Summary
The neuromuscular disorder spinal muscular atrophy (SMA), the most common inherited killer of infants, is caused by insufficient expression of survival motor neuron (SMN) protein. SMA therapeutics development efforts have focused on identifying strategies to increase SMN expression. We identified a long non-coding RNA (lncRNA) that arises from the antisense strand of SMN, SMN-AS1, which is enriched in neurons and transcriptionally represses SMN expression by recruiting the epigenetic Polycomb repressive complex-2. Targeted degradation of SMN-AS1 with antisense oligonucleotides (ASOs) increases SMN expression in patient-derived cells, cultured neurons, and the mouse central nervous system. SMN-AS1 ASOs delivered together with SMN2 splice-switching oligonucleotides additively increase SMN expression and improve survival of severe SMA mice. This study is the first proof-of-concept that targeting a lncRNA to transcriptionally activate SMN2 can be combined with SMN2 splicing modification to ameliorate SMA and demonstrates the promise of combinatorial ASOs for treatment of neurogenetic disorders.
Keywords: spinal muscular atrophy, SMN, long non-coding RNA, antisense oligonucleotides, SMA therapeutics
Graphical abstract
eTOC blurb
d’Ydewalle et al. identify a lncRNA that regulates SMN2 expression in a PRC2-dependent manner in neurons. Targeting SMN-AS1 in combination with SMN2 splicing modifiers has additive effects on SMN expression and improves the phenotype of severe SMA mice.
INTRODUCTION
The neuromuscular disease spinal muscular atrophy (SMA) is characterized by profound muscle weakness often within weeks or months after birth. It is caused by recessive mutations of the survival motor neuron 1 (SMN1) gene and retention of variable copy numbers of the highly homologous SMN2 gene (Lefebvre et al., 1995; Lorson et al., 1999). SMN2 contains a nucleotide substitution in a splice enhancer in exon 7 resulting in alternative splicing of SMN2-derived pre-mRNAs (Lorson et al., 1999; Monani et al., 1999a). Most mature SMN2 mRNAs lack exon 7 (Δ7-SMN2) and encode a truncated, unstable SMN protein resulting in reduced levels of functional SMN protein. All individuals affected by SMA have at least 1 copy of SMN2 and SMN2 copy number inversely correlates with disease severity (Lefebvre et al., 1997).
The identification of SMN2 as a genetic modifier of SMA prompted efforts to identify therapeutics that increase SMN2 expression (d'Ydewalle and Sumner, 2015). Splice-switching oligonucleotides (SSOs) and small molecules promote exon 7 inclusion in SMN2 pre-mRNAs and improve disease phenotypes in SMA mice (Hua et al., 2010; Hua et al., 2011; Naryshkin et al., 2014; Palacino et al., 2015). The SMN2 SSO Nusinersen is currently in phase III clinical trials in SMA patients (Chiriboga et al., 2016). While splicing modification is an elegant therapeutic strategy, it has a ceiling effect imposed by the amount of available SMN2 pre-mRNAs. This is particularly disadvantageous for individuals affected by the most common and severe type I SMA who have only 1 or 2 SMN2 copies but likely require the highest and most rapid induction of SMN. Another way to increase SMN levels is to increase SMN2 transcription (d'Ydewalle and Sumner, 2015). We and others showed that histone deacetylase (HDAC) inhibitors can activate the SMN2 promoter improving disease outcomes in SMA mice (Avila et al., 2007; Somers et al., 2013). HDAC inhibitors failed to show clinical efficacy in SMA patients likely due to low potency and specificity (Kissel et al., 2011; Swoboda et al., 2010). The identification of novel regulators of SMN transcription could be used to accentuate the effects of SMN2 splicing modulators.
Long non-coding RNAs (lncRNAs) have emerged as key regulators of protein-coding gene expression (Faghihi and Wahlestedt, 2009; Lee, 2012) with natural antisense transcripts (NATs), in particular, often affecting expression of their associated protein-coding genes (Carrieri et al., 2012; Faghihi et al., 2008; Mahmoudi et al., 2009). One mechanism by which lncRNAs regulate protein-coding gene expression is via binding of the Polycomb repressive complex-2 (PRC2) (Margueron and Reinberg, 2011). PRC2 binds to chromatin and di- and trimethylates Histone 3 at Lysine 27 (H3K27Me2/3), an epigenetic marker of repressed transcription (Margueron and Reinberg, 2011).
Many lncRNAs are enriched in the central nervous system (CNS) where they exhibit specific temporal and spatial expression patterns (Qureshi et al., 2010). They play roles in regulating neuronal cell fate specification and neurogenesis, synapse plasticity, neuronal homeostasis and behavior (Briggs et al., 2015; Meng et al., 2015; Ng et al., 2012; Qureshi et al., 2010; Ramos et al., 2015). Dysregulation of lncRNAs has been linked to neurodevelopmental and neurodegenerative disorders (Chamberlain and Brannan, 2001; Faghihi et al., 2008; Khalil et al., 2008). LncRNAs may also be promising therapeutic targets (Meng et al., 2015).
Here, we describe a neuronally-enriched NAT, SMN-AS1, which arises from a central region of intron 1 of the SMN genes. We show that SMN-AS1 can bind PRC2 and participate in recruiting it to the SMN transcription start site where it represses SMN expression. Knockdown of SMN-AS1 dissociates PRC2 from the SMN promoter and increases transcriptional activity of SMN in patient-derived cells, SMA neurons and a mouse model of severe SMA. Importantly, inducing SMN2 transcription by targeting SMN-AS1 in combination with SMN2 splice-switching oligonucleotides has additive effects on SMN expression and ameliorates SMA disease phenotypes in mice. Together, these studies provide proof-of-principle that SMN-AS1 is a clinically-relevant, novel therapeutic target for SMA.
RESULTS
Characterization of SMN-AS1
To identify putative non-coding RNAs associated with the human SMN1 and SMN2 genes, we utilized the UCSC Genome Browser to identify two expressed sequence tags (ESTs) (BC045789.1 and BC037819.1 of 1,613 and 1,618 base pairs length, respectively, isolated from human hypothalamus (Strausberg et al., 2002)), which align with 100% homology to the opposite DNA strand in a central region of intron 1 of the SMN genes (Figure 1A). Ensembl (www.ensembl.org) and the Basic local alignment search tool (BLAST) indicate these ESTs share no homology with other genomic regions. The ESTs co-align to epigenetic markers of active transcription in human embryonic stem cells (H1-ESCs) (Figure 1A), but not in human umbilical cord endothelial cells (HUVECs) suggesting that SMN-AS1 is expressed in a cell type-specific manner (Figure 1A). In Ensembl and AceView, these ESTs correspond to a single-exon antisense RNA, which we named SMN-AS1 (Figure 1A). A protein coding potential calculator (Kong et al., 2007) and an open reading frame finder tool (http://www.ncbi.nlm.nih.gov/orffinder/) indicated that SMN-AS1 has no protein-coding potential.
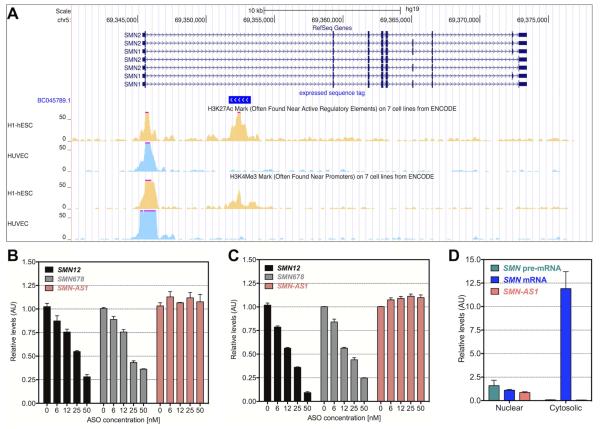
Figure 1. Identification of SMN-AS1.
(A) Genome browser views of human SMN1 and SMN2 on chromosome 5 are shown in 5′ to 3′-end with exons shown as blue boxes and introns as horizontal blue lines. The EST BC035789.1, shown in vivid blue, is transcribed from the opposite strand. ChIP-seq of the SMN loci for H3K27Ac and H3K4Me3 show active transcription co-aligning to the EST in H1-ESC cells (yellow tracks), but not in HUVEC cells (light blue tracks). (B,C) SMN mRNA levels measured at exon 1/2a boundary (SMN12) and at exon 6/7/8 boundary (SMN678) and SMN-AS1 levels in HeLa cells treated for 24 hours with ASO targeting SMN exon 1 (B) or SMN exon 2a (C). n = 4, mean + SEM. (D) SMN pre-mRNA, SMN mRNA and SMN-AS1 levels measured in the nuclear and cytoplasmic fraction of HeLa cells. n = 4, mean + SEM.
