; Teleman et al., 2005; Marr et al., 2007; Wessells et al., 2009). Alternative transcriptional regulatory mechanisms for 4E-BP and their biological significance have remained poorly characterized. Here, we show evidence that another pathway, mediated by GCN2 and ATF4, mediates the induction of 4E-BP transcription in response to the restriction of amino acids in the diet and during the development of specific tissues (summarized in Fig. 9). The specific data presented here include examination of 4E-BP protein through Western blot from starved larval extracts and examination of transcripts through quantitative PCR in cultured S2 cells, larvae, and adult tissues. Our new 4E-BP intron reporter, which responds to ATF4 activation, is widely expressed in Drosophila, indicating that ATF4 is a major mediator of 4E-BP induction during normal development as well as in response to dietary restriction of amino acids.
Figure 9.
Summary of the GCN2–ATF4–4E-BP pathway. Marked in red are the major findings reported in this work.
Our results also show that Drosophila gcn2 mutants have a shorter lifespan than wild-type controls when reared in food with low yeast content. These results are similar to what had been observed with mutants of C. elegans gcn2 (Rousakis et al., 2013) and yeast GCN4, an ATF4 equivalent gene in that organism (Steffen et al., 2008). The Drosophila gcn2 mutant phenotype is also similar to the reported phenotype of 4E-BP mutant flies (Tettweiler et al., 2005; Zid et al., 2009; Partridge et al., 2011). However, we have not examined through double-mutant analysis whether the two genes have a strictly linear genetic relationship in regulating lifespan. We point out that based on our current understanding, the two genes do not have a strictly linear relationship: GCN2–ATF4 has other transcriptional targets that also contribute to their phenotypes, and ATF4-independent regulatory inputs into 4E-BP exist, such as those mediated by FOXO and TOR. Thus, we speculate that the similar reported phenotypes of gcn2 and 4E-BP mutants on lifespan may be due to a broad effect of 4E-BP on other GCN2–ATF4 target gene expression, as 4E-BP’s target, eIF-4E, is thought to be involved in the expression of most eukaryotic genes.
Emerging evidence indicates that 4E-BP is not indiscriminate in the inhibition of general translation. For example, ribosome profiling studies in mammalian cultured cells have found that 4E-BP1’s effect on translation is highly selective, with some transcripts being highly sensitive to 4E-BP1 and others indifferent (Thoreen et al., 2012). Accordingly, it appears that 4E-BP activation would have cells shift their overall protein synthesis profile. The data in this study are consistent with that view. Specifically, we find that BiP and other stress-responsive transcripts score positive in the IRES assay and are resistant to suppression by 4E-BP. We note that mammalian BiP also reportedly has an IRES element in its 5′ UTR (Yang and Sarnow, 1997). Our finding that 4E-BP is a target of the UPR helps make sense of such an observation; IRES would help transcripts evade suppression by 4E-BP, whose expression level is high in stressed cells, allowing BiP to be expressed and help resolve stress. As 4E-BP activation results in a specific biological phenotype of enhanced stress resistance and lifespan extension (Tettweiler et al., 2005; Zid et al., 2009; Demontis and Perrimon, 2010), it appears that the proteome shift brought on by 4E-BP favors stress-responsive gene expression.
In Drosophila, 4E-BP is widely understood as a transcriptional target of FOXO. However, the role of FOXO in mediating the effects of dietary restriction of amino acids has been disputed (Giannakou et al., 2008; Min et al., 2008; Zid et al., 2009). Our own experiments presented in this paper show that the loss of foxo does not impair 4E-BP transcription, at least under conditions of amino acid restriction. Notably, we used a foxo mutant allele that is different from those used in the earlier studies on 4E-BP (Teleman et al., 2005; Tettweiler et al., 2005; Zid et al., 2009). Although the earlier studies had used foxo21/25 alleles with premature stop codons, recent studies indicate that full-length FOXO protein is still expressed in the foxo25 mutants (Nechipurenko and Broihier, 2012) and might still retain DNA-binding activity (Slack et al., 2011). Thus, it is possible that these alleles have neomorphic properties that may have led to results different from our current work. On the other hand, the foxo mutant allele that we used in this study has been validated to be a null allele (Slack et al., 2011; Nechipurenko and Broihier, 2012). We point out that our negative result with the foxo mutant is mostly related to the amino acid deprivation response and does not contradict FOXO’s known role in the induction of 4E-BP in other contexts.
In regards to the cellular response to amino acid deprivation, much focus had been placed on the TOR signaling pathway. It is interesting that the other amino acid–response pathway mediated by GCN2 leads to the transcriptional regulation of this TOR phosphorylation substrate. Our observation suggests that the two amino acid–responsive pathways work cooperatively.
Materials and methods
Fly strains
All Drosophila stocks were raised on standard cornmeal medium at 25°C. Genes were misexpressed in Drosophila eye imaginal discs using standard Gal4/UAS system (Brand and Perrimon, 1993). The following lines have been previously described: GMR-Gal4 (Hay et al., 1994), NP1-Gal4 (provided by E. Baehrecke, University of Massachusetts Medical School, Worcester, MA; Zaidman-Rémy et al., 2006), UAS-Rh-1G69D (Kang et al., 2012), UAS-dicer2 (Dietzl et al., 2007), thork13517 (4E-BP-lacZ) and thor2 (provided by D. Kimbrell, University of California, Davis, Davis, CA; Bernal and Kimbrell, 2000), foxoΔ94 (from L. Partridge, University College London, London, England, UK; Giannakou et al., 2008), and atf4crc1 and atf4R6 (provided by R. Hewes, University of Oklahoma, Norman, OK; Hewes et al., 2000). The gcn2FRT 12kb allele, which deleted the 12-kb region surrounding gcn2, was generated through the FLP-FRT–mediated chromosome deletion technique (Parks et al., 2004), using FRT elements within PBac{PB}CG1607c05674 and PBac{PB}CG11337c02097 lines. The deletion was confirmed through genomic PCR. One specific line (excision 47) was selected to derive all gcn2-specific mutant strains (see Molecular cloning) to ensure identical genetic background.
