Abstract
We describe a case of Gemella bergeriae endocarditis in a patient with a bicuspid aortic valve. Diagnosis was confirmed by sequencing of 16S rRNA genes in heart valve tissue. This is the first report of Gemella endocarditis confirmed by molecular detection of bacterial genes in heart valve tissue.
CASE REPORT
A 32-year-old Western European man visiting Canada, with a known history of asthma, hypercholesterolemia, and gastroesophageal reflux disease, presented at the hospital with a 1-month history of intermittent low-grade fever, chills, diaphoresis, fatigue, myalgia, lightheadedness, and anorexia and a 1-week history of worsening dyspnea and discomfort on the left side of his chest. He denied any rash, joint pain, or weight loss. He had recently traveled to Western Europe and South Africa. He was a cigarette smoker (10 packs/year). He occasionally used marijuana and rarely drank alcohol. He denied any current or past intravenous drug use. His medications prior to admission included omeprazole and salbutamol (the latter taken via an inhaler). He had no known medication allergies.
On physical examination, he was found to be alert and oriented, with a temperature of 38.6°C, a heart rate of 79 beats per minute, a respiratory rate of 18 breaths per minute, and normal oxygen saturation on room air. He had relatively normal dentition and no peripheral stigmata of infective endocarditis. Chest examination revealed normal breath sounds, while cardiac examination revealed the presence of a grade II/VI systolic ejection murmur at the apex with radiation to the carotid arteries, in addition to a grade I/VI diastolic murmur at the left lower sternal border. Laboratory investigations revealed a hemoglobin level of 129 g/liter, a leukocyte count of 19.1 × 109 leukocytes/liter with left shift, normal electrolyte and creatinine levels, and normal cardiac enzyme levels. Two sets of BacT/Alert FAN (BioMerieux Inc., Durham, N.C.) aerobic and anaerobic blood cultures were collected initially (day 0) and daily thereafter for several days. An urgent transesophageal echocardiogram was performed, demonstrating the presence of severe aortic regurgitation on a bicuspid valve, an aortic valve ring abscess, and a torn noncoronary cusp harboring multiple vegetations.
He was diagnosed with subacute infective endocarditis and started on intravenous ampicillin and gentamicin. Twenty-four hours later, he underwent ring abscess curettage and replacement of the aortic valve with a mechanical prosthesis. The resected aortic heart valve tissue was submitted for microbiological analysis. Gram staining of the tissue revealed moderate gram-positive cocci resembling streptococci (day 1). To facilitate a rapid microbiological diagnosis, DNA was extracted from the heart valve tissue using QIAmp DNA Blood Mini kits (QIAGEN Inc., Alameda, Calif.), and the bacterial 16S rRNA gene was then partially sequenced (days 3 and 4) using MicroSeq 500 kits and an ABI Prism 3100 genetic analyzer (Applied Biosystems, Foster City, Calif.). A GenBank BLAST search revealed a perfect and unique match of the 513-bp heart valve sequence (GenBank accession number AY632069) with that of a strain of Gemella bergeriae (GenBank accession number Y13365).
Just before the DNA sequencing results became available, cultures of blood and heart valve tissue became positive for gram-positive streptococci (day 2). The blood and heart valve isolates grew on chocolate, 5% sheep blood, and anaerobic (Brucella) blood agar media after 2 days of incubation but failed to grow on MacConkey agar. The isolates were beta-hemolytic, nonmotile, catalase negative, leucine aminopeptidase (LAP) positive, pyrrolidonlyarylamidase (PYR) negative, bacitracin resistant, arginine negative, esculin negative, mannitol negative, Voges-Proskauer negative, 6.5% NaCl intolerant, and failed to type with Lancefield group A, B, C, F, or G antiserum. The phenotypic identification (day 6) was consistent with Gemella spp. with the exception of the PYR result.
The patient required intubation for 3 days postoperatively, during which he developed acute renal failure requiring dialysis, which eventually improved. Treatment with ampicillin and gentamicin was continued, but he continued to have low-grade fever and diaphoresis. Oral rifampin was added to the antibiotic regimen after which the patient became afebrile. He was discharged from the hospital and sent home on these antibiotics and was monitored by the local home parenteral therapy program, although his planned return to Europe prompted atemporary switch of his antibiotics to intravenous ceftriaxone while in transit. The planned duration of antibiotic therapy was 4 to 6 weeks. Prior to discharge, he had developed second-degree heart block requiring temporary pacing.
The genus Gemella is comprised of catalase-negative, facultatively anaerobic, salt-intolerant, nonsatelliting, gram-positive cocci occurring in pairs (commonly with adjacent sides flattened), tetrads, and/or short chains (11). Six species, namely, Gemella haemolysans, Gemella morbillorum, Gemella bergeriae, Gemella sanguinis, Gemella palaticanis, and Gemella cuniculi are currently recognized (4-7, 11). G. palaticanis and G. cuniculi have been isolated only from animal sources (dog oropharynx and rabbit submandibular abscess, respectively) (6, 7). Only two species, G. haemolysans and G. morbillorum were recognized prior to 1998.
