Chung et al. show that the myomitokine GDF15 can act to modulate oxidative and lipolytic function in a non–cell-autonomous manner, thereby regulating systemic energy homeostasis in skeletal muscle-specific Crif1-deficient mice. This pathway may be a potential therapeutic target for preventing the onset of obesity and insulin resistance.
Abstract
Reduced mitochondrial electron transport chain activity promotes longevity and improves energy homeostasis via cell-autonomous and –non-autonomous factors in multiple model systems. This mitohormetic effect is thought to involve the mitochondrial unfolded protein response (UPRmt), an adaptive stress-response pathway activated by mitochondrial proteotoxic stress. Using mice with skeletal muscle–specific deficiency of Crif1 (muscle-specific knockout [MKO]), an integral protein of the large mitoribosomal subunit (39S), we identified growth differentiation factor 15 (GDF15) as a UPRmt-associated cell–non-autonomous myomitokine that regulates systemic energy homeostasis. MKO mice were protected against obesity and sensitized to insulin, an effect associated with elevated GDF15 secretion after UPRmt activation. In ob/ob mice, administration of recombinant GDF15 decreased body weight and improved insulin sensitivity, which was attributed to elevated oxidative metabolism and lipid mobilization in the liver, muscle, and adipose tissue. Thus, GDF15 is a potent mitohormetic signal that safeguards against the onset of obesity and insulin resistance.
Introduction
Mitochondrial function is critical for maintenance of local and systemic metabolic homeostasis. Preservation of normal mitochondrial function requires proteostasis, which is dynamically controlled by mitochondria-specific chaperones and proteases (Jensen and Jasper, 2014; Quirós et al., 2015). Mitochondrial proteotoxic stress, which is characterized by the accumulation of misfolded and unfolded proteins within mitochondrial compartments, activates a retrograde transcriptional response called the mitochondrial unfolded protein response (UPRmt). This adaptive response, which is aimed at reestablishing mitochondrial proteostasis, is conserved from worms to mammals. In multiple model organisms, genetic and pharmacological induction of the UPRmt exerts beneficial effects on lifespan, consistent with overall improvements in systemic metabolic homeostasis (Feng et al., 2001; Dillin et al., 2002; Durieux et al., 2011; Houtkooper et al., 2013; Owusu-Ansah et al., 2013). A recent study in Caenorhabditis elegans demonstrated that induction of the UPRmt promotes glycolytic metabolism, allowing subsequent recovery of mitochondrial function and oxidative phosphorylation; these results establish a precedent for the idea that UPRmt induction has metabolic consequences (Nargund et al., 2015). However, it remains to be determined whether induction of the UPRmt by genetic or pharmaceutical means exerts beneficial effects on systemic metabolic homeostasis in mammals. Moreover, in contrast to the situation in worm and fly, in mammals, the non–cell-autonomous signaling factors that communicate the state of the UPRmt between tissues, and thus influence organismal homeostasis, remain unknown.
Mitochondrial function in skeletal muscle is a critical determinant of systemic fuel homeostasis (Owusu-Ansah et al., 2013; Demontis et al., 2014). In humans, the mitochondrial content and oxidative function of skeletal muscle vary widely, and dysfunction of skeletal mitochondria is associated with abnormal nutrient handling typical of insulin resistance (Petersen et al., 2003; Morino et al., 2006; Jelenik and Roden, 2013). It remains unclear, however, whether muscle mitochondrial dysfunction results in insulin resistance or vice versa (Dumas et al., 2009; Samocha-Bonet et al., 2010). Investigations performed in animal models have ruled out a direct connection between deficiency of mitochondrial OxPhos in skeletal muscle and systemic insulin resistance. Indeed, studies of rodents with reduced mitochondrial function and mitochondrial stress in skeletal muscle revealed that these models are more insulin sensitive and thus protected against diet-induced obesity and diabetes, suggesting that dysfunctional mitochondria in a given tissue communicate with other tissues (Pospisilik et al., 2007; Han et al., 2011; Kim et al., 2013). Because the UPRmt is highly organ specific and depends on the duration or severity of OxPhos deficiency, the identification of the cell–non-autonomous mediators responsible for the interorgan communication of mitochondrial stress is a major challenge (Moisoi et al., 2009; Dogan et al., 2014; Wall et al., 2015). Furthermore, multiple cell–non-autonomous factors may interact to mediate systemic adaptations to organ-specific OxPhos dysfunction. FGF21 has been proposed to act as a mitokine that is released upon mitochondrial dysfunction caused by deficiencies in the respiratory chain or autophagy (Tyynismaa et al., 2010; Suomalainen et al., 2011; Nunnari and Suomalainen, 2012; Kim et al., 2013; Wall et al., 2015). However, a recent study suggested that FGF21 is dispensable for whole-body metabolic adaptations upon muscle mitochondrial stress (Ost et al., 2015). It remains unclear whether other mitokine factors are released upon mitochondrial dysfunction.
We wished to determine how tissue-specific activation of the UPRmt, in the context of altered mitochondrial proteostasis with reduced OxPhos activity, controls systemic energy homeostasis. To this end, we developed mice harboring skeletal muscle-specific loss of function (LOF) in the Crif1 gene, which encodes a newly defined protein of the large mitoribosomal subunit. Using these muscle-specific knockout (MKO) mice, we found that skeletal muscle-specific deficiency of Crif1 is sufficient to activate the UPRmt and stimulate production of mitokines that regulate systemic energy homeostasis. We identified growth differentiation factor 15 (GDF15; Bootcov et al., 1997; Yokoyama-Kobayashi et al., 1997) as a critical cell–non-autonomous factor that promotes oxidative function and lipolysis in liver and adipose tissues. We hypothesized that elevated production of GDF15 was responsible for the marked decrease of fat mass, resistance to obesity, and improved insulin sensitivity in MKO mice. Consistent with this hypothesis, administration of GDF15 to ob/ob mice promoted weight loss and increased insulin sensitivity. These findings provide robust evidence that the cell–non-autonomous factor GDF15 is a regulator of systemic energy homeostasis and is thus a potentially promising therapeutic agent for the treatment of obesity-associated insulin resistance.
Results
Skeletal MKO mice have reduced mitochondrial OxPhos function in skeletal muscle
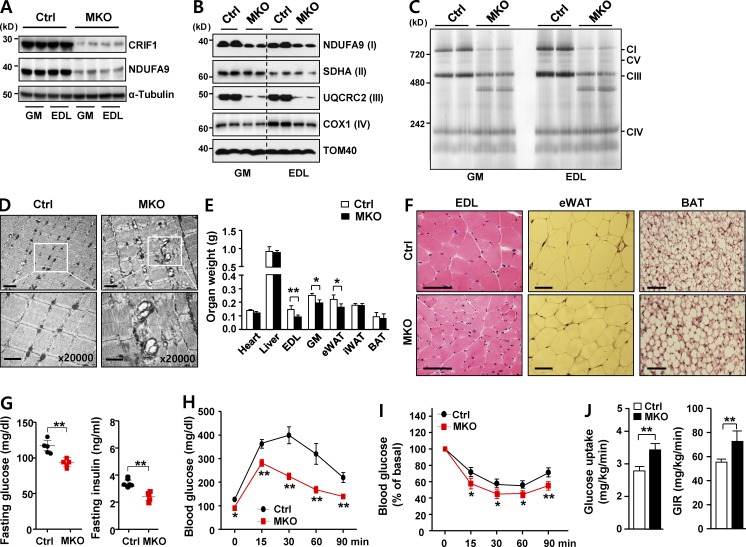
Mitochondrial ribosomes (mitoribosomes) are responsible for translation of OxPhos components encoded in the mitochondrial genome (O’Brien, 1971; Greber et al., 2015). CRIF1, a recently defined mitochondrial protein containing a long α-helix, is an integral component of the large subunit of the mammalian mitoribosome (Kim et al., 2012; Greber et al., 2014b). Homozygous Crif1-null mouse embryonic fibroblasts (MEFs) are profoundly deficient in translation and expression of respiratory chain subunits (Kim et al., 2012; Ryu et al., 2013). Consistent with this, disruption of the mouse Crif1 gene results in OxPhos deficiency in vivo (Kim et al., 2015). To assess the effects of muscle-specific Crif1 LOF on systemic energy homeostasis, we generated skeletal muscle–specific Crif1-knockout mice as an in vivo model of primary OxPhos deficiency. These mice were produced by crossing conditional Crif1 mice (Crif1flox/flox; hereafter, control; Kim et al., 2012) with mice expressing a Cre-recombinase gene under the control of the myosin light chain 1 fast promoter (Mlc1f-Cre; Bothe et al., 2000). Offspring from heterozygote intercrosses were born in a normal Mendelian ratio, and the MKO mice were healthy and viable. Expression of Crif1 in skeletal muscles was markedly lower in MKO mice than in control mice (Fig. 1 A). As expected, gastrocnemius muscle (GM) and extensor digitorum longus (EDL) in MKO mice contained reduced levels of three OxPhos subunits (NDUFA9, UQCRC2, and COX1; Fig. 1 B), as well as markedly reduced levels of OxPhos complexes I and III; moreover, subcomplex III (Fig. 1 C) had an abnormal appearance in blue native PAGE. In addition, electron microscopy revealed pronounced accumulation of abnormal, swollen mitochondria with disrupted cristae in GM of MKO mice (Fig. 1 D). MKO mice older than 13 wk exhibited overt features of mitochondrial myopathy, typified by lower muscle mass, myofibrillar disorganization with reduced fiber area, and reduced succinate dehydrogenase staining intensity (Fig. S1, A–C). In addition, MKO mice had reduced muscle strength in the grip test and latency period in the rotarod test, which was also associated with increased drop frequencies (Fig. S1 D). Thus, loss of Crif1 in skeletal muscle results in progressive functional and structural deterioration of the skeletal musculature.
Figure 1.
Impairment of OxPhos in MKO mice protects against insulin resistance. (A) Immunoblotting of CRIF1, NDUFA9 (a mitochondrial protein marker), and α-tubulin (a loading control) in gastrocnemius (GM) and extensor digitorum longus (EDL) muscle in chow-fed MKO and control (Ctrl) mice at 8 wk of age. Western blot (B) and blue native PAGE (C) of mitochondrial OxPhos complex subunits in GM and EDL muscle of Ctrl and MKO mice at 8 wk of age. (D) Mitochondrial morphology of Ctrl and MKO GM, visualized by electron microscopy. Bars: (top) 2 µm; (bottom) 1 µm. (E) Organ weights of 8-wk-old male Ctrl and MKO mice (n = 5 per group). (F) Representative images of hematoxylin and eosin staining of EDL, eWAT, and BAT in 8-wk-old male chow-fed Ctrl and MKO mice (n = 5 per group). Bars, 100 µm. (G) Overnight fasting glucose and insulin levels in MKO mice on a chow diet. (H) IPGTT and (I) IPITT in 8-wk-old male Ctrl and MKO mice fed a chow diet (n = 10 per group). (J) Hyperinsulinemic–euglycemic clamp. Glucose uptake and glucose infusion rate (GIR) in 8-wk-old male Ctrl and MKO mice on a chow diet (n = 5 per group). All data represent mean ± SEM. *, P < 0.05; **, P < 0.01. All blots are representative of more than three independent experiments.
MKO mice have elevated insulin sensitivity
To observe the impact of OxPhos deficiency in skeletal muscle on systemic energy homeostasis, we monitored body weight and food consumption in both control and MKO mice fed a standard chow diet. After 12 wk, food intake and weight gain did not significantly differ between control and MKO mice. After 13 wk, however, the MKO mice lost weight, and their muscle function progressively declined (Fig. S1, E and F).
To avoid the secondary effects of advanced muscle deterioration on metabolic parameters, we investigated the metabolic phenotype of 8-wk-old MKO mice. At that age, EDL, GM, and epididymal white adipose tissue (eWAT) mass was slightly lower in MKO mice than in controls (Fig. 1 E and 1F). However, whereas 8-wk-old MKO mice had reduced EDL muscle fiber mass, they had a normal distribution of central nuclei in muscle fibers, suggesting no apparent degenerative changes. The white adipose tissue (WAT) contained smaller adipocytes, whereas interscapular brown adipose tissue (BAT) contained markedly enlarged lipid droplets with unilocular deposition (Fig. 1 F). Fasting plasma glucose and insulin levels were also markedly reduced, although food consumption did not differ significantly from that of control mice (Figs. 1 G and S1 E). MKO mice exhibited improved glucose tolerance and insulin sensitivity in intraperitoneal glucose and insulin challenges (Fig. 1, H and I). Furthermore, glucose uptake and glucose infusion rate during a hyperinsulinemic–euglycemic clamp were higher in MKO mice (Fig. 1 J). Collectively, these findings indicated that OxPhos deficiency in MKO mice improved glucose tolerance and increased insulin sensitivity.