Molecular cloning
To make the rescue constructs, gcn2WT (as depicted in Fig. 3), a 20-kb genomic DNA covering the reference genome sequence from 31390122 to 31410205, was cloned from the CH322-20H13 bacterial artificial chromosome clone (Pacman Resources) into attB-P[acman]-ApR through the recombineering technology (Venken et al., 2006). To generate the gcn2 deletion mutants for the equivalent construct, we used a galK replacement method (Venken et al., 2006). gcn25′Δ rescue deletes 1,375 bp of sequences from exons 5 to 8 (which spans a sequence that encodes the kinase domain). gcn23′Δ deletes 1,040 bp of sequences encoding the tRNA binding domain from exons 8 to 9. gcn2Δ-1 rescue deletes from the ATG start codon of the first exon to the stop codon in the last exon. The following oligonucleotides were used to generate homology arms for cloning the GCN2 locus: GCN2WT_LA1_MluI-R: 5′-ACGCGTGCTATTGCCTTCTGGACCAT-3′; GCN2WT_RA1_MluI-F: 5′-ACGCGTGAGATGCTATTTATAAAC-3′; GCN2WT_RA1_Spe-R: 5′-ACTAGTCCAACATCAAATGCATAGA-3′; GCN2WT_LA2_SpeI-F: 5′-ACTAGTTTCTGCATTAAATCTCCGG-3′.
The following oligonucleotides were used to generate the GCN2 deletion constructs, as part of the galK replacement strategy: GCN2Δ-1 rescue_F: 5′-CACCCAGATGTAGCCAAGACTTATCCACTAACCGGAGATTTAATGCAGAACCTGTTGACAATTAATCATCGGCA-3′; GCN2Δ-1 rescue_R: 5′-ATCGGCTGGTCTACTAAGTAGTTCAGCCGATTCTATGCATTTGATGTTGGTCAGCACTGTCCTGCTCCTT-3′; GCN25′Δ rescue_F: 5′-GCACTGGATCCGCCATGCCGTACCAGATACCGACACTGGCCCTAAGCCAACCTGTTGACAATTAATCATCGGCA-3′; GCN25′Δ rescue_R: 5′-GCAACTAGGTTCTTGTAGGCCTTGCTCTGCGGATTAAGCGAGGGCGTGACGTCAGCACTGTCCTGCTCCTT-3′; GCN23′Δ rescue_F: 5′-TCGATGATATAGTATCCCTGAACCCAGTGATTGAGTTTGTTAAGGCTAAGCCTGTTGACAATTAATCATCGGCA-3′; GCN23′Δ rescue_R: 5′-GCCCGACAATCCTTAGCGTATTCCACACCAACAGCAGCTACCAGCTTGTCTCAGCACTGTCCTGCTCCTT-3′.
The derived constructs were inserted into a specific site at the chromosomal location 25C6 (available through Bestgene Inc., attP40, 25C6 site), using PhiC31 integrase-mediated site-specific transgenesis (Bischof et al., 2007). The resulting lines backcrossed to a laboratory w1118 strain for eight generations were then crossed into the background of gcn2FRT 12kb line (specifically excision 47) to generate gcn2-specific mutants. To generate the upstream 4E-BP/dsRed reporter (Fig. 1), a 1.6-kb intergenic sequence between thor (4E-BP) and pgant4 was amplified through genomic PCR and subcloned into the pRed H-stinger vector that has a dsRed reporter (Barolo et al., 2004). As for the 4E-BP intron reporter, the 0.42-kb of intron of thor (the sequence shown in Fig. 1 F) was similarly subcloned into the pRed H-stinger.
Immunohistochemistry and Western blots
All fluorescent images were obtained with LSM510 and LSM700 confocal microscopes (ZEISS), using ×20 objective lenses. The following antibodies were used: monoclonal anti–Rh-1 (1:500 for immunohistochemistry; Developmental Studies Hybridoma Bank, University of Iowa), mouse anti-profilin (1;1,000 for Western blot; Developmental Studies Hybridoma Bank, University of Iowa), anti-dsRed (1:500 for immunohistochemistry, 1:2,000 for Western blot; Takara Bio Inc.), rabbit anti-4E-BP(thor) antibody (Olson et al., 2013), guinea pig anti-BiP (Ryoo et al., 2007), anti-Hsp70 (Abcam), and rabbit anti-lacZ (1:2,000 for tissue-labeling; Molecular Probes) antibodies. Guinea pig anti-ATF4 was first used elsewhere (Kang et al., 2012), but the description of the antibody was omitted in that study. In brief, full-length His-tagged Drosophila ATF4 protein was expressed in Escherichia coli BL21 cells, purified, and injected into guinea pigs to generate a polyclonal antibody. After affinity purification against recombinant His-ATF4, the antibody was used at 1:100 dilution for Western blot using standard protocols.
