Abstract
A multiplex PCR for the simultaneous detection of five virulence genes (asa1, gelE, cylA, esp, and hyl) in enterococci was developed. The presence of these genes was investigated in 153 clinical and 118 fecal Enterococcus faecium isolates from inpatients at an increased risk of developing infections (such as patients in intensive care units and hematology wards) from 13 hospitals in eight European countries. Of the 271 E. faecium isolates, 135 were vancomycin resistant E. faecium (VREF) isolates and 136 were vancomycin susceptible E. faecium (VSEF) isolates. Susceptibilities to ampicillin, gentamicin, streptomycin, vancomycin, teicoplanin, ramoplanin, quinupristin-dalfopristin, and linezolid were tested by the microdilution method. Overall, the prevalence of esp was significantly higher (P = 0.03) in clinical VREF isolates (92%) than in fecal VREF isolates (73%). In Italy, the prevalence of esp was significantly higher (P = 0.02) in VREF isolates (91%) than in VSEF isolates (68%), whereas in the United Kingdom, hyl was significantly more prevalent (P = 0.01) in VREF isolates (71%) than in VSEF isolates (29%). No significant differences were found for the other countries. Pulsed-field gel electrophoresis was used to check the clonality among the strains tested and showed the spread of two center-specific (esp-positive) VREF clones in Italy and one center-specific (hyl-positive) clone in the United Kingdom. These clones were resistant to ampicillin, gentamicin, and streptomycin. The multiplex PCR reported in this study is a convenient and rapid method for the simultaneous detection of the virulence genes asa1, gelE, cylA, esp, and hyl in enterococci. Molecular analysis showed the intrahospital spread of esp-positive VREF clones (in Italy) and hyl-positive VREF clones (in the United Kingdom); the role of hyl remains to be elucidated.
Enterococci form part of the normal flora of both the human and the animal gastrointestinal tract. However, vancomycin-resistant enterococci have emerged as a major cause of nosocomial infections (28). Several virulence factors have been described in enterococci, for instance, aggregation substance (14), gelatinase (39), cytolysin (4), enterococcal surface protein (38), and, very recently, hyaluronidase (35). The first four virulence factors are found in Enterococcus faecalis, while the fourth and fifth virulence factors are specific for Enterococcus faecium.
Aggregation substance, encoded by asa1, which is carried on a plasmid, is a pheromone-inducible protein that enables the conjugative transfer of sex pheromone gene-containing plasmids through the clumping of one Enterococcus to another (14). As a virulence factor, aggregation substance increases bacterial adherence to renal tubular cells (26) and heart endocardial cells (18), augments internalization by intestinal epithelial cells (30), and has been shown to increase the valvular vegetation mass in an animal model of endocarditis (3).
Gelatinase, encoded by the chromosomal gelE, is an extracellular zinc endopeptidase that hydrolyzes collagen, gelatin, and small peptides (39) and that has been shown to exacerbate endocarditis in an animal model (17).
The production of cytolysin has also been shown to significantly worsen the severity of endocarditis (3) and endophthalmitis (24) in animal models as well as to contribute to the severity of enterococcal disease in humans (22). Cytolysin genes are carried on a plasmid or are integrated into the bacterial chromosome (23). Cytolysin consists of two components, lysin (L) and activator (A). The cytolysin operon consists of five genes, of which cylL1, cylL2, cylM, and cylB are relevant to the expression of component L, whereas cylA is necessary for the expression of component A (15, 21).
The enterococcal surface protein, encoded by the chromosomal esp, has an interesting structure that includes a central core consisting of distinct tandem repeat units. This central repeat region serves as a retractable arm, extending the N-terminal globular domain through the cell wall to the surface, which might facilitate immune evasion in case of immune deficiency (38). Enterococcal surface protein is associated with increased virulence (38), colonization and persistence in the urinary tract (37), and biofilm formation (41). A variant esp gene has recently been identified as a marker of highly prevalent vancomycin-resistant E. faecium (VREF) clones among hospitalized patients (44). However, the esp gene has also been detected in vancomycin-susceptible E. faecium (VSEF) isolates (45).
Recently, another virulence factor, hyaluronidase, was described in E. faecium (35). The E. faecium hyaluronidase, encoded by the chromosomal hyl, shows homology to the hyaluronidases previously described in Streptococcus pyogenes, Staphylococcus aureus, and Streptococcus pneumoniae (20), which are believed to contribute to invasion of the nasopharynx and pneumococcal pneumonia (2, 33).
Multiplex PCR is a rapid and convenient assay that allows simultaneous amplification of more than one locus in the same reaction and is used in both clinical and research laboratories (19). The purpose of this study was to develop a multiplex PCR for the detection of five potential virulence factors in enterococci and to investigate their presence in E. faecium isolates from eight European countries. Any possible correlation between these potential virulence factors, antibiotic resistance, and clonality was also explored.
(This work was presented in part at the 43rd Interscience Conference on Antimicrobial Agents and Chemotherapy, Chicago, Ill., 14 to 17 September 2003 [V. Vankerckhoven, T. Van Autgaerden, C. Vael, L. Lammens, S. Chapelle, R. Rossi, D. Jabes, and H. Goossens, Abstr. 43rd Intersci. Conf. Antimicrob. Agents Chemother., poster B-816, 2003].)
MATERIALS AND METHODS
Bacterial isolates.