MKO mice are protected from HFD-induced obesity and insulin resistance
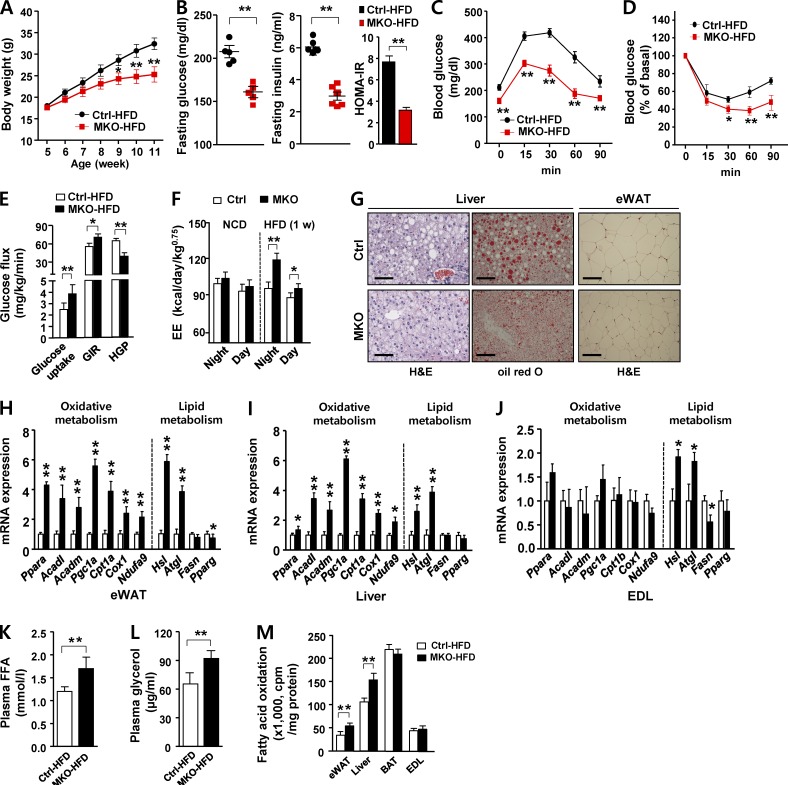
Given that MKO mice fed a standard chow diet were lean and had elevated glucose tolerance, we investigated weight gain on a high-fat diet (HFD). Remarkably, HFD-fed MKO (MKO-HFD) mice gained less weight than HFD-fed control (Ctrl-HFD) mice (Fig. 2 A). Dual-energy x-ray absorptiometry revealed that, at 11 wk of age, MKO-HFD mice had significantly lower fat mass than Ctrl-HFD mice (Fig. S2 A). Consistent with this, MKO-HFD mice had lower fasting glucose and insulin levels, concomitant with a decrease in insulin resistance index (Fig. 2 B), and also had higher glucose disposal rates than Ctrl-HFD mice in the intraperitoneal glucose tolerance test (IPGTT; Fig. 2 C) and intraperitoneal insulin tolerance test (IPITT; Fig. 2 D). To obtain further insight into whole-body and tissue-specific glucose metabolism in MKO-HFD mice, we performed a hyperinsulinemic–euglycemic clamp. Consistent with the results of the IPGTT and IPITT, MKO-HFD mice had higher whole-body glucose uptake and glucose infusion rate, suggesting that they had higher whole-body and peripheral insulin sensitivity (Fig. 2 E). In addition, hepatic glucose production was significantly lower in MKO-HFD mice than in Ctrl-HFD mice (Fig. 2 E).
Figure 2.
Improved metabolic parameters in MKO mice fed an HFD. (A) Body weight evolution of male Ctrl and MKO mice fed an HFD for 6 wk (n = 8 per group). (B) Fasting glucose, fasting insulin, and homeostatic model assessment-insulin resistance in male Ctrl and MKO mice fed an HFD for 6 wk (n = 5–6 per group). IPGTT (C) and IPITT (D) in male Ctrl and MKO mice fed an HFD for 6 wk (n = 6 per group). (E) Hyperinsulinemic–euglycemic clamp. Glucose uptake, glucose infusion rate (GIR) and hepatic glucose production (HGP) in male Ctrl and MKO mice fed an HFD for 6 wk (n = 5 per group). (F) Energy expenditure (EE) adjusted for body mass by ANOVA in 8-wk-old male Ctrl and MKO mice fed an HFD for 1 wk (n = 5 per group). (G) hematoxylin and eosin (H&E) and oil red O staining of liver (left) and eWAT (right) in male Ctrl and MKO mice fed an HFD for 6 wk (n = 5 per group). Bar, 100 µm. (H–J) Relative expression of genes related to β-oxidation (Ppara, Acadl, Acadm, Pgc1a, and Cpt1a/b), mitochondria (Cox1 and Ndufa9), and lipid metabolism (Hsl and Atgl, lipolysis; Fasn and Pparg, lipogenesis) in eWAT (H), liver (I), and EDL (J) in Ctrl and MKO mice fed an HFD for 6 wk (n = 5 per group). Plasma free fatty acid (K) and glycerol (L) concentrations in HFD-fed male Ctrl and MKO mice (n = 5 per group). (M) Ex vivo fatty acid oxidation (FAO) of infused 1-14C-palmitate in eWAT, liver, BAT, and EDL of HFD-fed Ctrl and MKO mice (n = 5 per group). All data represent mean ± SEM. *, P < 0.05; **, P < 0.01.
Energy expenditure, oxidative metabolism, and lipid catabolism are elevated in MKO mice
We next conducted indirect calorimetry after a short-term (1 wk) HFD to determine calorimetric parameters in the early dynamic phase of weight gain (i.e., before the late static phase reached after prolonged HFD feeding). Energy expenditure (EE) adjusted for body mass was significantly higher in MKO-HFD mice than in Ctrl-HFD mice (Figs. 2 F and S2 B), whereas spontaneous locomotor activity (Fig. S2 C) and food intake adjusted in the same manner did not differ between the two groups (not depicted). MKO-HFD mice exhibited markedly reduced accumulation of lipids, as assessed by oil red O staining, suggesting that these mice were protected from hepatic steatosis upon an HFD challenge. Furthermore, the adipocytes in eWAT were smaller in MKO-HFD mice than in Ctrl-HFD mice (Fig. 2 G).
To further characterize the elevated EE in MKO-HFD mice, we analyzed gene expression in adipose tissue, liver, and skeletal muscle. Genes involved in β-oxidation and mitochondrial OxPhos function were robustly induced in WAT and liver, but not in skeletal muscle (Fig. 2, H–J). Consistent with the up-regulation of lipolytic genes in eWAT, liver, and EDL (Fig. 2, H–J), fasting plasma free fatty acid and glycerol concentrations were significantly higher in MKO-HFD mice (Fig. 2, K and L), suggesting elevated in vivo lipolysis. Next, we performed an ex vivo fatty acid oxidation (FAO) assay using major metabolic organs such as adipose (eWAT and BAT), liver, and skeletal muscle. The rate of FAO in eWAT and liver was significantly higher in MKO-HFD mice than in Ctrl-HFD mice, whereas FAO in BAT and EDL did not differ between the two groups (Fig. 2 M). Collectively, the reduced adiposity and improved insulin sensitivity of MKO-HFD mice could be attributed to elevated lipolysis, β-oxidation, and EE. Collectively, these data show that reduced OxPhos function in skeletal muscle protected mice against HFD-induced adiposity and insulin resistance and was characterized by elevated lipolysis and oxidative metabolism in adipose tissue and liver.
Elevated circulating GDF15 in MKO mice
These robust phenotypic changes suggested that MKO mice expressed humoral factors involved in signaling from muscle to nonmuscle tissues. To identify the cell–non-autonomous factors secreted from skeletal muscle of MKO mice, we conducted a transcriptomic analysis by RNA sequencing (RNA-seq; Fig. 3 A), focusing on genes encoding secreted peptides (red dots).
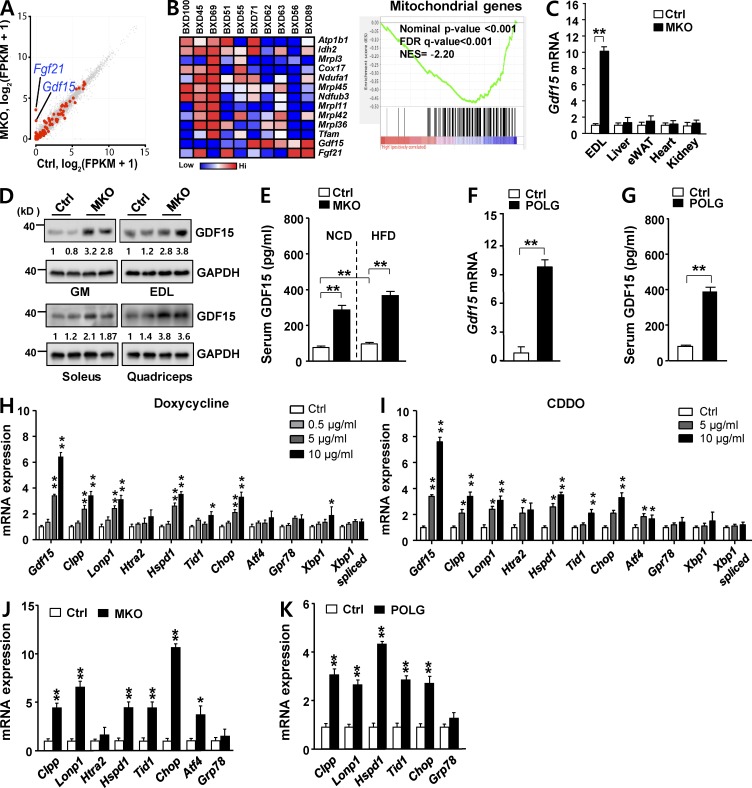
Figure 3.
GDF15 induction is associated with activation of the UPRmt in MKO and POLG mice. (A) Scatterplot of RNA-seq data, displaying transcript levels in EDL muscle of 8-wk-old male Ctrl (x axis) and MKO (y axis) mice. Gene sets encoding secretory proteins are represented in red. (B) Gene Set Enrichment Analysis was performed on skeletal muscle transcriptome data from BXD genetic reference population mice (Gdf15 expression in skeletal muscle transcriptomes of BXD mice on HFD; GeneNetwork accession numbers GN379 and GN380). The gene set encompassing mitochondria genes has an enrichment score of −0.477, a normalized enrichment score (NES) of −2.202, nominal P < 0.001, false discovery rate (FDR) q < 0.001, and family-wise error rate P < 0.001. (C) Gdf15 mRNA expression levels in EDL, liver, eWAT, heart, and kidney of 8-wk-old MKO mice. (D) GDF15 expression in GM, EDL, soleus, and quadriceps muscles in 8-wk-old male Ctrl and MKO mice fed a chow diet (n = 5 per group). (E) Serum GDF15 concentration in chow-fed 8-wk-old male Ctrl and MKO mice in the fasting state (n = 6 per group). (F and G) Gdf15 mRNA expression in GM (F) and serum GDF15 concentration (G) of 8-wk-old male POLG and control mice fed a chow diet (n = 5 per group). (H and I) Expression of Gdf15 and UPRmt genes after treatment of C2C12 myotubes with doxycycline (H) or CDDO (I) for 24 h. (J) Relative mRNA expression of UPRmt (Clpp, Lonp, Htra2, Hspd1, Tid1, and Chop) and ER stress (Atf4 and Grp78) genes in GM in 8-wk-old Ctrl and MKO mice. (K) Relative mRNA expression of UPRmt genes in GM of 8-wk-old control and POLG mice. All data represent mean ± SEM. *, P < 0.05; **, P < 0.01.
The mRNA levels of two secreted proteins, GDF15 and fibroblast growth factor 21 (FGF21), which are rarely expressed in wild-type skeletal muscle, were potently induced in OxPhos-deficient skeletal muscle (Fig. 3 A). Hence, we sought to determine whether the expression level of Gdf15 is negatively correlated with those of mitochondrial genes. To address this question, we performed gene set enrichment analysis (GSEA) using entire muscle transcriptomes from selected lines of the BXD mouse genetic reference population. In particular, we analyzed transcriptomes of individuals that exhibited either elevated (n = 5) or reduced (n = 5) expression of Gdf15 under HFD (Fig. 3 B, left). In this dataset, Gdf15 and the mitochondrial gene sets were clearly negatively correlated (enrichment score = −0.477, normalized enrichment score = −2.202, nominal P < 0.001, false discovery rate q < 0.001, and family-wise error rate P < 0.001; Fig. 3 B, right). However, Fgf21 and the mitochondrial gene set were not clearly correlated (Fig. 3 B, left). GDF15 expression is significantly induced in patients with mitochondrial diseases (Fujita et al., 2015). Consistent with this, Gdf15 mRNA and protein were markedly up-regulated in the GM, EDL, soleus, and quadriceps muscles of MKO mice (Fig. 3, C and D). Serum concentrations of GDF15 were also significantly higher in MKO mice (Fig. 3 E), suggesting that GDF15 produced in OxPhos-deficient skeletal muscles acts systemically as an endocrine signaling factor.