Cell culture and cell transfection
Drosophila S2 cells were grown in Schneider’s medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (Invitrogen). To grow S2 cells stably transfected with pMT-4E-BPLLAA, we used 5% fetal bovine serum, because of trace levels of metals in the serum that led to leaky expression of the MtnA promoter. Cells were transfected using Effectene (QIAGEN). To knock down genes, double-stranded RNA (dsRNA) was generated following the protocols of flyrnai.org. The following oligonucleotide sequences were used to generate T7-promoter–containing amplicons: Ire1-R 5′-TAATACGACTCACTATAGGGCAAAAGCAGAGCGAGAATG-3′; Ire1-S 5′-TAATACGACTCACTATAGGGTTAATGTCGCGATGCACAA-3′. xbp1-R 5′-TAATACGACTCACTATAGGGCAGCAGCACAACACCAGA-3′; xbp1-S 5′-TAATACGACTCACTATAGGGTGTGGGTTTCCATTTATCTTCA-3′ (517 bp); PERK-R 5′-TAATACGACTCACTATAGGGTGGCACAAGGAGGGGAAC-3′; PERK-S 5′-TAATACGACTCACTATAGGGGCACCACTGGACCTAGTAAA-3′ (495 bp); ATF4-R 5′-TAATACGACTCACTATAGGGGCGGTGTAGAGGATCGAAAG-3′; ATF4-S 5′-TAATACGACTCACTATAGGGCACTGTCCGATTTGCAGAAA-3′ (553 bp); FOXO(DRSC15463)-R 5′-TAATACGACTCACTATAGGGATGATGGACGGCTACGC-3′; FOXO(DRSC15463)-S 5′-TAATACGACTCACTATAGGG ATTGCTGGCCTTTGTACTG-3′.
For RNAi in cultured cells, we followed a previously described protocol (Ryoo et al., 2007), with two rounds of transfection at day 1 and 4 to enhance knockdown efficiency. The RNA was extracted for RT-PCR at day 9. To induce hyperactive 4E-BP, we used cells that were stably transfected with pMT-4E-BPLLAA, which is inducible by the addition of Cu2+.
Metabolic labeling with AHA
To assess the effect of 4E-BP overexpression, 4E-BPLLAA was induced in S2 cells stably transfected with pMT-4E-BPLLAA with 0.5mM copper sulfate for 2 h in methionine-free Schneider’s medium to enable AHA uptake. The cells were then labeled with 4 mM AHA (Anaspec) for 1 h. AHA-labeled proteins in the extracts were clicked to biotin-PEG4 alkyne (Invitrogen) using the manufacturer’s protocols. AHA-conjugated peptides were further enriched by incubation with Streptavidin-agarose (Thermo Fisher Scientific) followed by rigorous washing with PBS-Tween (0.5%). The bound fraction was collected by boiling the beads in sample buffer and analyzed by Western blotting with Streptavidin-IRDye 800 (Rockland Immunochemicals), guinea pig anti-BiP (1:1,000), mouse anti-Hsp70 (Abcam), mouse antiactin (EMD Millipore), and mouse antitubulin (Abcam).
BONCAT and proteomic analysis
For BONCAT analysis in adult flies, 2- to 3-d-old wild-type (gcn2WT rescue; gcn2FRT 12kb −/−) or gcn2 mutant male flies were fed with low-protein-content food with 0.25% YE for 2 d. The next day, flies were fed with low-protein-content food supplemented with 4 mM AHA. After 24 h, whole flies were extracted with a urea buffer (8 M urea, 200 mM Tris, pH 8.4, 4% CHAPS, 1 M NaCl, and protease inhibitor), sonicated to reduce viscosity, and centrifuged at 10,000 g for 5 min. Next, we used a Bradford assay to quantify the protein content. Equal amount of proteins from mutant and wild-type extracts were used as input. AHA-labeled proteins were cross-linked covalently to alkyne agarose beads using reagents provided by Click Chemistry Capture kit (Click Chemistry Tools). Beads were washed with a SDS wash buffer (1% SDS, 100 mM Tris, 250 mM NaCl, and 5 mM EDTA, pH 8.0), and proteins on beads were reduced with DTT at 70°C and alkylated with iodoacetamide at RT. To remove nonspecifically bound proteins, the beads then were washed with 5 × 2 ml SDS wash buffer, 10 × 2 ml urea buffer (8M urea and 100 mM Tris, pH 8.0), and 10 × 2 ml of 20% acetonitrile. Cross-linked proteins were digested with trypsin on-resin at 37°C overnight in digestion buffer (100 mM Tris, pH 8.0, 2 mM CaCl2, and 10% acetonitrile), and the tryptic peptides were desalted using Sep-PAC C-18 cartridge (WAT054955; Waters) and dried under vacuum in a SpeedVac.