Since SMN-AS1 is complementary to SMN pre-mRNA, we developed a strand-specific RT-qPCR assay to detect SMN-AS1. We measured SMN-AS1 expression in HeLa cells (not shown) and human embryonic kidney (HEK293T) cells, which are known to express a wide variety of lncRNAs (Werner and Ruthenburg, 2015)(Figure S1A). SMN-AS1 was expressed at low levels compared to SMN mRNA and other protein-coding genes, but at similar levels to other lncRNAs (Figure S1A) (Rinn et al., 2007; Smilinich et al., 1999). To confirm that SMN-AS1 is a distinct transcript, we utilized two chimeric phosphorothioate 2′-O-methoxyethyl (2′MOE)/DNA ASOs targeting either exon 1 or exon 2a of the SMN1 or SMN2 pre-mRNAs (Figure 1C,D). Both ASOs dose-dependently reduced SMN mRNA levels, but did not affect SMN-AS1 levels (Figure 1B–D). SMN-AS1 was predominantly expressed in the nucleus like SMN pre-mRNA, while SMN mRNA was in the cytoplasm (Figures 1D and S1B). SMN-AS1 does not contain sequences encoding polyadenylation signals and SMN-AS1 was not detected in poly(A)-enriched RNA consistent with other reported lncRNAs (Figure S1C) (Quinn and Chang, 2016; Yang et al., 2011). The SMN-AS1 locus, unlike SMN mRNA, is not conserved across vertebrates suggesting that SMN-AS1 is expressed only in humans (Figure S1D).
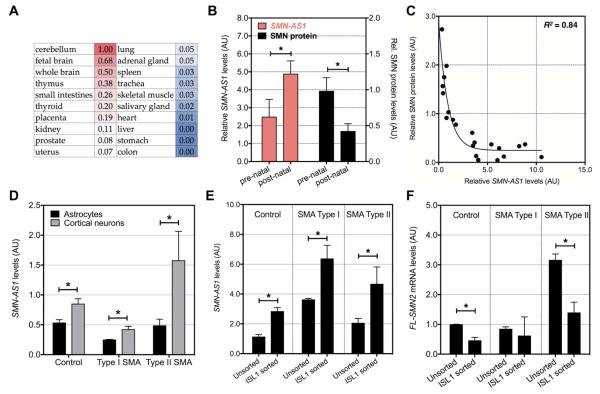
SMN-AS1 was particularly abundant in human cerebellum and brain (Figure 2A and S2A,B). To investigate the correlation between SMN-AS1 and SMN expression, we evaluated spinal cord samples (Figure 2B). SMN protein levels decreased between prenatal and early postnatal time points as previously described (Battaglia et al., 1997; Burlet et al., 1998), while SMN-AS1 levels increased (Figure 2B). A case-by-case correlation confirmed a robust inverse relationship (Figure 2C). SMN-AS1 was also enriched in neurons derived from induced pluripotent stem cells (iPSCs) isolated from control, type I SMA or type II SMA patients (Naryshkin et al., 2014; Ng et al., 2015). Expression levels were higher in cortical neurons compared to astrocytes and in purified Islet 1-positive motor neurons (Ng et al., 2015) compared to mixed motor neuron cultures (in which ~40% of cells are motor neurons)(Figures 2D-F and S2C). SMN-AS1 abundance varied in the different cell lines perhaps due to differences in neuronal differentiation rates and/or variability in FACS efficiency (Figure S2D). Nonetheless, within a cell line SMN-AS1 levels inversely correlated with SMN expression (Figure 2D,E).
Figure 2. SMN-AS1 expression in neuronal tissues and neurons.
(A) Heat map indicating relative SMN-AS1 levels in 20 different human tissues. (B) SMN-AS1 and SMN protein levels measured in human thoracic spinal cords at prenatal (n=12; age: 15 to 35 weeks of gestation) and postnatal (n=10; age: 1 day to 1 year) stages. Mean + SEM, * P < 0.05. (C) One-phase decay model indicating an inverse correlation (R2 = 0.84) between SMN-AS1 and SMN protein levels in human thoracic spinal cords. (D) SMN-AS1 levels in astrocytes and cortical neurons differentiated from human control and type I or type II SMA individuals. n = 3, mean + SEM, * P < 0.05. (E,F) SMN-AS1 levels (E) and FL-SMN2 mRNA levels (F) in mixed motor neuron cultures and sorted ISL1-positive motor neurons derived from human control and type I or type II SMA individuals. n = 3, mean + SEM, * P < 0.05.
SMN-AS1 negatively regulates SMN expression
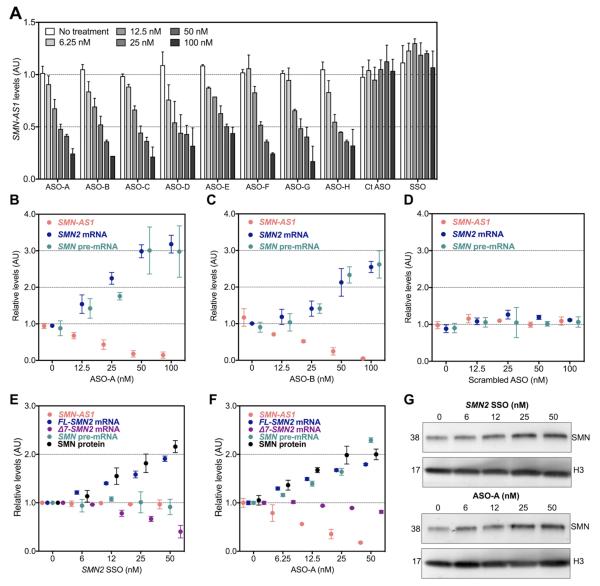
To investigate whether SMN-AS1 regulates SMN expression, we designed 78 individual chimeric 20-nucleotide phosphorothioate-modified 2′MOE/DNA ASOs complementary to SMN-AS1 tiling >90% of the transcript. We identified 8 ASOs that reduced SMN-AS1 levels in HeLa cells in a dose-dependent manner (Figure 3A). An ASO with a single nucleotide mismatch as well as an SMN2 SSO did not affect SMN-AS1 levels (Figure 3A). The 2 most effective ASOs dose-dependently increased full length (FL)-SMN2 mRNA levels while an ASO with a scrambled sequence did not (Figures 3B–D and S3A–C). This increase was consistent with transcriptional activation of SMN2 as SMN pre-mRNA levels also increased (Figures 3B–D and S3A–C). To further investigate the effects of SMN-AS1 on SMN, we expressed a reporter construct containing the SMN minimal promoter region (−163 bp upstream of the ATG start codon) coupled to a luciferase cDNA in HeLa cells (Figure S3D). Over-expression of SMN-AS1 reduced luciferase activity by ~54% and knockdown of SMN-AS1 increased luciferase activity by 55-74% (Figure S3D,E).
Figure 3. Antisense oligonucleotides targeting SMN-AS1 activate SMN expression.
(A) SMN-AS1 levels in HeLa cells treated for 1 day with various concentrations of ASOs targeting SMN-AS1 (ASO A-H) or control ASOs. Ct. ASO-1 = Control ASO-1 = single nucleotide mismatch ASO. SSO = SMN2 SSO. n = 4, mean + SEM. (B–D) SMN-AS1, SMN2 mRNA and SMN pre-mRNA levels in HeLa cells treated for 1 day with various concentrations of ASO-A (B), ASO-B (C), or scrambled ASO (D). UD = undetermined. n = 4, mean ± SEM. (E–G) SMN-AS1, FL-SMN2 mRNA, Δ7-SMN2 mRNA, SMN pre-mRNA and SMN protein levels in SMA fibroblasts treated for 3 days with various concentrations of SMN2 SSO (E) or SMN-AS1 ASO-A (F). n = 4, mean ± SEM. (G) Representative Western blots of SMN protein levels in SMA fibroblasts treated for 3 days with SMN2 SSO or SMN-AS1 ASO-A. The membranes were re-probed for a general loading control Histone-3 (H3). n = 3, mean ± SEM.
To verify that SMN-AS1 regulates SMN2 in SMA patient cells, we first confirmed SMN-AS1 expression in multiple fibroblast lines derived from SMA patients with variable SMN2 copy numbers (Figure S4A). We then transfected the GM03813 line with SMN-AS1 ASO-A or SMN2 SSO (Figure 3E-G). As expected, SMN2 SSO did not affect SMN-AS1 nor SMN pre-mRNA levels, but it increased FL-SMN2 mRNA and SMN protein levels, while Δ7-SMN2 mRNA levels decreased in a dose-dependent manner (Figure 3E,G). SMN-AS1 knockdown also augmented FL-SMN2 mRNA and SMN protein levels, but in contrast to SMN2 SSO, it also increased SMN pre-mRNA levels. There was little change in Δ7-SMN2 mRNA levels (Figure 3F) suggesting that SMN-AS1 ASOs may cause both transcriptional activation and some modulation of splicing leading to minimal net change in Δ7-SMN2 mRNA levels.
