Significance
In animal cells, cholesterol is essential for the assembly of membranes and production of steroid hormones and bile acids. Cells obtain cholesterol through receptor-mediated endocytosis of LDL into lysosomes. After lysosomal degradation, LDL-derived cholesterol must cross the lysosomal membrane to execute its functions. Lysosomal export is mediated by Niemann–Pick C1 (NPC1), a membrane protein. Here we show that the triazole drugs posaconazole and itraconazole inhibit lysosomal cholesterol export. A photoactivatable derivative of posaconazole cross-links to NPC1 in intact cells and to purified NPC1 in lipid nanodiscs. A point mutation in the membrane domain of NPC1 prevents lysosomal export of cholesterol and blocks posaconazole cross-linking.
Keywords: Niemann–Pick C disease, cholesterol transport, sterol-sensing domain, lipid nanodiscs, photoactivatable cross-linking
Abstract
Niemann–Pick C1 (NPC1), a membrane protein of lysosomes, is required for the export of cholesterol derived from receptor-mediated endocytosis of LDL. Lysosomal cholesterol export is reportedly inhibited by itraconazole, a triazole that is used as an antifungal drug [Xu et al. (2010) Proc Natl Acad Sci USA 107:4764–4769]. Here we show that posaconazole, another triazole, also blocks cholesterol export from lysosomes. We prepared P-X, a photoactivatable cross-linking derivative of posaconazole. P-X cross-linked to NPC1 when added to intact cells. Cross-linking was inhibited by itraconazole but not by ketoconazole, an imidazole that does not block cholesterol export. Cross-linking of P-X was also blocked by U18666A, a compound that has been shown to bind to NPC1 and inhibit cholesterol export. P-X also cross-linked to purified NPC1 that was incorporated into lipid bilayer nanodiscs. In this in vitro system, cross-linking of P-X was inhibited by itraconazole, but not by U18666A. P-X cross-linking was not prevented by deletion of the N-terminal domain of NPC1, which contains the initial binding site for cholesterol. In contrast, P-X cross-linking was reduced when NPC1 contained a point mutation (P691S) in its putative sterol-sensing domain. We hypothesize that the sterol-sensing domain has a binding site that can accommodate structurally different ligands.
The mechanisms by which lipids, such as cholesterol, are transported across membrane bilayers are poorly understood. Recent advances have begun to elucidate one such mechanism, the transport of cholesterol across the lysosome membrane. Mammalian cells supply themselves with cholesterol through the receptor-mediated endocytosis of cholesterol-carrying LDL (1). After delivery to lysosomes, the cholesterol is released from LDL and immediately bound by Niemann–Pick C2 (NPC2), a soluble cholesterol carrier. NPC2 delivers cholesterol to Niemann–Pick C1 (NPC1), a complex polytopic membrane protein of 1,278 amino acids embedded in the lysosome membrane (2). NPC1 transfers its cholesterol to the lysosomal membrane for delivery to other membranes, notably the plasma membrane and the endoplasmic reticulum (3, 4). Genetic defects in NPC2 or NPC1 cause a fatal disease in which multiple organs fail because of lysosomal cholesterol accumulation (2, 5).
The mechanism for NPC1 function is beginning to be unraveled through biochemical and structural studies (6–12). NPC1 has 13 transmembrane helices and three large domains that project into the lumen (13). Cholesterol-carrying NPC2 binds to the middle luminal domain (MLD), which positions the protein so that it can transfer its cholesterol to the N-terminal domain (NTD) (11). This transfer is facilitated by the formation of a channel between NPC2 and the NTD that allows cholesterol to slide from NPC2 to the NTD of NPC1 in a process known as a “hydrophobic handoff” (7).
A major unsolved problem is how cholesterol moves from the NTD of NPC1 to the membrane. As seen on cryoelectron microscopy and X-ray crystallography, the MLD of NPC1 is an extended structure that is predicted to penetrate through the thick glycocalyx that lines the lysosome membrane (10, 12). NPC2 binds to the far end of this rod-like structure, which is ∼60 Å from the membrane (11). The NTD accepts cholesterol at this point, and somehow must traverse the glycocalyx to deliver its cholesterol to the membrane domain of NPC1. This membrane domain contains five transmembrane helices, designated the sterol-sensing domain (SSD) (14). Within its transmembrane segments, the SSD contains a cleft large enough to accommodate cholesterol, which is postulated to be the site where cholesterol is inserted into the membrane (10).
Insight into the potential cholesterol-binding site in the NPC1 membrane domain has emerged from studies of U18666A, a derivative of androstenolone that blocks the export of cholesterol from lysosomes (15). U-X, a photoactivatable derivative of U18666A, was found to cross-link to NPC1 when added to cultured cells (16). U-X also was found to cross-link to mutant NPC1 containing two substitutions (P202A/F203A) that prevent cholesterol binding to the NTD (7), suggesting that U-X was not binding to the NTD. Instead, U-X cross-linking was abolished by the P691S mutation in the SSD that prevents cholesterol transport. These studies suggest that U18666A inhibits cholesterol transport by binding to the postulated cholesterol-binding site in the SSD (16).
In the present study, we used a photo-cross-linking approach to define the action of another class of inhibitors of cholesterol export—namely, a series of triazole compounds. Produced by pharmaceutical companies as antifungal drugs (17, 18), these compounds inhibit a variety of enzymes in sterol biosynthesis, showing higher potency against yeast enzymes vs. mammalian enzymes. It was recently demonstrated that one triazole, itraconazole, inhibits the export of LDL-derived cholesterol from lysosomes of endothelial cells in culture (19). Here we prepared a photoactivatable derivative of posaconazole, another triazole, and found that it cross-linked to NPC1 when added to cultured cells. Posaconazole also cross-linked to purified NPC1 that was incorporated into nanodisc lipid bilayers. Cross-linking of posaconazole was inhibited by itraconazole but not by ketoconazole, an imidazole that does not block cholesterol export from lysosomes. These findings indicate that the putative cholesterol-binding site in the SSD of NPC1 may have a relaxed specificity that permits the binding of many seemingly unrelated chemicals, including androstenone derivatives and triazoles.
Results
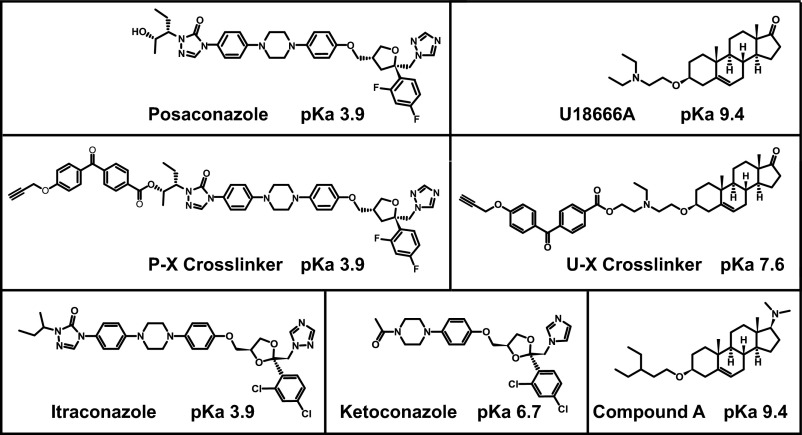
Fig. 1 shows the structures and acid dissociation constants (expressed as pKa values) of the compounds used in this study. Posaconazole and itraconazole are triazoles, and ketoconazole is an imidazole. U18666A and compound A are sterol derivatives. P-X and U-X are derivatives of posaconazole and U18666A, respectively, that were modified by attachment of a benzophenone moiety and an alkyne. The benzophenone allows UV light to induce cross-linking to the backbone atoms of proteins, and the alkyne permits the attachment of azide-containing fluorescent compounds through the use of click chemistry.