A total of 271 E. faecium isolates (153 clinical isolates and 118 fecal isolates) collected in 2001 from inpatients at increased risk of the development of infections (such as patients in intensive care units [ICUs] and hematology wards) from 13 hospitals in eight European countries (12 centers: Austria, n = 1; Belgium, n = 1; France, n = 2; Germany, n = 1; Italy, n = 3; The Netherlands, n = 1; Spain, n = 2; United Kingdom, n = 1) were included in this study. In total, 135 E. faecium isolates were VREF, whereas 136 were VSEF. Of the 271 strains, 115 (96 VREF isolates and 19 VSEF isolates) were isolated in Italy; 42 (28 VREF isolates and 14 VSEF isolates) were isolated in the United Kingdom; and 114 (11 VREF isolates and 103 VSEF isolates) were isolated in Austria, Belgium, France, Greece, Spain, and The Netherlands. The isolates have been identified in a previous study on ramoplanin susceptibility (16).
The reference strains used for the multiplex PCR were E. faecalis MMH594 (a generous gift from N. Shankar, Department of Medicinal Chemistry and Pharmaceutics, University of Oklahoma Health Sciences Center, Oklahoma City) (38) was used as a positive control strain for the detection of asa1, gelE, cylA, and esp; E. faecium C68 and C38 (both strains were generous gifts from L. Rice, Research and Medical Services, Louis Stokes Cleveland Veterans Affairs Medical Center, and Department of Medicine, Case Western Reserve University, Cleveland, Ohio) (35) were used as positive and negative control strains, respectively, for the detection of hyl (both controls were also positive for esp); and E. faecalis 217, an endocarditis isolate from The Netherlands, was used as a negative control for the detection of the virulence genes tested.
Furthermore, the multiplex PCR protocol was validated with 50 E. faecalis strains (35 clinical and 15 fecal strains), E. faecalis JH2-7 and E. faecalis OG1X (both strains were a generous gift from B. Murray, Center for Infectious Diseases, Department of Internal Medicine and Department of Microbiology and Molecular Genetics, University of Texas, Houston) (4), E. faecalis FA2-2 (a generous gift from N. Shankar) (38), E. faecium E-470 (a generous gift from R. Willems, University Medical Center Utrecht, Utrecht, The Netherlands) (44), and five E. faecium strains for which the virulence genes present were unknown to us (the five strains were sent by G. Werner from the Robert Koch Institute, Wernigerode, Germany, for use as quality control strains).
Oligonucleotide primers.
The five oligonucleotide primer pairs (Eurogentec, Seraing, Belgium) used to amplify the genes asa1, gelE, cylA, esp, and hyl and the expected amplicon sizes are listed in Table 1. Primers were based on published primer pairs for cylA (4) and esp (44), while primers for the detection of asa1 (GenBank accession number X17214) (14), gelE (GenBank accession number M37185) (39), and hyl (GenBank accession number AF544400) (35) were developed by using Primer3 (http://www.genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi). Primers were designed so that the PCR products were sufficiently different in size to be distinguishable by agarose gel electrophoresis. Primer specificity was checked by a search with the BLAST program, available through the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/).
TABLE 1.
PCR primers and products for the detection of virulence genes
| Gene | Virulence factor | Primer name | Oligonucleotide sequence (5′ to 3′) | Product size (bp) |
|---|---|---|---|---|
| asa1 | Aggregation substance | ASA 11 | GCACGCTATTACGAACTATGA | 375 |
| ASA 12 | TAAGAAAGAACATCACCACGA | |||
| gelE | Gelatinase | GEL 11 | TATGACAATGCTTTTTGGGAT | 213 |
| GEL 12 | AGATGCACCCGAAATAATATA | |||
| cylA | Cytolysin | CYT I | ACTCGGGGATTGATAGGC | 688 |
| CYT IIb | GCTGCTAAAGCTGCGCTT | |||
| esp | Enterococcal surface protein | ESP 14F | AGATTTCATCTTTGATTCTTGG | 510 |
| ESP 12R | AATTGATTCTTTAGCATCTGG | |||
| hyl | Hyaluronidase | HYL n1 | ACAGAAGAGCTGCAGGAAATG | 276 |
| HYL n2 | GACTGACGTCCAAGTTTCCAA |
Multiplex PCR.
All cultures were grown on Columbia agar (Becton Dickinson, Sparks, Md.) supplemented with 5% defibrinated horse blood and were incubated at 37°C. The template DNA was prepared by suspending one loopful of bacterial cells from an overnight culture in 1 ml of Milli Q water. The bacterial suspensions were heated for 5 min at 95°C and centrifuged to remove the debris. PCR was performed in a GeneAmp PCR System 9600 (Perkin-Elmer, Wellesley, Mass.). Each 50-μl PCR mixture consisted of 5 μl of bacterial suspension; a 0.1 μM concentration (each) of primers specific for asa1, gelE, and hyl; a 0.2 μM concentration (each) of primers specific for cylA and esp; 25 μl of HotStarTaq master mixture (Qiagen, Hilden, Germany), which consisted of 2.5 U of HotStarTaq DNA polymerase, 1.5 mM MgCl2, and 200 μM deoxynucleoside triphosphates; and an additional 1.0 mM MgCl2. An initial activation step at 95°C for 15 min, during which the HotStarTaq DNA polymerase is activated, was followed by 30 cycles of denaturation (94°C for 1 min), annealing (56°C for 1 min), and extension (72°C for 1 min), followed by one cycle consisting of 10 min at 72°C. After amplification, 25 μl of the amplicon was mixed with 5 μl of gel loading buffer (50% glycerol, 0.8 mg of bromophenol blue per ml) and electrophoresed in a 1.5% pronarose D1 gel (SphaeroQ, Burgos, Spain) for 1 h at 150 V in 0.5× TBE (Tris-borate-EDTA) containing 0.05 mg of ethidium bromide per liter (positive and negative controls were included in each set of amplifications). A 100-bp DNA ladder (Invitrogen, Merelbeke, Belgium) was used as a molecular size marker.
