Significance
Leaves are the primary source of photoassimilate in crop plants. A precise understanding of the genetic architecture underlying leaf morphology is critical to engineering climate-resilient crop varieties. An ideal cotton cultivar would produce a lower canopy of broad, normal leaves before transitioning to an upper canopy of highly lobed, okra leaves. Here we show that the major leaf shapes of cotton are controlled by the okra locus, which encodes an HD-Zip transcription factor Gossypium hirsutum LATE MERISTEM IDENTITY1-D1b (GhLMI1-D1b). Using gene silencing, we temporarily induced normal leaf formation in okra, thus validating the candidate gene and creating the leaf shape ideotype in cotton. This study, identifying a single locus responsible for cotton leaf shape, expands the genetic toolbox for breeders to produce superior cotton varieties.
Keywords: cotton, leaf shape, okra, gene cloning
Abstract
Leaf shape varies spectacularly among plants. Leaves are the primary source of photoassimilate in crop plants, and understanding the genetic basis of variation in leaf morphology is critical to improving agricultural productivity. Leaf shape played a unique role in cotton improvement, as breeders have selected for entire and lobed leaf morphs resulting from a single locus, okra (l-D1), which is responsible for the major leaf shapes in cotton. The l-D1 locus is not only of agricultural importance in cotton, but through pioneering chimeric and morphometric studies, it has contributed to fundamental knowledge about leaf development. Here we show that an HD-Zip transcription factor homologous to the LATE MERISTEM IDENTITY1 (LMI1) gene of Arabidopsis is the causal gene underlying the l-D1 locus. The classical okra leaf shape allele has a 133-bp tandem duplication in the promoter, correlated with elevated expression, whereas an 8-bp deletion in the third exon of the presumed wild-type normal allele causes a frame-shifted and truncated coding sequence. Our results indicate that subokra is the ancestral leaf shape of tetraploid cotton that gave rise to the okra allele and that normal is a derived mutant allele that came to predominate and define the leaf shape of cultivated cotton. Virus-induced gene silencing (VIGS) of the LMI1-like gene in an okra variety was sufficient to induce normal leaf formation. The developmental changes in leaves conferred by this gene are associated with a photosynthetic transcriptomic signature, substantiating its use by breeders to produce a superior cotton ideotype.
Leaf shape is spectacularly diverse (1–3). This diversity reflects evolutionary processes—either adaptive or neutral—that manifest through changes in developmental programming, environmental plasticity, or the interaction thereof (4). As the primary sources of photoassimilate in the world’s major crops, the role of leaves—their shapes, the constraints morphology places on other physiologically relevant features, and the contributions of leaf shape to canopy and plant architecture—whether directly or indirectly selected upon during domestication, is an indisputably important consideration when discussing agricultural productivity. Although much is known about the developmental genetic basis of leaf morphology, only a handful of genes modifying leaf shapes in crops or responsible for natural variation among species have been identified (5–10).
Cotton (Gossypium spp.) is the world’s most important source of natural fiber as well as a leading oilseed crop. The cultivated cottons (Gossypium hirsutum and Gossypium barbadense) are allotetraploid species (2n = 4x = 52, AADD) formed from the hybridization of diploids Gossypium arboreum (2n = 2x = 26, AA) and Gossypium raimondii (2n = 2x = 26, DD) (11). Remarkable phenotypic diversity exists for leaf shape in cotton, widely ranging from entire (lacking dissection) to deeply lobed across both diploids and polyploids (12–14). Leaf shape in Gossypium is an important agronomic trait that affects plant and canopy architecture, yield, stress tolerance, and other production attributes (15). Among crops, leaf shape in cotton is unique; in recent history, breeders used a single locus, okra, to purposefully alter leaf shape among cotton cultivars (15, 16). The four major leaf shapes of cotton: normal, subokra, okra, and superokra (Fig. 1A) are semidominant and allelomorphic at the l-D1 (okra) locus (15–21), whereas laciniate, similar in morphology to okra, maps to the orthologous diploid A-genome locus (l-A1) (22). Beyond agriculture, the okra locus is also of historical importance to leaf development. Not only was it used for one of the first comprehensive morphometric descriptions of leaves (12, 23), but pioneering studies creating okra chimeras determined the contributions of different cell layers to leaf shape (24, 25).
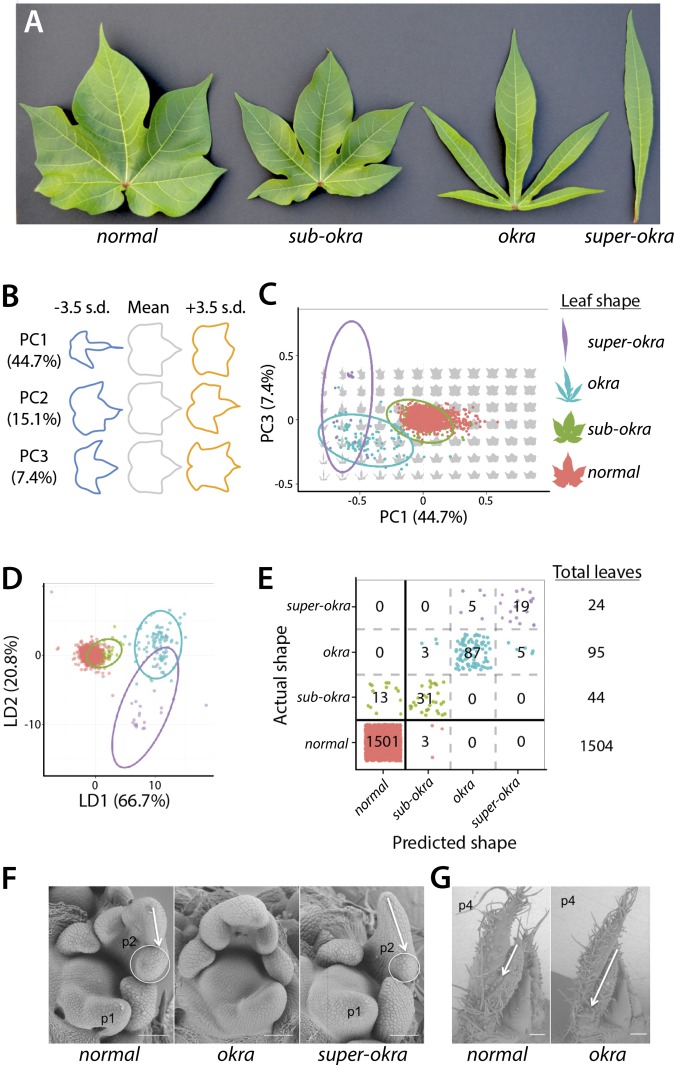
Fig. 1.
Morphometric analysis of leaf shapes conferred by the subokra, okra, and superokra mutations. (A, Left to Right) Leaves representative of normal, subokra, okra, and superokra leaf shape phenotypes. (B) Eigenleaves representative of leaf shapes found ±3.5 SDs along each Principal Component (PC) axis calculated from the harmonic series of an EFD analysis. (C) Percent variance explained by each PC is provided. PC1 and PC3 (PC2 not shown, as it explains asymmetric shape variance) separate normal and various okra leaf shape classes. Also provided are 95% confidence ellipses. (D) LDA maximizes the discrimination of normal and okra leaf shape classes. LD1 and LD2 are shown, and the percent separation between phenotypic classes of leaves is indicated. Also provided are 95% confidence ellipses. (E) A confusion matrix, showing actual versus predicted leaf shapes, constructed using linear discriminants. Leaf shape alone discriminants a majority of normal from subokra, okra, and superokra leaf types. (F) SEMs of the SAM, P1, and P2 leaf primordia for normal, okra, and superokra shoot apices. Note the displaced lobe in the P2 of superokra relative to normal. (G) SEM of normal and okra shoot apices showing a more pronounced leaf lobe present by the P4 stage of leaf primordium development in okra relative to normal. Colors, normal, red; okra, blue; subokra, green; superokra, purple. [Scale bar, 100 μm (F) and 200 μm (G).]
The okra allelic series confers increasingly lobed leaf shapes from subokra to okra with the proximal lobes in mature superokra reduced to a single linear blade (Fig. 1A). The characteristic shape of these four leaf morphs can be used both qualitatively and quantitatively (12, 23) to distinguish among their alleles (Fig. 1 B–E). Classical development studies involving okra revealed the underlying factor acted early in leaf development in all tissue layers (L1, L2, L3) and cell autonomously (24, 25). Scanning Electron Micrographs (SEMs) of the shoot apical meristem show that the deeply lobed phenotype of superokra is apparent by the P2 stage of leaf development (Fig. 1F), whereas the less severe okra manifests by the P4 stage (Fig. 1G).
The l-D1 locus was placed on the short arm of chromosome 15-D1 (Chr15) using cytogenetics (26, 27) and confirmed by quantitative trait loci (QTL) mapping (28–30). The l-D1 locus was localized to a 5.4 cM interval near the telomere of Chr15 (31), and shuttle mapping using the laciniate gene (l-A2L) from G. arboreum further reduced the candidate region to 112 kb and 10 genes (22). Mapping and genomic targeting indicated two putative paralogous genes on Chr15 as the possible candidate genes for the l-D1 locus (31, 32). Here, we report the identification of a LATE MERISTEM IDENTITY1 (GhLMI1-D1b) gene, encoding an HD-Zip transcription factor, as the major determinant of leaf shape variation at the l-D1 locus in cotton.
Results
The Okra Locus Explains a Majority of Leaf Shape Variation in Cultivated Cotton.