Gemella spp. are commonly found as residents of the oropharyngeal, gastrointestinal, and/or urogenital bacterial flora of humans and other animals (11). They have been implicated in a wide variety of infectious illnesses, usually in patients with chronic medical conditions or compromised immune systems. Human infections caused by Gemella spp. include endocarditis, septic arthritis, osteomyelitis, spondylodiscitis, empyema, lung abscess, foreign device infections, meningitis, and septic shock with or without documented bacteremia (3-5, 8, 9, 11-13).
Endocarditis is one of the most commonly recognized infections associated with Gemella spp. To date, approximately 35 cases, including ours, of endocarditis associated with Gemella spp. have been reported in the literature (1-5, 8, 12, 13). Of these cases, 15 have been attributed to G. morbillorum, 14 have been attributed to G. haemolysans, 4 have been attributed to G. bergeriae, and 2 have been attributed to G. sanguinis (1-5, 8, 12, 13). No reports prior to ours have confirmed the presence of Gemella spp. directly from heart valve tissue either phenotypically or by molecular methods.
The identification of Gemella isolates represents a challenge to clinical laboratories. Manual or commercial phenotypic methods may result in misidentification of Gemella spp. as viridans group streptococci or other related organisms and vice versa (11, 13).
Gemella spp. are usually PYR positive, LAP positive, 6.5% NaCl intolerant, esculin negative, and vancomycin sensitive (11). Cells of G. haemolysans are typically arranged in pairs, tetrads, or small clusters, while those of other Gemella spp. are arranged in pairs and chains, although cells of G. palaticanis may also form clusters (4-7, 11). Occasionally, Gemella spp. may stain gram negative, particularly in the case of G. haemolysans (11).
The phenotypic profile of blood and heart valve isolates of our patient, based on microscopic, colonial, and biochemical traits, matched that expected for Gemella spp., except for a discrepant PYR result. However, the results of sequencing the 16S rRNA gene in heart valve tissue from our patient were completely concordant with the known 16S rRNA genetic profile (in the public GenBank database) of a previously characterized strain of G. bergeriae.
The increasing availability and affordability of molecular methods, such as DNA sequencing, have facilitated the identification of uncommon and/or phenotypically difficult-to-identify microbial pathogens encountered in the clinical microbiology laboratory. The 16S rRNA gene (approximately 1,500 bp), which is ubiquitous in all eubacteria, has served as the principal target of bacterial sequence-based identification protocols. Each unique bacterial species has a distinctive 16S rRNA gene sequence profile (signature); hence, the signatures of unknown bacteria can be compared to those found in publicly or commercially available sequence databases to determine whether the organism belongs to a known species.
Gemella bergeriae was first described in 1998 by Collins and coworkers (4) and was named after Ulrich Berger in recognition of his contributions to the microbiology of gemellae. These investigators used 16S rRNA gene sequence analysis and an extensive repertoire of biochemical tests to characterize six strains of previously undescribed gram-positive, catalase-negative, facultatively anaerobic cocci recovered from blood cultures of hospitalized patients (half of whom were diagnosed with subacute bacterial endocarditis) (4), leading to their proposal of a new Gemella species, G. bergeriae, to which these strains belonged. These strains were phenotypically similar to ours except for the PYR result, although one strain in the report by Collins and coinvestigators (4) was found to be weakly positive for this enzyme, and it is known that other Gemella spp. may have weakly positive or apparently negative PYR results, depending on the test inoculum size (unpublished observations).
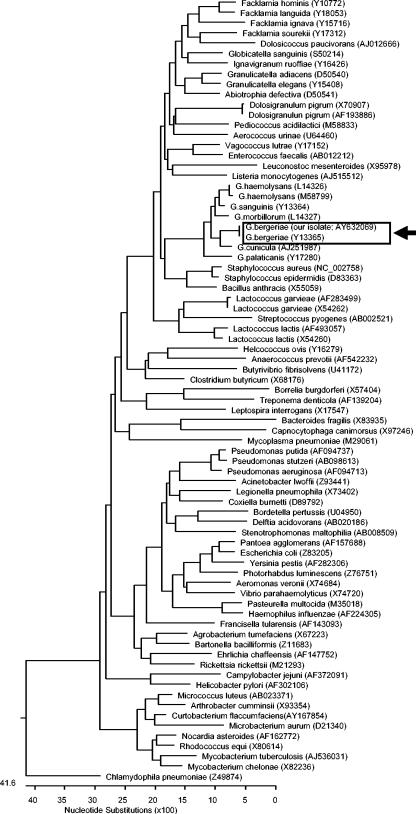
G. bergeriae displays the closest phylogenetic relationships with other members of the genus Gemella (Fig. 1). Recently, the species name was changed to G. bergeri to conform to proper taxonomical nomenclature, although the former name is still used in the Manual of Clinical Microbiology (11). Our report highlights the importance of the use of molecular methods for the identification of microorganisms directly from human specimens. This case of Gemella endocarditis is the first in the world literature to be confirmed by 16S rRNA gene sequencing directly from heart valve tissue. Other reports of endocarditis associated with Gemella spp. diagnosed using 16S rRNA gene sequencing have involved characterization of blood culture isolates from patients with suspected endocarditis. It has been suggested that molecular diagnosis of infective endocarditis be considered an additional major criterion in Duke's classification system for this disease, particularly for culture-negative cases (10).