SMN-AS1 represses SMN expression in differentiating neurons
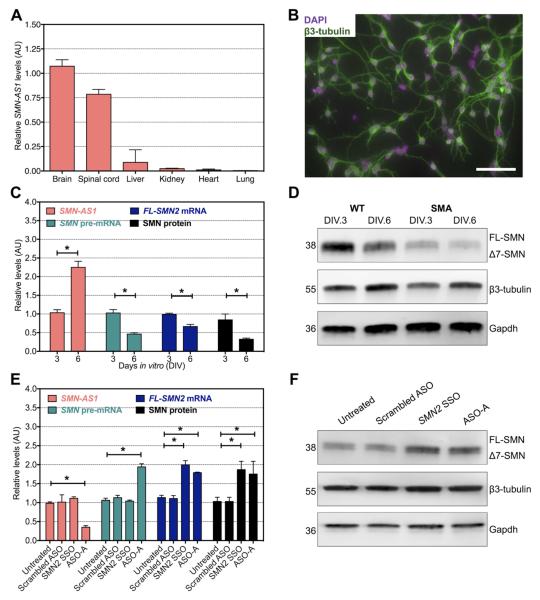
Severe SMA mice harbor the human SMN2 gene including a ~3.4 kb portion of the human SMN2 promoter, its exons and introns including the SMN-AS1 locus in intron 1 (Le et al., 2005). Like in humans, SMN-AS1 was predominantly expressed in the CNS in these mice (Figures 1 and 4A). We cultured primary cortical neurons with approximately 95% purity as assessed by immunocytochemical analyses of the neuronal markers β3-tubulin, Map2 and SMI32 (Figure 4B and not shown). SMN-AS1 levels increased 2.4-fold between day 3 and day 6 in vitro (DIV.3 and DIV.6, respectively) correlating inversely with a 50% decrease in FL-SMN2 mRNA and 57% decrease in SMN protein expression (Figure 4C,D). This inverse expression pattern was also observed in undifferentiated compared to differentiated human neuroblastoma cells and is consistent with the changes observed in developing human spinal cord (Figures 2 and S4B,C). Treatment of neurons with either SMN-AS1 ASO-A or ASO-B by free uptake starting at DIV.3 resulted in a 65% decrease in SMN-AS1 levels by DIV.6 and a 68% and 72% increase of FL-SMN2 mRNA and SMN protein levels, respectively (Figures 4E,F and S4D). These effects were comparable to those observed with an equimolar concentration of a SMN2 SSO (Figure 4E,F).
Figure 4. SMN-AS1 regulates SMN expression in primary SMA neurons.
(A) SMN-AS1 levels in brain, spinal cord, liver, kidney, heart and lungs of P10 SMA mice. n = 3, mean + SEM. (B) Fluorescence micrograph of primary cortical neurons stained for the neuronal marker β3-tubulin (green) and 4′,6-diamidino-2-phenylindole (DAPI, magenta) on DIV.3. Scale bar = 20 μm. (C) SMN-AS1, SMN pre-mRNA, FL-SMN2 mRNA and SMN protein levels in primary SMA cortical neurons on DIV.3 and DIV.6. n = 3, mean + SEM. * P < 0.05. (D) Representative Western blot of SMN protein levels in primary SMA cortical neurons on DIV.3 and DIV.6. The membranes were re-probed for neuronal marker β3-tubulin and a general loading control Gapdh. (E) SMN-AS1, SMN pre-mRNA, FL-SMN2 mRNA and SMN protein levels in primary SMA cortical neurons treated on DIV.3 for 3 days with 5 μM of scrambled ASO, SMN2 SSO or SMN AS1 ASO-A. (F). Representative Western blot of SMN protein levels in primary SMA neurons treated for 3 days with 5 μM of scrambled ASO, SMN2 SSO or SMN-AS1 ASO-A. The membrane was re-probed for the neuronal marker β3-tubulin and a general loading control Gapdh. n = 3, mean + SEM. * P < 0.05.
SMN-AS1 recruits PRC2 to the SMN promoter
Many nuclear lncRNAs negatively regulate expression of associated protein-coding genes by recruiting PRC2 to their promoter regions (Margueron and Reinberg, 2011; Wang and Chang, 2011). PRC2 contains the scaffolding proteins Embryonic ectoderm development (EED) and Suppressor of Zeste-12 (SUZ12) as well as the enzymatic component Enhancer of Zeste-2 (EZH2) and other proteins (Lee et al., 2006; Plath et al., 2003). To determine whether SMN-AS1 can bind to PRC2, we performed RNA binding immunoprecipitation (RIP) experiments using HEK293T cells (Figure 5A,B). When either EED or SUZ12 were immunoprecipitated, SMN-AS1 was detectable at levels comparable to HOTAIR, a lncRNA known to bind PRC2 (Figure 5A,B) (Rinn et al., 2007). We also immunoprecipitated SMN and snRNP70, another splicing protein with RNA binding capacity. SMN-AS1 and HOTAIR were undetectable in these precipitated fractions, but U1 snRNA, a small non-coding RNA known to bind to SMN and snRNP70, was present at robust levels (Figure S5A). Knockdown of SMN-AS1 in HEK293T cells resulted in reduced binding of SMN-AS1 to PRC2 (Figure S5B,C).
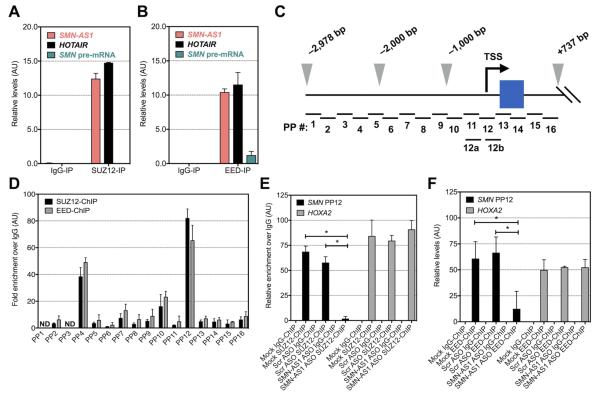
Figure 5. SMN-AS1 recruits PRC2 to SMN promoter to repress transcription.
(A,B) SMN-AS1, HOTAIR and SMN pre-mRNA levels following SUZ12-RIP (A) or EED-RIP (B) in HEK293T cells. The negative control IgG-RIP was set to 0. (C) Schematic of the SMN promoters spanning a region from −2,987 bp to +737 bp relative to the ATG start codon (+1 bp) of exon 1 (blue box). The TSS is indicated with a black arrow and the 2,000 and −1,000 bp marks are indicated with grey arrowheads. PP = Primer Pairs; see extended experimental procedures for details. (D) Relative enrichment of amplicons with indicated PP following SUZ12- and EED-ChIP in HEK293T cells. The negative control IgG-ChIP was set to 0. ND = not determined. n = 3, mean + SEM. (E,F) Relative enrichment of the PP12 amplicon following SUZ12-ChIP (E) or EED-ChIP (F) in HEK293T cells treated for 3 days with vehicle (mock), 250 nM scrambled (Scr) ASO or 250 nM SMN-AS1 ASO-A. n = 3, Mean + SEM. * P < 0.05.
EZH2 di- and tri-methylates Histone-3 at Lysine-27 (H3K27Me2/3), an epigenetic marker for repressed transcription found in promoter regions (Cao et al., 2002). To evaluate whether PRC2 can bind the promoter region of the SMN genes, we performed chromatin immunoprecipitations (ChIPs) in HEK293T cells with 16 primer sets spanning −3 kb to +700 bp of the SMN promoters relative to the ATG start codon (Figure 5C). We were unable to discriminate the SMN1 and SMN2 promoters as their sequences are nearly identical (Boda et al., 2004; Echaniz-Laguna et al., 1999; Monani et al., 1999b). EED- and SUZ12 ChIP indicated that PRC2 can bind the SMN promoters near the transcription start site (TSS) and this was associated with enriched H3K27Me3 marks in this region (Figures 5D and S4E). RNA polymerase-2 (POLR2A) ChIP showed increased occupancy of POLR2A downstream of the SMN TSS (Figure S4C). SMN-AS1 knockdown reduced binding of SUZ12 and EED to the SMN promoters indicating that PRC2 recruitment is at least in part SMN-AS1-dependent (Figures 5E,F). This was also associated with decreased H3K27Me3 levels and increased POLR2A occupancy near the TSS (Figure S5F,G). PRC2 binding to and H3K27Me3 levels in the HOXA2 proximal promoter, a known PRC2 target gene, was not affected by the ASO treatment (Figures 5E,F and S5F,G). Together, these data indicate that SMN-AS1 can bind PRC2 and that PRC2 is recruited to the SMN promoter to repress transcription by regulating H3K27Me3 levels near the SMN TSS.
PRC2 represses SMN expression in neurons
PRC2, and EZH2 in particular, play well established roles in maintaining pluripotency of stem cells and in controlling cell proliferation (Piunti et al., 2014; Rao et al., 2015). The role of EZH2 in differentiating neurons or the developing CNS is less well established. We confirmed Ezh2 expression in the CNS by Western blot analysis of brain (not shown) and spinal cord at embryonic and early postnatal time points in wild type (WT) and SMA mice, and loss of expression in adult stages (Figure S6A). Immunohistochemical analyses of neonatal WT and SMA spinal cords indicated that Ezh2 is expressed in motor neurons (Figure 6A). Ezh2 was also expressed in primary cortical neurons isolated from SMA mice (Figure S6B). To explore if Ezh2 regulates SMN expression in neurons, we transfected SMA cortical neurons with 2 ASOs targeting mouse Ezh2. Both Ezh2 ASOs significantly reduced Ezh2 mRNA and Ezh2 protein levels and increased SMN protein expression to similar levels achieved with SMN1-AS1 ASO treatment (Figure 6B,C).
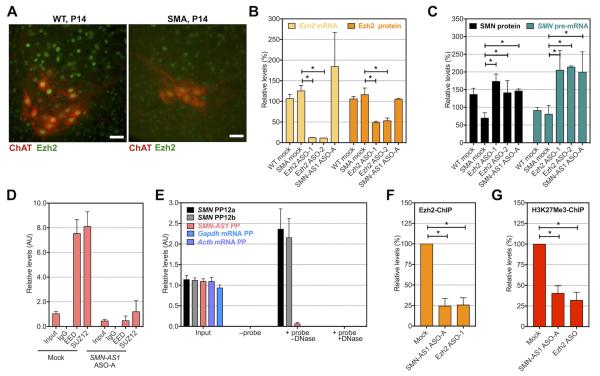
Figure 6. SMN-AS1 recruits PRC2 to SMN promoter in neurons.