To determine the quantitative extent that the okra locus is responsible for controlling leaf shape in cultivated cotton, we morphometrically analyzed 1,504 leaves from 420 cultivated cotton lines (Dataset S1). The eigenleaves (representations of shape variance) resulting from a Principal Component Analysis (PCA) of the harmonic series of Elliptical Fourier Descriptors (EFDs) describe shape features associated with linear versus palmately lobed leaf types (PC1) and pronounced distal lobing (PC3), in addition to fluctuating asymmetry (PC2) (Fig. 1B). PC1 and PC3 (in addition to other PCs not shown) separate normal from subokra, okra, and superokra leaf types and explain the majority of shape variance in the cotton accessions analyzed (Fig. 1C). A Linear Discriminant Analysis (LDA) performed to distinguish cotton accessions by leaf shape led to the following correct percentage of assignments to phenotypic class by EFDs alone: 99.8% normal (1,501 of 1,504 cases), 70.5% subokra (31 of 44 cases), 91.6% okra (87 of 95 cases), and 79.2% superokra (19 of 24 cases) (Fig. 1 D and E). The classification became 99% correct (1,651 of 1,667 cases) if only two classes were formed: normal and non-normal (inclusive of subokra, okra, and superokra) (Fig. 1E).
Our results indicate that okra (l-D1), a monogenic locus, is quantitatively responsible for the majority of leaf shape variance in cotton, the alleles of which can be discriminated from each other at high correct classification rates using shape information alone. A strongly monogenic basis for leaf shape in cotton is in contrast to a polygenic basis for leaf shape described in other crops (7, 33). Our results are consistent with classical morphometric work describing the profound role the l-D1 locus plays in determining cotton leaf shape (12, 23).
Fine Mapping of the l-D1 Locus in a Large F2 Population.
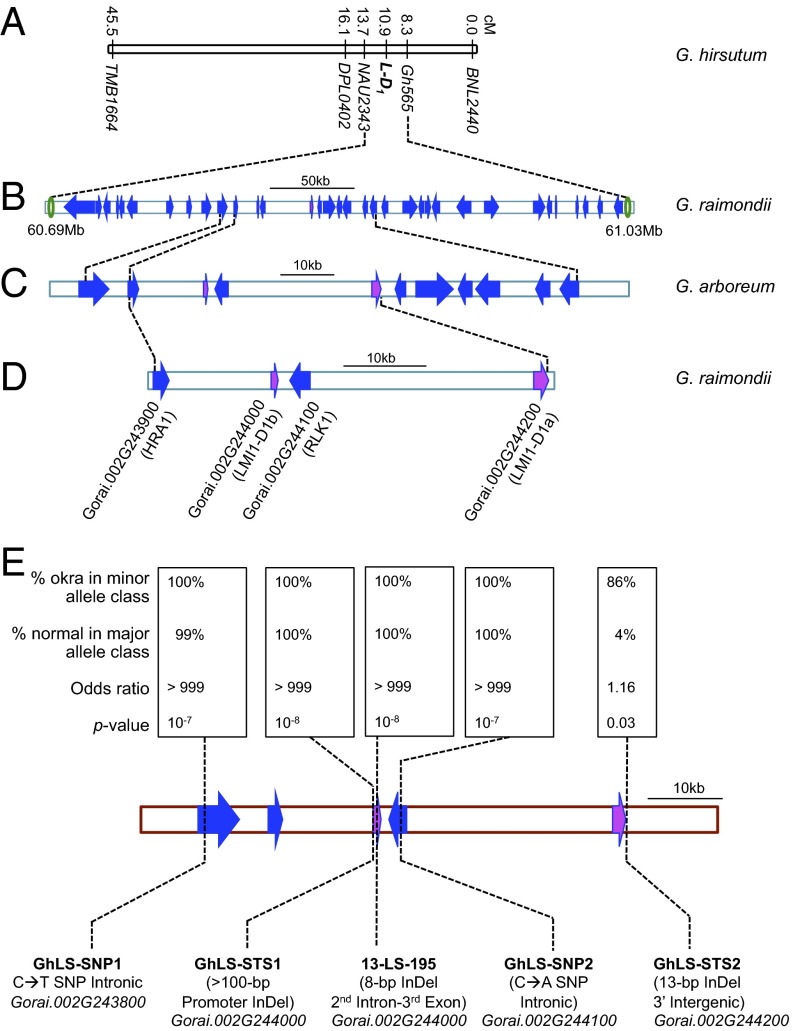
A total of 1,027 F2 plants from the cross NC05AZ21 × NC11-2100 showed the expected phenotypic ratio for single gene inheritance of okra leaf shape (SI Appendix, Table S1). Genotyping with the codominant flanking simple sequence repeats (SSRs) of l-D1 (Gh565 and DPL0402) identified 122 recombinants and produced a genetic map (Fig. 2A) similar to the preliminary mapping (31). Furthermore, a sequence-tagged site (STS) marker (13-LS-195) that had previously cosegregated with leaf shape phenotype (31) continued to do so in the present fine mapping population, confirming the tight linkage of this marker to the leaf shape locus (l-D1) in cotton. The resulting 337-kb, 34-gene candidate interval (Fig. 2B) was further resolved to 112 kb and 10 genes using orthologous mapping of the homeologous laciniate gene of the diploid cotton G. arboreum with the molecular markers closely linked to the l-D1 locus (22) (Fig. 2C).
Fig. 2.
Genetically resolving the l-D1 locus in Upland cotton. (A) Genetic mapping of l-D1 locus based on biparental mapping. (B) Orthologous mapping of the l-D1 locus to the sequenced D genome donor (G. raimondii) chromosome 2 (337 kb, 34 putative genes) (30), and (C) shuttle mapping using the orthologous laciniate (l-A2) locus from diploid donor species G. arboreum (112 kb, 10 putative genes) (21). (D) Fine mapping of l-D1 locus using an association mapping panel and two sets (BC8 and BC3) of isogenic lines (52 kb, putative four genes). (E) Association analysis statistics, adjusting for population structure for variants within candidate gene region of l-D1 locus.
Fine Mapping Using Association Mapping and Isogenic Lines.
To supplement cosegregating STS marker 13-LS-195, three additional markers (GhLS-STS1, GhLS-SNP2, and GhLS-STS2) were developed using G. raimondii sequence information (34) within the 10-gene candidate orthologous region (SI Appendix, Fig. S1). An additional SNP marker (GhLS-SNP1) was also designed despite the fact this gene had already been excluded by the orthologous mapping of l-D1 locus with the laciniate gene of diploid cotton G. arboreum (Fig. 2C).
We then used association mapping to narrow down the candidate genomic region using the newly developed markers. Of the five markers used to genotype the diversity panel of 538 tetraploid cotton accessions with diverse leaf shapes, three markers (GhLS-STS1, 13-LS-195, and GhLS-SNP2) showed complete association with leaf shape, whereas GhLS-STS2 showed no association between the two most common leaf shapes, normal and okra (SI Appendix, Table S2). The lack of association of this STS marker with leaf shape was sufficient to reduce the 10-gene, 112-kb candidate region (Fig. 2C) to a 4-gene, 52-kb region between Gorai.002G243900 and Gorai.002G244200 (Fig. 2D and SI Appendix, Table S2).
Population structure was estimated in a subset of 404 lines of the 538-member panel using SSR markers distributed throughout the genome. Association tests of variants in the candidate gene region and of SSRs throughout the genome confirmed that the four remaining candidate genes showed very strong and significant (P < 10−7) association with leaf shape after correcting for population structure (Fig. 2E and Dataset S2). A few SSR markers also had significant associations, which may have occurred by chance when rare allele classes were observed in okra types (SI Appendix, Fig. S2 and Dataset S2). After fitting the most significant candidate gene marker as a covariate and retesting the background markers for associations, no markers outside the candidate gene interval were significant (SI Appendix, Fig. S2 and Dataset S2).
Association mapping of leaf shape with the above novel markers was also performed using two sets of near isogenic lines. We confirmed that genes conferring the okra phenotype in the mapping parent NC05AZ21 and okra isogenic line (LA213-okra) are allelic by phenotyping their F1 (SI Appendix, Fig. S3) and an F2 population. Genotyping of two sets of isolines (BC8 in Stoneville 213 and BC3 in Stoneville 7A backgrounds) with the four markers showed a similar marker pattern as observed in the association mapping (SI Appendix, Tables S2 and S3), confirming the resolution of the candidate region to four genes and 52 kb.
Thus, characterizing the association-mapping panel and isolines with novel markers reduced the candidate genomic region to 52 kb containing four genes. Of the four genes, Gorai.002G244200 (hereafter GhLMI1-D1a) and Gorai.002G244000 (hereafter GhLMI1-D1b) are paralogues coding for HD-Zip transcription factors with 71.2% protein similarity. Their homologs were implicated in flowering time and leaf complexity in Arabidopsis (8, 9, 35). Of the remaining two putative genes, Gorai002G244100 (hereafter GhRLK1) is a serine/threonine protein kinase, whereas Gorai.002G243900 (hereafter GhHRA1) is a trihelix transcription factor.
Expression Analysis of Leaf Shape Gene Candidates.
