Significance
Pancreatic ductal adenocarcinoma cancer (PDAC) cells have an exceptional propensity to invade nerves via pronounced crosstalk between nerves and cancer cells, but the mechanisms of this early neural invasion are yet unknown. By using genetically engineered mouse models, we show that in the precursor stage PDAC induces the generation of ready-to-use nerves for dissemination by secreting the chemokine CXCL12 that attracts glia (Schwann) cells of nerves. This migration of glia cells to cancerous cells at this very early stage intriguingly attenuates cancer-associated pain via downregulation of pain-associated targets in Schwann cells and via suppression of central glia. Hence, malignant transformed cells seem to disguise cancer-associated symptoms (such as pain) actively and thereby delay the early diagnosis of cancer.
Keywords: Schwann cells, pancreatic cancer, CXCL12, CXCR4, CXCR7
Abstract
Pancreatic ductal adenocarcinoma (PDAC) cells (PCC) have an exceptional propensity to metastasize early into intratumoral, chemokine-secreting nerves. However, we hypothesized the opposite process, that precancerous pancreatic cells secrete chemokines that chemoattract Schwann cells (SC) of nerves and thus induce ready-to-use routes of dissemination in early carcinogenesis. Here we show a peculiar role for the chemokine CXCL12 secreted in early PDAC and for its receptors CXCR4/CXCR7 on SC in the initiation of neural invasion in the cancer precursor stage and the resulting delay in the onset of PDAC-associated pain. SC exhibited cancer- or hypoxia-induced CXCR4/CXCR7 expression in vivo and in vitro and migrated toward CXCL12-expressing PCC. Glia-specific depletion of CXCR4/CXCR7 in mice abrogated the chemoattraction of SC to PCC. PDAC mice with pancreas-specific CXCL12 depletion exhibited diminished SC chemoattraction to pancreatic intraepithelial neoplasia and increased abdominal hypersensitivity caused by augmented spinal astroglial and microglial activity. In PDAC patients, reduced CXCR4/CXCR7 expression in nerves correlated with increased pain. Mechanistically, upon CXCL12 exposure, SC down-regulated the expression of several pain-associated targets. Therefore, PDAC-derived CXCL12 seems to induce tumor infiltration by SC during early carcinogenesis and to attenuate pain, possibly resulting in delayed diagnosis in PDAC.
The nervous system has recently been discovered to react to even the earliest stages of carcinogenesis by supplying growing tumors with nerves and with multiple modulations of tumor innervation, which constitute cancer-associated neuropathy (1–3). Pancreatic ductal adenocarcinoma (PDAC) features a pronounced neuropathy with an exceptionally high frequency of tumor cell penetration into nerves, i.e., neural invasion (NI), reaching 100% in retrospective case series (4). The pathogenesis of NI holds major translational relevance, because NI is independently associated with a dismal overall survival, local recurrence, and severe neuropathic pain during the already highly lethal course of PDAC (5, 6). Classically, cancer cells are assumed to penetrate into nerves actively and to use them as paths of dissemination (6, 7). However, we recently reported that peripheral glia cells, i.e., Schwann cells (SC), become activated in PDAC and suppress pain sensation (8). Importantly, they emerge around the premalignant precursor lesions of different human and murine gastrointestinal cancers and correlate to the frequency of NI (2). This observation led us to consider the possibility that tumor-derived chemoattractants such as chemokines may recruit neuro-glial cells very early during carcinogenesis.
The chemokine CXCL12, also known as “stromal-derived factor 1 alpha” (SDF-1α), is a CXC-class chemokine that has been shown to be widely expressed in several well-vascularized mammalian tissues and cancers and is known to regulate homing, proliferation, and survival of bone marrow-derived hematopoietic stem cells and stromal cells via its cognate receptor CXCR4 (9–11). Moreover, CXCR4 was reported to be overexpressed, to influence cancer cell proliferation and metastasis, and to modulate the tumor microenvironment in more than 25 types of human cancers including PDAC, prostate, breast, and ovarian cancer and melanoma (9, 12). Recently, CXCL12 was shown to exert its effect through an alternative receptor, CXCR7 (RDC-1). Intriguingly, CXCR7 is able to heterodimerize with CXCR4 and to fine-tune CXCR4 function (9, 13).
