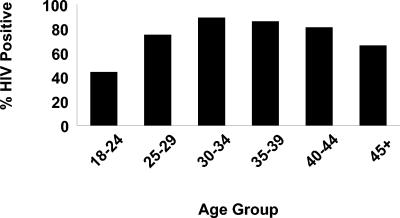Abstract
We examined the pattern of tuberculosis (TB) transmission (i.e., reactivation versus recent transmission) and the impact of human immunodeficiency virus (HIV) infection in Harare, Zimbabwe. Consecutive adult smear-positive pulmonary TB patients presenting to an urban hospital in Harare were enrolled. A detailed epidemiological questionnaire was completed, and tests for HIV type 1 and CD4 cell counts were performed for each patient. Molecular fingerprinting of the genomic DNA recovered from cultures of sputum was performed by two molecular typing methods: spacer oligonucleotide typing (spoligotyping) and analysis of variable number of tandem DNA repeats (VNTRs). A cluster was defined as isolates from two or more patients that shared the same spoligotype pattern or the same VNTR pattern, or both. DNA suitable for typing was recovered from 224 patients. The prevalence of HIV infection was 79%. Of 187 patient isolates (78.6%) typed by both spoligotyping and analysis of VNTRs, 147 were identified as part of a cluster by both methods. By spoligotyping alone, 84.1% of patient isolates were grouped into 20 clusters. The cluster size was generally <8 patient isolates, although three large clusters comprised 68, 25, and 23 patient isolates. A total of 89.4% of the patient isolates grouped into 12 clusters defined by analysis of VNTRs, with 2 large clusters consisting of 127 and 13 patient isolates, respectively. Thirty-six percent of patient isolates with a shared spoligotype and 17% with a shared VNTR pattern were geographically linked within Harare, but they were not linked on the basis of the patient's home district. In a multivariate analysis, there were no independent predictors of clustering, including HIV infection status. Comparison with the International Spoligotype database (Pasteur Institute, Pointe à Pitre, Guadeloupe) demonstrated that our three largest spoligotype clusters are well recognized and ubiquitous in Africa. In this epidemiologically well characterized urban population with a high prevalence of HIV infection, we identified a very high level of strain clustering, indicating substantial ongoing recent TB transmission. Geographic linkage could be detected in a proportion of these clusters. A small group of actively circulating strains accounted for most of the cases of TB transmission.
Tuberculosis (TB) has reemerged as an important public health problem in Zimbabwe. Following a stabilization of the TB rates in the early 1980s at approximately 50 per 100,000 population, since 1987 there has been a dramatic increase to 250 per 100,000 population, which is largely attributable to the human immunodeficiency virus (HIV) epidemic in sub-Saharan Africa (9, 28).
Strain-specific markers for the differentiation of Mycobacterium tuberculosis complex strains are reliable tools for the identification of specific strains and are useful for epidemiological studies of TB. The most extensively used differentiation method is restriction fragment length polymorphism (RFLP) typing, which uses the insertion sequence IS6110 to differentiate clinical isolates (39). However, this technique is technically demanding and expensive and requires viable bacterial growth, and so it may not be optimal for large epidemiological studies in resource-poor settings. New PCR-based typing methods such as spacer oligonucleotide typing (spoligotyping), based on polymorphisms in the direct repeat locus, require less DNA and have been increasingly used for the rapid typing of isolates. However, spoligotyping used alone is less discriminatory than RFLP typing and tends to overestimate the number of epidemiological links, so the use of a second method for confirmation of the results is recommended (22). Fingerprinting based on the variable number of tandem DNA repeats (VNTRs) is a further highly reproducible rapid typing method (15) that yields a high level of cluster discrimination comparable to that of RFLP analysis when it is used as a second-line test with spoligotyping (13, 26).
Use of conventional epidemiological data in combination with these DNA techniques can help answer important questions about the epidemiology of TB in large populations by distinguishing between newly acquired and reactivated disease (1, 4, 6, 7, 16, 23, 32, 37). Patients whose isolates have identical patterns (i.e., a cluster) are likely to have been infected recently and can be targeted for epidemiological investigation to identify a chain of transmission, whereas patients whose isolates demonstrate unique patterns are likely to have reactivation of a latent infection. Most previous studies that have used these DNA fingerprinting techniques have focused on the tracing of TB outbreaks within a hospital or other high-risk environments and the dissemination of multidrug-resistant strains, in which isolates from epidemiologically related patients are usually identical (8, 10, 11, 14). Although it was previously thought that 90% of TB cases in the developed world resulted from the endogenous reactivation of latent infection, population-based RFLP studies conducted in the United States and Western Europe, areas with a relatively low incidence of TB, show that recent infection accounts for up to half of the cases among both HIV-infected and HIV-negative patients in urban areas (1, 4, 6, 7, 16, 23, 32, 37). These studies also showed that only a fraction of the main TB transmission routes are disclosed by classical contact-tracing practices.
In Africa, where TB is endemic, it has generally been considered that most cases of TB in HIV-infected and HIV-negative patients result from reactivation (9). However, there are few data to support this, as few detailed molecular epidemiological studies have been undertaken in countries with high rates of both TB and HIV infection. It has been shown that despite the more limited strain diversity in sub-Saharan Africa, there is still adequate strain variability among M. tuberculosis isolates from patients living in developing countries to permit the application of molecular approaches to the tracking of TB transmission (24, 25). Our objectives were, first, to examine the pattern of TB transmission (i.e., the relative frequency of reactivation versus that of recent infection) and the role of HIV infection using two molecular typing methods, spoligotyping and analysis of VNTRs, in tandem with epidemiological data and, second, to identify the risk factors for recent transmission in a well-characterized cohort of smear-positive TB patients in Harare, Zimbabwe.
(This study was presented in part at the 9th Conference on Retroviruses and Opportunistic Infections, Seattle, Wash., 24 to 28 February 2002 [P. Easterbrook, A. Gibson, S. Murad, A. Ferguson, P. Mason, A. Ndudza, L. Mbengeranwa, and F. Drobniewski, Abstr. 9th Conf. Retrovir. Opportunistic Infect., abstract 621 W, 2002].)
MATERIALS AND METHODS
A total of 516 consecutive adult smear-positive pulmonary TB patients presenting to the Beatrice Road TB Hospital in Harare, Zimbabwe, between May and October 1997 were enrolled in the study. The Beatrice Road TB Hospital is the main referral center for TB and other infectious diseases in Harare and captures the majority of TB cases in the city, which has a population of about 1.25 million. Data for each patient were collected by using a standardized questionnaire and pro forma by a trained research nurse and included demographic data (age, gender, occupation, marital status, number of children, rural home district, present address and all previous residence addresses over the last year in Harare, household size, number of rooms and household crowding level [i.e., number of dwellers per number of rooms], medical history [past history of TB and treatment, M. bovis BCG vaccination, and history of other medical conditions, e.g., diabetes], the history of TB in the household within the last 5 years, and smoking and alcohol consumption). A chest X ray was obtained for each patient, and a comprehensive physical examination focused on the detection of clinical features suggestive of HIV disease. Informed consent to perform HIV testing was obtained from all patients, and the study was approved by the Medical Research Council Ethics Committee of Zimbabwe.