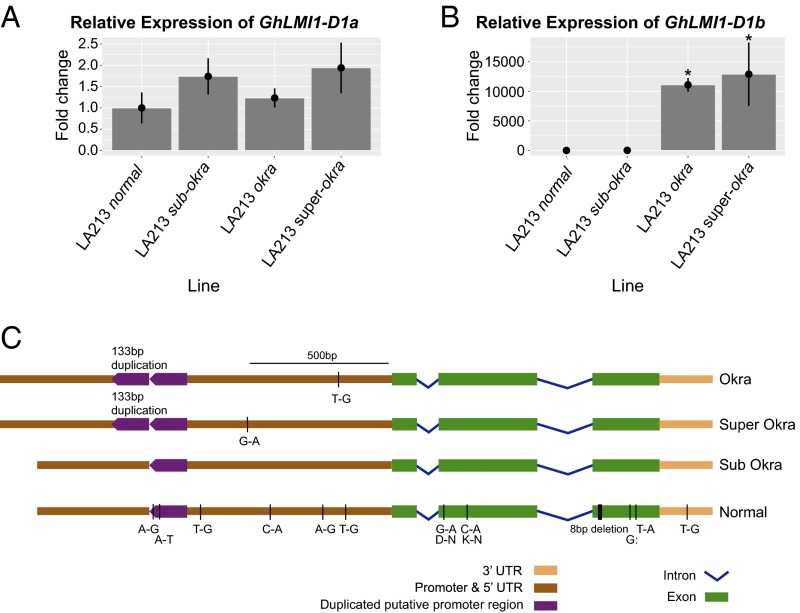
Expression analysis was performed to help identify the causal gene among the four remaining candidates. Semiquantitative expression analysis revealed that neither GhHRA1 nor GhRLK1 were expressed in young leaf tissue (SI Appendix, Fig. S4). Based on this observation together with their lack of sequence polymorphisms from novel STS marker development, we eliminated these two genes from consideration. GhLMI1-D1a was similarly expressed among leaf shapes, whereas GhLMI1-D1b expression was detected only in okra and superokra (SI Appendix, Fig. S4). Quantitative (q)RT-PCR confirmed the equivalent expression of GhLMI1-D1a among leaf shapes, whereas GhLMI1-D1b was up-regulated in okra and superokra compared with normal and subokra (Fig. 3 A and B). This indicated that up-regulation of GhLMI1-D1b in okra and superokra may be responsible for these leaf shapes. Congruent with these findings, RNA sequencing of okra and normal plastochron 2 (P2) samples indicated that GhLMI1-D1b is ∼15-fold enriched in okra relative to normal samples and is the only significantly differentially expressed gene (false discovery rate ≤ 0.05) within the 52-kb candidate interval (SI Appendix, Fig. S5 and Dataset S3).
Fig. 3.
Nucleotide polymorphisms of GhLMI1-D1b gene and expression analysis of candidate genes using qRT-PCR among different leaf shapes. (A and B) Relative transcript levels of leaf shape candidate genes (GhLMI1-D1a and GhLMI1-D1b) among isolines (n = 3) at ∼90 d after planting. There is no difference in the expression of GhLMI1-D1a among leaf shapes but a significant increase of GhLMI1-D1b expression in okra and superokra. Error bars represent the SD of the fold change. (B) Asterisks represent statistically significant differences as determined by unpaired t tests at P < 0.05. (C) Nucleotide polymorphisms of GhLMI1-D1b gene among four major leaf shapes of tetraploid cotton. The 133-bp tandem duplicated region in the putative promoter was found only in okra and superokra and in parallel with elevated gene expression. The 8-bp deletion was found only in normal and causes a frameshift mutation and premature stop codon. All other polymorphisms are SNPs with unknown effect on the gene expression and protein function. Subokra is set as the standard to which the other three leaf shapes are compared.
Sequencing of GhLMI1-Like Genes.
To identify DNA polymorphisms among leaf shapes, GhLMI1-D1a and GhLMI1-D1b were sequenced in 20 tetraploid cotton varieties (SI Appendix, Table S4), and comparisons were made relative to subokra (Fig. 3C and SI Appendix, Fig. S6). Sequence analysis of GhLMI1-D1a identified two variants (SI Appendix, Fig. S6). Variant 1 was found in two normal and all five okra and subokra lines. Variant 2 was found in the remaining three normal and all five superokra lines. The 13-bp InDel (GhLS-STS2), which showed no association with leaf shape in the association analysis (SI Appendix, Table S2), is placed ∼100 bp from the end of the 3′ UTR. Although this polymorphism lies outside GhLMI1-D1a, the proximity, lack of major polymorphisms, and identical expression among leaf shapes (SI Appendix, Figs. S4 and S5) provide evidence against GhLMI1-D1a as the candidate gene underlying l-D1.
Sequence analysis of GhLMI1-D1b identified two prominent polymorphisms among leaf shapes. First, a 133-bp tandem duplication located ∼800-bp upstream of the translation start site was unique to okra and superokra (Fig. 3C) and may explain the altered expression of GhLMI1-D1b seen earlier (Fig. 3B). The second notable polymorphism was an 8-bp deletion in the third exon of GhLMI1-D1b found only in normal (Fig. 3C). The exonic location of this deletion was confirmed through Sanger sequencing of GhLMI1-D1b okra cDNA. Translation of the resulting normal coding sequence (CDS) produces a frameshift 156 aa into the protein and a premature stop codon truncating the protein from 228 aa to 178 aa (see Fig. 5A). Neither of the conserved functional domains of an HD-Zip transcription factor appear directly impacted by the deletion. However, the frameshift introduces multiple leucines that may disturb the characteristic spacing of the leucine zipper and alter protein–protein interactions (see Fig. 5A).
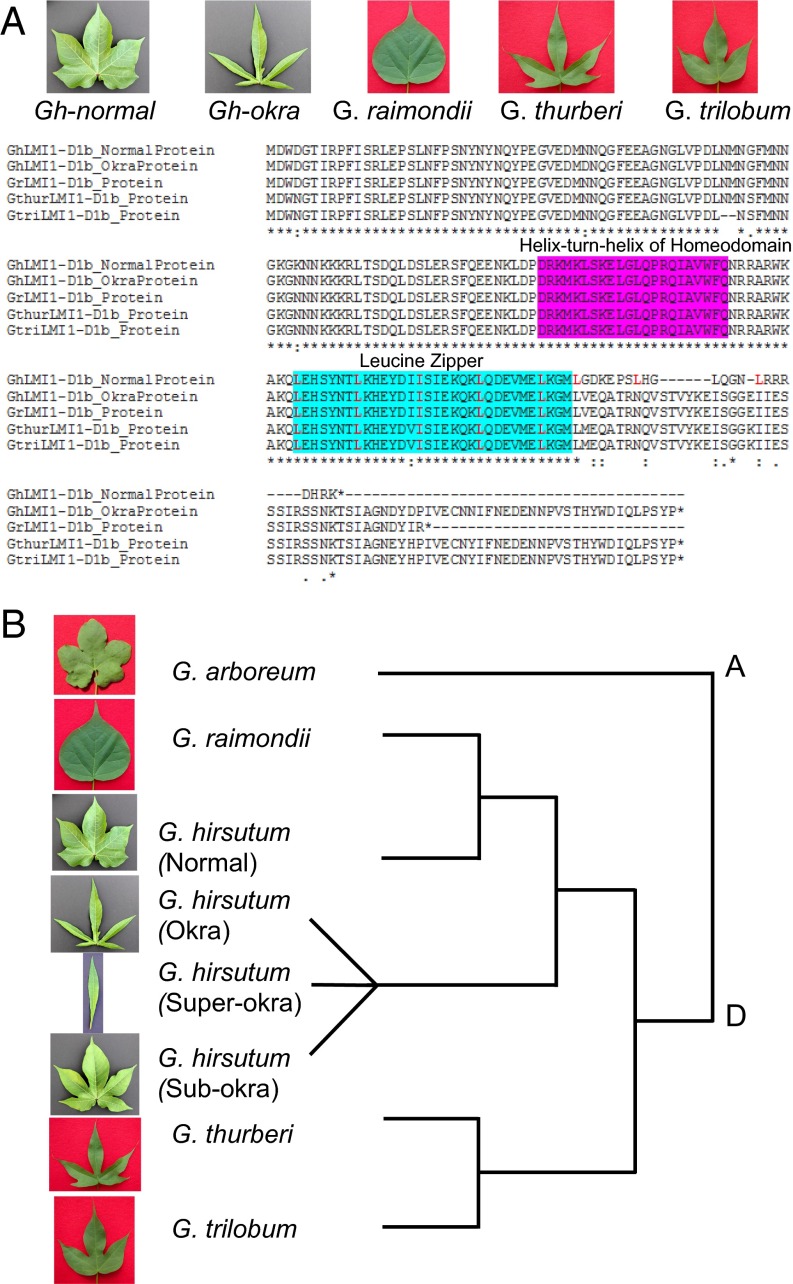
Fig. 5.
Functional prediction and phylogenetic analysis of LMI1-like genes among different cotton leaf shapes. (A) Amino acid translations in the Gossypium LMI1-D1b alleles. Normal in G. hirsutum and simple leaf in G. raimondii show truncated proteins while G. hirsutum-okra, G. trilobum and G. thurbeii with dissected leaves code for functional LMI1-D1b protein. Frameshift mutation resulting from 8bp deletion in normal introduces additional leucines (L) at 7 amino acid intervals (highlighted in red) that may interfere with the functionality of this domain. (B) Phylogenetic analysis showing the close relationship between okra, subokra, and superokra but not to G. thurberi or G. trilobum. Conversely, normal CDS appears more closely related to G. raimondii.
Both major polymorphisms in GhLMI1-D1b were covered by markers used in genotyping the association-mapping panel and the isogenic lines. The promoter duplication was assayed by the marker GhLS-STS1, whereas the 8-bp exonic deletion was underscored by the marker 13-LS-195. Combined with the gene expression differences, the complete association of these markers with leaf shape phenotype (SI Appendix, Tables S2 and S3) provided strong evidence that modifications to GhLMI1-D1b are responsible for the various leaf shapes of cotton.
Functional Characterization of GhLMI1-D1b Using Virus-Induced Gene Silencing (VIGS).