FIG. 1.
Phylogenetic tree showing the 16S rRNA relationships of our G. bergeriae isolate with various Gemella spp. and other medically important bacteria. The tree was constructed on the basis of the first 500 nucleotides of the 16S rRNA gene and was constructed by Clustal W analysis. The sequences were obtained from the GenBank database, and their nucleotide sequence accession numbers are shown in parentheses. Chlamydophila pneumoniae was used as the outgroup to root the tree. The locations within the tree of the G. bergeriae sequence from our patient (GenBank accession number AY632069) along with one previously published G. bergeriae sequence (GenBank accession number Y13365) are shown boxed with an arrow.
REFERENCES
- 1.Al Soub, H., S. S. El-Shafie, A. L. Al-Khal, and A. M. Salam. 2003. Gemella morbillorum endocarditis. Saudi Med. J. 24:1135-1137. [PubMed] [Google Scholar]
- 2.Benes, J., D. Picha, M. Kabelkova, O. Dzupova, B. Horova, and A. Gabrielova. 2002. Infective endocarditis caused by unusual gram-positive pathogens. Folia Microbiol. (Prague) 47:737-741. [DOI] [PubMed] [Google Scholar]
- 3.Brouqui, P., and D. Raoult. 2001. Endocarditis due to rare and fastidious bacteria. Clin. Microbiol. Rev. 14:177-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins, M. D., R. A. Hutson, E. Falsen, B. Sjoden, and R. R. Facklam. 1998. Gemella bergeriae sp. nov., isolated from human clinical specimens. J. Clin. Microbiol. 36:1290-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins, M. D., R. A. Hutson, E. Falsen, B. Sjoden, and R. R. Facklam. 1998. Description of Gemella sanguinis sp. nov., isolated from human clinical specimens. J. Clin. Microbiol. 36:3090-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins, M. D., M. Rodriguez Jovita, G. Foster, B. Sjoden, and E. Falsen. 1999. Characterization of a Gemella-like organism from the oral cavity of a dog: description of Gemella palaticanis sp. nov. Int. J. Syst. Bacteriol. 49:1523-1526. [DOI] [PubMed] [Google Scholar]
- 7.Hoyles, L., G. Foster, E. Falsen, and M. D. Collins. 2000. Characterization of a Gemella-like organism isolated from an abscess of a rabbit: description of Gemella cunicula sp. nov. Int. J. Syst. E vol. Microbiol. 50:2037-2041. [DOI] [PubMed] [Google Scholar]
- 8.La Scola, B., and D. Raoult. 1998. Molecular identification of Gemella species from three patients with endocarditis. J. Clin. Microbiol. 36:866-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martha, B., M. Duong, M. Buisson, M. Grappin, L. Piroth, P. Chavenet, and H. Portier. 2003. Acute Gemella haemolysans spondylodiscitis in an immunocompetent patient. Presse Med. 32:1273-1275. [PubMed] [Google Scholar]
- 10.Millar, B., J. Moore, P. Mallon, J. Xu, M. Crowe, R. Mcclurg, D. Raoult, J. Earle, R. Hone, and P. Murphy. 2001. Molecular diagnosis of infective endocarditis—a new Duke's criterion. Scand. J. Infect. Dis. 33:673-680. [DOI] [PubMed] [Google Scholar]
- 11.Ruoff, K. 2003. Aerococcus, Abiotrophia, and other infrequently isolated aerobic catalase-negative, gram-positive cocci, p. 434-444. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed. American Society for Microbiology, Washington, D.C.
- 12.Shukla, S. K., T. Tak, R. C. Haselby, C. S. McCauley, Jr., and K. D. Reed. 2002. Second case of infective endocarditis caused by Gemella sanguinis. Wis. Med. J. 101:37-39. [PubMed] [Google Scholar]
- 13.Woo, P. C. Y., S. K. P. Lau, A. M. Y. Fung, S. K. Chiu, R. W. H. Yung, and K. Y. Yuen. 2003. Gemella bacteremia characterized by 16S ribosomal RNA gene sequencing. J. Clin. Microbiol. 56:690-693. [DOI] [PMC free article] [PubMed] [Google Scholar]