Fig. 1.
Chemical structures of compounds used in this study. pKa values denote the negative logarithm of the acid dissociation constant.
The ability of various compounds to block the NPC1-mediated export of cholesterol from lysosomes was tested using a cholesterol re-esterification assay (20). In this assay, cultured CHO-7 cells are deprived of cholesterol to induce the synthesis of LDL receptors. The cells are then incubated with FCS, which contains LDL. If NPC1 is functional, then LDL-derived cholesterol is exported from the lysosome and re-esterified in the endoplasmic reticulum through the action of acyl-CoA:cholesterol acyltransferase (ACAT). To monitor the re-esterification, we pulse-labeled the cells with [14C]oleate and measured the synthesis of cholesteryl [14C]oleate. To ensure that any inhibition was not caused by diminished uptake and lysosomal degradation of LDL, we routinely monitored the uptake and degradation of 125I-labeled LDL (20). No such inhibition was seen in experiments with posaconazole, itrconazole, or U18666A.
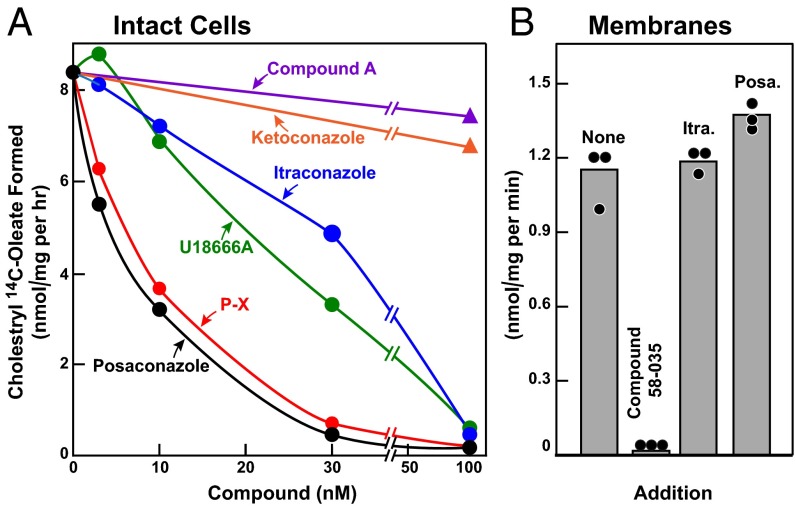
Fig. 2A shows that posaconazole and P-X are potent inhibitors of cholesterol export from lysosomes, as reflected in the cholesterol re-esterification assay in intact CHO-7 cells, with 50% inhibition occurring at around 7 nM. U18666A and itraconazole were somewhat less potent (50% inhibition at 24 and 34 nM, respectively), whereas ketoconazole and compound A were inactive at concentrations up to 100 nM.
Fig. 2.
Triazole-mediated inhibition of cholesterol esterification in CHO-7 cells. On day 0, CHO-7 cells were set up in medium A with 5% LPDS at a density of 2.5 × 105 cells/60-mm dish. (A) [14C]Oleate incorporation into cholesteryl [14C]oleate in intact monolayers. On day 2, the medium was switched to fresh medium containing 10 µM sodium compactin and 50 µM sodium mevalonate. On day 3, the medium was switched to medium B containing 5% FCS, 10 µM compactin, 50 µM mevalonate, and various concentrations of the indicated compound. After a 3-h incubation, each cell monolayer was incubated for 2 h with 0.2 mM sodium [14C]oleate (18,412 dpm/nmol). The cells were then harvested for measurement of their cholesteryl [14C]oleate and [14C]triglyceride contents, as described in SI Materials and Methods. Each value is the average of duplicate incubations. The inhibitory constant (Ki) values (denoting the concentration at which each compound inhibited cholesterol esterification by 50%) were 6.6 nM for posaconazole, 7 nM for P-X, 24 nM for U18666A, 34 nM for itraconazole, and >100 nM for both ketoconazole and compound A. Values for incorporation of [14C]oleate into [14C]triglycerides in cells treated with 100 nM of all six compounds ranged from 70 to 81 nmol/h per 1 mg of protein. (B) ACAT activity in membranes. On day 3, cells were harvested, and a 105g membrane fraction was assayed for ACAT activity as described in SI Materials and Methods. Itraconazole (Itra.) and posaconaczole (Posa.) were added at final concentrations of 0.3 µM. Sandoz 58–035 (ACAT inhibitor) was added at a final concentration of 10 µM. Each bar represents the mean of triplicate incubations.
To exclude the possibility that posaconazole and itraconazole were inhibiting ACAT activity directly, we measured the effects of these compounds in an in vitro enzyme assay using a 105g membrane fraction from CHO-7 cells (Fig. 2B). Neither triazole inhibited ACAT activity, in contrast to the marked inhibitory effect of a known ACAT inhibitor, Sandoz 58–035 (21). These data suggest that the inhibitory effect of triazoles on cholesterol re-esterification is mediated by inhibition of cholesterol export and not by direct inhibition of ACAT.
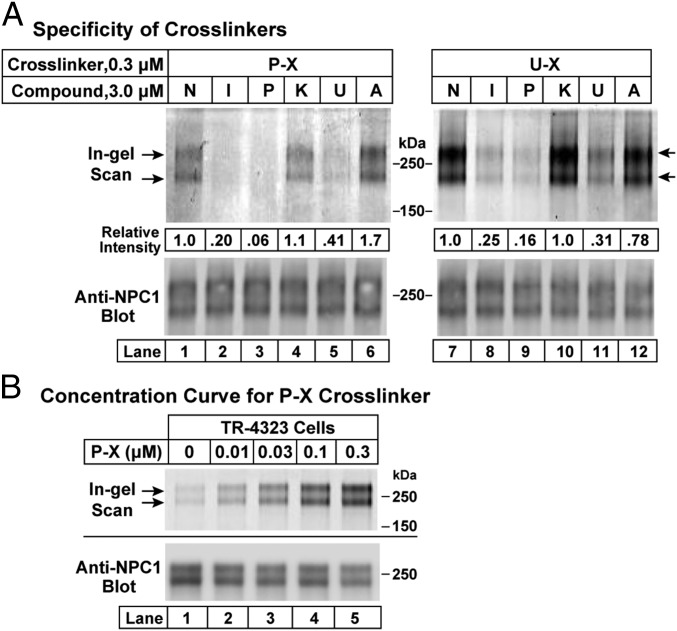
To measure the cross-linking of P-X and U-X to NPC1, we added the compounds to CHO-7 cells and then exposed the cells to UV light. The cells were disrupted and NPC1 was immunoprecipitated. The immunoprecipitated protein was incubated with Alexa Fluor 532 azide under conditions that catalyzed its attachment to the alkyne on P-X and U-X. After SDS/PAGE, the fluorescent proteins were visualized in situ with a fluorescence detector (Fig. 3A, Upper). The proteins were then transferred to nitrocellulose and incubated with an antibody to NPC1 (Fig. 3A, Lower). The results indicate that P-X at a concentration of 0.3 µM was cross-linked to bands corresponding to NPC1 (Fig. 3A, Left). Fluorescent labeling was reduced when the cell incubations were conducted in the presence of a 10-fold excess (i.e., 3 µM) of itraconazole, posaconazole, or U18666A, but not with ketoconazole or compound A. A similar pattern of competition was seen when U-X was the cross-linker (Fig. 3A, Right). These results indicate that P-X and U-X bind to NPC1, and that the binding site also recognizes itraconazole and U18666A, but not ketoconazole or compound A. NPC1 appears as a doublet in the SDS/PAGE gels of Fig. 3A. This doublet is due to differential glycosylation, with the upper band reduced to the lower band when the protein is treated with PNGase F (8). In other experiments when we ran the gels for a shorter time (75 min, compared with 120 min in Fig. 3), the two bands were much closer together (Figs. 3B and 4–6).