(A) Fluorescence micrographs of ventral spinal cord isolated from a P14 WT (left panel) or SMA (right panel) mouse stained with ChAT (red) and Ezh2 (green). Scale bar = 50 μm. (B) Ezh2 mRNA and Ezh2 protein levels in untreated WT cortical neurons or SMA cortical neurons untreated or treated for 3 days with 5 μM Ezh2 ASO-1 or Ezh2 ASO-2, or 5 μM of SMN-AS1 ASO A. n = 3, mean + SEM. * P < 0.05. (C) SMN protein and SMN pre-mRNA levels in untreated WT cortical neurons or SMA cortical neurons untreated or treated on DIV.3 for 3 days with 5 μM Ezh2 ASO-1 or Ezh2 ASO-2, or 5 μM of SMN-AS1 ASO-A. n = 3, mean + SEM. * P < 0.05. (D) SMN-AS1 levels following SUZ12-RIP and EED-RIP in primary SMA cortical neurons treated on DIV.3 for 3 days with vehicle (mock) or 5 μM SMN-AS1 ASO-A. The negative control IgG-RIP was set to 0. n = 3, mean + SEM. (E) Relative enrichment of PP12a, PP12b or SMN-AS1 amplicons following SMN-AS1 ChIRP with biotinylated ASOs in primary SMA cortical neurons on DIV.6. Gapdh and Actb mRNA primers were used as negative control. n = 3, mean + SEM. (F,G) Relative enrichment of PP12 amplicon following Ezh2-ChIP (F) and H3K27Me3-ChIP (G) in primary SMA cortical neurons treated on DIV.3 for 3 days with vehicle (mock), 5 μM SMN-AS1 ASO-A or 5 μM Ezh2 ASO-1. n = 3, mean + SEM. * P < 0.05.
We confirmed that SMN-AS1 can bind to PRC2 in SMA neurons using RIP. Knockdown of SMN-AS1 reduced SMN-AS1 levels and abolished PRC2:SMN-AS1 binding (Figure 6D). SMN-AS1 can also bind PRC2 in vivo in WT and SMA mouse brain (Figure S6C). To determine whether SMN-AS1 directly binds to the SMN promoter in the TSS region, we performed chromatin isolation by RNA purification (ChIRP) in cultured neurons (Figure 6E). We modified our 8 lead SMN-AS1ASOs by replacing 10 core nucleotides with 2′MOE nucleotides and adding a 3′-end biotin label. We validated that these biotinylated ASOs pulled down SMN-AS1 (Figure S6D). Two separate primer sets amplifying the SMN TSS region confirmed that SMN-AS1 binds directly to the SMN proximal promoter region (Figure 6E). A DNase treatment of the precipitated material confirmed that we amplified proximal SMN promoter DNA (and not duplexed 5′UTR SMN mRNA). The SMN-AS1 DNA locus was not amplified indicating that SMN-AS1 does not bind its complementary DNA in this setting (Figure 6E).
We next evaluated whether Prc2 binds to SMN2 in primary neurons. Suz12 (not shown), Ezh2 and its associated repressive marker H3K27Me3 localize to the SMN2 proximal promoter (Figure 6F,G). Knockdown of either Ezh2 or SMN-AS1 reduced Ezh2 binding and decreased H3K27Me3 levels in the SMN TSS region (Figure 6F,G). We conclude that in neurons SMN-AS1 and Prc2 are recruited to the SMN proximal promoter, and that SMN-AS1 knockdown alleviates PRC2 binding and de-represses SMN transcriptional activity.
SMN-AS1 is a therapeutic target for SMA
As a primary goal of SMA therapeutics is to increase SMN levels, targeted knockdown of SMN-AS1 could represent a novel SMA therapeutic strategy. Severe SMA mice were injected subcutaneously with SMN-AS1 ASO-A (400 mg/kg) at P1 and P3 as ASOs are CNS penetrant at this age (Hua et al., 2011). At P10, there was a ~69 % and ~51% reduction of SMN-AS1 in brain and spinal cord, respectively and increased SMN2 pre-mRNA, FL-SMN2 mRNA and SMN protein levels (Figures 7A–D and Figure S7A). Despite a moderate increase of SMN expression in the CNS, survival, body weight and righting reflex were not improved compared to saline-treated SMA mice (Figure 7E–G).
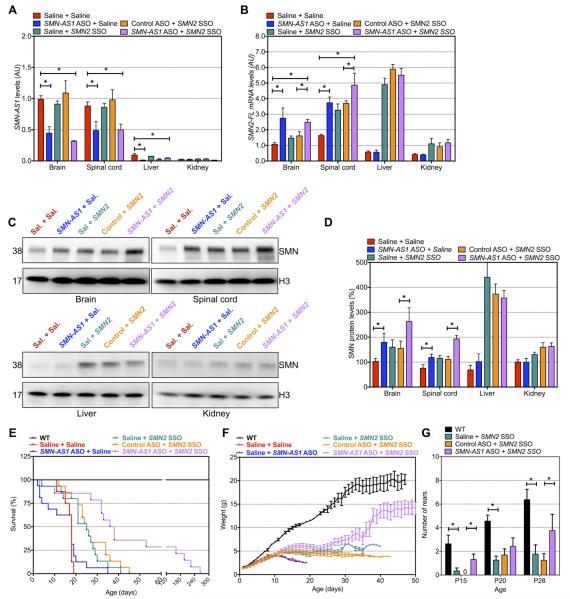
Figure 7. Targeting SMN-AS1 in SMA mice increases SMN expression.
(A–D) SMN-AS1 (A), FL-SMN2 mRNA (B) and SMN protein (C,D) levels in brain, spinal cord, liver and kidney at P10. n = 3, mean + SEM. * P < 0.05. (C) Representative Western blots of data presented in C, D. (E) Kaplan-Meier curve for treated SMA mice. WT mice are shown as a reference. n = 15 mice per group. (F) Daily body weight measurements of treated SMA mice. mean ± SEM. (G) Number of rears in 60 seconds for treated animals. Mean + SEM. * P < 0.05.
Severe SMA mice have dysfunction of several organ systems of uncertain relevance to the human disease (Bevan et al., 2010; Gombash et al., 2015). These systemic impairments may account for the limited phenotypic improvement previously observed with neuronal-specific genetic rescue of SMN (Gogliotti et al., 2012; Martinez et al., 2012). Given the neuronal enrichment of SMN-AS1 and the potential to miss meaningful therapeutic effects because of systemic abnormalities in SMA mice, we evaluated the effects of combined systemic administration of a low, sub-optimal dose (50 mg/kg) of SMN2 SSO together with a high dose (400 mg/kg) of SMN-AS1 ASO. SMN2 SSO alone or in combination with a control ASO increased SMN expression to the same magnitude as the SMN-AS1 ASO alone in the brain and spinal cord (Figure 7B–D). SMN2 SSO also increased FL-SMN2 mRNA and SMN protein levels in other organs while SMN-AS1 ASO alone did not (Figure 7B–D). When combined, SMN2 SSO and SMN-AS1 ASO had additive effects on FL-SMN2 mRNA and SMN protein levels in the CNS (Figure 7C,D).
Combination therapy also improved survival of SMA mice. While saline-treated mice had a median survival of 18 days and low dose SMN2 SSO-treated mice had a median survival time of 25 days (Figure 7E), combining SMN2 SSO with SMN-AS1 ASO increased median survival to 37 days with 4 out of 15 (~25%) mice surviving more than 120 days (Figure 7E). SMA mice receiving combination ASOs were heavier than SMN2 SSO alone treated mice (Figure 7F). Combination-treated mice also had improved motor behavior and in some tests animals receiving both SMN-AS1 ASO and SMN2 SSO performed as well as untreated WT control animals (Figure 7G and Figure S7B,C). Taken together, these data show that reducing SMN-AS1 levels in combination with modulating SMN2 splicing ameliorates SMA disease manifestations.
DISCUSSION
The neuromuscular disorder SMA is caused by insufficient expression levels of SMN protein resulting in profound muscle weakness and often premature death. Antisense oligonucleotides and small molecules that modulate SMN2 splicing are in clinical trials; however, their efficacy could be accentuated by parallel induction of SMN2 transcription. Identification of SMN2 transcriptional activators has been limited by inadequate understanding of the mechanisms that regulate the SMN genes particularly in neurons. In this study, we identified a neuronally-enriched NAT, SMN-AS1, whose expression levels increase during neuronal differentiation inversely correlating with SMN. We show that SMN-AS1 contributes to the recruitment of the epigenetic chromatin modifier PRC2 to the SMN2 promoter and that SMN-AS1 knockdown dissociates PRC2 from the SMN promoter thus increasing SMN expression in neurons and in the CNS. We also provide proof-of-principle that SMN-AS1 is a clinically relevant, novel therapeutic target by demonstrating that SMN-AS1 ASOs together with SMN2 SSOs show combinatorial therapeutic effects in SMA mice. Together these data reveal a novel mechanism regulating SMN expression in neurons, validate a new SMA therapeutic strategy that can be combined with SMN2 splicing modification, and highlight the potential of combinatorial ASOs targeting distinct gene regulatory mechanisms.
