Novel approaches in mammalian synthetic biology are advancing the study of cellular processes, regulatory networks, and multicellular interactions. Mathur et al. describe how the design of sophisticated genetic components supported by quantitative standards and computational tools will continue to expand the impact of synthetic biology on cell biology research.
Abstract
Synthetic biology is advancing the design of genetic devices that enable the study of cellular and molecular biology in mammalian cells. These genetic devices use diverse regulatory mechanisms to both examine cellular processes and achieve precise and dynamic control of cellular phenotype. Synthetic biology tools provide novel functionality to complement the examination of natural cell systems, including engineered molecules with specific activities and model systems that mimic complex regulatory processes. Continued development of quantitative standards and computational tools will expand capacities to probe cellular mechanisms with genetic devices to achieve a more comprehensive understanding of the cell. In this study, we review synthetic biology tools that are being applied to effectively investigate diverse cellular processes, regulatory networks, and multicellular interactions. We also discuss current challenges and future developments in the field that may transform the types of investigation possible in cell biology.
Introduction
Synthetic biology has pioneered transformative approaches that are affecting how scientists tackle key questions in mammalian cell biology. Synthetic biology techniques have wide-ranging applicability and commonly make use of genetic devices, or collections of genetic elements encoding particular functions, for probing key cellular mechanisms. Early success focused on engineered transcription-based regulatory systems primarily in bacteria. More recently, new endeavors have shifted to mammalian gene regulatory processes to allow flexible, precise, and comprehensive control over gene expression and cellular development. Novel and more complex genetic devices have been used to probe cellular mechanisms, including alternative splicing, RNAi, and epigenetics. Moreover, the ability to modulate integrated and complex regulatory networks involved in cell signaling, cell communication, cell cycle, and differentiation has been achieved. This review focuses on key areas of inquiry in cell biology research that are enabled by mammalian synthetic biology approaches, the challenges that exist in effectively using these approaches, and how this area of research is likely to develop over the next few years.
Advancing cell biology research with engineered genetic devices
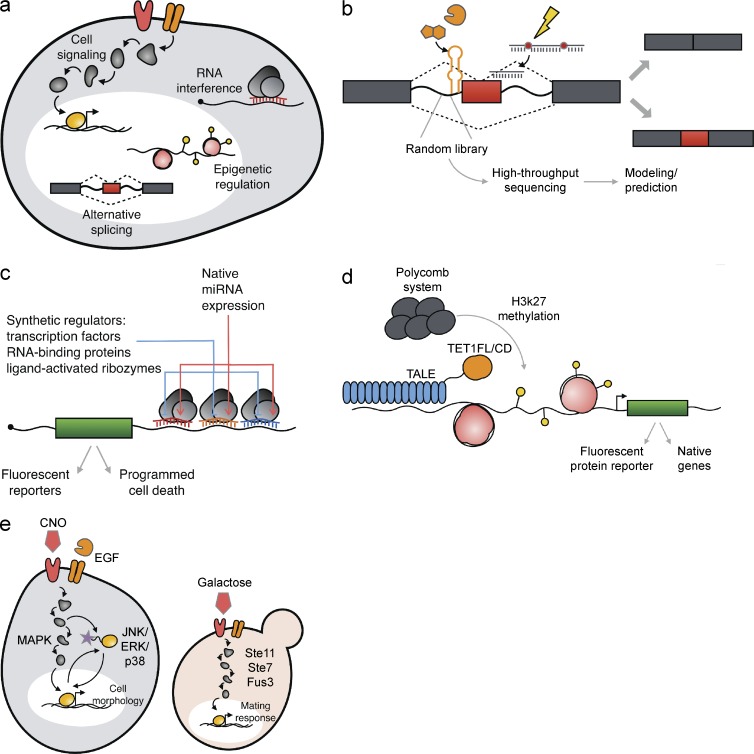
Genetic devices have been used to gain insight into cellular mechanisms with an emphasis on introducing precise perturbations to complex biological networks for studying impacts on cellular behavior. We begin by discussing mechanisms of mammalian synthetic biology approaches that are distinct from those used for interrogating prokaryotic systems (Fig. 1 a). We then discuss specific areas of mammalian cell biology that have used synthetic biology techniques to advance fundamental understanding.
Figure 1.
Tools and approaches for studying the molecular mechanisms of mammalian cells. (a) Mammalian synthetic biology enables the study of a variety of cellular mechanisms, including alternative splicing, RNAi, epigenetics, and signaling pathways within complex networks. (b) Approaches to precisely modulate alternative splicing via light-responsive splice switching oligonucleotides, ligand-responsive splicing devices, and the assessment and prediction of splicing patterns through high-throughput screening of synthetic libraries. (c) RNAi-based devices leverage synthetic regulators, including transcription factors, RNA-binding proteins, and ligand-activated ribozymes, for classifying cells based on miRNA expression and regulating cell fate. (d) Epigenetic tools that activate silenced loci with human Polycomb chromatin protein for increased transcription of a senescence locus and transcription activator–like effector (TALE)–TET1 fusions for locus-specific demethylation of endogenous genes. (e) Engineered cell-signaling components, such as G-protein–coupled receptors, GEFs, and MAPKs, that direct cellular response to regulate specific cell morphology and the mating response.
Tools and approaches for studying molecular mechanisms in mammalian cells
Alternative splicing
Synthetic biology is advancing the design of molecular tools that enable the precise and conditional modulation of splicing activity to alter protein sequence, diversity, and ultimately cellular behavior. In particular, functional nucleic acids have been used to modulate splicing patterns in response to diverse classes of molecules, thereby increasing the capacity to modify splicing patterns based on changing conditions in the cellular environment. In early examples, an RNA aptamer to the small molecule theophylline was shown to impart conditional control over splicing of a target gene via sequestration of key canonical splicing sequences, such as the branchpoint sequence and 3′ splice site (Gusti et al., 2008; Kim et al., 2008). In a subsequent study, RNA aptamers to cellular proteins (p50, p65, and β-catenin) were placed in intronic regions to control alternative splicing that modulated target gene expression in response to activation of the associated cellular signaling pathways (Fig. 1 b; Culler et al., 2010a). This genetic device conditionally altered cellular fate by linking activation of the nuclear factor κB and Wnt signaling pathways to expression of the drug-responsive herpes simplex virus type 1 thymidine kinase gene for cellular apoptosis.
In addition to molecular inputs, light has been used to achieve precise conditional control over splicing via functional oligonucleotides. Specifically, light-removable groups and photocleavable backbone linkers were positioned on synthetic splice-switching oligonucleotides for optochemical control of splicing with high spatial and temporal resolution (Hemphill et al., 2015). Additionally, alternative splicing has been harnessed to regulate other molecular mechanisms, such as RNAi. In one example, functional siRNA molecules were expressed within synthetic introns to differentially control siRNA silencing (Greber et al., 2008).
