Abstract
Vibrio cholerae, the causative agent of cholera, is a natural inhabitant of the aquatic ecosystem. We examined a unique collection of V. cholerae clinical and environmental isolates of widespread geographic distribution recovered over a 60-year period to determine their evolutionary genetic relationships based on analysis of two housekeeping genes, malate dehydrogenase (mdh) and a chaperonin (groEL). In addition, the phylogenetic distribution of 12 regions associated with virulence was determined. Comparative sequence analysis of mdh revealed that all V. cholerae O1 and O139 serogroup isolates belonged to the same clonal lineage. Single-strand conformational polymorphism (SSCP) analysis of these O1 and O139 strains at groEL confirmed the presence of an epidemic clonal complex. Of the 12 virulence regions examined, only three regions, Vibrio seventh pandemic island 1 (VSP-I), VSP-II, and RS1, were absent from all classical V. cholerae isolates. Most V. cholerae El Tor biotype and O139 serogroup isolates examined encoded all 12 virulence regions assayed. Outside of V. cholerae O1/O139 serogroup isolates, only one strain, VO7, contained VSP-I. Two V. cholerae El Tor isolates, GP155 and 2164-78, lacked both VSP-I and VSP-II, and one El Tor isolate, GP43, lacked VSP-II. Five non-O1/non-O139 serogroup isolates had an mdh sequence identical to that of the epidemic O1 and O139 strains. These isolates, similar to classical strains, lack both VSP-I and VSP-II. Four of the 12 virulence regions examined were found to be present in all isolates: hlyA, pilE, MSHA and RTX. Among non-O1/non-O139 isolates, however, the occurrence of the additional eight regions was considerably lower. The evolutionary relationships and multilocus virulence gene profiles of V. cholerae natural isolates indicate that consecutive pandemic strains arose from a common O1 serogroup progenitor through the successive acquisition of new virulence regions.
Vibrio cholerae is a natural inhabitant of the aquatic environment and is found associated with shellfish and crustaceans (12, 21, 29, 48). V. cholerae is the causative agent of the diarrheal disease cholera, and humans are the only known animal host. V. cholerae is an extracellular pathogen of the small intestine and causes significant human disease and death, particularly on the Indian subcontinent. A recent study has shown that the human host may contribute significantly to cholera epidemics, since passage through the human intestine was shown to induce a hyperinfectious state, which was perpetuated in the natural environment after release (43). Of the 200 O-antigen serogroups so far identified among V. cholerae isolates, only two serogroups, O1 and O139, are known to cause epidemic and pandemic cholera (34). The V. cholerae O1 serogroup can be further divided into two biotypes of epidemiological relevance, classical and El Tor, based on minor phenotypic differences. The first cholera pandemic, which began in 1817 in Asia, and subsequent pandemics, were probably caused by the classical biotype. In 1961, the seventh and present pandemic began, which was caused by the El Tor biotype (34). In 1992, for the first time in the recorded history of cholera a novel O-serogroup, O139 emerged to cause epidemic cholera (1). Significantly, exposure to O1 serogroup cholera does not protect against O139 cholera (45). The El Tor strain reemerged to overtake the O139 serogroup as the major cause of cholera by 1996 (23). However, the O139 serogroup is still present on the Indian subcontinent and, in some areas, is the predominant cause of cholera (23). Interestingly, several studies have proposed that the origin of the serogroup O139 strain was an El Tor strain that obtained the O139 biosynthesis genes (as well as the SXT element and a capsule) via antigenic switching from a donor strain (3, 44, 60, 65). Recently, it has been proposed based on comparative sequence analysis that an O22 serogroup maybe a possible donor for the O139 serogroup (17, 67). Sporadic cholera outbreaks caused by V. cholerae non-O1 and non-O139 isolates have been documented; for example, in 1968 in Sudan there was a cholera outbreak caused by an O37 serogroup isolate (21, 68).
The evolutionary genetic relationships among V. cholerae strains have been examined by multilocus enzyme electrophoresis (2, 11, 19, 20, 54, 58), single locus sequence analysis (7, 37, 38, 59), and multilocus sequence analysis of housekeeping genes (9, 36, 40). These analyses have given conflicting results regarding the ancestry of O1 serogroup classical and El Tor biotype strains. Several studies suggest that at least three pathogenic clones exist, consisting of classical and El Tor biotype strains and U.S. Gulf Coast strains (37, 63). Others have suggested that the three pathogenic clones are very closely related (2, 9, 18). In all studies, the epidemic V. cholerae isolates form a lineage separate from nonepidemic strains.
Although the O-antigen is a major protective antigen in V. cholerae virulence and probably plays a role in host colonization, the two major virulence factors of V. cholerae are cholera toxin (CT), the main cause of the explosive rice watery diarrhea, and toxin-coregulated pilus (TCP), the main intestinal colonization factor (61). The ctxAB genes, which encode CT, are integral components of a novel filamentous phage CTXφ (64), and the TCP biosynthesis genes are encoded on the Vibrio pathogenicity island (hereafter designated VPI-1) (35). A number of studies have found that the two main virulence factors, CT and TCP, are predominately associated with V. cholerae O1 and O139 serogroup strains and are only occasionally found in nonepidemic isolates (5, 7, 8, 10, 13, 25, 40, 47, 49, 50, 55).
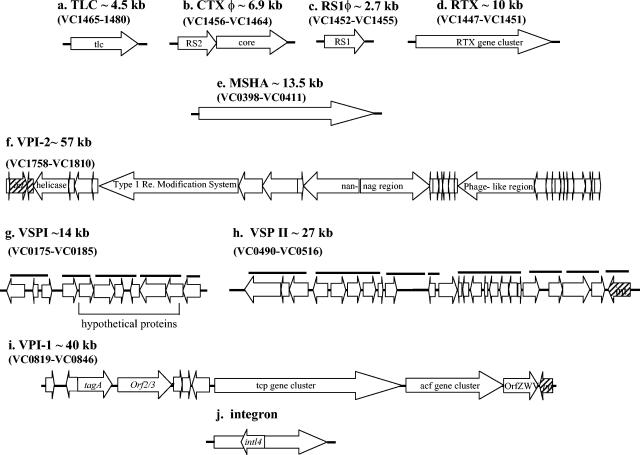
Among V. cholerae El Tor isolates, CTXφ is flanked by an additional filamentous phage RS1φ that is required for CTXφ production (16, 22) (Fig. 1). The CTX prophage is also flanked by the toxin-linked cryptic plasmid (TLC), whose role in pathogenesis is unknown (56) (Fig. 1). A number of other gene clusters have also been identified that are found predominantly among epidemic V. cholerae isolates: the RTX toxin gene cluster (42), the mannose-sensitive hemolysin agglutination pilin (MSHA) (32), VPI-2 (31), hemolysin, and PilE pilin (28, 30) (Fig. 1). Recently, comparative genomic studies which used a V. cholerae DNA microarray among 11 epidemic isolates identified two regions, Vibrio seventh pandemic island I (VSP-I), encompassing VC0175 to VC0185, and VSP-II, encompassing VC0490 to VC0497, that were found exclusively among El Tor biotype isolates (18). The role of VSP-I and VSP-II in V. cholerae virulence remains undetermined.
FIG. 1.
Schematic representation of nine regions that are associated with pathogenesis in V. cholerae. The positions and directions of transcription of the open reading frames are indicated by the directions of the arrows. The black bold horizontal lines indicate the positions of the PCR primers.
To elucidate the steps and significance of virulence gene acquisition in the evolution of V. cholerae it is essential to know the underlying phylogenetic relationships among strains. In this study we examined a unique collection of 64 V. cholerae and 5 Vibrio mimicus isolates to determine their evolutionary genetic relationships and multilocus virulence gene profiles to elucidate the steps involved in the emergence of epidemic isolates. Our results show that V. cholerae serogroup O1 classical and El Tor biotype strains encompass a single epidemic clonal complex and that differences between biotype strains arose through the acquisition of additional virulence regions by El Tor isolates. The emergence of epidemic V. cholerae O139 serogroup strains was not a unique occurrence in the history of cholera, since V. cholerae O37 and O8 serogroup isolates phylogenetically cluster with O1 and O139 serogroup isolates, indicating antigenic switching.
MATERIALS AND METHODS
Bacterial isolates.
A total of 64 V. cholerae isolates were examined in this study (Table 1). The 64 V. cholerae isolates belonged to 19 different serogroups, 3 isolates had no serogroup designation, and 23 isolates belonged to serogroup O1, of which 6 isolates were of the classical biotype and 15 isolates were of the El Tor biotype. The O139 serogroup was represented by 13 isolates, the O37 serogroup was represented by 7 isolates, and the O8 and O141 serogroups each were represented by 2 isolates. There were 14 serogroups represented by a single V. cholerae isolate. The V. cholerae isolates were recovered from six continents (North and South America, Asia, Europe, Australia, and Africa) over a 60-year period (1937 to 2000) (Table 1). In addition, our study also included five V. mimicus isolates, four O115 and one O41 serogroup isolates. Of the 69 strains examined, 51 were clinical isolates and 12 were environmental isolates (Table 1). All Vibrio strains were grown in Luria-Bertani (LB) broth and stored at −70°C in LB broth with 20% (vol/vol) glycerol.
TABLE 1.
Strains used in this study
| Strain | Serogroup (biotype) | Source | Place of isolation | Yr of isolation |
|---|---|---|---|---|
| V. cholerae | ||||
| Sixth pandemic classical strains | ||||
| O395 | O1 (classical) | Clinical | India | 1964 |
| 569B | O1 (classical) | Clinical | India | 1948 |
| C1 | O1 (classical) | Unknown | Unknown | 1955 |
| C14 | O1 (classical) | Environmental | Unknown | 1973 |
| CA401 | O1 (classical) | Clinical | India | 1953 |
| GP12 | O1 (classical) | Unknown | India | 1971 |
| Seventh pandemic El Tor strains | ||||
| GP33 | O1 (El Tor) | Unknown | Union of Soviet Socialist Republics | 1971 |
| GP43 | O1 (El Tor) | Unknown | Australia | 1972 |
| GP155 | O1 (El Tor) | Unknown | Australia | 1979 |
| N16961 | O1 (El Tor) | Clinical | Bangladesh | 1975 |
| 3038 | O1 (El Tor) | Clinical | Vietnam | Unknown |
| F1873 | O1 (El Tor) | Clinical | Rwanda | 1994 |
| F1875 | O1 (El Tor) | Clinical | Goma, Zaire | 1994 |
| F1939 | O1 (El Tor) | Clinical | Rwanda | 1994 |
| SM115 | O1 (El Tor) | Clinical | Bahrain | 1978 |
| 2125-98 | O1 (El Tor) | Clinical | Bangladesh | 1998 |
| 2164-78 | O1 (El Tor) | Environmental | U.S. Gulf Coast | 1978 |
| Pre-seventh pandemic El Tor strains | ||||
| RV79 | O1 (El Tor) | Clinical | Indonesia | 1937 |
| C5 | O1 (El Tor) | Unknown | Indonesia | 1957 |
| Nontoxigenic El Tor strains | ||||
| 2740-80 | O1 (El Tor) | Environmental | U.S. Gulf Coast | 1980 |
| 468-83 | O1 (El Tor) | Clinical | United States | 1983 |
| O1 serogroup strains of unknown biotype | ||||
| 1528-79 | O1 | Environmental | Louisiana | 1979 |
| 917-84 | O1 | Clinical | Georgia (United States) | 1984 |
| O139 serogroup | ||||
| 36054-98 | O139 | Clinical | Bangladesh | 1998 |
| SG20 | O139 | Clinical | Calcutta, India | Unknown |
| AS207 | O139 | Clinical | Calcutta, India | 1996 |
| AS209 | O139 | Clinical | Calcutta, India | 1996 |
| AS212 | O139 | Clinical | Calcutta, India | 1996 |
| AS213 | O139 | Clinical | Calcutta, India | 1996 |
| AS231 | O139 | Clinical | Calcutta, India | 1996 |
| AS259 | O139 | Clinical | Calcutta, India | 1996 |
| AS260 | O139 | Clinical | Calcutta, India | 1996 |
| 35636-97 | O139 | Clinical | Bangladesh | 1997 |
| MO2 | O139 | Clinical | Madras, India | 1992 |
| MO10 | O139 | Clinical | Madras, India | 1992 |
| MO45 | O139 | Clinical | Madras, India | 1992 |
| Non-O1 and non-O139 serogroup strains | ||||
| 151 | O37 | Environmental | Mexico | 1998 |
| CO476 | O37 | Clinical | Calcutta, India | 1994 |
| VO7 | O37 | Environmental | Varanasi, India | 1988 |
| CO130 | O37 | Environmental | Calcutta, India | 1993 |
| V45 | Rough | Clinical | Bangladesh | 1978 |
| V46 | O141 | Clinical | United States | 1978 |
| V47 | O141 | Clinical | United States | 1984 |
| V52 | O37 | Clinical | Sudan | 1968 |
| V53 | O37 | Clinical | Sudan | 1968 |
| V54 | O8 | Clinical | Thailand | Unknown |
| SG3 | O32 | Clinical | Calcutta, India | 1992-1993 |
| SG6 | O45 | Clinical | Calcutta, India | 1992-1993 |
| SG7 | O56 | Clinical | Calcutta, India | 1992-1993 |
| SG8 | O37 | Clinical | Calcutta, India | 1992-1993 |
| SG9 | O38 | Clinical | Calcutta, India | Unknown |
| SG10 | O69 | Clinical | Calcutta, India | 1992-1993 |
| SG14 | O54 | Clinical | Calcutta, India | 1992-1993 |
| AM112 | O39 | Clinical | Calcutta, India | 1996 |
| VIG1613 | O12 | Clinical | Peru | Unknown |
| DK59 | O70 | Environmental | Germany | 1994 |
| DK67 | O74 | Environmental | Korea | 1994 |
| DK71 | O66 | Environmental | Germany | 1994 |
| SCE4 | O8 | Environmental | India | 1997 |
| SCE188 | O44 | Environmental | India | 1997 |
| 208 | O11 | Clinical | Thailand | 1998 |
| C43 | NAG | Clinical | Unknown | Unknown |
| 9581 | O41 | Clinical | India | 1990 |
| 9582 | Rough | Clinical | India | 1990 |
| V. mimicus | ||||
| PT5 | O115 | Clinical | Bangladesh | 1985 |
| PT48 | O115 | Clinical | Bangladesh | 1985 |
| 9583 | O115 | Clinical | United States | 1980 |
| 523-80 | O115 | Clinical | United States | 1980 |
| 531-90 | O41 | Clinical | Japan | 1990 |
DNA isolation.
Chromosomal DNA was extracted from each V. cholerae and V. mimicus isolate by using the G-nome DNA isolation kit from Bio 101 (Vista, Calif.). Briefly, a single colony of each isolate was inoculated into 3 ml of LB broth and incubated overnight at 37°C with shaking at 150 rpm. The bacterial cells were pelleted at 3,000 rpm for 5 min, the supernatant was discarded, and the pellet brought to a final volume of 1.85 ml in cell suspension solution. The cells were lysed and treated with RNase and protease. DNA was extracted with Tris-EDTA buffer and ethanol and resuspended in Tris-EDTA buffer.
PCR amplification and nucleotide sequencing.
PCR primers to amplify the chromosomal housekeeping gene malate dehydrogenase (mdh) were designed from the mdh sequence of V. cholerae strain N16961 (26). The following PCR cycle was used to amplify the mdh gene for each isolate: an initial denaturation step at 96°C for 1 min followed by 30 cycles of denaturation at 94°C for 30 s, 30 s of primer annealing at 53.9°C, and 1.5 min of primer extension at 72°C. The primer pair mdh1-mdh2 amplified an 892-bp fragment, representing 84% of the mdh gene. PCR products were purified with the QIAquick PCR purification kit (Qiagen, Hilden, Germany) in accordance with the manufacturer's instructions. After purification, an aliquot of 10 μl was used as a sequencing template. The mdh gene sequences were determined in both directions by MWG-Biotech based on the dye deoxy terminator method.
Phylogenetic analyses.
The mdh gene sequences were aligned by using the CLUSTALW multiple-sequence alignment program (27). From the mdh sequence alignments, a 648-bp region was further analyzed by using the Molecular Evolutionary Genetics Analysis (MEGA) suite of programs, version 2.1 (39). Phylogenetic gene trees were constructed by the neighbor-joining method with the Jukes-Cantor distance method (33, 57). Bootstrap values were calculated for 1,000 trees. The proportions of synonymous (silent) substitutions per synonymous site (Ds) and nonsynonymous (replacement) substitutions per nonsynonymous site (Dn) were calculated.
PCR-SSCP.
In conjunction with mdh sequencing, an additional 25 V. cholerae O1 and O139 serogroup strains were analyzed to confirm sequence identity at this locus within these two serogroups by PCR-single-strand conformational polymorphism analysis (PCR-SSCP), a simple and rapid method to determine point mutations within genes. Two oligonucleotide primers, mdh1 and mdh2 (Table 2), were used to amplify an 892-bp PCR product, which was then restricted with HindIII (Roche Molecular Biochemicals, East Sussex, United Kingdom) at 37°C to generate two fragments. Then 5 μl of the restricted DNA was mixed with 5 μl of denaturation buffer (5 mM EDTA, 0.05% bromophenol blue, and xylene cyanole in formamide), and the mixture was incubated at 95°C for 8 min. The sample was then placed directly in ice for 10 min before being loaded onto a nondenaturing 8% polyacrylamide gel. Samples (8 μl) were run at 100 V for 2 h. As a control, 4 μl of undenatured digested DNA (mdh gene) was used.
TABLE 2.