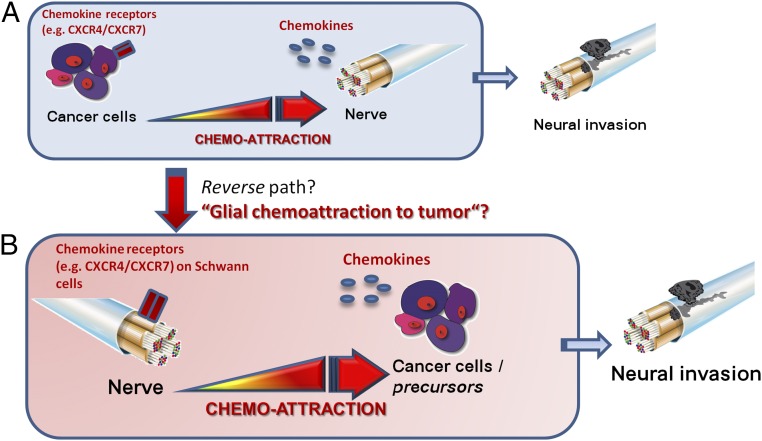
In the present study we hypothesized that the CXCL12/CXCR4/CXCR7 axis may mediate the chemoattraction of SC to cancer cells, thereby initiating NI and its impact on cancer dissemination (Fig. 1). Here we show that PDAC cells (PCC) harbor CXCL12, which attracts SC of peripheral nerves via CXCR4 and CXCR7. In two glia-specific murine CXCR4- and CXCR7-KO models, specific depletion of CXCR4 or CXCR7 abrogated the chemoattraction of SC to tumor cells. Correspondingly, pancreas-specific ablation of CXCL12 signaling in mice that develop PDAC disrupted the chemoattraction of glia cells to PDAC precursor lesions and resulted in increased abdominal mechanosensitivity. Accordingly, PDAC patients with pain exhibit less neural CXCR4- and CXCR7. Together with the multiple effects of CXCL12 on the transcription of pain-associated targets in activated SC, these observations collectively suggest a link between chemokine-mediated SC activation, early tumor dissemination, and the suppression of pain sensation in cancer.
Fig. 1.
The reverse pathogenesis of NI in cancer. (A) The classical theory of NI in cancer dictates that cancer cells that express chemokine receptors are chemoattracted to nerves that secrete chemokines. Cancer cells then actively invade nerves in the cancer-hosting organ and use the nerves as longitudinal paths for their spread. (B) In the present study we propose that a reverse direction of this interaction might also be possible based on the expression profile of nerves and especially SC for chemokine receptors and on the multitude of chemokines that are known to be secreted by cancer cells.
Results
SC in Pancreatic Nerves Express CXCR4 and CXCR7, and PCC Are a Source of CXCL12 in PDAC.