PFGE.
The clonal distribution among VREF isolates was studied as described previously (16) by pulsed-field gel electrophoresis (PFGE) by the method of Descheemaeker et al. (7). Briefly, bacterial cells from an overnight culture were imbedded in low-melting-point preparative agarose (Bio-Rad Laboratories, Nazareth, Belgium). After cell wall and protein digestion, the plugs were digested overnight with 30 U of SmaI (MBI Fermentas, St. Leon-Rot, Germany) at 25°C. PFGE was performed with a 1% agarose gel by using a CHEF Mapper apparatus (Bio-Rad Laboratories) in 0.5× TBE buffer at 14°C and 6 V/cm. For separation, a linearly ramped switching time from 5 to 35 s was applied for 24 h. The gels were stained with ethidium bromide to detect the DNA band profiles, and the image was digitized with a Gel Doc 1000 system (Bio-Rad Laboratories). Conversion, normalization, and further analysis of the DNA band patterns were performed with GelCompar software (version 4.0b; Applied Maths, Kortrijk, Belgium), as described previously (34). The similarity between PFGE patterns was evaluated by use of the Dice coefficient and was observed visually by the detection of a maximum of three clearly visible bands.
Antimicrobial susceptibility testing.
The antimicrobial susceptibilities of all strains were tested as described previously (16). They were tested for their susceptibilities to ampicillin (Sigma Chemical Co., St. Louis, Mo.), gentamicin (Sigma), streptomycin (Sigma), vancomycin (Sigma), teicoplanin (Gruppo Lepetit, Milan, Italy), ramoplanin (Vicuron Pharmaceuticals, Gerenzano, Italy), quinupristin-dalfopristin (Synercid; commercial preparation of Aventis, Milan, Italy), and linezolid (Zyvox; commercial preparation of Pharmacia, Milan, Italy) by the microdilution method, according to the guidelines of NCCLS (29). The genes responsible for resistance to vancomycin (vanA, vanB, vanC1, and vanC2) were detected by PCR, as described previously (9).
Statistical analysis.
Chi-square analysis of contingency tables and Fisher's exact test were used for statistical analysis. A P value <0.05 was considered statistically significant.
RESULTS
Development of multiplex PCR.
Template DNA was prepared by using cell suspensions of only a few colonies since thicker cell suspensions, as described by other investigators (25, 31), increased the amplicon intensity, which has also been noticed by others (13, 32). Although at first the primer concentrations used for the detection of each gene were the same, increasing the primer concentrations for cylA and esp helped visualize the expected PCR products much more consistently. Moreover, a higher concentration of Mg2+ was needed to optimize the intensities of the band patterns generated.
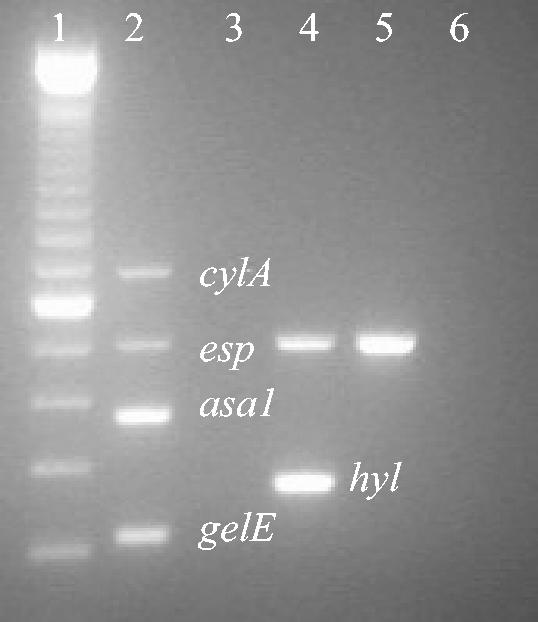
As shown in Fig. 1, the PCR product of the expected size for each of the four control strains was observed. The E. faecalis positive control strain, strain MMH594 (Fig. 1, lane 2), yielded four gene-specific products (asa1, gelE, cylA, and esp), while no products were generated from the E. faecalis negative control strain, strain 217 (Fig. 1, lane 3). The E. faecium positive control strain for hyl, strain C68 (Fig. 1, lane 4), yielded two gene-specific products (esp and hyl), while only esp was generated from the E. faecium negative control strain for hyl, strain C38 (Fig. 1, lane 5). Each multiplex PCR assay was performed with a negative control containing all the reagents but no DNA template (Fig. 1, lane 6).
FIG. 1.
Multiplex PCR of control strains. Lane 1, molecular marker (100 bp); lane 2, E. faecalis MMH594 (positive control for asa1, gelE, cylA, and esp); lane 3, E. faecalis 217 (negative control for asa1, gelE, cylA, and esp); lane 4, E. faecium C68 (positive control for hyl; also positive for esp); lane 5, E. faecium C38 (negative control for hyl; positive for esp); lane 6, negative control (no DNA added).
The 53 E. faecalis strains contained one, two, three, all, or none of the virulence genes tested for in this study, which was identical to the results generated by uniplex PCRs for each of the virulence genes tested for. Upon comparison of the two primer sets specific for esp reported so far (38, 44), only those described by Willems et al. (44) could consistently detect esp in two of six E. faecium isolates.