PCR primers used in this study
| Primer | Sequence (5′-3′) | Predicted PCR product size (bp) | Annealing temp (°C) | Reference or source |
|---|---|---|---|---|
| Housekeeping gene primers | ||||
| mdh1 | ATGAAAGTCGCTGTTATT | 892 | 53.9 | 7 |
| mdh2 | GTATCTAACATGCCATCC | |||
| groEL1A | GATCCATATGGCTGCTAAAGACGTACG | 1,600 | 58 | This study |
| groEL1B | CTAGGTCGACTTACATCATGCGGCCCATGC | |||
| Virulence gene primers | ||||
| VSP-1 | ||||
| VC0175F | TGGATGCTCTCTTCTTCA | 2,834 | 52 | This study |
| VC0175R | CGCTCACTCACTAATACCGAG | |||
| VC0178F | AGAGGCTTGTTTACTATCAG | 2,053 | 50 | This study |
| VC0178R | ATCGGTACTGTCAGGGCT | |||
| VC0180F | GGATGAGCAAATACAGCTAAC | 2,283 | 50 | This study |
| VC0180R | CTAGGAAGAATTTTATCGGC | |||
| VC0183F | CAGTAAGAGTGTAGCGTGCC | 3,389 | 52 | This study |
| VC0183R | CCTGCACATCGAGATGC | |||
| VC0185F | AGGAGGCGTGTAAGTCATAGC | 1,110 | 55 | This study |
| VC0185R | AGACCACGAATACCTGCTCC | |||
| MSHA | ||||
| msha398F | GGAACGTGGCACAAATG | 3,000 | 49 | This study |
| msha398R | TGACGTAAGTGAGCCGC | |||
| msha400F | AAGATGAAATCGGGTTG | 2,212 | 45 | This study |
| msha400R | TATCTGGCGACGCTTGC | |||
| msha403F | GAACCGATTTATCTGTAGGAG | 3,874 | 51 | This study |
| msha403R | TGACCGCCATTATCTGATAC | |||
| msha406F | CGAGTATTAAGGTACTGAAGG | 596 | 50 | This study |
| msha406R | ATCGGTCAGCTTGATCG | |||
| HlyA | ||||
| VC0489F | AGATCAACTACGATCAAGCC | 1,677 | 54.2 | This study |
| VC0489R | AGAGGTTGCTATGCTTTCTAC | |||
| VSP-II | ||||
| VC0490F | CGTGAAGGGATATAGGAG | 2,337 | 49.6 | This study |
| VC0490R | TGCAGTTGTTGAATGGAC | |||
| VC0493F | AATGCTTCTCAGGGGGGTCTT | 3,600 | 57 | This study |
| VC0493R | CGCTCTTCTTTCCACGCTTCA | |||
| VC0498F | AGGTGGTATCGGGCTGGT | 4,140 | 58 | This study |
| VC0498R | TGCGGCTGGAATGGAGTCTG | |||
| VC0502F | TCATCAGTTAGCACACGAAC | 476 | 52 | This study |
| VC0502R | GCTATCGTTATACTTGGCG | |||
| VC0504F | CAGCAAAGGCGGAAGAGGTAG | 3,240 | 55 | This study |
| VC0504R | AGCCCGAAATGAATCCCAAAA | |||
| VC0512F | CAGTGGCTTCGCAGAGGA | 3,900 | 53 | This study |
| VC0512R | CCCTCCACTGCTATTCCG | |||
| VC0514F | TTATGATCCAAGGAGTAGGG | 2,089 | 52 | This study |
| VC0514R | AGGCTGAAAAACAACTTGAG | |||
| VC0516F | GTTTTCTGCGTTGTTCGAG | 965 | 52 | This study |
| VC0516R | TCCTGATGTCTCTCTTGCCG | |||
| VC0517F | CCCACTTCTTCCAGAGTATG | 1,753 | 54.3 | This study |
| VC0517R | CGCAGTCACAGCTTAAACAAC | |||
| VPI-1 | ||||
| tcpH1 | AGCCGCCTAGATAGTCTGTG | 2,176 | 51.7 | 51 |
| tcpA4 | TCGCCTCCAATAATCCGAC | |||
| toxt1 | AGGAGATGGAAGTGGTGTG | 1,055 | 48.7 | 53 |
| toxt2 | CTTGGTGCTACATTCATGG | |||
| acfB1 | GATGAAAGAACAGGAGAGA | 1,180 | 49 | 53 |
| acfB2 | CAGCAACCACAGCAAAACC | |||
| PilE | ||||
| PilEF | CATACCTTTTGAGCATCGAC | 3,087 | 50 | This study |
| PilER | GTGGCAAGAAGGACTCG | |||
| RTX | ||||
| rtxA1 | GCGATTCTCAAAGAGATGC | 1,366 | 53.8 | 42 |
| rtxA2 | CACTCATTCCGATAACCAC | |||
| RS1φ | ||||
| rstC1 | AACAGCTACGGGCTTATTC | 238 | 52.4 | 66 |
| rstC2 | TGAGTTGCGGATTTAGGC | |||
| CTXφ | ||||
| rstA1 | ACTCGATACAAACGCTTCTC | 1,009 | 53.7 | 66 |
| rstA2 | AGAATCTGGAAGGTTGAGTG | |||
| orfU | CGTCACACCAGTTACTTTTCG | 1,072 | 54.5 | 62 |
| orfU | AGAATGTACGCCATCGC | |||
| zot1 | GGCTTAAACCTTGAACGC | 1,036 | 54.7 | 24 |
| zot2 | AACCCCGTTTCACTTCTAC | |||
| ctxA1 | AGTCAGGTGGTCTTATGCC | 1,037 | 51.2 | This study |
| ctxB2 | TTGCCATACTAATTGCGG | |||
| tlc3 | GGGAATGTTGAGTTCTCAGTG | 1,548 | 55.5 | 56 |
| tlc4 | GTTGCGAAGTGGATTTTGTG | |||
| intl4:3 | CCTTCATTGGATCACTCG | 597 | 51.9 | This study |
| intl4:4 | GACGGAAAAAGATAGTGCC |
In addition, all epidemic V. cholerae O1 and O139 serogroup isolates were examined at the groEL locus by PCR-SSCP analysis. Primer pair groEL1A and groEL1B, designed from V. cholerae genome sequence (26) were used to PCR amplify a 1.6-kb band from 20 V. cholerae O1 and O139 serogroup isolates and 2 O37 serogroup isolates. PCR products were digested with BstYI at 60°C to generate four restriction bands. Restricted DNA (10 μl) was denatured as described above in 10 μl of denaturation buffer and electrophoresed at 175 V for 6 h.
After electrophoresis, the 8% polyacrylamide gels were silver stained with a DNA silver staining kit (Pharmacia Biotech). Briefly, the silver staining procedure was as follows. The gels were first fixed in 10% acetic acid for approximately 30 min at room temperature and washed with deionized water three times for 2 min. Color impregnation lasted for 20 min at room temperature. The gel was then washed for 5 to 10 s with deionized water, followed by color development for 6 min with a color development solution. The color reaction was stopped, and the bands were fixed. The gel was air dried for approximately 2 h. SSCP profiles were interpreted visually.
PCR analysis.
PCR was used to assay 64 V. cholerae and 5 V. mimicus isolates for the presence of 12 regions associated with V. cholerae virulence. Of the 12 virulence regions examined, 10 regions were comprised of three or more genes (VSP-I, MSHA pilin, VSP-II, VPI-1, Repeat in toxin [RTX], RS1φ, CTXφ, TLC, VPI-2, and class 1 integron) and 2 loci were single gene regions (hlyA and pilE). Of the 10 virulence gene clusters examined, 7 are associated with mobile genetic elements (Fig. 1). A total of 31 primer pairs were used to determine the distribution of the 12 regions among the 69 Vibrio isolates (Table 2). Five primer pairs were used to assay for the presence of VSP-I, four primer pairs were used to assay for MSHA, nine primer pairs were used to assay for VSP-II, three primer pairs were used to assay for VPI-1, four primer pairs were used to assay for the presence of CTXφ, and one primer pair (each) was used to assay for the presence of pilE, hlyA, RTX, RS1φ, TLC, and intl4 (Table 2). Gene fragments were amplified from chromosomal DNA isolated from the 64 V. cholerae strains and the 5 V. mimicus strains. PCR was performed in a 20-μl reaction mixture by using the following cycles: an initial denaturation step at 96°C for 1 min followed by 30 cycles of denaturation at 94°C for 30 s, 30 s of primer annealing at 45 to 58°C, and 1 to 4 min of primer extension at 72°C (Table 2).
Southern blot analysis.
To confirm negative PCR results, Southern hybridization analysis was carried out. DNA from each strain of interest was digested with the restriction enzyme EcoRI (Roche Molecular Biochemicals) and separated by electrophoresis in 0.6% (wt/vol) 1× Tris-borate-EDTA agarose. Separated DNA fragments were transferred to a nitrocellulose membrane for Southern hybridization. A single DNA probe was generated for each of the 12 regions by PCR amplification with V. cholerae strain N16961 as a template and labeled with horseradish peroxidase to verify the absence of a particular gene. Southern hybridization was carried out by using the enhanced chemiluminescence direct nucleic acid labeling and detection system according to the manufacturer's instructions (Amersham Pharmacia Biotech). In all experiments, V. cholerae strain N16961 was used as a positive control.
RESULTS
Genetic variation at the mdh locus among V. cholerae isolates.
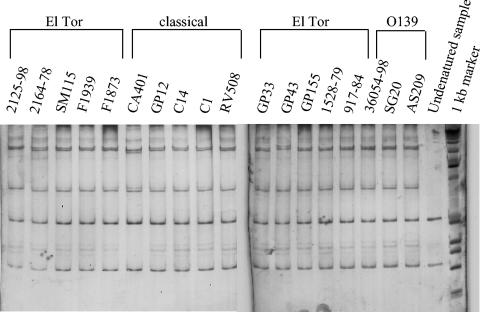
To determine the evolutionary genetic relationships among our collection of V. cholerae isolates, we analyzed a 648-bp region of the housekeeping gene malate dehydrogenase (mdh) from 36 V. cholerae isolates and 5 V. mimicus isolates. Previous studies have shown that comparative nucleotide sequence analysis of the mdh locus is a reliable indicator of overall genetic relationships between strains (6). Within the 648-bp region from the 36 V. cholerae strains examined, there was a total of 44 polymorphic sites, which included two amino acid replacement sites (Table 3; Fig. 2). Of the 44 polymorphic sites, 22 were phylogenetically informative (at least two or more sequences contained the polymorphism) (Table 3). The average pairwise difference for the 36 V. cholerae mdh sequences was 1.03%, with a maximum pairwise difference of 4.61% observed between V. cholerae strain DK71, an environmental O66 serogroup strain from Germany, and V. cholerae strain 9581, a clinical O41 serogroup isolate from India. Eight epidemic V. cholerae O1 El Tor and O139 serogroup strains examined had identical mdh sequences which differed from classical biotype strains at a single site. Three V. cholerae O37 serogroup strains, V52, V53, and CO130, one O8 serogroup strain, V54, and one rough strain, V45, had mdh sequences identical to the El Tor O1 and O139 serogroup mdh sequence. Among the 23 V. cholerae non-O1 and non-O139 isolates examined at the mdh locus, there were a total of 43 polymorphic sites, which resulted in 42 synonymous polymorphic sites and 1 nonsynonymous polymorphic site (Table 3). An additional 25 V. cholerae O1 and O139 serogroup isolates were examined for sequence variation at the mdh locus by PCR-SSCP analysis (Table 1) (Fig. 3). The sensitivity of PCR-SSCP tends to decrease with increasing fragment length, therefore the 892-bp amplicon was digested with HindIII to generate shorter fragments before PCR-SSCP analysis. Undenatured, digested mdh DNA of the V. cholerae strain produced two HindIII restricted bands of ∼500 and ∼300 bp. Denatured, digested mdh DNA produced 11 HindIII-restricted bands for all V. cholerae strains tested. Of the 25 strains analyzed, 23 exhibited PCR-SSCP profile 1, 1 classical strain, CA401, exhibited PCR-SSCP profile 2, and strain GP43 exhibited profile 3 (Fig. 3). There is a minor difference in the banding pattern of the three profiles, which could have resulted from a single nucleotide substitution. Overall, the mdh sequence and PCR-SSCP analyses indicate that the epidemic V. cholerae isolates at the mdh locus are highly homologous.