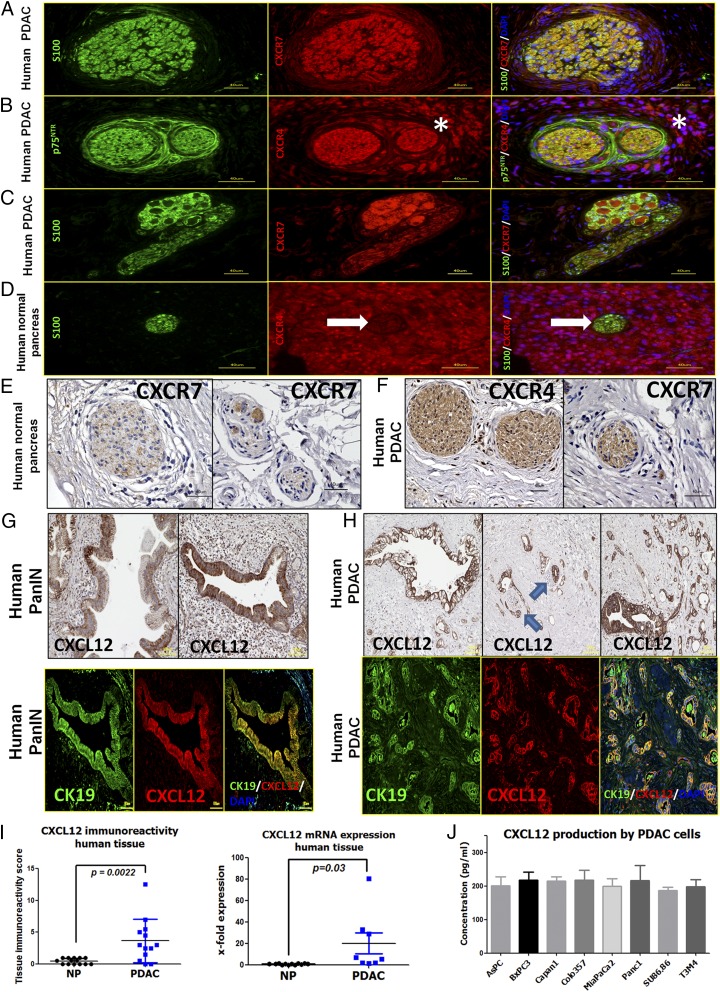
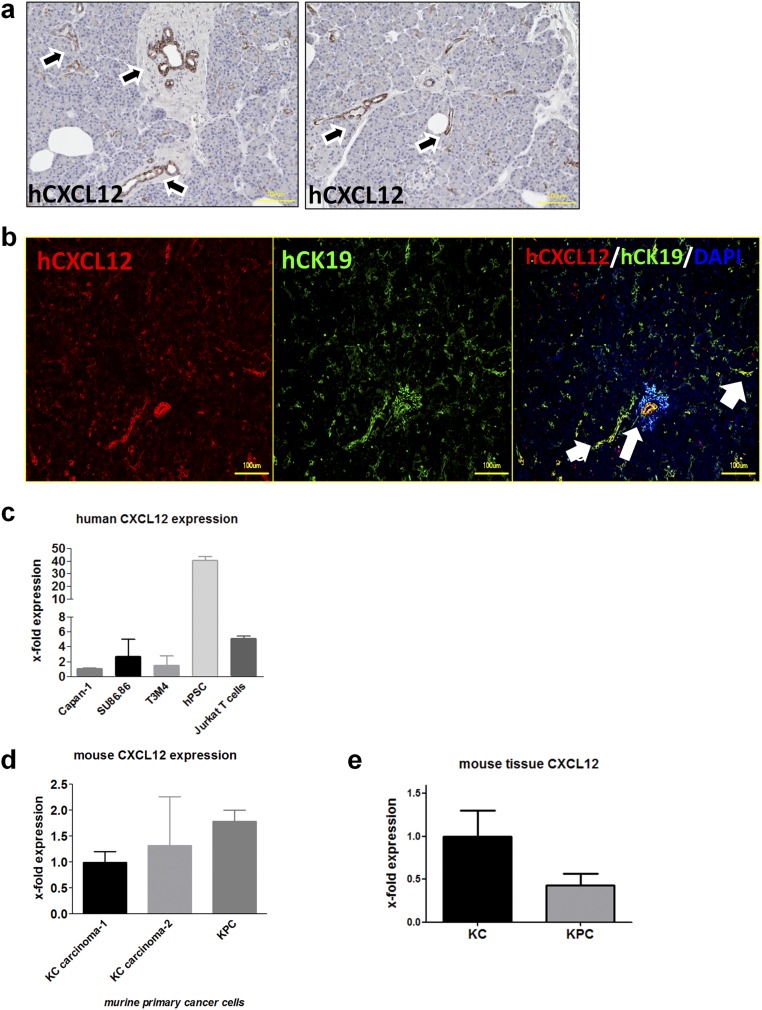
The emergence of SC around the precursor lesions or cancer cells in PDAC (2) suggests that SC may express chemokine receptors that can respond to the chemokines secreted from cancer cells or their precursors (Fig. 1). Previous studies suggested overexpression of the CXCL12 receptor CXCR4 in PDAC (14, 15). Furthermore, CXCL12 was previously detected on tumor-associated fibroblasts (16) and on FAP+ fibroblasts (17), and its receptor CXCR4 was detected on pancreatic stellate cells (18) and also on endothelial cells (14). However, no studies have focused on the distribution of CXCL12/CXCR4/CXCR7 in pancreatic nerves. In double-immunolabeling experiments using the SC marker S100, we detected the prominent and highly frequent presence of CXCR4 and CXCR7 not only in immune cells and cancer cells but also in SC of nerves within PDAC tissues (Fig. 2 A–C). CXCR4 or CXCR7 also was detectable in nerves within in the normal pancreas (NP) (Fig. 2 D–F and Tables S1–S6). In the corresponding immunolabeling experiments, CXCL12 was prominently up-regulated in the precursor pancreatic intraepithelial neoplasia (PanIN) lesions and PCC compared with NP and colocalized with the PCC marker cytokeratin-19 (CK19) (Fig. 2 G and H). In NP, CXCL12 was expressed in intrapancreatic ducts (Fig. S1 and Table S3), whereas in PDAC it was detected within the extracellular matrix (ECM), i.e., stroma, and in endothelium as well as in cancer cells (Table S3). Because of this prominent CXCL12 immunoreactivity detected in cancer precursors and cancer cells, the mean immunoreactivity score for CXCL12 in PDAC tissues (3.7 ± 3.4) was remarkably greater than in NP (0.46 ± 0.46) (Fig. 2I). We next performed ELISA to quantify the levels CXCL12 in lysates of eight different human PDAC cell lines. All eight PCC lines contained prominent levels of CXCL12 (average CXCL12 content = 207.5 ± 12.2 pg/mL) (Fig. 2J). In a comparative analysis of mRNA expression, human PDAC cell lines exhibited CXCL12 expression comparable to that of human Jurkat T cells, and the highest CXCL12 expression was observed in human