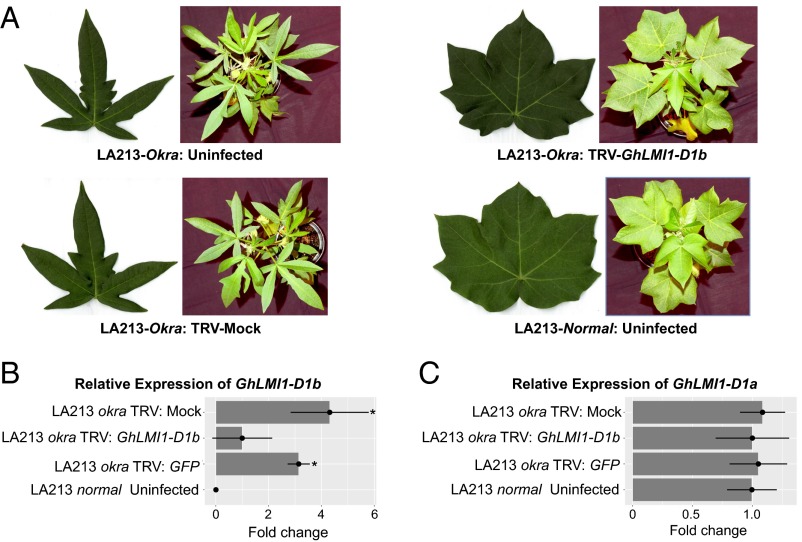
As expression analysis indicated the elevated expression of GhLMI1-D1b could be responsible for okra leaf shape (Fig. 3B), we hypothesized that silencing of GhLMI1-D1b in okra would reduce transcript levels and confer a simpler leaf shape. A 461-bp fragment of GhLMI1-D1bOkra encompassing 268 bp of the CDS and 193 bp of the 3′ UTR was used in VIGS. Including the 3′ UTR sequence was expected to minimize off-target silencing of other GhLMI1-like genes, especially GhLMI1-D1a. Because VIGS is environmentally sensitive (36) and can fluctuate over time (37), a TRV:CHLI treatment that blocks chlorophyll production was used as a visible marker to monitor and verify viral infection. Silencing of GhLMI1-D1b in an okra isoline led to a pronounced reduction in leaf lobing compared with uninfected and negative controls and a brief period of normal leaf production (Fig. 4A and SI Appendix, Fig. S7). Silencing of GhLMI1-D1b was eventually overcome, leading to a reversion to okra in a timeframe similar to the loss of silencing seen in the TRV:CHLI positive control (SI Appendix, Fig. S7).
Fig. 4.
Functional characterization of GhLMI1-D1b using VIGS. (A) Representative sixth true leaf and 4-wk-old plants from VIGS experiment showing the reversion to normal leaf shape in the GhLMI1-D1b silencing treatment. (B) Relative transcript levels of candidate genes in the GhLMI1-D1b silenced and control LA213-Okra plants (n = 3) confirmed the effective knockdown of GhLMI1-D1b. Asterisks represent statistically significant differences as determined by unpaired t tests at *P < 0.05. (C) Transcript levels of GhLMI1-D1a were unaffected by VIGS treatment, confirming silencing was specific to GhLMI1-D1b.
The level of GhLMI1-D1b transcript was substantially reduced in the TRV:GhLMI1-D1b treatment compared with the negative controls (Fig. 4B). This proved that knocking down the GhLMI1-D1b transcript through VIGS was sufficient to induce normal leaf formation in an okra variety. Furthermore, expression of GhLMI1-D1a was unaffected by VIGS (Fig. 4C), demonstrating specificity to GhLMI1-D1b. Thus, phenotyping and transcript profiling of TRV:GhLMI1-D1b leaves confirmed that altered expression of GhLMI1-D1b was responsible for the okra leaf shape of cotton.
Comparative Sequence Alignments of LMI1-Like Genes in Diploid and Tetraploid Cotton.
Outside of the promoter duplication, subokra, okra, and superokra GhLMI1-D1b were identical except for one promoter SNP unique to both okra and superokra (Fig. 3C). Comparative sequence alignments confirmed the close sequence relatedness of the subokra, okra, and superokra GhLMI1-D1b alleles (Fig. 5B). However, normal GhLMI1-D1b was considerably different from the other alleles, with six unique promoter SNPs and two SNPs in the second exon, both of which cause amino acid changes. Normal also carried a 1-bp deletion in the third exon, an additional third exon SNP, and a SNP in the 3′ region (Fig. 3C).
The 1-bp deletion would also cause a frameshift and premature stop codon if not for the preceding 8-bp deletion. Interestingly, the simple-leaved D genome donor G. raimondii (Fig. 5A) also carried this 1-bp deletion. Normal leaf GhLMI1-D1b also shares many of its other polymorphisms with Gorai.002G244000, including four out of the six promoter SNPs, the G→A SNP in the second exon, the T→A SNP in the third exon, and the 3′ UTR SNP. Comparative sequence alignments indicated that normal leaf GhLMI1-D1b CDSs show higher sequence similarity to the G. raimondii LMI1-D1b gene than to the alleles in other tetraploids with variable leaf shapes (Fig. 5B). GaLMI1-A1b from moderately lobed Gossypium aboreum is predicted to produce a full-length LMI1-like protein that is identical in length to those produced by the non-normal leaf shapes of tetraploid cotton.
To further assess the variability of LMI1-D1b genes in Gossypium, GhLMI1-D1b was Sanger sequenced in two additional D genome diploids, Gossypium thurberi and Gossypium trilobum. Both of these species have highly lobed leaves similar to G. hirsutum okra (Fig. 5A). Both of the G. thurberi and G. trilobum LMI1-D1b genes were full-length and similar to those found in the G. hirsutum leaf shape mutants and G. arboreum. Thus, although only a handful of species from the large Gossypium genus have been analyzed, there exists a trend that species with lobed leaves carry full-length LMI1-D1b genes whereas those with entire leaves produce truncated LMI1-1b genes (Fig. 5A). Extending beyond G. hirsutum, this provides evidence for a broad role of LMI1-1b genes in controlling the diversity of leaf shapes seen throughout Gossypium.
Transcriptome-Wide Changes Accompanying GhLMI1-D1b.
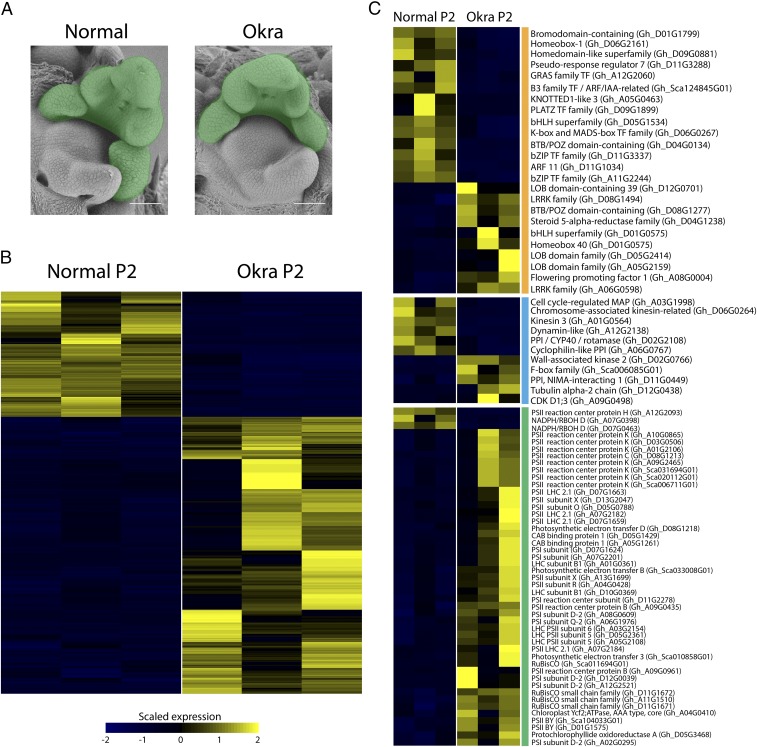
The morphological effects of okra leaf shape are evident by the P4 leaf primordia stage (Fig. 1G). To analyze gene expression changes preceding observable changes in leaf morphology, we used RNA-Seq to determine transcriptional changes at the P2 stage of leaf development (Fig. 6A). Our analyses uncovered 401 differentially expressed genes (false discovery rate ≤ 0.05) at the P2 stage between okra and normal BC8 isolines (Fig. 6B and Dataset S4). Gene Ontology (GO) enrichment analyses for this gene set indicated that microtubule and kinesin processes are down-regulated in okra relative to normal samples, whereas photosynthesis GO categories are highly enriched in okra relative to normal samples (SI Appendix, Fig. S8 and Dataset S6).
Fig. 6.
Transcriptomic comparison of okra versus normal leaf shapes in P2 stage primordia. (A) SEMs of normal and okra shoot apices. (Scale bar, 100 µm.) P2 stage leaf primordia (highlighted in green) were hand-dissected and processed for Illumina RNA-sequencing. (B) Heatmap visualization of the 401 genes that are significantly differentially expressed (false discovery rate ≤ 0.05) between normal and okra P2 samples (Datasets S4–S6). Scaled and centered reads per million values for normal and okra biological replicates are plotted in the left three columns and right three columns, respectively. Yellow indicates up-regulated and blue indicates down-regulated expression values. (C) Gene expression differences between normal and okra P2 samples for three functional categories are highlighted—developmental regulators in orange, cell cycle in blue, and photosynthesis-related transcripts in green.
To gain further insight into the molecular basis for the okra leaf phenotype, we examined differentially expressed genes that fall into three functional categories—photosynthesis-related transcripts, cell-cycle proteins, and putative developmental regulators (Fig. 6C). Congruent with the findings from our GO analyses, nearly all of the differentially expressed photosynthesis-related genes are up-regulated in the okra (44 genes) relative to the normal (three genes) transcriptomes. We were unable to identify any strong cell-cycle candidates from our differential gene expression analyses (Fig. 6C). However, our investigation of putative developmental genes that are associated with increased leaf complexity in okra revealed several transcripts with homology to known leaf development regulators (e.g., BTB/POZ domain-containing, Homeodomain-containing, and GRAS family transcription factors). Notably, three Lateral Organ Boundary (LOB) genes (Gh_D12G0701, Gh_D05G2414, and Gh_A05G2159) are significantly up-regulated in okra relative to normal P2 samples.