Laboratory methods.
HIV infection status was determined by an enzyme-linked immunosorbent assay (Dupont, Wilmington, Del.). CD4 cell counts were measured by flow cytometry (FACScan; Becton Dickinson, Paramus, N.J.). Three smear-positive sputum samples from each patient were sent to the TB reference laboratory in Bulawayo, Zimbabwe, by the usual transport system and were cultured on Lowenstein-Jensen medium. Cultures and drug susceptibility testing were performed for all 502 patients with microbiologically confirmed cases of TB. Isolates were categorized into drug-sensitive and single-drug- or multidrug-resistant strains.
Molecular typing was performed at the National Mycobacterial Reference Unit, Health Protection Agency, London, United Kingdom, after transport of the cultures by overnight courier from Zimbabwe. Spoligotyping was performed with genomic DNA extracted from cultures of sputum by a standard phenol-chloroform method (43). The spacers between the direct repeats in the target region were amplified by using two 18-nucleotide primers (primer 5′-CCAAGAGGGGACGGAAAC-3′ and biotinylated primer 5′-GGTTTTGGGTCTGACGAC-3′). The PCR products were then hybridized to a Biodyne C membrane (Isogen Bioscience, Maarsen, The Netherlands). This membrane contains immobilized synthetic oligomeric spacer sequences derived from the direct-repeat region of M. tuberculosis H37Rv and M. bovis BCG. Hybridized DNA was detected by using an enhanced chemiluminesence kit (Amersham International plc, Little Chalfont, United Kingdom), with exposure to X-ray film producing a pattern or profile reminiscent of a bar code.
Analysis of VNTRs was performed as described previously (15), but with the following modifications: DNA was extracted from the cultures as described above. PCR was performed in a total volume of 20 μl containing 11 μl of HotStarTaq DNA polymerase (Qiagen, Chichester, United Kingdom) and 10 to 100 ng of DNA sample. An initial denaturation at 94°C for 15 min was followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 1 min, and extension at 72°C for 2 min, followed by a final extension at 72°C for 10 min. An aliquot (10 μl) from each reaction tube was run on a 2% Nusieve GTG Agarose gel (Flowgen, Leicestershire, United Kingdom). Molecular weight standards (100-bp ladder [Promega, Southampton, United Kingdom] and a 20-bp ladder [Sigma-Aldrich, Dorset, United Kingdom]) were loaded every eight lanes. The gel was stained with ethidium bromide and visualized under UV light. Once the size of the PCR product was determined, the number of copies for each exact tandem repeats (ETR) (A to E) was determined by a previously described method (15) and a five-digit number representing the allele profiles was created.
Statistical analysis.
Patient isolates were categorized into one of two groups according to whether the spoligotype or VNTR pattern was unique or was identical to that for another sample (i.e., a cluster). A cluster was defined as a group of two or more patient isolates that shared the same spoligotype or VNTR pattern. Clusters were considered false positive if the sample was processed in the microbiology laboratory on the same day that a specimen with a positive smear from another patient with the same typing pattern was processed. We also reanalyzed the data by assuming that each cluster of n patients comprised one source case and that all the other cases in the clusters (i.e., n − 1) were due to recently acquired disease through active transmission (32). Therefore, a minimum estimate of the proportion of cases of TB caused by recent transmission was calculated as (number of clustered patients − number of clusters)/total number of patients. Patient isolates contained within clusters and those with unique patterns were compared by chi-square tests or Fisher's exact test and by Mann-Whitney U tests for categorical and continuous variables, respectively. Risk factors for clustering identified by univariate analysis (P < 0.2) were then included in a multivariate logistic regression model to identify independent risk factors for clustering, with clustered and nonclustered being the dependent outcomes and with a cluster defined as two or more patient isolates that shared the same (i) spoligotype and VNTR patterns, (ii) the same spoligotype pattern, and (iii) the same VNTR pattern. Since the overall rate of clustering may not always be informative, we also examined whether certain Harare districts and rural home districts were associated with a higher frequency of overall clustering and with particular clusters identified by spoligotyping or analysis of VNTRs using a binomial probability test and one-sided P values after exclusion of those districts in which only one study patient was recorded and exclusion of those patients within a district who were part of a cluster but from another district. For each patient within a cluster, the location of the patient's home was recorded by using the Harare Street Atlas. Individuals living within 0.5 km of a patient who was part of the same cluster were considered part of a geographically based cluster. Data were analyzed by using STATA software (version 7.0; STATA Corporation, College Station, Tex.). Taxatron software (Institut Pasteur, Paris, France) was used to calculate the molecular weights of the hybridizing bands and to compare the isolates with those in the International Spoligotype database.
RESULTS
Study population.
A total of 516 patients with smear-positive TB who presented to the Beatrice Road TB Hospital from May to October 1997 were enrolled in the study. Complete data were available for 502 patients, and these patients were included in the analysis. Of the 502 patients, 371 (74%) were HIV positive and 65% were male. The median age at presentation was 31 years (interquartile range [IQR], 27 to 40 years). Among the 502 patients, 204 (41%) gave a history of recent household contact with TB and 44 (9%) had a past history of TB. There was a striking difference in the prevalence of HIV infection by age (Fig. 1). For the HIV-positive patients, the median CD4 cell count at the time of presentation was 150 × 106/liter (IQR, 64 to 291/liter). Drug resistance was identified in isolates from only 17 (4%) of 458 patients.
FIG. 1.
Prevalence of HIV-1 infection among TB patients by age group.
Spoligotyping and VNTR patterns.
Useable DNA was recovered from 224 of the 228 individual cultures available for 502 (44.6%) patients. A significant number of the remaining cultures were extensively contaminated with bacteria or yeasts, which led to degradation of the medium. Spoligotyping was performed on 214 samples (after the exclusion of 10 contaminated samples), and analysis of VNTRs was performed on 198 samples (after the exclusion of 3 contaminated samples). There were no statistically significant differences in the characteristics of the 187 patients from whom DNA was available for typing compared to those of the 317 patients from whom DNA was not available. Of the 187 patients with available spoligotype and VNTR typing data, 147 (78.6%) were identified as part of a cluster on the basis of both methods, with an additional 20 identified as part of a cluster on the basis of analysis of VNTRs alone and 12 identified as a cluster on the basis of spoligotyping alone.