pancreatic stellate cells (Fig. S1C). In cancer cells isolated from murine PDAC models [i.e., KC (p48-Cre;LSL-KrasG12D) and KPC (p48-Cre;LSL-KrasG12D; trp53lox/lox) mice], the p53-deficient KPC cancer cells tended to have higher CXCL12 expression (Fig. S1D). On the other hand, the tissue expression of CXCL12 tended to be higher in 12- to 20-wk-old KC mice that have not yet progressed to overt invasive cancer than in KPC mice with overt cancer (Fig. S1E), indicating PanIN lesions as a potential major source of CXCL12.
Fig. 2.
SC in human PDAC harbor the chemokine receptors CXCR4 and CXCR7, whereas PanIN lesions and established human PCC lines contain CXCL12. (A–C) Coimmunolabeling of nerves in PDAC tissues with the SC markers S100/p75NTR demonstrates the presence of CXCR4 and CXCR7 in nerves (A and B) and also in intrapancreatic ganglia (C). The asterisks indicate the immunostained perineural inflammatory cells. (D and E) In normal human pancreas the chemokine receptors are only faintly detectable in SC. The white arrow points to the nerve embedded in the exocrine tissue. (F) However, several nerves in PDAC tissues exhibited immunoreactivity for CXCR4 and CXCR7. (G and H) Immunolabeling of PDAC tissues against CXCL12 and colabeling with the PCC marker CK19 revealed PanIN lesions (G) and PCC (H) as the major source of this chemokine in PDAC. The blue arrows point to nerves that are invaded by CXCL12-expressing cancer cells. (I) Accordingly, the mean immunoreactivity of PDAC tissues for CXCL12 (Left) and the mean tissue CXCL12 mRNA expression (Right) was prominently greater in PDAC than in NP. For the mRNA expression, the levels of NP tissues were normalized to 1 (Mann–Whitney u test). (J) Human PCC lines synthesize substantial amounts of CXCL12 as detected via ELISA. Experiments were repeated three times. (Scale bars: A–F, 40 μm; G–H, 100 μm.)
Table S1.
Patient characteristics: NP and PDAC
| Sex | Differentiation | UICC grade | |||||||||
| Tissue | No. patients | Male | Female | Median age, y | Good | Moderate | Poor | Ib | IIa | IIb | III |
| NP | 12 | 7 | 5 | 69 | |||||||
| PDAC | 25 | 17 | 8 | 63.5 | 1 | 13 | 11 | 2 | 2 | 20 | 1 |
Table S6.