Dynamic expression of SMN and SMN-AS1
Prior studies have reported that SMN levels are developmentally regulated (Battaglia et al., 1997; Burlet et al., 1998). Our data confirms a decrease of SMN levels between prenatal and early postnatal periods in human spinal cord as well as a 2-fold reduction in SMN levels during differentiation of cultured neurons. Motor neurons may be particularly dependent on high SMN levels for normal maturation and/or maintenance during perinatal development and SMA disease onset often occurs during this critical period. In SMA mice, neonatal knockdown of SMN is required to recapitulate disease pathology (Kariya et al., 2014), while early postnatal restoration of SMN is essential to ameliorate disease manifestations (Lutz et al., 2011). These studies highlight the importance of this temporal window, yet the mechanisms that regulate SMN expression during this period have not been well defined. SMN-AS1 is developmentally regulated in the spinal cord in an inverse pattern to SMN suggesting that it represses SMN during neuronal maturation and CNS development. Many lncRNAs have been shown to regulate the expression of protein-coding genes in a temporally and spatially restricted manner (Herriges et al., 2014). The NAT SOX2OT, for example, increases during aging (Barry et al., 2015) repressing the neural pluripotency marker SOX2. Our data raise the question of how SMN-AS1 expression itself is controlled. In some cases, NATs and protein coding genes are inversely regulated by direct transcriptional interference (Shearwin et al., 2005). Given the active co-expression of SMN and SMN-AS1, it may be more likely that each are regulated by distinct transcription factors (Alam et al., 2014). Another question to be answered in future investigations is whether SMN-AS1 expression levels modify SMA disease severity. These experiments are challenging, however, as they likely require determining expression levels specifically in motor neurons of multiple SMA patients.
The interaction between SMN-AS1 and the SMN genes
Many nuclear lncRNAs regulate gene expression by associating with chromatin (Faghihi and Wahlestedt, 2009; Wang and Chang, 2011). Direct binding of NATs to complementary DNA is a straightforward way to explain how lncRNAs interact with their target. This mode of action has been proposed for lncRNAs including MEG3, which forms RNA-DNA triplexes in GA-rich sequences of TGF-β pathway genes (Mondal et al., 2015). Our ChIRP data suggest that, at least in primary cortical neurons, SMN-AS1 does not principally bind to its DNA locus, but rather to the SMN TSS region corresponding to a local hotspot of PRC2 recruitment. Sequence alignment of SMN-AS1 and the SMN promoters indicates ~70% sequence homology between the 3′ end of SMN-AS1 (1376 – 1613 nt) and a region −0.6 kb upstream of the ATG start codon. This suggests that this 3′ sequence of SMN-AS1 could mediate SMN promoter binding. The site of action of lncRNAs may also be determined by chromatin tertiary structure (Vance and Ponting, 2014). Although separated by 7 kb, it is plausible that the SMN-AS1 locus and the proximal SMN promoter are in close vicinity due to ‘DNA looping’ and that SMN-AS1 functions locally near its site of synthesis. Our studies have not determined whether SMN-AS1 can regulate SMN expression only in cis or also in trans (Faghihi and Wahlestedt, 2009). SMN-AS1 is likely transcribed from both the SMN1 and SMN2 genes and our data from SMA mouse neurons and tissues, which harbor only the human SMN2 gene, indicate that SMN-AS1 can repress the SMN locus from which it arises. Whether SMN-AS1 can also regulate a neighboring SMN locus in trans will require further study.
PRC2 contributes to the regulation of SMN expression in neurons
PRC2 has traditionally been associated with genomic imprinting (i.e., silencing of alleles in a parent-of-origin dependent manner), X-chromosome inactivation and irreversible silencing of protein-coding genes by trimethylating H3K27 in their promoter regions (Mager et al., 2003; Margueron and Reinberg, 2011; Pinter et al., 2012; Simon et al., 2013). In contrast, our data indicate that PRC2 regulates SMN expression despite ongoing SMN transcriptional activity. These data are consistent with a model in which PRC2 together with the repressive H3K27Me3 mark co-exist with and are balanced by markers of active transcription resulting in a basal level of SMN expression, which can be further modulated. Our observations are consistent with other work demonstrating that EZH2 partially inhibits SLIT2 expression in a variety of cancers and that SLIT2 expression can be increased by interfering with EZH2 function (Yu et al., 2010). Furthermore in hippocampal neurons, it has been shown that neuronal stimulation can de-repress Bdnf expression by displacement of Ezh2 from the Bdnf promoters (Palomer et al., 2016) and that Ezh2 knockdown can de-repress Runx2/p57 (Aguilar et al., 2016). Knockdown of specific lncRNAs, even in fully differentiated cells, has been shown to displace PRC2 thus de-repressing gene transcription (Gonzalez et al., 2015; Li et al., 2013).
The reversibility of PRC2-induced repression of SMN may also imply that both repressive and activating complexes can be recruited to the SMN promoter. Previous work by our group and others has implicated several other epigenetic modifiers in the regulation of SMN expression including histone deacetylases (Avila et al., 2007; Kernochan et al., 2005) and methyl-CpG binding protein 2 (MECP2) (Hauke et al., 2009). The recruitment of these epigenetic modifiers are not mutually exclusive and can co-occupy the SMN promoter to maintain steady-state SMN expression levels. Since several chromatin modifiers likely co-regulate SMN transcription, this raises the question whether SMN-AS1 recruits only PRC2, or whether it also tethers other epigenetic complexes to the SMN2 promoter. Future ChIRP experiments followed by mass spectrometry of the precipitated proteins could identify additional interactors of SMN-AS1, which would provide further insights regarding the neuronal regulation of SMN expression.
Although knockdown of SMN-AS1 increased SMN pre-mRNA and FL-SMN mRNA levels, it unexpectedly did not significantly increase Δ7-SMN2 mRNA levels. This suggests that suppression of SMN-AS1 results both in transcriptional activation of SMN2 as well as in some modulation of SMN2 exon 7 splicing with little net change in Δ7-SMN2 mRNA levels. Transcriptional activation can alter RNA polymerase II elongation rates with consequent changes in splicing both in vitro and in vivo (Fong et al., 2014; de la Mata et al., 2003). Alternatively, it is possible that SMN-AS1 knockdown not only alters the epigenetic state of the SMN promoter region, but also downstream regions with subsequent effects on exon 7 splicing as has been described for other genes (Ameyar-Zazoua et al., 2012; Luco et al., 2010).
SMN-AS1 as therapeutic target for SMA?
ASOs can be used to target cytoplasmic or nuclear RNA molecules thus modulating splicing, 5′ capping, polyadenylation, protein binding, or protein translation. They can also be used to direct degradation of target RNAs by RNase H (Corey, 2007; Kole et al., 2012). The potential of ASOs as a therapeutic for neurological and other genetic disorders has been studied extensively in vitro and in vivo. (Cheng et al., 2015; Donnelly et al., 2013; Lagier-Tourenne et al., 2013; Meng et al., 2015; Schoch et al., 2016). This is particularly true for SMA, in which SMN2 SSOs are able to ameliorate disease in SMA mouse models by influencing SMN2 splicing in the nucleus (Hua et al., 2010; 2008; Passini et al., 2011; Sahashi et al., 2013). Importantly, although penetration of ASOs into the adult CNS is low after systemic delivery as has been shown by others (Hua et al., 2008), clinical trials in patients with amyotrophic lateral sclerosis and SMA have indicated that intrathecal administration of ASOs is tolerated and safe (Chiriboga et al., 2016; Miller et al., 2013).
In an effort to determine whether SMN-AS1 could be a novel therapeutic target, we identified ASOs that degrade SMN-AS1 and dose-dependently activate SMN expression. When administered alone, SMN-AS1 ASO did not improve the survival of severe SMA mice. This is not entirely unexpected as the SMN1-AS1 ASO sequences and chemistries have not yet been optimized for in vivo targeting efficiency as has been done for SMN2 SSOs (Hua et al., 2010). In addition, the expression of SMN-AS1 and induction of SMN expression after SMN-AS1 knockdown is quite restricted to neurons and prior neuronal-specific genetic rescue experiments in severe SMA mice showed only modest phenotypic improvements (Gogliotti et al., 2012; Martinez et al., 2012; Paez-Colasante et al., 2013). These studies highlight that in SMA mouse models there are abnormalities of multiple other organ systems (Bevan et al., 2010; Biondi et al., 2012; Gombash et al., 2015; Heier et al., 2010) which are an important determinant of SMA mouse survival, but are of uncertain relevance to the human disease. Although systemic administration of high dose SMN2 SSO alone substantially improves the survival of SMA mice (Hua et al., 2011), this is likely because of restoration of SMN levels peripherally (Hua et al., 2015). Given that SMN-AS1 may specifically down-regulate SMN expression during perinatal periods, it is also possible that earlier intervention during this temporal window would be more efficacious.
Importantly, when combined with suboptimal doses of SMN2 SSO, the disease modifying effects of SMN-AS1 ASOs were evident with improved motor behavior, weights and survival in severe SMA mice. Our study shows that SMN-AS1 can be a therapeutic target, and also demonstrates the potential of combinatorial ASOs targeting distinct molecular regulators of a single gene. Combining ASOs targeting two different genes with traditional chemotherapy has been proposed for cancer therapeutics (Biroccio et al., 2003; Zupi et al., 2005), but this is the first example to our knowledge of two ASOs used in combination for a neurological disorder. Like others, we found that dosing of multiple ASOs was well tolerated in SMA mice (Hua et al., 2015; Zupi et al., 2005). Although the clinical development of combination ASOs will present new challenges, they hold the promise of additive or synergistic therapeutic effects for multiple neurogenetic diseases.
EXPERIMENTAL PROCEDURES
All mice in this study were maintained according to protocols approved by the Institutional Animal Care and Use Committee at Johns Hopkins University School of Medicine. De-identified human tissue samples were collected at autopsy following parental or patient informed consent in strict observance of the legal and institutional ethical regulations. Protocols were approved by the Institutional Review Board at the Johns Hopkins University School of Medicine.
Antisense oligonucleotide synthesis
We synthesized and purified all chemically modified oligonucleotides as described in Swayze et al., 2007. The 2′-O-methoxyethyl (2′MOE) gapmer ASOs are 20 nucleotides in length, wherein the central gap segment comprising of ten 2′-deoxynucleotides are flanked on the 5′ and 3′ wings by five 2′-MOE modified nucleotides. Internucleoside linkages are purely phosphorothioate (in vitro) or interspersed with phosphodiester (in vivo), and all cytosine residues are 5′-methylcytosines. The sequences of the ASOs used in this study are listed in supplemental experimental procedures.
Detailed experimental procedures for cell cultures, pluripotent stem cell cultures, primary cortical neuron cultures, Western blotting, RNA isolation and RT-qPCR, ChIP, RIP and ChIRP experiments, and ASO administration and behavioral assessment of the SMA mice are described in supplemental materials.
Supplementary Material
Highlights.
SMN-AS1 is the first long non-coding RNA identified that is associated with SMN;
SMN-AS1 recruits PRC2 to the SMN2 promoter repressing its transcription in neurons;
SMN-AS1 knockdown increases SMN expression in vitro and in vivo;
Combining SMN-AS1 ASOs with SMN2 splicing ASO attenuates disease in SMA mice.
ACKNOWLEDGEMENTS
We thank all the individuals and their families for providing tissue samples. We thank Dr. Thomas Crawford (Johns Hopkins University School of Medicine) for his help with collecting human tissues. We thank Dr. Loyal Goff (Johns Hopkins University School of Medicine) and Dr. Mohammad Ali Faghihi (University of Miami Miller School of Medicine) for fruitful discussions. Human tissue was received from the NIH Neuro-BioBank at the University of Maryland Brain and Tissue Bank. We are grateful to Dr. Amal Dakka and Dr. Nikolai Naryshkin (PTC Therapeutics) for providing primer sequences. This work was supported by the SMA Research Team (C.J.S.), the Muscular Dystrophy Association (C.dY.) and the American Association of Neuromuscular and Electrodiagnostic Medicine (C.dY.), FightSMA, Inc. (C.dY.) and the Gwendolyn Strong Foundation (C.dY.), and NINDS R01NS096770 (C.J.S.). This publication was made possible by the Johns Hopkins Institute for Clinical and Translational Research (ICTR) which is funded in part by Grant Number UL1 TR 001079 from the National Center for Advancing Translational Sciences (NCATS) a component of the National Institutes of Health (NIH), and NIH Roadmap for Medical Research. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of the Johns Hopkins ICTR, NCATS or NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
Conceptualization, C.dY. and C.J.S.; Methodology, C.dY., D.M.R., and C.J.S.; Formal Analysis, C.dY., D.M.R., and N.J.P.; Investigation: C.dY., D.M.R., and N.J.P.; Resources: L.L.R., F.R., and C.F.B.; Writing – Original Draft, C.dY.; Writing – Review and Editing, C.dY., D.M.R., K.L., A.J.W., L.L.R, F.R., C.F.B. and C.J.S.; Visualization: C.dY. and D.M.R.; Supervision, C.dY. and C.J.S.; Project Administration, C.dY. and C.J.S.; Funding Acquisition, C.dY. and C.J.S.
REFERENCES
- Aguilar R, Bustos FJ, Saez M, Rojas A, Allende ML, van Wijnen AJ, van Zundert B, Montecino M. Polycomb PRC2 complex mediates epigenetic silencing of a critical osteogenic master regulator in the hippocampus. Biochim. Biophys. Acta. 2016;1859:1043–1055. doi: 10.1016/j.bbagrm.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam T, Medvedeva YA, Jia H, Brown JB, Lipovich L, Bajic VB. Promoter analysis reveals globally differential regulation of human long non-coding RNA and protein-coding genes. PLoS ONE. 2014;9:e109443. doi: 10.1371/journal.pone.0109443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameyar-Zazoua M, Rachez C, Souidi M, Robin P, Fritsch L, Young R, Morozova N, Fenouil R, Descostes N, Andrau J-C, Mathieu J, Hamiche A, Ait-Si-Ali S, Muchardt C, Batsché E, Harel-Bellan A. Argonaute proteins couple chromatin silencing to alternative splicing. Nat. Struct. Mol. Biol. 2012;19:998–1004. doi: 10.1038/nsmb.2373. [DOI] [PubMed] [Google Scholar]
- Avila AM, Burnett BG, Taye AA, Gabanella F, Knight MA, Hartenstein P, Cizman Z, Di Prospero NA, Pellizzoni L, Fischbeck KH, Sumner CJ. Trichostatin A increases SMN expression and survival in a mouse model of spinal muscular atrophy. J. Clin. Invest. 2007;117:659–671. doi: 10.1172/JCI29562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry G, Guennewig B, Fung S, Kaczorowski D, Weickert CS. Long Non-Coding RNA Expression during Aging in the Human Subependymal Zone. Front. Neurol. 2015;6:45. doi: 10.3389/fneur.2015.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia G, Princivalle A, Forti F, Lizier C, Zeviani M. Expression of the SMN gene, the spinal muscular atrophy determining gene, in the mammalian central nervous system. Hum. Mol. Genet. 1997;6:1961–1971. doi: 10.1093/hmg/6.11.1961. [DOI] [PubMed] [Google Scholar]
- Bevan AK, Hutchinson KR, Foust KD, Braun L, McGovern VL, Schmelzer L, Ward JG, Petruska JC, Lucchesi PA, Burghes AHM, et al. Early heart failure in the SMNDelta7 model of spinal muscular atrophy and correction by postnatal scAAV9-SMN delivery. Hum. Mol. Genet. 2010;19:3895–3905. doi: 10.1093/hmg/ddq300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biondi O, Lopes P, Desseille C, Branchu J, Chali F, Ben Salah A, Pariset C, Chanoine C, Charbonnier F. Physical exercise reduces cardiac defects in type 2 spinal muscular atrophy-like mice. J. Physiol. 2012;590:5907–5925. doi: 10.1113/jphysiol.2012.238196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biroccio A, Leonetti C, Zupi G. The future of antisense therapy: combination with anticancer treatments. Oncogene. 2003;22:6579–6588. doi: 10.1038/sj.onc.1206812. [DOI] [PubMed] [Google Scholar]
- Boda B, Mas C, Giudicelli C, Nepote V, Guimiot F, Levacher B, Zvara A, Santha M, LeGall I, Simonneau M. Survival motor neuron SMN1 and SMN2 gene promoters: identical sequences and differential expression in neurons and non-neuronal cells. Eur. J. Hum. Genet. 2004;12:729–737. doi: 10.1038/sj.ejhg.5201217. [DOI] [PubMed] [Google Scholar]
- Briggs JA, Wolvetang EJ, Mattick JS, Rinn JL, Barry G. Mechanisms of Long Non-coding RNAs in Mammalian Nervous System Development, Plasticity, Disease, and Evolution. Neuron. 2015;88:861–877. doi: 10.1016/j.neuron.2015.09.045. [DOI] [PubMed] [Google Scholar]
- Burlet P, Huber C, Bertrandy S, Ludosky MA, Zwaenepoel I, Clermont O, Roume J, Delezoide AL, Cartaud J, Munnich A, et al. The distribution of SMN protein complex in human fetal tissues and its alteration in spinal muscular atrophy. Hum. Mol. Genet. 1998;7:1927–1933. doi: 10.1093/hmg/7.12.1927. [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, Pesce E, Ferrer I, Collavin L, Santoro C, et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491:454–457. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- Chamberlain SJ, Brannan CI. The Prader-Willi syndrome imprinting center activates the paternally expressed murine Ube3a antisense transcript but represses paternal Ube3a. Genomics. 2001;73:316–322. doi: 10.1006/geno.2001.6543. [DOI] [PubMed] [Google Scholar]
- Cheng H-M, Chern Y, Chen I-H, Liu C-R, Li S-H, Chun SJ, Rigo F, Bennett CF, Deng N, Feng Y, et al. Effects on murine behavior and lifespan of selectively decreasing expression of mutant huntingtin allele by supt4h knockdown. PLoS Genet. 2015;11:e1005043. doi: 10.1371/journal.pgen.1005043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiriboga CA, Swoboda KJ, Darras BT, Iannaccone ST, Montes J, De Vivo DC, Norris DA, Bennett CF, Bishop KM. Results from a phase 1 study of nusinersen (ISIS-SMN(Rx)) in children with spinal muscular atrophy. Neurology. 2016;86:890–897. doi: 10.1212/WNL.0000000000002445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey DR. RNA learns from antisense. Nature Chemical Biology. 2007;3:8–11. doi: 10.1038/nchembio0107-8. [DOI] [PubMed] [Google Scholar]