Approaches that leverage high-throughput, quantitative assays to characterize large numbers of sequences for splicing activity can enhance our understanding of the splicing code. High-throughput in vivo screens have been developed that allow for the identification of sequence characteristics of splicing regulatory elements, thereby enabling the development of new tools for splicing control. Researchers have systematically examined randomized nucleotide libraries to identify cis-acting splicing regulatory elements that confer changes in splicing patterns. In one study, unique intronic splicing regulatory elements were identified using a high-throughput in vivo screening platform (Culler et al., 2010b). Bioinformatics analyses and functional characterization revealed novel consensus motifs with intrinsic and combinatorial regulatory activity and indicated multiple splicing regulatory factors affect regulation at an intronic regulatory element. Another study corroborated that the activity of individual cis-acting elements is determined by a unique set of regulatory factors present in the cellular context (Wang et al., 2013). By testing groups of intronic silencing elements known to also exhibit exonic silencing and enhancer activities, researchers found that many proteins bound combinations of the regulatory elements and that each element was recognized by multiple proteins. These results revealed distinct patterns of context-dependent splicing activity and enabled the identification of new splicing regulators. Finally, novel high-throughput approaches have been used to assess splicing patterns of synthetic minigenes to gain mechanistic insight and substantially expand our understanding of the splicing code. Researchers recently built a synthetic dataset comprising approximately two million synthetic alternatively spliced minigenes and assayed splicing patterns, which allowed them to discern a possible universal mechanism for alternative exon recognition (Rosenberg et al., 2015). This research can serve to support the discovery of novel mechanistic trends, as the model trained using the synthetic dataset was also highly predictive of alternative splicing events in native mammalian genes.
RNAi
Over the years, synthetic biologists have focused on modifying cellular behavior in predictable and quantifiable ways with RNAi-based genetic devices that are modular, reversible, and tunable. RNAi-based devices can be used to silence various mRNA target sequences and mediate gene expression. Researchers harnessed RNAi early in synthetic biology by developing a tunable mammalian control system that coupled a TetR-regulated promoter to shRNA, such that the addition of an inducer molecule turned off expression of the shRNA and increased target gene expression (Deans et al., 2007). Control systems that leverage RNAi silencing to modify gene expression serve to elucidate gene expression threshold requirements for specific cellular phenotypes (e.g., BAX expression in cellular apoptosis) and modulate biological processes. A newer study focused on developing a quantitative model for the relationship between RNAi activity exhibited by miRNAs and target gene expression (Bloom et al., 2014). The researchers initially assessed several functional parameters, such as mRNA half-life and miRNA target-site number, to develop a model for correlating miRNA and gene expression levels. The model was extended to account for protein-responsive miRNAs and thus predict the relationship among protein input, miRNA concentration, and target gene expression levels. This quantitative understanding enabled the development of a noninvasive sensor for measuring the nuclear protein concentration of β-catenin induced by the Wnt signaling pathway.
Several synthetic RNAi-based control systems have been developed to regulate complex behaviors, including homeostasis and cellular detection. A genetic device that can form the foundation of homeostasis in mammalian cells was built using a negative-feedback control system comprised of synthetic miRNA regulators that were controlled by protein-responsive ribozymes (Bloom et al., 2015). In another example, an miRNA sensor containing an RNA-binding protein-based repressor, L7Ae, recognized the miRNA expression profile indicative of HeLa cells to specifically kill HeLa cells in a mixed HeLa-HEK293 cell population (Fig. 1 c; Wroblewska et al., 2015). The sensor comprised two transcripts, one of which detected a miRNA expressed highly in HeLa cells (e.g., miR-21) and encoded L7Ae. A second transcript recognized miRNAs expressed at low levels in HeLa [e.g., miR-141, miR-142(3p), and miR-146a], contained a binding site for L7Ae, and encoded the BAX gene to induce apoptosis. Therefore, upon sensing high versus low miR-21 in the cellular environment, HeLa cells underwent apoptosis or maintained cell survival, respectively. Similar genetically encoded miRNA detection systems have been used to recover specific cell types. An engineered transcript encoding a fluorescent reporter and miRNA target site was designed to detect miRNAs expressed in cardiomyocytes to isolate cardiomyocytes differentiated from human induced pluripotent stem cells (hiPSCs) via cell sorting (Miki et al., 2015). The detection system was further modified to control the expression of the apoptosis inducer Bim in response to a specified miRNA profile to enrich for cardiomyocytes without cell sorting. This approach proved to be broadly applicable, as miR-126, miR-122-5p, and miR-375 were used to enrich endothelial cells, hepatocytes, and insulin-producing cells differentiated from hiPSCs, respectively. In this manner, synthetic RNAi-based control systems have been shown to support the noninvasive assessment of expression profiles for the interrogation and analysis of increasingly complex cellular behaviors.
Epigenetic regulation
Synthetic biologists have examined chromatin-based interactions to expand our understanding of epigenetics as the key roles of these regulatory systems in mammalian biological processes have become apparent. In a recent study, researchers constructed a synthetic transcription factor that interacts with chromatin to reactivate silenced loci in human cells (Haynes and Silver, 2011). Specifically, the human Polycomb chromatin protein and homologues from nonmammalian cells were used to recognize the repressive trimethyl-histone H3 lysine 27 (H3K27me3) signal (Fig. 1 d). When expressed in U2OS osteosarcoma cells, these synthetic transcription factors led to increased transcription of a senescence locus, thereby reducing cell proliferation. Locus-specific control of chromatin state has also been demonstrated by the removal of DNA methylation from specific CpG sites (Maeder et al., 2013). Transcription activator–like effectors were fused to the catalytic domains of ten-eleven translocation 1 (TET1), which oxidizes mC to hmC and thereby initiates demethylation to produce locus-specific demethylation and activation at three endogenous genes (KLF4, RHOXF2, and HBB) in three human cell lines. These TET1 fusion proteins provide a tool for assessing the functional significance of specific CpG methylation marks on endogenous loci. In another example, chromatin regulation has been applied to maintain relative protein levels using an engineered epigenetic switch comprising two antibiotic-inducible transcriptional regulatory systems that repress each other's expression in CHO-K1 cells (Kramer et al., 2004).
Although chromatin regulators have been leveraged extensively in synthetic biology to alter gene expression, precise measurements of changes in epigenetic state in individual cells remain unclear. Recently, the dynamics of four silencing chromatin regulators that enact distinct modifications, including DNA methylation, histone deacetylation, and histone methylation, were observed with time-lapse microscopy via coupling to a fluorescent reporter protein (Bintu et al., 2016). The system enabled the study of epigenetic regulation dynamics, including activity over different time scales with individual transition stages and the generation of distinct epigenetic states at a single-cell level. These novel methods permit systematic functional studies of chromatin modifications that lay the foundation for investigating combinatorial and spatiotemporal layers of chromatin regulation in cell biology.
Signaling pathways
Synthetic biology provides new tools for introducing precise, controlled perturbations into signal transduction pathways to analyze both information flow through the pathway and downstream biological behavior. Several innovative strategies have been used to further understand prominent cell signaling components, such as G-protein–coupled receptors that sense extracellular molecules to activate signaling pathways, guanine nucleotide exchange factors (GEFs) that activate GTPases for signal transduction, and MAPKs that direct cellular response to regulate specific cellular function (Fig. 1 e).
G-protein–coupled receptors were engineered by one group to direct migration of cells along the gradient of a small-molecule drug, clozapine-N-oxide (CNO), which does not interact with native signaling pathways (Park et al., 2014). The synthetic receptor was engineered into a variety of cell types, including neutrophils, T lymphocytes, keratinocytes, and endothelial cells, to achieve CNO-responsive migration, thus broadly enabling cell migration studies. Another research team developed synthetic GEFs (based on a family of GEFs with a highly modular structure) by systematically reprogramming them with new regulatory domains such that their activity was controlled by protein kinase A (Yeh et al., 2007). These synthetic GEFs were subsequently used to generate protein kinase A–dependent changes in cell morphology with modifications to the cytoskeleton.