Peptide separation was performed using Dionex UltiMate 3000 RSLCnano system (Thermo Fisher Scientific). Tryptic peptides from bead column were reconstituted using 0.1% formic acid and separated on a 50-cm Easy-Spray column with a 75-µm inner diameter packed with 2 µm C18 resin (Thermo Scientific) over 120 min (300 nl/min) using a 0 to 45% acetonitrile gradient in 0.1% formic acid at 50°C. The liquid chromatography was coupled to a Q Exactive mass spectrometer with a nano-electrospray ionization source. Mass spectra were acquired in a data-dependent mode with an automatic switch between a full scan with five data-dependent tandem mass spectrometry (MS/MS) scans. The target value for the full scan MS spectra was 3,000,000 with a maximum injection time of 120 ms and a resolution of 70,000 at m/z 400. The ion target value for MS/MS was set to 1,000,000 with a maximum injection time of 120 ms and a resolution of 17,500 at m/z 400. Dynamic exclusion of repeated peptides was applied for 20 s. Resulting raw files were processed using MaxQuant (version 1.5.2.8) for identification with the database of Drosophila melanogaster (2014.12.18 version, 20049 entries; UniProt). Samples from biological duplication for each set were loaded as each experiment. The search parameters were set as default including cysteine carbamidomethylation as a fixed modification, N-terminal acetylation, methionine oxidation phosphoserine, phosphothreonine, and phosphotyrosine as variable modifications with two miscleavages. Peptide identification was based on a search with an initial mass deviation of the precursor ion of up to 10 ppm, and the allowed fragment mass deviation was set to 20 ppm. The peptide-spectrum match false discovery rate, protein false discovery rate, and the site decoy fraction were set to 0.01. The minimal scores for unmodified and modified peptides were 0 and 40, respectively. The minimal delta scores for unmodified and modified peptides were 0 and 17, respectively. The minimum of unique and razor peptides for identification was set to 1. For the label-free quantitation, “LFQ method” was chosen with minimum ratio count to be 1.5. LFQ intensity from each group were tested with a t test to find statistically differential expressed proteins, and proteins showing P < 0.05 were chosen for gene ontology analysis using DAVID bioinformatics tool (https://david.ncifcrf.gov).
Nutrient restriction on Drosophila larvae
Crosses were performed in cages with apple-juice plates (25% [vol/vol] apple juice, 1.25% [wt/vol] sucrose, and 2.5% [wt/vol] agar) supplemented with active yeast paste. Larvae were collected ∼47–49 h after egg laying and transferred to standard cornmeal food (5.9% wt/vol glucose, 6.6% cornmeal, 1.2% baker’s yeast, and 0.7% agar in water) or to nutrient restricted medium (5% sucrose and 1% agar in PBS) for 4 or 18 h at 25°C.
Lifespan analysis
Newly hatched flies were collected within 18 h at 18°C and transferred to YE food (8.6% cornmeal, 5% sucrose, 0.46% agar, 1% acid mix, and YE; 212750 Bacto Yeast Extract; BD; Zid et al., 2009) at a density of ∼100 flies in a cage. Flies were transferred to fresh food every 2 d and death scored on those days at 25°C.
Analysis of retinal degeneration
ninaEG69D/+ flies had Rh1-GFP in the background that fluorescently marked their photoreceptors. Those with regular array of photoreceptors showed deep pseudopupil pattern of GFP. The loss of such green fluorescent pseudopupil was used as an indication of retinal degeneration in live flies. In addition to the knock down of the indicated genes through the combined action of GMR-Gal4 and Rh1-Gal4, all flies expressed an RNAi line against the white gene to eliminate eye pigments, which may otherwise affect the course of retinal degeneration. These flies were selected and reared in indicated food vials (15–20 flies in each vial) in permanent light at 25°C. The vials were changed frequently to avoid mixing the flies with eventual progeny. The quantification of pseudopupils was performed on a pad under blue fluorescent light after anaesthetizing the flies with CO2.
Bicistronic assay
5′ UTRs of selected UPR genes were amplified by PCR and cloned into a previously described biscistronic reporter plasmid containing the renilla and firefly luciferase open reading frames (Marr et al., 2007). Drosophila S2 cells were maintained in Schneiders medium supplemented with 10% FBS. Cells were split 48 h before beginning the experiment. On the day of the experiment cells were counted and diluted to 1X10E6 per milliliter, 0.5 ml of cells was added to each well of a 24-well plate, and the cells were allowed to attach. A transfection cocktail was prepared containing 1 µg expression construct (either pACdsRED or pAC4EBP) and 0.2 µg of the respective bicistronic reporter plasmid in 150 µl EC buffer. 8 µl enhancer was added, followed by 5-min incubation at RT. Then, 18 µl Effectene was added, followed by 15-min incubation at RT. Finally, 1.2 dml complete media was added to the cocktail. Media was removed from the cells and replaced by 400 µl of the transfection cocktail. Renilla and firefly luciferase was measured 48 h later using a Promega dual luciferase assay. Each condition was assayed in triplicate. The following oligonucleotides were used to subclone the 5′ UTRs : MB910: 5′-CTCCATGGCTCATTACCCCAACTCTTCTAG-3′, Xbp1 5′ UTR for (NncoI); MB911: 5′-CTCCATGGGTGCCATGTTTAACTGGTTC-3′, Xbp1 5′ UTR REV (NcoI); MB912: 5′-CTCCATGGAAGATAATTCACCATCTGGTAGC-3′, HRD1 5′UTR for (NcoI); MB913: 5′-GCGCCATGGAAACGGACGATAAGAG-3′, HRD1 5′ UTR REV (NcoI); MB914: 5′-CTCCATGGATTCCAACACTCATTACCGTTCC-3′, ATF4 5′ UTR forward (NcoI); MB915: 5′-GGCAACTGCAAGCTCTCCATGGTT-3′, ATF4 5′ UTR REV (NcoI); MB916: 5′-CTCCATGGAAATTACTATTATAGTATATATTG-3′, ATF6 5′ UTR for (NcoI); MB917: 5′-GATGTCCATGGCTAATCGGCCATAG-3′, ATF6 5′UTR rev (NcoI); MB918: 5′-CTCCATGGCAGCTGGTCACACTGAGGGTGC-3′, EDEM2 5′ UTR for (NcoI); MB919: 5′-CTCCATGGTGCTGGCTATCTGGAGACTAC-3′, EDEM2 5′ UTR rev (NcoI); MB920: 5′-TACCATGGACTTCATATTGAAGATCTC-3′, Hsc3 reverse (NcoI); MB921: 5′-CTCCATGGAGTTGCCGGGCAGTTAGCCATTGG-3′, Hsc-RC for (NcoI); MB922: 5′-CTCCATGGATCGACACGTCAGTGGTAGGTGAGAG-3′, Hsc-RB for (NcoI); MB923: 5′-CTCCATGGAAAAACGTCAGATAAGCAGCAGCCAC-3′, Hsc-RD for (NcoI); MB924: 5′-CTCCATGGAAAAACGTCAGATAAGCAGCAGCC-3′, Hsc-RA for (NcoI).