Multiplex PCR survey for virulence factors among E. faecium isolates.
We tested 271 E. faecium strains for the presence of five virulence factors. The genes asa1, gelE, and cylA were not detected. Of all 271 (VREF and VSEF) strains, 176 (65%) were positive for esp and 45 (17%) were positive for hyl. The prevalence of esp was significantly higher (P = 0.04) among clinical VREF isolates (24 of 26 [92%]) than among fecal VREF isolates (80 of 109 [73%]). The esp gene was significantly more prevalent (P < 0.0001) among the VREF isolates than among the VSEF isolates: 104 of 135 (77%) VREF isolates versus 72 of 136 (53%) VSEF isolates. A significant difference in the prevalence of esp (P = 0.02) was found among the 115 Italian isolates (96 VREF isolates and 19 VSEF isolates): 87 of 96 (91%) VREF isolates versus 13 of 19 (68%) VSEF isolates possessed this gene. No significant difference in the prevalence of esp was observed between United Kingdom VSEF isolates and VREF isolates. The esp gene was significantly more common (P < 0.0001) among ampicillin-resistant VREF isolates (93 of 109 [85%]) than among ampicillin-susceptible VREF isolates (11 of 26 [42%]). Among the esp-positive VREF isolates, ampicillin resistance was seen in 93 of 104 (89%), of which 82 of 87 (94%) were Italian esp-positive VREF isolates and 9 of 15 (60%) were esp-positive United Kingdom VREF isolates.
The hyl gene was found in 7 of 26 (27%) of the clinical VREF isolates, whereas it was found in 15 of 109 (14%) of the fecal VREF isolates (P = 0.1). No significant difference in the prevalence of hyl was seen between the Italian VSEF and VREF isolates. A significant difference (P = 0.01) in the prevalence of hyl was observed among the 42 United Kingdom isolates (28 VREF isolates and 14 VSEF isolates): 20 of 28 (71%) VREF isolates versus 4 of 14 (29%) VSEF isolates were found to be positive for this gene; however, no significant difference in the prevalence of hyl was observed between the Italian VREF and VSEF isolates. The hyl gene was significantly more common (P < 0.0001) among ampicillin-resistant VREF isolates (16 of 109 [15%]) than among ampicillin-susceptible VREF isolates (6 of 26 [2%]). Ampicillin resistance was seen in 16 of 22 (73%) of the hyl-positive VREF isolates, of which the only Italian hyl-positive VREF isolates and 14 of 20 (70%) United Kingdom hyl-positive VREF isolates were resistant.
Of the 114 isolates (11 VREF isolates and 103 VSEF isolates) collected from Austria, Belgium, France, Greece, Spain, and The Netherlands, esp was detected in 51 (45%) isolates (2 of 11 [18%] VREF isolates and 49 of 103 [48%] VSEF isolates) and hyl was present in 19 (17%) isolates (1 of 11 [9%] VREF isolates and 18 of 103 [18%] VSEF isolates).
PFGE typing, susceptibility testing, and the presence of virulence determinants.
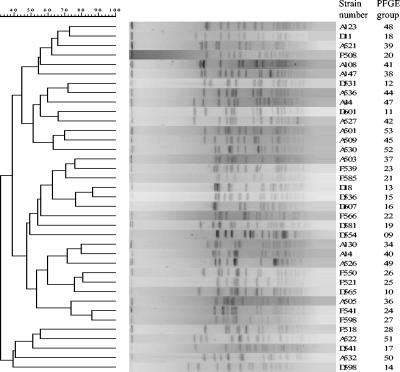
PFGE groups were defined as described previously (40). A dendrogram is shown in Fig. 2. PFGE revealed the spread of two center-specific esp-positive VREF clones in Italy (referred to as centers D and F below) and one center-specific hyl-positive VREF clone in the United Kingdom (referred to as center A below); these clones were resistant to ampicillin, gentamicin, and streptomycin. In center D in Italy, among the 58 VREF isolates collected, PFGE revealed 11 groups, all of which were of the VanA phenotype (Table 2). Except for the isolates belonging to PFGE groups 9 and 17, all isolates were positive for esp, of which group 11 represented the largest number of isolates. In center F in Italy, among the 36 VREF isolates collected, PFGE revealed 9 groups, all of which were of the VanA phenotype (Table 2). The isolates of PFGE group 20 were positive for esp and hyl, while the isolates of PFGE groups 25, 26, 27, and 28, of which group 26 contained the largest number of isolates, were positive only for esp. The other isolates (PFGE groups 21, 22, 23, and 24) did not show the presence of any virulence factors. All of the Italian isolates were highly resistant to ampicillin, gentamicin, and streptomycin. In center A in the United Kingdom, among the 28 VREF isolates collected, 24 of the VanA phenotype and 4 of the VanB phenotype, PFGE revealed 17 groups, of which group 49 contained the largest number of isolates (Table 2). The isolates of PFGE groups 34, 39, 40, 41, 47, 48, 49, 50, 51, and 52 were positive for esp and hyl; the isolates of PFGE group 37 were positive only for hyl; and the isolates of PFGE groups 38 and 53 were positive only for esp. No virulence factors were detected in the other groups (groups 36, 42, 44, and 45). All the United Kingdom isolates were resistant to ampicillin, and of these, 19 and 21 isolates also showed resistance to gentamicin and streptomycin, respectively. One Italian strain of PFGE group 16 and one United Kingdom strain of PFGE group 49 showed esp positivity, while the other isolates of the same group did not (Table 2). Finally, one strain was resistant to linezolid (8 mg/liter); this strain was isolated in the United Kingdom, belonged to PFGE group 49, and was positive for hyl. Ramoplanin was active against all strains of VREF, with an MIC at which 90% of isolates are inhibited of 0.5 mg/liter for the clinical isolates.