Subcellular/Nuclear Localization of GhLMI1-D1b.
Transient expression of 35S::GhLMI1-D1bOkra-GFP and pUBQ10::GhLMI1-D1bOkra-GFP in Nicotiana benthamiana was used to determine the subcellular localization of GhLMI1-D1bOkra. GhLMI1-D1bOkra-GFP was detected in the nucleus of transformed tobacco plants (SI Appendix, Fig. S9) in a pattern that colocalized with the nuclear marker RFP-Histone2B (SI Appendix, Fig. S9). Protein fusion experiments showed that GhLMI1-D1bOkra-GFP localized to the nucleus, consistent with the classification of LMI1-like genes as transcription factors (8, 9, 35).
Discussion
Leaf shape varies dramatically across plant evolution and in response to the environment. Understanding the genetic architecture of leaf shape is critical for harnessing its variation to modify plant physiology and improve agronomic profitability (2, 4). Using a diverse array of genomic and molecular tools, we established that the multiallelic, major leaf shape locus l-D1 of cotton is governed by GhLMI1-D1b encoding an HD-Zip transcription factor.
LMI1-like genes, in particular their duplication and regulatory diversification, have recently been proposed as evolutionary hotspots for leaf shape diversity in model plants (8, 9). The evidence presented here indicates that the major leaf shapes of cultivated cotton are controlled by the same pathway. The near-tandem duplication of LMI1-like genes in Gossypium (Fig. 2D) are unique from those previously described, as the Malvales and Brassicales diverged before duplications observed in the Brassicaceae (8, 9). Therefore, the separate LMI1-like duplication event in Gossypium indicates convergent evolution and strengthens the evidence that LMI1-like genes are evolutionary hot spots for modifying leaf shape (8, 9).
Sequence analyses of GhLMI1-D1b in a set of 20 cultivars elucidated two major polymorphisms among different leaf shapes at the l-D1 locus (Fig. 3C and SI Appendix, Tables S2–S4). First, an 8-bp deletion near the beginning of the third exon was found only in normal GhLMI1-D1b. This deletion results in a frameshift and premature stop codon in the predicted normal GhLMI1-D1b protein that may interfere with the function of the leucine zipper motif (Fig. 5A). Additionally, the C-terminal truncation may impact protein stability by removing residues necessary for proper folding. The second cosegregating polymorphism was a 133-bp tandem duplication ∼800 bp upstream of the GhLMI1-D1b translation start site (Fig. 3C and SI Appendix, Table S2). This duplication was present in all superokra and okra alleles but absent from all normal and subokra alleles. Furthermore, expression of GhLMI1-D1b was up-regulated in plants with okra and superokra leaf shapes (Fig. 3B). This finding is consistent with previous reports that promoter modifications to LMI1-like genes alter expression and leaf shape in model plants (8, 9). Finally, the down-regulation of GhLMI1-D1b by VIGS in a cotton accession with the okra leaf phenotype strongly reduced leaf lobing and resulted in the production of normal leaves (Fig. 4A).
Based on comparative sequence analysis, it appears that normal originated in G. raimondii LMI1-D1b (Fig. 5B). Owing to their shared 1-bp deletion, frameshift, and premature stop codon, this protein may already have been non- or partially functional. The subsequent 8-bp deletion, which causes an earlier frameshift and premature stop codon, would be expected to even further compromise the protein function. Sequence analysis indicated the other three leaf shape alleles are not derivatives of G. raimondii LMI1-D1b. This ancestral allele may have derived from a lobed D genome diploid, although likely not G. thurberi or G. trilobum (Fig. 5B). Once the subokra allele developed within the D genome, it likely gave rise to okra via a single duplication of 133 bp in the promoter region. The origin of this promoter duplication is currently unclear, but it may have arisen via unequal crossing over or replication slippage or due to transposable elements. Consistent with the expression results and gene silencing results obtained here, this promoter duplication led to overexpression of GhLMI1-D1b and an increase in the degree of leaf lobing and complexity. The finding of only a single additional promoter SNP in the entire GhLMI1-D1b genic region between subokra and okra indicates this event happened relatively recently. Only two promoter SNPs differentiate superokra from okra (Fig. 3C), consistent with the knowledge that superokra originated from okra within the last 90 y (15). Phylogenetic analysis involving sequences from all Gossypium species would establish the evolution and adaptive significance of LMI1 genes in cotton.
To address the molecular basis of the okra phenotype, we compared gene expression between okra and normal BC8 isolines using RNA-Seq at the P2 stage of leaf development (Fig. 6 and Dataset S4). GO enrichment analyses for the set of differentially expressed genes indicate that microtubule and kinesin processes are down-regulated in okra relative to normal samples, suggesting a cellular basis for the observed changes in leaf morphology (Dataset S5), whereas photosynthesis GO categories are highly enriched in okra relative to normal samples (SI Appendix, Fig. S8 and Dataset S6). This finding is in line with previous transcriptomic analyses from tomato shoot apices that revealed a positive correlation between photosynthetic-related gene expression and genetic changes in leaf complexity (33). The direction of this correlation is the same in okra (i.e., photosynthetic gene expression is positively correlated with increased leaf dissection), implying a broad connection between leaf morphology and the capacity for photosynthesis. Previous work from Cardamine hirsuta demonstrated that REDUCED COMPLEXITY (RCO), a homolog of GhLMI1-D1b, promotes leaf dissection by inhibiting cell division in the sinuses of young leaf primordia (8). We were unable to identify any strong cell-cycle candidates from our differential gene expression analyses (Fig. 6C). One possible explanation for this discrepancy is that cell-cycle repression in okra occurs at a later developmental stage or is not manifest through transcriptomic changes. Among our list of differentially expressed developmental genes between okra and normal leaves are several putative developmental regulators, which may be associated with leaf complexity such as BTB/POZ domain-containing, Homeodomain-containing, and GRAS family transcription factors. Additionally, three LOB genes, which are known to delineate organ boundaries, including the formation of lobes and serrations (38), are significantly up-regulated in okra relative to normal P2 samples (Gh_D12G0701, Gh_D05G2414, and Gh_A05G2159), suggesting that these genes may act downstream of GhLMI1-D1b to pattern the okra phenotype.
Okra is an exceptional mutation affecting leaf development. It inspired an early quantitative framework for leaf shape across evolution and development (12, 23) and, through chimeric studies, revealed some insights into the morphogenetic and cellular basis of leaf morphology (24, 25). In two separate instances within the Brassicaceae, the homologs of GhLMI1-D1b have been implicated in evolutionary shifts in leaf morphology (8, 9). That the okra locus confers a monogenic basis to most of the leaf shape variance in cotton, the mechanisms for which have been studied in other model organisms, and that it is implicated in the productivity of a major crop demonstrate a unique intersection between agriculture and the evolutionary and developmental basis of leaf morphology.
Materials and Methods
Plant Material.
To fine-map the l-D1 locus, we used parental accessions NC05AZ21 and NC11-2100 (TX-2324; PI607650) (SI Appendix, Fig. S10), which were used previously in the preliminary mapping study (31). A total of 1,027 F2 plants derived from the original Okra (NC05AZ21) × Normal (NC11-2100) cross were used to identify recombinants for fine-mapping the l-D1 locus. A 538-member diversity panel was used for association mapping and genome-wide association studies. This panel consisted of the 384-member cotton diversity panel (39), plus 154 wild and landrace accessions, all of which were obtained from USDA Cotton Germplasm Collection (Dataset S1). Two sets of isolines were used in fine-mapping and/or in gene expression and VIGS studies, a BC8 set that included all four leaf shapes in the Stoneville 213 background (40) and a BC3 pair of normal and okra in the Stoneville 7A background (41).
Morphometric Analysis.
Four leaves from each of the accessions in the cotton diversity panel were sampled from a field in Central Crops Research Station, Clayton, North Carolina mid-August 2015. Leaves were arranged on a scanner (Epson Workforce DS-50000) with a ruler, and the abaxial side of the leaf was scanned. Files were named by the order they were scanned and appended with a field number corresponding to genotype information. In ImageJ (42), the “Make Binary,” “Fill Holes,” “Open,” “Close-,” and “Image Inverter” functions were used to convert leaves to binary, polished objects. Individual binary leaves were then manually selected using the “Wand” tool and copied and pasted into individual files named by genotype. Binary leaf silhouettes were converted to chain code using the program SHAPE (43, 44). The nef code file from SHAPE (.nef file) was then imported into the Momocs package in R (45–48) using the NEF2COE function. Individual leaf contours in the Coe object were assigned phenotype factor levels of normal, subokra, okra, and superokra and harmonics isolated for subsequent analyses. The PC.contrib() and pca() functions in Momocs were used to visualize eigenleaves and perform PCA (respectively) on harmonics, and the morpho.space() function was used to visualize the morphospace (47). LDA on harmonic coefficients was performed using the lda function in conjunction with the MASS package (49). The predict function (stats package) and table function (base package) were used to reallocate leaves by their predicted phenotypic class. R package ggplot2 (50) was used for all data visualizations unless indicated otherwise.
Cryo-SEM of Okra and WT SEMs.
For comparisons of P2, vegetative shoot apices were hand-dissected from 4-wk-old normal, okra, and superokra BC8 isolines (40) to expose the shoot apical meristem and the two most recently initiated leaf primordia: P1 and P2. Apices were affixed to SEM stubs using cryo-glue, frozen in liquid nitrogen, and viewed using a Hitachi TM-1000 tabletop scanning electron microscope. Image contrast adjustment and scale addition were done in Fiji (fiji.sc/).
Association Mapping.