Histology scores for CXCR7 immunostaining
| Tissue | Acini | Ducts | Islets | Vessels | Nerves | Ganglion | Cancer | ECM | Immune cells | IHC score |
| NP | ||||||||||
| BP1576 | 0.5 | 0.0 | 0.0 | 0.0 | 0.0 | X | X | 0.0 | 0.0 | 0.0 |
| HD1516-2 | 10.5 | 1.0 | 0.0 | 0.0 | 0.5 | X | X | 0.5 | 0.5 | 2.5 |
| HD256-1 | 0.5 | 0.0 | 0.0 | 0.0 | 1.0 | X | X | 0.0 | 0.0 | 1.0 |
| HD1303 | 0.5 | 0.0 | 0.0 | 0.0 | 0.5 | X | X | 0.0 | 0.0 | 0.5 |
| BP442 | 0.5 | 0.0 | 0.0 | 0.0 | 0.5 | 1.0 | X | 0.0 | 1.0 | 2.5 |
| HD388-6 | 0.5 | 0.0 | 0.0 | 0.5 | 0.5 | 0.5 | X | 0.0 | 0.0 | 1.5 |
| HD1768-1 | 0.5 | 0.0 | 0.0 | 0.0 | 0.5 | X | X | 1.0 | 0.0 | 1.5 |
| BN1005 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | X | X | 0.0 | 0.0 | 0.0 |
| HD846-1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | X | X | 0.0 | 0.0 | 0.0 |
| HD1611-2 | 0.5 | 0.0 | 0.0 | 0.0 | 0.5 | X | X | 0.0 | 0.0 | 0.0 |
| Average | 1.4 | 0.1 | 0.0 | 0.1 | 0.4 | 0.8 | X | 0.2 | 0.2 | 3.0 |
| PDAC | ||||||||||
| HD282-1 | X | X | X | 0.0 | 1 | X | 0.5 | 0.0 | 0.0 | 1.5 |
| HD1650-1 | X | X | X | 0.0 | 0 | X | 0.5 | 0.5 | 1.0 | 2.0 |
| BP2422 | 0.0 | 0.5 | X | 0.0 | 0 | X | 0.5 | 1.0 | 2.0 | 4.0 |
| 3824/09 | 0.0 | 0.5 | 0.0 | 0.0 | 0 | 0.0 | 0.0 | 0.5 | 1.0 | 2.0 |
| HD644-1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.5 | X | 0.5 | 1.0 | 2.0 | 4.0 |
| HD502-1 | 0.0 | 0.0 | 0.0 | 0.0 | 0 | X | 0.0 | 0.0 | 0.0 | 0.0 |
| HD599-1 | X | X | X | 0.0 | 0 | X | 0.0 | 0.0 | 0.0 | 0.0 |
| HD176-1 | X | X | X | 0.0 | 0.5 | X | 0.0 | 0.5 | 1.0 | 2.0 |
| HD545-1 | X | X | X | 0.0 | 0 | X | 1.0 | 1.0 | 2.0 | 4.0 |
| HD562-1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.5 | X | 0.0 | 0.5 | 1.0 | 2.0 |
| HD1642-1 | 0.0 | 0.0 | 0.0 | 0.0 | 1 | X | 0.5 | 1.5 | 3.0 | 6.0 |
| HD151 | X | X | X | 0.0 | 0 | X | 0.0 | 0.0 | 0.0 | 0.0 |
| HD746-1 | X | X | X | 0.0 | 1.5 | X | 1.5 | 3.0 | 6.0 | 12.0 |
| HD484-1 | X | X | X | 0.0 | 0 | X | 1.5 | 1.5 | 3.0 | 6.0 |
| HD257-1 | X | X | X | 0.5 | 0.5 | X | 1.0 | 2.0 | 4.0 | 8.0 |
| HD263 | X | X | X | 0.0 | 0 | 2.0 | 0.0 | 2.0 | 4.0 | 8.0 |
| