Monachino et al. review recent developments in single-molecule biophysical approaches and the cell biological advances they allow.
Abstract
Single-molecule manipulation and imaging techniques have become important elements of the biologist’s toolkit to gain mechanistic insights into cellular processes. By removing ensemble averaging, single-molecule methods provide unique access to the dynamic behavior of biomolecules. Recently, the use of these approaches has expanded to the study of complex multiprotein systems and has enabled detailed characterization of the behavior of individual molecules inside living cells. In this review, we provide an overview of the various force- and fluorescence-based single-molecule methods with applications both in vitro and in vivo, highlighting these advances by describing their applications in studies on cytoskeletal motors and DNA replication. We also discuss how single-molecule approaches have increased our understanding of the dynamic behavior of complex multiprotein systems. These methods have shown that the behavior of multicomponent protein complexes is highly stochastic and less linear and deterministic than previously thought. Further development of single-molecule tools will help to elucidate the molecular dynamics of these complex systems both inside the cell and in solutions with purified components.
Introduction
Single-molecule approaches are transforming our understanding of cell biology. In the context of the living cell, proteins are found in various states of structural conformation and association in complexes, with the transitioning between states occurring in a seemingly chaotic fashion. Observing molecular properties at the single-molecule level allows characterization of subpopulations, the visualization of transient intermediates, and the acquisition of detailed kinetic information that would otherwise be hidden by the averaging over an ensemble of stochastically behaving constituents. Although the field is rapidly evolving, and many technical challenges still exist, methods to visualize individual proteins in purified systems, henceforth referred to as in vitro, contribute to a tremendous gain in mechanistic insight into many cellular processes. However, the comparatively low complexity of such in vitro experiments does not necessarily represent the physiology of the cell. Development of single-molecule tools has begun to enable the visualization of complex biochemical reactions with great resolution in the dynamic and crowded environment of the cell. In vitro single-molecule studies on reconstituted systems of high complexity are informing on how these systems may behave in a cellular environment, and live-cell single-molecule imaging is providing pictures of increasing clarity about the physiological relevance of pathways observed in vitro. This interplay between in vitro and in vivo assays will play a major role in future studies, with bottom-up and top-down approaches required to fill the gaps.
In this review, we provide an overview of the state of the field and discuss the main classes of single-molecule methods that have found applications in in vitro and in vivo studies. In particular, we describe the principles of both force- and fluorescence-based single-molecule methods, and we highlight how these approaches have increased our understanding of molecular machineries. Using recent work, we illustrate both the advances in methodology and new insights into the dynamic behavior of complex systems that they provide. To guide our review of the main technological developments and the biological breakthroughs they have allowed, in the context of what seems like an overwhelming amount of examples and applications, we focus on studies of the molecular motors that carry cellular cargo and the multiprotein complex involved in DNA replication, the replisome. Our focus on these studies merely represents an attempt to illustrate the methodological possibilities—the reader is advised to consult the many other excellent sources and reviews that discuss the use of single-molecule tools in other fields and systems.
Push, pull, poke, and prod: Mechanical single-molecule techniques
The folding of proteins into functional structures, the manner with which they undergo conformational transitions, and their interactions between binding partners are all complex processes that are strictly ruled by the shape of the free-energy landscapes describing the thermodynamics of the system. Theoretically, there is a huge number of possible 3D conformations that a one-dimensional sequence of amino acids can assume, each characterized by a specific free energy. However, a protein assumes only those states that minimize the free energy, with preference for the absolute minimum. Thus, the number of possible protein conformations is limited to very few, if not only one (Onuchic et al., 1997). The application of forces to these systems introduces well-defined changes to the energetics and enables a precise interrogation of the relevant interactions and processes. Single-molecule mechanical techniques have been developed to use small forces to controllably manipulate individual biomolecules so that molecular mechanisms can be investigated at a level of detail inaccessible with conventional ensemble-averaged assays. In this paper, we focus on three main classes of these methods: atomic force microscopy (AFM), optical tweezers (OT), and magnetic tweezers (MT). Each of these techniques works in a different force regimen, with these three techniques together covering a range from femto-Newtons (fN) to nano-Newtons (nN), providing experimental access to forces that are relevant to biochemical processes and reactions. More comprehensive reviews on each technique and applications can be found elsewhere (Greenleaf et al., 2007; Neuman and Nagy, 2008; Müller and Dufrêne, 2011; de Souza, 2012; Dulin et al., 2013; Robinson and van Oijen, 2013; Ando, 2014; Ando et al., 2014; Blehm and Selvin, 2014; Whited and Park, 2014; Lyubchenko and Shlyakhtenko, 2016).
AFM
AFM is a scanning probe microscopy technique that allows visualization of the surface topography of a sample at subnanometer resolution. It uses an atomically sharp tip on the free end of a projecting arm (called cantilever) to measure the height (z axis) at a specific (x,y) position (Fig. 1 A). In biological imaging applications, AFM is typically used in the so-called tapping mode with the cantilever oscillating at a frequency close to its mechanical resonance. In this way, interactions with the surface can be detected with great sensitivity without the tip in constant contact with the sample, thus eliminating dragging and frictional effects during the (x,y) scan and avoiding distortion of image data. Ultimately, the tapping mode helps to preserve the integrity of the soft biological sample and allows the visualization of biomolecules for periods up to hours (Santos et al., 2013).
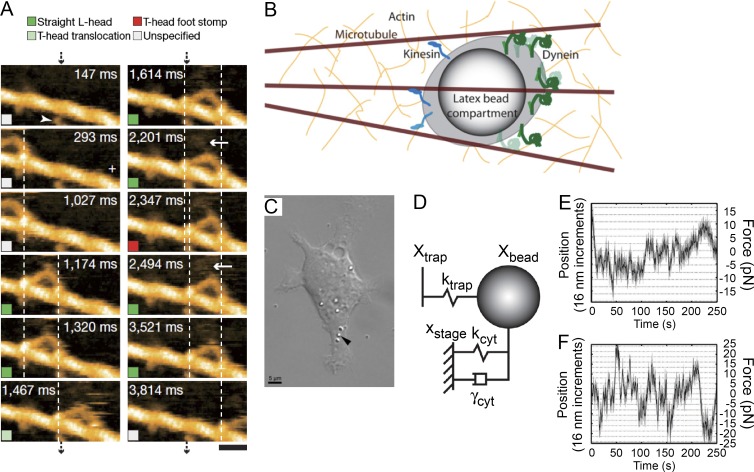
Figure 1.
Single-molecule approaches. (A) AFM. A tip is attached to a cantilever, with deflection of the tip or changes in its resonance frequency reporting on proximity to features on a cellular surface. By raster scanning the sample, an image of the 3D shape can be formed with subnanometer resolution. (B) OT. A functionalized bead is introduced into the cell. The bead is trapped and manipulated by a focused laser beam. (C) MT. Magnetic beads that specifically interact with a substrate of interest are introduced into the cell. By applying a magnetic field, the beads can be rotated or translated, thereby introducing a force to the system. (D) Fluorescence microscopy. Substrates of interest are labeled with a fluorescent tag. Their fluorescence is detected on a sensitive camera, allowing real-time visualization of spatiotemporal dynamics. (E) PAINT. This technique works by labeling a substrate that interacts transiently with a receptor. A low concentration of fluorescent ligands is introduced in the extracellular medium such that at a constant rate, receptors in the membrane are being visualized by short-lived fluorophore immobilization during the imaging sequence. (F and G) smFRET. (F) Two substrates of interest are labeled with two specific fluorescent tags (a donor–acceptor FRET pair). The emission of the donor tag spectrally overlaps with the absorption of the acceptor dye. The donor transfers its energy to the acceptor in a distance-dependent manner (FRET). An interaction between the two substrates will give a FRET signal, providing a dynamic observation of molecular interactions. (G) A molecule of interest is labeled with a FRET pair at known positions, one with a donor and the other with an acceptor. A change in the conformation of the substrate can be observed as a change in the FRET efficiency.