To enable the assessment of complex mammalian MAPK cascades, minimal mammalian MAPK cascades were reconstructed in yeast as a novel approach to understand how changes inherent and external to the cascade would modify cell signaling in an insulated environment (O’Shaughnessy et al., 2011). By measuring the activation profile of extracellular signal–regulated kinase (ERK) in the cascade, researchers determined that kinase concentrations could broadly tune activation dynamics and generate ultrasensitivity in the cascade. Experiments have recently also been conducted to measure single-cell activity dynamics of kinases within signaling networks, such as JNK and nuclear factor-κB activation (Regot et al., 2014). This approach led researchers to new insights into the impact of p38 on ERK signaling by measuring JNK, p38, and ERK activities simultaneously in single cells using a reporter that converted a phosphorylation event into a nucleocytoplasmic translocation event.
Beyond assessing components involved in signaling, synthetic biologists developed scaffolding proteins for spatial and temporal organization of molecules to assemble molecular components and rewire signaling cascades. Scaffolds have been shown to tether partner molecules together via short peptide motifs that serve as interaction domains to precisely control signaling behaviors. For example, a synthetic MAPK pathway derived from the ERK–MAPK pathway was shown to mediate signaling within 293T cells (Ryu and Park, 2015). Peptide tags for the PDZ domain were used to recruit kinases in the ERK pathway to a PDZ domain-based scaffold. Upon activation by EGF, a peptide-tagged ERK1 was phosphorylated, thereby demonstrating the ability of the synthetic scaffold to redirect a phosphorylation cascade. Engineered versions of cell signaling components generated by synthetic biology approaches refine our understanding of mammalian signaling pathways by redirecting information flow in novel yet precise ways.
Tools and approaches for studying gene networks
Regulatory feedback/feedforward motifs
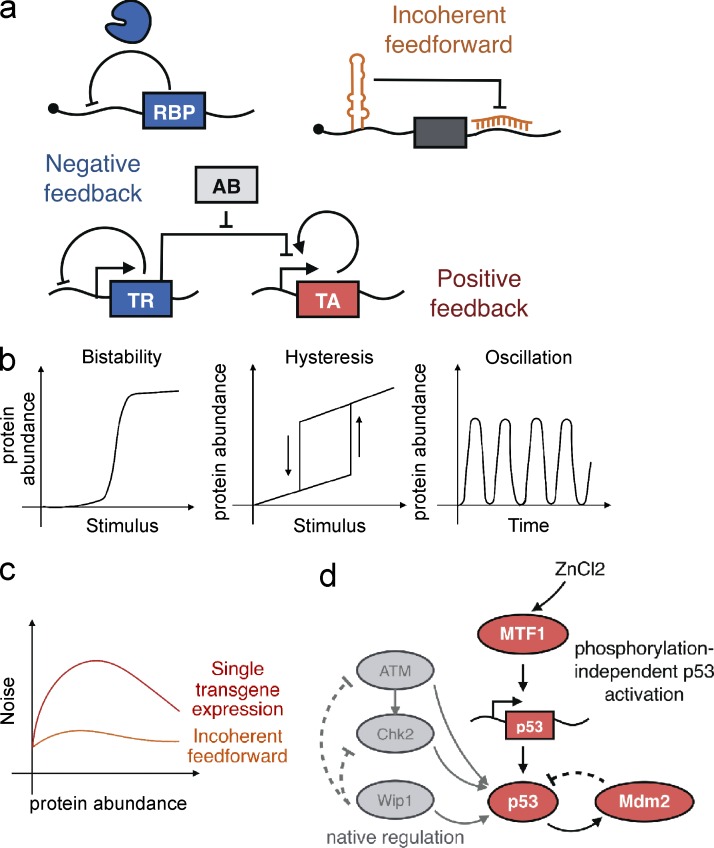
Biological networks are often composed of recurring motifs to regulate gene expression and organize other aspects of multicomponent cellular activity. Examples of such motifs are found in transcriptional, posttranscriptional, signaling, and neural networks from simple prokaryotes to complex mammalian cells. For instance, feedforward and feedback loops (Fig. 2 a) are common regulatory motifs in natural systems. By reconstructing such motifs, synthetic biology has supported an improved quantitative understanding of their mechanism and utility.
Figure 2.
Tools and approaches for studying gene networks. (a) Biological networks in natural systems are often composed of recurring motifs, including positive and negative feedback, and incoherent feedforward loops that regulate gene expression and organize multicomponent cellular activity. (b) Positive-feedback control enables signal bistability and hysteresis in response to a stimulus, whereas oscillations arise in networks that combine positive-feedback with negative-feedback loops. (c) Incoherent feedforward systems have superior buffering capacity against variability, higher expression levels, and lower intrinsic noise when compared with single transgene expression levels. (d) To study complex regulatory systems, a p53 activator-Mdm2 repressor synthetic-hybrid oscillator was stimulated with nonnatural inputs and feedback loops to affect the tightly controlled frequency of the oscillations in response to DNA damage. AB, antibiotic; RBP, RNA-binding protein; TA, transcriptional activator; TR, transcriptional repressor.
Positive-feedback control enables signal amplification, hysteresis, and bistability (Fig. 2 b), which are present in many cell fate–determining processes including chemotaxis, myogenesis, and cell cycle (Ferrell, 2002; Brandman and Meyer, 2008). An early demonstration of a hysteretic switch in mammalian cells was achieved with a synthetic transactivator that activated its own expression and was tunably repressed via a transcriptional repressor (Kramer and Fussenegger, 2005). This engineered system consisted of well-studied components enabling precise mathematical modeling and testing of mechanistic sufficiency. Congruent with modeling parameters, it was observed that hysteresis was observed only at comparable intracellular concentrations of the transrepressor and transactivator.
Negative-feedback loops can reduce the noise in a biological system and increase the responsiveness of the system to changes (Alon, 2007). Using a set of synthetic transcriptional repressors, researchers compared the expression noise from a simple negative regulation system composed of a dox-inducible LacI repressor to that from a system in which autoregulatory negative feedback of the repressor was implemented via transcriptional modulation (Shimoga et al., 2013). The system encoding the negative-feedback loop displayed significantly reduced levels of total transcriptional noise in mammalian cells, whereas the negative regulation system increased intrinsic transcriptional noise. In another example, a feedback control system based on modulation of protein translation was engineered, in which an RNA-binding protein, L7Ae, exerted negative feedback by binding to a motif within the 5′ untranslated region of its own transcript (Stapleton et al., 2012). This approach could be used to study natural RNA-binding proteins, such as the L30 protein, S15 protein, and U1A protein, that exert feedback control on translation (Boelens et al., 1993; Dabeva and Warner, 1993; Philippe et al., 1993).
Posttranscriptional processes have also been used to build feedback and feedforward motifs in mammalian cells. An incoherent feedforward system (Fig. 2 c) was explored in a synthetic RNAi-mediated gene network and found to buffer cells against gene-dosage variability (Bleris et al., 2011). In this study, transcriptional and posttranscriptional feedforward loops were compared, and the latter were shown to lead to higher gene expression, lower intrinsic noise, and improved adaptation to variable plasmid levels.