TABLE 3.
Sequence variation at the mdh locus among V. cholerae and V. mimicus strains
| No. of strains | Fragment size (bp) | Total no. of sites
|
ds ± SE | dn ± SE | ||||
|---|---|---|---|---|---|---|---|---|
| Polymorphic | Synonymous | Nonsynonymous | Informative | Singleton | ||||
| Within V. cholerae | ||||||||
| 36 | 648 | 44 | 42 | 2 | 22 | 22 | 0.038 ± 0.008 | 0.0004 ± 0.0003 |
| Within O1/O139 serogroups | ||||||||
| 13 | 648 | 9 | 8 | 1 | 2 | 7 | 0.008 ± 0.003 | 0.0006 ± 0.0006 |
| Within non-O1/non-O139 serogroups | ||||||||
| 23 | 648 | 43 | 42 | 1 | 21 | 22 | 0.049 ± 0.010 | 0.0002 ± 0.0002 |
| Within V. mimicus | ||||||||
| 5 | 648 | 7 | 6 | 1 | 0 | 7 | 0.014 ± 0.006 | 0.001 ± 0.001 |
| Between V. cholerae and V. mimicus | ||||||||
| 41 | 648 | 91 | 87 | 4 | 81 | 10 | 0.144 ± 0.017 | 0.003 ± 0.001 |
FIG. 2.
Polymorphic sites within the mdh gene of V. cholerae and V. mimicus isolates. N16961 represents 3038, F1875, RV79, AS207, MO10, MO45, CO130, V45, V52, V53, and V54. The numbering of the polymorphic sites (vertical format) is from the first position of the sequence segment. The position within the codon for each polymorphic site is shown below the sequences. Asterisks indicate polymorphic sites that gave rise to an amino acid change.
FIG. 3.
PCR-SSCP profiles of mdh fragments after HindIII digestion of 18 strains (25 strains profiled, 7 strains not shown). PCR-SSCP profiles are as follows. Profile 1, 2125-98, 2164-78, SM115, F1939, F1873, GP12, C14, C1, RV508, GP33, GP155, 1528-79, 917-84, 36054-98, SG20, and AS209 (AS212, AS213, AS231, AS259, AS260, 35636-97, and MO2 are not shown); profile 2, CA401; profile 3, GP43.
Genetic variation at the mdh locus between V. cholerae and V. mimicus isolates.
Analysis of the mdh sequence from the five clinical V. mimicus isolates identified seven polymorphic sites, six synonymous polymorphic sites, and one nonsynonymous site among these isolates (Table 3). Clinical V. mimicus O115 serogroup strains PT5, PT48, 9583, and 523-80 all had identical mdh sequences, which differed from strain 531-90, a clinical O41 serogroup isolate recovered in Japan in 1990. Comparative nucleotide sequence analysis of the mdh locus between V. cholerae and V. mimicus isolates revealed a total of 91 polymorphic nucleotide sites, of which 81 sites were phylogenetically informative (Table 3). Of the 91 polymorphic sites, 45 were unique to V. mimicus isolates and resulted in two amino acid replacements (Fig. 2). The average pairwise difference for the 36 V. cholerae and 5 V. mimicus mdh sequences was 3.4%, the maximum difference of 12.03% was between the V. mimicus isolates and V. cholerae isolates, which is similar to the divergence between Escherichia coli and Salmonella enterica serovar Typhimurium isolates at the mdh locus.
Genetic variation at groEL.
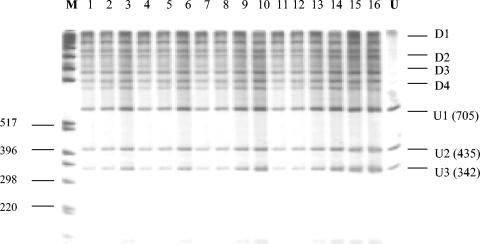
To elucidate further the relationships between V. cholerae O1 serogroup isolates, we examined 5 classical, 10 El Tor, 5 O139, and 2 O37 isolates by PCR-SSCP analysis at an additional locus, groEL. One of the most widely used techniques to localize mutations is PCR-SSCP, which is capable of detecting almost 100% of mutations. Alteration of the nucleotide sequence of the molecule by as little as a single base can reshape the secondary structure, with consequent changes in electrophoretic mobilities through a gel (52). The 1.6-kb groEL PCR amplicon was digested with BstYI, which resulted in four bands of 705, 435, 342, and 153 bp. Denatured, digested groEL DNA produced 11 bands representing profile 1 for all strains examined (Fig. 4). As can be seen from Fig. 4, V. cholerae classical and El Tor biotype strains gave identical banding patterns at groEL, indicating a lack of polymorphic sites in this gene among these isolates. In addition, V. cholerae O37 serogroup isolates V52 and V53 were also examined by PCR-SSCP at the groEL locus and, as expected, gave an identical banding pattern to the epidemic isolates, again confirming a common origin (data not shown). Taken together, the mdh sequence analysis and PCR-SSCP analyses at the mdh and groEL loci indicate that the V. cholerae O1 classical and El Tor biotypes and O139 serogroup strains are a highly homologous group of isolates representing a single clonal lineage.
FIG. 4.
PCR-SSCP profiles of groEL restriction fragments after BstYI digestion from three V. cholerae serogroups separated on a 0.8% nondenaturing polyacrylamide gel. The strains and respective serogroups are as follows: lanes 1 to 8, N16961, 2740-80, F1875, 3038, GP33, 2125-98, 2164-78, C5, O1 serogroup, biotype El Tor; lanes 9 to 12, CA401, O395, 569B, C14, O1 serogroup, biotype classical; lanes 13 to 15, MO10, MO45, 36054-98, O139 serogroup; lane 16, V52, O37 serogroup; lane M, Gibco-BRL 1-kb marker (with sizes in base pairs noted on the left); lane U, undenatured groEL restriction fragments from N16961. D1 to D4, single-stranded band pairs of the denatured 705-, 435-, 342-, and 153-bp BstYI groEL restriction fragments; U1 to U3, renatured double-stranded BstYI groEL restriction fragments (with sizes in base pairs noted in parentheses). The smaller 153- and 112-bp fragments were too faint to be presented here.
Evolutionary genetic relationships among V. cholerae natural isolates.
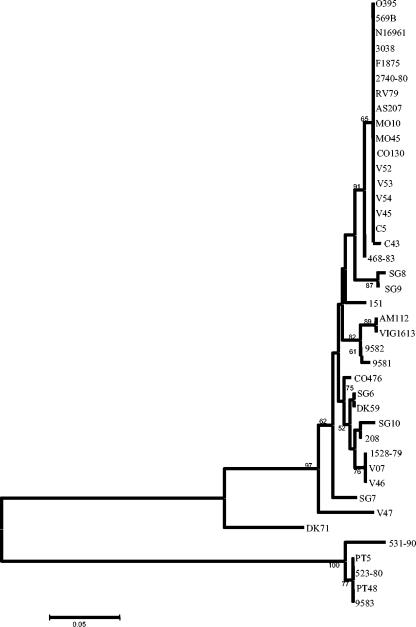
From the 36 V. cholerae and 5 V. mimicus mdh sequences, we constructed a neighbor-joining tree based on synonymous polymorphic sites, which are sites in a codon predicted to not result in amino acid replacements and are therefore not under selective pressure (Fig. 5). The mdh gene tree groups V. cholerae O1 classical and El Tor isolates and O139 serogroup isolates together to form an epidemic clone complex (Fig. 5). Interestingly, several V. cholerae non-O1 and non-O139 serogroup isolates also clustered with this epidemic clone complex: three toxigenic V. cholerae serogroup O37 strains, V52, V53, isolated in Sudan in 1968, and CO130, isolated in India in 1993, and one toxigenic O8 serogroup strain, V54, recovered in Thailand. In addition, V. cholerae strain V45, a rough isolate clustered with the epidemic clone, as well as a nonagglutinable strain, C43, and a nontoxigenic clinical O1 serogroup strain, 468-83, isolated on the U.S. Gulf Coast in 1983 (Fig. 5). Comparative sequence analysis also demonstrates that strains of the same serogroup may belong to two or more widely divergent lineages (Fig. 5). Thus, for example, of the seven V. cholerae O37 serogroup strains examined, the remaining four strains (SG8, 151, CO476, and VO7), which are nontoxigenic clinical and environmental isolates (SG8, CO476, and VO7 were isolated in India and 151 was isolated in Mexico), were found on four separate branches of the mdh gene tree, indicating their diverse evolutionary origins (Fig. 5). A similar picture emerges from the analysis of two O141 strains (V46 and V47) examined; they are also found on divergent branches of the mdh tree, suggesting that serogroup designation is not an indicator of overall relatedness but represents lateral gene transfer of the O-antigen among strains. Of the remaining 17 V. cholerae non-O1 and non-O139 isolates examined at the mdh locus, strains SG7, V47, and DK71 formed the most divergent branches. The non-O1 and non-O139 serogroup strains formed separate lineages from the epidemic strains but in general are closely related to one another, hence, the very small branch lengths. Two clinical V. cholerae strains, AM112, an O39 serogroup isolate from India, and VIG1613, an O12 serogroup isolate from Peru, clustered together, indicating identity. In addition, an O45 serogroup strain, SG6, from India and an O70 serogroup strain, DK59, from Germany clustered together, as did strains VO7, V46, and 1528-79 (Fig. 5). These data suggest the occurrence of clones of wide geographic distribution.