Elevated GDF15 expression is associated with activation of the UPRmt in MKO and POLG mice
We next asked whether the induction of GDF15 was a general consequence of mitochondrial stress in the muscle, using both in vivo (i.e., the polymerase gamma mutator [POLG] mouse) and in vitro (i.e., C2C12 myotubes) models. First, we analyzed young (8-wk-old) POLG mice, which carry a missense mutation that abolishes POLG proofreading activity, resulting in oxidative damage in skeletal muscle (Trifunovic et al., 2004; Kolesar et al., 2014; Wall et al., 2015). As with MKO mice, Gdf15 mRNA and serum GDF15 levels were significantly higher in POLG mice than in control mice (Fig. 3, F and G). Next, we chemically induced mitochondrial stress in cultured C2C12 myotubes using doxycycline and the triterpenoid 2-cyano-3,12-dioxooleana-1,9-dien-28-oic acid (CDDO), both of which trigger the UPRmt (Bernstein et al., 2012; Houtkooper et al., 2013). Expression of the intrinsic mitochondrial proteases Clpp and Lonp1, the mitochondrial chaperones Hspd1 and Tid1, and Chop, a transcription factor that mediates the UPRmt (Horibe and Hoogenraad, 2007), was robustly up-regulated in C2C12 myotubes treated with doxycycline and CDDO, indicative of mitochondrial stress (Fig. 3, H and I). Under the same conditions, Gdf15 was strongly induced, indicating that GDF15 expression in skeletal muscle reflects activation of the UPRmt. These observations led us to analyze the UPRmt in MKO and POLG mice. Skeletal muscles from both strains expressed high levels of UPRmt transcripts such as Clpp, Lonp1, Hspd1, and Tid1 (Fig. 3, J and K). We also confirmed that Gdf15 induction was restricted to muscle: Gdf15 transcript levels in liver, eWAT, heart, and kidney did not differ between MKO and control mice (Fig. 3 C). Thus, activation of the UPRmt in skeletal muscle of POLG and MKO mice was associated with elevated GDF15 expression.
CHOP-dependent induction of GDF15 in MKO skeletal muscle upon UPRmt activation
GDF15 expression is regulated by various transcriptional pathways, dependent on the cellular and signaling context (Ago and Sadoshima, 2006; Wang et al., 2014). Expression of Gdf15 is generally low in quiescent cells but is rapidly induced by stress stimuli that activate diverse signaling pathways (Vaňhara et al., 2012; Wang et al., 2013).
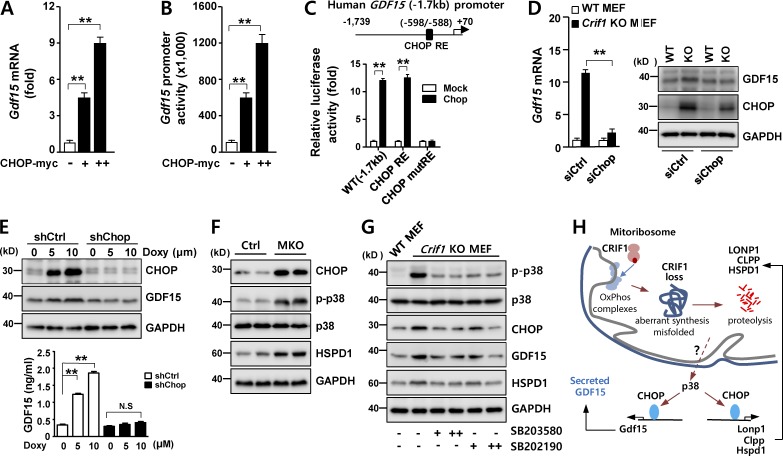
Based on our observations that GDF15 expression is associated with the UPRmt, we next explored the potential involvement of transcription factors such as the ATFs and CHOP, which mediate stress responses in mammalian cells (Zhao et al., 2002; Horibe and Hoogenraad, 2007; Pellegrino et al., 2014). In C2C12 myoblasts, Gdf15 promoter activity was not altered by overexpression of ATF4, ATF5, or ATF6 (Fig. S3, A–C). In contrast to the ATFs, overexpression of CHOP strongly induced both Gdf15 mRNA levels and promoter activity (Fig. 4, A and B). Moreover, reporter assays revealed the functional importance of putative CHOP-responsive elements (CHOP REs) in the human GDF15 promoter (Fig. 4 C). siRNA-mediated knockdown of Chop suppressed Gdf15 expression in Crif1-deficient MEF cells (Fig. S3, D and E; and Fig. 4 D), demonstrating that CHOP induces GDF15 in these cells. Moreover, we developed a stable C2C12 cell line harboring knockdown of Chop by lentivirus-mediated transduction of Chop-specific shRNA. In C2C12 shChop cells, CHOP was not induced after doxycycline treatment, and expression and secretion of GDF15 were not stimulated (Fig. 4 E). This observation confirmed that CHOP is required for UPRmt-induced expression of GDF15 in vitro. Consistent with this result, CHOP was highly expressed in skeletal muscle of MKO mice (Fig. 4 F).
Figure 4.
CHOP-dependent induction of GDF15 during the UPRmt. (A) Gdf15 mRNA expression in C2C12 myoblasts transfected with either an empty (mock) or myc-tagged CHOP expression vector (CHOP-myc). (B) Luciferase activity of human GDF15 promoter in C2C12 myoblasts transfected with the CHOP-myc vector. (C) Luciferase activity of reporters with deletions or point mutations of the human GDF15 promoter in C2C12 myoblasts transfected with either an empty (mock) or CHOP expression vector for 36 h. (D) GDF15 expression in Crif1-deficient MEFs transfected with control siRNA (siCtrl) or Chop siRNA (siChop) for 48 h. Expression of CHOP was determined by Western blotting. (E) GDF15 expression (top) and secretion (bottom) in CHOP-knockdown cells using lentivirus-delivered CHOP-specific shRNA. 24-h doxycycline treatment induced GDF15 in stable shCtrl cells, but not in shChop cells. (F) Immunoblotting of CHOP, HSPD1, and phospho–p38 T180/Y182 expression in MKO mice. (G) A p38-specific inhibitor blocks p38-CHOP–induced GDF15 expression in Crif1-deficient MEF cells. GAPDH is used as loading control. (H) A schematic model showing the UPRmt pathways governing GDF15 expression in Crif1-deficient skeletal muscle. All data represent mean ± SEM. **, P < 0.01. All results are representative of more than three independent experiments.
CHOP, a member of the C/EBP family of transcription factors, is phosphorylated by p38 MAPK; this modification increases the transcriptional activity of CHOP (Wang and Ron, 1996). Given that skeletal muscle in MKO mice contained high levels of phosphorylated p38 kinase (Fig. 4 F), we hypothesized that the p38–CHOP pathway could regulate Gdf15 expression under mitochondrial stress. To validate the influence of p38 kinase–CHOP on GDF15 expression, we treated Crif1-deficient MEF cells with two different p38 kinase inhibitors, SB203580 and SB202190, resulting in attenuation of Gdf15 and Chop expression (Fig. 4 G). These findings indicate that p38 kinase is a key regulator of CHOP-dependent Gdf15 transcription in Crif1-deficient skeletal muscle (Fig. 4 H).
GDF15 increases lipolysis and FAO
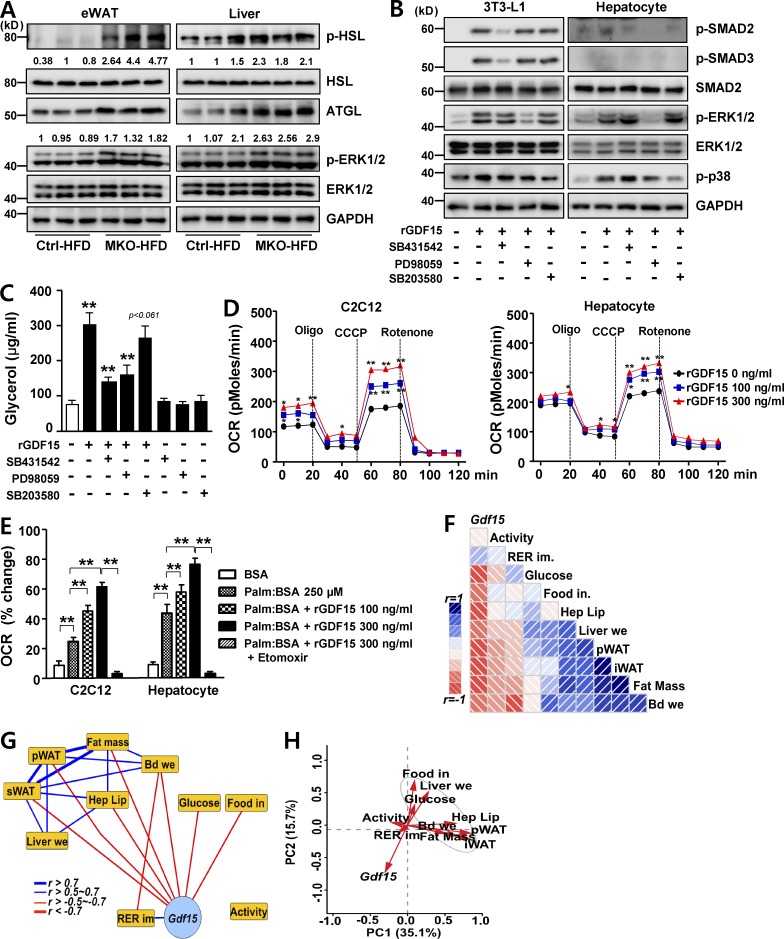
Because the circulating level of GDF15 was elevated in MKO mice, we next investigated its roles in metabolism and how they contribute to the phenotypes of MKO mice. Higher levels of phosphorylated hormone-sensitive lipase (HSL S660) and ERK1/2 suggested that MKO mice had elevated lipolytic activities in eWAT and liver (Fig. 5 A). To determine whether the higher lipolytic activities in MKO mice were related to the increased activity of GDF15, we monitored phosphorylation of SMAD, ERK1/2, and p38 after recombinant GDF15 (rGDF15) treatment in differentiated 3T3-L1 adipocytes and primary cultured mouse hepatocytes. rGDF15-induced phosphorylation of SMAD2 and SMAD3 was inhibited by the chemical inhibitor SB431542, a potent and selective inhibitor of TGF-β reversible inhibitor receptor kinases. Moreover, rGDF15-induced phosphorylation of ERK1/2 and p38 was inhibited by PD98059 and SB203580, respectively (Fig. 5 B). rGDF15 significantly increased the amount of glycerol released into the media by 3T3-L1 adipocytes after 3 h. Moreover, the lipolytic effect of GDF15 was suppressed by TGF-β reversible inhibitor and ERK inhibitors (Fig. 5 C). Based on these findings, we conclude that MKO-HFD mice are protected from obesity via activation of TGF-β reversible inhibitor receptor kinases by GDF15.
Figure 5.
GDF15 induces lipolysis and FAO. (A) Immunoblotting for ATGL, phosphorylated HSL-Ser660 (pHSL), HSL, phospho-ERK1/2 (pERK1/2), and GAPDH in eWAT or liver of male Ctrl and MKO mice fed an HFD for 6 wk (n = 5–6 per group). (B) Immunoblotting to detect SMAD (phospho-SMAD2, phospho-SMAD3, and SMAD2), phospho-ERK1/2, and phospho-p38 in differentiated 3T3-L1 adipocytes and primary cultured hepatocytes after exposure to 100 ng/ml rGDF15 in the absence or presence of 5 µM SB431542 (ALK4/5/7 inhibitor), 25 µM PD98059 (ERK1/2 inhibitor), and 10 µM SB203580 (p38 inhibitor). GAPDH was used as a loading control. (C) In vitro lipolysis assay. Glycerol content was measured in differentiated 3T3-L1 adipocytes treated with 300 ng/ml rGDF15 in the absence or presence of SB431542, PD98059, and SB203580. (D) Oxygen consumption rate (OCR) measured in C2C12 myoblasts (left) and primary cultured hepatocytes (right) treated with the indicated concentrations of rGDF15. CCCP, carbonyl cyanide m-chlorophenyl hydrazine; Oligo, oligomycin. (E) Fatty acid oxidation (FAO) in C2C12 myoblasts and primary hepatocytes using palmitate-BSA as a substrate after pretreatment with different concentrations of rGDF15 in the presence or absence of 40 µM of the CPT1 inhibitor etomoxir. (F and G) Corrgram (F) and interaction network (G) showing correlations between Gdf15 mRNA levels in skeletal muscle of HFD-fed mice and the indicated clinical phenotypes across different mice strains of the BXD genetic reference population. Activity, locomotor activity for 24 h (counts per hour; n = 52); RER im, RER improvement (ratio; n = 49); Glucose, serum glucose level (millimoles per liter); Food in, food intake (grams) in 24 h (n = 32); Hep Lip, hepatic lipid contents normalized to total protein; Liver we, liver weight (grams; n = 32); pWAT, peritoneal adipose tissue weight (grams); sWAT, subcutaneous fat weight (grams; n = 32); Fat Mass, total white adipose weight (grams; n = 32); and Bd We, body weight gain (grams; n = 32). In the corrgram (F), the depth of shading at the correlation matrices indicates the magnitude of the correlation. Positive and negative correlations are represented in blue and red, respectively. In the interaction network (G), positive Pearson’s correlation coefficients are represented as blue and red lines (r = 0.5–0.7 [normal], 0.7–1.0 [bold], and −1.0 to −0.7 [bold], −0.7 to −0.5 [normal], respectively). (H) Principal-component analysis plot of the expression of Gdf15 and selected metabolic phenotype in the BXD genetic reference population, based on Pearson’s correlation coefficients. All data represent mean ± SEM. *, P < 0.05; **, P < 0.01. All results are representative of more than three independent experiments.