Oscillatory gene networks
Genetic oscillators such as the circadian rhythm are found across all kingdoms of life and coordinate the expression of numerous genes (Reppert and Weaver, 2002). Oscillatory gene networks combine positive-feedback amplification modules and negative-feedback repressors with a time delay to achieve periodically changing gene expression and/or cellular phenotype (Novák and Tyson, 2008). Inspired by work in bacteria (Elowitz and Leibler, 2000), early efforts reconstructed an oscillatory gene network in mammalian cells to display the circadian rhythm (Chilov and Fussenegger, 2004). Researchers designed a circadian oscillator by expressing transcription activators CLOCK and BMAL1, which bind to a synthetic promoter to induce the expression of a reporter protein, and transcription repressors PER1 and CRY2 (Chilov and Fussenegger, 2004). In turn, PER1 and CRY2 repressed transcription from the synthetic promoter, forming an autoregulatory negative-feedback loop. In a subsequent study, researchers replaced the CLOCK and BMAL1 activators with a synthetic tetracycline transactivator (tTA) activator and used amplified repression via siRNA-mediated silencing to generate autoregulatory feedback (Tigges et al., 2010). Modeling corroborated the dynamics of the low-frequency (∼26 h) oscillating system, providing insights into the accuracy and sufficiency of each system component. The results from this study contrast with an analogous oscillator (Tigges et al., 2009), in which antisense RNA was used in place of siRNA. Without the amplifying effect in the negative-feedback loop, the synthetic oscillator gene network displayed highly variable periodicity and amplitude depending on plasmid dosage and reporter protein stability.
In addition to demonstrating our understanding of the oscillatory-feedback mechanism, synthetic negative-feedback and oscillatory networks have been leveraged to characterize the dynamics of network subcomponents. By varying the intron length of a transcriptional repressor in a synthetic transcription factor–based autoinhibitory feedback network, one study effectively introduced variable time delay into the system (Swinburne et al., 2008), resulting in significant variations in the period of the oscillatory system. In this example, the responsiveness of the engineered oscillator enabled the study of how intron length affects transcriptional and alternative splicing rates. Synthetic oscillatory networks can potentially further our understanding of gene expression dynamics in cellular development, in which transcriptional time delay may influence the early gene activation program.
Synthetic gene networks embedded within natural regulatory networks have also been used to perturb a subset of the biological network as a means to examine complex regulatory systems. In one example, researchers built a p53 activator–Mdm2 repressor synthetic-hybrid oscillator (Toettcher et al., 2010) to probe the p53-controlled oscillations that are observed during γ-irradiation–induced DNA damage (Fig. 2 d). By stimulating the p53–Mdm2 pathways with nonnatural inputs and feedback loops, specific characteristics of p53 oscillations were altered, including amplitude, frequency, and damping. Guided by computational modeling, the researchers found that treatment with the Mdm2 inhibitor Nutlin3A could affect the tightly controlled frequency of the oscillations in response to DNA damage. Synthetic biology has provided tools in the form of transcriptional and posttranscriptional regulators to test our understanding of oscillatory gene networks with mathematical modeling of each isolable component.
Tools and approaches for studying multicellular behavior
Cell-to-cell communication
Cell-to-cell communication is critical for the exchange of information between specialized cells within multicellular organisms. Synthetic biology techniques enable the assessment of the underlying complex regulatory networks of multiple interacting genes that can otherwise be difficult to isolate and interrogate. To date, synthetic cell–cell communication systems have been based on either distant, diffusion-based transfer of signaling molecules or local, contact-based signaling through engineered Notch pathways.
Synthetic mammalian cell–cell communication systems have commonly been based on sender cells secreting a signaling molecule that diffuses to receiver cells to elicit a gene expression response. In one study, researchers engineered sender cells to express nitric oxide (NO) synthase, which converts L-arginine to L-citrulline and NO (Wang et al., 2008). The secreted NO diffused to receiver cells, where it activated guanylyl cyclase to produce cyclic guanosine monophosphate to induce reporter protein expression. This NO sender–receiver signaling system displayed quorum-sensing behavior, similar to bacterial systems with autoinducers. In a separate study, MDCK cells were engineered to express a sender–receiver system in 3D cell culture, in which sender cells produced hepatocyte growth factor, which diffused through the collagen matrix to receiver cells (Carvalho et al., 2014). Hepatocyte growth factor concentration gradients were created from sender cells over millimeter distances and induced tubule formation along the gradient in receiver cells. Both approaches model natural morphogen-dependent cellular communication and can be used to study this mechanism of cellular communication while minimizing cross talk with native host pathways.
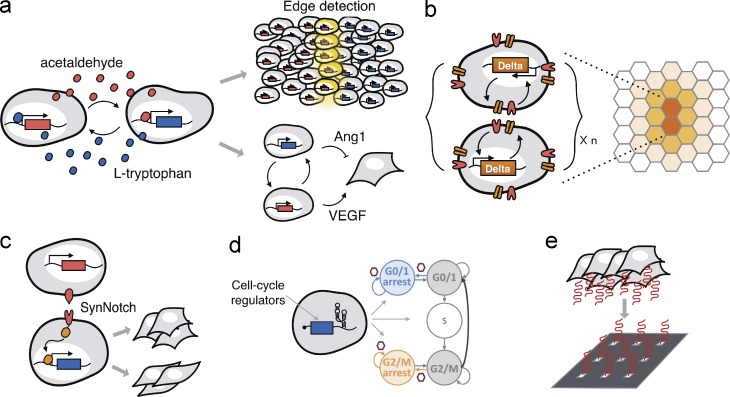
Researchers have also built two-way communication systems that mimic natural intercellular communication. In one example, sender cells were engineered to produce L-tryptophan, and receiver cells that expressed an L-tryptophan–inducible synthetic gene network in turn expressed alcohol dehydrogenase to convert ethanol in the media to acetaldehyde (Fig. 3 a), which ultimately activated a reporter gene network in the sender cells (Bacchus et al., 2012). The two-way communication module was used to coordinate the expression of VEGF and Ang1 to mimic the natural process of blood vessel formation. More recently, an L-tryptophan and interleukin-4 two-way communication system was engineered to detect the boundary between two cell populations (Kolar et al., 2015). This synthetic edge detection system may allow for the study of population border formation, (e.g., in tissue generation) by controlling the border sharpness through tuning the sensitivity of individual receiver cells and controlling diffusion rates of the signaling molecules.
Figure 3.
Tools and approaches for studying multicellular behavior. (a) A two-way communication system was engineered in which sender cells produce L-tryptophan to induce the production of acetaldehyde in receiver cells, thus activating the sender cells to coordinate the expression of VEGF and Ang1 for blood vessel formation; a similar two-way communication system was applied to engineer a mammalian cell-edge detector. (b) Engineered trigger Delta cells induce the expression of Delta in the adjacent co-cultured Notch-tTA–expressing cells to propagate the induction to adjacent cells, demonstrating the mechanistic sufficiency of this Delta-Notch positive-feedback loop in cell–cell communication. (c) Synthetic notch receptors generate novel cell–cell contact signaling pathways in which both the extracellular and intracellular domains can be exchanged. (d) Cell cycle regulators achieve conditional and reversible cell cycle arrest in the G1/0 and G2/M phases using small molecule-responsive ribozyme switches, enabling the study of cell cycle–dependent phenotypes and cell cycle misregulation. (e) Tunable control of cell-to-cell contact during cellular differentiation with interaction arrays using DNA-programmed adhesion to study the dynamics of stem cell fate decisions in response to competing juxtacrine signals.