FIG. 5.
Neighbor-joining tree constructed by the Jukes-Cantor method with the nucleotide sequences of mdh gene fragments of V. cholerae strains. Construction and bootstrapping of the trees were carried out with the MEGA suite of programs. One thousand bootstrap replicates were performed for each analysis, and bootstrap values are given at the nodes.
We also identified two strains, 9581 and 9582, which were originally designated V. mimicus but clustered with V. cholerae non-O1 and non-O139 serogroup isolates on the mdh gene tree (Fig. 5). To determine the species designation of these isolates, we performed two biochemical tests previously used to differentiate V. cholerae and V. mimicus isolates: the Voges-Proskauer and corn oil tests. As expected, strains 9581 and 9582 were positive for both tests, similar to the control V. cholerae strains tested, indicating that these strains are indeed V. cholerae.
Evolutionary genetic relationships between V. cholerae and V. mimicus.
As expected, the five V. mimicus isolates formed a separate divergent branch from the V. cholerae isolates on the mdh gene tree. Four V. mimicus isolates, PT5, PT48, 9583, and 523-80, clustered together, and strain 531-90 formed a separate divergent lineage (Fig. 5).
Presence of virulence regions in V. cholerae and V. mimicus.
In total, 64 V. cholerae strains were examined for the presence of 12 regions associated with virulence in V. cholerae by PCR assays with 31 primer pairs (Table 2). Of the six classical biotype strains assayed by PCR, all strains contained the same nine regions, MSHA, hlyA, VPI-1, pilE, RTX, CTXφ, TLC, VPI-2, and intl4 and lacked RS1φ, VSP-I, and VSP-II (Table 4). The 15 El Tor biotype strains analyzed were divided into three groups based on the year of isolation and the presence of ctxAB (Table 1). Of the 11 toxigenic seventh pandemic strains examined by PCR analysis, 7 strains contained all 12 virulence regions examined. Two toxigenic El Tor strains recovered from Australia in the 1970s, GP155 and GP43, lacked VSP-I and VSP-II and VSP-II, respectively, by PCR and Southern blot analyses (Table 4). In addition, strain GP33 lacked TLC by PCR and Southern blot analyses. PCR analysis with six primer pairs (Table 2) showed that the VSP-II region was larger than previously documented (18) and encompassed an additional 19.4-kb region from VC0498 to VC0516. From our PCR analysis, we estimate that the VSP-II region is an ∼27-kb region encompassing VC0490 to VC0516 (Fig. 1). PCR assays indicated that VC0489 marked the 5′ flanking region and was found in all V. cholerae strains examined. Similarly, PCR assays with primer pair VC0517F-VCO517R showed that VC0517 marked the 3′ flanking region of VSP-II and was present in all V. cholerae strains examined. An environmental toxigenic El Tor isolate 2164-78, recovered in the United States in 1978, was shown not to contain VSP-I, VSP-II, and TLC by PCR and Southern blot analyses. A clinical pre-seventh pandemic El Tor strain RV79 isolated in Indonesia in 1937 lacked only three of the virulence regions examined: VSP-I, VSP-II, and VPI-2 (Table 4). The El Tor strain C5 isolated 20 years later in Indonesia contained 11 of the virulence regions with only RS1φ missing. Two nontoxigenic El Tor isolates, 468-83 and 2740-80, isolated in the United States in the early 1980s both lacked VSP-I, VSP-II, RS1φ, and CTXφ; additionally, strain 468-83 did not contain VPI-1 and TLC (Table 4). Two V. cholerae O1 serogroup strains of unknown biotype, 1528-79 and 917-84, were examined. Neither contained VSP-I, VSP-II, nor RS1φ; strain 1528-79 also lacked VPI-1, CTXφ, TLC, and VPI-2 (Table 4). The 13 V. cholerae O139 serogroup strains examined yielded positive PCR bands for all 12 virulence regions. However, as previously shown, there was a partial deletion of VPI-2 from 12 of the 13 O139 strains examined (31). These 12 O139 serogroup strains only contained a 20-kb 3′ region of VPI-2. Strain MO2 isolated in India in 1992 contained the entire VPI-2 region (Table 4) (31).
TABLE 4.
Distribution of 12 regions associated with virulence among V. cholerae natural isolates as determined by PCR analysisa
| Strain type and name | VSP-I (0175-0185) | MSHA (0398-0411) | hlyA (0489) | VSP-II (0490-0516) | VPI-I (0819-0847) | PilE (0857) | RTX (1447-1451) | RS1 (1452-1455) | CTXφ (1456-1464) | TLC (1465-1480) | VPI-2 (1758-1809) | intl4 (vca0291) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| O1 classical | ||||||||||||
| O395 | − | + | + | − | + | + | + | − | + | + | + | + |
| 569B | − | + | + | − | + | + | + | − | + | + | + | + |
| C1 | − | + | + | − | + | + | + | − | + | + | + | + |
| C14 | − | + | + | − | + | + | + | − | + | + | + | + |
| CA401 | − | + | + | − | + | + | + | − | + | + | + | + |
| GP12 | − | + | + | − | + | + | + | − | + | + | + | + |
| O1 E1 Tor | ||||||||||||
| GP33 | + | + | + | + | + | + | + | + | + | − | + | + |
| GP43 | + | + | + | − | + | + | + | + | + | + | + | + |
| GP155 | − | + | + | − | + | + | + | + | + | + | + | + |
| N16961 | + | + | + | + | + | + | + | + | + | + | + | + |
| 3038 | + | + | + | + | + | + | + | + | + | + | + | + |
| F1873 | + | + | + | + | + | + | + | + | + | + | + | + |
| F1875 | + | + | + | + | + | + | + | + | + | + | + | + |
| F1939 | + | + | + | + | + | + | + | + | + | + | + | + |
| SM115 | + | + | + | + | + | + | + | + | + | + | + | + |
| 2125-98 | + | + | + | + | + | + | + | + | + | + | + | + |
| 2164-78 | − | + | + | − | + | + | + | + | + | − | + | + |
| Pre-seventh pandemic E1 Tor strains | ||||||||||||
| RV79 | − | + | + | − | + | + | + | + | + | + | − | + |
| C5 | + | + | + | + | + | + | + | − | + | + | + | + |
| Nontoxigenic El Tor strains | ||||||||||||
| 2740-80 | − | + | + | − | + | + | + | − | − | + | + | + |
| 468-83 | − | + | + | − | − | + | + | − | − | − | + | + |
| O1 strains of unknown biotype | ||||||||||||
| 1528-79 | − | + | + | − | − | + | + | − | − | − | − | + |
| 917-84 | − | + | + | − | + | + | + | − | + | + | + | + |
| O139 | ||||||||||||
| 36054-98 | + | + | + | + | + | + | + | + | + | + | +* | + |
| SG20 | + | + | + | + | + | + | + | + | + | + | +* | + |
| AS207 | + | + | + | + | + | + | + | + | + | + | +* | + |
| AS209 | + | + | + | + | + | + | + | + | + | + | +* | + |
| AS212 | + | + | + | + | + | + | + | + | + | + | +* | + |
| AS213 | + | + | + | + | + | + | + | + | + | + | +* | + |
| AS231 | + | + | + | + | + | + | + | + | + | + | +* | + |
| AS259 | + | + | + | + | + | + | + | + | + | + | +* | + |
| AS260 | + | + | + | + | + | + | + | + | + | + | +* | + |
| 35636-97 | + | + | + | + | + | + | + | + | + | + | +* | + |
| MO2 | + | + | + | + | + | + | + | + | + | + | + | + |
| MO10 | + | + | + | + | + | + | + | + | + | + | +* | + |
| MO45 | + | + | + | + | + | + | + | + | + | + | +* | + |
| Non-O1/non-O139 strains | ||||||||||||
| 151 | − | + | + | − | + | + | + | − | + | − | + | + |
| CO476 | − | + | + | − | − | + | + | − | − | + | − | − |
| VO7 | + | + | + | − | + | + | + | − | − | + | − | + |
| CO130 | − | + | + | − | + | + | + | + | + | + | − | + |
| V45 | − | + | + | − | + | + | + | − | + | + | + | + |
| V46 | − | + | + | − | + | + | + | + | + | − | + | + |
| V47 | − | + | + | − | + | + | + | + | + | − | + | + |
| V52 | − | + | + | − | + | + | + | + | + | + | + | + |
| V53 | − | + | + | − | + | + | + | + | + | + | + | + |
| V54 | − | + | + | − | + | + | + | + | + | + | + | + |
| SG3 | − | + | + | − | − | + | + | + | + | − | − | + |
| SG6 | − | + | + | − | − | + | + | − | − | + | − | + |
| SG7 | − | + | + | − | − | + | + | − | − | + | − | + |
| SG8 | − | + | + | − | − | + | + | − | − | + | − | + |
| SG9 | − | + | + | − | − | + | + | − | − | − | − | + |
| SG10 | − | + | + | − | − | + | + | + | − | + | − | + |
| SG14 | − | + | + | − | − | + | + | + | − | + | + | + |
| AM112 | − | + | + | − | − | + | + | − | − | + | + | + |
| VIG1613 | − | + | + | − | − | + | + | − | − | − | + | + |
| DK59 | − | + | + | − | − | + | + | − | − | − | − | + |
| DK67 | − | + | + | − | − | + | + | − | − | − | − | + |
| DK71 | − | + | + | − | − | + | + | − | − | − | + | + |
| SCE4 | − | + | + | − | + | + | + | − | − | − | + | + |
| SCE188 | − | + | + | − | + | + | + | + | + | + | + | + |
| 208 | − | + | + | − | + | + | + | + | + | − | − | + |
| C43 | − | + | + | − | − | + | + | − | − | − | + | + |
| 9581 | − | + | + | − | + | + | + | − | − | − | + | + |
| 9582 | − | + | + | − | + | + | + | − | − | − | + | + |
The numbers in parentheses. refer to the genetic organization, e.g., 0175 is VC0175. Asterisks denote strains that only contain the phage-like region of VP1-2; the nan-nag region and restriction modification system are absent. +, present; −, absent.