Next, we explored the effects of GDF15 on mitochondrial respiration in C2C12 myoblasts and cultured primary hepatocytes. In myoblasts and hepatocytes, rGDF15 treatment increased basal, ATP-coupled, and maximal respiratory rates in a dose-dependent manner (Fig. 5 D). Consistent with this, we found that pretreatment of rGDF15 increased FAO both in myoblasts and hepatocytes (Fig. 5 E). The oxygen consumption rate (OCR) reverted to basal levels upon treatment with the CPT1 inhibitor etomoxir, reflecting the fact that GDF15 promotes FAO and mitochondrial respiration (Fig. 5 E).
In light of these findings regarding the impact of GDF15 on systemic energy homeostasis, we next investigated whether Gdf15 also impacts the same metabolic pathways in a complex systems genetics model, the BXD mouse genetic reference population (Andreux et al., 2012; Wu et al., 2014). Consistent with our observations in MKO mice, HFD-fed BXD mice with higher expression of Gdf15 transcripts in skeletal muscle exhibited more favorable metabolic phenotypes than those expressing lower levels of Gdf15. Total, peritoneal, and subcutaneous fat mass, liver lipid content and weight, body weight gain, food intake, and serum glucose level were mostly positively correlated with one another (Pearson’s R > 0.5, represented as blue boxes and edges in Fig. 5, F and G, respectively), whereas those phenotypes were negatively correlated with Gdf15 expression (R < −0.5, displayed as red boxes and edges in Fig. 5, F and G, respectively). The improvement in the respiratory exchange ratio (RER) was also positively correlated with Gdf15 expression, but not significantly. In contrast, spontaneous locomotor activity was not significantly correlated with Gdf15 expression (Fig. 5, F and G). Furthermore, principal-component analysis confirmed that Gdf15 expression in skeletal muscles of HFD-fed BXD strains was negatively correlated with peritoneal and subcutaneous fat mass, liver lipid content and weight, body weight gain, food intake, and serum glucose level (Fig. 5 H).
GDF15 determines metabolic phenotypes of MKO mice
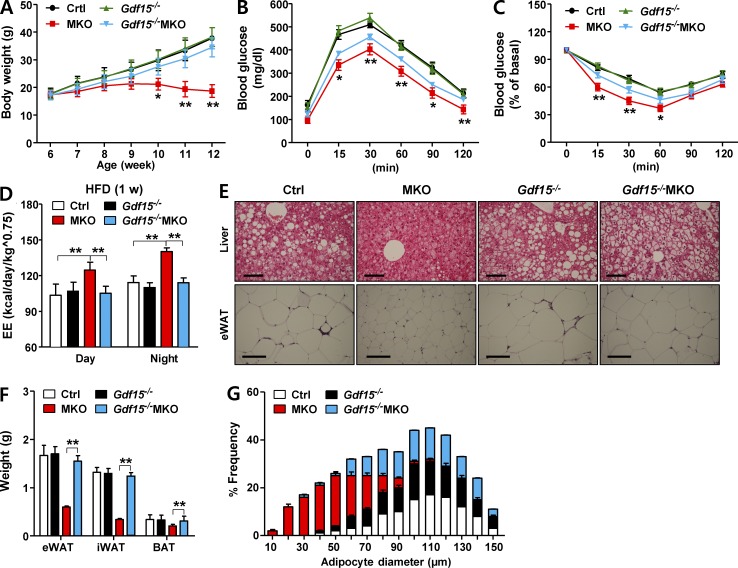
Given that GDF15 is secreted at high levels from skeletal muscle of MKO mice, we investigated the in vivo role of GDF15 in determining metabolic phenotypes of MKO mice. To this end, we observed the metabolic phenotypes of double Gdf15−/−MKO-knockout mice, which were generated by crossing whole-body Gdf15-deficient (Gdf15−/−) mice with MKO mice. Several phenotypes of MKO mice, including reduced body weight and improved glucose tolerance, were attenuated in Gdf15−/−MKO mice (Fig. 6, A–C), and the elevated EE (adjusted by body weight) in HFD-fed MKO mice was not observed in HFD-fed Gdf15−/−MKO mice (Fig. 6 D). In addition, Gdf15−/−MKO mice exhibited prominent histological phenotypes of steatosis, which were absent in MKO mice fed an HFD (Fig. 6 E). Moreover, the reductions in fat weight and the number of small adipocytes in eWAT observed in MKO mice were absent in Gdf15−/−MKO mice, and the corresponding parameters reverted to the levels in control mice (Fig. 6, F and G). Thus, protection against obesity and improved insulin sensitivity were mostly determined by the cell–non-autonomous factor GDF15, which is induced in skeletal muscle in MKO mice under mitochondrial stress.
Figure 6.
The metabolic phenotypes of Gdf15−/−MKO double knockouts are attenuated relative to those of MKO mice. (A–C) Time course of changes in body weight (n = 6–7 per group; A), IPGTT (n = 5–6 per group; B), and IPITT (n = 5–6 per group; C) in male Gdf15+/+Crif1f/f (Ctrl), Gdf15−/−Crif1f/f (Gdf15−/−), Gdf15+/+MKO (MKO), and Gdf15−/−MKO mice fed an HFD for 6 wk. The mice were fed an HFD from 6 to 12 wk of age, after which all metabolic analyses were performed. (D) Energy expenditure (EE) adjusted for body mass by analysis of variance in 8-wk-old male Ctrl and MKO mice fed an HFD for 1 wk (n = 5 per group). (E–G) Hematoxylin and eosin (H&E) staining of liver and eWAT (E), fat weight (F), and adipocyte diameter (G) in male Ctrl, Gdf15−/−, MKO, and Gdf15−/−MKO mice fed an HFD for 6 wk (n = 5 per group). Bar, 100 µm. All data represent means ± SEM. *, P < 0.05; **, P < 0.01. MKO vs. Gdf15−/−MKO, (n = 5−7 per group).
GDF15 reduces body weight and improves glucose tolerance in ob/ob mice
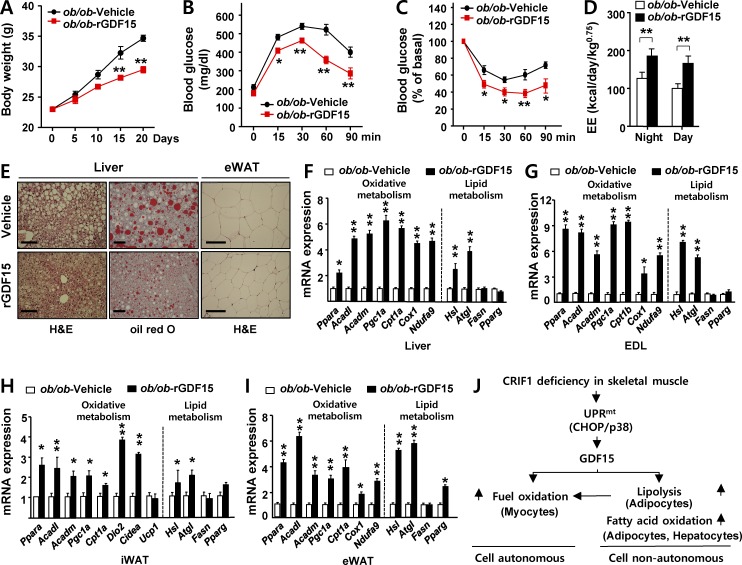
Although previous studies revealed that GDF15 exerts both prolongevity and protective effects on obesity in transgenic models (Macia et al., 2012; Chrysovergis et al., 2014; Wang et al., 2014), the pharmacological effects of GDF15 on obesity and insulin sensitivity remained to be elucidated. To investigate the protective effects of GDF15 on weight gain and insulin resistance, we intraperitoneally injected ob/ob mice three times per week with 0.5 mg/kg rGDF15 or vehicle for 3 wk, beginning at 6 wk of age. Injection of rGDF15 decreased body weight gain relative to vehicle-treated controls (Fig. 7 A), independent of food intake and locomotor activity (Fig. S4, A and B). In comparison with vehicle-treated ob/ob mice, GDF15-treated ob/ob mice had a higher glucose disposal rate in response to intraperitoneal glucose challenge and insulin stimulation (Fig. 7, B and C). Furthermore, oxygen consumption (VO2), carbon dioxide production (VCO2), and EE were significantly higher in rGDF15-treated ob/ob mice than in control mice (Fig. S4, C–E; and Fig. 7 D). In addition, hepatic steatosis and eWAT lipid droplet size were markedly reduced in rGDF15-treated ob/ob mice (Fig. 7 E). As with MKO mice, the expression of lipolytic genes in liver, EDL, inguinal white adipose tissue, and eWAT was robustly induced (Fig. 7, F–I). Transcripts of genes involved in FAO and mitochondrial OxPhos were also consistently up-regulated in WAT and liver (Fig. 7, F–I). Collectively, these data indicate that administration of rGDF15 decreased adiposity and insulin resistance and stimulated lipolysis and oxidative metabolism, reminiscent of the metabolic phenotypes observed in MKO mice.
Figure 7.
Elevated insulin sensitivity and lipid catabolism in ob/ob mice injected with GDF15. (A) Body weight of ob/ob mice treated with recombinant GDF15 (rGDF15). ob/ob mice were injected intraperitoneally with rGDF15 (0.5 mg/kg, red) or vehicle (black) for 3 wk every other day (n = 5 per group). IPGTT (B) and IPITT (C) in ob/ob mice administered rGDF15 or vehicle for 3 wk every other day (n = 5 per group). (D) Energy expenditure (EE) of ob/ob-rGDF15 or ob/ob-vehicle mice was analyzed by indirect calorimetry over a period of 12 h light/12 h dark cycles. EE adjusted for body mass was compared by ANOVA between ob/ob mice treated with rGDF15 for 3 wk and those treated with vehicle (n = 5 per group). (E) Representative images of hematoxylin and eosin and oil red O staining of liver (left) and eWAT (right) in ob/ob-rGDF15 or ob/ob-vehicle mice (n = 5 per group). Bars, 100 µm. (F–I) Relative expression of genes related to β-oxidation (Ppara, Acadl, Acadm, Pgc1a, and Cpt1a/b) mitochondria (Cox1 and Ndufa9), thermogenesis (Dio2, Cidea, and Ucp1), and lipid metabolism (Hsl and Atgl, lipolysis; Fasn and Pparg, lipogenesis) in liver (F), EDL (G), inguinal white adipose tissue (iWAT; H), and eWAT (I) in ob/ob-rGDF15 or ob/ob-vehicle mice (n = 5 per group). (J) Model of Gdf15 induction upon OxPhos dysfunction. All data represent mean ± SEM. *, P < 0.05; **, P < 0.01.
Discussion
Crif1, an essential mitoribosome protein required for the synthesis and insertion of mitochondria-encoded OxPhos polypeptides in the inner mitochondrial membrane, is abundantly expressed in skeletal muscle (Kim et al., 2012; Greber et al., 2014a). Loss of Crif1 function leads to OxPhos dysfunction caused by both aberrant synthesis and defective insertion of OxPhos proteins (Ryu et al., 2013; Kim et al., 2015). Consistent with this, we found that skeletal muscle deficiency of Crif1 induced progressive mitochondrial OxPhos dysfunction and activation of UPRmt in muscle, culminating in advanced muscular dystrophy and sarcopenia. Accordingly, to exclude potential confounding effects of advanced skeletal muscle and adipose tissue dysfunction on metabolism, we investigated metabolic phenotypes in young MKO mice at 8 wk of age, a time when control and MKO mice did not exhibit apparent phenotypic differences.
Although many studies have reported that primary defects in mitochondrial function in mouse skeletal muscle protect from obesity and insulin resistance, the molecular mechanisms and factors underlying the control of systemic energy homeostasis remain to be identified (Wredenberg et al., 2006; Pospisilik et al., 2007; Quintens et al., 2013). Reduced mitochondrial oxidative phosphorylation in mice with AIF LOF in skeletal muscle improves glucose tolerance and insulin sensitivity while reducing fat mass (Pospisilik et al., 2007). Cell-autonomous activation of AMP kinase in skeletal muscle with impaired OxPhos function is regarded as a potential explanation for the up-regulation of catabolic processes such as glucose uptake and FAO (Towler and Hardie, 2007). Our data indicate that GDF15 is a previously unrecognized circulating factor that regulates systemic energy homeostasis and protects against diet-induced obesity and insulin resistance in the context of selective muscle mitochondrial dysfunction and activation of the UPRmt.
Furthermore, our results show that p38 kinase activation of CHOP is required for the transcriptional activation of Gdf15 expression in Crif1-deficient cells. This finding indicates that CHOP plays a critical role in the mitohormetic response, typified here by the induction of GDF15 to promote metabolic adaptation. Consistent with this role in mitochondrial stress signaling, CHOP is an adaptive response gene that is induced by a variety of other stress signals, including hypoxia and cytokines (Zheng et al., 2012; Schönthal, 2013; Dong et al., 2014). Thus, our study identifies CHOP as essential for the adaptive GDF15 response in mice with electron transport chain–incompetent mitochondria and activated UPRmt in skeletal muscle. Notably, patients with mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes have high serum levels of GDF15; accordingly, this factor was proposed as a more sensitive and specific biomarker than FGF21 for diagnosis and therapeutic monitoring of mitochondrial diseases (Fujita et al., 2015).