FIG. 2.
Dendrogram of PFGE patterns.
TABLE 2.
Vancomycin-resistant E. faecium strains in centers D and F in Italy and center A in the United Kingdom
| Center/country | PFGE group | No. of isolates | Ward of isolation | No. of specimens of the following typea:
|
VanA or VanB phenotype | Detection of:
|
||
|---|---|---|---|---|---|---|---|---|
| Clinical | Fecal | esp | hyl | |||||
| D/Italy | 9 | 1 | ICU | 1 | VanA | − | − | |
| D/Italy | 10 | 1 | ICU | 1 | VanA | + | − | |
| D/Italy | 11 | 30 | ICU | 7 (3) | 23 | VanA | + | − |
| D/Italy | 12 | 7 | ICU | 1 (1) | 6 | VanA | + | − |
| D/Italy | 13 | 7 | ICU | 2 (2) | 5 | VanA | + | − |
| D/Italy | 14 | 1 | ICU | 1 | VanA | + | − | |
| D/Italy | 15 | 1 | ICU | 1 | VanA | + | − | |
| D/Italy | 16 | 4 | ICU | 1 | 3 | VanA | +b | − |
| D/Italy | 17 | 1 | ICU | 1 | VanA | − | − | |
| D/Italy | 18 | 1 | Surgery | 1 | VanA | + | − | |
| D/Italy | 19 | 4 | ICU | 1 | 3 | VanA | + | − |
| F/Italy | 20 | 1 | Medicine | 1 | VanA | + | + | |
| F/Italy | 21 | 1 | Medicine | 1 | VanA | − | − | |
| F/Italy | 22 | 1 | Outpatient | 1 | VanA | − | − | |
| F/Italy | 23 | 1 | Hematology | 1 | VanA | − | − | |
| F/Italy | 24 | 1 | Hematology | 1 | VanA | − | − | |
| F/Italy | 25 | 9 | Medicine, n = 6; urology, n = 1; hematology, n = 1; surgery, n = 1 | 1 | 8 | VanA | + | − |
| F/Italy | 26 | 18 | Medicine, n = 9; hematology, n = 8; surgery, n = 1 | 2 (1) | 16 | VanA | + | − |
| F/Italy | 27 | 1 | Surgery | 1 | VanA | + | − | |
| F/Italy | 28 | 3 | Hematology | 1 (1) | 2 | VanA | + | − |
| A/United Kingdom | 34 | 1 | ICU | 1 | VanA | + | + | |
| A/United Kingdom | 36 | 1 | Pediatrics | 1 | VanA | − | − | |
| A/United Kingdom | 37 | 1 | Pediatrics | 1 | VanA | − | + | |
| A/United Kingdom | 38 | 1 | Pediatrics | 1 (1) | VanA | + | − | |
| A/United Kingdom | 39 | 1 | Hematology | 1 | VanB | + | + | |
| A/United Kingdom | 40 | 1 | ICU | 1 | VanA | + | + | |
| A/United Kingdom | 41 | 1 | Medicine | 1 | VanB | + | + | |
| A/United Kingdom | 42 | 1 | Pediatrics | 1 | VanA | − | − | |
| A/United Kingdom | 44 | 1 | Hematology | 1 | VanA | − | − | |
| A/United Kingdom | 45 | 1 | Hematology | 1 | VanA | − | − | |
| A/United Kingdom | 47 | 2 | Urology, n = 1; solid organ transplant, n = 1 | 2 | VanA | + | + | |
| A/United Kingdom | 48 | 1 | Medicine | 1 (1) | VanA | + | + | |
| A/United Kingdom | 49 | 9 | Hematology | 9 | VanA | +c | + | |
| A/United Kingdom | 50 | 1 | Hematology | 1 | VanB | + | + | |
| A/United Kingdom | 51 | 1 | Hematology | 1 | VanB | + | + | |
| A/United Kingdom | 52 | 1 | Hematology | 1 | VanA | + | + | |
| A/United Kingdom | 53 | 3 | Hematology | 3 | VanA | + | − | |
Values in parentheses are the number of isolates from blood.
One of four isolates.
One of nine isolates.
DISCUSSION
A multiplex PCR developed for the simultaneous detection of enterococcal genes that encode for aggregation substance (asa1), gelatinase (gelE), cytolysin (cylA), enterococcal surface protein (esp), and hyaluronidase (hyl) has not been described before. The multiplex protocol for these five genes provides a reliable and rapid alternative to phenotypic testing and uniplex PCRs. Moreover, the use of 5 μl of a heat-treated bacterial suspension as the DNA template is a time-saving step compared to the amount of time required for DNA preparation, increasing the feasibility of the technique.
We surveyed European E. faecium isolates for the presence of these genes. The asa1, gelE, and cylA genes were not detected in any of the 271 E. faecium isolates, which is in agreement with the results reported by other investigators who also tested E. faecium strains for the presence of one or more of these genes (4, 8, 10, 12, 36). However, Eaton and Gasson (10) found one gelE-positive E. faecium isolate without phenotypic gelatinase activity, and Elsner et al. (12) found asa1 among 13% of clinical E. faecium isolates, but they used hybridization.