- d'Ydewalle C, Sumner CJ. Spinal Muscular Atrophy Therapeutics: Where do we Stand? Neurotherapeutics. 2015;12:303–316. doi: 10.1007/s13311-015-0337-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, Zhang P-W, Pham JT, Haeusler AR, Heusler AR, Mistry NA, Vidensky S, Daley EL, Poth EM, Hoover B, et al. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80:415–428. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echaniz-Laguna A, Miniou P, Bartholdi D, Melki J. The Promoters of the Survival Motor Neuron Gene (SMN) and Its Copy (SMNc) Share Common Regulatory Elements. Am. J. Hum. Genet. 1999;64:1365–1370. doi: 10.1086/302372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat. Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat. Rev. Mol. Cell Biol. 2009;10:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong N, Kim H, Zhou Y, Ji X, Qiu J, Saldi T, Diener K, Jones K, Fu X-D, Bentley DL. Pre-mRNA splicing is facilitated by an optimal RNA polymerase II elongation rate. Genes Dev. 2014;28:2663–2676. doi: 10.1101/gad.252106.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogliotti RG, Quinlan KA, Barlow CB, Heier CR, Heckman CJ, Didonato CJ. Motor neuron rescue in spinal muscular atrophy mice demonstrates that sensory-motor defects are a consequence, not a cause, of motor neuron dysfunction. J. Neurosci. 2012;32:3818–3829. doi: 10.1523/JNEUROSCI.5775-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gombash SE, Cowley CJ, Fitzgerald JA, Iyer CC, Fried D, McGovern VL, Williams KC, Burghes AHM, Christofi FL, Gulbransen BD, et al. SMN deficiency disrupts gastrointestinal and enteric nervous system function in mice. Hum. Mol. Genet. 2015;24:3847–3860. doi: 10.1093/hmg/ddv127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez I, Munita R, Agirre E, Dittmer TA, Gysling K, Misteli T, Luco RF. A lncRNA regulates alternative splicing via establishment of a splicing-specific chromatin signature. Nat. Struct. Mol. Biol. 2015;22:370–376. doi: 10.1038/nsmb.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauke J, Riessland M, Lunke S, Eyüpoglu IY, Blümcke I, El-Osta A, Wirth B, Hahnen E. Survival motor neuron gene 2 silencing by DNA methylation correlates with spinal muscular atrophy disease severity and can be bypassed by histone deacetylase inhibition. Hum. Mol. Genet. 2009;18:304–317. doi: 10.1093/hmg/ddn357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heier CR, Satta R, Lutz C, Didonato CJ. Arrhythmia and cardiac defects are a feature of spinal muscular atrophy model mice. Hum. Mol. Genet. 2010;19:3906–3918. doi: 10.1093/hmg/ddq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herriges MJ, Swarr DT, Morley MP, Rathi KS, Peng T, Stewart KM, Morrisey EE. Long noncoding RNAs are spatially correlated with transcription factors and regulate lung development. Genes Dev. 2014;28:1363–1379. doi: 10.1101/gad.238782.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y, Liu YH, Sahashi K, Rigo F, Bennett CF, Krainer AR. Motor neuron cell-nonautonomous rescue of spinal muscular atrophy phenotypes in mild and severe transgenic mouse models. Genes Dev. 2015;29:288–297. doi: 10.1101/gad.256644.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y, Sahashi K, Hung G, Rigo F, Passini MA, Bennett CF, Krainer AR. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev. 2010;24:1634–1644. doi: 10.1101/gad.1941310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y, Sahashi K, Rigo F, Hung G, Horev G, Bennett CF, Krainer AR. Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature. 2011;478:123–126. doi: 10.1038/nature10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y, Vickers TA, Okunola HL, Bennett CF, Krainer AR. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am. J. Hum. Genet. 2008;82:834–848. doi: 10.1016/j.ajhg.2008.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariya S, Obis T, Garone C, Akay T, Sera F, Iwata S, Homma S, Monani UR. Requirement of enhanced Survival Motoneuron protein imposed during neuromuscular junction maturation. J. Clin. Invest. 2014;124:785–800. doi: 10.1172/JCI72017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kernochan LE, Russo ML, Woodling NS, Huynh TN, Avila AM, Fischbeck KH, Sumner CJ. The role of histone acetylation in SMN gene expression. Hum. Mol. Genet. 2005;14:1171–1182. doi: 10.1093/hmg/ddi130. [DOI] [PubMed] [Google Scholar]
- Khalil AM, Faghihi MA, Modarresi F, Brothers SP, Wahlestedt C. A novel RNA transcript with antiapoptotic function is silenced in fragile X syndrome. PLoS ONE. 2008;3:e1486. doi: 10.1371/journal.pone.0001486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissel JT, Scott CB, Reyna SP, Crawford TO, Simard LR, Krosschell KJ, Acsadi G, Elsheik B, Schroth MK, D'Anjou G, et al. SMA CARNI-VAL TRIAL PART II: A Prospective, Single-Armed Trial of L-Carnitine and Valproic Acid in Ambulatory Children with Spinal Muscular Atrophy. PLoS ONE. 2011;6:e21296. doi: 10.1371/journal.pone.0021296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kole R, Krainer AR, Altman S. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat. Rev. Drug Discov. 2012;11:125–140. doi: 10.1038/nrd3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Zhang Y, Ye Z-Q, Liu X-Q, Zhao S-Q, Wei L, Gao G. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35:W345–9. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR. A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell. 2003;12:525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Baughn M, Rigo F, Sun S, Liu P, Li H-R, Jiang J, Watt AT, Chun S, Katz M, et al. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E4530–9. doi: 10.1073/pnas.1318835110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TT, Pham LT, Butchbach MER, Zhang HL, Monani UR, Coovert DD, Gavrilina TO, Xing L, Bassell GJ, Burghes AHM. SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum. Mol. Genet. 2005;14:845–857. doi: 10.1093/hmg/ddi078. [DOI] [PubMed] [Google Scholar]
- Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K-I, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre S, Bürglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- Li L, Liu B, Wapinski OL, Tsai M-C, Qu K, Zhang J, Carlson JC, Lin M, Fang F, Gupta RA, et al. Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell Rep. 2013;5:3–12. doi: 10.1016/j.celrep.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl. Acad. Sci. U.S.A. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz CM, Kariya S, Patruni S, Osborne MA, Liu D, Henderson CE, Li DK, Pellizzoni L, Rojas J, Valenzuela DM, et al. Postsymptomatic restoration of SMN rescues the disease phenotype in a mouse model of severe spinal muscular atrophy. J. Clin. Invest. 2011;121:3029–3041. doi: 10.1172/JCI57291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager J, Montgomery ND, de Villena FP-M, Magnuson T. Genome imprinting regulated by the mouse Polycomb group protein Eed. Nat. Genet. 2003;33:502–507. doi: 10.1038/ng1125. [DOI] [PubMed] [Google Scholar]
- Mahmoudi S, Henriksson S, Corcoran M, Méndez-Vidal C, Wiman KG, Farnebo M. Wrap53, a natural p53 antisense transcript required for p53 induction upon DNA damage. Mol. Cell. 2009;33:462–471. doi: 10.1016/j.molcel.2009.01.028. [DOI] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez TL, Kong L, Wang X, Osborne MA, Crowder ME, Van Meerbeke JP, Xu X, Davis C, Wooley J, Goldhamer DJ, et al. Survival motor neuron protein in motor neurons determines synaptic integrity in spinal muscular atrophy. J. Neurosci. 2012;32:8703–8715. doi: 10.1523/JNEUROSCI.0204-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L, Ward AJ, Chun S, Bennett CF, Beaudet AL, Rigo F. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature. 2015;518:409–412. doi: 10.1038/nature13975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller TM, Pestronk A, David W, Rothstein J, Simpson E, Appel SH, Andres PL, Mahoney K, Allred P, Alexander K, et al. An antisense oligonucleotide against SOD1 delivered intrathecally for patients with SOD1 familial amyotrophic lateral sclerosis: a phase 1, randomised, first-in-man study. Lancet Neurol. 2013;12:435–442. doi: 10.1016/S1474-4422(13)70061-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani R, Wakamatsu A, Tanaka N, Yoshida H, Tochigi N, Suzuki Y, Oonishi T, Tani H, Tano K, Ijiri K, et al. Identification and characterization of novel genotoxic stress-inducible nuclear long noncoding RNAs in mammalian cells. PLoS ONE. 2012;7:e34949. doi: 10.1371/journal.pone.0034949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modarresi F, Faghihi MA, Lopez-Toledano MA, Fatemi RP, Magistri M, Brothers SP, van der Brug MP, Wahlestedt C. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat. Biotechnol. 2012;30:453–459. doi: 10.1038/nbt.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monani UR, Lorson CL, Parsons DW, Prior TW, Androphy EJ, Burghes AH, McPherson JD. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum. Mol. Genet. 1999a;8:1177–1183. doi: 10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- Monani UR, McPherson JD, Burghes AH. Promoter analysis of the human centromeric and telomeric survival motor neuron genes (SMNC and SMNT) Biochim. Biophys. Acta. 1999b;1445:330–336. doi: 10.1016/s0167-4781(99)00060-3. [DOI] [PubMed] [Google Scholar]