In this study, we also showed that plasma concentrations of FGF21 were elevated in MKO mice (Fig. S3, G and H). This effect may be linked to the transcriptional activation of Fgf21 in the wake of higher ATF4 expression resulting from Crif1 deficiency in skeletal muscle (Fig. 3 J; Kim et al., 2013). FGF21, a marker of mitochondrial diseases, is a hormone that mediates the starvation-induced response in humans and mouse models (Tyynismaa et al., 2010; Kim et al., 2013). FGF21 induced in POLG mice, a model of mitochondrial disease and premature aging, provides metabolic benefits by increasing fat metabolism and improving mitochondrial function, particularly when the mice are fed an HFD (Wall et al., 2015). Collectively, the metabolic phenotypes of MKO mice, characterized by elevated glucose tolerance, lipolysis, and FAO, can be attributed to the convergent metabolic actions of both GDF15 and FGF21. However, the precise cell–non-autonomous role of FGF21 remains poorly understood (Ost et al., 2015). Skeletal muscle–specific UCP1 transgenic mice, a model of mitochondrial uncoupling, exhibit metabolic adaptation associated with robust induction of FGF21 in muscle. Strikingly, in double-knockout mice generated by crossing UCP1 transgenic with Fgf21-knockout mice, FGF21 played a negligible role in improvements in obesity resistance, glycemic control, and hepatic lipid homeostasis related to muscle mitochondrial stress. These observations challenge the idea that FGF21 is the key metabolic mediator of mitochondrial stress adaptation during muscle mitochondrial disease. Consistent with results obtained in UCP1 transgenic and Fgf21-knockout mice, double Fgf21−/−MKO–knockout mice did not exhibit significant metabolic differences related to body weight, glucose tolerance, or insulin sensitivity (unpublished data). Based on these observations, we propose that GDF15 is a critical humoral factor that is released from skeletal muscle fibers upon mitochondrial stress to promote reorganization and adaptation of systemic metabolism. Thus, GDF15 can be considered as a true myomitokine.
Consistent with the up-regulation of lipolytic genes and FAO in MKO mice, in vivo administration of rGDF15 increased the expression of genes involved in lipolysis and FAO in adipose tissue and liver. This finding suggests that GDF15 mobilizes free fatty acids from adipose deposits. Because GDF15 administration increased both free fatty acid mobilization and oxidation, elevated GDF15 levels may be responsible for the metabolic benefits observed in MKO mice. The metabolic phenotypes in MKO mice appear to mimic the phenotype of transgenic mouse models expressing human GDF15, which is characterized by elevated expression of lipolytic and thermogenic genes (Chrysovergis et al., 2014). Based on our results, rGDF15 activates activin receptor–like kinase (ALK)–mediated MAPK and SMAD 2/3 phosphorylation in adipose tissue and liver. The various members of the TGF-β ligand family, including GDF15, TGF-β, and activins, play diverse roles in mouse and human models of obesity and systemic glucose tolerance. Although they share common signaling pathways mediated by ALK isoforms, each member has specific effects on the regulation of metabolic tissues. For example, the activins are multifunctional growth and differentiation factors. In the pancreas, these proteins stimulate the differentiation of β cells and secretion of insulin, enabling the cells to respond by taking up glucose efficiently (Ueda, 2000). In contrast, in liver, skeletal muscle, and WAT, activins negatively regulate organogenesis, leading to impaired insulin sensitivity (Rebbapragada et al., 2003; Dani, 2013). In macrophages, activins induce a phenotypic switch from the M1 to M2 phenotypes, thereby diminishing inflammation. Because adipose inflammation is closely associated with insulin resistance and the onset of type 2 diabetes, activins may improve insulin resistance via their antiinflammatory activity (Hashimoto and Funaba, 2011). The molecular pathways by which GDF15 exerts the observed pharmacological effects on obesity and diabetes remain to be determined.
Previous studies showed that controlled treatment with mitochondrial uncouplers such as 2,4-dinitrophenol or niclosamide ethanolamine salt decreases insulin resistance and hepatic steatosis in various animal models, suggesting that mild mitochondrial uncoupling can improve metabolic profiles (Perry et al., 2013, 2015; Tao et al., 2014). Although the authors of those studies attributed the beneficial metabolic effects to futile cycling, they did not investigate the induction of UPRmt or mitokine action. A separate study in U2OS and HEK293 cells revealed that GDF15 was induced by the addition of carbonyl cyanide m-chlorophenyl hydrazine (CCCP) in an in vitro model, establishing a precedent for GDF15 induction in response to mild mitochondrial uncoupling (Gleyzer and Scarpulla, 2011). These findings suggest that GDF15 is a mitokine intermediary partially responsible for the metabolic benefits reported in previous studies. In support of this idea, our in vivo experiments demonstrated that GDF15 protected against obesity and improved insulin sensitivity in ob/ob mice. Accordingly, we speculate that GDF15 or its analogues could be useful in the management of obesity-related diseases, in particular type 2 diabetes. Although we provided in vivo evidence that administration of rGDF15 could successfully protect from obesity and improve insulin sensitivity in animal models, a full understanding of the precise molecular mechanism of GDF15 requires further study.
Various clinical studies have shown a positive correlation between GDF15 and obesity, type 2 diabetes mellitus, and resultant comorbidities such as cardiovascular complications (Kempf et al., 2012; Dominguez-Rodriguez et al., 2014). Patients with obesity and insulin resistance may exhibit impaired mitochondrial function (Pagel-Langenickel et al., 2010; Hesselink et al., 2016). We observed expression and secretion of GDF15 in both adipocytes (unpublished data) and skeletal myocytes, which have altered mitochondrial OxPhos function. Therefore, we suspect that induction of GDF15 in obesity is the result of compensation for muscle mitochondrial dysfunction. Thus, elevated GDF15 levels may reflect a physiological response aimed at reestablishing metabolic homeostasis, albeit insufficiently. This may explain why administration of rGDF15 to obese mice improved their metabolic phenotype. It should also be noted that multiple studies have reported that GDF15 plays a protective role in these diseases (Kempf et al., 2006, 2011; Xu et al., 2006; Preusch et al., 2013), consistent with our observations. Clinical studies will be needed to establish the therapeutic value of GDF15 in the context of human metabolic diseases.
In summary, our study revealed that OxPhos dysfunction in skeletal muscle increases the production of GDF15, which we classify as a novel myomitokine. GDF15 is an essential transmitter of systemic mitohormetic adaptation and redistribution of energy stores during OxPhos dysfunction (Fig. 7 J). Our findings lay the foundation for the identification of mitokines and their potential therapeutic applications in the treatment of obesity and insulin resistance.
Materials and methods
Mice
Floxed Crif1 (Crif1f/f) mice were generated as previously described (Kwon et al., 2008). Crif1f/f mice were bred with Mlc1f-Cre and GDF15-null (Gdf15−/−) mice (provided by S.J. Burden, New York University, New York, NY, and S. Lee, Johns Hopkins University School of Medicine, Baltimore, MD) to generate MKO and Gdf15−/−MKO mice, respectively. POLG mice were maintained as described previously (Wall et al., 2015). The HFD, which contained 60% fat, was purchased from Research Diets Inc. (D12492). Mice were maintained in a controlled environment (12 h light/12 h dark cycle; humidity 50–60%; ambient temperature 22 ± 2°C) and fed a normal chow diet except as noted. All experimental procedures were conducted in accordance with the Institutional Animal Care and Use Committee of the Chungnam National University School of Medicine (Daejeon, Korea). The 5-wk-old male ob/+ control mice and ob/ob C57BL/6J Lep(−/−) mice were purchased from Envigo. All mice were of the C57BL/6 background.
rGDF15 protein and reagents
Purified recombinant human GDF15 containing an N-terminal 6-His tag was purchased from R&D Systems (957-GD/CF). Lyophilized protein was reconstituted with sterile 4 mM HCl and stored at −70°C. SB431542, PD98059, SB203580, SB202190, oligomycin, CCCP, etomoxir, and all other reagents were purchased from Sigma-Aldrich, unless otherwise noted.
Measurement of the OCR
The OCR was measured in using a Seahorse XF-24 analyzer (Seahorse Bioscience). Cells were seeded at 20,000–50,000 cells per well 18 h before the analysis. On the day before the experiment, the sensor cartridge was placed into the calibration buffer supplied by Seahorse Bioscience and incubated at 37°C in a non-CO2 incubator. Immediately before measurement, cells were washed and incubated at 37°C with media lacking sodium bicarbonate. Three readings were taken after each addition of mitochondrial inhibitor, but before injection of the subsequent inhibitor. 2 µg/ml oligomycin, 5 µM CCCP, and 2 µM rotenone were used as mitochondrial inhibitors. The OCR was automatically calculated and recorded by the sensor cartridge and Seahorse XF-24 software.
Serum biochemical measurements
Blood was collected from the heart under general anesthesia. Samples were centrifuged at 10,000 rpm for 5 min, and the supernatants were collected. Serum insulin (Alpco Diagnostics), FGF21 (R&D Systems), and GDF15 (R&D Systems) levels were measured by ELISA using kits obtained from the indicated suppliers. Biochemical analyses, including determination of free fatty acid and total cholesterols, were performed using a Hitachi 7180 auto analyzer and Wako reagents (Wako Pure Chemical Industries). Plasma glycerol levels were measured using the Free Glycerol Determination kit (Sigma-Aldrich).
Isolation of mitochondria and blue native PAGE
For mitochondrial isolation and blue native PAGE, cells or tissues were homogenized in isolation buffer B (210 mM mannitol, 70 mM sucrose, 1 mM EGTA, and 5 mM Hepes, pH 7.2) with a Teflon-glass homogenizer. The homogenate was centrifuged at 600 g for 5 min at 4°C. Mitochondrial fractions were washed with buffer B and resuspended in the same buffer. Mitochondria were either used immediately or stored at −80°C for later use. For blue native PAGE, the mitochondrial fraction was analyzed using the Native PAG Novex Bis-Tris Gel system (Invitrogen) with 0.5% (wt/vol) n-dodecyl-β-d-maltoside to determine the content of the mitochondrial OxPhos complex. After electrophoresis, proteins were transferred to a polyvinylidene fluoride membrane and incubated with anti-OxPhos antibody cocktail (45-8099 and 45-7999; Invitrogen). After incubation in diluted primary antibody, the membrane was washed and visualized using the Western Breeze Chromogenic Western Blot Immunodetection kit (Invitrogen).
Cell culture and isolation of primary hepatocytes
Wild-type and Crif1-deficient MEF cells were grown in DMEM supplemented with 10% FBS, 1% penicillin streptomycin (Invitrogen), 100 µg/ml pyruvate, and 50 µg/ml uridine (Sigma-Aldrich). 3T3-L1 cells were maintained in DMEM supplemented with 10% bovine calf serum (Invitrogen). After 48 h, cells were differentiated with 0.5 mM IBMX, 1 mM dexamethasone, 10 mg/ml insulin, and 10% FBS (Ryu et al., 2013). C2C12 cells were cultured in DMEM containing 10% FBS. Myoblasts were differentiated with DMEM supplemented with 2% horse serum. Mouse primary hepatocytes were prepared from 8-wk-old male C57BL/6 mice using the collagenase perfusion method (0.8 mg/ml collagenase; Chung et al., 2015). Isolated cells were further purified through a Percoll density gradient and plated in culture plates at an appropriate density in Medium 199 (Sigma-Aldrich). All cell lines used in this study were regularly tested for mycoplasma contamination using the MycoAlert PLUS kit (Lonza).
Plasmid constructs
The pcDNA-HA human ATF4, pcDNA-HA human ATF5, pcDNA-HA human ATF6, and pcDNA-human CHOP-myc expression vectors and the pGL3B-human GDF15 (−1,739/+70) luciferase reporter constructs were provided by H.S. Choi (Chungnam National University, Daejeon, Korea) and Y. Moon (Pusan National University, Busan, Korea). pGL3B-GDF15 deletion (CHOP RE [−670 bp]) and pGL3B-CHOP mutRE reporter constructs were constructed by inserting a PCR fragment of the human GDF15 promoter into KpnI–XhoI-digested pGL3Basic (Promega). The nucleotide sequence of the 5′-flanking region of the GDF15 promoter was scanned, and one putative CHOP binding site (GDF15 [−598 to −588; 5′-TGCAATAACA-3′]) was identified based on the nucleotide sequence. To create a CHOP mutRE point mutant reporter, the 5′-TGCAATAACA-3′ sequence (−598 to −588) of the CHOP RE (−670-bp) vector was mutated to 5′-AAAAAAAACA-3′ by DpnI-based site-directed mutagenesis (Agilent Technologies). Nucleotide sequences of all plasmids were confirmed by automatic sequencing.