By using spoligotyping alone, 54 distinct spoligotypes were identified and a total of 180 patient isolates (84.1%) were grouped into 20 clusters (or 15 clusters on the basis of the n − 1 approach, as 10 patient isolates were in clusters with only 2 isolates). The cluster size was generally less than 8 isolates (Table 1); however, three large clusters comprised 68, 25, and 23 patient isolates, respectively, and these accounted for 50% of the isolates. In comparison, there were 33 distinct strains by analysis of VNTRs, and 177 patient isolates (89.4%) grouped in 12 clusters defined by analysis of VNTRs (or 10 clusters on the basis of the n − 1 approach, as 4 patient isolates were in clusters with only 2 isolates), with 2 large clusters comprising 127 isolates (which accounted for 64.1% of the isolates in the study) and 13 patients. The minimum estimate for the proportion of TB cases due to recent transmission was 74.8% [(180 − 20)/214, i.e., (number of clustered patients − number of clusters)/total number of patients] for spoligotyping and 83.3% [(177 − 12)/198] for analysis of VNTRs.
TABLE 1.
Clustering results obtained by analysis of VNTRs as a first-line test followed by subclustering by spoligotyping and obtained by spoligotyping as a first-line test followed by subclustering by analysis of VNTRs
| Primary typing method and cluster size (no. of isolates) | Allele designation by analysis of VNTRs | No. of subclusters by:
|
|
|---|---|---|---|
| Spoligotyping | Analysis of VNTRsa | ||
| Analysis of VNTRs | |||
| 5 | 3233*3 | 3 | |
| 4 | 2123*2 | 2 | |
| 4 | 2123*3 | 2 | |
| 3 | 2143*1 | 2 | |
| 5 | 2143*2 | 4 | |
| 127 | 2143*3 | 23 | |
| 2 | 4243*5 | ||
| 13 | 3243*3 | 6 | |
| 2 | 2243*2 | 2 | |
| 6 | 2243*3 | 3 | |
| 3 | 2343*3 | 2 | |
| 3 | 3233*2 | 3 | |
| Spoligotyping | |||
| 4 | —b (NT = 3) | ||
| 2 | — (NT = 1) | ||
| 68 | 8 (NT = 7) | ||
| 2 | — (NT = 4) | ||
| 23 | 4 (NT = 3) | ||
| 3 | 2 | ||
| 3 | — (NT = 2) | ||
| 5 | 2 | ||
| 8 | — | ||
| 4 | 2 (NT = 1) | ||
| 2 | 2 | ||
| 2 | — | ||
| 3 | — | ||
| 2 | — (NT = 1) | ||
| 7 | — | ||
| 25 | 11 (NT = 3) | ||
| 4 | — | ||
| 3 | 2 | ||
| 7 | |||
| 3 | |||
Expressions in parentheses represent numbers of samples not tested (NT) because a strain type was not available by analysis of VNTRs for some isolates analyzed by spoligotyping.
no subclusters identified.
Analysis of VNTRs was less discriminatory as a first-line fingerprinting method (Table 1). Eleven of the 12 (91.7%) clusters identified by analysis of VNTRs could be further subdivided by spoligotyping into distinct subtypes, with each cluster containing from 2 to 23 subtypes. Conversely, only 8 of 20 (40%) clusters obtained by spoligotyping were further subdivided by analysis of VNTRs. Within the three largest clusters obtained by spoligotyping, analysis of VNTRs identified 8, 11, and 4 additional unique strains, respectively. However, a corresponding fingerprint obtained by analysis of VNTRs was not available for five distinct spoligotypes, so the discriminatory power of analysis of VNTRs may have been underestimated.
Risk factors for clustering.
To identify risk factors for recent infection with M. tuberculosis, the 180 patients whose isolates were in spoligotype-defined clusters were compared with the 34 patients whose isolates were not in clusters. Univariate analysis (Table 2) showed that patients whose isolates were in clusters were less likely to be male (P = 0.02), less likely to harbor a drug-resistant isolate (P = 0.08), and less likely to have a history of TB (P = 0.08) but were more likely to have never smoked (P = 0.004) than patients whose isolates had unique DNA patterns. Factors not associated with clustering were age, HIV infection status, household crowding level, history of TB in the household in the last 5 years, number of symptoms suggestive of HIV (i.e., chronic diarrhea of >1 month, recurrent pneumonia, wasting syndrome, herpes zoster rash, and persistent genital ulcers), presence of a BCG scar, and CD4 cell count. We found similar results when we compared the 177 patients whose isolates were in clusters identified by analysis of VNTRs and the 21 patients whose isolates were not clustered and when we compared the 147 patients whose isolates were defined as part of a cluster on the basis of both spoligotyping and analysis of VNTRs and 8 patients whose isolates were not clustered, except that gender was no longer statistically significant in these analyses. In a multivariate analysis, we found no independent predictors of clustering. We also performed a similar analysis to establish risk factors for membership in a large cluster (≥6, ≥12, and ≥20 patients with a shared strain). We found that a history of TB in the household within the last 5 years was significantly less common among those within a cluster comprising six or more patient isolates with a shared spoligotype and VNTR pattern (30.2%) than in patients whose isolates comprised a cluster with less than six isolates (60.7%; P = 0.003), and the CD4 cell count was significantly higher in the former group than in the latter group (320 × 106 and 240 × 106 cells/liter, respectively; P = 0.02). In a multivariate analysis, a history of TB in the household in the last 5 years (odd ratio = 0.24; 95% confidence interval = 0.09 to 0.68; P = 0.007) and infection with a drug-resistant isolate (odds ratio = 0.05; 95% confidence interval = 0.003 to 0.95; P = 0.05) were inversely associated with the presence of clustering.
TABLE 2.
Characteristics of clustered and nonclustered patients determined by spoligotypinga
| Characteristic | Value for group
|
P value | |
|---|---|---|---|
| Cluster of ≥2 isolates (n = 180) | Noncluster (n = 34) | ||
| Median age (yr [IQR]) | 30 (26-38.7) | 31 (26.7-37.2) | 0.71 |
| No. male (%) | 110 (61.1) | 28 (82.3) | 0.02 |
| No. (%) HIV-1 seropositive | 138 (77.1) | 30 (88.2) | 0.17 |
| No. (%) resistant with isolates to one or more anti-TB drug | 4 (2.2) | 3 (9.1) | 0.08 |
| No. (%) with household crowding level of:b | 0.30 | ||
| <1 | 19 (10.8) | 4 (12.1) | |
| 1-2 | 29 (16.5) | 9 (27.3) | |
| >2 | 128 (72.7) | 20 (60.6) | |
| No. (%) who ever smoked | 59 (32.8) | 20 (58.8) | 0.004 |
| No. (%) with: | |||
| BCG scar | 161 (89.9) | 32 (94.1) | 0.75 |
| TB in household in last 5 year | 65 (36.7) | 12 (35.3) | 0.89 |
| Past history of TB | 11 (6.1) | 5 (14.7) | 0.08 |
| Symptoms of HIV infectionc | 0.70 | ||
| 1 symptom | 53 (29.6) | 12 (35.3) | |
| ≥2 symptoms | 44 (24.6) | 9 (26.5) | |
| Harare districtsd | 0.47 | ||
| Median CD4 count (106/liter [IQR]) | 232.5 (87-443) | 262.5 (125-396) | 0.83 |
Comparison of clusters defined by two or more isolates by analysis of VNTRs (n = 177) and noncluster (n = 21) and those defined as a cluster by both spoligotyping and analysis of VNTRs (n = 147) versus noncluster (n = 8) yielded similar results, except that gender was no longer statistically significant.