RT-PCR and quantitative real-time RT-PCR
Total RNA was isolated using TRIzol Reagent (Invitrogen), and 200 ng total RNA was used for reverse transcription with the SuperScript First-Strand Synthesis System (Invitrogen). The following primer sequences were used: Thor-F 5′-GCTAAGATGTCCGCTTCACC-3′; Thor-R 5′-CCTCCAGGAGTGGTGGAGTA-3′; CG16974-F 5′-CTTAACTGCGAGCATGTGGA-3′; CG16974-R 5′-CGTGGAAGCTCAGTCAACAA-3′; Thor175F 5′-CAGCTAAGATGTCCGCTTCA-3′; Thor260R 5′-ATCCGAGATGACAACCTTCC-3′; Rp49-F 5′-AGATCGTGAAGAAGCGCACCAAG-3′; Rp49-R 5′-CACCAGGAACTTCTTGAATCCGG-3′; PERK857F 5′-GCCAGCTGCTGTATGAATGT-3′; PERK936R 5′-CCTAATGGTGTCGTCGATTG-3′; gcn2_1644F 5′-CCCTGGTGGAGAGTTTGATGC-3′; gcn2_1708R 5′-GTTACACTTGTCTACAAAGTCG-3′; ATF4_586F 5′-TCGCAAAAGTTGGTTAAACG-3′; ATF4673R 5′-TCCGTAGGATTCAACTGCTG-3′; Foxo340F 5′-ACCGGCAAAATCAACAATTT-3′; Foxo416R 5′-TGTTTGCAATGGGAAATAGC-3′.
Online supplemental material
Figure S1 shows the knockdown efficiency of the RNAi lines in the developing fat body. Figure S2 shows that overexpression of atf4, but not foxo, induces 4E-BP intron reporter. Figure S3 shows a comparison of the expression patterns between 4E-BP-lacZ, 4E-BP upstream dsRed, and 4E-BP intron dsRed. Figure S4 shows 4E-BP upstream dsRed reporter in the foxo mutant background. Figure S5 shows Drosophila ovaries in gcn2−/− flies. Table S1 is a summary of changes in lifespan and their statistical significance in adult flies reared with or without dietary restriction. Table S2 shows the relative fold change of newly synthesized peptides in control versus gcn2 mutant flies. Table S3 shows GO enrichment by the DAVID functional annotation tool.
Supplementary Material
Acknowledgments
We thank the Bloomington Stock Center for reagents and Joong-Jean Park for technical advice.
This work was supported by grants from the National Institutes of Health (R01EY020866 to H.D. Ryoo and NS050276 and RR027990 to T.A. Neubert); the March of Dimes Foundation (to H.D. Ryoo); the Korean Health Technology research and development project, the Ministry of Health and Welfare, South Korea (H13C1821); and the National Research Foundation of Korea (NRF-2015R1C1A2A01051560; to M.-J. Kang).
The authors declare no competing financial interests.
Author contributions: M.-J. Kang and H.D. Ryoo conceived the project. M.-J. Kang, D. Vasudevan, K. Kang, K. Kim, J.-E. Park, N. Zhang, M.T. Marr, and H.D. Ryoo performed experiments and analyzed data. X. Zeng and T.A. Neubert provided guidance for recombineering and BONCAT/proteomics. M.-J. Kang and H.D. Ryoo wrote the paper, incorporating suggestions from all authors.