AFM was initially limited to the imaging of static structures, but the last decade has seen the introduction of even smaller cantilevers (Walters et al., 1996) and improvements in the image acquisition rate, making it possible to scan surfaces at high speed (high-speed AFM [HS-AFM]). HS-AFM is one of the few techniques so far that allows observation of biological molecules at both subnanometer and sub–100-ms resolution. This technical breakthrough has enabled real-time observation of molecular processes, such as the movement of motor proteins along cytoskeletal filaments, and has allowed the direct study of relationships between structural and dynamic properties of biochemical reactions, at the single-molecule level, with one single technique (Ando et al., 2008). This powerful and quite unique ability of HS-AFM to relate structure to function was highlighted in a hallmark study in which the walking of myosin V on actin was imaged (Fig. 2 A; Kodera et al., 2010). Not only did the high-speed imaging visualize the hand-over-hand mechanism of myosin V translocation, but the authors of this study were also able to explain the mechanism in structural terms. They showed that the forward movement of the myosin is a purely mechanical process related to the accumulation of tension in the leading head. Recently, a further technical improvement has allowed imaging of large fields of view at high speed and visualization of biochemical reactions occurring on the outer surfaces of cells (Watanabe et al., 2013). In vivo biological imaging with AFM offers several advantages over other techniques with high spatial resolution such as scanning EM. In particular, AFM does not require dehydration steps and can provide topographic images with nanometer resolution under physiological conditions (Essmann et al., 2016). These aspects position AFM as a technique with great potential to provide unique insight in various areas of cell biology such as membrane structure and dynamics, cell division, growth, and morphology. Finally, there have been attempts to bring AFM inside cells (Müller and Dufrêne, 2011), opening to the use of its high spatial and temporal resolution to observe fundamental cellular processes inside the cell itself.
Figure 2.
Force-based measurements on motor proteins. (A) Myosin V walking on actin was directly observed using high-speed AFM. The acquisition times are indicated on each frame. Bar, 30 nm. (A is adapted with permission from Kodera et al. [2010].) (B–F) The in vivo transport of intracellular cargoes and the associated forces were measured with OT. (B) Cartoon describing the experiment. Multiple copies of the motor proteins dynein and kinesin carry along microtubules a bead that has been internalized by the cell. The bead was optically trapped and its movement tracked. (C) Picture of a mouse macrophage cell with internalized polystyrene beads (arrowhead is pointing at one of the beads). (D) Diagram indicating the various contributions experienced by the bead because of the trapping force and viscous drag experienced inside the cytoplasm. (E and F) Example trajectories tracking the displacement of the bead with respect to the beam focus in living cells. (B–F are adapted from Hendricks et al. [2012].)
In addition to its topographic imaging applications, AFM is a powerful tool to perform force spectroscopy on single molecules in the 10 pN to 10 nN range. In this application, the tip of the AFM is used to capture one end of a biomolecule that is bound to a surface at its other end, apply a stretching force to it by moving the cantilever away from the surface, and thus unfolding it with a precise and controllable force (Li et al., 2002; Alegre-Cebollada et al., 2014). This approach makes it possible to probe the molecular interactions that stabilize the protein in a specific conformation. The alternative conformations of proteins, when subjected to mechanical forces inside the cell, can then be revealed (Alegre-Cebollada et al., 2014). Finally, by using different loading rates, researchers can model the kinetics of transitions and obtain details of the free-energy landscape controlling the various structural transitions (Whited and Park, 2014). An early example of AFM-based force spectroscopy involved the unfolding of the integral membrane protein bacteriorhodopsin out of archaeal purple membranes (Oesterhelt et al., 2000). Further, the role of ligands in stabilizing biomolecular structures can be assessed and quantified by mechanical unfolding. The interaction between a ligand and a protein affects the free-energy landscape of the system and potentially yields different unfolding profiles as a function of the ligand (Zocher et al., 2012). This approach is not limited to answer fundamental questions about cellular mechanisms, but also benefits applied research. For instance, researchers have been able to study in vivo membrane protein–ligand interactions to facilitate drug development (Zhang et al., 2012).
OT
In OT (also called optical traps), a tightly focused laser beam is diffracted by a dielectric particle, resulting in a force that traps the particle nearby the focus of the laser. At the same time, by changing the position of the focus, it is possible to move the particle, just as if the laser beam were a pair of tweezers. By tethering one end of a molecule of interest to the bead and the other end either to a surface or to a second trapped particle, a stretching force can be applied to the molecule in the 0.1–100 pN range. The applied force can be modulated by either changing the tightness of the trap or by moving the position of the particle with respect to the beam focus (Fig. 1 B). Tracking of the 3D displacement of the trapped particle allows measurements with subnanometer spatial resolution and submillisecond time resolution. Thanks to such precision, this technology has, for instance, enabled the visualization of the motion of motor proteins such as kinesins and dyneins along microtubules (Svoboda et al., 1993; Mallik et al., 2004), myosins along actin, and nucleic-acid enzymes along DNA (Abbondanzieri et al., 2005; Johnson et al., 2007).
Anytime lasers are used, photo damage to biological samples is a reason of concern. In the case of OT, this problem is minimized because biological samples are almost transparent to the near-infrared wavelengths of the lasers that are typically used to trap particles (Neuman et al., 1999). This compatibility with cellular specimens, combined with recently developed sophisticated force-calibration techniques (Blehm and Selvin, 2014; Jun et al., 2014), allows the use of OT in vivo and opens the possibility of studying the same biological system both in vitro and in vivo. Such hybrid approaches will be key in filling the gap between the mechanistic understanding obtained from in vitro reconstituted systems and biochemical reactions that occur in a cellular environment. This strategy has been very successful already in the characterization of the motor proteins kinesins, dyneins, and myosins (Holzbaur and Goldman, 2010; Blehm and Selvin, 2014; Bhabha et al., 2016). The Xie group played a pioneering role in the development and use of OT in vivo at the submillisecond time resolution needed to observe organelle transport (Nan et al., 2008; Sims and Xie, 2009). They reported that, in living human lung cancer cells, cargoes carried by kinesins make individual steps of 8 nm, whereas those carried by dyneins make individual steps of 8, 12, 16, 20, and 24 nm, providing new insight into the cooperative effects of multiple dyneins carrying the same cargo (Nan et al., 2008). They also observed that kinesins and dyneins both have a stall force of ∼7 to 8 pN (Sims and Xie, 2009). In a study by the Goldman group (Hendricks et al., 2012), it was shown that the force exerted by individual motors is the same both in vivo (in mouse macrophage cells) and in vitro. These researchers suggest, however, that the viscoelastic cell environment and the presence of cytoskeletal networks favor motor binding. By comparing in vitro with in vivo experiments, they propose that, in living macrophages, cargo is carried by as many as 12 dyneins and up to 3 kinesins in a tug-of-war mechanism (Fig. 2, B–F). A study by the Selvin laboratory (Blehm et al., 2013) characterized the transport of lipid vesicles and phagocytosed polystyrene beads in A549 human epithelial cells and in Dictyostelium discoideum, allowing them to propose that a single kinesin is sufficient to carry the cargo toward the periphery of the cell, whereas two to three dyneins are needed to transport the cargo toward the center. During outward motion, dyneins act as a drag on the kinesin–cargo translocation by pulling the cargo in the opposite direction. During inward motion, the kinesin is still bound to the cargo but not to the microtubule and therefore does not obstruct the action of the dynein (Blehm et al., 2013).
MT
MT are conceptually similar to OT: a magnetic field is used to trap a superparamagnetic bead that is bound to one end of the molecule of interest (Fig. 1 C). MT can apply forces between fractions of pN up to several hundreds of pN, depending on the experimental design. Importantly, unlike optical traps, MT can apply torque by making use of the fact that magnetic beads act as a dipole with a preferred orientation in the external magnetic field. By applying bright-field illumination and using the interference patterns of the individual beads to provide information on their position with respect to the focal plane, the movement of the beads can be tracked with nanometer resolution. The large homogeneity of magnetic fields allows tracking of hundreds of beads simultaneously, a throughput difficult or impossible to achieve with OT. Moreover, magnetic fields are very selective for the magnetic particles and, therefore, do not interfere with the biological system under study, making MT ideal for in vivo investigations. The downside of this approach, compared with OT, is the difficulty of combining high forces with 3D control over the magnetic bead. In vivo MT experiments have been reported (de Vries et al., 2005), but more development is needed for the method to be used as an alternative to OT.