Contact-dependent cellular communication underlies many developmental processes. One prominent example is the Delta–Notch signaling pathway, in which Delta ligand binds to Notch resulting in the cleavage and detachment of the Notch intracellular domain from the membrane, which ultimately enters the nucleus to up-regulate target genes. Researchers have engineered a synthetic Notch intracellular domain in the form of a heterologous transcription factor tTA to express fluorescent reporter genes in MDCK and CHO cells to isolate the effects that individual receptors have on the Notch signaling pathways (Matsuda et al., 2012). To simulate cell–cell communication, Trigger Delta cells were engineered to express Delta, which induced the release of the synthetic tTA in the adjacent co-cultured Notch-tTA–expressing cells. Signal propagation was achieved when the synthetic tTA was used to express another transcriptional activator TRE-GV for signal amplification before the expression of Delta. Subsequently, the next adjacent cells are induced by Delta and so forth. Although the synthetic system required a two-step amplification to achieve propagating levels of Notch-tTA and Delta expression (Fig. 3 b), this study demonstrated the mechanistic sufficiency of this Delta-Notch positive-feedback loop. Researchers also built upon this synthetic system by replacing the transcriptional activator with a transcriptional repressor rTS to demonstrate lateral inhibition and a minimal mechanism for cell-type bifurcation (Matsuda et al., 2015). In a recent study, researchers found that sensing and response behaviors could also be modified with customizable extracellular and intracellular domains in synthetic notch receptors to generate novel cell–cell contact signaling pathways (Morsut et al., 2016; Fig. 3 c).
Cellular development and differentiation
Cells undergo growth and division during normal cellular development. Dynamic regulation implemented by genetic devices has been used to examine both cell cycle progression and proliferation. Reversible, drug-responsive control of cell cycle arrest in the G1/0 and G2/M phases was recently demonstrated in U2OS cells (Fig. 3 d; Wei and Smolke, 2015). Cell cycle controllers composed of small molecule-responsive ribozyme switches conditionally and reversibly arrested up to ∼80% of a cell population in a target phase of the cell cycle, with a durable response from the G0/1 control device lasting several weeks. Such methods to reversibly interrogate cell cycle enable the assessment of cell cycle–dependent phenotypes and cell cycle misregulation. Similarly, controlled proliferation to enforce control over cell growth has been demonstrated in mammalian synthetic biology. In one study, researchers placed p27Kip1, a Cdk inhibitor that binds G1-cyclin/CDK complexes and inhibits transition from the G1 into the S phase of the cell cycle, under a tetracycline-regulated expression system for reversible and conditional control over proliferation (Cachat et al., 2014). In a separate example, small molecule-responsive ribozyme switches were used to provide stringent gene expression control and modulate T cell growth rate (Chen et al., 2010). These advancements in tools to control cell cycle and cellular proliferation support cell biology studies that look to understand processes essential to cellular development.
Novel gene expression devices enable unprecedented spatiotemporal control and dynamic resolution for the study of mammalian cell differentiation and self-organization. In one recent example, angiogenesis was precisely orchestrated in chicken embryos via red light–inducible human VEGF (Müller et al., 2013). The system can be reversibly and rapidly toggled between transcriptionally on or off states using alternating cycles of light. Through precise spatiotemporal control, illumination with red light (660 nm) showed definite and dense neovascularization and angiogenesis. In another recent example, an array platform (Fig. 3 e) that allowed tunable control over cell-to-cell contact during differentiation was used to analyze the dynamics of adult neural stem cell fate decisions in response to competing juxtacrine signals (Chen et al., 2016). Primary cortical astrocytes were engineered to express EfnB2, Delta-like 1, or both, and the distribution of neuronal stem cell fates that arose from conflicting signals was measured in vivo. A potential signaling hierarchy between Delta-like 1 and ephrin-B2 ligands was uncovered, as neural stem cells adopt the Delta-like 1 phenotype of stem cell maintenance upon simultaneous presentation of both signals. A third study recently demonstrated that self-organization can be achieved with an engineered pulse induction of GATA-binding protein 6 expression to direct the differentiation of a single autologous hiPSC source into three germ layers (Guye et al., 2016). Codifferentiation of progenitor populations triggered spatially and temporally organized differentiation of hiPSC as a complex function of GATA-binding protein 6 levels and tissue context over a 2-wk period. The ability to affect cellular differentiation and self-organization with synthetic biology approaches highlights their utility for exploring complex processes.
Challenges in applying synthetic biology tools and approaches
Although mammalian synthetic biology has advanced the investigation of molecular and cell biology in numerous examples, several challenges remain that limit effective and widespread adoption of these approaches. Cellular noise in a synthetic system can arise from the stochasticity of low molecule count, transcriptional bursting, translational effects, silencing effects, chromatin remodeling, and other unpredictable variables in a cellular environment (Murphy et al., 2010). A recent study investigated cellular variability and found that there is an inherent specificity and sensitivity tradeoff in a synthetic transcriptional program for cell death–targeting cancer cells and other similar cell state classifiers (Morel et al., 2016). Such variability in tool or system performance may need to be mitigated to improve stringency in control, especially in more complex feedback and cell–cell interaction studies. To build more robust genetic systems, researchers may need to incorporate greater complexity into the design of synthetic circuits, such as developing more effective feedback control, similar to native noise-filtering capacities in cells. It has been proposed that more sophisticated modeling and quantitative analysis methods can be used to understand and manage the noise in engineered systems, in which existing tools are unable to mitigate these effects (Rekhi and Qutub, 2013). Epigenetic silencing of transgenes can also place a limit on the time of use in longitudinal studies, such as those involving stem cell development and differentiation (Ramunas et al., 2007). Although the recent development of minimal plasmids devoid of bacterial DNA in their backbones, or minicircles, significantly reduces silencing (Kay et al., 2010), there remains a need for better insulators to enable the construction of more complex synthetic biology tools and systems involving many components for long-term observations (Tolmachov et al., 2013).
There is also a lack of tools that can sense and manipulate different aspects of cell physiology. Given its roots in bacterial transcriptional gene networks, synthetic biology has yet to develop a comprehensive set of tools that are suitable for interrogating mammalian cell morphology, cellular mechanics, intracellular organelles, and trafficking and compartmentalization. To increase the available toolset, high-throughput functional discovery methods will likely be used, analogous to a recent transcriptional repressor–mining study in prokaryotes (Stanton et al., 2014). Similarly, improved computational methods may significantly expand the diversity of small molecule dependent modulators of protein stability (Feng et al., 2015). In addition, the development of new light-inducible effectors will enable exquisite temporal-spatial control in synthetic mammalian gene networks (Kramer et al., 2003; Lohmueller et al., 2012; Slomovic and Collins, 2015) and protein–protein interactions (Zhou et al., 2012). Finally, increased dialog and collaboration between synthetic and cell biologists will facilitate the development of new tools.
Future developments in mammalian synthetic biology
Advances in synthetic biology have generated numerous approaches with broad utility for studying mammalian cell biology, as researchers have increasingly focused on building tools to control key regulatory processes unique to mammalian cells, such as epigenetic and alternative splicing regulation. The impact of these tools will be bolstered by the development of quantitative standards for measuring the activity of biological components to support their reproducibility and portability across cellular contexts. Novel computational tools will systematically enhance the design and predictability of genetic devices to advance the study of cell biology. Such developments will underpin the broad utility of mammalian synthetic biology tools for continued assessment and observation of complex biological phenomena, such as memory.