Knockdown of Chop expression by shRNA or siRNA and luciferase assay
Mouse shChop (NM_007837.2-501s1c1) and nontarget shRNA control (SHC001) in lentiviral vector pLKO.1 were purchased from Sigma-Aldrich. To generate stably expressing shRNA targeting shChop cells, the lentiviral vector was cotransfected into 293T (Takara Bio Inc.) cells with virus packaging mix (Sigma-Aldrich) using the Lipofectamine PLUS (Invitrogen) reagent. Chop expression levels were analyzed by RT-PCR and Western blot analyses. Chop siRNA (siChop) and control (siCtrl) were purchased from Invitrogen (siChop, 5′-CCAGGAAACGAAGAGGAAGAAUCAAdTdT-3′; siCtrl, 5′-GCAUCAAGGUGAACUUCAAdTdT-3′). Crif1-deficient or wild-type MEFs were cultured in six-well plates and transfected with 100 nM siCtrl or siChop using Lipofectamine 2000 (Invitrogen). For luciferase assays, C2C12 myoblasts were plated in 24-well culture plates and transfected with 100 ng of a human GDF15 (−1,750/+70) promoter luciferase reporter and 30 ng Renilla reporter vector, along with 100–300 ng of the indicated expression plasmids using Lipofectamine PLUS. pcDNA3 empty vector was added so that the same total amount of plasmid DNA was used per transfection. After transfection for the indicated times, cells were lysed, and luciferase assays were performed using a luciferase assay kit (Promega). The transfection efficiency was normalized against Renilla luciferase activity. All assays were performed at least in triplicate.
Hyperinsulinemic–euglycemic clamping
Hyperinsulinemic–euglycemic clamping was performed as previously described (Ryu et al., 2013). In brief, after an overnight fast, a 2-h hyperinsulinemic–euglycemic clamp was performed in 8-wk-old male MKO mice and age-matched littermate controls fed a chow diet. In addition, MKO and control mice fed an HFD for 6 wk starting at 5 wk of age were subjected to a hyperinsulinemic–euglycemic clamp at 11 wk of age. The insulin clamp began with a primed-continuous infusion of 0.3 U/kg insulin. Blood samples were collected at 20-min intervals for immediate measurement of plasma glucose concentrations, and 20% glucose was infused at variable rates to maintain glucose at basal concentrations (∼120 mg/dl). Basal and insulin-stimulated whole-body glucose uptake was estimated with a continuous infusion of 3H-glucose (PerkinElmer) for 2 h before clamping (0.05 µCi/min) and throughout clamping (0.1 CuI/min), respectively. At 75 min after the start of the clamp, 2-deoxy-d-1-14C glucose (PerkinElmer) was injected with a Hamilton syringe to measure insulin-stimulated glucose uptake. To determine 3H-glucose and 2-deoxy-d-1-14C concentrations, plasma samples were deproteinized with ZnSO4 and Ba(OH)2, dried, resuspended in water, and counted in scintillation fluid for detection of 3H and 14C.
Histological analysis
All tissue samples were obtained from 8-wk-old male Ctrl and MKO mice, Ctrl and MKO mice fed an HFD for 6 wk, or GDF15- and vehicle-treated ob/ob mice. Samples for light microscopy were fixed in 4% PFA for 1 h. Paraffin embedding, sectioning, and hematoxylin and eosin and oil red O staining were performed according to standard protocols.
Transmission electron microscopy
Tissue samples from mice were fixed in 1% (wt/vol) glutaraldehyde at 4°C and then washed five times with 0.1 M cacodylate buffer, pH 7.2, at 4°C. Washed tissues were fixed for 1 h at 4°C with 1% (wt/vol) OsO4 in 0.1 M cacodylate buffer, pH 7.2, containing 0.1% (wt/vol) CaCl2. Samples were dehydrated by ethanol series and propylene oxide treatment, and then embedded in Embed-812 (Electron Microscopy Sciences). The resin was then polymerized at 60°C for 36 h. Tissues were sectioned using an EM UC6 ultramicrotome (Leica Biosystems) and stained with 4% (wt/vol) uranyl acetate and citrate. Specimens were observed on a JEM ARM 1300S high-voltage electron microscope (JEOL).
Western blot analysis
Western blot analysis was performed according to standard methods using commercially available antibodies. Primary antibodies against mitochondrial OXPHOS complex subunits (NDUFA9, SDHA, UQCRC2, and COX1) were purchased from Invitrogen. Anti-CRIF1 (sc-374122) and anti-GDF15 (sc-66905) were purchased from Santa Cruz Biotechnology, Inc. Anti–phospho-p44/42 MAPK (Erk1/2; Thr202/Tyr204; 4370), anti–p44/42 MAPK (9102), anti–phospho SMAD2 (Ser465/467; 3108), anti-SMAD2 (5339), anti–phospho-SMAD3 (Ser423/425; 9520), anti-SMAD3 (9523), anti-ATGL (2138), anti-phospho HSL (Ser660; 4126), anti-HSL (4107), anti-CHOP (2895), anti-p38 MAPK (9212), anti–phospho-p38 MAPK (Thr180/Tyr182; 9211), and anti-GAPDH (2118) were obtained from Cell Signaling Technology. Anti-LONP1 (ab103809), anti-CLPP (ab124822), and anti-HSPD1 (ab46798) were purchased from Abcam. Images were scanned on an ODYSSEY instrument and quantitated using Image Studio Software (LI-COR Biosciences).
Isolation of RNA and analysis by real-time PCR
Total RNA was isolated using TRIzol reagent (Thermo Fisher Scientific), and cDNA was prepared from total RNA using M-MLV reverse transcription and oligo-dT primers (Invitrogen). The resultant cDNA was amplified on a 7500 Fast Real-Time PCR System (Applied Biosystems). Real-time PCR was performed in triplicate with individual time-matched vehicle-treated or control mice using 2× SYBR Green Mix (Applied Biosystems). All quantitative calculations were performed using the ΔΔCT method. All mouse primer sequences are listed in Table S1.
RNA-seq data and analysis
Total RNA was prepared from EDL muscle of 8-wk-old male and MKO mice using TRIzol (Invitrogen) reagent, and poly(A) RNA was extracted with Dynabeads Oligo(dT)25 (Ambion). Sequencing libraries were prepared using the Epicentre ScriptSeq v2 and then subjected to 100-nt paired-end sequencing on an Illumina HiSeq2000 (Illumina). The libraries were quantitated by qPCR according to the qPCR Quantification Protocol Guide, and quality control was performed on an Agilent Technologies 2100 Bioanalyzer. Reads were processed from the sequencer and aligned to the Mus musculus reference genome (mm10) using TopHat v2.0.13 (Trapnell et al., 2009). The reference genome and annotation data were downloaded from the UCSC Genome Browser (http://genome.uscs.edu). Gene annotation information was used to run TopHat with the “-G” option. After alignment of reads to the genome, Cufflinks v2.2.1 (Trapnell et al., 2010) was used to assemble aligned reads into transcripts and estimate their abundance. To correct sequence expression count bias, the “–max-bundle-frags 50000000” option was used. The transcript counts in isoform level were calculated, and relative transcript abundances were measured in FPKM (fragments per kilobase of exon per million fragments mapped) using Cufflinks, as well as gene level expression values. Raw data were calculated as FPKM of each gene in each sample using the Cufflinks software. Genes with more than two zero FPKM values were excluded. To facilitate log2 transformation, 1 was added to each FPKM value of filtered genes. Filtered data were log2-transformed and subjected to quantile normalization. The resultant values were used for DEG analysis. A fold change of two or more was deemed significant.
IPGTT and insulin tolerance tests
For the IPGTT, mice were fasted for 16 h, and then 1 or 2 g glucose per kilogram of body weight was injected into the intraperitoneal cavity. Blood glucose levels were measured at 0, 15, 30, 60, 90, and 120 min using a glucometer (ACCU-CHEK; Roche). The insulin tolerance test was performed by measuring blood glucose after 6 h of fasting followed by intraperitoneal injection of 0.75 U/kg insulin (Humalog).
Indirect calorimetry
Mice fed either standard chow or an HFD were placed into individual metabolic chambers, with free access to food and water. VO2, VCO2, EE (calculated according to the following formula: 1.44 × VO2 × (3.815 + 1.232 × respiratory quotient)), and ambulatory activity were determined by indirect calorimetry using a Physical cage system (Oxylet; Panlab). VO2 and VCO2 were measured over a 24-h period. Data were analyzed using METABOLISM software, version 2.2. (Oxylet; Panlab). Mean values during light and dark periods were calculated. Student’s t test was used to calculate p-values. Data distribution was assumed to be normal, but this was not formally tested.
In vitro lipolysis assay
3T3L1 cells were seeded in 96-well plates at 103 cells per well and allowed to differentiate for 5 d, after which the medium was replaced with Krebs-Ringer bicarbonate buffer supplemented with 25 mM Hepes and 1% BSA. Samples were treated at 37°C for 3 h with vehicle or recombinant GDF15 (R&D Systems) at a final concentration of 100–300 ng/ml. Glycerol released from adipose tissues was measured using the Free Glycerol Reagent (Sigma-Aldrich) and on a SpectraMax Gemini spectrophotometer (Molecular Devices).
Ex vivo FAO assay
Fresh tissues (eWAT, liver, and BAT, 100–300 mg) were immediately placed in ice-cold medium containing 250 mM sucrose, 1 mM EDTA, and 10 mM Tris-HCl, pH 7.4. Tissues were rinsed to remove any blood and then homogenized in a glass homogenization tube with a motor-driven Teflon pestle. Diluted tissue homogenates were preincubated with a 95% O2–5% CO2 mixture at 30°C for 15 min. Reaction mixture (100 mM sucrose, 10 mM Tris-HCl, 5 mM KH2PO4, 0.2 mM EDTA, 7.5% fatty acid–free BSA, 80 mM KCl, 1 mM MgCl2, 2 mM L-carnitine, 0.1 mM malate, 0.05 mM coenzyme A, 1 mM DTT, and 2 mM ATP) was then added to the preincubated tissue homogenates. The substrate was 0.5 mM [1-14C] 0.5 µCi palmitate with 0.7% BSA. After 60 min of incubation at 37°C, 4 N sulfuric acid was injected to stop the reaction. CO2 produced during the 1-h incubation was trapped with 2 M sodium hydroxide. After acid treatment, the reaction supernatants and 14CO2 trap were transferred to scintillation fluid, and 14C radioactivity was counted using a Tri-Carb 2900TR by liquid scintillation analyzer (PerkinElmer).
Bioinformatic analyses
An unbiased bioinformatic analysis (GSEA; http://www.broadinstitute.org/gsea) was performed using skeletal muscle transcriptome data from BXD genetic reference population mice as described in previous studies (Subramanian et al., 2005; Ryu et al., 2014). In brief, Gdf15 expression was analyzed in skeletal muscle transcriptomes of both normal chow- and HFD-fed BXD mice (GeneNetwork accession numbers GN379 and GN380). Next, the transcriptomes of 10 HFD-fed BXD mouse lines exhibiting the largest alterations (highlighted in red and blue, Fig. 3 B) in Gdf15 expression were subjected to GSEA. Heat maps (Fig. 3 B, left) were built using GENE-E (The Broad Institute; http://www.broadinstitute.org/cancer/software/GENE-E/). Correlation and principal-component analyses (Fig. 5, F–H) were performed using the appropriate functions and packages in R (http://www.r-project.org). The depth of shading at the correlation matrices (corrgram) indicates the magnitude of the correlation (Pearson’s R). Positive and negative correlations are represented in blue and red, respectively. All raw data used in GSEA are available on the GeneNetwork website (http://www.genenetwork.org). Parameters for all assays were as follows. Activity: locomotor activity for 24 h (counts per hour), standard housing cage at 23 wk of age, HFD, males (n = 52, GeneNetwork record ID 17714). RER imp: RER improvement (ratio) after 10 d of voluntary exercise, treadmill at 10° incline, chow diet, males (n = 49, GeneNetwork record ID 17749). Glucose: serum glucose level (millimoles per liter) at 70 d in males fed an HFD from 4 wk (n = 32, GeneNetwork record ID 15045). Food in: food intake [g] in 24 h at 16 wk of age, metabolic cage, chow diet, males (n = 36, 24 h at 16 wk of age, metabolic cage 17633). Hep Lip: hepatic lipid (ratio of total lipid to total protein) measured by Fourier transform infrared spectroscopy in liver at 140 d in males fed an HFD from 4 wk (n = 31, GeneNetwork record ID 15095). Liver we: liver weight (grams) at 29 wk of age, chow diet, males (n = 50, GeneNetwork record ID 17767). pWAT: peritoneal adipose tissue weight (grams) at 20 wk in females and males fed an HFD from 4 wk (n = 32, GeneNetwork record ID 14981). sWAT: residual subcutaneous fat weight (grams) at 20 wk in females and males on HFD feeding from 4 wk (n = 32, GeneNetwork record ID 14992). Fat mass: total white adipose weight (grams) at 20 wk in the whole body in females and males on HFD feeding from 4 wk (n = 32, GeneNetwork record ID 14987). Bd We: body weight gain (grams) between 9 and 10 wk in females and males fed an HFD from 4 wk (n = 32, GeneNetwork record ID 14952).