Fig. 3.
(A) Specificity of cross-linking of P-X (Left) and U-X (Right) to NPC1 in intact cells. On day 0, CHO-7 cells were set up at 1.6 × 106 cells per 150-mm dish in 25 mL of medium A with 5% LPDS as described in Materials and Methods. On day 3, each monolayer received 25 µL of DMSO containing either 0.3 µM P-X or U-X in the presence of 3 µM of one of the following potential competitors: none (N), itraconazole (I), posaconazole (P), ketoconazole (K), U18666A (U), or compound A (A). After incubation for 1 h at 37 °C, cells were exposed to UV light and harvested, after which cell lysates were incubated with 1 µg/mL anti–NPC-1 antibody. The solutions were applied to protein A/G agarose beads, washed, and eluted as described in Materials and Methods. (B) Cross-linking at different concentrations of P-X. TR-4323 cells overexpressing NPC1 were set up as above. On day 3, each monolayer received 25 µL of DMSO containing the indicated amount of P-X. After incubation for 1 h at 37 °C, cells were exposed to UV light and harvested, after which cell lysates were incubated with 25 µL of anti-FLAG M2 magnetic beads, washed, and eluted as described in Materials and Methods. In A and B, eluted proteins were subjected to fluorescent labeling using click chemistry, followed by 8% SDS/PAGE and in-gel fluorescence scanning (Upper) or immunoblotting with 0.5 µg/mL anti-NPC1 (Lower). Quantification of bands in the upper lanes in A was carried out as described in Materials and Methods. “Relative intensity” values of 1.0 in A refer to samples (lanes 1 and 7) that did not receive competitor compound.
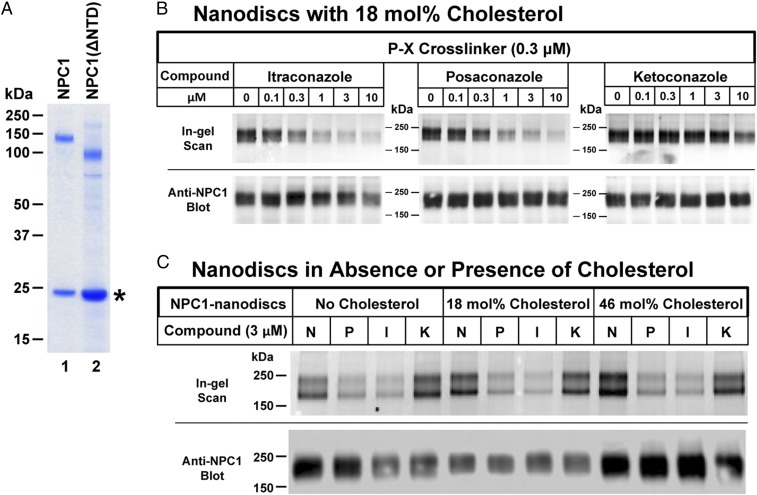
Fig. 4.
Specificity of cross-linking of P-X to purified NPC1 reconstituted in nanodiscs with and without cholesterol. (A) SDS/PAGE of purified and reconstituted WT NPC1 and NPC1(ΔNTD) used for cross-linking. Proteins were purified and reconstituted as described in Materials and Methods and then subjected to a 4–12% Bis-Tris gradient gel. The amount of protein was normalized so that similar amounts of full-length NPC1 and NPC1(ΔNTD) (∼0.6 µg) were loaded onto the gel. An asterisk denotes the position of migration of MSP1D1. (B and C) Each reaction contained, in a final volume of 100 µL of buffer A, 0.3 µM P-X and 0.1 µg of NPC1 nanodiscs prepared with the indicated concentration of cholesterol and the indicated concentration of one of the following: none (N), itraconazole (I), posaconazole (P), or ketoconazole (K). Nanodiscs with varying amounts of cholesterol were prepared as described in Materials and Methods. After incubation for 1 h at 37 °C, each mixture was subjected to UV cross-linking and fluorescent labeling. Proteins were subjected to 8% SDS/PAGE, followed by in-gel fluorescence scanning (Upper) or immunoblotting with 0.5 µg/mL anti-NPC1 (Lower).
Fig. 6.
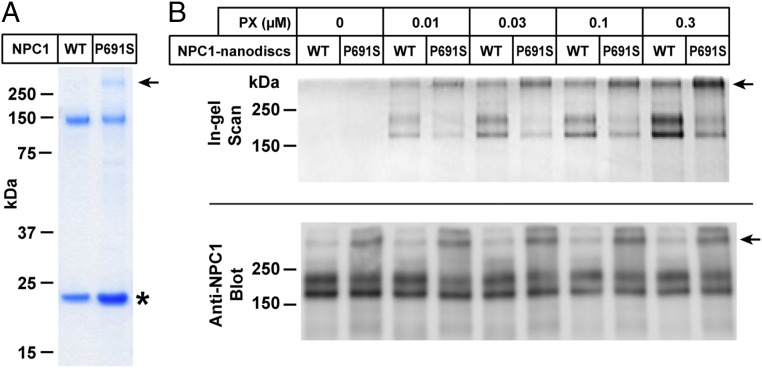
Cross-linking of P-X to purified NPC1(P691S) in nanodiscs. (A) SDS/PAGE of purified and reconstituted WT NPC1 and mutant NPC1(P691S) used for cross-linking. Proteins were purified and reconstituted as described in Materials and Methods and then subjected to a 4–12% Bis-Tris gradient gel. The amount of protein was normalized so that similar amounts of WT and mutant NPC1 proteins (∼0.3 µg) were loaded onto the gel. The asterisk denotes the position of migration of MSP1D1. The arrow denotes aggregated NPC1(P691S) nanodiscs that did not enter gel. (B) Cross-linking of purified proteins. Each reaction contained, in a final volume of 100 µL of buffer A, 0.1 µg of either NPC1 nanodiscs or NPC1(P691S) nanodiscs, and the indicated amount of P-X. After incubation for 1 h at 37 °C, the mixtures were subjected to UV cross-linking and fluorescent labeling. Proteins were subjected to 8% SDS/PAGE, followed by in-gel fluorescence scanning (Upper) or immunoblotting with 0.5 µg/mL anti-NPC1 (Lower).