Calculated from number of dwellers/number of rooms in household.
Chronic diarrhea for >1 month, recurrent pneumonia, wasting syndrome, herpes zoster rash, and persistent genital ulcers.
Eleven districts: Budiriro, Chitungwiza, Dzivaresekwa, Glen Norah, Glen View, Higfield, Kambuzuma, Mbara, Mufakose, Mabunku, and Kuwadzana.
Geographic mapping of clusters identified by spoligotyping and analysis of VNTRs.
Overall, we found no evidence for a significantly higher frequency of clustering within certain Harare districts of residence or on the basis of the patients' rural home districts in Zimbabwe. However, when we repeated the analysis according to the individual clusters identified by spoligotyping and analysis of VNTRs, 7 of 20 (35%) strains that clustered by spoligotyping and 2 of 12 (16.7%) strains that clustered by analysis of VNTRs had a higher frequency in certain Harare districts (but not the rural home districts) compared to the frequency of the strain cluster in the overall study population, although the differences did not generally attain statistical significance (Table 3). Isolates in one large cluster identified by spoligotyping (n = 68), which accounted for 37.8% of all isolates, were found at a higher frequency (>50%) in five districts, while a further cluster of 23 isolates (12.8% of the study population) was found among >40% of patients in three districts. Five other smaller clusters identified by spoligotyping were also disproportionately represented in certain Harare districts. A similar picture emerged with 2 of the 12 clusters identified by analysis of VNTRs. The largest cluster identified by analysis of VNTRs (n = 127), which accounted for 64.1% of all isolates, was found at a higher frequency (>80%) in five districts, although this did not attain statistical significance. A further small cluster of four patients identified by analysis of VNTRs was found among patients from only two districts. When we mapped the locations of the patients' homes on a map of Harare, we found that the vast majority of the patients in patient clusters within a particular district resided within 0.5 km of each other.
TABLE 3.
Geographic clusters within Harare districts for 7 of 20 clusters identified by spoligotyping and 2 of 12 clusters identified by analysis of VNTRs
| Typing method and cluster code | No. of patients in cluster | Frequency (%) in overall study population | Harare district | No. of study patients in cluster in district/no. of study patients in district | Frequency (%) of cluster isolate in district | P value |
|---|---|---|---|---|---|---|
| Spoligotyping | ||||||
| 777777606060771 | 68 | 37.8 | Glen View | 10/19 | 52.6 | 0.14 |
| Highfield | 13/23 | 56.5 | 0.06 | |||
| Kuwadzana | 4/7 | 57.1 | 0.25 | |||
| Mbare | 12/26 | 46.1 | 0.25 | |||
| Waterfalls | 2/2 | 100 | 0.14 | |||
| 777777607760771 | 23 | 12.8 | Chitungwiza | 2/5 | 40 | 0.13 |
| Dzivaresekwa | 2/5 | 40 | 0.13 | |||
| Kambuzuma | 2/3 | 66.7 | 0.05 | |||
| 777777677760771 | 5 | 2.8 | Glen View | 2/19 | 10.5 | 0.11 |
| 777777604060731 | 7 | 3.9 | Mufukose | 3/13 | 23.1 | 0.01 |
| 777777777760731 | 25 | 13.9 | Mufakose | 4/13 | 30.8 | 0.09 |
| 777777606060631 | 4 | 2.2 | Highfield | 2/23 | 9.0 | 0.09 |
| 777777606060731 | 7 | 3.9 | Braeside | 2/11 | 18.2 | 0.07 |
| Analysis of VNTRs | ||||||
| 2123*2 | 4 | 2.0 | Highfield | 2/26 | 7.7 | 0.10 |
| Glen View | 2/22 | 9.1 | 0.07 | |||
| 2143*3 | 127 | 64.1 | Arcadia | 2/2 | 100 | 0.41 |
| Budiriro | 8/9 | 88.9 | 0.11 | |||
| Glen View | 16/22 | 72.7 | 0.26 | |||
| Harare | 2/2 | 100 | 0.41 | |||
| Kambuzuma | 3/4 | 75 | 0.54 | |||
| Mabvuku | 6/7 | 85.7 | 0.22 | |||
| Southerton | 3/3 | 100 | 0.26 |
Relationships between isolates identified by spoligotyping (Table 4).
TABLE 4.