Statistical analysis
Statistical analyses were performed using Stat Graph Prism 5 (GraphPad). Data are reported as mean ± SEM. All data from animal studies were analyzed by two-way repeated-measures analysis of variance followed by Bonferroni’s correction for multiple comparison, one-way analysis of variance followed by Tukey post hoc test, or two-tailed Student’s t test. A p-value <0.05 was considered statistically significant.
Online supplemental material
Fig. S1 shows muscle phenotypes and metabolic parameters of MKO mice, demonstrating that Crif1 LOF leads to sarcopenia and reduced muscle function after 12 wk. Fig. S2 shows that MKO mice fed an HFD exhibited elevated EE and reduced muscle and fat mass. Fig. S3 shows that GDF15 promoter activity is independent of ATF4/5/6 in Crif1-deficient MEF cells. GDF15 and FGF21 were induced in Crif1-deficient MEF cells and MKO mice. Fig. S4 shows that rGDF15 treatment in ob/ob mice leads to elevated EE. Table S1 shows a list of primer sets used for quantitative RT-PCR analyses.
Supplementary Material
Acknowledgments
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Science, ICT and Future Planning, Korea (NRF-2015R1A2A1A13000951 and NRF-2014M3A9D8034464). H.K. Chung was supported by an NRF grant from the Ministry of Education, Korea (NRF-2013R1A1A2058889). J. Auwerx was supported by grants from the École Polytechnique Fédérale de Lausanne, the Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (31003A-140780), the Aging X program of the Swiss Initiative for Systems Biology (51RTP0-151019), the Krebsliga Schweiz (KFS-3082-02-2013), and the National Institutes of Health (R01AG043930). R.M. Evans is an Investigator of the Howard Hughes Medical Institute at the Salk Institute and March of Dimes Chair Foundation in Molecular and Developmental Biology and is supported by the National Institutes of Health (grants DK057978, DK090962, HL088093, HL105278, and ES010337), the Glenn Foundation for Medical Research, and the Leona M. and Harry B. Helmsley Charitable Trust (grant 2012-PG-MED002).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- ALK
- activin receptor–like kinase
- BAT
- brown adipose tissue
- Ctrl
- control
- EDL
- extensor digitorum longus
- EE
- energy expenditure
- eWAT
- epididymal white adipose tissue
- FAO
- fatty acid oxidation
- FPKM
- fragments per kilobase of exon per million fragments mapped
- GDF
- growth differentiation factor
- GM
- gastrocnemius muscle
- GSEA
- gene set enrichment analysis
- HFD
- high-fat diet
- IPGTT
- intraperitoneal glucose tolerance test
- IPITT
- intraperitoneal insulin tolerance test
- LOF
- loss of function
- MEF
- mouse embryonic fibroblast
- mitoribosome
- mitochondrial ribosome
- MKO
- muscle-specific knockout
- OCR
- oxygen consumption rate
- POLG
- polymerase gamma mtDNA mutator
- RE
- responsive element
- RER
- respiratory exchange ratio
- RNA-seq
- RNA sequencing
- VCO2
- carbon dioxide production
- VO2
- oxygen consumption
- WAT
- white adipose tissue
References
- Ago T., and Sadoshima J.. 2006. GDF15, a cardioprotective TGF-beta superfamily protein. Circ. Res. 98:294–297. 10.1161/01.RES.0000207919.83894.9d [DOI] [PubMed] [Google Scholar]
- Andreux P.A., Williams E.G., Koutnikova H., Houtkooper R.H., Champy M.F., Henry H., Schoonjans K., Williams R.W., and Auwerx J.. 2012. Systems genetics of metabolism: the use of the BXD murine reference panel for multiscalar integration of traits. Cell. 150:1287–1299. 10.1016/j.cell.2012.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein S.H., Venkatesh S., Li M., Lee J., Lu B., Hilchey S.P., Morse K.M., Metcalfe H.M., Skalska J., Andreeff M., et al. 2012. The mitochondrial ATP-dependent Lon protease: a novel target in lymphoma death mediated by the synthetic triterpenoid CDDO and its derivatives. Blood. 119:3321–3329. 10.1182/blood-2011-02-340075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bootcov M.R., Bauskin A.R., Valenzuela S.M., Moore A.G., Bansal M., He X.Y., Zhang H.P., Donnellan M., Mahler S., Pryor K., et al. 1997. MIC-1, a novel macrophage inhibitory cytokine, is a divergent member of the TGF-beta superfamily. Proc. Natl. Acad. Sci. USA. 94:11514–11519. 10.1073/pnas.94.21.11514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothe G.W., Haspel J.A., Smith C.L., Wiener H.H., and Burden S.J.. 2000. Selective expression of Cre recombinase in skeletal muscle fibers. Genesis. 26:165–166. [DOI] [PubMed] [Google Scholar]
- Chrysovergis K., Wang X., Kosak J., Lee S.H., Kim J.S., Foley J.F., Travlos G., Singh S., Baek S.J., and Eling T.E.. 2014. NAG-1/GDF-15 prevents obesity by increasing thermogenesis, lipolysis and oxidative metabolism. Int. J. Obes. 38:1555–1564. 10.1038/ijo.2014.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H.K., Kim Y.K., Park J.H., Ryu M.J., Chang J.Y., Hwang J.H., Lee C.H., Kim S.H., Kim H.J., Kweon G.R., et al. 2015. The indole derivative NecroX-7 improves nonalcoholic steatohepatitis in ob/ob mice through suppression of mitochondrial ROS/RNS and inflammation. Liver Int. 35:1341–1353. 10.1111/liv.12741 [DOI] [PubMed] [Google Scholar]
- Dani C. 2013. Activins in adipogenesis and obesity. Int. J. Obes. 37:163–166. 10.1038/ijo.2012.28 [DOI] [PubMed] [Google Scholar]
- Demontis F., Patel V.K., Swindell W.R., and Perrimon N.. 2014. Intertissue control of the nucleolus via a myokine-dependent longevity pathway. Cell Reports. 7:1481–1494. 10.1016/j.celrep.2014.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillin A., Hsu A.L., Arantes-Oliveira N., Lehrer-Graiwer J., Hsin H., Fraser A.G., Kamath R.S., Ahringer J., and Kenyon C.. 2002. Rates of behavior and aging specified by mitochondrial function during development. Science. 298:2398–2401. 10.1126/science.1077780 [DOI] [PubMed] [Google Scholar]
- Dogan S.A., Pujol C., Maiti P., Kukat A., Wang S., Hermans S., Senft K., Wibom R., Rugarli E.I., and Trifunovic A.. 2014. Tissue-specific loss of DARS2 activates stress responses independently of respiratory chain deficiency in the heart. Cell Metab. 19:458–469. 10.1016/j.cmet.2014.02.004 [DOI] [PubMed] [Google Scholar]
- Dominguez-Rodriguez A., Abreu-Gonzalez P., and Avanzas P.. 2014. Usefulness of growth differentiation factor-15 levels to predict diabetic cardiomyopathy in asymptomatic patients with type 2 diabetes mellitus. Am. J. Cardiol. 114:890–894. 10.1016/j.amjcard.2014.06.020 [DOI] [PubMed] [Google Scholar]
- Dong B., Zhou H., Han C., Yao J., Xu L., Zhang M., Fu Y., and Xia Q.. 2014. Ischemia/reperfusion-induced CHOP expression promotes apoptosis and impairs renal function recovery: the role of acidosis and GPR4. PLoS One. 9:e110944 10.1371/journal.pone.0110944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas J.F., Simard G., Flamment M., Ducluzeau P.H., and Ritz P.. 2009. Is skeletal muscle mitochondrial dysfunction a cause or an indirect consequence of insulin resistance in humans? Diabetes Metab. 35:159–167. 10.1016/j.diabet.2009.02.002 [DOI] [PubMed] [Google Scholar]
- Durieux J., Wolff S., and Dillin A.. 2011. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 144:79–91. 10.1016/j.cell.2010.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Bussière F., and Hekimi S.. 2001. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev. Cell. 1:633–644. 10.1016/S1534-5807(01)00071-5 [DOI] [PubMed] [Google Scholar]
- Fujita Y., Ito M., Kojima T., Yatsuga S., Koga Y., and Tanaka M.. 2015. GDF15 is a novel biomarker to evaluate efficacy of pyruvate therapy for mitochondrial diseases. Mitochondrion. 20:34–42. 10.1016/j.mito.2014.10.006 [DOI] [PubMed] [Google Scholar]
- Gleyzer N., and Scarpulla R.C.. 2011. PGC-1-related coactivator (PRC), a sensor of metabolic stress, orchestrates a redox-sensitive program of inflammatory gene expression. J. Biol. Chem. 286:39715–39725. 10.1074/jbc.M111.291575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber B.J., Boehringer D., Leibundgut M., Bieri P., Leitner A., Schmitz N., Aebersold R., and Ban N.. 2014a The complete structure of the large subunit of the mammalian mitochondrial ribosome. Nature. 515:283–286. 10.1038/nature13895 [DOI] [PubMed] [Google Scholar]
- Greber B.J., Boehringer D., Leitner A., Bieri P., Voigts-Hoffmann F., Erzberger J.P., Leibundgut M., Aebersold R., and Ban N.. 2014b Architecture of the large subunit of the mammalian mitochondrial ribosome. Nature. 505:515–519. 10.1038/nature12890 [DOI] [PubMed] [Google Scholar]
- Greber B.J., Bieri P., Leibundgut M., Leitner A., Aebersold R., Boehringer D., and Ban N.. 2015. Ribosome. The complete structure of the 55S mammalian mitochondrial ribosome. Science. 348:303–308. 10.1126/science.aaa3872 [DOI] [PubMed] [Google Scholar]
- Han D.H., Hancock C.R., Jung S.R., Higashida K., Kim S.H., and Holloszy J.O.. 2011. Deficiency of the mitochondrial electron transport chain in muscle does not cause insulin resistance. PLoS One. 6:e19739 10.1371/journal.pone.0019739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto O., and Funaba M.. 2011. Activin in glucose metabolism. Vitam. Horm. 85:217–234. 10.1016/B978-0-12-385961-7.00011-1 [DOI] [PubMed] [Google Scholar]
- Hesselink M.K., Schrauwen-Hinderling V., and Schrauwen P.. 2016. Skeletal muscle mitochondria as a target to prevent or treat type 2 diabetes mellitus. Nat. Rev. Endocrinol. 12:633–645. 10.1038/nrendo.2016.104 [DOI] [PubMed] [Google Scholar]
- Horibe T., and Hoogenraad N.J.. 2007. The chop gene contains an element for the positive regulation of the mitochondrial unfolded protein response. PLoS One. 2:e835 10.1371/journal.pone.0000835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper R.H., Mouchiroud L., Ryu D., Moullan N., Katsyuba E., Knott G., Williams R.W., and Auwerx J.. 2013. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 497:451–457. 10.1038/nature12188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelenik T., and Roden M.. 2013. Mitochondrial plasticity in obesity and diabetes mellitus. Antioxid. Redox Signal. 19:258–268. 10.1089/ars.2012.4910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen M.B., and Jasper H.. 2014. Mitochondrial proteostasis in the control of aging and longevity. Cell Metab. 20:214–225. 10.1016/j.cmet.2014.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempf T., Eden M., Strelau J., Naguib M., Willenbockel C., Tongers J., Heineke J., Kotlarz D., Xu J., Molkentin J.D., et al. 2006. The transforming growth factor-beta superfamily member growth-differentiation factor-15 protects the heart from ischemia/reperfusion injury. Circ. Res. 98:351–360. 10.1161/01.RES.0000202805.73038.48 [DOI] [PubMed] [Google Scholar]
- Kempf T., Zarbock A., Widera C., Butz S., Stadtmann A., Rossaint J., Bolomini-Vittori M., Korf-Klingebiel M., Napp L.C., Hansen B., et al. 2011. GDF-15 is an inhibitor of leukocyte integrin activation required for survival after myocardial infarction in mice. Nat. Med. 17:581–588. 10.1038/nm.2354 [DOI] [PubMed] [Google Scholar]
- Kempf T., Guba-Quint A., Torgerson J., Magnone M.C., Haefliger C., Bobadilla M., and Wollert K.C.. 2012. Growth differentiation factor 15 predicts future insulin resistance and impaired glucose control in obese nondiabetic individuals: results from the XENDOS trial. Eur. J. Endocrinol. 167:671–678. 10.1530/EJE-12-0466 [DOI] [PubMed] [Google Scholar]
- Kim K.H., Jeong Y.T., Oh H., Kim S.H., Cho J.M., Kim Y.N., Kim S.S., Kim D.H., Hur K.Y., Kim H.K., et al. 2013. Autophagy deficiency leads to protection from obesity and insulin resistance by inducing Fgf21 as a mitokine. Nat. Med. 19:83–92. 10.1038/nm.3014 [DOI] [PubMed] [Google Scholar]
- Kim S.J., Kwon M.C., Ryu M.J., Chung H.K., Tadi S., Kim Y.K., Kim J.M., Lee S.H., Park J.H., Kweon G.R., et al. 2012. CRIF1 is essential for the synthesis and insertion of oxidative phosphorylation polypeptides in the mammalian mitochondrial membrane. Cell Metab. 16:274–283. 10.1016/j.cmet.2012.06.012 [DOI] [PubMed] [Google Scholar]