The association mapping population consisted of the 384-member elite cotton diversity panel (39) and 154 wild and landrace accessions of tetraploid cotton, some of which were sensitive to long-day photoperiod conditions. Of this population, a diversity panel of 447 photoperiod insensitive lines were grown under summer field conditions in Clayton, North Carolina, whereas the 91 photoperiod-sensitive accessions were grown in 10-inch single pots in the greenhouse under short-day conditions. All plants were phenotyped as described previously (31).
A total of 47 STS markers were designed from the 10-gene candidate region (Fig. 2C). Three were polymorphic and run on the association mapping panel. Additionally, two SNPs were converted into a Kompetitive Allele Specific PCR (KASP) assay and analyzed on the population at the Eastern Regional Small Grains Genotyping Laboratory. Marker locations are summarized in SI Appendix, Fig. S1, and primer sequences are provided in SI Appendix, Table S5. DNA isolation and genotyping using SSRs and STS markers were done as described previously (31). All of the primer pairs were synthesized by Integrated DNA Technologies.
To verify that the associations between leaf shape and the candidate genes tested were not due to population structure, 149 multiallelic SSR markers distributed throughout the genome were also analyzed on the 384-line diversity panel as well as 42 of the additional lines with okra leaf shape from the larger set tested above. Multiallelic SSR genotypes were converted to numeric allele content scores (0, 1, or 2) using the “Expand Multiallelic Genotypes” option in JMP Genomics version 8 (SAS Institute). PCA was used to estimate population structure of the diversity panel. After initial analysis of population structure, it was clear that 17 wild accessions were distinct outliers along the first principal component axis (which accounted for 23% of the marker variation). In addition, only one line exhibited subokra phenotype and one line exhibited superokra phenotype. The wild accessions and sub- and superokra types were excluded from further association analyses, resulting in 54 SSR markers being monomorphic in the remaining sample of lines. These SSRs were excluded from further analysis, and PCA was performed again on the remaining sample of 404 lines and 95 SSR markers, which included 36 okra types and 368 normal leaf shape types.
All markers were then tested for association with the binary trait okra versus normal leaf shape using a logistic regression model in the PROC LOGISTIC procedure in SAS software version 9.4 (SAS Institute). Population structure was controlled by including the first three principal components (explaining 9% of variation in marker profiles) as covariates in the model. To deal with complete and quasicomplete separation observed in some markers, we used the FIRTH option available in SAS software, which produces finite parameter estimates by means of penalized maximum likelihood estimation (51).
In the initial scan, several candidate gene region markers and several SSRs had significant associations with leaf shape. Inspection of the marker data revealed that genotypes at significant SSRs were correlated with the candidate gene region marker genotypes. Therefore, we performed a second scan of all SSR markers in which the most significant candidate gene variant (GhLS-STS1) was included as an additional covariate in the model.
Semiquantitative Expression Analysis.
Total RNA was collected from three field-grown plants [n (number of replications) = 3] each of six varieties: NC05AZ21 (okra), NC11-2100 (normal), LA213-63 (normal recurrent parent), LA213 Sea Island Leaf (subokra), LA213 okra, and LA213 superokra. Samples were taken ∼90 d after planting. Leaves were taken at the earliest possible time they could reliably be distinguished from the shoot apical meristem without the help of any equipment. At this time point, leaves were ∼30–50 mm in length from tip to base.
RNA was isolated using the Spectrum Plant Total RNA Kit (Sigma-Aldrich) following the manufacturer’s instructions. Total RNA was converted to cDNA using the ImProm-II Reverse Transcription System (Promega) per the manufacturer’s instructions. cDNA was then used as template in 50 µL PCR reactions and visualized on 3% (wt/vol) HiRes agarose (GeneMate BioExpress). GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE (GAPDH) was used as the reference gene. Primers used in semiquantitative expression analysis are provided in SI Appendix, Table S6.
Real-Time Quantitative PCR Analysis of GhLMI1-Like Gene Expression.
cDNA from LA213 isolines in the preceding section was used in 25 µL Real-Time qPCR reactions with Power SYBR Green PCR Master Mix (Applied Biosystems) and the ABI 7300 Real-Time PCR System (Applied Biosystems). The positive control/reference gene was UBI14 (52). Technical replicates were run in triplicate. Ct values were analyzed using the ΔΔCt method (53). Fold changes [=2^(–ΔΔCt)] (53) and their SDs were plotted using Microsoft Excel. Unpaired t tests were calculated between isolines or VIGS experimental treatments to detect significant changes in gene expression. Primers used in RT-qPCR are provided in SI Appendix, Table S6.
Sequencing of GhLMI1-Like Genes.
Sanger sequencing was used to obtain the genomic DNA sequence of GhLMI1-D1a and GhLMI1-D1b in 20 different tetraploid Gossypium accessions listed in SI Appendix, Table S4. Sequencing was performed at the North Carolina State University Genomic Sciences Laboratory. Genome-specific primers for PCR were developed by aligning homeologous sequences from G. raimondii and G. arboreum and targeting differences between the two donor diploid genomes (SI Appendix, Tables S6–S8). Genome specificity of the primers was confirmed by analyzing amplification in a panel of diploid species from both genomes (SI Appendix, Table S9). Nested PCR was necessary for the 5′ end of GhLMI1-D1a. A complete list of the primers used for Sanger Sequencing can be found in SI Appendix, Table S10. Sequencing results were analyzed and assembled using Sequencher 5.2.3 (Gene Codes) with additional alignments performed using Clustal Omega Multiple Sequence Alignment (www.ebi.ac.uk/Tools/msa/). Publicly available predictions for Gorai.002G244200 and Gorai.002G244000 (Phytozome, https://phytozome.jgi.doe.gov/) were used to determine the exon/intron structure of GhLMI1-like genes with expansions to a stop codon when necessary. Protein translations were performed using the ExPASy Translate Tool (web.expasy.org/translate/). Renderings of GhLMI1-like genes were drawn with fancyGENE (bio.ieo.eu/fancygene/) and redrawn to scale in Microsoft PowerPoint.
VIGS of GhLMI1-D1b in LA213 Okra.
All enzymes used in the construction of the TRV silencing vectors were supplied by New England Biolabs. Kits with ‘Zymo’ in the name were supplied by Zymo Research. To construct TRV2:GhLMI1-D1b, a 461-bp fragment of the GhLMI1-D1b gene was amplified from cDNA derived from LA213 Okra. This 461-bp silencing fragment of GhLMI1-D1b included the last 29 bp of the second exon, the entire 239 bp of the third exon, and the first 193 bp of the proposed 3′ UTR. This fragment, along with the pYL156 (TRV2) vector (obtained from the Arabidopsis Biological Resource Center), was then digested with the restriction enzyme Acc65I overnight according to the manufacturer’s instructions. Following the digestion, the restriction enzyme was inactivated by placing the reactions at 65 °C for 20 min. Digested vector DNA was then dephosphorylated with Antarctic Phosphatase per the manufacturer’s instructions. Both digested vector and GhLMI1-D1b fragment were separated on a 0.8% agarose gel, excised, and purified using the Zymoclean Gel DNA Recovery Kit and ligated using T4 DNA ligase. When combined with the TRV1 vector of the bipartite TRV VIGS system, this treatment was named TRV:GhLMI1-D1b.
In addition to the TRV:GhLMI1-D1b experimental treatment, two negative controls were used as before (54). TRV:Mock consisted of only an empty TRV2 vector, which does not support the spread of infection due to the absence of TRV1 and thereby controls for effects of the inoculation process. TRV:GFP, consisting of TRV1 plus TRV2 containing a silencing fragment for GFP, is capable of spreading throughout the plant. However, cotton lacks an endogenous GFP gene so that potential effects of infection only could be monitored. Because silencing from TRV can fluctuate over time (37) and TRV VIGS is very sensitive to temperature and humidity (36), a TRV:CHLI treatment that blocks chlorophyll production was used as a visible marker to ensure that environmental conditions were suitable for VIGS and to time the phenotyping of LMI knockdowns.
Two of the VIGS control constructs, TRV:GFP and TRV:ChlI, were produced by digesting the TRV2 plasmid pYL156 with Acc65I. The ends of the digested vector were blunted using DNA Polymerase I, Large (Klenow) Fragment and dephosphorylated with Antarctic Phosphatase. A 499-bp GFP fragment flanked with StuI restriction sites was amplified from transgenic cotton carrying the mGFP5-ER transgene (55) using the primers mGFPerF: 5′-ATA AGG CCT GTG ATG CAA CAT ACG GAA AAC-3′ and mGFPerR2: 5′-ATT AGG CCT AGG TAA TGG TTG TCT GGT AAA AG-3′. The GFP PCR products were desalted using the Zymo DNA Clean and Concentrator kit. The cleaned PCR product was then digested with StuI. A 501-bp blunt-ended fragment of the cotton ChlI gene was digested out of pJRT.CLCrVA.009 (55) using MscI, gel-purified, and extracted from the agarose gel using the Zymoclean Gel DNA Recovery Kit. Blunt-ended GFP and ChlI gene fragments were ligated into the TRV2 vector using T4 DNA ligase.
All vector constructs were transformed into DH10-beta competent cells (New England Biolabs) and plated on Luria–Bertani agar plates containing 50 µg/mL each of kanamycin and gentamicin. Transformants were screened for insert direction using the following primers: TRV2MCSF, 5′-CTT AGA TTC TGT GAG TAA GGT TAC C-3′ and mGFPerR2, 5′ ATT AGG CCT AGG TAA TGG TTG CT GGT AAA AG 3′ for TRVGFP; and TRV2MCSF, 5′-CTT AGA TTC TGT GAG TAA GGT TAC C-3′, and GhChlIR, 5′ GCT TGG CCA ATC AAA CCG TGC TCT TT-3′ for TRVChlI. Positive clones were confirmed by sequencing.