Twenty-eight V. cholerae non-O1 and non-O139 serogroup strains were assayed by PCR for the presence of 12 virulence regions (Table 4). None of the 28 strains examined by PCR and Southern blot analyses contained VSP-II, and only one strain, VO7, an environmental O37 serogroup isolate from India, contained VSP-I (Fig. 6). Only four regions MSHA, hlyA, pile, and RTX were present in all 28 V. cholerae non-O1 and non-O139 serogroup strains. The intl4 region was absent in strain CO476. The three O37 serogroup strains, V52, V53, and CO130, which form part of the epidemic clone complex, lacked VSP-I and VSP-II; CO130 also lacked VPI-2. Similarly, strain V54 only lacked VSP-I and VSP-II, as did strain V45, which also lacked RS1φ (Table 4).
FIG. 6.
PCR amplification of gene VC0180, which is part of VSP-I. (A) Lane 1, molecular size ladder; lanes 2 to 29, PCR amplicon with primer pair VC0180F-VC0180R. (B) Lane 1, molecular size ladder; lanes 2 to 27, PCR amplicon with primer pair VC0180F-VC0180R.
Among the five V. mimicus strains investigated, all gave negative PCR results for eight regions assayed: VSP-I, MSHA, hlyA, VSP-II, pilE, RTX, TLC, and intl4. Four V. mimicus strains, PT5, PT48, 9583, and 523-80, contained both VPI-1 and CTXφ; two strains (PT5 and PT48) also contained RS1φ.
DISCUSSION
In this study we show that clinical epidemic V. cholerae O1, O139, and O37 serogroup isolates form a highly uniform clone and that the emergence of the sixth and seventh cholera pandemic strains resulted from the successive acquisition of virulence regions.
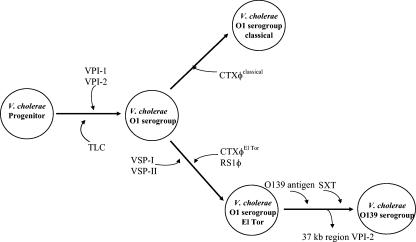
Genotypic and phenotypic analysis of two pre-seventh pandemic isolates, RV79 and C5, isolated in Indonesia in 1937 and 1957, respectively, give some interesting insights into a possible scenario for the evolution of epidemic isolates. El Tor strain RV79 is identical to other O1 serogroup strains at mdh and groEL and lacks only 3 of the 12 virulence regions, VSP-I, VSP-II, and VPI-2, examined in this study. El Tor strain C5, similar to RV79, is identical to other O1 serogroup strains, isolated 20 years later lacks only 1 of the 12 regions examined, RS1φ. Since classical biotype strains were still circulating in the human population prior to the emergence of the seventh cholera pandemic El Tor strain, the question arises as to whether El Tor seventh pandemic isolates arose from a classical progenitor strain via the acquisition of RS1φ, VSP-I, and VSP-II or, alternatively, whether they arose from an RV79 and C5 progenitor-like isolate. The most parsimonious scenario (one requiring the least number of steps) is that an O1 isolate acquired CTXφ, VSP-I, VSP-II, and RS1φ (Fig. 7).
FIG. 7.
Hypothetical evolutionary scenario for the emergence of epidemic V. cholerae O1 and O139 isolates. From the left, the parsimonious evolutionary steps beginning with the V. cholerae progenitor to the right to the contemporary states are indicated. The model begins with a V. cholerae ancestor that most resembles the present day V. cholerae isolates in metabolic functions. From this ancestral state, V. cholerae natural isolates diverged from one another through mutation. The V. cholerae O1 serogroup strains appear to have only arisen once and given rise to the highly successful epidemic classical and El Tor biotype isolates through the independent acquisition of CTXφ by these isolates. The V. cholerae El Tor biotype, which is responsible for the present seventh cholera pandemic, acquired additional virulence regions including RS1φ, VSP-I, and VSP-II. The V. cholerae O139 serogroup, which arose in 1992, acquired a new O-antigen as well as the SXT element.
In 1968 there was a large outbreak of cholera in Sudan caused by an O37 serogroup isolate (68). Interestingly, Bik and colleagues (4) determined by IS1004 fingerprinting that this O37 serogroup strain from Sudan is closely related to classical O1 strains and may have acquired the O37 biosynthesis genes via lateral gene transfer. Beltran et al. (2) confirmed the identity of the O37 strain to O1 strains by multilocus enzyme electrophoresis analysis and identified an additional O37 serogroup strain from India that was similar to O1 classical strains. Recently, an analysis of the O-antigen biosynthesis region also identified an O37 serogroup strain from India that had an O1 serogroup core genome (41). In our study, two V. cholerae O37 serogroup isolates (isolated in Sudan in 1968) had mdh sequences and PCR-SSCP profiles for groEL identical to those of the O1 and O139 strains, indicating that these have an O1 serogroup core genome. These strains likely arose by modification of an O1 strain similar to the emergence of the O139 serogroup clone as previously suggested by Bik et al. (4). Based on multilocus virulence gene profiles of the O37 and O8 serogroups, it is likely that strains V45, V52, V53, and V54 arose from a classical-like progenitor, since they lack only VSP-I and VSP-II; in addition, V45 lacks RS1φ, similar to classical strains. Interestingly, a recent study with the infant mouse cholera model has shown that several non-O1 and non-O139 serogroup isolates, including O37 serogroup strains are efficient intestinal colonizers (8). Morris et al. (46) also demonstrated that a non-O1 and non-O139 V. cholerae strain was capable of causing severe diarrheal disease in humans.
In contrast to the data for O1 and O139 serogroup strains, which all belong to a single unique epidemic clone, our analyses indicate that strains from the same serogroup can belong to divergent lineages and that strains with different serogroup designations can belong to the same lineage, which is expected for regions, such as the O-antigen, that can be acquired by lateral gene transfer (2, 4, 9, 37, 41, 59).
Multilocus virulence gene profile analysis demonstrates the cooccurrence of several virulence regions among V. cholerae isolates (Table 5). For example, with the exception of SG3, CTXφ was only found in strains containing VPI-1, which is to be expected, since it encodes the CTXφ receptor TCP (64). In addition, RS1φ was mainly present in isolates that also contained CTXφ, which again is to be expected, since recent data suggest that both elements require each other for transfer (22). Two strains, SG10 and SG14, however, contained only RS1φ. This observation may be explained by a recent finding that described an alternative mechanism for RS1φ transfer via a novel filamentous phage named KSF-1φ (23). Consistent with previous studies, we found that all classical strains examined in this study lacked RS1φ, VSP-I, and VSP-II and that these three regions are all present in El Tor strains (Table 4) (14, 18). Dziejman et al. (18) recently found VSP-I and VSP-II present only in El Tor isolates. In our study, we found VSP-I in an O37 serogroup strain and both VSP-I and VSP-II were absent from several El Tor isolates. Furthermore, Dziejman and colleagues found that VSP-II encompassed open reading frames VC0490 to VC0497; however, we found that VSP-II spanned a larger region encompassing VC0490 to VC0516. Nonetheless, consistent with their results, we found that VSP-II is confined to seventh pandemic strains.
TABLE 5.
Cooccurrence of virulence regions among V. cholerae
| Serogroup (biotype) | Total no. of strains | No. of strains containing:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| VSP-I | VSP-II | VPI-I | PilE | RTX | RS1 | CTXφ | TLC | VPI-2 | ||
| O1 (classical) | 6 | 0 | 0 | 6 | 6 | 6 | 0 | 6 | 6 | 6 |
| O1 (El Tor) | 15 | 10 | 9 | 14 | 15 | 15 | 12 | 13 | 12 | 14 |
| O1 | 2 | 0 | 0 | 1 | 2 | 2 | 0 | 1 | 1 | 1 |
| O139 | 13 | 13 | 13 | 13 | 13 | 13 | 13 | 13 | 13 | 13a |
| Non-O1/non-O139 | 28 | 1 | 0 | 14 | 28 | 28 | 11 | 11 | 14 | 16 |
These strains only contain the phage-like region of VP1-2; the nan-nag region and restriction modification system are absent. MO2 is an exception, it contains the entire VPI-2.
Four of the virulence regions, MSHA, hlyA, pilE, and RTX, were present in all V. cholerae isolates and absent from all V. mimicus strains examined, indicating that these regions were acquired after V. cholerae and V. mimicus diverged from their most recent common ancestor. Among the V. mimicus isolates examined, only three regions, VPI-1, CTXφ, and TLC, were present. Previous studies of VPI-1 and CTXφ have indicated recent interspecies lateral transfer between V. cholerae and V. mimicus, suggesting that transfer of virulence factors among isolates is an ongoing process.