Footnotes
Abbreviations used:
- AHA
- azidohomoalanine
- BONCAT
- bio-orthogonal noncanonical amino acid tagging
- dsRNA
- double-stranded RNA
- GO
- gene ontology
- IRES
- internal ribosome entry site
- ISC
- intestinal stem cell
- MS
- mass spectrometry
- Rh
- rhodopsin
- SE
- standard error
- TU
- tunicamycin
- UPR
- unfolded protein response
- YE
- yeast extract
References
- Armstrong A.R., Laws K.M., and Drummond-Barbosa D.. 2014. Adipocyte amino acid sensing controls adult germline stem cell number via the amino acid response pathway and independently of Target of Rapamycin signaling in Drosophila. Development. 141:4479–4488. 10.1242/dev.116467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird T.D., Palam L.R., Fusakio M.E., Willy J.A., Davis C.M., McClintick J.N., Anthony T.G., and Wek R.C.. 2014. Selective mRNA translation during eIF2 phosphorylation induces expression of IBTKα. Mol. Biol. Cell. 25:1686–1697. 10.1091/mbc.E14-02-0704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barolo S., Castro B., and Posakony J.W.. 2004. New Drosophila transgenic reporters: insulated P-element vectors expressing fast-maturing RFP. Biotechniques. 36:436–440: 442. [DOI] [PubMed] [Google Scholar]
- Bernal A., and Kimbrell D.A.. 2000. Drosophila Thor participates in host immune defense and connects a translational regulator with innate immunity. Proc. Natl. Acad. Sci. USA. 97:6019–6024. 10.1073/pnas.100391597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J., Maeda R.K., Hediger M., Karch F., and Basler K.. 2007. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA. 104:3312–3317. 10.1073/pnas.0611511104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjordal M., Arquier N., Kniazeff J., Pin J.P., and Léopold P.. 2014. Sensing of amino acids in a dopaminergic circuitry promotes rejection of an incomplete diet in Drosophila. Cell. 156:510–521. 10.1016/j.cell.2013.12.024 [DOI] [PubMed] [Google Scholar]
- Brand A.H., and Perrimon N.. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 118:401–415. [DOI] [PubMed] [Google Scholar]
- Chakrabarti S., Liehl P., Buchon N., and Lemaitre B.. 2012. Infection-induced host translational blockage inhibits immune responses and epithelial renewal in the Drosophila gut. Cell Host Microbe. 12:60–70. 10.1016/j.chom.2012.06.001 [DOI] [PubMed] [Google Scholar]
- Colley N.J., Cassill J.A., Baker E.K., and Zuker C.S.. 1995. Defective intracellular transport is the molecular basis of rhodopsin-dependent dominant retinal degeneration. Proc. Natl. Acad. Sci. USA. 92:3070–3074. 10.1073/pnas.92.7.3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis F., and Perrimon N.. 2010. FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell. 143:813–825. 10.1016/j.cell.2010.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever T.E., Feng L., Wek R.C., Cigan A.M., Donahue T.F., and Hinnebusch A.G.. 1992. Phosphorylation of initiation factor 2 alpha by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell. 68:585–596. 10.1016/0092-8674(92)90193-G [DOI] [PubMed] [Google Scholar]
- Dieterich D.C., Link A.J., Graumann J., Tirrell D.A., and Schuman E.M.. 2006. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT). Proc. Natl. Acad. Sci. USA. 103:9482–9487. 10.1073/pnas.0601637103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K.C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S., et al. . 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 448:151–156. 10.1038/nature05954 [DOI] [PubMed] [Google Scholar]
- Fristrom J.W. 1965. Development of the morphological mutant cryptocephal of Drosophila melanogaster. Genetics. 52:297–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakou M.E., Goss M., and Partridge L.. 2008. Role of dFOXO in lifespan extension by dietary restriction in Drosophila melanogaster: not required, but its activity modulates the response. Aging Cell. 7:187–198. 10.1111/j.1474-9726.2007.00362.x [DOI] [PubMed] [Google Scholar]
- Hadorn E., and Gloor H.. 1943. Cryptocephal, ein spat wirkender Leftalfaktor bei Drosophila melanogaster. Rev. Suisse Zool. 50:256–261. [Google Scholar]
- Han J., Back S.H., Hur J., Lin Y.H., Gildersleeve R., Shan J., Yuan C.L., Krokowski D., Wang S., Hatzoglou M., et al. . 2013. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat. Cell Biol. 15:481–490. 10.1038/ncb2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding H.P., Zhang Y., and Ron D.. 1999. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 397:271–274. 10.1038/16729 [DOI] [PubMed] [Google Scholar]
- Harding H.P., Novoa I., Zhang Y., Zeng H., Wek R., Schapira M., and Ron D.. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell. 6:1099–1108. 10.1016/S1097-2765(00)00108-8 [DOI] [PubMed] [Google Scholar]
- Harding H.P., Zhang Y., Zeng H., Novoa I., Lu P.D., Calfon M., Sadri N., Yun C., Popko B., Paules R., et al. . 2003. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell. 11:619–633. 10.1016/S1097-2765(03)00105-9 [DOI] [PubMed] [Google Scholar]
- Hay B.A., Wolff T., and Rubin G.M.. 1994. Expression of baculovirus P35 prevents cell death in Drosophila. Development. 120:2121–2129. [DOI] [PubMed] [Google Scholar]
- Hernández G., Vázquez-Pianzola P., Sierra J.M., and Rivera-Pomar R.. 2004. Internal ribosome entry site drives cap-independent translation of reaper and heat shock protein 70 mRNAs in Drosophila embryos. RNA. 10:1783–1797. 10.1261/rna.7154104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewes R.S., Schaefer A.M., and Taghert P.H.. 2000. The cryptocephal gene (ATF4) encodes multiple basic-leucine zipper proteins controlling molting and metamorphosis in Drosophila. Genetics. 155:1711–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A.G. 2014. The scanning mechanism of eukaryotic translation initiation. Annu. Rev. Biochem. 83:779–812. 10.1146/annurev-biochem-060713-035802 [DOI] [PubMed] [Google Scholar]