The experiment shown in Fig. 3A was carried out in parental CHO-7 cells at a P-X concentration of 0.3 µM. To determine the lowest concentration at which P-X cross-linking to NPC1 could be detected, we first produced a stably transfected cell line of CHO-7 cells that overexpress human NPC1. When these TR-4323 cells were incubated with various amounts of P-X, cross-linking to NPC1 was detected at concentrations as low as 10 nM (Fig. 3B).
To study the binding of the triazoles to purified NPC1, we prepared a cDNA encoding full-length NPC1 with a streptactin tag at the C terminus. The cDNA was inserted into a BacMam vector that was then transduced into human HEK293S GnTI− cells as described previously (10). Cell membranes were solubilized with n-dodecyl-β-d-maltopyranoside (DDM), and the tagged NPC1 was purified on a column containing Strep-Tactin resin. The purified protein was incorporated into lipid nanodiscs that contained a 9:2 molar ratio of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine and cholesterol together with MSP1D1, a membrane scaffold protein, as described in Materials and Methods. Fig. 4A, lane 1, shows that the nanodiscs contained pure NPC1 (Upper) and MSP1D1 (Lower), as determined by SDS/PAGE and Coomassie blue staining.
Nanodiscs containing purified NPC1 were incubated with 0.3 µM P-X in the absence or presence of potential competitors. After exposure to UV light and attachment of Alexa Fluor 532, the protein was subjected to SDS/PAGE (Fig. 4B). In the absence of competitors, P-X was cross-linked to NPC1. Increasing amounts of itraconazole or posaconazole competed for cross-linking, but ketoconazole had much less effect. Quantification of this cross-linking as determined by fluorescence scanning of the bands (22) showed that itraconazole and posaconazole competed with half-maximal concentrations of ∼1 µM, whereas ketoconazole was more than 10-fold less potent. As shown in Fig. 4B, Bottom, all lanes had similar amounts of NPC1 as determined by immunoblotting.
Our standard procedure for preparing nanodiscs includes 18 mol% cholesterol. To determine whether this cholesterol influences P-X cross-linking, we prepared nanodiscs with no cholesterol and with 46 mol% cholesterol. As shown in Fig. 4C, varying the cholesterol content had no effect on P-X cross-linking.
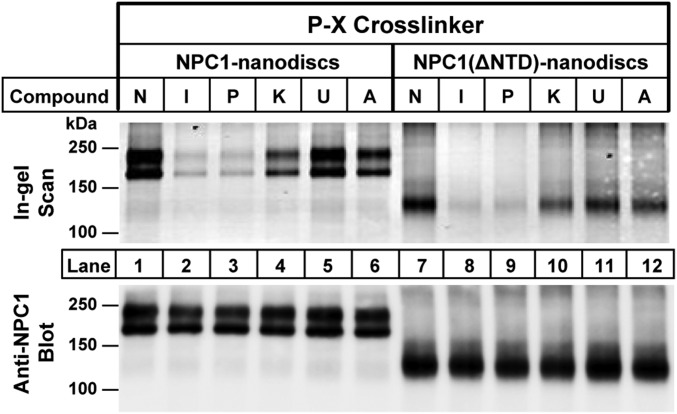
In previous studies with U-X (16), we showed that cross-linking to NPC1 was not prevented by a point mutation in the NTD that abolishes cholesterol binding (7), indicating that U-X cross-linking is not dependent on the cholesterol-binding site in the NTD. To further test this hypothesis, we prepared a plasmid encoding NPC1 with a deletion of the NTD. The mutant protein, designated NPC1(ΔNTD), was purified (Fig. 4A, lane 2) and incorporated into nanodiscs in the same fashion as full-length NPC1. Fig. 5 shows that P-X was cross-linked to NPC1(ΔNTD) in the same fashion as full-length NPC1. Moreover, cross-linking was competed by itraconazole and posaconazole, but not by ketoconazole or compound A. Fig. 5 also shows that U18666A did not compete with P-X for cross-linking to NPC1 in nanodiscs. A possible explanation for this unexpected finding (observed in multiple experiments) is provided in Discussion.
Fig. 5.
Cross-linking of P-X to purified NPC1(ΔNTD) reconstituted in nanodiscs. Each reaction contained, in a final volume of 100 µL of buffer A, 0.1 µg of either NPC1 nanodiscs or NPC1(ΔNTD) nanodiscs, 0.3 μM P-X, and 3 μM of one of the following: none (N), itraconazole (I), posaconazole (P), ketoconazole (K), U18666A (U), or compound A (A). After incubation for 1 h at 37 °C, the mixtures were subjected to UV cross-linking and fluorescent labeling. Proteins were subjected to 8% SDS/PAGE, followed by in-gel fluorescence scanning (Upper) or immunoblotting with 0.5 µg/mL anti-NPC1 (Lower).
In previous studies of intact cells, we observed reduced cross-linking of U-X to the P691S mutant of NPC1 (16). This mutation occurs in transmembrane helix 5, which is part of the SSD, and it may possibly alter the rigidity of the putative sterol-binding cleft (10). The mutant protein moves to lysosomes, but it does not transport cholesterol (23, 24). Moreover, it showed reduced susceptibility to cross-linking when a radiolabeled photoactivated derivative of cholesterol was added to cultured cells (25). Fig. 6A shows a Coomassie blue-stained SDS/PAGE gel of purified wild type (WT) and mutant NPC1(P691S) proteins incorporated into nanodiscs. Fig. 6B shows reduced cross-linking to the P691S mutant compared with WT in nanodiscs incubated with varying concentrations of P-X. Whereas P-X was cross-linked to WT NPC1 at 0.01 µM, it showed much less cross-linking to the P691S mutant at every concentration tested up to 0.3 µM. We observed that a small fraction of the P691S mutant protein migrated near the top of the SDS/PAGE gel, indicating a tendency toward aggregation. To ensure a valid comparison, we equalized the amount of nonaggragated WT and mutant proteins in the nanodiscs, as determined by blotting with anti-NPC1 (Fig. 6B, Bottom).
Discussion
In this work we have defined properties of the SSD of NPC1, and described approaches to studying this region in the future. We showed that posaconazole and itraconazole, two antifungal triazoles, inhibited cholesterol export from lysosomes of mammalian cells in culture. P-X, a photoactivatable derivative of posaconazole, was cross-linked to NPC1 when exposed to UV light after incubation with cultured cells. In addition, we prepared pure NPC1, incorporated it into lipid bilayer nanodiscs, and showed that P-X could be cross-linked to the pure protein. The specificity of the cross-linking reaction was confirmed by data showing that cross-linking was inhibited competitively by excess posaconazole and itraconazole, but not by ketoconazole, a related compound that contains an imidazole rather than a triazole moiety.
Two lines of evidence suggest that P-X cross-links to the membrane domain of NPC1. First, in the nanodisc assay with purified proteins, P-X cross-linking was not affected by deletion of the NTD, which is the initial cholesterol-binding site in NPC1 (26). Second, P-X binding was markedly reduced when the purified NPC1 contained the P691S mutation in the membranous SSD. This mutation has been shown to prevent the cross-linking of U-X, a photoactivatable derivative of U18666A that also blocks cholesterol export from lysosomes (16). Considered together, these data indicate that U18666A, posaconazole, and itraconazole may all bind to a common site that may lie in the SSD. This site is required for the function of NPC1 in transporting LDL-derived cholesterol from lysosomes.