The esp gene was detected in 65% of E. faecium isolates, in accordance with the findings of other studies (10, 11), which identified the esp gene in about 80% of E. faecium strains. However, this is in contrast to the findings of Shankar et al. (38), who reported the absence of esp in E. faecium. We detected the esp gene in a significantly higher number (P < 0.0001) of VREF strains (77%) than VSEF strains (53%). Previous studies on the incidence of esp in VREF and VSEF are contradictory: some studies (44) indicated a higher prevalence of esp among VREF strains than among VSEF strains, other studies showed the opposite (5, 8, 27), whereas again in others an equal distribution of the esp gene was found among VREF and VSEF strains (35, 45). Interestingly, we found that 91% of the Italian VREF strains harbored the esp gene. This is in contrast to the results of Baldassarri et al. (1), who found this gene in 33% of clinical VSEF isolates. The prevalence of esp in the present study was significantly higher (P = 0.04) in the clinical VREF isolates (92%) than in the fecal VREF isolates (73%). This is in agreement with the results of Rice et al. (35), who also reported a higher prevalence of esp among clinical isolates compared with that among fecal isolates. These results suggest a possible role of esp in the pathogenicity of enterococci.
Analysis of the clonality E. faecium strains harboring the esp gene showed that 5 of 9 clonal types (56%) from center D in Italy, 8 of 11 clonal types (73%) from center F in Italy, and 11 of 19 clonal types (58%) from center A in the United Kingdom were esp positive. The clonal distribution of E. faecium strains harboring esp has also been reported by Coque et al. (5, 6), while Willems et al. (44) described a larger number of esp-positive clones and, thus, reported a distribution of esp more heterogeneous than that found in our study.
Deviating results were found for one Italian strain of PFGE group 16 and one United Kingdom strain of group 49. Both strains were found to be esp positive, while other strains belonging to the same group were esp negative. This difference might be explained by the fact that the restriction enzyme used for PFGE, SmaI, does not recognize restriction sites in the esp gene. Moreover, SmaI generates segments of kilobase pairs, while the multiplex PCR generates segments of base pairs, and because of the difference in the lengths of the fragments generated by PFGE and PCR, it is unlikely that the virulence gene will be observed by PFGE. Our findings could be further explored by using a second restriction enzyme, which might be able to detect the differences. According to Waar et al. (43), E. faecalis isolates are clonal if they reveal a similarity of ≥90% by amplified fragment length polymorphism analysis and an identical pattern of virulence factors. On the basis of these conclusions, the clones reported in our study may have to be reclassified in a different PFGE cluster.
The hyl gene was detected in 17% of the 271 E. faecium isolates collected in eight European countries, which is in contrast to the findings of Rice et al. (35), who detected hyl in only 3% of the European clinical isolates. We found the hyl gene among 16% of the 135 VREF isolates and 17% of the 136 VSEF isolates. Rice et al. (35) found the hyl gene only in European VREF isolates and in none of the European VSEF isolates included in their study. Moreover, our study showed that the hyl gene is even more prevalent among the United Kingdom VREF isolates (71%) than among the U.S. VREF isolates (39%) described by Rice et al. (35).
The esp and hyl genes were significantly more common among ampicillin-resistant VREF isolates than among ampicillin-susceptible VREF isolates, which is in accordance with the findings of other studies (5, 6, 27, 42).
Finally, we found that 34 of 45 (76%) hyl-positive strains were also esp positive, which is in accordance with the findings of Rice et al. (35), who also described the combined presence of hyl and esp in >90% of the strains that they tested. On the contrary, Coque et al. (6) found only 4% of their isolates to be positive for esp and hyl.
In conclusion, the multiplex PCR developed and described herein is a convenient and rapid method for the simultaneous detection of five potential virulence genes, asa1, gelE, cylA, esp, and hyl, in enterococci. Molecular analysis showed the intrahospital spread of esp-positive VREF clones (in Italy) and of hyl-positive VREF clones (in the United Kingdom); the role of hyl remains to be elucidated.
Acknowledgments
This work was supported by a grant from the European Commission (QLRT-2001-01273) and Vicuron Pharmaceuticals.
REFERENCES
- 1.Baldassarri, L., L. Bertuccini, M. G. Ammendolia, G. Gherardi, and R. Creti. 2001. Variant esp gene in vancomycin-sensitive Enterococcus faecium. Lancet 357:1802. [DOI] [PubMed] [Google Scholar]
- 2.Berry, A. M., and J. C. Paton. 2000. Additive attenuation of virulence of Streptococcus pneumoniae by mutation of the genes encoding pneumolysin and other putative pneumococcal virulence proteins. Infect. Immun. 68:133-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chow, J. W., L. A. Thal, M. B. Perri, J. A. Vazquez, S. M. Donabedian, D. B. Clewell, and M. J. Zervos. 1993. Plasmid-associated hemolysin and aggregation substance production contribute to virulence in experimental enterococcal endocarditis. Antimicrob. Agents Chemother. 37:2474-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coque, T. M., J. E. Patterson, J. M. Steckelberg, and B. E. Murray. 1995. Incidence of hemolysin, gelatinase, and aggregation substance among enterococci isolated from patients with endocarditis and other infections and from feces of hospitalized and community-based persons. J. Infect. Dis. 171:1223-1229. [DOI] [PubMed] [Google Scholar]
- 5.Coque, T. M., R. Willems, R. Canton, R. Del Campo, and F. Baquero. 2002. High occurrence of esp among ampicillin-resistant and vancomycin-susceptible Enterococcus faecium clones from hospitalized patients. J. Antimicrob. Chemother. 50:1035-1038. [DOI] [PubMed] [Google Scholar]
- 6.Coque, T. M., R. Willems, J. Fortún, J. Top, S. Diz, R. Canton, E. Loza, and F. Baquero. 2003. Population structure of Spanish Enterococcus faecium isolates causing bacteremia: evolution, diversity of isolates with variable antibiotic resistance and virulence profiles and clinical outcomes, abstr. K-1111. In Abstracts of the 43rd Interscience Conference on Antimicrobial Agents and Chemotherapy. American Society for Microbiology, Washington, D.C.