- Hu C., Pang S., Kong X., Velleca M., and Lawrence J.C. Jr. 1994. Molecular cloning and tissue distribution of PHAS-I, an intracellular target for insulin and growth factors. Proc. Natl. Acad. Sci. USA. 91:3730–3734. 10.1073/pnas.91.9.3730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J.C., Jaruga E., Repnevskaya M.V., and Jazwinski S.M.. 2000. An intervention resembling caloric restriction prolongs life span and retards aging in yeast. FASEB J. 14:2135–2137. [DOI] [PubMed] [Google Scholar]
- Jünger M.A., Rintelen F., Stocker H., Wasserman J.D., Végh M., Radimerski T., Greenberg M.E., and Hafen E.. 2003. The Drosophila forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J. Biol. 2:20 10.1186/1475-4924-2-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M., Powers R.W. III, Steffen K.K., Westman E.A., Hu D., Dang N., Kerr E.O., Kirkland K.T., Fields S., and Kennedy B.K.. 2005. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 310:1193–1196. 10.1126/science.1115535 [DOI] [PubMed] [Google Scholar]
- Kang K., Ryoo H.D., Park J.E., Yoon J.H., and Kang M.J.. 2015. A Drosophila reporter for the translational activation of ATF4 marks stressed cells during development. PLoS One. 10:e0126795 10.1371/journal.pone.0126795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M.J., Chung J., and Ryoo H.D.. 2012. CDK5 and MEKK1 mediate pro-apoptotic signalling following endoplasmic reticulum stress in an autosomal dominant retinitis pigmentosa model. Nat. Cell Biol. 14:409–415. 10.1038/ncb2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapahi P., Zid B.M., Harper T., Koslover D., Sapin V., and Benzer S.. 2004. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr. Biol. 14:885–890. 10.1016/j.cub.2004.03.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klass M.R. 1977. Aging in the nematode Caenorhabditis elegans: major biological and environmental factors influencing life span. Mech. Ageing Dev. 6:413–429. 10.1016/0047-6374(77)90043-4 [DOI] [PubMed] [Google Scholar]
- Kurada P., and O’Tousa J.E.. 1995. Retinal degeneration caused by dominant rhodopsin mutations in Drosophila. Neuron. 14:571–579. 10.1016/0896-6273(95)90313-5 [DOI] [PubMed] [Google Scholar]
- Malzer E., Daly M.L., Moloney A., Sendall T.J., Thomas S.E., Ryder E., Ryoo H.D., Crowther D.C., Lomas D.A., and Marciniak S.J.. 2010. Impaired tissue growth is mediated by checkpoint kinase 1 (CHK1) in the integrated stress response. J. Cell Sci. 123:2892–2900. 10.1242/jcs.070078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malzer E., Szajewska-Skuta M., Dalton L.E., Thomas S.E., Hu N., Skaer H., Lomas D.A., Crowther D.C., and Marciniak S.J.. 2013. Coordinate regulation of eIF2α phosphorylation by PPP1R15 and GCN2 is required during Drosophila development. J. Cell Sci. 126:1406–1415. 10.1242/jcs.117614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak S.J., Yun C.Y., Oyadomari S., Novoa I., Zhang Y., Jungreis R., Nagata K., Harding H.P., and Ron D.. 2004. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 18:3066–3077. 10.1101/gad.1250704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marr M.T. II, D’Alessio J.A., Puig O., and Tjian R.. 2007. IRES-mediated functional coupling of transcription and translation amplifies insulin receptor feedback. Genes Dev. 21:175–183. 10.1101/gad.1506407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCay C.M., Crowell M.F., and Maynard L.A.. 1935. The effect of retarded growth upon the length of lifespan and upon the ultimate body size. J. Nutr. 10:63–79. [PubMed] [Google Scholar]
- Min K.J., Yamamoto R., Buch S., Pankratz M., and Tatar M.. 2008. Drosophila lifespan control by dietary restriction independent of insulin-like signaling. Aging Cell. 7:199–206. 10.1111/j.1474-9726.2008.00373.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miron M., Lasko P., and Sonenberg N.. 2003. Signaling from Akt to FRAP/TOR targets both 4E-BP and S6K in Drosophila melanogaster. Mol. Cell. Biol. 23:9117–9126. 10.1128/MCB.23.24.9117-9126.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechipurenko I.V., and Broihier H.T.. 2012. FoxO limits microtubule stability and is itself negatively regulated by microtubule disruption. J. Cell Biol. 196:345–362. 10.1083/jcb.201105154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson C.M., Donovan M.R., Spellberg M.J., and Marr M.T. II. 2013. The insulin receptor cellular IRES confers resistance to eIF4A inhibition. eLife. 2:e00542 10.7554/eLife.00542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palam L.R., Baird T.D., and Wek R.C.. 2011. Phosphorylation of eIF2 facilitates ribosomal bypass of an inhibitory upstream ORF to enhance CHOP translation. J. Biol. Chem. 286:10939–10949. 10.1074/jbc.M110.216093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks A.L., Cook K.R., Belvin M., Dompe N.A., Fawcett R., Huppert K., Tan L.R., Winter C.G., Bogart K.P., Deal J.E., et al. . 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36:288–292. 10.1038/ng1312 [DOI] [PubMed] [Google Scholar]
- Partridge L., Green A., and Fowler K.. 1987. Effects of egg-production and of exposure to males on female survival in Drosophila melanogaster. J. Insect Physiol. 33:745–749. 10.1016/0022-1910(87)90060-6 [DOI] [Google Scholar]
- Partridge L., Alic N., Bjedov I., and Piper M.D.. 2011. Ageing in Drosophila: the role of the insulin/Igf and TOR signalling network. Exp. Gerontol. 46:376–381. 10.1016/j.exger.2010.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pause A., Belsham G.J., Gingras A.C., Donzé O., Lin T.A., Lawrence J.C. Jr., and Sonenberg N.. 1994. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 371:762–767. 10.1038/371762a0 [DOI] [PubMed] [Google Scholar]