High-throughput methods, such as flow cytometry and next-generation sequencing, underscore the need for a comprehensive set of standards and benchmarks for mammalian cellular components to improve cellular measurement in biology. Significant steps have been taken to increase our quantitative understanding of core promoters that enable precise regulation of gene expression in mammalian cells (Ede et al., 2016). In a recent study, a selection of core promoters was shown to produce a range of gene expression levels and achieve a gradient of fold-inductions spanning two orders of magnitude. Standardization of programmable transcription factors across mammalian cell types (Chavez et al., 2016; Lebar and Jerala, 2016) and the generation of quantitative benchmarks for promoter and transcription factor pairs will allow researchers to achieve a quantifiable understanding of component properties, such as potency and activity, to support their usage in a variety of cellular environments.
Sustained development of novel computational tools and automation will support the creation of advanced genetic devices for which designs are informed by complex models and large datasets. Systematic bioinformatics approaches have been used to assess transcriptomic profiles in representative cellular states to identify key genetic motifs and rationally build synthetic promoters with predicted expression levels before validating their activities in vivo (Cheng and Alper, 2016). Computational design will also be increasingly used in more complex situations, such as minimizing cross talk while rewiring signaling pathways. In a recent example, orthogonal protein–protein interfaces were computationally designed, allowing key signaling proteins to be activated exclusively by an engineered cognate partner in vivo (Kapp et al., 2012). Such computational workflows will enable the construction of robust and reliable biological components with defined properties within the cell.
Together, quantitative standards and computational tools will extend the repertoire of tools to study increasingly complex processes such as biological memory. Synthetic biology can provide new approaches for identifying and tracing lineage in cellular subpopulations, thereby enabling the tracking of developmental progression. Recently, a genetic device was shown to facilitate heritable memory of hypoxia and DNA damage in mammalian cell populations (Burrill et al., 2012). CRISPR/Cas9 has also been used to introduce diverse mutations into precise genomic locations that accumulate over cell divisions (McKenna et al., 2016). Researchers demonstrated the accumulation of hundreds to thousands of uniquely altered DNA sequences in zebrafish, from which lineage relationships were inferred based on shared mutations. We expect that this approach will ultimately be adapted to create synthetic memory in mammalian systems. Developments and extensions of genetic devices in mammalian synthetic biology will continue to serve as key approaches for providing new insight into developmental processes in cell biology.
Conclusion
Tools and approaches in mammalian synthetic biology are enabling researchers to probe molecular and cellular processes unique to mammalian systems and build model synthetic networks to study cellular mechanisms, complex regulatory networks, and phenotype. Synthetic biology has also enabled researchers to achieve greater quantitative understanding of intracellular biomolecular interactions as well as multicellular behavior. Future developments in applying synthetic biology to study the cell will likely address current limitations because of noise associated with genetic devices and increase the diverse repertoire of tools available in mammalian synthetic biology. In addition, with improved quantitative standards for characterizing genetic devices and sophistication in computational modeling and automation, tools such as synthetic memory will open up new areas of inquiry in complex developmental processes in cell biology.
Acknowledgments
The authors thank S. Rudina, J. Payne, M. Kaplan, B. Kotopka, and C. Dorosin for valuable comments on the manuscript.
This work was supported by the National Institutes of Health, National Science Foundation, and Human Frontier Science Program (grants to C.D. Smolke); Stanford Bio-X Institute, National Science Foundation, and Siebel Scholars Foundation (graduate student fellowships to M. Mathur); and Agency for Science, Technology and Research (graduate student fellowship to J.S. Xiang).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- CNO
- clozapine-N-oxide
- ERK
- extracellular signal–regulated kinase
- GEF
- guanine nucleotide exchange factor
- hiPSC
- human induced pluripotent stem cell
- NO
- nitric oxide
- TET1
- ten-eleven translocation 1
- tTA
- tetracycline transactivator
References
- Alon U. 2007. Network motifs: Theory and experimental approaches. Nat. Rev. Genet. 8:450–461. 10.1038/nrg2102 [DOI] [PubMed] [Google Scholar]
- Bacchus W., Lang M., El-Baba M.D., Weber W., Stelling J., and Fussenegger M.. 2012. Synthetic two-way communication between mammalian cells. Nat. Biotechnol. 30:991–996. 10.1038/nbt.2351 [DOI] [PubMed] [Google Scholar]
- Bintu L., Yong J., Antebi Y.E., McCue K., Kazuki Y., Uno N., Oshimura M., and Elowitz M.B.. 2016. Dynamics of epigenetic regulation at the single-cell level. Science. 351:720–724. 10.1126/science.aab2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleris L., Xie Z., Glass D., Adadey A., Sontag E., and Benenson Y.. 2011. Synthetic incoherent feedforward circuits show adaptation to the amount of their genetic template. Mol. Syst. Biol. 7:519 10.1038/msb.2011.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom R.J., Winkler S.M., and Smolke C.D.. 2014. A quantitative framework for the forward design of synthetic miRNA circuits. Nat. Methods. 11:1147–1153. 10.1038/nmeth.3100 [DOI] [PubMed] [Google Scholar]
- Bloom R.J., Winkler S.M., and Smolke C.D.. 2015. Synthetic feedback control using an RNAi-based gene-regulatory device. J. Biol. Eng. 9:5 10.1186/s13036-015-0002-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boelens W.C., Jansen E.J., van Venrooij W.J., Stripecke R., Mattaj I.W., and Gunderson S.I.. 1993. The human U1 snRNP-specific U1A protein inhibits polyadenylation of its own pre-mRNA. Cell. 72:881–892. 10.1016/0092-8674(93)90577-D [DOI] [PubMed] [Google Scholar]
- Brandman O., and Meyer T.. 2008. Feedback loops shape cellular signals in space and time. Science. 322:390–395. 10.1126/science.1160617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrill D.R., Inniss M.C., Boyle P.M., and Silver P.A.. 2012. Synthetic memory circuits for tracking human cell fate. Genes Dev. 26:1486–1497. 10.1101/gad.189035.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cachat E., Liu W., Hohenstein P., and Davies J.A.. 2014. A library of mammalian effector modules for synthetic morphology. J. Biol. Eng. 8:26 10.1186/1754-1611-8-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho A., Menendez D.B., Senthivel V.R., Zimmermann T., Diambra L., and Isalan M.. 2014. Genetically encoded sender-receiver system in 3D mammalian cell culture. ACS Synth. Biol. 3:264–272. 10.1021/sb400053b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez A., Tuttle M., Pruitt B.W., Ewen-Campen B., Chari R., Ter-Ovanesyan D., Haque S.J., Cecchi R.J., Kowal E.J.K., Buchthal J., et al. 2016. Comparison of Cas9 activators in multiple species. Nat. Methods. 13:563–567. 10.1038/nmeth.3871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Bremer A.W., Scheideler O.J., Na Y.S., Todhunter M.E., Hsiao S., Bomdica P.R., Maharbiz M.M., Gartner Z.J., and Schaffer D.V.. 2016. Interrogating cellular fate decisions with high-throughput arrays of multiplexed cellular communities. Nat. Commun. 7:10309 10.1038/ncomms10309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.Y., Jensen M.C., and Smolke C.D.. 2010. Genetic control of mammalian T-cell proliferation with synthetic RNA regulatory systems. Proc. Natl. Acad. Sci. USA. 107:8531–8536. 10.1073/pnas.1001721107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J.K., and Alper H.S.. 2016. Transcriptomics-guided design of synthetic promoters for a mammalian system. ACS Synth. Biol. 10.1021/acssynbio.6b00075 [DOI] [PubMed] [Google Scholar]