- 7.Descheemaeker, P., C. Lammens, B. Pot, P. Vandamme, and H. Goossens. 1997. Evaluation of arbitrarily primed PCR analysis and pulsed-field gel electrophoresis of large genomic DNA fragments for identification of enterococci important in human medicine. Int. J. Syst. Bacteriol. 47:555-561. [DOI] [PubMed] [Google Scholar]
- 8.Dupre, I., S. Zanetti, A. M. Schito, G. Fadda, and L. A. Sechi. 2003. Incidence of virulence determinants in clinical Enterococcus faecium and Enterococcus faecalis isolates collected in Sardinia (Italy). J. Med. Microbiol. 52:491-498. [DOI] [PubMed] [Google Scholar]
- 9.Dutka-Malen, S., S. Evers, and P. Courvalin. 1995. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J. Clin. Microbiol. 33:1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eaton, T. J., and M. J. Gasson. 2001. Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl. Environ. Microbiol. 67:1628-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eaton, T. J., and M. J. Gasson. 2002. A variant enterococcal surface protein Esp(fm) in Enterococcus faecium; distribution among food, commensal, medical, and environmental isolates. FEMS Microbiol. Lett. 216:269-275. [DOI] [PubMed] [Google Scholar]
- 12.Elsner, H. A., I. Sobottka, D. Mack, M. Claussen, R. Laufs, and R. Wirth. 2000. Virulence factors of Enterococcus faecalis and Enterococcus faecium blood culture isolates. Eur. J. Clin. Microbiol. Infect. Dis. 19:39-42. [DOI] [PubMed] [Google Scholar]
- 13.Feng, P., and S. R. Monday. 2000. Multiplex PCR for detection of trait and virulence factors in enterohemorrhagic Escherichia coli serotypes. Mol. Cell. Probes 14:333-337. [DOI] [PubMed] [Google Scholar]
- 14.Galli, D., F. Lottspeich, and R. Wirth. 1990. Sequence analysis of Enterococcus faecalis aggregation substance encoded by the sex pheromone plasmid pAD1. Mol. Microbiol. 4:895-904. [DOI] [PubMed] [Google Scholar]
- 15.Gilmore, M. S., R. A. Segarra, and M. C. Booth. 1990. An HlyB-type function is required for expression of the Enterococcus faecalis hemolysin/bacteriocin. Infect. Immun. 58:3914-3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goossens, H., D. Jabes, R. Rossi, C. Lammens, G. Privitera, and P. Courvalin. 2003. European survey of vancomycin-resistant enterococci in at-risk hospital wards and in vitro susceptibility testing of ramoplanin against these isolates. J. Antimicrob. Chemother. 51(Suppl. 3):iii5-iii12. [DOI] [PubMed] [Google Scholar]
- 17.Gutschik, E., S. Moller, and N. Christensen. 1979. Experimental endocarditis in rabbits. 3. Significance of the proteolytic capacity of the infecting strains of Streptococcus faecalis. Acta Pathol. Microbiol. Scand. Sect. B 87:353-362. [PubMed] [Google Scholar]
- 18.Guzman, C. A., C. Pruzzo, G. LiPira, and L. Calegari. 1989. Role of adherence in pathogenesis of Enterococcus faecalis urinary tract infection and endocarditis. Infect. Immun. 57:1834-1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henegariu, O., N. A. Heerema, S. R. Dlouhy, G. H. Vance, and P. H. Vogt. 1997. Multiplex PCR: critical parameters and step-by-step protocol. BioTechniques 23:504-511. [DOI] [PubMed] [Google Scholar]
- 20.Hynes, W. L., and S. L. Walton. 2000. Hyaluronidases of gram-positive bacteria. FEMS Microbiol. Lett. 183:201-207. [DOI] [PubMed] [Google Scholar]
- 21.Ike, Y., D. B. Clewell, R. A. Segarra, and M. S. Gilmore. 1990. Genetic analysis of the pAD1 hemolysin/bacteriocin determinant in Enterococcus faecalis: Tn917 insertional mutagenesis and cloning. J. Bacteriol. 172:155-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ike, Y., H. Hashimoto, and D. B. Clewell. 1987. High incidence of hemolysin production by Enterococcus (Streptococcus) faecalis strains associated with human parenteral infections. J. Clin. Microbiol. 25:1524-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jett, B. D., M. M. Huycke, and M. S. Gilmore. 1994. Virulence of enterococci. Clin. Microbiol. Rev. 7:462-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jett, B. D., H. G. Jensen, R. E. Nordquist, and M. S. Gilmore. 1992. Contribution of the pAD1-encoded cytolysin to the severity of experimental Enterococcus faecalis endophthalmitis. Infect. Immun. 60:2445-2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kariyama, R., H. Kuman, A. M. Hammerum, F. M. Aarestrup, and L. B. Jensen. 2001. Identification of a Tn1546-like (type 2) element in vancomycin-resistant Enterococcus faecium isolated from hospitalized patients in Japan. Antimicrob. Agents Chemother. 45:992-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kreft, B., R. Marre, U. Schramm, and R. Wirth. 1992. Aggregation substance of Enterococcus faecalis mediates adhesion to cultured renal tubular cells. Infect. Immun. 60:25-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leavis, H. L., R. Willems, J. Top, E. Spalburg, E. M. Mascini, A. C. Fluit, A. Hoepelman, A. J. de Neeling, and M. J. Bonten. 2003. Epidemic and non-epidemic multidrug-resistant Enterococcus faecium. Emerg. Infect. Dis. 9:1108-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moellering, R. C., Jr. 1992. Emergence of Enterococcus as a significant pathogen. Clin. Infect. Dis. 14:1173-1176. [DOI] [PubMed] [Google Scholar]
- 29.National Committee for Clinical Laboratory Standards. 2002. Performance standards for antimicrobial susceptibility testing; twelfth informational supplement, M100-S12, vol. 22, no. 11. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 30.Olmsted, S. B., G. M. Dunny, S. L. Erlandsen, and C. L. Wells. 1994. A plasmid-encoded surface protein on Enterococcus faecalis augments its internalization by cultured intestinal epithelial cells. J. Infect. Dis. 170:1549-1556. [DOI] [PubMed] [Google Scholar]
- 31.Pass, M. A., R. Odedra, and R. M. Batt. 2000. Multiplex PCRs for identification of Escherichia coli virulence genes. J. Clin. Microbiol. 38:2001-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patel, R., J. R. Uhl, P. Kohner, M. K. Hopkins, and F. R. Cockerill, III. 1997. Multiplex PCR detection of vanA, vanB, vanC-1, and vanC-2/3 genes in enterococci. J. Clin. Microbiol. 35:703-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Polissi, A., A. Pontiggia, G. Feger, M. Altieri, H. Mottl, L. Ferrari, and D. Simon. 1998. Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect. Immun. 66:5620-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pot, B., P. Vandamme, and K. Kersters. 1994. Analysis of electrophoretic whole-organism protein fringerprints, p. 493-521. In M. Goodfellow and A. G. O'Donnell (ed.), Modern microbial methods. Chemical methods in prokaryotic systematics. John Wiley & Sons Ltd., Chichester, United Kingdom.
- 35.Rice, L. B., L. Carias, S. Rudin, C. Vael, H. Goossens, C. Konstabel, I. Klare, S. R. Nallapareddy, W. Huang, and B. E. Murray. 2003. A potential virulence gene, hylEfm, predominates in Enterococcus faecium of clinical origin. J. Infect. Dis. 187:508-512. [DOI] [PubMed] [Google Scholar]
- 36.Semedo, T., S. M. Almeida, P. Martins, M. F. Silva Lopes, J. J. Figueiredo Marques, R. Tenreiro, and M. T. Barreto Crespo. 2003. Comparative study using type strains and clinical and food isolates to examine hemolytic activity and occurrence of the cyl operon in enterococci. J. Clin. Microbiol. 41:2569-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shankar, N., C. V. Lockatell, A. S. Baghdayan, C. Drachenberg, M. S. Gilmore, and D. E. Johnson. 2001. Role of Enterococcus faecalis surface protein Esp in the pathogenesis of ascending urinary tract infection. Infect. Immun. 69:4366-4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shankar, V., A. S. Baghdayan, M. M. Huycke, G. Lindahl, and M. S. Gilmore. 1999. Infection-derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infect. Immun. 67:193-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Su, Y. A., M. C. Sulavik, P. He, K. K. Makinen, P. L. Makinen, S. Fiedler, R. Wirth, and D. B. Clewell. 1991. Nucleotide sequence of the gelatinase gene (gelE) from Enterococcus faecalis subsp. liquefaciens. Infect. Immun. 59:415-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toledo-Arana, A., J. Valle, C. Solano, M. J. Arrizubieta, C. Cucarella, M. Lamata, B. Amorena, J. Leiva, J. R. Penades, and I. Lasa. 2001. The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Appl. Environ. Microbiol. 67:4538-4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vael, C., V. Vankerckhoven, S. Chapelle, and H. Goossens. 2001. Detection of esp gene in non-epidemic vancomycin-sensitive Enterococcus faecium isolates in association with ampicillin resistance, abstr. 967. In Abstracts of the 41st Interscience Conference on Antimicrobial Agents and Chemotherapy. American Society for Microbiology, Washington, D.C.
- 43.Waar, K., A. B. Muscholl-Silberhorn, R. J. Willems, M. J. Slooff, H. J. Harmsen, and J. E. Degener. 2002. Genogrouping and incidence of virulence factors of Enterococcus faecalis in liver transplant patients differ from blood culture and fecal isolates. J. Infect. Dis. 185:1121-1127. [DOI] [PubMed] [Google Scholar]
- 44.Willems, R. J., W. Homan, J. Top, M. Santen-Verheuvel, D. Tribe, X. Manzioros, C. Gaillard, C. M. Vandenbroucke-Grauls, E. M. Mascini, E. van Kregten, J. D. van Embden, and M. J. Bonten. 2001. Variant esp gene as a marker of a distinct genetic lineage of vancomycin-resistant Enterococcus faecium spreading in hospitals. Lancet 357:853-855. [DOI] [PubMed] [Google Scholar]
- 45.Woodford, N., M. Soltani, and K. J. Hardy. 2001. Frequency of esp in Enterococcus faecium isolates. Lancet 358:584. [DOI] [PubMed] [Google Scholar]