An unexpected finding in the present study was the failure of U18666A to block the cross-linking of P-X to purified NPC1 in nanodiscs (Fig. 6B). This failure occurred even though U18666A did block cross-linking of P-X to NPC1 when added to intact cells (Fig. 3). A possible explanation lies in the pKa values shown in Fig. 1. U18666A is a basic compound that may concentrate in the acidic environment of lysosomes. This concentration may enhance the apparent potency of U18666A when added to intact cells. Posaconazole and itraconazole are acidic compounds that should not concentrate in lysosomes (27). U18666A appears to be equally potent to itraconazole in intact cells where U18666A reaches high intralysosomal concentrations, but it would be much less potent when added to nanodiscs in vitro.
Our current findings indicate that the triazoles inhibit lysosomal cholesterol export by binding to the membrane domain of NPC1, but they do not reveal whether these compounds compete for binding to the same site as cholesterol or whether they bind to another site that inhibits cholesterol binding allosterically. Arguing against a single binding site is the observation that P-X cross-linking is not inhibited even when the nanodiscs contain 46 mol% of cholesterol (Fig. 4C). This argument is not conclusive, however. It is possible that posaconazole and cholesterol bind to a common site but the nanodisc cholesterol is not in an orientation that allows access to this site. It is conceivable that cholesterol binding requires delivery by the NTD, which could open a channel in the SSD (11) in much the same way as NPC2 does when it delivers cholesterol to the NTD (7). Somehow, U18666A and the triazoles may gain access to the SSD without requiring opening by the NTD.
The alternative possibility is that the membrane domain of NPC1 contains an allosteric regulatory site to which U18666A and the triazoles bind. If such an allosteric site exists, then its ability to bind must depend on the configuration of proline 691. The P691S mutation in the SSD disrupts cholesterol export (16, 23, 24), triazole binding (Fig. 6), and cross-linking of photocholesterol (25), an observation that suggests a single binding site. A solution to this conundrum may emerge from more detailed structural studies of the binding site(s) for cholesterol, triazoles, and U18666A in the membrane domain of NPC1.
Materials and Methods
Chemical Compounds for Cross-Linking.
Itraconazole, ketoconazole, and U18666A were obtained from Sigma-Aldrich, and posaconazole was purchased from eNovation Chemicals. U-X and compound A were synthesized as described previously (16). P-X was synthesized as described in SI Materials and Methods. All compounds were dissolved in DMSO and stored as 10 mM stock solutions in multiple aliquots at −80 °C.
Nanodisc Reconstitution of Purified NPC1.
A cDNA encoding full-length human NPC1 (GI: 83305902) with a C-terminal StrepTactin epitope tag (WSHPQFEK) (28) was cloned into pEG BacMam (29). The baculovirus was transduced into HEK293S GnTI− cells (American Type Culture Collection; CRL-3022). At 50 h postinfection, the cells were disrupted by sonication in buffer containing 20 mM Hepes at pH 7.0, 150 mM NaCl, 1 mM phenylmethyl sulfonyl fluoride, and 5 μg/mL each of leupeptin and aprotinin. After low-speed centrifugation, the resulting supernatant was incubated with 1% (wt/vol) DDM (final concentration) for 2 h at 4 °C, after which the mixture was centrifuged again, and the supernatant was loaded onto Strep-Tactin resin (IBA). After two washes with buffer containing 20 mM Hepes at pH 7.0, 150 mM NaCl, and 0.02% DDM, the protein was eluted with the same buffer (but with 5 mM desthiobiotin) and then concentrated to ∼5 mg/mL. For all experiments except those shown in Fig. 4B, purified NPC1 was incorporated into lipid nanodiscs containing 18 mol% cholesterol as described previously (30).
To form the lipid stock solution, 9 mg of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (Avanti) and 1 mg of cholesterol (Avanti) dissolved in chloroform were dried by argon stream, and residual chloroform was further removed by vacuum desiccation for overnight. Lipids were rehydrated in 0.5 mL of buffer containing 20 mM Hepes pH 7.0, 150 mM NaCl, and 25 mM sodium cholate, and then sonicated in a water bath, resulting in a clear lipid stock solution at 20 mg/mL. Purified NPC1 protein (∼5 mg/mL) in 0.02% DDM was mixed with membrane scaffold protein MSP1D1 (30) and the lipid stock solution at a molar ratio of 1:10:100 and then incubated at 4 °C for 1 h. To remove excess detergent, Bio-Beads SM2 (0.7 g per 1 mL mixture; Bio-Rad) were added, followed by incubation at 4 °C for 4 h with constant rotation. The Bio-Beads were removed by centrifugation, and the resulting supernatant was applied to a Superdex 200 size-exclusion column (GE Healthcare) in buffer containing 20 mM Hepes pH 7.0 and 150 mM NaCl. The peak corresponding to NPC1 reconstituted in lipid nanodiscs was collected for cross-linking assays.
Two mutant versions of human NPC1, one in which the NTD (amino acids 24–260) was deleted and the other containing an amino acid substitution (P691S) (23, 24), were expressed, purified, and incorporated into nanodiscs as described above. The resulting proteins are referred to as NPC1(ΔNTD) nanodiscs and NPC1(P691S) nanodiscs.
Immunoprecipitation of Cross-Linked NPC1.
For immunoprecipitation of P-X or U-X binding proteins, parental CHO-7 or mutant TR-4323 cells overexpressing NPC1 were set up on day 0 in 25 mL of medium A with 5% lipoprotein-deficient serum (LPDS) at a density of 1.6 × 106 cells per 150-mm dish. On day 3, each monolayer received a direct addition of DMSO (final concentration, 0.2%) containing the indicated cross-linker and various competitor compounds as described in the figure legends. After incubation for 1 h at 37 °C in the dark and without a change of media, the cells were irradiated for 10 min at room temperature under 306-nm UV light (Atlanta Light Bulb Co.) in a UV Stratalinker 2400 apparatus (Stratagene). All subsequent operations were carried out at 4 °C. Cells were scraped from the dishes, and each cell suspension was subjected to centrifugation at 1 × 103 × g for 5 min. The pellet was washed once with PBS and then solubilized with 1 mL of buffer containing 50 mM Tris⋅HCl pH 7.5, 150 mM NaCl, 1% (wt/vol) DDM, and Protease Inhibitor Mixture. Each solubilized lysate was passed through a 22.5-gauge needle 10 times, rotated for 1 h, and clarified by centrifugation at 1.5 × 106 × g for 30 min. The supernatant was then transferred to a fresh tube.
CHO-7 cell lysates were incubated with monoclonal anti-NPC1 as described in the figure legends. After rotating for 1 h, 100 μL of Protein A/G PLUS Agarose Immunoprecipitation Reagent (beads) were added, followed by rotation overnight, and centrifugation at 1 × 103 × g for 5 min. The pelleted beads were washed five times with 1 mL of wash buffer (50 mM Tris⋅HCl 7.5, 150 mM NaCl, and 0.02% DDM), suspended in 1 mL of wash buffer mixed with 1% SDS, and then boiled for 5 min, after which the mixture was clarified by centrifugation for 5 min at 1 × 103 × g. TR-4323 cell lysates were incubated with anti-FLAG M2 magnetic beads for 1 h, after which the beads were applied to a magnet, washed three times with 1 mL of wash buffer, and eluted with 100 µL of wash buffer containing 15 µg of 3× FLAG peptide.