- Kim Y.K., Joung K.H., Ryu M.J., Kim S.J., Kim H., Chung H.K., Lee M.H., Lee S.E., Choi M.J., Chang J.Y., et al. 2015. Disruption of CR6-interacting factor-1 (CRIF1) in mouse islet beta cells leads to mitochondrial diabetes with progressive beta cell failure. Diabetologia. 58:771–780. 10.1007/s00125-015-3506-y [DOI] [PubMed] [Google Scholar]
- Kolesar J.E., Safdar A., Abadi A., MacNeil L.G., Crane J.D., Tarnopolsky M.A., and Kaufman B.A.. 2014. Defects in mitochondrial DNA replication and oxidative damage in muscle of mtDNA mutator mice. Free Radic. Biol. Med. 75:241–251. 10.1016/j.freeradbiomed.2014.07.038 [DOI] [PubMed] [Google Scholar]
- Kwon M.C., Koo B.K., Moon J.S., Kim Y.Y., Park K.C., Kim N.S., Kwon M.Y., Kong M.P., Yoon K.J., Im S.K., et al. 2008. Crif1 is a novel transcriptional coactivator of STAT3. EMBO J. 27:642–653. 10.1038/sj.emboj.7601986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macia L., Tsai V.W., Nguyen A.D., Johnen H., Kuffner T., Shi Y.C., Lin S., Herzog H., Brown D.A., Breit S.N., and Sainsbury A.. 2012. Macrophage inhibitory cytokine 1 (MIC-1/GDF15) decreases food intake, body weight and improves glucose tolerance in mice on normal and obesogenic diets. PLoS One. 7:e34868 10.1371/journal.pone.0034868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moisoi N., Klupsch K., Fedele V., East P., Sharma S., Renton A., Plun-Favreau H., Edwards R.E., Teismann P., Esposti M.D., et al. 2009. Mitochondrial dysfunction triggered by loss of HtrA2 results in the activation of a brain-specific transcriptional stress response. Cell Death Differ. 16:449–464. 10.1038/cdd.2008.166 [DOI] [PubMed] [Google Scholar]
- Morino K., Petersen K.F., and Shulman G.I.. 2006. Molecular mechanisms of insulin resistance in humans and their potential links with mitochondrial dysfunction. Diabetes. 55(Suppl. 2):S9–S15. 10.2337/db06-S002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargund A.M., Fiorese C.J., Pellegrino M.W., Deng P., and Haynes C.M.. 2015. Mitochondrial and nuclear accumulation of the transcription factor ATFS-1 promotes OXPHOS recovery during the UPR(mt). Mol. Cell. 58:123–133. 10.1016/j.molcel.2015.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J., and Suomalainen A.. 2012. Mitochondria: in sickness and in health. Cell. 148:1145–1159. 10.1016/j.cell.2012.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien T.W. 1971. The general occurrence of 55 S ribosomes in mammalian liver mitochondria. J. Biol. Chem. 246:3409–3417. [PubMed] [Google Scholar]
- Ost M., Coleman V., Voigt A., van Schothorst E.M., Keipert S., van der Stelt I., Ringel S., Graja A., Ambrosi T., Kipp A.P., et al. 2015. Muscle mitochondrial stress adaptation operates independently of endogenous FGF21 action. Mol. Metab. 5:79–90. 10.1016/j.molmet.2015.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu-Ansah E., Song W., and Perrimon N.. 2013. Muscle mitohormesis promotes longevity via systemic repression of insulin signaling. Cell. 155:699–712. 10.1016/j.cell.2013.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagel-Langenickel I., Bao J., Pang L., and Sack M.N.. 2010. The role of mitochondria in the pathophysiology of skeletal muscle insulin resistance. Endocr. Rev. 31:25–51. 10.1210/er.2009-0003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino M.W., Nargund A.M., Kirienko N.V., Gillis R., Fiorese C.J., and Haynes C.M.. 2014. Mitochondrial UPR-regulated innate immunity provides resistance to pathogen infection. Nature. 516:414–417. 10.1038/nature13818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry R.J., Kim T., Zhang X.M., Lee H.Y., Pesta D., Popov V.B., Zhang D., Rahimi Y., Jurczak M.J., Cline G.W., et al. 2013. Reversal of hypertriglyceridemia, fatty liver disease, and insulin resistance by a liver-targeted mitochondrial uncoupler. Cell Metab. 18:740–748. 10.1016/j.cmet.2013.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry R.J., Zhang D., Zhang X.M., Boyer J.L., and Shulman G.I.. 2015. Controlled-release mitochondrial protonophore reverses diabetes and steatohepatitis in rats. Science. 347:1253–1256. 10.1126/science.aaa0672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen K.F., Befroy D., Dufour S., Dziura J., Ariyan C., Rothman D.L., DiPietro L., Cline G.W., and Shulman G.I.. 2003. Mitochondrial dysfunction in the elderly: possible role in insulin resistance. Science. 300:1140–1142. 10.1126/science.1082889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pospisilik J.A., Knauf C., Joza N., Benit P., Orthofer M., Cani P.D., Ebersberger I., Nakashima T., Sarao R., Neely G., et al. 2007. Targeted deletion of AIF decreases mitochondrial oxidative phosphorylation and protects from obesity and diabetes. Cell. 131:476–491. 10.1016/j.cell.2007.08.047 [DOI] [PubMed] [Google Scholar]
- Preusch M.R., Baeuerle M., Albrecht C., Blessing E., Bischof M., Katus H.A., and Bea F.. 2013. GDF-15 protects from macrophage accumulation in a mouse model of advanced atherosclerosis. Eur. J. Med. Res. 18:19 10.1186/2047-783X-18-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintens R., Singh S., Lemaire K., De Bock K., Granvik M., Schraenen A., Vroegrijk I.O., Costa V., Van Noten P., Lambrechts D., et al. 2013. Mice deficient in the respiratory chain gene Cox6a2 are protected against high-fat diet-induced obesity and insulin resistance. PLoS One. 8:e56719 10.1371/journal.pone.0056719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirós P.M., Langer T., and López-Otín C.. 2015. New roles for mitochondrial proteases in health, ageing and disease. Nat. Rev. Mol. Cell Biol. 16:345–359. 10.1038/nrm3984 [DOI] [PubMed] [Google Scholar]
- Rebbapragada A., Benchabane H., Wrana J.L., Celeste A.J., and Attisano L.. 2003. Myostatin signals through a transforming growth factor beta-like signaling pathway to block adipogenesis. Mol. Cell. Biol. 23:7230–7242. 10.1128/MCB.23.20.7230-7242.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu D., Jo Y.S., Lo Sasso G., Stein S., Zhang H., Perino A., Lee J.U., Zeviani M., Romand R., Hottiger M.O., et al. 2014. A SIRT7-dependent acetylation switch of GABPβ1 controls mitochondrial function. Cell Metab. 20:856–869. 10.1016/j.cmet.2014.08.001 [DOI] [PubMed] [Google Scholar]
- Ryu M.J., Kim S.J., Kim Y.K., Choi M.J., Tadi S., Lee M.H., Lee S.E., Chung H.K., Jung S.B., Kim H.J., et al. 2013. Crif1 deficiency reduces adipose OXPHOS capacity and triggers inflammation and insulin resistance in mice. PLoS Genet. 9:e1003356 10.1371/journal.pgen.1003356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samocha-Bonet D., Heilbronn L.K., Lichtenberg D., and Campbell L.V.. 2010. Does skeletal muscle oxidative stress initiate insulin resistance in genetically predisposed individuals? Trends Endocrinol. Metab. 21:83–88. 10.1016/j.tem.2009.09.008 [DOI] [PubMed] [Google Scholar]
- Schönthal A.H. 2013. Pharmacological targeting of endoplasmic reticulum stress signaling in cancer. Biochem. Pharmacol. 85:653–666. 10.1016/j.bcp.2012.09.012 [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomalainen A., Elo J.M., Pietiläinen K.H., Hakonen A.H., Sevastianova K., Korpela M., Isohanni P., Marjavaara S.K., Tyni T., Kiuru-Enari S., et al. 2011. FGF-21 as a biomarker for muscle-manifesting mitochondrial respiratory chain deficiencies: a diagnostic study. Lancet Neurol. 10:806–818. 10.1016/S1474-4422(11)70155-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao H., Zhang Y., Zeng X., Shulman G.I., and Jin S.. 2014. Niclosamide ethanolamine-induced mild mitochondrial uncoupling improves diabetic symptoms in mice. Nat. Med. 20:1263–1269. 10.1038/nm.3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towler M.C., and Hardie D.G.. 2007. AMP-activated protein kinase in metabolic control and insulin signaling. Circ. Res. 100:328–341. 10.1161/01.RES.0000256090.42690.05 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., and Salzberg S.L.. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 25:1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., and Pachter L.. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28:511–515. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifunovic A., Wredenberg A., Falkenberg M., Spelbrink J.N., Rovio A.T., Bruder C.E., Bohlooly-Y M., Gidlöf S., Oldfors A., Wibom R., et al. 2004. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 429:417–423. 10.1038/nature02517 [DOI] [PubMed] [Google Scholar]
- Tyynismaa H., Carroll C.J., Raimundo N., Ahola-Erkkilä S., Wenz T., Ruhanen H., Guse K., Hemminki A., Peltola-Mjøsund K.E., Tulkki V., et al. 2010. Mitochondrial myopathy induces a starvation-like response. Hum. Mol. Genet. 19:3948–3958. 10.1093/hmg/ddq310 [DOI] [PubMed] [Google Scholar]
- Ueda Y. 2000. Activin A increases Pax4 gene expression in pancreatic beta cell lines. FEBS Lett. 480:101–105. 10.1016/S0014-5793(00)01914-1 [DOI] [PubMed] [Google Scholar]
- Vaňhara P., Hampl A., Kozubík A., and Souček K.. 2012. Growth/differentiation factor-15: prostate cancer suppressor or promoter? Prostate Cancer Prostatic Dis. 15:320–328. 10.1038/pcan.2012.6 [DOI] [PubMed] [Google Scholar]
- Wall C.E., Whyte J., Suh J.M., Fan W., Collins B., Liddle C., Yu R.T., Atkins A.R., Naviaux J.C., Li K., et al. 2015. High-fat diet and FGF21 cooperatively promote aerobic thermogenesis in mtDNA mutator mice. Proc. Natl. Acad. Sci. USA. 112:8714–8719. 10.1073/pnas.1509930112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.Z., and Ron D.. 1996. Stress-induced phosphorylation and activation of the transcription factor CHOP (GADD153) by p38 MAP Kinase. Science. 272:1347–1349. 10.1126/science.272.5266.1347 [DOI] [PubMed] [Google Scholar]
- Wang X., Baek S.J., and Eling T.E.. 2013. The diverse roles of nonsteroidal anti-inflammatory drug activated gene (NAG-1/GDF15) in cancer. Biochem. Pharmacol. 85:597–606. 10.1016/j.bcp.2012.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Chrysovergis K., Kosak J., Kissling G., Streicker M., Moser G., Li R., and Eling T.E.. 2014. hNAG-1 increases lifespan by regulating energy metabolism and insulin/IGF-1/mTOR signaling. Aging (Albany, N.Y.). 6:690–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wredenberg A., Freyer C., Sandström M.E., Katz A., Wibom R., Westerblad H., and Larsson N.G.. 2006. Respiratory chain dysfunction in skeletal muscle does not cause insulin resistance. Biochem. Biophys. Res. Commun. 350:202–207. 10.1016/j.bbrc.2006.09.029 [DOI] [PubMed] [Google Scholar]
- Wu Y., Williams E.G., Dubuis S., Mottis A., Jovaisaite V., Houten S.M., Argmann C.A., Faridi P., Wolski W., Kutalik Z., et al. 2014. Multilayered genetic and omics dissection of mitochondrial activity in a mouse reference population. Cell. 158:1415–1430. 10.1016/j.cell.2014.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Kimball T.R., Lorenz J.N., Brown D.A., Bauskin A.R., Klevitsky R., Hewett T.E., Breit S.N., and Molkentin J.D.. 2006. GDF15/MIC-1 functions as a protective and antihypertrophic factor released from the myocardium in association with SMAD protein activation. Circ. Res. 98:342–350. 10.1161/01.RES.0000202804.84885.d0 [DOI] [PubMed] [Google Scholar]
- Yokoyama-Kobayashi M., Saeki M., Sekine S., and Kato S.. 1997. Human cDNA encoding a novel TGF-beta superfamily protein highly expressed in placenta. J. Biochem. 122:622–626. 10.1093/oxfordjournals.jbchem.a021798 [DOI] [PubMed] [Google Scholar]
- Zhao Q., Wang J., Levichkin I.V., Stasinopoulos S., Ryan M.T., and Hoogenraad N.J.. 2002. A mitochondrial specific stress response in mammalian cells. EMBO J. 21:4411–4419. 10.1093/emboj/cdf445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Zheng X., Wang X., Ma Z., Gupta Sunkari V., Botusan I., Takeda T., Björklund A., Inoue M., Catrina S.B., et al. 2012. Acute hypoxia induces apoptosis of pancreatic β-cell by activation of the unfolded protein response and upregulation of CHOP. Cell Death Dis. 3:e322 10.1038/cddis.2012.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.