Recent developments in bright, laser-based illumination sources, improvements in complementary metal-oxide semiconductor camera speeds, and the introduction of graphics processing unit–based calculation have made it possible to acquire bead images and track them in real time at kHz rates. These methods have made it possible for MT experiments to achieve subnanometer and submillisecond resolution and have enabled the observation of in vitro processes in real time at high spatiotemporal resolution (Dulin et al., 2015). The combination of force and torque provided by MT has proven to be ideally suited to study DNA conformations and the activity of DNA-binding proteins. For example, it has revealed important mechanistic aspects of proteins involved in DNA replication. Studies investigating primer extension with the T7 polymerase and Escherichia coli DNA polymerase I (Pol I) produced a model in which DNA synthesis is rate-limited by conformational changes involving multiple bases on the template strand (Maier et al., 2000). Using MT to study helicase activity of the T4 bacteriophage and its coupling to partner proteins in the replisome, such as the primase and the polymerase, provided new insight into how the replisome is assembled onto DNA and how DNA replication is initiated. These experiments visualized how the synthesis of an RNA primer on the lagging strand results in the formation of loops of single-stranded DNA (ssDNA), a phenomenon that later was shown to occur in other replication systems (Manosas et al., 2009; Pandey et al., 2009; Duderstadt et al., 2016). A study of the interplay between the T4 phage helicase and its DNA polymerase activities revealed that replication is faster than the unwinding by the helicase or synthesis by the polymerase as individual activities. Because the physical interaction between the two proved to be very weak, such synergies suggest an important role for ratchet-type mechanisms in speeding up reactions that consist of both reversible and irreversible steps (Manosas et al., 2012). Recent studies on replication termination demonstrate the strength of mechanical approaches in their ability to apply external forces to rationalize mechanistic aspects of findings originally made in vivo. By using MT to exert different levels of force to the E. coli Tus–Ter replication fork barrier in vitro and by observing its lifetime on DNA, a pathway describing barrier formation was proposed that reconciled previous structural, biochemical, and microbiology studies (Berghuis et al., 2015).
Summarizing, it is clear that the various experimental platforms to apply mechanical force to individual molecules represent a powerful toolbox, each method with its own strengths and weaknesses. AFM combines high-resolution microscopy with force manipulation, with high time resolution. First, a biological sample is imaged, and then a specific part of it is directly probed. Therefore, it can provide structural, dynamic, and force information all from a single platform. OT and MT, instead, offer only force manipulation, but they can follow dynamics up to 100 times faster than AFM, thus granting access to short-lived states. Furthermore, both OT and MT can probe soft biological samples with virtually no damage at all. In the case of OT, this aspect has resulted in a mature tool for in vivo investigations, allowing mechanical manipulation inside the cell.
What you see is what you get: Imaging techniques
Fluorescence imaging
Mechanical single-molecule techniques allow the precise measurement of force and energy changes and have, therefore, been invaluable to studies on protein folding, DNA stability, and protein–DNA interactions. In this section, we describe single-molecule fluorescence imaging methods, approaches that take a more passive approach than force-based methods in that they are based on the visualization of mechanically unperturbed, fluorescently tagged molecules. Single-molecule fluorescence imaging methods are especially powerful in the visualization of molecular associations, copy numbers, conformational changes in biomolecules, and enzymatic activity, often in real time. By using a fluorescence microscope equipped with a laser source to excite the fluorescent tag and a sensitive camera to detect its fluorescence emission, a single fluorophore can be imaged with high spatiotemporal precision (10s of nanometers within 10s of milliseconds). Labeling with such fluorophores, therefore, allows direct, real-time observation of a system of interest (Fig. 1 D). The first single-molecule fluorescence experiment was performed in 1990 under cryogenic conditions (Orrit and Bernard, 1990). These low temperatures were necessary to increase the stability and lifetime of the fluorophores. Only 5 y later, the increase in the quality of optics and photon detectors allowed the first room-temperature single-molecule experiment to be performed, showing individual ATP turnovers by myosin (Funatsu et al., 1995). The limited stability and lifetime of fluorophores impose significant challenges on the use of fluorescent tags to follow the dynamics of individual biomolecules, as they affect the quality of the signal and the duration of the experiment. Furthermore, the fluorophores need to be able to be specifically linked to a biomolecule of interest. Through the development of new fluorophores and photo-stabilizing compounds (Dave et al., 2009; Ha and Tinnefeld, 2012), the brightness, stability, and lifetime of fluorescent probes have increased significantly. Current efforts are directed toward improving the compounds that confer increased photostability to reduce their toxic effects and potential interference with the system of interest (van der Velde et al., 2016). Another key challenge in single-molecule fluorescence imaging experiments is the optical diffraction limit, giving rise to a lower limit of the smallest detection volume achievable. At high concentrations, this limitation results in a total number of fluorophores in the detection volume that is too large to allow single-molecule detection. As a result, single-molecule fluorescence-imaging tools were originally only useful at low nanomolar concentrations. Initial methodological advances were mainly made in the area of molecular motors, like DNA-based polymerases, myosins, and kinesins (Peterman et al., 2004), in part because the tight binding of these systems to their templates allows their study at very low concentrations. Over the past decade, developments in fluorophore stability and imaging techniques have increased the useful concentration range for single-molecule imaging by ∼10,000-fold. These developments have expanded the variety and complexity of systems probed by single-molecule fluorescence tools tremendously.
Total internal reflection fluorescence (TIRF)
One of the first methods introduced to increase the useful concentration range of single-molecule fluorescence imaging was TIRF microscopy. In TIRF microscopy (Axelrod et al., 1984), an evanescent wave excites only those molecules in an ∼100-nm thin layer above a glass–water interface (van Oijen, 2011). Though TIRF can be used to study molecular and cellular phenomena at any liquid–solid interface (such as transport on membranes), it has proven to be most useful in single-molecule microscopy. The reduction of the excited volume as a result of the thin evanescent wave results in an increase of the signal-to-background ratio that allows high-contrast imaging of single molecules up to a concentration of ∼10s of nM. A good example of the application of TIRF microscopy in single-molecule studies is the mechanism of DNA replication. Applying TIRF imaging to purified and fluorescently labeled replication proteins acting on surface-tethered and flow-stretched DNA molecules, the dynamic behavior of bacteriophage T7 polymerases within replisomes was visualized during DNA synthesis. Though it was previously assumed that polymerases are stably bound to the replication fork, it was demonstrated that the polymerases in fact rapidly exchange with those in solution (Geertsema et al., 2014). TIRF microscopy has also allowed the real-time visualization of in vitro reconstituted eukaryotic replication-origin firing. It was shown that the helicase motor domains Mcm2–7 bind as double hexamers preferentially at a native origin sequence and that single Mcm2–7 hexamers propagate bidirectionally, monotonically, and processively as constituents of active replisomes (Duzdevich et al., 2015). For kinesins, TIRF microscopy has been used to work out a longstanding mechanistic controversy on their walking mechanism. By labeling a single head of dimeric kinesin with a fluorophore and localizing the position of the dye, it was observed that a single kinesin head moves in alternating steps of 16.6 and 0 nm. This observation proves that kinesins take steps in a hand-over-hand mechanism and not an inchworm mechanism (Yildiz et al., 2004).
In vivo, near-TIRF microscopy has been used to examine the replisome stoichiometry and architecture in living cells. Using fully functional fluorescent derivatives of E. coli replisome components expressed from their endogenous promoters, it was shown that active replisomes contain three molecules of the replicative polymerase Pol III core, rather than the historically accepted two (Fig. 3, A–F; Reyes-Lamothe et al., 2010). The mutagenic polymerase Pol V, one of the players in the bacterial SOS response to DNA damage, was recently visualized at the single-molecule level in live E. coli cells. It was shown that Pol V is, beyond the known regulatory mechanisms at the transcriptional and posttranslational level, subject to a novel form of spatial regulation, in which it is transiently sequestered at the inner cell membrane (Robinson et al., 2015). Movement of kinesins and dyneins has been observed inside living cells using fluorescence imaging with one-nanometer accuracy (FIONA). GFP-tagged peroxisomes in cultured Drosophila melanogaster S2 cells were located within 1.5 nm in 1.1 ms. Surprisingly, dyneins and kinesins do not work against each other during peroxisome transport in vivo. Rather, multiple kinesins or multiple dyneins work together, producing up to 10 times the speed previously reported in in vitro measurements (Kural et al., 2005).
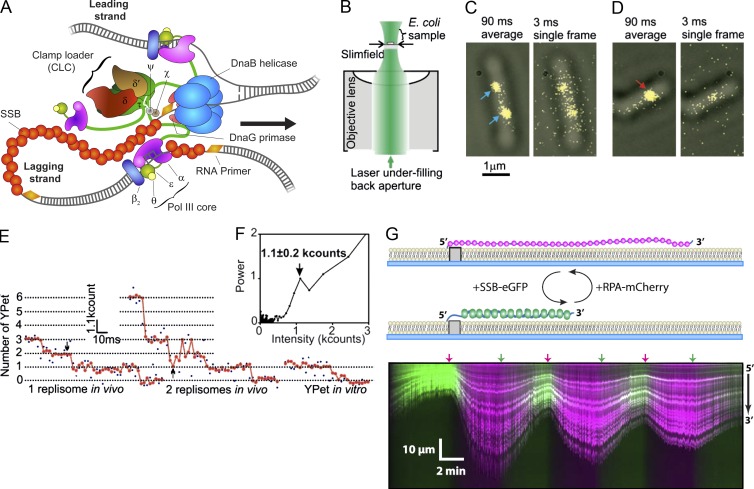
Figure 3.
Fluorescence imaging of DNA replication. (A) Schematic representation of the E. coli DNA-replication machinery. Coordinated unwinding of parental double-stranded DNA and synthesis of two daughter duplexes is catalyzed by a large multiprotein complex, the replisome, built up from 12 different proteins and held together by a large number of weak and strong protein–protein and protein–DNA interactions. (B–F) Quantitative characterization of the number of polymerases per replisome in living E. coli using single-molecule slim-field microscopy. (B) Laser light is focused on the back aperture of the microscope objective, generating an intense Gaussian field at the sample just large enough to image a single E. coli cell. (C and D) Overlay of bright-field images of cells (gray) and 90-ms frame-averaged fluorescence images (yellow) of fluorescently labeled polymerases (ε-YPet). The blue arrows point at replisomes with three polymerases and the red arrow indicates a replisome with six polymerases. (E) Raw (blue) and filtered (red) intensity for a putative single (left panel) and double (middle panel) replisome spot were compared with the intensity of a single surface-immobilized YPet in vitro (right panel). Combined with the Fourier spectral analysis to find the brightness of a single YPet (F), these data show that the in vivo steps were integer multiples of the intensity of a single YPet molecule and replisomes contain a mean of three polymerases. (B–F are adapted with permission from Reyes-Lamothe et al. (2010).) (G) Two-color fluorescence imaging of the concentration-dependent exchange of ssDNA binding proteins on ssDNA. A microfluidic flow cell with ssDNA curtains was alternatingly injected with RPA-mCherry (magenta) and E. coli ssDNA binding protein (SSB)-EGFP (green). The exchange is evident by the change in color of the fluorescence and length of the ssDNA. Arrows placed above the kymograph indicate the time points of the injections. (G is adapted from Gibb et al. [2014].)
Local activation of dye (LADye), photoactivation, diffusion, and excitation (PhADE), and point accumulation for imaging in nanoscale topography (PAINT)
To reduce the background fluorescence even further and enable the visualization of individual labeled molecules at physiologically relevant concentrations, techniques have been introduced that rely on photoactivatable tags. In PhADE (Loveland et al., 2012), a protein of interest is fused to a photoactivatable protein and introduced to its surface-immobilized substrate. After photoactivation of the protein near the surface, rapid diffusion of the unbound proteins away from the detection volume reduces background fluorescence, whereupon the bound molecules are imaged. This method allowed the visualization of the micrometer-scale movement of replication forks, the spatiotemporal pattern of replication initiation along individual DNA molecules, and the dynamics of individual proteins at replication forks in undiluted cellular extracts (Loveland et al., 2012). The drawback of this technique is the need for photoactivatable proteins. In an alternative method, LADye (Geertsema et al., 2015) relies on the labeling of proteins with inorganic fluorophores that are chemically darkened (Vaughan et al., 2012). Only those proteins bound to their substrate are selectively activated, via a short-distance energy-transfer mechanism. Although the chemicals used to darken the fluorophores could potentially alter the behavior of the system, this approach has already allowed the observation of the sequence-independent interaction of interferon-inducible protein 16 with DNA and the sliding via diffusion of adenovirus protease on DNA in the presence of very high, micromolar concentrations of protein (Geertsema et al., 2015). PhADE and LADye have increased the useful concentration of proteins in in vitro single-molecule experiments to levels closer to in vivo conditions than ever before, thereby providing new insight into the behavior of DNA-interacting proteins at physiologically relevant concentrations.
The concentrations of most proteins inside living cells are well above the concentration limit that allows visualization using conventional single-molecule imaging methods (Lewis et al., 2016). Therefore, similar techniques to reduce background fluorescence are used in vivo. In PAINT (Sharonov and Hochstrasser, 2006), the objects to be imaged are continuously targeted based on many cycles of transient association by fluorescent probes present in the solution, rather than having the fluorescent probe stably bound to the objects. As a result, a fluorescent signal appears as a diffraction-limited spot on the object when a label briefly binds to it and is momentarily immobilized (Fig. 1 E). This method was used to track endogenous AMPA glutamate receptors on living neurons, revealing high receptor densities and reduced diffusion in synapses (Giannone et al., 2010).
Single-molecule fluorescence resonance energy transfer (smFRET)
FRET is the distance-dependent nonradiative energy transfer between two fluorescent molecules that occurs when the emission spectrum of one fluorophore overlaps with the absorption spectrum of the other. Measuring the FRET efficiency allows the visualization of changes in the distance between fluorophores between ∼1 and 10 nm (Ha, 2001). By attaching two fluorophores with the appropriate spectral properties to two molecules of interest, association events and relative movements can be observed through smFRET (Fig. 1 F). By labeling a protein with two fluorophores at known positions within the protein, conformational changes and dynamics within a single molecule can be detected (Fig. 1 G). Since the initial development of the method (Ha et al., 1996), smFRET has rapidly evolved as an experimental platform to answer fundamental questions in all aspects of cellular biochemistry. For example, by labeling the two heads of a kinesin with a FRET pair, it was shown that the kinesin waits for ATP in a one-head–bound state and makes brief transitions to a two-head–bound intermediate as it walks along the microtubule (Mori et al., 2007).
Further, smFRET has allowed the direct observation of the conformational dynamics of single amino-acid transporters during substrate transport (Erkens et al., 2013; Akyuz et al., 2015). Also, smFRET studies revealed the real-time dynamics of the conformational change of the β2 clamp, the processivity factor in the DNA replication machinery, during loading onto DNA. The distance between the clamp and DNA was monitored by attaching a red Cy5 acceptor fluorophore to β2 and a green Cy3 donor fluorophore to the DNA. Three successive FRET states were seen, corresponding to closure of the clamp, followed by clamp release from its loader, and diffusion on the DNA (Cho et al., 2014).
To enable in vivo fluorescence imaging, proteins are traditionally genetically fused to a fluorescent protein. The spectral properties and poor photostability of these fluorescent proteins, however, make their use in smFRET very challenging. Therefore, observing smFRET in living cells requires new labeling, internalization, and imaging strategies. Significant progress in all these areas has been made in the last decade (Sustarsic and Kapanidis, 2015). Fluorescently labeled DNA was internalized in living E. coli cells using heat shock (Fessl et al., 2012). By electroporating a large fragment of DNA polymerase I (Klenow fragment), doubly labeled on the fingers and thumb domains, FRET was measured between internalized, immobile Klenow fragment molecules. This study shows that the distance between the two domains is preserved in live cells (Crawford et al., 2013).
Cryo-EM
Perhaps the most rapidly developing single-molecule imaging technique is cryo-EM. In cryo-EM, rapid freezing techniques (vitrification) provide immobilization of biological samples embedded in amorphous ice, preserving the structure of the samples in their native state. Using EM, these biological structures can be resolved down to the atomic level. The ability to obtain near-atomic resolution structures using cryo-EM was initially shown almost three decades ago (Henderson et al., 1990). By now, cryo-EM is a firmly established tool to gain structural information on both purified and cellular systems. Recent developments in both sample preparation and detection techniques have given access to resolutions as high as 2.2 Å for proteins as small as ∼100 kD (Cheng, 2015; Fernandez-Leiro and Scheres, 2016; Merk et al., 2016). 8- to 9-Å resolution structures of four different states of kinesins bound to microtubules allowed precise docking of a kinesin crystal structure into the map. With this information, structural rearrangements that occur upon binding of the kinesin motor domain to the microtubules could be identified (Sindelar and Downing, 2010). The structure of the Saccharomyces cerevisiae helicase, CMG, was determined by cryo-EM at a resolution of 3.7–4.8 Å, hinting toward a new unwinding mechanism. In this mechanism, two domains of the helicase move in a pumpjack-like motion to translocate on DNA (Yuan et al., 2016). 8-Å resolution structures of DNA-bound and DNA-free states of the E. coli polymerase complex revealed previously unknown interactions, thereby shedding light on different operational modes of the polymerase (Fernandez-Leiro et al., 2015).
Cryo-EM and fluorescence microscopy are now being combined into correlative light EM (CLEM; Sartori et al., 2007). This combination of techniques uses fluorescence microscopy to guide the search for specific features and locate areas worth recording and examining by cryo-EM. Fluorescence imaging can furthermore provide valuable information about local variations in ice thickness, ice crystal contamination, or other defects that could affect cryo-EM data quality. In live CLEM, proteins in a living cell are first observed using fluorescence microscopy, followed by the observation of cellular structures, such as organelles or membranes, using cryo-EM in the same cell (Kobayashi et al., 2016). With the combination of these two techniques, dynamic events can be observed in specific cellular structures. This potentially makes live CLEM a powerful method to provide functional and structural understanding of dynamic and complex events, such as nuclear envelope formation (Haraguchi et al., 2008).
Summarizing, single-molecule fluorescence imaging methods and fluorescence tagging strategies have matured to the point at which they can almost routinely be used to visualize biological processes, often in real time. Methods that allow the detection of individual molecules in high-concentration, crowded environments, combined with advances in specific and selective fluorescent labeling, pave the way to a precise interrogation of molecular processes inside living cells. Combined with the advent of cryo-EM methods, in particular those that visualize cellular structures, we are now able to visualize the dynamics of individual proteins inside a living cell with access to the structural properties of their immediate environment. These methods will enable the field to study more and more complex systems in increasingly physiologically relevant environments.
Two’s company, three’s a crowd: Multiprotein complexes in crowded environments
All molecular processes that support cellular activity arise from an intricate network of macromolecular interactions that take place in complex, crowded environments. It is therefore of fundamental importance to decipher this “molecular sociology” (Robinson et al., 2007; Mahamid et al., 2016) ideally by direct visualization. The great advances that have been made in single-molecule techniques are emphasizing a view of dynamic multiprotein systems that is not linear and deterministic, but highly stochastic (van Oijen and Dixon, 2015). In this review, we compared in vitro and in vivo experiments on cytoskeletal motors. It is clear that increasing the complexity of the system, for example by having multiple kinesins and dyneins acting on the same cargo, changes their dynamics. We also described the dynamic behavior of the DNA-replication machinery (replisome) during DNA synthesis. The composition of the replisome has previously been shown to be very stable and highly resistant to dilution (Debyser et al., 1994; Georgescu et al., 2011; Tanner et al., 2011). Single-molecule studies on the bacteriophage T7 and E. coli replisomes demonstrated that the composition of the replisome is in fact highly dynamic when operating in an environment with replisomal components present in solution, with proteins binding and unbinding extremely rapidly (Loparo et al., 2011; Geertsema et al., 2014). This suggests a mechanism in which, in a low-concentration condition, a protein remains stably bound to a complex, while being exchanged rapidly in the presence of competing protein at high concentration. Such a perhaps counterintuitive concentration-dependent dissociative mechanism has recently also been reported for replication protein A (RPA) in S. cerevisiae (Gibb et al., 2014). Using DNA curtains and fluorescently labeled RPA, it was shown that RPA remains bound to ssDNA for long periods of time when free protein is absent from solution. In contrast, RPA rapidly dissociates from ssDNA when free RPA or free E. coli ssDNA binding protein is present in solution, allowing rapid exchange between the free and bound states (Fig. 3 G). Further, in a study on the binding and unbinding kinetics of DNA transcription regulators in living E. coli cells, the kinetics of dissociation from chromosomal recognition sites was shown to be concentration dependent (Chen et al., 2015).
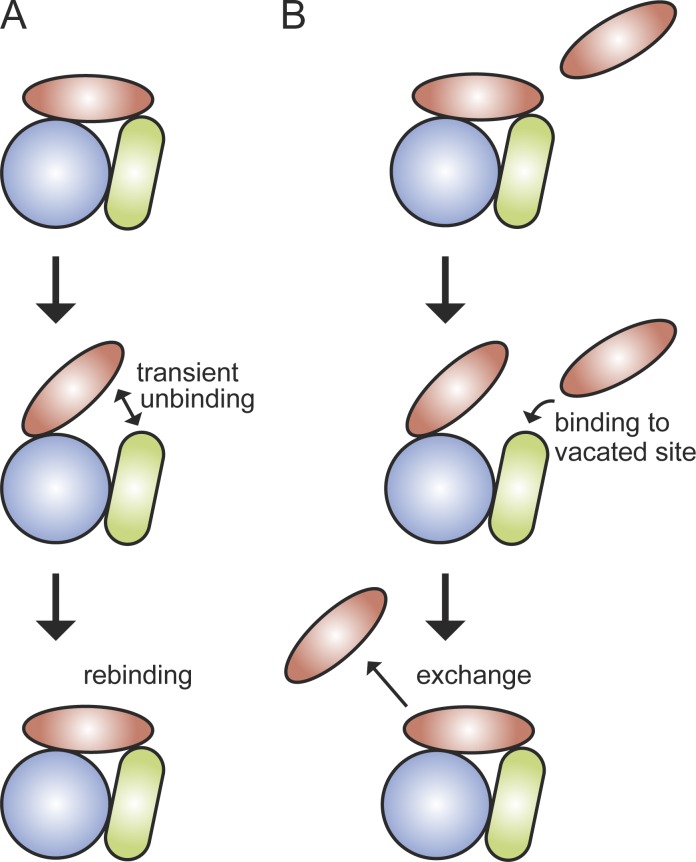
The apparent paradox between stability under high dilution and plasticity at high concentrations can be rationalized through a network of many weak interactions (Fig. 4). Under dilute conditions, stochastic, transient disruptions of any one of the interactions within a protein complex will not result in dissociation of the protein, as it is held to the complex via the other bonds, and the interaction would be rapidly reform (Fig. 4 A). Under more physiologically relevant protein concentrations, however, a protein can bind at a transiently vacated binding site and consequently compete out the original protein (Fig. 4 B; Geertsema and van Oijen, 2013). This phenomenon obeys fundamental chemical and thermodynamic principles and can be mathematically described (Sing et al., 2014; Åberg et al., 2016). This multisite exchange mechanism would allow components of multiprotein complexes to be easily replaced. In the case of the replisome, for example, this mechanism may represent a pathway through which a defective polymerase can easily be replaced, thereby insuring replication with a high fidelity. Furthermore this concentration-dependent exchange could provide easy access to other potential binding partners, like repair polymerases (Sutton, 2010). The upregulation of these repair polymerases will increase their copy number and stimulate the dissociation of Pol III through the multisite exchange mechanism, thereby guaranteeing fast DNA repair.
Figure 4.
Stability versus plasticity. (A) Under dilute conditions, transient disruption of any one of the weak interactions holding a complex together would be followed by its rapid reformation, preventing complete dissociation of the protein from the complex. This rapid microscopic reassociation would allow a protein to remain stably bound to the complex. (B) If, however, there are competing proteins in close proximity to the complex, one of these can bind at a transiently vacated binding site and consequently be at a sufficiently high local concentration to compete out the original protein.
Outlook
Single-molecule tools have enabled experimental access to the dynamic behavior of complex biomolecular systems under physiologically relevant conditions. An important next direction is to further develop the single-molecule methods to study larger, more complex systems. The in vitro use of force- and fluorescence-based tools described in this review has matured to a point at which the complexity of the systems under study seems limitless. Single-molecule studies of complex biochemical systems have already significantly changed our view of the dynamic behavior of molecular systems. The role of stochastic processes in how biological macromolecules move and interact with one another has significant impact on how biochemical processes are controlled. Instead of deterministic pathways, multiprotein complexes seem to perform their tasks by choosing from a multitude of pathways, each made possible by the constellation of weak and strong interactions that hold such a complex together. Applications of these tools within cells are still comparatively limited, however, in their ability to monitor structural and functional properties in real time at the single-molecule level. Further development of these tools and new labeling approaches are needed to further elucidate the molecular gymnastics of these complexes in vivo and bridge the gap between in vitro studies and observations inside living cells.
Acknowledgments
The authors would like to thank Dr. Slobodan Jergic, Dr. Andrew Robinson, and Mr. Jacob Lewis for their contributions to the figures.
E. Monachino and L.M. Spenkelink acknowledge support from the Netherlands Foundation for Fundamental Research on Matter (12CMCE03). A.M. van Oijen would like to acknowledge support by the Australian Research Council (DP150100956 and FL140100027).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- AFM
- atomic force microscopy
- CLEM
- correlative light EM
- HS-AFM
- high-speed atomic force microscopy
- LADye
- local activation of dye
- MT
- magnetic tweezers
- OT
- optical tweezers
- PAINT
- point accumulation for imaging in nanoscale topography
- PhADE
- photoactivation, diffusion, and excitation
- Pol
- DNA polymerase
- RPA
- replication protein A
- smFRET
- single-molecule fluorescence resonance energy transfer
- ssDNA
- single-stranded DNA
References
- Abbondanzieri E.A., Greenleaf W.J., Shaevitz J.W., Landick R., and Block S.M.. 2005. Direct observation of base-pair stepping by RNA polymerase. Nature. 438:460–465. 10.1038/nature04268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Åberg C., Duderstadt K.E., and van Oijen A.M.. 2016. Stability versus exchange: A paradox in DNA replication. Nucleic Acids Res. 44:4846–4854. 10.1093/nar/gkw296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akyuz N., Georgieva E.R., Zhou Z., Stolzenberg S., Cuendet M.A., Khelashvili G., Altman R.B., Terry D.S., Freed J.H., Weinstein H., et al. 2015. Transport domain unlocking sets the uptake rate of an aspartate transporter. Nature. 518:68–73. 10.1038/nature14158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alegre-Cebollada J., Kosuri P., Giganti D., Eckels E., Rivas-Pardo J.A., Hamdani N., Warren C.M., Solaro R.J., Linke W.A., and Fernández J.M.. 2014. S-glutathionylation of cryptic cysteines enhances titin elasticity by blocking protein folding. Cell. 156:1235–1246. 10.1016/j.cell.2014.01.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando T. 2014. High-speed AFM imaging. Curr. Opin. Struct. Biol. 28:63–68. 10.1016/j.sbi.2014.07.011 [DOI] [PubMed] [Google Scholar]
- Ando T., Uchihashi T., and Fukuma T.. 2008. High-speed atomic force microscopy for nano-visualization of dynamic biomolecular processes. Prog. Surf. Sci. 83:337–437. 10.1016/j.progsurf.2008.09.001 [DOI] [Google Scholar]
- Ando T., Uchihashi T., and Scheuring S.. 2014. Filming biomolecular processes by high-speed atomic force microscopy. Chem. Rev. 114:3120–3188. 10.1021/cr4003837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod D., Burghardt T.P., and Thompson N.L.. 1984. Total internal reflection fluorescence. Annu. Rev. Biophys. Bioeng. 13:247–268. 10.1146/annurev.bb.13.060184.001335 [DOI] [PubMed] [Google Scholar]
- Berghuis B.A., Dulin D., Xu Z.Q., van Laar T., Cross B., Janissen R., Jergic S., Dixon N.E., Depken M., and Dekker N.H.. 2015. Strand separation establishes a sustained lock at the Tus-Ter replication fork barrier. Nat. Chem. Biol. 11:579–585. 10.1038/nchembio.1857 [DOI] [PubMed] [Google Scholar]
- Bhabha G., Johnson G.T., Schroeder C.M., and Vale R.D.. 2016. How dynein moves along microtubules. Trends Biochem. Sci. 41:94–105. 10.1016/j.tibs.2015.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blehm B.H., and Selvin P.R.. 2014. Single-molecule fluorescence and in vivo optical traps: How multiple dyneins and kinesins interact. Chem. Rev. 114:3335–3352. 10.1021/cr4005555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blehm B.H., Schroer T.A., Trybus K.M., Chemla Y.R., and Selvin P.R.. 2013. In vivo optical trapping indicates kinesin’s stall force is reduced by dynein during intracellular transport. Proc. Natl. Acad. Sci. USA. 110:3381–3386. (published erratum appears in Proc. Natl. Acad. Sci. USA. 2013. 110:9613) 10.1073/pnas.1219961110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T.Y., Santiago A.G., Jung W., Krzemiński Ł., Yang F., Martell D.J., Helmann J.D., and Chen P.. 2015. Concentration- and chromosome-organization-dependent regulator unbinding from DNA for transcription regulation in living cells. Nat. Commun. 6:7445 10.1038/ncomms8445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y. 2015. Single-particle cryo-EM at crystallographic resolution. Cell. 161:450–457. 10.1016/j.cell.2015.03.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho W.K., Jergic S., Kim D., Dixon N.E., and Lee J.B.. 2014. Loading dynamics of a sliding DNA clamp. Angew. Chem. Int. Ed. Engl. 53:6768–6771. 10.1002/anie.201403063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford R., Torella J.P., Aigrain L., Plochowietz A., Gryte K., Uphoff S., and Kapanidis A.N.. 2013. Long-lived intracellular single-molecule fluorescence using electroporated molecules. Biophys. J. 105:2439–2450. 10.1016/j.bpj.2013.09.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave R., Terry D.S., Munro J.B., and Blanchard S.C.. 2009. Mitigating unwanted photophysical processes for improved single-molecule fluorescence imaging. Biophys. J. 96:2371–2381. 10.1016/j.bpj.2008.11.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debyser Z., Tabor S., and Richardson C.C.. 1994. Coordination of leading and lagging strand DNA synthesis at the replication fork of bacteriophage T7. Cell. 77:157–166. 10.1016/0092-8674(94)90243-7 [DOI] [PubMed] [Google Scholar]
- de Souza N. 2012. Pulling on single molecules. Nat. Methods. 9:873–877. 10.1038/nmeth.2149 [DOI] [PubMed] [Google Scholar]
- de Vries A.H.B., Krenn B.E., van Driel R., and Kanger J.S.. 2005. Micro magnetic tweezers for nanomanipulation inside live cells. Biophys. J. 88:2137–2144. 10.1529/biophysj.104.052035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duderstadt K.E., Geertsema H.J., Stratmann S.A., Punter C.M., Kulczyk A.W., Richardson C.C., and van Oijen A.M.. 2016. Simultaneous real-time imaging of leading and lagging strand synthesis reveals the coordination dynamics of single replisomes. Mol. Cell. 64:1–13. 10.1016/j.molcel.2016.10.028 [DOI] [PubMed] [Google Scholar]
- Dulin D., Lipfert J., Moolman M.C., and Dekker N.H.. 2013. Studying genomic processes at the single-molecule level: Introducing the tools and applications. Nat. Rev. Genet. 14:9–22. 10.1038/nrg3316 [DOI] [PubMed] [Google Scholar]
- Dulin D., Cui T.J., Cnossen J., Docter M.W., Lipfert J., and Dekker N.H.. 2015. High spatiotemporal-resolution magnetic tweezers: Calibration and applications for DNA dynamics. Biophys. J. 109:2113–2125. 10.1016/j.bpj.2015.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duzdevich D., Warner M.D., Ticau S., Ivica N.A., Bell S.P., and Greene E.C.. 2015. The dynamics of eukaryotic replication initiation: origin specificity, licensing, and firing at the single-molecule level. Mol. Cell. 58:483–494. 10.1016/j.molcel.2015.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkens G.B., Hänelt I., Goudsmits J.M.H., Slotboom D.J., and van Oijen A.M.. 2013. Unsynchronised subunit motion in single trimeric sodium-coupled aspartate transporters. Nature. 502:119–123. 10.1038/nature12538 [DOI] [PubMed] [Google Scholar]
- Essmann C.L., Elmi M., Shaw M., Anand G.M., Pawar V.M., and Srinivasan M.A.. 2016. In-vivo high resolution AFM topographic imaging of Caenorhabditis elegans reveals previously unreported surface structures of cuticle mutants. Nanomedicine (Lond.). 13:183–189. 10.1016/j.nano.2016.09.006 [DOI] [PubMed] [Google Scholar]
- Fernandez-Leiro R., and Scheres S.H.W.. 2016. Unravelling biological macromolecules with cryo-electron microscopy. Nature. 537:339–346. 10.1038/nature19948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Leiro R., Conrad J., Scheres S.H.W., and Lamers M.H.. 2015. cryo-EM structures of the E. coli replicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease and τ. eLife. 4:e11134 10.7554/eLife.11134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fessl T., Adamec F., Polívka T., Foldynová-Trantírková S., Vácha F., and Trantírek L.. 2012. Towards characterization of DNA structure under physiological conditions in vivo at the single-molecule level using single-pair FRET. Nucleic Acids Res. 40:e121 10.1093/nar/gks333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funatsu T., Harada Y., Tokunaga M., Saito K., and Yanagida T.. 1995. Imaging of single fluorescent molecules and individual ATP turnovers by single myosin molecules in aqueous solution. Nature. 374:555–559. 10.1038/374555a0 [DOI] [PubMed] [Google Scholar]
- Geertsema H.J., and van Oijen A.M.. 2013. A single-molecule view of DNA replication: the dynamic nature of multi-protein complexes revealed. Curr. Opin. Struct. Biol. 23:788–793. 10.1016/j.sbi.2013.06.018 [DOI] [PubMed] [Google Scholar]
- Geertsema H.J., Kulczyk A.W., Richardson C.C., and van Oijen A.M.. 2014. Single-molecule studies of polymerase dynamics and stoichiometry at the bacteriophage T7 replication machinery. Proc. Natl. Acad. Sci. USA. 111:4073–4078. 10.1073/pnas.1402010111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geertsema H.J., Schulte A.C., Spenkelink L.M., McGrath W.J., Morrone S.R., Sohn J., Mangel W.F., Robinson A., and van Oijen A.M.. 2015. Single-molecule imaging at high fluorophore concentrations by local activation of dye. Biophys. J. 108:949–956. 10.1016/j.bpj.2014.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgescu R.E., Kurth I., and O’Donnell M.E.. 2011. Single-molecule studies reveal the function of a third polymerase in the replisome. Nat. Struct. Mol. Biol. 19:113–116. 10.1038/nsmb.2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannone G., Hosy E., Levet F., Constals A., Schulze K., Sobolevsky A.I., Rosconi M.P., Gouaux E., Tampé R., Choquet D., and Cognet L.. 2010. Dynamic superresolution imaging of endogenous proteins on living cells at ultra-high density. Biophys. J. 99:1303–1310. 10.1016/j.bpj.2010.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb B., Ye L.F., Gergoudis S.C., Kwon Y., Niu H., Sung P., and Greene E.C.. 2014. Concentration-dependent exchange of replication protein A on single-stranded DNA revealed by single-molecule imaging. PLoS One. 9:e87922 10.1371/journal.pone.0087922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenleaf W.J., Woodside M.T., and Block S.M.. 2007. High-resolution, single-molecule measurements of biomolecular motion. Annu. Rev. Biophys. Biomol. Struct. 36:171–190. 10.1146/annurev.biophys.36.101106.101451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha T. 2001. Single-molecule fluorescence resonance energy transfer. Methods. 25:78–86. 10.1006/meth.2001.1217 [DOI] [PubMed] [Google Scholar]
- Ha T., and Tinnefeld P.. 2012. Photophysics of fluorescent probes for single-molecule biophysics and super-resolution imaging. Annu. Rev. Phys. Chem. 63:595–617. 10.1146/annurev-physchem-032210-103340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha T., Enderle T., Ogletree D.F., Chemla D.S., Selvin P.R., and Weiss S.. 1996. Probing the interaction between two single molecules: fluorescence resonance energy transfer between a single donor and a single acceptor. Proc. Natl. Acad. Sci. USA. 93:6264–6268. 10.1073/pnas.93.13.6264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi T., Kojidani T., Koujin T., Shimi T., Osakada H., Mori C., Yamamoto A., and Hiraoka Y.. 2008. Live cell imaging and electron microscopy reveal dynamic processes of BAF-directed nuclear envelope assembly. J. Cell Sci. 121:2540–2554. 10.1242/jcs.033597 [DOI] [PubMed] [Google Scholar]
- Henderson R., Baldwin J.M., Ceska T.A., Zemlin F., Beckmann E., and Downing K.H.. 1990. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J. Mol. Biol. 213:899–929. 10.1016/S0022-2836(05)80271-2 [DOI] [PubMed] [Google Scholar]
- Hendricks A.G., Holzbaur E.L., and Goldman Y.E.. 2012. Force measurements on cargoes in living cells reveal collective dynamics of microtubule motors. Proc. Natl. Acad. Sci. USA. 109:18447–18452. 10.1073/pnas.1215462109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzbaur E.L., and Goldman Y.E.. 2010. Coordination of molecular motors: From in vitro assays to intracellular dynamics. Curr. Opin. Cell Biol. 22:4–13. 10.1016/j.ceb.2009.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D.S., Bai L., Smith B.Y., Patel S.S., and Wang M.D.. 2007. Single-molecule studies reveal dynamics of DNA unwinding by the ring-shaped T7 helicase. Cell. 129:1299–1309. 10.1016/j.cell.2007.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun Y., Tripathy S.K., Narayanareddy B.R., Mattson-Hoss M.K., and Gross S.P.. 2014. Calibration of optical tweezers for in vivo force measurements: How do different approaches compare? Biophys. J. 107:1474–1484. 10.1016/j.bpj.2014.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S., Iwamoto M., and Haraguchi T.. 2016. Live correlative light-electron microscopy to observe molecular dynamics in high resolution. Microscopy (Oxf.). 65:296–308. 10.1093/jmicro/dfw024 [DOI] [PubMed] [Google Scholar]
- Kodera N., Yamamoto D., Ishikawa R., and Ando T.. 2010. Video imaging of walking myosin V by high-speed atomic force microscopy. Nature. 468:72–76. 10.1038/nature09450 [DOI] [PubMed] [Google Scholar]
- Kural C., Kim H., Syed S., Goshima G., Gelfand V.I., and Selvin P.R.. 2005. Kinesin and dynein move a peroxisome in vivo: A tug-of-war or coordinated movement? Science. 308:1469–1472. 10.1126/science.1108408 [DOI] [PubMed] [Google Scholar]
- Lewis J.S., Jergic S., and Dixon N.E.. 2016. The E. coli DNA replication fork. Enzymes. 39:31–88. 10.1016/bs.enz.2016.04.001 [DOI] [PubMed] [Google Scholar]
- Li H., Linke W.A., Oberhauser A.F., Carrion-Vazquez M., Kerkvliet J.G., Lu H., Marszalek P.E., and Fernandez J.M.. 2002. Reverse engineering of the giant muscle protein titin. Nature. 418:998–1002. 10.1038/nature00938 [DOI] [PubMed] [Google Scholar]
- Loparo J.J., Kulczyk A.W., Richardson C.C., and van Oijen A.M.. 2011. Simultaneous single-molecule measurements of phage T7 replisome composition and function reveal the mechanism of polymerase exchange. Proc. Natl. Acad. Sci. USA. 108:3584–3589. 10.1073/pnas.1018824108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loveland A.B., Habuchi S., Walter J.C., and van Oijen A.M.. 2012. A general approach to break the concentration barrier in single-molecule imaging. Nat. Methods. 9:987–992. 10.1038/nmeth.2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyubchenko Y.L., and Shlyakhtenko L.S.. 2016. Imaging of DNA and protein-DNA complexes with atomic force microscopy. Crit. Rev. Eukaryot. Gene Expr. 26:63–96. 10.1615/CritRevEukaryotGeneExpr.v26.i1.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahamid J., Pfeffer S., Schaffer M., Villa E., Danev R., Cuellar L.K., Förster F., Hyman A.A., Plitzko J.M., and Baumeister W.. 2016. Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science. 351:969–972. 10.1126/science.aad8857 [DOI] [PubMed] [Google Scholar]
- Maier B., Bensimon D., and Croquette V.. 2000. Replication by a single DNA polymerase of a stretched single-stranded DNA. Proc. Natl. Acad. Sci. USA. 97:12002–12007. 10.1073/pnas.97.22.12002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallik R., Carter B.C., Lex S.A., King S.J., and Gross S.P.. 2004. Cytoplasmic dynein functions as a gear in response to load. Nature. 427:649–652. 10.1038/nature02293 [DOI] [PubMed] [Google Scholar]
- Manosas M., Spiering M.M., Zhuang Z., Benkovic S.J., and Croquette V.. 2009. Coupling DNA unwinding activity with primer synthesis in the bacteriophage T4 primosome. Nat. Chem. Biol. 5:904–912. 10.1038/nchembio.236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manosas M., Spiering M.M., Ding F., Croquette V., and Benkovic S.J.. 2012. Collaborative coupling between polymerase and helicase for leading-strand synthesis. Nucleic Acids Res. 40:6187–6198. 10.1093/nar/gks254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merk A., Bartesaghi A., Banerjee S., Falconieri V., Rao P., Davis M.I., Pragani R., Boxer M.B., Earl L.A., Milne J.L.S., and Subramaniam S.. 2016. Breaking cryo-EM resolution barriers to facilitate drug discovery. Cell. 165:1698–1707. 10.1016/j.cell.2016.05.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T., Vale R.D., and Tomishige M.. 2007. How kinesin waits between steps. Nature. 450:750–754. 10.1038/nature06346 [DOI] [PubMed] [Google Scholar]