The purified proteins from the CHO-7 and TR-4323 lysates were then tagged with Alexa Fluor 532 by means of copper-catalyzed azide-alkyne cycloaddition (click chemistry) (31, 32). The reactions were carried out by mixing an aliquot of each SDS lysate (43 µL), in a final volume of 50 µL, with 3 µL of 1.7 mM Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (TBTA), 2 µL of 50 mM CuSO4, 1 µL of 50 mM Tris(2-carboxyethyl)phosphine (TCEP), and 1 µL of 1.25 mM Alexa Fluor 532 azide. The mixture was incubated at room temperature for 1 h.
To visualize the fluorescent-labeled proteins by in-gel fluorescence, 20-µL aliquots of each sample were mixed with 5× loading dye and subjected to 8% SDS/PAGE, after which the gels were scanned with a Typhoon image scanner (GE Healthcare; filter setting: excitation, 533 nM; emission, 555 nm; high sensitivity). After scanning, the proteins were transferred to nitrocellulose, blotted with rabbit monoclonal anti-NPC1, and visualized by chemiluminescence using goat anti-rabbit IgG conjugated to horseradish peroxidase, as described below. Quantification of fluorescent-tagged NPC1 bands in SDS/PAGE was carried out using ImageJ (22).
UV Cross-Linking of P-X to NPC1 in Nanodiscs.
Each reaction contained, in a final volume of 100 µL of buffer A (50 mM Hepes pH 7.5 and 150 mM NaCl), 0.1 µg of WT NPC1 or NPC1(ΔNTD) reconstituted in nanodiscs, 0.3 µM P-X, and various amounts of itraconazole, posaconazole, or ketoconazole as indicated in the figure legends. After incubation for 1 h at 37 °C, the mixture was subjected to UV cross-linking for 10 min. After the addition of SDS (final concentration 1%), the cross-linked P-X was tagged with Alexa Fluor 532 using click chemistry as described above. An aliquot of each sample was subjected to 8% SDS/PAGE, and fluorescent-labeled proteins were visualized by in-gel fluorescence and quantified by ImageJ as described above. After scanning the gel, the proteins were transferred to nitrocellulose and blotted with rabbit monoclonal anti-NPC1.
Reproducibility.
Each experiment was repeated at least three times, with similar results obtained.
Additional information on materials, synthesis of posaconazole P-X cross-linker, cell culture, generation of TR-4323 cells, cholesterol esterification assays, and immunoblot analysis is provided in SI Materials and Methods.
SI Materials and Methods
Materials.
We obtained Dulbecco’s PBS, desthiobiotin, CuSO4, NaCl, FCS, SDS, 3× FLAG peptide, anti-FLAG M2 magnetic beads, and Sandoz 58–035 (ACAT inhibitor) from Sigma-Aldrich; Tris⋅HCl from Affymetrix; Hepes from Alfa Aesar; [1-14C]oleic acid (55 mCi/mmol) and [1-14C]oleoyl CoA (54 mCi/mmol) from PerkinElmer; Protein A/G PLUS-Agarose Immunoprecipitation Reagent from Santa Cruz Biotechnology; cOmplete EDTA-Free Protease Inhibitor Mixture (1 tablet per 50 mL of buffer) from Roche Diagnostics; DDM from Anatrace; Alexa Fluor 532 azide from Joseph M. Ready, University of Texas Southwestern Medical Center; and TBTA, TCEP, and Bolt 4–12% Bis-Tris Plus gradient gels from Thermo Fisher Scientific. All tissue culture supplies were obtained from sources as described previously (33). Stock solutions of sodium mevalonate and sodium compactin (34) and cholesterol/methyl-β-cyclodextrin (33) were prepared as described previously. Newborn calf LPDS (d < 1.215 g/mL) was prepared by ultracentrifugation as described previously (20).
Synthesis of Posaconazole Cross-Linker (P-X).
To a solution of posaconazole (70 mg, 0.1 mmol) and 4-(4-(prop-2-yn-1-yloxy)benzoyl)benzoic acid (42 mg, 0.12 mmol) in CH2Cl2 (3 mL) was added 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (28 mg, 0.12 mmol) and 4-(dimethylamino)pyridine (18 mg, 0.12 mmol) at room temperature. The solution was stirred for 12 h at room temperature, followed by dilution with EtOAc (30 mL) and washing with brine (3 × 3 mL). The organic layer was dried (Na2SO4) and concentrated. The residue was purified by flash chromatography (silica gel; CH2Cl2:MeOH = 15:1) to provide P-X (69 mg, 0.071 mmol; 71%). P-X was dissolved in DMSO and stored as a 10 mM stock solution in multiple aliquots at −80 °C.
1H NMR (400 MHz, CDCl3) δ 8.13 (d, J = 8.4 Hz, 2H), 8.10 (s, 1H), 7.78 (d, J = 8.8 Hz, 2H), 7.77 (s, 1H), 7.72 (d, J = 8.4 Hz, 2H), 7.57 (s, 1H), 7.38 (dt, J = 8.8, 6.4 Hz, 1H), 7.24 (d, J = 9.2 Hz, 2H), 7.03 (d, J = 8.8 Hz, 2H), 6.95 (d, J = 9.2 Hz, 2H), 6.94 (d, J = 9.2 Hz, 2H), 6.89–6.77 (m, 2H), 6.76 (d, J = 9.2 Hz, 2H), 5.41(dq, J = 7.6, 6.4 Hz, 1H), 4.76 (d, J = 2.4 Hz, 2H), 4.64 (d, J = 14.4 Hz, 1H), 4.50 (d, J = 14.4 Hz, 1H), 4.37 (ddd, J = 10.8, 7.6, 4.0 Hz, 1H), 4.10 (dd, J = 8.8, 7.6 Hz, 1H), 3.77 (dd, J = 8.8, 6.4 Hz, 1H), 3.69 (dd, J = 8.8, 5.6 Hz, 1H), 3.60 (dd, J = 9.2, 7.6 Hz, 1H), 3.42–3.28 (m, 4H), 3.26–3.14 (m, 4H), 2.67–2.47 (m, 2H), 2.56 (t, J = 2.4 Hz, 1H), 2.07 (dd, J = 12.8, 8.4 Hz, 1H), 2.04–1.94 (m, 1H), 1.94–1.80 (m, 1H), 1.45 (d, J = 6.4 Hz, 3H), 0.95 (t, J = 7.6 Hz, 3H); 13C NMR (100 MHz, CDCl3) δ 194.67, 164.89, 162.80 (dd, J = 48.3, 12.0 Hz, 1C), 161.33, 159.02 (dd, J = 46.3, 12.0 Hz, 1C), 153.34, 153.15, 151.08, 150.45, 145.46, 144.61, 141.88, 134.60, 132.81, 132.50, 130.24, 129.61, 129.45, 128.57 (dd, J = 9.5, 5.6 Hz, 1C), 125.60, 125.35 (dd, J = 13.0, 3.6 Hz, 1C), 123.60, 118.74, 116.60, 115.15, 114.58, 111.31 (dd, J = 20.6, 3.4 Hz, 1C), 104.62 (dd, J = 25.8, 25.8 Hz, 1C), 84.02 (d, J = 4.1 Hz, 1C), 77.65, 76.39, 72.50, 70.73, 68.91, 60.37, 55.89, 50.87, 48.93, 38.81, 37.41 (d, J = 3.3 Hz, 1C), 22.39, 17.54, 10.55; ESI MS: [M+H]+ calcd. for C54H53F2N8O7 963.4, found 963.4; [α]D: -66.3 (c = 0.32, CHCl3); Calculated pKa: 3.9.