- Mondal T, Subhash S, Vaid R, Enroth S, Uday S, Reinius B, Mitra S, Mohammed A, James AR, Hoberg E, et al. MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA-DNA triplex structures. Nat. Commun. 2015;6:7743. doi: 10.1038/ncomms8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narver HL, Kong L, Burnett BG, Choe DW, Bosch-Marcé M, Taye AA, Eckhaus MA, Sumner CJ. Sustained improvement of spinal muscular atrophy mice treated with trichostatin A plus nutrition. Ann. Neurol. 2008;64:465–470. doi: 10.1002/ana.21449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naryshkin NA, Weetall M, Dakka A, Narasimhan J, Zhao X, Feng Z, Ling KKY, Karp GM, Qi H, Woll MG, et al. Motor neuron disease. SMN2 splicing modifiers improve motor function and longevity in mice with spinal muscular atrophy. Science. 2014;345:688–693. doi: 10.1126/science.1250127. [DOI] [PubMed] [Google Scholar]
- Ng S-Y, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012;31:522–533. doi: 10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S-Y, Lin L, Soh BS, Stanton LW. Long noncoding RNAs in development and disease of the central nervous system. Trends Genet. 2013;29:461–468. doi: 10.1016/j.tig.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Ng S-Y, Soh BS, Rodriguez-Muela N, Hendrickson DG, Price F, Rinn JL, Rubin LL. Genome-wide RNA-Seq of Human Motor Neurons Implicates Selective ER Stress Activation in Spinal Muscular Atrophy. Cell Stem Cell. 2015;17:569–584. doi: 10.1016/j.stem.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oskoui M, Levy G, Garland CJ, Gray JM, O'Hagen J, De Vivo DC, Kaufmann P. The changing natural history of spinal muscular atrophy type 1. Neurology. 2007;69:1931–1936. doi: 10.1212/01.wnl.0000290830.40544.b9. [DOI] [PubMed] [Google Scholar]
- Paez-Colasante X, Seaberg B, Martinez TL, Kong L, Sumner CJ, Rimer M. Improvement of neuromuscular synaptic phenotypes without enhanced survival and motor function in severe spinal muscular atrophy mice selectively rescued in motor neurons. PLoS ONE. 2013;8:e75866. doi: 10.1371/journal.pone.0075866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacino J, Swalley SE, Song C, Cheung AK, Shu L, Zhang X, Van Hoosear M, Shin Y, Chin DN, Keller CG, et al. SMN2 splice modulators enhance U1-pre-mRNA association and rescue SMA mice. Nat. Chem. Biol. 2015;11:511–517. doi: 10.1038/nchembio.1837. [DOI] [PubMed] [Google Scholar]
- Palomer E, Carretero J, Benvegnù S, Dotti CG, Martin MG. Neuronal activity controls Bdnf expression via Polycomb de-repression and CREB/CBP/JMJD3 activation in mature neurons. Nat. Commun. 2016;7:11081. doi: 10.1038/ncomms11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passini MA, Bu J, Richards AM, Kinnecom C, Sardi SP, Stanek LM, Hua Y, Rigo F, Matson J, Hung G, et al. Antisense oligonucleotides delivered to the mouse CNS ameliorate symptoms of severe spinal muscular atrophy. Sci. Transl. Med. 2011;3:72ra18–72ra18. doi: 10.1126/scitranslmed.3001777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinter SF, Sadreyev RI, Yildirim E, Jeon Y, Ohsumi TK, Borowsky M, Lee JT. Spreading of X chromosome inactivation via a hierarchy of defined Polycomb stations. Genome Res. 2012;22:1864–1876. doi: 10.1101/gr.133751.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piunti A, Rossi A, Cerutti A, Albert M, Jammula S, Scelfo A, Cedrone L, Fragola G, Olsson L, Koseki H, et al. Polycomb proteins control proliferation and transformation independently of cell cycle checkpoints by regulating DNA replication. Nat. Commun. 2014;5:3649. doi: 10.1038/ncomms4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, la Cruz, de CC, Otte AP, Panning B, Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- Qureshi IA, Mattick JS, Mehler MF. Long non-coding RNAs in nervous system function and disease. Brain Res. 2010;1338:20–35. doi: 10.1016/j.brainres.2010.03.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos AD, Andersen RE, Liu SJ, Nowakowski TJ, Hong SJ, Gertz CC, Salinas RD, Zarabi H, Kriegstein AR, Lim DA. The long noncoding RNA Pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Cell Stem Cell. 2015;16:439–447. doi: 10.1016/j.stem.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao RA, Dhele N, Cheemadan S, Ketkar A, Jayandharan GR, Palakodeti D, Rampalli S. Ezh2 mediated H3K27me3 activity facilitates somatic transition during human pluripotent reprogramming. Sci. Rep. 2015;5:8229. doi: 10.1038/srep08229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahashi K, Ling KKY, Hua Y, Wilkinson JE, Nomakuchi T, Rigo F, Hung G, Xu D, Jiang Y-P, Lin RZ, et al. Pathological impact of SMN2 mis-splicing in adult SMA mice. EMBO Mol. Med. 2013;5:1586–1601. doi: 10.1002/emmm.201302567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch KM, De Vos SL, Miller RL, Chun SJ, Norrbom M, Wozniak DF, Dawson HN, Bennett CF, Rigo F, Miller TM. Increased 4R-Tau Induces Pathological Changes in a Human-Tau Mouse Model. Neuron. 2016;90:941–947. doi: 10.1016/j.neuron.2016.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearwin KE, Callen BP, Egan JB. Transcriptional interference--a crash course. Trends Genet. 2005;21:339–345. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MD, Pinter SF, Fang R, Sarma K, Rutenberg-Schoenberg M, Bowman SK, Kesner BA, Maier VK, Kingston RE, Lee JT. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature. 2013;504:465–469. doi: 10.1038/nature12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smilinich NJ, Day CD, Fitzpatrick GV, Caldwell GM, Lossie AC, Cooper PR, Smallwood AC, Joyce JA, Schofield PN, Reik W, et al. A maternally methylated CpG island in KvLQT1 is associated with an antisense paternal transcript and loss of imprinting in Beckwith-Wiedemann syndrome. Proc. Natl. Acad. Sci. U.S.A. 1999;96:8064–8069. doi: 10.1073/pnas.96.14.8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers E, Riessland M, Schreml J, Wirth B, Gillingwater TH, Parson SH. Increasing SMN levels using the histone deacetylase inhibitor SAHA ameliorates defects in skeletal muscle microvasculature in a mouse model of severe spinal muscular atrophy. Neurosci. Lett. 2013;544:100–104. doi: 10.1016/j.neulet.2013.03.052. [DOI] [PubMed] [Google Scholar]
- Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, Wagner L, Shenmen CM, Schuler GD, Altschul SF, et al. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl. Acad. Sci. U.S.A. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swayze EE, Siwkowski AM, Wancewicz EV, Migawa MT, Wyrzykiewicz TK, Hung G, Monia BP, Bennett CF. Antisense oligonucleotides containing locked nucleic acid improve potency but cause significant hepatotoxicity in animals. Nucleic Acids Res. 2007;35:687–700. doi: 10.1093/nar/gkl1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swoboda KJ, Scott CB, Crawford TO, Simard LR, Reyna SP, Krosschell KJ, Acsadi G, Elsheik B, Schroth MK, D'Anjou G, et al. SMA CARNI-VAL trial part I: double-blind, randomized, placebo-controlled trial of L-carnitine and valproic acid in spinal muscular atrophy. PLoS ONE. 2010;5:e12140. doi: 10.1371/journal.pone.0012140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance KW, Ponting CP. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 2014;30:348–355. doi: 10.1016/j.tig.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CH, Finkel RS, Bertini ES, Schroth M, Simonds A, Wong B, Aloysius A, Morrison L, Main M, Crawford TO, et al. Consensus statement for standard of care in spinal muscular atrophy. Presented at the Journal of child neurology; SAGE Publications; 2007. pp. 1027–1049. [DOI] [PubMed] [Google Scholar]
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner MS, Ruthenburg AJ. Nuclear Fractionation Reveals Thousands of Chromatin-Tethered Noncoding RNAs Adjacent to Active Genes. Cell Rep. 2015;12:1089–1098. doi: 10.1016/j.celrep.2015.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Duff MO, Graveley BR, Carmichael GG, Chen L-L. Genomewide characterization of non-polyadenylated RNAs. Genome Biol. 2011;12:R16. doi: 10.1186/gb-2011-12-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C, Wang J, Zhang H, Zhou S, Qian T, Ding F, Gu X, Yu B. Long non-coding RNA uc.217 regulates neurite outgrowth in dorsal root ganglion neurons following peripheral nerve injury. Eur. J. Neurosci. 2015;42:1718–1725. doi: 10.1111/ejn.12966. [DOI] [PubMed] [Google Scholar]
- Yu B, Zhou S, Hu W, Qian T, Gao R, Ding G, Ding F, Gu X. Altered long noncoding RNA expressions in dorsal root ganglion after rat sciatic nerve injury. Neurosci. Lett. 2013;534:117–122. doi: 10.1016/j.neulet.2012.12.014. [DOI] [PubMed] [Google Scholar]
- Yu J, Cao Q, Wu L, Dallol A, Li J, Chen G, Grasso C, Cao X, Lonigro RJ, Varambally S, et al. The neuronal repellent SLIT2 is a target for repression by EZH2 in prostate cancer. Oncogene. 2010;29:5370–5380. doi: 10.1038/onc.2010.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zupi G, Scarsella M, Semple SC, Mottolese M, Natali PG, Leonetti C. Antitumor efficacy of bcl-2 and c-myc antisense oligonucleotides in combination with cisplatin in human melanoma xenografts: relevance of the administration sequence. Clin. Cancer Res. 2005;11:1990–1998. doi: 10.1158/1078-0432.CCR-04-1284. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.