Frequency of spoligotypes in Harare, Zimbabwe
| Octal codea | No. of isolates observed in Harare | International Spoligotype database code | Octal codea | No. of isolates observed in Harare | International Spoligotype database codeb | |
|---|---|---|---|---|---|---|
| 000000000003771 | 4 | 1 | ||||
| 077777606060771 | 1 | 816 | ||||
| 100775747413771 | 1 | Not seen | ||||
| 457347607760671 | 1 | Not seen | ||||
| 477777607760771 | 4 | 753 | ||||
| 477777777760771 | 3 | 804 | ||||
| 517347606060661 | 1 | 79 | ||||
| 557163777760671 | 1 | Not seen | ||||
| 557757606060771 | 1 | 85 | ||||
| 577347606060631 | 1 | Not seen | ||||
| 577717606060731 | 1 | Not seen | ||||
| 577756777760601 | 1 | Not seen | ||||
| 577757604040731 | 1 | Not seen | ||||
| 577757606060731 | 1 | Not seen | ||||
| 577757606060771 | 1 | 87 | ||||
| 577777603760771 | 1 | Not seen | ||||
| 601777606060731 | 1 | Not seen | ||||
| 601777606060771 | 1 | Not seen | ||||
| 637775777763770 | 1 | Not seen | ||||
| 677777607760771 | 2 | 20 | ||||
| 700046677760671 | 1 | Not seen | ||||
| 700076777760771 | 1 | 92 | ||||
| 700775747473771 | 1 | Not seen | ||||
| 700777747413771 | 2 | 129 | ||||
| 703377400001771 | 3 | 21 | ||||
| 730377600003771 | 1 | Not seen | ||||
| 737417606060731 | 2 | 805 | ||||
| 737777607030771 | 1 | Not seen | ||||
| 737777677670371 | 1 | Not seen | ||||
| 757757606060771 | 1 | Not seen | ||||
| 757777606060771 | 1 | Not seen | ||||
| 757777777413731 | 2 | 806 | ||||
| 770077606060731 | 3 | 807 | ||||
| 773777606060731 | 1 | Not seen | ||||
| 774017606060731 | 1 | Not seen | ||||
| 774777777760771 | 1 | 801 | ||||
| 777357606060731 | 1 | Not seen | ||||
| 777357607760631 | 1 | Not seen | ||||
| 777377607760771 | 1 | 81 | ||||
| 777417606060731 | 9 | 184 | ||||
| 777737774020771 | 1 | 218 | ||||
| 777737777760731 | 1 | 73 | ||||
| 777737777760771 | 4 | 37 | ||||
| 777747606060771 | 2 | 808 | ||||
| 777757606060731 | 2 | 82 | ||||
| 777757777760671 | 1 | 83 | ||||
| 777773604060731 | 1 | 84 | ||||
| 777774606060731 | 1 | Not seen | ||||
| 777775606060771 | 3 | 809 | ||||
| 777775770020771 | 1 | Not seen | ||||
| 777776777760771 | 1 | 119 | ||||
| 777777414020731 | 1 | Not seen | ||||
| 777777602060771 | 1 | 810 | ||||
| 777777604000171 | 1 | Not seen | ||||
| 777777604060731 | 8 | 811 | ||||
| 777777606060571 | 3 | 812 | ||||
| 777777606060631 | 5 | 813 | ||||
| 777777606060631 | 1 | Not seen | ||||
| 777777606060671 | 4 | 814 | ||||
| 777777606060731 | 9 | 815 | ||||
| 777777606060771 | 68 | 59 | ||||
| 777777607760731 | 1 | 60 | ||||
| 777777607760771 | 23 | 42 | ||||
| 777777674063771 | 1 | Not seen | ||||
| 777777676060771 | 1 | Not seen | ||||
| 777777677760771 | 6 | 291 | ||||
| 777777747473771 | 1 | Not seen | ||||
| 777777757760771 | 1 | 44 | ||||
| 777777757763771 | 1 | Not seen | ||||
| 777777773413731 | 1 | Not seen | ||||
| 777777774020731 | 2 | 62 | ||||
| 777777774020771 | 1 | 47 | ||||
| 777777774060771 | 1 | Not seen | ||||
| 777777777413771 | 1 | 236 | ||||
| 777777777420731 | 1 | 817 | ||||
| 777777777473771 | 1 | Not seen | ||||
| 777777777600771 | 1 | 780 | ||||
| 777777777760671 | 1 | 245 | ||||
| 777777777760771 | 25 | 53 |
Octal codes were determined by the protocol of Dale et al. (7a).
Obtained from the Pasteur Institute, Pointe à Pitre, Guadeloupe.
Linkage with the International Spoligotype database (Pasteur Institute, Pointe à Pitre, Guadeloupe) demonstrated that our three largest spoligotype cluster types (International Spoligotype Database code numbers 59, 53, and 42) are all well recognized and ubiquitous in Africa. All clusters between types 804 and 815 have not yet been described elsewhere and could be considered a result of the specific evolution of type 59. Type 1 is the Beijing/W strain, which was represented by the isolates from four patients in the study population, none of whom were geographically linked, although two of the patients originated from the same rural home district in Chinhoyi, which is just north of Harare. Two of the patients infected with the Bejing/W strain also gave a history of a household contact with TB within the last 10 years, but none harbored a drug-resistant isolate.
DISCUSSION
We have used spoligotyping and VNTR typing of an epidemiologically well characterized patient population to describe the current pattern of TB transmission in Harare, Zimbabwe, a country in which HIV and TB are endemic. On the basis of shared spoligotype and VNTR patterns, more than three-quarters of the strains were potentially linked, suggesting a high level of recent TB transmission in Harare. This is substantially higher than the estimates of recent transmission reported from other studies. The proportions of new cases due to recent infection have been estimated in cities and countries where the rates of endemic TB are low and have been found to be 38 to 41% in New York City (1, 14); 32% in Baltimore, Md. (6); 40% in San Francisco, Calif. (32); 28% in Berne, Switzerland (16); 46% in Amsterdam, The Netherlands (37); 28% in France (36); and 38% in Seville, Spain (31). Fewer molecular epidemiological studies of TB transmission have been undertaken in countries where HIV and TB are endemic; and estimates range from 25% of 84 TB patients in Honduras (30); 32% of 239 patients in San Paulo, Brazil, of whom approximately half were HIV infected (12); 33% of 51 patients in Guadeloupe (33); 41% in a study of 41 HIV-positive TB patients from Nairobi, Kenya (17); 42% of 301 acid-fast bacillus smear-positive patients from Botswana (69% were HIV seropositive) (27); 45% of 38 patients in Havana, Cuba (29); 45% of 246 patients (20) and 50% of 371 patients (42) in South Africa; 62% of patients in Tunisia (24); and 53.6% of 28 TB patients in a small study conducted in Harare in 1995 (25). Our high rate of clustering suggesting recent transmission probably reflects the high prevalence of HIV infection (74%) in our urban population and the relatively young age (median age, 31 years) of our urban population living in generally overcrowded, high-density housing. HIV infection increases susceptibility to exogenous TB infection (3), and in turn, primary TB is more likely to develop in the first 12 months following acquisition (3). Among HIV-infected patients living in New York City and San Francisco, more than 60% of new TB cases were the result of recent transmission (1, 32). These studies also found that the frequency of strains with clustered patterns was significantly higher among seropositive patients (1, 32), although this has not been a consistent finding (20, 27, 38).
The significance of clustering in population-based studies is controversial (18). While epidemiological data from urban areas strongly suggest that clustering indicates recent transmission of TB, in geographically stable rural populations, clustering may result from the simultaneous reactivation of infection acquired from the same source in the distant past (7) or from the predominance of certain well-conserved ancestral strains in the population, which would result in an overestimate of recent transmission. For example, in a study from Malawi and Kenya, strains from a widespread area were found to have identical DNA fingerprints, but no apparent epidemiological links could be identified (21). We found that approximately half of our M. tuberculosis isolates that clustered by spoligotyping and that 70% of our M. tuberculosis isolates that clustered by analysis of VNTRs belonged to just a few major families of clustered patterns. A recent study of strains from countries with a high prevalence of M. tuberculosis infection has also suggested that the majority of circulating strains in a particular locality belong to a limited number of families (24), although Warren et al. (41) found a high degree of strain diversity in Cape Town, South Africa, where the incidence of TB is high. However, despite this, there was still substantial diversity in our typing patterns, with 54 different strains detected among our study population by spoligotyping and 33 different strains detected by analysis of VNTRs, suggesting that the chance occurrence of identical patterns among unrelated cases would be unusual. Furthermore, with 8 (35%) of 20 clusters identified by spoligotyping and 2 (17%) of the 12 clusters identified by analysis of VNTRs, there was a trend toward a higher rate of clustering within certain Harare districts, but statistical confirmation was limited by the small sample size. Although we did not undertake formal contact tracing to establish the presence of specific epidemiological links between these geographically based clusters, we found that almost all patients within these clusters lived within 0.5 km of each other. Importantly, we did not identify any clustering according to rural home district, which argues against the possibility that these clustered strains reflect reactivation of strains acquired from a common source in the past. Overall, these observations suggest that cases of TB caused by strains with identical spoligotype or VNTR patterns in our study are due to recently transmitted disease but that the incidence of TB in Harare is strongly influenced by a relatively small subset of actively circulating strains.