Culture Media.
Medium A is a 1:1 mixture of Ham’s F-12 medium and DMEM containing 2.5 mM l-glutamine. Medium B is l-glutamine–free DMEM. All media contained 100 U/mL penicillin and 100 µg/mL streptomycin sulfate unless noted otherwise in the figure legends.
Cell Culture.
Stock cultures of hamster CHO-7 cells (a clone of CHO-K1 cells selected for growth in LPDS) (35), 10–3 cells (mutant CHO-K1 cells that lack detectable NPC1 mRNA) (36), and TR-4323 (a clone of mutant 10–3 cells stably transfected with a plasmid encoding human NPC1 (see below) were maintained in monolayer culture at 37 °C in an 8.8% CO2 incubator.
Generation of TR-4323 Cells Expressing Recombinant Human NPC1.
The 10–3 cells, a stable line of mutant CHO-K1 cells that lack detectable NPC1 (36), were set up on day 0 at a density of 4 × 105 cells per 100-mm dish in medium A with 5% LPDS. On day 2, cells were transfected with 1 µg of pCMV-NPC1-3× Flag-TEV-2× StrepTactin (16) using FuGENE HD transfection reagent according to the manufacturer’s instructions. At 24 h after transfection, cells were switched to the foregoing medium supplemented with 700 µg/mL Zeocin for selection. Fresh medium was added every 2–3 d until colonies formed at ∼15 d. Individual colonies were isolated with cloning cylinders, and expression of NPC1-Flag-TEV-StrepTactin was assessed by immunoblot analysis with anti-NPC1 antibody. Cells from single colonies were cloned by limiting dilution, maintained in medium A with 5% LPDS containing 500 µg/mL Zeocin, and are hereinafter referred to as TR-4323 cells.
Cholesterol Esterification Assay in Intact Cells.
The rate of incorporation of [14C]oleate into cholesteryl [14C]oleate and [14C]triglycerides by monolayers of CHO-7 cells was measured as described previously (20). In brief, at 16 h before the experiments, cells were switched to medium containing LPDS supplemented with compactin and mevalonate. On the day of the experiment, cells were incubated for 3 h with fresh medium containing FCS as a source of cholesterol-containing lipoproteins. Cells were then pulse-labeled for 2 h with sodium [14C]oleate-albumin complex as described in the figure legends. The cells were then washed, and the lipids were extracted in hexane:isopropanol (3:2), separated on a silica gel G thin-layer chromatogram (developed in heptane:ethylether:acetic acid, 90:30:1), and quantified by scintillation counting. The amounts of cholesteryl [14C]oleate and [14C]triglycerides formed are expressed as nanomoles formed per hour per milligram of cell protein.
ACAT Activity in Membranes.
ACAT activity in membranes from CHO-7 cell extracts was assayed by measuring the rate of conversion of [1-14C]oleoyl CoA to cholesteryl [14C]oleoate as described previously (21) with several modifications. A 105g membrane pellet was resuspended in 25 mM Tris⋅HCl at pH 7.5, homogenized by 10–15 passages through an 18-gauge needle. The membranes (100 µg protein) were incubated in a final volume of 0.3 mL of 25 mM Tris⋅HCl at pH 7.5 containing 2 mg/mL BSA and 50 µM cholesterol/methyl-β-cyclodextrin in the presence of various compounds (added in 1 µl of DMSO) as indicated in Fig. 2B. After incubation for 20 min at 37 °C, each reaction received a direct addition of [1-14C]oleoyl CoA (final concentration, 50 µM; 7,789 dpm/nmol), and the incubations were continued for 1 h at 37 °C. Reactions were terminated by the addition of 2 mL of hexane/isopropanol (3:2, vol:vol) and processed for cholesteryl [14C]oleate formation as described above.
SDS/PAGE and Immunoblot Analysis.
In all experiments except those depicted in Fig. 3A, SDS/PAGE gels were run as follows: Denatured protein samples and marker proteins (250–37 kDa) were loaded onto 8% gels, to which 100 V was applied for 75 min. Gels were subsequently cut below the 50-kDa marker before fluorescent scanning. These procedures ensured that free Alexa Fluor 532 would not contaminate the gels. The gel in Fig. 3A was run at 100 V for 120 min. After electrophoresis, the proteins were transferred to nitrocellulose filters for immunoblot analysis. The filters were incubated at room temperature with 1 µg/mL rabbit monoclonal IgG against amino acids 1,261–1,278 of human NPC1 (Abcam). Bound antibody was visualized by chemiluminescence (SuperSignal West Pico Chemiluminescent Substrate; Thermo Scientific) after incubation with a 1:5,000 dilution of goat anti-rabbit IgG conjugated to horseradish peroxidase (Jackson ImmunoResearch). Images were scanned using an Odyssey FC Imager (dual-mode imaging system; 2-min integration time) and analyzed using Image Studio version 5.0 (LI-COR).
Acknowledgments
We thank our colleagues Yansong Gao, Donald Anderson, and Ting Han for helpful suggestions; Lisa Beatty, Shomanike Head, and Lucie Batte for invaluable help with tissue culture; and Jessica Proulx for excellent technical assistance. This work was supported by grants from the National Institutes of Health (HL20948, to J.L.G. and M.S.B.) and the Robert A. Welch Foundation (I-1422, to J.K.D.B.). X.L. is supported by funds from Howard Hughes Medical Institute (Günter Blobel, Investigator), and is a Gordon and Betty Moore Foundation Fellow of Life Sciences Research Foundation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1619571114/-/DCSupplemental.