Two-week-old okra seedlings were agro-inoculated as described previously (56). In each experimental replicate, five plants were inoculated per treatment with TRV:Mock, TRV:GFP, and TRV:GhLMI1-D1b. Additionally, at least two plants in each replication were inoculated with TRV:CHLI. Plants were grown under a 26/22 °C day/night cycle. Starting at 3 wk postinoculation, all plants were photographed weekly. At 4 wk postinoculation, leaves were collected randomly from three of the five plants (n = 3) in the TRV:GhLMI1-D1b, TRV:GFP, and TRV:Mock treatments for expression analysis as described above. Three experimental replicates of the VIGS experiments were performed with consistent results. All primers used in the VIGS experiment are listed in SI Appendix, Table S6.
Comparative Sequence Analysis of LMI1-Like Genes in Gossypium.
In addition to sequencing GhLMI1-like genes in tetraploid cotton, LMI1-D1b was sequenced in lobed d-genome diploid species G. thurberi (PIs 530766 and 530789) and G. trilobum (PI 530967). Sequences of LMI1-A1b and LMI1-D1b, pulled from G. arboreum and G. raimondii, respectively, were obtained from https://www.cottongen.org/. Alignment and comparative sequence analysis were performed using Clustal Omega. Helix-turn-helix prediction was carried out using https://npsa-prabi.ibcp.fr/cgi-bin/primanal_hth.pl. Leucine zipper was predicted using 2zip.molgen.mpg.de/.
GhLMI1-D1b cDNA isolated from the okra leaf mapping population parent NC05AZ21 was sequenced via Sanger sequencing as described previously. Analysis, assembly, and alignment of the sequence were as described previously for the genomic sequencing of the LMI1-like genes. Extraction of RNA and conversion to cDNA are described in Semiquantitative Expression Analysis. Alignment of cDNA sequence to gDNA sequence confirmed the predicted exon/intron structure.
RNA-Seq Transcriptome Analysis.
Sample harvesting and RNA preparation for RNA-seq.
Three biological replicates of P2, corresponding to leaf 8, were hand-dissected directly into ice-cold acetone from normal and okra shoot apices. Ten individuals were pooled for each biological replicate. Acetone was removed from the samples and replaced with extraction buffer from the PicoPure RNA Isolation Kit (ThermoFisher Scientific). RNA was isolated following the manufacturer’s protocol with the optional on-column DNase treatment. The RNA integrity was assessed by running the samples on an Agilent RNA 6000 Pico Chip (Agilent Technologies). The Clontech SMARTer cDNA synthesis kit (Clontech) was used to amplify 10 ng of total RNA into polyA tail-enriched dscDNA. A total of 150 ng of dscDNA was fragmented for 17 min with Fragmentase (New England Biolabs) and processed into Illumina sequencing libraries using the NEBNext Ultra DNA Library Prep Kit for Illumina (New England Biolabs). Illumina libraries were quantified with a Nanodrop and pooled to a final concentration of 20 nM. The pooled libraries were sequenced for single-end 100-bp reads on an Illumina HiSEq. 2500 at the Washington University in St. Louis School of Medicine Genome Technology Access Center (https://gtac.wustl.edu/).
Bioinformatic processing of RNA-seq data.
Illumina adapters and low-quality bases were trimmed using Trimmomatic (57) with the following default parameters: LEADING: 3 TRAILING: 3 SLIDINGWINDOW: 4:15 MINLEN: 36. Trimmed reads were aligned to the G. hirsutum AD1_NBI genome and to the G. raimondii (D5) BGI-CGP Genome v1.0 (https://www.cottongen.org/data/download/genome) (58, 59) to produce Sequence Alignment/Map (SAM) files using the HISAT2 alignment software (60) with the following alignment option: –dta-cufflinks. SAM files were converted into compressed and sorted Binary Alignment/Map (BAM) files using Samtools –view, -bSh, and -sort-sort commands, respectively (samtools.sourceforge.net/) (61). Reads mapping to annotated genes for the AD1_NBI and D5 BGI-CGP genomes were extracted using StringTie (https://ccb.jhu.edu/software/stringtie/) (62). Genes with less than one count per million across at least three samples were discarded from the analysis. Significantly differentially expressed genes (FDR adjusted P value ≤ 0.05) were identified by performing a pairwise comparison between normal and okra P2 samples in edgeR version 3.0 (https://bioconductor.org/packages/3.0/bioc/html/edgeR.html) (63, 64).
GO enrichment.
The R package TopGO (65) was used to test for GO category enrichment in differentially expressed genes between okra and normal P2 transcriptomes. The Fisher’s Exact Test (P value ≤ 0.05) was used to identify significantly enriched GO categories in gene sets that are significantly differentially expressed between okra and normal P2 samples.
Colocalization of GhLMI1-D1bOkra-GFP in N. benthamiana.
Fluorescent protein fusions of GhLMI1-D1b were generated by Gateway cloning (Life Technologies). The GhLMI1-D1bOkra CDS without the STOP codon was amplified from okra cDNA using primers GhLMI1-D1b-Okra-TOPO-F (5′-CACCATGGATTGGGATGGCACCATTCGACCCTTT-3′) and GhLMI1-D1b-Okra-STOP-R (5′-GGGATAAGAAGGGAGTTGAA-3′) and cloned into pENTR/D/TOPO (Life Technologies) to generate pENTR::GhLMI1-D1b-Okra-stop. Recombination with LR Clonase (Invitrogen) was carried out between pENTR::GhLMI1-D1b-Okra-stop and the following destination vectors: pGWB5 (66), pGWB8 (66), and pUBQ10-C-GFP. pUBQ10-C-GFP is a modified version of pUBC-GFP (67) but includes the full pUBQ10 promoter. The following constructs were obtained and confirmed by sequencing: 35S::GhLMI1-D1b-GFP and pUBQ10::GhLMI1-D1b-GFP. Both constructs were introduced by transient Agrobacterium transformation into N. benthamiana plants carrying a RFP-Histone2B marker (68) for colocalization analysis.
A Zeiss LSM 710 confocal microscope with a 40× water objective (1.1 N.A.) was used to image fluorescence protein fusions. The excitation/emission wavelengths during acquisition were 488 nm/492–570 nm for GFP and 561 nm/588–696 nm for RFP.
Supplementary Material
Acknowledgments
We appreciate the technical help by Jared Smith and Sharon Williamson of United States Department of Agriculture-Agricultural Research Service and Cathy Herring of North Carolina Department of Agriculture as well as Drs. Richard Percy and James Frelichowski of the United States Department of Agriculture-National Cotton Germplasm Collection for supplying the cotton accessions used in this research. Cotton Incorporated supported this work through its core funding and PhD Fellowship program. For additional support of this research, we thank the North Carolina Cotton Growers Association Inc. and National Science Foundation Grants IOS-1238014 (to J.B.H.) and IOS-1025947 (to C.H.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KY293601 to KY293643).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1613593114/-/DCSupplemental.
References
- 1.Bell AD, Bryan A. Plant Form: An Illustrated Guide to Flowering Plant Morphology. Timber; London: 2008. [Google Scholar]
- 2.Nicotra AB, et al. The evolution and functional significance of leaf shape in the angiosperms. Funct Plant Biol. 2011;38(7):535–552. doi: 10.1071/FP11057. [DOI] [PubMed] [Google Scholar]
- 3.Tsukaya H. Leaf development. Arabidopsis Book. 2013;11:e0163. doi: 10.1199/tab.0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chitwood DH, Sinha NR. Evolutionary and environmental forces sculpting leaf development. Curr Biol. 2016;26(7):R297–R306. doi: 10.1016/j.cub.2016.02.033. [DOI] [PubMed] [Google Scholar]
- 5.Kimura S, Koenig D, Kang J, Yoong FY, Sinha N. Natural variation in leaf morphology results from mutation of a novel KNOX gene. Curr Biol. 2008;18(9):672–677. doi: 10.1016/j.cub.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 6.Piazza P, et al. Arabidopsis thaliana leaf form evolved via loss of KNOX expression in leaves in association with a selective sweep. Curr Biol. 2010;20(24):2223–2228. doi: 10.1016/j.cub.2010.11.037. [DOI] [PubMed] [Google Scholar]
- 7.Tian F, et al. Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat Genet. 2011;43(2):159–162. doi: 10.1038/ng.746. [DOI] [PubMed] [Google Scholar]
- 8.Vlad D, et al. Leaf shape evolution through duplication, regulatory diversification, and loss of a homeobox gene. Science. 2014;343(6172):780–783. doi: 10.1126/science.1248384. [DOI] [PubMed] [Google Scholar]
- 9.Sicard A, et al. Repeated evolutionary changes of leaf morphology caused by mutations to a homeobox gene. Curr Biol. 2014;24(16):1880–1886. doi: 10.1016/j.cub.2014.06.061. [DOI] [PubMed] [Google Scholar]
- 10.Jöst M, et al. The INDETERMINATE DOMAIN protein BROAD LEAF1 limits barley leaf width by restricting lateral proliferation. Curr Biol. 2016;26(7):903–909. doi: 10.1016/j.cub.2016.01.047. [DOI] [PubMed] [Google Scholar]
- 11.Wendel JF, Brubaker C, Alvarez I, Cronn R, Stewart JM. Evolution and natural history of the cotton genus. In: Paterson AH, editor. Genetics and Genomics of Cotton. Springer-Verlag; New York: 2009. pp. 3–19. [Google Scholar]
- 12.Hammond D. The expression of genes for leaf shape in Gossypium hirsutum L. and Gossypium arboreum L. I. The expression of gene for leaf shape in Gossypium hirsutum L. Am J Bot. 1941;28(2):124–137. [Google Scholar]
- 13.Hutchinson JB, Silow RA, Stephens SG. Evolution of Gossypium. Oxford Univ Press; London: 1947. [Google Scholar]
- 14.Saunders JH. The Wild Species of Gossypium. Oxford Univ Press; London: 1961. [Google Scholar]
- 15. Andres RJ, Bowman DT, Jones DC, Kuraparthy V, Major leaf shapes of cotton: Genetics and agronomic effects in crop production. J Cotton Sci, in press.