Various studies have identified multiple factors associated with TB due to recent infection, including young age (1, 30, 32); HIV infection status (1, 32); drug resistance (1, 12); and membership in ethnic minority populations, drug and alcohol use, and homelessness (1, 14, 16, 32, 37). The only risk factor for clustering in a recent molecular epidemiological study from Botswana comparable to our study was prior imprisonment (27). In our multivariate analysis, infection with an isolate with a pattern found to be part of a cluster was not associated with any particular demographic or clinical characteristics, including HIV infection. However, in the analysis of risk factors for membership in a large cluster (six or more strains), unique or nonclustered strains were significantly associated with a history of TB in the household in the last 5 years, and there was a borderline association with drug resistance. The lack of an association between HIV seropositivity and clustering has been reported in other studies (20, 27, 38, 42), and in our study it most likely reflects the very high HIV type 1 (HIV-1) seroprevalence in the population. Our explanation of the association between a household history of TB and infection with a nonclustered strain rather than a clustered strain is that residence within the various high-density districts of Harare is likely to be the more dominant factor in recent exposure and transmission and that reactivation of TB acquired from a household contact, often years previously, would result in a unique strain rather than a clustered strain. Finally, since we enrolled patients with new pulmonary cases of TB only over a 6-month period, it is unlikely that we would have been able to identify a link with a case from many years earlier.
Comparison with the International Spoligotype database demonstrated that our three largest spoligotype clusters are well recognized and ubiquitous in Africa. Type 59 is found almost exclusively in Africa and was described in a previous study in Harare (25). Types 53 and 42 are ubiquitous, with a trend for type 42 to be overrepresented in Latin America and Mediterranean countries, but probably also in Africa. We identified the presence of the Beijing/W genotype strain in four patients, none of whom were geographically or epidemiologically linked. Strains of the Beijing/W genotype family have had a strong impact on the TB epidemics in Asia, Vietnam, and the former republics of the USSR (40) and increasingly in other geographic regions (19). They have also been associated with large outbreaks, some involving multidrug resistance (19). The occurrence of the Beijing/W strain in Vietnam has been correlated with young age, suggesting recent transmission (2), and its presence in four patients in our study population is likely to represent imported transmission.
The spoligotyping and analysis of VNTR methods together confirmed the presence of a cluster of strains in 78.6% of patients, whereas 84.1% clustering was found on the basis of spoligotyping alone and 89.4% clustering was found on the basis of analysis of VNTRs alone. This high level of agreement between unlinked genetic markers is consistent with a clonal population structure of circulating M. tuberculosis strains (26). Analysis of VNTRs was less discriminatory as a first-line fingerprinting method: 11 of the 12 clusters identified by analysis of VNTRs could be further subdivided into distinct subtypes by spoligotyping, whereas only 8 of the 20 clusters identified by spoligotyping could be further subdivided by analysis of VNTRs. However, five spoligotypes did not have a corresponding VNTR fingerprint, so the discriminatory power of analysis of VNTRs may have been underestimated. Overall, the use of spoligotyping in tandem with analysis of VNTRs provided additional cluster discrimination and is comparable to that achieved by IS6110 RFLP typing (13, 26). More recently, a promising new molecular typing method based on 12 minsatellite-like loci containing VNTRs of genetic elements, called mycobacterial interspersed repetitive units (MIRUs), has been developed (35). The resolution obtained by analysis of MIRUs-VNTRs is close to that of typing by RFLP analysis, and it is more discriminatory than spoligotyping or analysis of VNTRs alone (34). The level of clustering in our study may therefore have been less if the isolates had been typed by either the RFLP method or a two-PCR-based genotyping strategy of spoligotyping with MIRUs.
Our study has three main limitations affecting the generalizability of the data. First, we obtained DNA from only half of the patients enrolled in the study. However, the characteristics of the patients from whom isolates were and were not used for spoligotyping were similar, so this is unlikely to have biased our results. Similarly, although our study population was enrolled from only one hospital in Harare, the majority of TB patients in the city are seen at this hospital, and we therefore believe that the patients that we enrolled are representative of the TB patients in Harare. We also enrolled only smear-positive patients, and given that smear-negative but culture-positive cases can account for considerable TB transmission, our study may provide an incomplete assessment of transmission within the community (5). However, by confining our analysis to smear-positive patients, we could identify the most efficient sources of TB transmission. Finally, we excluded laboratory cross-contamination as a likely reason for the clustering, as none of the samples with identical patterns were processed on the same day.
The study has several major implications for TB control. First, if chemoprophylaxis is to be used, it must be lifelong to cover the constant risk of exposure and not just short term to cover reactivation. Similarly, there may be little value in prolonging standard treatment to reduce the likelihood of relapse if the majority of cases are due to reinfection. Second, we assume that clustering occurs either because the contact is immunocompromised and progresses rapidly to disease after exposure or because the index case patient delays seeking medical attention, resulting in the potential for many secondary cases. If this is the case, then an increased emphasis should be placed on the early identification of cases through enhanced public health education campaigns, together with stricter implementation of the traditional methods of TB control, including treatment until cure.
Acknowledgments
Financial support was provided by the Royal College of Physicians of London and a travel grant from the Wellcome Trust.