- Preston A.M., and Hendershot L.M.. 2013. Examination of a second node of translational control in the unfolded protein response. J. Cell Sci. 126:4253–4261. 10.1242/jcs.130336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O., Marr M.T., Ruhf M.L., and Tjian R.. 2003. Control of cell number by Drosophila FOXO: downstream and feedback regulation of the insulin receptor pathway. Genes Dev. 17:2006–2020. 10.1101/gad.1098703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A., Zhou Z., Tang M.L., Meller S., Chen J., Bellen H., and Kimbrell D.A.. 1996. Identification of immune system and response genes, and novel mutations causing melanotic tumor formation in Drosophila melanogaster. Genetics. 143:929–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousakis A., Vlassis A., Vlanti A., Patera S., Thireos G., and Syntichaki P.. 2013. The general control nonderepressible-2 kinase mediates stress response and longevity induced by target of rapamycin inactivation in Caenorhabditis elegans. Aging Cell. 12:742–751. 10.1111/acel.12101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo H.D., Domingos P.M., Kang M.J., and Steller H.. 2007. Unfolded protein response in a Drosophila model for retinal degeneration. EMBO J. 26:242–252. 10.1038/sj.emboj.7601477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack C., Giannakou M.E., Foley A., Goss M., and Partridge L.. 2011. dFOXO-independent effects of reduced insulin-like signaling in Drosophila. Aging Cell. 10:735–748. 10.1111/j.1474-9726.2011.00707.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N., and Hinnebusch A.G.. 2009. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 136:731–745. 10.1016/j.cell.2009.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen K.K., MacKay V.L., Kerr E.O., Tsuchiya M., Hu D., Fox L.A., Dang N., Johnston E.D., Oakes J.A., Tchao B.N., et al. . 2008. Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell. 133:292–302. 10.1016/j.cell.2008.02.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teleman A.A., Chen Y.W., and Cohen S.M.. 2005. 4E-BP functions as a metabolic brake used under stress conditions but not during normal growth. Genes Dev. 19:1844–1848. 10.1101/gad.341505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettweiler G., Miron M., Jenkins M., Sonenberg N., and Lasko P.F.. 2005. Starvation and oxidative stress resistance in Drosophila are mediated through the eIF4E-binding protein, d4E-BP. Genes Dev. 19:1840–1843. 10.1101/gad.1311805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen C.C., Chantranupong L., Keys H.R., Wang T., Gray N.S., and Sabatini D.M.. 2012. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 485:109–113. 10.1038/nature11083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken K.J., He Y., Hoskins R.A., and Bellen H.J.. 2006. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 314:1747–1751. 10.1126/science.1134426 [DOI] [PubMed] [Google Scholar]
- Wang L., Ryoo H.D., Qi Y., and Jasper H.. 2015. PERK limits Drosophila lifespan by promoting intestinal stem cell proliferation in response to systemic and local ER stress. PLoS Genet. 11:e1005220 10.1371/journal.pgen.1005220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.C., Bohmann D., and Jasper H.. 2005. JNK extends life span and limits growth by antagonizing cellular and organism-wide responses to insulin signaling. Cell. 121:115–125. 10.1016/j.cell.2005.02.030 [DOI] [PubMed] [Google Scholar]
- Wek R.C., Jackson B.M., and Hinnebusch A.G.. 1989. Juxtaposition of domains homologous to protein kinases and histidyl-tRNA synthetases in GCN2 protein suggests a mechanism for coupling GCN4 expression to amino acid availability. Proc. Natl. Acad. Sci. USA. 86:4579–4583. 10.1073/pnas.86.12.4579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessells R., Fitzgerald E., Piazza N., Ocorr K., Morley S., Davies C., Lim H.Y., Elmen L., Hayes M., Oldham S., and Bodmer R.. 2009. d4eBP acts downstream of both dTOR and dFoxo to modulate cardiac functional aging in Drosophila. Aging Cell. 8:542–552. 10.1111/j.1474-9726.2009.00504.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S., Ishihara H., Yamada T., Tamura A., Usui M., Tominaga R., Munakata Y., Satake C., Katagiri H., Tashiro F., et al. . 2008. ATF4-mediated induction of 4E-BP1 contributes to pancreatic beta cell survival under endoplasmic reticulum stress. Cell Metab. 7:269–276. 10.1016/j.cmet.2008.01.008 [DOI] [PubMed] [Google Scholar]
- Yang Q., and Sarnow P.. 1997. Location of the internal ribosome entry site in the 5′ non-coding region of the immunoglobulin heavy-chain binding protein (BiP) mRNA: evidence for specific RNA-protein interactions. Nucleic Acids Res. 25:2800–2807. 10.1093/nar/25.14.2800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidman-Rémy A., Hervé M., Poidevin M., Pili-Floury S., Kim M.S., Blanot D., Oh B.H., Ueda R., Mengin-Lecreulx D., and Lemaitre B.. 2006. The Drosophila amidase PGRP-LB modulates the immune response to bacterial infection. Immunity. 24:463–473. 10.1016/j.immuni.2006.02.012 [DOI] [PubMed] [Google Scholar]
- Zhang W., Hietakangas V., Wee S., Lim S.C., Gunaratne J., and Cohen S.M.. 2013. ER stress potentiates insulin resistance through PERK-mediated FOXO phosphorylation. Genes Dev. 27:441–449. 10.1101/gad.201731.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zid B.M., Rogers A.N., Katewa S.D., Vargas M.A., Kolipinski M.C., Lu T.A., Benzer S., and Kapahi P.. 2009. 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell. 139:149–160. 10.1016/j.cell.2009.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinke I., Schütz C.S., Katzenberger J.D., Bauer M., and Pankratz M.J.. 2002. Nutrient control of gene expression in Drosophila: microarray analysis of starvation and sugar-dependent response. EMBO J. 21:6162–6173. 10.1093/emboj/cdf600 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.