Initially, a V. cholerae O1 serogroup strain first acquired the pathogenic island VPI-1, which encodes TCP, an essential colonization factor and the receptor for CTXφ. This proposition is supported by the near sequence identity between classical and El Tor biotype strains across most of the VPI-1 region (36). The hypervariability documented at the tcpA gene is likely the result of positive Darwinian selection in this region (8). A second pathogenic island, VPI-2, which encodes genes involved in restriction modification and N-acetyl neuraminic acid utilization, is found predominantly among O1 and O139 epidemic V. cholerae isolates (31) and was most likely present in an O1 serogroup strain that gave rise to classical and El Tor biotype strains. Following the acquisition of VPI-1 and VPI-2 by an O1 serogroup progenitor strain, classical and El Tor biotype isolates emerged and diverged from one another through the acquisition of VSP-I, VSP-II, and RS1φ. Studies based on comparative nucleotide sequence analysis of CTXφ genes indicate that this region was acquired independently in classical and El Tor biotype isolates (7). The V. cholerae classical biotype was responsible for the sixth cholera pandemic, which began in 1899, and presumably previous cholera pandemics. V. cholerae El Tor biotype isolates, which are responsible for the ongoing seventh cholera pandemic, which began in 1961, acquired at least three regions in addition to CTXφ: RS1φ, which facilitates CTXφ production, and VSP-I and VSP-II, whose roles in V. cholerae virulence are unknown (18). The V. cholerae O139 strains that emerged in 1992 were derived from an El Tor progenitor by O-antigen switching likely facilitated by bacteriophages as well as the acquisition of a novel CTXφ and SXT constin (4, 15, 66).
Since the beginning of the modern era of cholera pandemics, all epidemic V. cholerae isolates appear to have a highly conserved core genome onto which additional DNA was added via lateral transfer, facilitating pathogenesis. In addition, the sequence identity at mdh and groEL among V. cholerae epidemic O1, O139, and O37 isolates suggests that these strains have emerged recently, evolutionarily speaking, which is also indicated by the fact that humans are the only known animal hosts for V. cholerae.
Acknowledgments
We thank Frits Mooi and Matthew Waldor for V. cholerae isolates.
The research in E.F.B.'s laboratory is funded by the Higher Education Authority PRTLI-3 grant and an Enterprise Ireland basic research grant.
REFERENCES
- 1.Albert, M. J., A. K. Siddique, M. S. Islam, A. S. Faruque, M. Ansaruzzaman, S. M. Faruque, and R. B. Sack. 1993. Large outbreak of clinical cholera due to Vibrio cholerae non-O1 in Bangladesh. Lancet 341:704. [DOI] [PubMed] [Google Scholar]
- 2.Beltran, P., G. Delgado, A. Navarro, F. Trujillo, R. K. Selander, and A. Cravioto. 1999. Genetic diversity and population structure of Vibrio cholerae. J. Clin. Microbiol. 37:581-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berche, P., C. Poyart, E. Abachin, H. Lelievre, J. Vandepitte, A. Dodin, and J. M. Fournier. 1994. The novel epidemic strain O139 is closely related to the pandemic strain O1 of Vibrio cholerae. J. Infect. Dis. 170:701-704. [DOI] [PubMed] [Google Scholar]
- 4.Bik, E., R. Gouw, and F. Mooi. 1996. DNA fingerprinting of Vibrio cholerae strains with a novel insertion sequence element: a tool to identify epidemic strains. J. Clin. Microbiol. 34:1453-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd, E. F., A. J. Heilpern, and M. K. Waldor. 2000. Molecular analysis of a putative CTXφ precursor and evidence for independent acquisition of distinct CTXφs by toxigenic Vibrio cholerae. J. Bacteriol. 182:5530-5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd, E. F., K. Nelson, F.-S. Wang, T. S. Whittam, and R. K. Selander. 1994. Molecular genetic basis of allelic polymorphism in malate dehydrogenase (mdh) in natural populations of Escherichia coli and Salmonella enterica. Proc. Natl. Acad. Sci. USA 91:1280-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyd, E. F., K. L. Moyer, L. Shi, and M. K. Waldor. 2000. Infectious CTXφ and the Vibrio pathogenicity island prophage in Vibrio mimicus: evidence for recent horizontal transfer between V. mimicus and V. cholerae. Infect. Immun. 68:1507-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyd, E. F., and M. K. Waldor. 2002. Evolutionary and functional analyses of variants of the toxin-coregulated pilus protein TcpA from toxigenic Vibrio cholerae non-O1/non-O139 serogroup isolates. Microbiology 148:1655-1666. [DOI] [PubMed] [Google Scholar]
- 9.Byun, R., L. D. Elbourne, R. Lan, and P. R. Reeves. 1999. Evolutionary relationships of pathogenic clones of Vibrio cholerae by sequence analysis of four housekeeping genes. Infect. Immun. 67:1116-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chakraborty, S., A. K. Mukhopadhyay, R. K. Bhadra, A. N. Ghosh, R. Mitra, T. Shimada, S. Yamasaki, S. M. Faruque, Y. Takeda, R. R. Colwell, and G. B. Nair. 2000. Virulence genes in environmental strains of Vibrio cholerae. Appl. Environ. Microbiol. 66:4022-4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, F., G. M. Evins, W. L. Cook, R. Almeida, N. Hargrett-Bean, and K. Wachsmuth. 1991. Genetic diversity among toxigenic and nontoxigenic Vibrio cholerae O1 isolated from the Western Hemisphere. Epidemiol. Infect. 107:225-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colwell, R. R. 1996. Global climate and infectious disease: the cholera paradigm. Science 274:2025-2031. [DOI] [PubMed] [Google Scholar]
- 13.Dalsgaard, A., O. Serichantalergs, A. Forslund, W. Lin, J. Mekalanos, E. Mintz, T. Shimada, and J. G. Wells. 2001. Clinical and environmental isolates of Vibrio cholerae serogroup O141 carry the CTX phage and the genes encoding the toxin-coregulated pili. J. Clin. Microbiol. 39:4086-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis, B. M., K. E. Moyer, E. F. Boyd, and M. K. Waldor. 2000. CTX prophages in classical biotype Vibrio cholerae: functional phage genes but dysfunctional phage genomes. J. Bacteriol. 182:6992-6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis, B. M., H. H. Kimsey, W. Chang, and M. K. Waldor. 1999. The Vibrio cholerae O139 calcutta CTXΦ is infectious and encodes a novel repressor. J. Bacteriol. 181:6779-6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis, B. M., H. H. Kimsey, A. V. Kane, and M. K. Waldor. 2002. A satellite phage-encoded antirepressor induces repressor aggregation and cholera toxin gene transfer. EMBO J. 21:4240-4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dumontier, S., and P. Berche. 1998. Vibrio cholerae O22 might be a putative source of exogenous DNA resulting in the emergence of the new strain of Vibrio cholerae O139. FEMS Microbiol. Lett. 164:91-98. [DOI] [PubMed] [Google Scholar]
- 18.Dziejman, M., E. Balon, D. Boyd, C. M. Fraser, J. F. Heidelberg, and J. J. Mekalanos. 2002. Comparative genomic analysis of Vibrio cholerae: genes that correlate with cholera endemic and pandemic disease. Proc. Natl. Acad. Sci. USA 99:1556-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evins, G. M., D. N. Cameron, J. G. Wells, K. D. Greene, T. Popovic, S. Giono-Cerezo, I. K. Wachsmuth, and R. V. Tauxe. 1995. The emerging diversity of the electrophoretic types of Vibrio cholerae in the Western Hemisphere. J. Infect. Dis. 172:173-179. [DOI] [PubMed] [Google Scholar]
- 20.Farfan, M., D. Minana, M. C. Fuste, and J. G. Loren. 2000. Genetic relationships between clinical and environmental Vibrio cholerae isolates based on multilocus enzyme electrophoresis. Microbiology 146:2613-2626. [DOI] [PubMed] [Google Scholar]
- 21.Faruque, S. M., M. J. Albert, and J. J. Mekalanos. 1998. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 62:1301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faruque, S. M., Asadulghani, M. Kamruzzaman, R. K. Nandi, A. N. Ghosh, G. B. Nair, J. J. Mekalanos, and D. A. Sack. 2002. RS1 element of Vibrio cholerae can propagate horizontally as a filamentous phage exploiting the morphogenesis genes of CTXphi. Infect. Immun. 70:163-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Faruque, S. M., D. A. Sack, R. B. Sack, R. R. Colwell, Y. Takeda, and G. B. Nair. 2003. Emergence and evolution of Vibrio cholerae O139. Proc. Natl. Acad. Sci. USA 100:1304-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fasano, A., B. Baudry, D. W. Pumplin, S. S. Wasserman, B. D. Tall, J. Ketley, and J. B. Kaper. 1991. Vibrio cholerae produces a second enterotoxin, which affects intestinal tight junctions. Proc. Natl. Acad. Sci. USA 88:5242-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghosh, C., R. K. Nandy, S. K. Dasgupta, G. B. Nair, R. H. Hall, and A. C. Ghose. 1997. A search for cholera toxin (CT), toxin coregulated pilus (TCP), the regulatory element ToxR and other virulence factors in non-O1/non-O139 Vibrio cholerae. Microb. Pathog. 22:199-208. [DOI] [PubMed] [Google Scholar]
- 26.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, and O. White. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 12:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higgins, D. G., J. D. Thompson, and T. J. Gibson. 1996. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 266:383-402. [DOI] [PubMed] [Google Scholar]
- 28.Hobbs, M., and J. S. Mattick. 1993. Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol. Microbiol. 10:233-243. [DOI] [PubMed] [Google Scholar]
- 29.Huq, A., R. R. Colwell, R. Rahman, A. Ali, M. A. Chowdhury, S. Parveen, D. A. Sack, and E. Russek-Cohen. 1990. Detection of Vibrio cholerae O1 in the aquatic environment by fluorescent-monoclonal antibody and culture methods. Appl. Environ. Microbiol. 56:2370-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iredell, J. R., and P. A. Manning. 1994. The toxin-co-regulated pilus of Vibrio cholerae O1: a model for type 4 pilus biogenesis? Trends Microbiol. 2:187-192. [DOI] [PubMed] [Google Scholar]
- 31.Jermyn, W. S., and E. F. Boyd. 2002. Characterization of a novel Vibrio pathogenicity island (VPI-2) encoding neuraminidase (nanH) among toxigenic Vibrio cholerae isolates. Microbiology 148:3681-3693. [DOI] [PubMed] [Google Scholar]
- 32.Jonson, G., J. Holmgren, and A. M. Svennerholm. 1991. Identification of a mannose-binding pilus on Vibrio cholerae El Tor. Microb. Pathog. 11:433-441. [DOI] [PubMed] [Google Scholar]
- 33.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules. Academic Press, Inc., New York, N.Y.