REFERENCES
- 1.Alland, D., G. E. Kalkut, A. R. Moss, R. A. McAdam, J. A. Hahn, W. Bosworth, E. Drucker, and B. R. Bloom. 1994. Transmission of tuberculosis in New York City. An analysis by DNA fingerprinting and conventional epidemiologic methods. N. Engl. J. Med. 330:1710-1716. [DOI] [PubMed] [Google Scholar]
- 2.Anh, D. D., M. W. Van Borgdorff, L. N. Van, N. T. Lan, T. van Gorkom, K. Kremer, and D. van Soolingen. 2000. Mycobacterium tuberculosis Beijing genotype emerging in Vietnam. Emerg. Infect. Dis. 6:302-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnes, P. F., A. B. Bloch, P. T. Davidson, and D. E. Snider, Jr. 1991. Tuberculosis in patients with human immunodeficiency virus infection. N. Engl. J. Med. 324:1644-1650. [DOI] [PubMed] [Google Scholar]
- 4.Barnes, P. F., Z. Yang, S. Preston-Martin, J. M. Pogoda, B. E. Jones, M. Otaya, K. D. Eisenach, L. Knowles, S. Harvey, and M. D. Cave. 1997. Patterns of tuberculosis transmission in central Los Angeles. JAMA 278:1159-1163. [PubMed] [Google Scholar]
- 5.Behr, M. A., S. A. Warren, H. Salamon, P. C. Hopewell, A. Ponce de Leon, C. L. Daley, and P. M. Small. 1999. Transmission of Mycobacterium tuberculosis from AFB smear-negative patients. Lancet 353:444-449. [DOI] [PubMed] [Google Scholar]
- 6.Bishai, W. R., N. M. Graham, S. Harrington, D. S. Pope, N. Hooper, J. Astemborski, L. Sheely, D. Vlahov, G. E. Glass, and R. E. Chaisson. 1998. Molecular and geographic patterns of tuberculosis transmission after 15 years of directly observed therapy. JAMA 280:1679-1684. [DOI] [PubMed] [Google Scholar]
- 7.Braden, C. R., G. L. Templeton, M. D. Cave, S. Valway, I. M. Onorato, K. G. Castro, D. Moers, Z. Yang, W. W. Stead, and J. H. Bates. 1997. Interpretation of restriction fragment length polymorphism analysis of Mycobacterium tuberculosis isolates from a state with a large rural population. J. Infect. Dis. 175:1446-1452. [DOI] [PubMed] [Google Scholar]
- 7a.Dale, J. W., D. Brittain, A. A. Cataldi, D. Cousins, J. T. Crawford, J. Driscoll, H. Heersma, T. Lillebaek, T. Quitugua, N. Rastogi, R. A. Skuce, C. Sola, D. Van Soolingen, and V. Vincent. 2001. Spacer oligonucleotide typing of bacteria of the Mycobacterium tuberculosis complex: recommendations for standardised nomenclature. Int. J. Tuberc. Lung Dis. 5:216-269. [PubMed] [Google Scholar]
- 8.Daley, C. L., P. M. Small, G. F. Schecter, G. K. Schoolnik, R. A. McAdam, W. R. Jacobs, Jr., and P. C. Hopewell. 1992. An outbreak of tuberculosis with accelerated progression among persons infected with human immunodeficiency virus: an analysis using restriction fragment length polymorphisms. N. Engl. J. Med. 326:231-235. [DOI] [PubMed] [Google Scholar]
- 9.DeCock, K. M., B. Soro, I. M. Coulibaly, and S. B. Lucas. 1992. Tuberculosis and HIV infection in sub-Saharan Africa. JAMA 268:1581-1587. [DOI] [PubMed] [Google Scholar]
- 10.Dooley, S. W., M. E. Villarino, M. Lawrence, L. Salinas, S. Amil, J. V. Rullan, W. R. Jarvis, A. B. Bloch, and G. M. Cauthen. 1992. Nosocomial transmission of tuberculosis in a hospital unit for HIV-infected patients. JAMA 267:2632-2634. [PubMed] [Google Scholar]
- 11.Edlin, B. R., J. I. Tokars, M. H. Grieco, J. T. Crawford, J. Williams, E. M. Sordillo, K. R. Ong, J. O. Kilburn, S. W. Dooley, and K. G. Castro. 1992. An outbreak of multidrug-resistant tuberculosis among hospitalized patients with the acquired immunodeficiency syndrome. N. Engl. J. Med. 326:1514-1521. [DOI] [PubMed] [Google Scholar]
- 12.Ferrazoli, L., M. Palaci, L. R. Marques, L. F. Jamal, J. B. Afiune, E. Chimara, M. C. Martins, M. A. Silva Telles, C. A. Oliveira, M. C. Palhares, D. T. Spada, and L. W. Riley. 2000. Transmission of tuberculosis in an endemic urban setting in Brazil. Int. J. Tuberc. Lung Dis. 4:18-25. [PubMed] [Google Scholar]
- 13.Filliol, I., S. Ferdinand, L. Negroni, C. Sola, and N. Rastogi. 2000. Molecular typing of Mycobacterium tuberculosis based on variable number of tandem DNA repeats used alone and in association with spoligotyping. J. Clin. Microbiol. 38:2520-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedman, C. R., M. Y. Stoeckle, B. N. Kreiswirth, W. D. Johnson, Jr., S. M. Manoach, J. Berger, K. Sathianathan, A. Hafner, and L. W. Riley. 1995. Transmission of multidrug-resistant tuberculosis in a large urban setting. Am. J. Respir. Crit. Care Med. 152:355-359. [DOI] [PubMed] [Google Scholar]
- 15.Frothingham, R., and W. A. Meeker-O'Connell. 1998. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem repeats. Microbiology 144:1189-1196. [DOI] [PubMed] [Google Scholar]
- 16.Genewein, A., A. Telenti, C. Bernasconi, C. Mordasini, S. Weiss, A. M. Maurer, H. L. Rieder, K. Schopfer, and T. Bodmer. 1993. Molecular approach to identifying route of transmission of tuberculosis in the community. Lancet 342:841-844. [DOI] [PubMed] [Google Scholar]
- 17.Gilks, C. F., P. Godfrey-Faussett, B. I. Batchelor, J. C. Ojoo, S. J. Ojoo, R. J. Brindle, J. Paul, J. Kimari, M. C. Bruce, J. Bwayo, F. A. Plummer, and D. A. Warrell. 1997. Recent transmission of tuberculosis in a cohort of HIV-1 infected female sex workers in Nairobi, Kenya. AIDS 11:911-918. [DOI] [PubMed] [Google Scholar]
- 18.Glynn, J. R., J. Bauer, A. S. de Boer, M. W. Borgdorff, P. E. Fine, P. Godfrey-Faussett, and E. Vynnycky. 1999. Interpreting DNA fingerprint clusters of Mycobacterium tuberculosis. Int. J. Tuberc. Lung Dis. 3:1055-1060. [PubMed] [Google Scholar]
- 19.Glynn, J. R., J. Whiteley, P. J. Bifani, K. Kremer, and D. van Soolingen. 2002. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis. Emerg. Infect. Dis. 8:843-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Godfrey-Faussett, P., P. Sonnenberg, S. C. Shearer, M. C. Bruce, C. Mee, L. Morris, and J. Murray. 2000. Tuberculosis control and molecular epidemiology in a South African gold-mining community. Lancet 356:1066-1071. [DOI] [PubMed] [Google Scholar]
- 21.Godfrey-Faussett, P., and N. G. Stoker. 1992. Aspects of tuberculosis in Africa. Genetic fingerprinting for clues to the pathogenesis of tuberculosis. Trans. R. Soc. Trop. Med. Hyg. 86:472-475. [DOI] [PubMed] [Google Scholar]