- Chilov D., and Fussenegger M.. 2004. Toward construction of a self-sustained clock-like expression system based on the mammalian circadian clock. Biotechnol. Bioeng. 87:234–242. 10.1002/bit.20143 [DOI] [PubMed] [Google Scholar]
- Culler S.J., Hoff K.G., and Smolke C.D.. 2010a Reprogramming cellular behavior with RNA controllers responsive to endogenous proteins. Science. 330:1251–1255. 10.1126/science.1192128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culler S.J., Hoff K.G., Voelker R.B., Berglund J.A., and Smolke C.D.. 2010b Functional selection and systematic analysis of intronic splicing elements identify active sequence motifs and associated splicing factors. Nucleic Acids Res. 38:5152–5165. 10.1093/nar/gkq248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabeva M.D., and Warner J.R.. 1993. Ribosomal protein L32 of Saccharomyces cerevisiae regulates both splicing and translation of its own transcript. J. Biol. Chem. 268:19669–19674. [PubMed] [Google Scholar]
- Deans T.L., Cantor C.R., and Collins J.J.. 2007. A tunable genetic switch based on RNAi and repressor proteins for regulating gene expression in mammalian cells. Cell. 130:363–372. 10.1016/j.cell.2007.05.045 [DOI] [PubMed] [Google Scholar]
- Ede C., Chen X., Lin M.-Y., and Chen Y.Y.. 2016. Quantitative analyses of core promoters enable precise engineering of regulated gene expression in mammalian cells. ACS Synth. Biol. 5:395–404. 10.1021/acssynbio.5b00266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz M.B., and Leibler S.. 2000. A synthetic oscillatory network of transcriptional regulators. Nature. 403:335–338. 10.1038/35002125 [DOI] [PubMed] [Google Scholar]
- Feng J., Jester B.W., Tinberg C.E., Mandell D.J., Antunes M.S., Chari R., Morey K.J., Rios X., Medford J.I., Church G.M., et al. 2015. A general strategy to construct small molecule biosensors in eukaryotes. eLife. 4:10606 10.7554/eLife.10606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell J.E., Jr 2002. Self-perpetuating states in signal transduction: Positive feedback, double-negative feedback and bistability. Curr. Opin. Cell Biol. 14:140–148. 10.1016/S0955-0674(02)00314-9 [DOI] [PubMed] [Google Scholar]
- Greber D., El-Baba M.D., and Fussenegger M.. 2008. Intronically encoded siRNAs improve dynamic range of mammalian gene regulation systems and toggle switch. Nucleic Acids Res. 36:e101 10.1093/nar/gkn443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusti V., Kim D.-S., and Gaur R.K.. 2008. Sequestering of the 3′ splice site in a theophylline-responsive riboswitch allows ligand-dependent control of alternative splicing. Oligonucleotides. 18:93–99. 10.1089/oli.2007.0107 [DOI] [PubMed] [Google Scholar]
- Guye P., Ebrahimkhani M.R., Kipniss N., Velazquez J.J., Schoenfeld E., Kiani S., Griffith L.G., and Weiss R.. 2016. Genetically engineering self-organization of human pluripotent stem cells into a liver bud-like tissue using Gata6. Nat. Commun. 7:10243 10.1038/ncomms10243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes K.A., and Silver P.A.. 2011. Synthetic reversal of epigenetic silencing. J. Biol. Chem. 286:27176–27182. 10.1074/jbc.C111.229567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemphill J., Liu Q., Uprety R., Samanta S., Tsang M., Juliano R.L., and Deiters A.. 2015. Conditional control of alternative splicing through light-triggered splice-switching oligonucleotides. J. Am. Chem. Soc. 137:3656–3662. 10.1021/jacs.5b00580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapp G.T., Liu S., Stein A., Wong D.T., Reményi A., Yeh B.J., Fraser J.S., Taunton J., Lim W.A., and Kortemme T.. 2012. Control of protein signaling using a computationally designed GTPase/GEF orthogonal pair. Proc. Natl. Acad. Sci. USA. 109:5277–5282. 10.1073/pnas.1114487109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay M.A., He C.-Y., and Chen Z.-Y.. 2010. A robust system for production of minicircle DNA vectors. Nat. Biotechnol. 28:1287–1289. 10.1038/nbt.1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.-S., Gusti V., Dery K.J., and Gaur R.K.. 2008. Ligand-induced sequestering of branchpoint sequence allows conditional control of splicing. BMC Mol. Biol. 9:23 10.1186/1471-2199-9-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolar K., Wischhusen H.M., Müller K., Karlsson M., Weber W., and Zurbriggen M.D.. 2015. A synthetic mammalian network to compute population borders based on engineered reciprocal cell-cell communication. BMC Syst. Biol. 9:97 10.1186/s12918-015-0252-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer B.P., and Fussenegger M.. 2005. Hysteresis in a synthetic mammalian gene network. Proc. Natl. Acad. Sci. USA. 102:9517–9522. 10.1073/pnas.0500345102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer B.P., Weber W., and Fussenegger M.. 2003. Artificial regulatory networks and cascades for discrete multilevel transgene control in mammalian cells. Biotechnol. Bioeng. 83:810–820. 10.1002/bit.10731 [DOI] [PubMed] [Google Scholar]
- Kramer B.P., Viretta A.U., Daoud-El-Baba M., Aubel D., Weber W., and Fussenegger M.. 2004. An engineered epigenetic transgene switch in mammalian cells. Nat. Biotechnol. 22:867–870. 10.1038/nbt980 [DOI] [PubMed] [Google Scholar]
- Lebar T., and Jerala R.. 2016. Benchmarking of TALE- and CRISPR/dCas9-based transcriptional regulators in mammalian cells for the construction of synthetic genetic circuits. ACS Synth. Biol. 5:1050–1058. 10.1021/acssynbio.5b00259 [DOI] [PubMed] [Google Scholar]
- Lohmueller J.J., Armel T.Z., and Silver P.A.. 2012. A tunable zinc finger-based framework for Boolean logic computation in mammalian cells. Nucleic Acids Res. 40:5180–5187. 10.1093/nar/gks142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder M.L., Angstman J.F., Richardson M.E., Linder S.J., Cascio V.M., Tsai S.Q., Ho Q.H., Sander J.D., Reyon D., Bernstein B.E., et al. 2013. Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins. Nat. Biotechnol. 31:1137–1142. 10.1038/nbt.2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M., Koga M., Nishida E., and Ebisuya M.. 2012. Synthetic signal propagation through direct cell-cell interaction. Sci. Signal. 5:ra31 10.1126/scisignal.2002764 [DOI] [PubMed] [Google Scholar]
- Matsuda M., Koga M., Woltjen K., Nishida E., and Ebisuya M.. 2015. Synthetic lateral inhibition governs cell-type bifurcation with robust ratios. Nat. Commun. 6:6195 10.1038/ncomms7195 [DOI] [PubMed] [Google Scholar]