- 34.Kaper, J. B., J. G. Morris, Jr., and M. M. Levine. 1995. Cholera. Clin. Microbiol. Rev. 8:48-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karaolis, D. K., J. A. Johnson, C. C. Bailey, E. C. Boedeker, J. B. Kaper, and P. R. Reeves. 1998. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc. Natl. Acad. Sci. USA 95:3134-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karaolis, D. K., R. Lan, J. B. Kaper, and P. R. Reeves. 2001. Comparison of Vibrio cholerae pathogenicity islands in sixth and seventh pandemic strains. Infect. Immun. 69:1947-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karaolis, D. K., R. Lan, and P. R. Reeves. 1995. The sixth and seventh cholera pandemics are due to independent clones separately derived from environmental, nontoxigenic, non-O1 Vibrio cholerae. J. Bacteriol. 177:3191-3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kotetishvili, M., O. C. Stine, Y. Chen, A. Kreger, A. Sulakvelidze, S. Sozhamannan, and J. G. Morris, Jr. 2003. Multilocus sequence typing has better discriminatory ability for typing Vibrio cholerae than does pulsed-field gel electrophoresis and provides a measure of phylogenetic relatedness. J. Clin. Microbiol. 41:2191-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 40.Li, M., M. Kotetishvili, Y. Chen, and S. Sozhamannan. 2003. Comparative genomic analyses of the Vibrio pathogenicity island and cholera toxin prophage regions in nonepidemic serogroup strains of Vibrio cholerae. Appl. Environ. Microbiol. 69:1728-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li, M., T. Shimada, J. G. Morris, Jr., A. Sulakvelidze, and S. Sozhamannan. 2002. Evidence for the emergence of non-O1 and non-O139 Vibrio cholerae strains with pathogenic potential by exchange of O-antigen biosynthesis regions. Infect. Immun. 70:2441-2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin, W., K. J. Fullner, R. Clayton, J. A. Sexton, M. B. Rogers, K. E. Calia, S. B. Calderwood, C. Fraser, and J. J. Mekalanos. 1999. Identification of a Vibrio cholerae RTX toxin gene cluster that is tightly linked to the cholera toxin prophage. Proc. Natl. Acad. Sci. USA 96:1071-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Merrell, D. S., S. M. Butler, F. Qadri, N. A. Dolganov, A. Alam, M. B. Cohen, S. B. Calderwood, G. K. Schoolnik, and A. Camilli. 2002. Host-induced epidemic spread of the cholera bacterium. Nature 417:642-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mooi, F. R., and E. M. Bik. 1997. The evolution of epidemic Vibrio cholerae strains. Trends Microbiol. 5:161-165. [DOI] [PubMed] [Google Scholar]
- 45.Morris, J. G., Jr., G. E. Losonsky, J. A. Johnson, C. O. Tacket, J. P. Nataro, P. Panigrahi, and M. M. Levin. 1995. Clinical and immunologic characteristics of Vibrio cholerae O139 Bengal infection in North American volunteers. J. Infect. Dis. 171:903-908. [DOI] [PubMed] [Google Scholar]
- 46.Morris, J. G., Jr., T. Takeda, B. D. Tall, G. A. Losonsky, S. K. Bhattacharya, B. D. Forrest, B. A. Kay, and M. Nishibuchi. 1990. Experimental non-O group 1 Vibrio cholerae gastroenteritis in humans. J. Clin. Investig. 85:697-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mukhopadhyay, A. K., S. Chakraborty, Y. Takeda, G. B. Nair, and D. E. Berg. 2001. Characterization of VPI pathogenicity island and CTXphi prophage in environmental strains of Vibrio cholerae. J. Bacteriol. 183:4737-4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nair, G. B., B. L. Sarkar, S. P. De, M. K. Chakraborti, R. K. Bhadra, and S. C. Pal. 1988. Ecology of Vibrio cholerae in the freshwater environs of Calcutta, India. Microb. Ecol. 15:203-216. [DOI] [PubMed] [Google Scholar]
- 49.Nandi, B., R. K. Nandy, A. C. Vicente, and A. C. Ghose. 2000. Molecular characterization of a new variant of toxin-coregulated pilus protein (TcpA) in a toxigenic non-O1/non-O139 strain of Vibrio cholerae. Infect. Immun. 68:948-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Novais, R. C., A. Coelho, C. A. Salles, and A. C. Vicente. 1999. Toxin-co-regulated pilus cluster in non-O1, non-toxigenic Vibrio cholerae: evidence of a third allele of pilin gene. FEMS Microbiol. Lett. 171:49-55. [DOI] [PubMed] [Google Scholar]
- 51.Ogierman, M. A., E. Voss, C. Meaney, R. Faast, S. R. Attridge, and P. A. Manning. 1996. Comparison of the promoter proximal regions of the toxin-co-regulated tcp gene cluster in classical and El Tor strains of Vibrio cholerae O1. Gene 170:9-16. [DOI] [PubMed] [Google Scholar]
- 52.Orita, M., H. Iwahana, H. Kanazawa, K. Hayashi, and T. Sekiya. 1989. Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc. Natl. Acad. Sci. USA 86:2766-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O'Shea, Y. A., and E. F. Boyd. 2002. Mobilization of the Vibrio pathogenicity island between Vibrio cholerae isolates mediated by CP-T1 generalized transduction. FEMS Microbiol. Lett. 214:153-157. [DOI] [PubMed] [Google Scholar]
- 54.Popovic, T., C. A. Bopp, O. Olsvik, and K. Wachsmuth. 1993. Epidemiologic application of a standardized ribotype scheme for V. cholerae O1. Clin. Microbiol. 31:2474-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rivera, I. N., J. Chun, A. Huq, R. B. Sack, and R. R. Colwell. 2001. Genotypes associated with virulence in environmental isolates of Vibrio cholerae. Appl. Environ. Microbiol. 67:2421-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rubin, E. J., W. Lin, J. J. Mekalanos, and M. K. Waldor. 1998. Replication and integration of a Vibrio cholerae cryptic plasmid linked to the CTX prophage. Mol. Microbiol. 28:1247-1254. [DOI] [PubMed] [Google Scholar]
- 57.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 58.Salles, C. A., and H. Momen. 1991. Identification of Vibrio cholerae by enzyme electrophoresis. Trans. R. Soc. Trop. Med. Hyg. 85:544-547. [DOI] [PubMed] [Google Scholar]
- 59.Stine, O. C., S. Sozhamannan, Q. Gou, S. Zheng, J. G. Morris, Jr., and J. A. Johnson. 2000. Phylogeny of Vibrio cholerae based on recA sequence. Infect. Immun. 68:7180-7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stroeher, U. H., G. Parasivam, B. K. Dredge, and P. A. Manning. 1997. Novel Vibrio cholerae O139 genes involved in lipopolysaccharide biosynthesis. J. Bacteriol. 179:2740-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taylor, R. K., V. L. Miller, D. B. Furlong, and J. J. Mekalanos. 1987. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc. Natl. Acad. Sci. USA 84:2833-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trucksis, M., J. E. Galen, J. Michalski, A. Fasano, and J. B. Kaper. 1993. Accessory cholera enterotoxin (Ace), the third toxin of a Vibrio cholerae virulence cassette. Proc. Natl. Acad. Sci. USA 90:5267-5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wachsmuth, I. K., G. M. Evins, P. I. Fields, O. Olsvik, T. Popovic, C. A. Bopp, J. G. Wells, C. Carrillo, and P. A. Blake. 1993. The molecular epidemiology of cholera in Latin America. J. Infect. Dis. 167:621-626. [DOI] [PubMed] [Google Scholar]
- 64.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910-1914. [DOI] [PubMed] [Google Scholar]
- 65.Waldor, M. K., and J. J. Mekalanos. 1994. Vibrio cholerae O139 specific gene sequences. Lancet 343:1366. [DOI] [PubMed] [Google Scholar]
- 66.Waldor, M. K., E. J. Rubin, G. D. Pearson, H. Kimsey, and J. J. Mekalanos. 1997. Regulation, replication, and integration functions of the Vibrio cholerae CTXΦ are encoded by region RS2. Mol. Microbiol. 24:917-926. [DOI] [PubMed] [Google Scholar]
- 67.Yamasaki, S., S. Garg, G. B. Nair, and Y. Takeda. 1999. Distribution of Vibrio cholerae O1 antigen biosynthesis genes among O139 and other non-O1 serogroups of Vibrio cholerae. FEMS Microbiol. Lett. 179:115-121. [DOI] [PubMed] [Google Scholar]
- 68.Zinnaka, Y., and C. C. Carpenter. 1972. An enterotoxin produced by noncholera vibrios. Johns Hopkins Med. J. 131:403-411. [PubMed] [Google Scholar]