- 22.Goguet de la Semoniere, Y. O., H. M. Li, G. Torrea, A. Bunschoten, J. van Embden, and B. Gicquel. 1997. Evaluation of spoligotyping in a study of the transmission of Mycobacterium tuberculosis. J. Clin. Microbiol. 35:2210-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayward, A. C., S. Goss, F. Drobniewski, N. Saunders, R. J. Shaw, M. Goyal, A. Swan, A. Uttley, A. Pozniak, J. Grace-Parker, and J. M. Watson. 2002. The molecular epidemiology of tuberculosis in inner London. Epidemiol. Infect. 128:175-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hermans, P. W. M., F. Messadi, H. Guebrexabher, D. van Soolingen, P. E. de Haas, H. Heersma, H. de Neeling, A. Ayoub, F. Portaels, and D. Frommel. 1995. Analysis of the population structure of Mycobacterium tuberculosis in Ethiopia, Tunisia, and The Netherlands: usefulness of DNA typing for global tuberculosis epidemiology. J. Infect. Dis. 171:1504-1513. [DOI] [PubMed] [Google Scholar]
- 25.Heyderman, R. S., M. Goyal, P. Roberts, S. Ushewokunze, S. Zizhou, B. G. Marshall, R. Makombe, J. D. Van Embden, P. R. Mason, and R. J. Shaw. 1998. Pulmonary tuberculosis in Harare, Zimbabwe: analysis by spoligotyping. Thorax 53:346-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kremer, K., D. van Soolingen, R. Frothingham, W. H. Haas, P. W. Hermans, C. Martin, P. Palittapongarnpim, B. B. Plikaytis, L. W. Riley, M. A. Yakrus, J. M. Musser, and J. D. van Embden. 1999. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 37:2607-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lockman, S., J. D. Sheppard, C. Braden, M. J. Mwasekaga, C. L. Woodley, T. A. Kenyon, N. J. Binkin, M. Steinman, F. Montsho, M. Kesupile-Reed, C. Hirschfeldt, M. Notha, T. Moeti, and J. W. Tappero. 2001. Molecular and conventional epidemiology of Mycobacterium tuberculosis in Botswana: a population-based prospective study of 301 pulmonary tuberculosis patients. J. Clin. Microbiol. 39:1042-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ministry of Health and Child Welfare, Department of Epidemiology and Disease Control. 1994. Tuberculosis annual report, Zimbabwe. Ministry of Health and Child Welfare, Harare, Zimbabwe.
- 29.Montoro, E., J. Valdivia, and S. Cardoso Leao. 1998. Molecular fingerprinting of Mycobacterium tuberculosis isolates obtained in Havana, Cuba, by IS6110 restriction fragment length polymorphism analysis and by the double-repetitive element PCR method. J. Clin. Microbiol. 36:3099-3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pineda-Garcia, L. A., A. Ferreira, and S. E. Hoffner. 1997. DNA fingerprinting of Mycobacterium tuberculosis strains from patients with pulmonary tuberculosis in Honduras. J. Clin. Microbiol. 35:2393-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Safi, H., J. Aznar, and J. C. Palomares. 1997. Molecular epidemiology of Mycobacterium tuberculosis strains isolated during a 3-year period (1993 to 1995) in Seville, Spain. J. Clin. Microbiol. 35:2472-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Small, P. M., P. C. Hopewell, S. P. Singh, A. Paz, J. Parsonnet, D. C. Ruston, G. F. Schecter, C. L. Daley, and G. K. Schoolnik. 1994. The epidemiology of tuberculosis in San Francisco: a population-based study using conventional and molecular methods. N. Engl. J. Med. 330:1703-1709. [DOI] [PubMed] [Google Scholar]
- 33.Sola, C. L., K. S. Horgen, S. Goh, and N. Rastogi. 1997. Molecular fingerprinting of Mycobacterium tuberculosis on a Caribbean island with IS6110 and DRr probes. J. Clin. Microbiol. 35:843-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sola, C. L., I. Filliol, E. Legrand, S. Lesjean, C. Locht, P. Supply, and N. Rastogi. 2002. Genotyping of the Mycobacterium tuberculosis complex using MIRUs: association with VNTR and spoligotyping for molecular epidemiology and evolutionary genetics. Infect. Genet. Evol. 3:125-133. [DOI] [PubMed] [Google Scholar]
- 35.Supply, P., S. Lesjean, E. Savine, K. Kremer, D. van Soolingen, and C. Locht. 2001. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J. Clin. Microbiol. 39:3563-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Torrea, G., C. Offredo, M. Simonet, B. Gicquel, P. Berche, and C. Pierre-Audigier. 1996. Evaluation of tuberculosis transmission in a community by 1 year of systematic typing of Mycobacterium tuberculosis clinical isolates. J. Clin. Microbiol. 34:1043-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Deutekom, H., J. J. J. Gerritsen, D. van Soolingen, E. J. van Ameijden, J. D. van Embden, and R. A. Coutinho. 1997. A molecular epidemiological approach to studying the transmission of tuberculosis in Amsterdam. Clin. Infect. Dis. 25:1071-1077. [DOI] [PubMed] [Google Scholar]
- 38.Van Soolingen, D., M. W. Borgdorff, P. E. de Haas, M. M. Sebek, J. Veen, M. Dessens, K. Kremer, and J. D. van Embden. 1999. Molecular epidemiology of tuberculosis in The Netherlands: a nationwide study from 1993 through 1997. J. Infect. Dis. 180:726-736. [DOI] [PubMed] [Google Scholar]
- 39.Van Soolingen, D., P. W. M. Hermans, P. E. W. de Haas, D. R. Soll, and J. D. van Embden. 1991. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J. Clin. Microbiol. 29:2578-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Soolingen, D., L. Qian, P. E. de Haas, J. T. Douglas, H. Traore, F. Portaels, H. Z. Qing, D. Enkhsaikan, P. Nymadawa, and J. D. van Embden. 1995. Predominance of a single genotype of Mycobacterium tuberculosis in countries of east Asia. J. Clin. Microbiol. 33:3234-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Warren, R., J. Hauman, N. Beyers, M. Richardson, H. S. Schaaf, P. Donald, and P. van Helden. 1996. Unexpectedly high strain diversity of Mycobacterium tuberculosis in a high incidence community. S. Afr. Med. J. 86:45-49. [PubMed] [Google Scholar]
- 42.Wilkinson, D., M. Pillay, J. Cramp, G. R. Davies, and A. W. Sturm. 1997. Molecular epidemiology and transmission dynamics of Mycobacterium tuberculosis in rural Africa. Trop. Med. Int. Health 2:747-753. [DOI] [PubMed] [Google Scholar]
- 43.Wilson, S. A., S. Gross, and F. Drobniewski. 1998. Evaluation of strategies for molecular fingerprinting for their use in the routine work of a Mycobacterium reference unit. J. Clin. Microbiol. 36:3385-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]