- 16.Stephens SG. A genetic survey of leaf shape in new world cottons – A problem in critical identification of alleles. J Genet. 1945;46(2):313–330. [Google Scholar]
- 17.Shoemaker DN. A study of leaf characters in cotton hybrids. J Hered. 1909;5(1):116–119. [Google Scholar]
- 18.McClendon CA. Mendelian inheritance in cotton hybrids. Georgia Experiment Station Bulletin. 1912;99:139–228. [Google Scholar]
- 19.Harland SC. The genetics of Gossypium. Bibliographica Genetica. 1932;9:107–182. [Google Scholar]
- 20.Hutchinson JB, Silow RA. Gene symbols for use in cotton genetics. J Hered. 1939;30(10):461–464. [Google Scholar]
- 21.Green JM. Sub-okra, a new leaf shape in Upland cotton. J Hered. 1953;44(6):229–232. [Google Scholar]
- 22.Kaur B, Andres RJ, Kuraparthy V. Major leaf shape genes, laciniate in diploid cotton and okra in polyploid Upland cotton, map to an orthologous genomic region. Crop Sci. 2016;56(3):1095–1105. [Google Scholar]
- 23.Hammond D. The expression of genes for leaf shape in Gossypium hirsutum L. and Gossypium arboreum L. II. The expression of gene for leaf shape in Gossypium arboreum L. Am J Bot. 1941;28(2):138–150. [Google Scholar]
- 24.Dolan L, Poethig RS. Genetic analysis of leaf development in cotton. Development. 1991;113:39–46. [PubMed] [Google Scholar]
- 25.Dolan L, Poethig R. The OKRA leaf shape mutation in cotton is active in all cell layers of the leaf. Am J Bot. 1998;85(3):322–327. [PubMed] [Google Scholar]
- 26.Endrizzi JE, Brown MS. Identification of monosomes for six chromosomes in Gossypium hirsutum. Am J Bot. 1964;51(2):117–120. [Google Scholar]
- 27.Endrizzi JE, Kohel RJ. Use of telosomes in mapping three chromosomes in cotton. Genetics. 1966;54(2):535–550. doi: 10.1093/genetics/54.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang C, Wright RJ, Woo SS, DelMonte TA, Paterson AH. QTL analysis of leaf morphology in tetraploid Gossypium (cotton) Theor Appl Genet. 2000;100(3):409–418. [Google Scholar]
- 29.Song XL, Guo WZ, Han ZG, Zhang TZ. Quantitative trait loci mapping of leaf morphological traits and chlorophyll content in cultivated tetraploid cotton. J Integr Plant Biol. 2005;47(11):1382–1390. [Google Scholar]
- 30.Lacape JM, et al. Mapping QTLs for traits related to phenology, morphology, and yield components in an inter-specific Gossypium hirsutum x G. barbadense cotton RIL population. Field Crops Res. 2013;144:256–267. [Google Scholar]
- 31.Andres RJ, Bowman DT, Kaur B, Kuraparthy V. Mapping and genomic targeting of the major leaf shape gene (L) in Upland cotton (Gossypium hirsutum L.) Theor Appl Genet. 2014;127(1):167–177. doi: 10.1007/s00122-013-2208-4. [DOI] [PubMed] [Google Scholar]
- 32.Zhu QH, et al. Integrated mapping and characterization of the gene underlying the okra leaf trait in Gossypium hirsutum L. J Exp Bot. 2016;67(3):763–774. doi: 10.1093/jxb/erv494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chitwood DH, et al. A quantitative genetic basis for leaf morphology in a set of precisely defined tomato introgression lines. Plant Cell. 2013;25(7):2465–2481. doi: 10.1105/tpc.113.112391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paterson AH, et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature. 2012;492(7429):423–427. doi: 10.1038/nature11798. [DOI] [PubMed] [Google Scholar]
- 35.Saddic LA, et al. The LEAFY target LMI1 is a meristem identity regulator and acts together with LEAFY to regulate expression of CAULIFLOWER. Development. 2006;133(9):1673–1682. doi: 10.1242/dev.02331. [DOI] [PubMed] [Google Scholar]
- 36.Fu DQ, et al. Enhancement of virus-induced gene silencing in tomato by low temperature and low humidity. Mol Cells. 2006;21(1):153–160. [PubMed] [Google Scholar]
- 37.Senthil-Kumar M, Mysore KS. Virus-induced gene silencing can persist for more than 2 years and also be transmitted to progeny seedlings in Nicotiana benthamiana and tomato. Plant Biotechnol J. 2011;9(7):797–806. doi: 10.1111/j.1467-7652.2011.00589.x. [DOI] [PubMed] [Google Scholar]
- 38.Majer C, Hochholdinger F. Defining the boundaries: Structure and function of LOB domain proteins. Trends Plant Sci. 2011;16(1):47–52. doi: 10.1016/j.tplants.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 39.Tyagi P, et al. Genetic diversity and population structure in the US Upland cotton (Gossypium hirsutum L.) Theor Appl Genet. 2014;127(2):283–295. doi: 10.1007/s00122-013-2217-3. [DOI] [PubMed] [Google Scholar]
- 40.Kennedy CW, Smith WC, Jones JE. Effect of early season square removal on three leaf types of cotton. Crop Sci. 1986;26(1):139–145. [Google Scholar]
- 41.Andries JA, Jones JE, Sloane LW, Marshall JG. Effects of okra leaf shape on boll rot, yield, and other important characters of Upland cotton, Gossypium hirsutum L. Crop Sci. 1969;9(6):705–710. [Google Scholar]
- 42.Abràmoff MD, Magalhães PJ, Ram SJ. Image processing with ImageJ. Biophoton Int. 2004;11(7):36–42. [Google Scholar]
- 43.Iwata H, Niikura S, Matsuura S, Takano Y, Ukai Y. Evaluation of variation of root shape of Japanese radish (Raphanus sativus L.) based on image analysis using elliptic Fourier descriptors. Euphytica. 1998;102(2):143–149. [Google Scholar]
- 44.Iwata H, Ukai Y. SHAPE: A computer program package for quantitative evaluation of biological shapes based on elliptic Fourier descriptors. J Hered. 2002;93(5):384–385. doi: 10.1093/jhered/93.5.384. [DOI] [PubMed] [Google Scholar]
- 45.Claude J. Morphometrics with R. Springer; New York: 2008. [Google Scholar]
- 46.Claude J. Log-shape ratios, Procrustes superimposition, elliptic Fourier analysis: Three worked examples in R. Hystrix. 2013;24(1):94–102. [Google Scholar]
- 47.Sankaran K, Holmes S. structSSI: Simultaneous and Selective Inference for Grouped or Hierarchically Structured Data. J Stat Softw. 2014;59(13):1–21. doi: 10.18637/jss.v059.i13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. R Development Core Team (2011) R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, Austria), www.R-project.org.
- 49.Venables WN, Ripley BD. Modern Applied Statistics with S. 4th Ed Springer; New York: 2002. [Google Scholar]
- 50.Wickham H. 2009. ggplot2: Elegant Graphics for Data Analysis. (Springer-Verlag, New York)
- 51.Firth D. Bias reduction of maximum likelihood estimates. Biometrika. 1993;80(1):27–38. [Google Scholar]
- 52.Artico S, Nardeli SM, Brilhante O, Grossi-de-Sa MF, Alves-Ferreira M. Identification and evaluation of new reference genes in Gossypium hirsutum for accurate normalization of real-time quantitative RT-PCR data. BMC Plant Biol. 2010;10:49. doi: 10.1186/1471-2229-10-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 54.Tuttle JR, Idris AM, Brown JK, Haigler CH, Robertson D. Geminivirus-mediated gene silencing from Cotton leaf crumple virus is enhanced by low temperature in cotton. Plant Physiol. 2008;148(1):41–50. doi: 10.1104/pp.108.123869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sunilkumar G, Mohr L, Lopata-Finch E, Emani C, Rathore KS. Developmental and tissue-specific expression of CaMV 35S promoter in cotton as revealed by GFP. Plant Mol Biol. 2002;50(3):463–474. doi: 10.1023/a:1019832123444. [DOI] [PubMed] [Google Scholar]
- 56.Tuttle JR, Haigler CH, Robertson D. Method: Low-cost delivery of the cotton leaf crumple virus-induced gene silencing system. Plant Methods. 2012;8(1):27. doi: 10.1186/1746-4811-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang T, et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol. 2015;33(5):531–537. doi: 10.1038/nbt.3207. [DOI] [PubMed] [Google Scholar]
- 59.Wang K, et al. The draft genome of a diploid cotton Gossypium raimondii. Nat Genet. 2012;44(10):1098–1103. doi: 10.1038/ng.2371. [DOI] [PubMed] [Google Scholar]
- 60.Kim D, Langmead B, Salzberg SL. HISAT: A fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11(9):1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou X, Lindsay H, Robinson MD. Robustly detecting differential expression in RNA sequencing data using observation weights. Nucleic Acids Res. 2014;42(11):e91. doi: 10.1093/nar/gku310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Alexa A, Rahnenführer J. 2016 Gene Set Enrichment Analysis with topGO. https://www.bioconductor.org/packages/3.3/bioc/vignettes/topGO/inst/doc/topGO.pdf.
- 66.Nakagawa T, et al. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng. 2007;104(1):34–41. doi: 10.1263/jbb.104.34. [DOI] [PubMed] [Google Scholar]
- 67.Grefen C, et al. A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J. 2010;64(2):355–365. doi: 10.1111/j.1365-313X.2010.04322.x. [DOI] [PubMed] [Google Scholar]
- 68.Martin K, et al. Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J. 2009;59(1):150–162. doi: 10.1111/j.1365-313X.2009.03850.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.