References
- 1.Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232(4746):34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- 2.Pentchev PG. Niemann–Pick C research from mouse to gene. Biochim Biophys Acta. 2004;1685(1-3):3–7. doi: 10.1016/j.bbalip.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 3.Das A, Brown MS, Anderson DD, Goldstein JL, Radhakrishnan A. Three pools of plasma membrane cholesterol and their relation to cholesterol homeostasis. eLife. 2014;3(e02882):1–16. doi: 10.7554/eLife.02882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iaea DB, Maxfield FR. Cholesterol trafficking and distribution. Essays Biochem. 2015;57:43–55. doi: 10.1042/bse0570043. [DOI] [PubMed] [Google Scholar]
- 5.Vance JE, Karten B. Niemann–Pick C disease and mobilization of lysosomal cholesterol by cyclodextrin. J Lipid Res. 2014;55(8):1609–1621. doi: 10.1194/jlr.R047837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Infante RE, et al. NPC2 facilitates bidirectional transfer of cholesterol between NPC1 and lipid bilayers, a step in cholesterol egress from lysosomes. Proc Natl Acad Sci USA. 2008;105(40):15287–15292. doi: 10.1073/pnas.0807328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwon HJ, et al. Structure of N-terminal domain of NPC1 reveals distinct subdomains for binding and transfer of cholesterol. Cell. 2009;137(7):1213–1224. doi: 10.1016/j.cell.2009.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang ML, et al. Identification of surface residues on Niemann–Pick C2 essential for hydrophobic handoff of cholesterol to NPC1 in lysosomes. Cell Metab. 2010;12(2):166–173. doi: 10.1016/j.cmet.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deffieu MS, Pfeffer SR. Niemann–Pick type C 1 function requires lumenal domain residues that mediate cholesterol-dependent NPC2 binding. Proc Natl Acad Sci USA. 2011;108(47):18932–18936. doi: 10.1073/pnas.1110439108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X, et al. Structure of human Niemann–Pick C1 protein. Proc Natl Acad Sci USA. 2016;113(29):8212–8217. doi: 10.1073/pnas.1607795113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X, Saha P, Li J, Blobel G, Pfeffer SR. Clues to the mechanism of cholesterol transfer from the structure of NPC1 middle lumenal domain bound to NPC2. Proc Natl Acad Sci USA. 2016;113(36):10079–10084. doi: 10.1073/pnas.1611956113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gong X, et al. Structural insights into the Niemann–Pick C1 (NPC1)-mediated cholesterol transfer and Ebola infection. Cell. 2016;165(6):1467–1478. doi: 10.1016/j.cell.2016.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davies JP, Ioannou YA. Topological analysis of Niemann–Pick C1 protein reveals that the membrane orientation of the putative sterol-sensing domain is identical to those of 3-hydroxy-3-methylglutaryl-CoA reductase and sterol regulatory element binding protein cleavage-activating protein. J Biol Chem. 2000;275(32):24367–24374. doi: 10.1074/jbc.M002184200. [DOI] [PubMed] [Google Scholar]
- 14.Nohturfft A, Brown MS, Goldstein JL. Sterols regulate processing of carbohydrate chains of wild-type SREBP cleavage-activating protein (SCAP), but not sterol-resistant mutants Y298C or D443N. Proc Natl Acad Sci USA. 1998;95(22):12848–12853. doi: 10.1073/pnas.95.22.12848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liscum L. Pharmacological inhibition of the intracellular transport of low-density lipoprotein-derived cholesterol in Chinese hamster ovary cells. Biochim Biophys Acta. 1990;1045(1):40–48. doi: 10.1016/0005-2760(90)90201-8. [DOI] [PubMed] [Google Scholar]
- 16.Lu F, et al. Identification of NPC1 as the target of U18666A, an inhibitor of lysosomal cholesterol export and Ebola infection. eLife. 2015;4(e12177):1–16. doi: 10.7554/eLife.12177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peyton LR, Gallagher S, Hashemzadeh M. Triazole antifungals: A review. Drugs Today (Barc) 2015;51(12):705–718. doi: 10.1358/dot.2015.51.12.2421058. [DOI] [PubMed] [Google Scholar]
- 18.Nagappan V, Deresinski S. Reviews of anti-infective agents: Posaconazole, a broad-spectrum triazole antifungal agent. Clin Infect Dis. 2007;45(12):1610–1617. doi: 10.1086/523576. [DOI] [PubMed] [Google Scholar]
- 19.Xu J, Dang Y, Ren YR, Liu JO. Cholesterol trafficking is required for mTOR activation in endothelial cells. Proc Natl Acad Sci USA. 2010;107(10):4764–4769. doi: 10.1073/pnas.0910872107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldstein JL, Basu SK, Brown MS. Receptor-mediated endocytosis of low-density lipoprotein in cultured cells. Methods Enzymol. 1983;98:241–260. doi: 10.1016/0076-6879(83)98152-1. [DOI] [PubMed] [Google Scholar]
- 21.Metherall JE, Ridgway ND, Dawson PA, Goldstein JL, Brown MS. A 25-hydroxycholesterol-resistant cell line deficient in acyl-CoA:cholesterol acyltransferase. J Biol Chem. 1991;266(19):12734–12740. [PubMed] [Google Scholar]
- 22.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watari H, et al. Niemann-Pick C1 protein: Obligatory roles for N-terminal domains and lysosomal targeting in cholesterol mobilization. Proc Natl Acad Sci USA. 1999;96(3):805–810. doi: 10.1073/pnas.96.3.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ko DC, Gordon MD, Jin JY, Scott MP. Dynamic movements of organelles containing Niemann–Pick C1 protein: NPC1 involvement in late endocytic events. Mol Biol Cell. 2001;12(3):601–614. doi: 10.1091/mbc.12.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohgami N, et al. Binding between the Niemann–Pick C1 protein and a photoactivatable cholesterol analog requires a functional sterol-sensing domain. Proc Natl Acad Sci USA. 2004;101(34):12473–12478. doi: 10.1073/pnas.0405255101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Infante RE, et al. Purified NPC1 protein, II: Localization of sterol binding to a 240-amino acid soluble luminal loop. J Biol Chem. 2008;283(2):1064–1075. doi: 10.1074/jbc.M707944200. [DOI] [PubMed] [Google Scholar]
- 27.de Duve C, et al. Commentary: Lysosomotropic agents. Biochem Pharmacol. 1974;23(18):2495–2531. doi: 10.1016/0006-2952(74)90174-9. [DOI] [PubMed] [Google Scholar]
- 28.Schmidt TGM, Skerra A. The Strep-tag system for one-step purification and high-affinity detection or capturing of proteins. Nat Protoc. 2007;2(6):1528–1535. doi: 10.1038/nprot.2007.209. [DOI] [PubMed] [Google Scholar]
- 29.Goehring A, et al. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat Protoc. 2014;9(11):2574–2585. doi: 10.1038/nprot.2014.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ritchie TK, et al. Chapter 11: Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 2009;464:211–231. doi: 10.1016/S0076-6879(09)64011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kolb HC, Finn MG, Sharpless KB. Click chemistry: Diverse chemical function from a few good reactions. Angew Chem Int Ed Engl. 2001;40(11):2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 32.Sapkale P, Sahu M, Chaudhari M, Patil PR. Trends of click synthesis: A review. Int J Pharm Pharm Sci. 2014;6:99–103. [Google Scholar]
- 33.Das A, Goldstein JL, Anderson DD, Brown MS, Radhakrishnan A. Use of mutant 125I-perfringolysin O to probe transport and organization of cholesterol in membranes of animal cells. Proc Natl Acad Sci USA. 2013;110(26):10580–10585. doi: 10.1073/pnas.1309273110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown MS, Faust JR, Goldstein JL, Kaneko I, Endo A. Induction of 3-hydroxy-3-methylglutaryl coenzyme A reductase activity in human fibroblasts incubated with compactin (ML-236B), a competitive inhibitor of the reductase. J Biol Chem. 1978;253(4):1121–1128. [PubMed] [Google Scholar]
- 35.Metherall JE, Goldstein JL, Luskey KL, Brown MS. Loss of transcriptional repression of three sterol-regulated genes in mutant hamster cells. J Biol Chem. 1989;264(26):15634–15641. [PubMed] [Google Scholar]
- 36.Wojtanik KM, Liscum L. The transport of low-density lipoprotein-derived cholesterol to the plasma membrane is defective in NPC1 cells. J Biol Chem. 2003;278(17):14850–14856. doi: 10.1074/jbc.M300488200. [DOI] [PubMed] [Google Scholar]