- Müller D.J., and Dufrêne Y.F.. 2011. Force nanoscopy of living cells. Curr. Biol. 21:R212–R216. 10.1016/j.cub.2011.01.046 [DOI] [PubMed] [Google Scholar]
- Nan X., Sims P.A., and Xie X.S.. 2008. Organelle tracking in a living cell with microsecond time resolution and nanometer spatial precision. ChemPhysChem. 9:707–712. 10.1002/cphc.200700839 [DOI] [PubMed] [Google Scholar]
- Neuman K.C., and Nagy A.. 2008. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods. 5:491–505. 10.1038/nmeth.1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman K.C., Chadd E.H., Liou G.F., Bergman K., and Block S.M.. 1999. Characterization of photodamage to Escherichia coli in optical traps. Biophys. J. 77:2856–2863. 10.1016/S0006-3495(99)77117-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oesterhelt F., Oesterhelt D., Pfeiffer M., Engel A., Gaub H.E., and Müller D.J.. 2000. Unfolding pathways of individual bacteriorhodopsins. Science. 288:143–146. 10.1126/science.288.5463.143 [DOI] [PubMed] [Google Scholar]
- Onuchic J.N., Luthey-Schulten Z., and Wolynes P.G.. 1997. Theory of protein folding: The energy landscape perspective. Annu. Rev. Phys. Chem. 48:545–600. 10.1146/annurev.physchem.48.1.545 [DOI] [PubMed] [Google Scholar]
- Orrit M., and Bernard J.. 1990. Single pentacene molecules detected by fluorescence excitation in a p-terphenyl crystal. Phys. Rev. Lett. 65:2716–2719. 10.1103/PhysRevLett.65.2716 [DOI] [PubMed] [Google Scholar]
- Pandey M., Syed S., Donmez I., Patel G., Ha T., and Patel S.S.. 2009. Coordinating DNA replication by means of priming loop and differential synthesis rate. Nature. 462:940–943. 10.1038/nature08611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterman E.J.G., Sosa H., and Moerner W.E.. 2004. Single-molecule fluorescence spectroscopy and microscopy of biomolecular motors. Annu. Rev. Phys. Chem. 55:79–96. 10.1146/annurev.physchem.55.091602.094340 [DOI] [PubMed] [Google Scholar]
- Reyes-Lamothe R., Sherratt D.J., and Leake M.C.. 2010. Stoichiometry and architecture of active DNA replication machinery in Escherichia coli. Science. 328:498–501. 10.1126/science.1185757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson A., and van Oijen A.M.. 2013. Bacterial replication, transcription and translation: Mechanistic insights from single-molecule biochemical studies. Nat. Rev. Microbiol. 11:303–315. 10.1038/nrmicro2994 [DOI] [PubMed] [Google Scholar]
- Robinson A., McDonald J.P., Caldas V.E., Patel M., Wood E.A., Punter C.M., Ghodke H., Cox M.M., Woodgate R., Goodman M.F., and van Oijen A.M.. 2015. Regulation of mutagenic DNA polymerase V activation in space and time. PLoS Genet. 11:e1005482 10.1371/journal.pgen.1005482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson C.V., Sali A., and Baumeister W.. 2007. The molecular sociology of the cell. Nature. 450:973–982. 10.1038/nature06523 [DOI] [PubMed] [Google Scholar]
- Santos S., Barcons V., Christenson H.K., Billingsley D.J., Bonass W.A., Font J., and Thomson N.H.. 2013. Stability, resolution, and ultra-low wear amplitude modulation atomic force microscopy of DNA: Small amplitude small set-point imaging. Appl. Phys. Lett. 103:063702 10.1063/1.4817906 [DOI] [Google Scholar]
- Sartori A., Gatz R., Beck F., Rigort A., Baumeister W., and Plitzko J.M.. 2007. Correlative microscopy: Bridging the gap between fluorescence light microscopy and cryo-electron tomography. J. Struct. Biol. 160:135–145. 10.1016/j.jsb.2007.07.011 [DOI] [PubMed] [Google Scholar]
- Sharonov A., and Hochstrasser R.M.. 2006. Wide-field subdiffraction imaging by accumulated binding of diffusing probes. Proc. Natl. Acad. Sci. USA. 103:18911–18916. 10.1073/pnas.0609643104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims P.A., and Xie X.S.. 2009. Probing dynein and kinesin stepping with mechanical manipulation in a living cell. ChemPhysChem. 10:1511–1516. 10.1002/cphc.200900113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sindelar C.V., and Downing K.H.. 2010. An atomic-level mechanism for activation of the kinesin molecular motors. Proc. Natl. Acad. Sci. USA. 107:4111–4116. 10.1073/pnas.0911208107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sing C.E., Olvera de la Cruz M., and Marko J.F.. 2014. Multiple-binding-site mechanism explains concentration-dependent unbinding rates of DNA-binding proteins. Nucleic Acids Res. 42:3783–3791. 10.1093/nar/gkt1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sustarsic M., and Kapanidis A.N.. 2015. Taking the ruler to the jungle: Single-molecule FRET for understanding biomolecular structure and dynamics in live cells. Curr. Opin. Struct. Biol. 34:52–59. 10.1016/j.sbi.2015.07.001 [DOI] [PubMed] [Google Scholar]
- Sutton M.D. 2010. Coordinating DNA polymerase traffic during high and low fidelity synthesis. Biochim. Biophys. Acta. 1804:1167–1179. 10.1016/j.bbapap.2009.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda K., Schmidt C.F., Schnapp B.J., and Block S.M.. 1993. Direct observation of kinesin stepping by optical trapping interferometry. Nature. 365:721–727. 10.1038/365721a0 [DOI] [PubMed] [Google Scholar]
- Tanner N.A., Tolun G., Loparo J.J., Jergic S., Griffith J.D., Dixon N.E., and van Oijen A.M.. 2011. E. coli DNA replication in the absence of free β clamps. EMBO J. 30:1830–1840. 10.1038/emboj.2011.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Velde J.H.M., Oelerich J., Huang J., Smit J.H., Aminian Jazi A., Galiani S., Kolmakov K., Guoridis G., Eggeling C., Herrmann A., et al. 2016. A simple and versatile design concept for fluorophore derivatives with intramolecular photostabilization. Nat. Commun. 7:10144 10.1038/ncomms10144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oijen A.M. 2011. Single-molecule approaches to characterizing kinetics of biomolecular interactions. Curr. Opin. Biotechnol. 22:75–80. 10.1016/j.copbio.2010.10.002 [DOI] [PubMed] [Google Scholar]
- van Oijen A.M., and Dixon N.E.. 2015. Probing molecular choreography through single-molecule biochemistry. Nat. Struct. Mol. Biol. 22:948–952. 10.1038/nsmb.3119 [DOI] [PubMed] [Google Scholar]
- Vaughan J.C., Jia S., and Zhuang X.. 2012. Ultrabright photoactivatable fluorophores created by reductive caging. Nat. Methods. 9:1181–1184. 10.1038/nmeth.2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters D.A., Cleveland J.P., Thomson N.H., Hansma P.K., Wendman M.A., Gurley G., and Elings V.. 1996. Short cantilevers for atomic force microscopy. Rev. Sci. Instrum. 67:3583 10.1063/1.1147177 [DOI] [Google Scholar]
- Watanabe H., Uchihashi T., Kobashi T., Shibata M., Nishiyama J., Yasuda R., and Ando T.. 2013. Wide-area scanner for high-speed atomic force microscopy. Rev. Sci. Instrum. 84:053702 10.1063/1.4803449 [DOI] [PubMed] [Google Scholar]
- Whited A.M., and Park P.S.. 2014. Atomic force microscopy: A multifaceted tool to study membrane proteins and their interactions with ligands. Biochim. Biophys. Acta. 1838:56–68. 10.1016/j.bbamem.2013.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz A., Tomishige M., Vale R.D., and Selvin P.R.. 2004. Kinesin walks hand-over-hand. Science. 303:676–678. 10.1126/science.1093753 [DOI] [PubMed] [Google Scholar]
- Yuan Z., Bai L., Sun J., Georgescu R., Liu J., O’Donnell M.E., and Li H.. 2016. Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation. Nat. Struct. Mol. Biol. 23:217–224. 10.1038/nsmb.3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Wu G., Song C., Li Y., Qiao H., Zhu P., Hinterdorfer P., Zhang B., and Tang J.. 2012. Single molecular recognition force spectroscopy study of a luteinizing hormone-releasing hormone analogue as a carcinoma target drug. J. Phys. Chem. B. 116:13331–13337. 10.1021/jp306882r [DOI] [PubMed] [Google Scholar]
- Zocher M., Fung J.J., Kobilka B.K., and Müller D.J.. 2012. Ligand-specific interactions modulate kinetic, energetic, and mechanical properties of the human β2 adrenergic receptor. Structure. 20:1391–1402. 10.1016/j.str.2012.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]