- McKenna A., Findlay G.M., Gagnon J.A., Horwitz M.S., Schier A.F., and Shendure J.. 2016. Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science. 353:aaf7907 10.1126/science.aaf7907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki K., Endo K., Takahashi S., Funakoshi S., Takei I., Katayama S., Toyoda T., Kotaka M., Takaki T., Umeda M., et al. 2015. Efficient detection and purification of cell populations using synthetic microRNA switches. Cell Stem Cell. 16:699–711. 10.1016/j.stem.2015.04.005 [DOI] [PubMed] [Google Scholar]
- Morel M., Shtrahman R., Rotter V., Nissim L., and Bar-Ziv R.H.. 2016. Cellular heterogeneity mediates inherent sensitivity-specificity tradeoff in cancer targeting by synthetic circuits. Proc. Natl. Acad. Sci. USA. 113:8133–8138. 10.1073/pnas.1604391113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morsut L., Roybal K.T., Xiong X., Gordley R.M., Coyle S.M., Thomson M., and Lim W.A.. 2016. Engineering customized cell sensing and response behaviors using synthetic Notch receptors. Cell. 164:780–791. 10.1016/j.cell.2016.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller K., Engesser R., Metzger S., Schulz S., Kämpf M.M., Busacker M., Steinberg T., Tomakidi P., Ehrbar M., Nagy F., et al. 2013. A red/far-red light-responsive bi-stable toggle switch to control gene expression in mammalian cells. Nucleic Acids Res. 41:e77 10.1093/nar/gkt002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K.F., Adams R.M., Wang X., Balázsi G., and Collins J.J.. 2010. Tuning and controlling gene expression noise in synthetic gene networks. Nucleic Acids Res. 38:2712–2726. 10.1093/nar/gkq091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák B., and Tyson J.J.. 2008. Design principles of biochemical oscillators. Nat. Rev. Mol. Cell Biol. 9:981–991. 10.1038/nrm2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shaughnessy E.C., Palani S., Collins J.J., and Sarkar C.A.. 2011. Tunable signal processing in synthetic MAP kinase cascades. Cell. 144:119–131. 10.1016/j.cell.2010.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.S., Rhau B., Hermann A., McNally K.A., Zhou C., Gong D., Weiner O.D., Conklin B.R., Onuffer J., and Lim W.A.. 2014. Synthetic control of mammalian-cell motility by engineering chemotaxis to an orthogonal bioinert chemical signal. Proc. Natl. Acad. Sci. USA. 111:5896–5901. 10.1073/pnas.1402087111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe C., Eyermann F., Bénard L., Portier C., Ehresmann B., and Ehresmann C.. 1993. Ribosomal protein S15 from Escherichia coli modulates its own translation by trapping the ribosome on the mRNA initiation loading site. Proc. Natl. Acad. Sci. USA. 90:4394–4398. 10.1073/pnas.90.10.4394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramunas J., Montgomery H.J., Kelly L., Sukonnik T., Ellis J., and Jervis E.J.. 2007. Real-time fluorescence tracking of dynamic transgene variegation in stem cells. Mol. Ther. 15:810–817. [DOI] [PubMed] [Google Scholar]
- Regot S., Hughey J.J., Bajar B.T., Carrasco S., and Covert M.W.. 2014. High-sensitivity measurements of multiple kinase activities in live single cells. Cell. 157:1724–1734. 10.1016/j.cell.2014.04.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rekhi R., and Qutub A.A.. 2013. Systems approaches for synthetic biology: A pathway toward mammalian design. Front. Physiol. 4:285 10.3389/fphys.2013.00285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reppert S.M., and Weaver D.R.. 2002. Coordination of circadian timing in mammals. Nature. 418:935–941. 10.1038/nature00965 [DOI] [PubMed] [Google Scholar]
- Rosenberg A.B., Patwardhan R.P., Shendure J., and Seelig G.. 2015. Learning the sequence determinants of alternative splicing from millions of random sequences. Cell. 163:698–711. 10.1016/j.cell.2015.09.054 [DOI] [PubMed] [Google Scholar]
- Ryu J., and Park S.-H.. 2015. Simple synthetic protein scaffolds can create adjustable artificial MAPK circuits in yeast and mammalian cells. Sci. Signal. 8:ra66 10.1126/scisignal.aab3397 [DOI] [PubMed] [Google Scholar]
- Shimoga V., White J.T., Li Y., Sontag E., and Bleris L.. 2013. Synthetic mammalian transgene negative autoregulation. Mol. Syst. Biol. 9:670 10.1038/msb.2013.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slomovic S., and Collins J.J.. 2015. DNA sense-and-respond protein modules for mammalian cells. Nat. Methods. 12:1085–1090. 10.1038/nmeth.3585 [DOI] [PubMed] [Google Scholar]
- Stanton B.C., Nielsen A.A.K., Tamsir A., Clancy K., Peterson T., and Voigt C.A.. 2014. Genomic mining of prokaryotic repressors for orthogonal logic gates. Nat. Chem. Biol. 10:99–105. 10.1038/nchembio.1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton J.A., Endo K., Fujita Y., Hayashi K., Takinoue M., Saito H., and Inoue T.. 2012. Feedback control of protein expression in mammalian cells by tunable synthetic translational inhibition. ACS Synth. Biol. 1:83–88. 10.1021/sb200005w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinburne I.A., Miguez D.G., Landgraf D., and Silver P.A.. 2008. Intron length increases oscillatory periods of gene expression in animal cells. Genes Dev. 22:2342–2346. 10.1101/gad.1696108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tigges M., Marquez-Lago T.T., Stelling J., and Fussenegger M.. 2009. A tunable synthetic mammalian oscillator. Nature. 457:309–312. 10.1038/nature07616 [DOI] [PubMed] [Google Scholar]
- Tigges M., Dénervaud N., Greber D., Stelling J., and Fussenegger M.. 2010. A synthetic low-frequency mammalian oscillator. Nucleic Acids Res. 38:2702–2711. 10.1093/nar/gkq121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toettcher J.E., Mock C., Batchelor E., Loewer A., and Lahav G.. 2010. A synthetic-natural hybrid oscillator in human cells. Proc. Natl. Acad. Sci. USA. 107:17047–17052. 10.1073/pnas.1005615107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolmachov O.E., Subkhankulova T., and Tolmachova T.. 2013. Silencing of transgene expression: A gene therapy perspective. In Gene Therapy—Tools and Potential Applications. F. Martin-Molina, editor. InTech, Rijeka, Croatia: 49–68. [Google Scholar]
- Wang W.-D., Chen Z.-T., Kang B.-G., and Li R.. 2008. Construction of an artificial intercellular communication network using the nitric oxide signaling elements in mammalian cells. Exp. Cell Res. 314:699–706. 10.1016/j.yexcr.2007.11.023 [DOI] [PubMed] [Google Scholar]
- Wang Y., Xiao X., Zhang J., Choudhury R., Robertson A., Li K., Ma M., Burge C.B., and Wang Z.. 2013. A complex network of factors with overlapping affinities represses splicing through intronic elements. Nat. Struct. Mol. Biol. 20:36–45. 10.1038/nsmb.2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei K.Y., and Smolke C.D.. 2015. Engineering dynamic cell cycle control with synthetic small molecule-responsive RNA devices. J. Biol. Eng. 9:21 10.1186/s13036-015-0019-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wroblewska L., Kitada T., Endo K., Siciliano V., Stillo B., Saito H., and Weiss R.. 2015. Mammalian synthetic circuits with RNA binding proteins for RNA-only delivery. Nat. Biotechnol. 33:839–841. 10.1038/nbt.3301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh B.J., Rutigliano R.J., Deb A., Bar-Sagi D., and Lim W.A.. 2007. Rewiring cellular morphology pathways with synthetic guanine nucleotide exchange factors. Nature. 447:596–600. 10.1038/nature05851 [DOI] [PubMed] [Google Scholar]
- Zhou X.X., Chung H.K., Lam A.J., and Lin M.Z.. 2012. Optical control of protein activity by fluorescent protein domains. Science. 338:810–814. 10.1126/science.1226854 [DOI] [PMC free article] [PubMed] [Google Scholar]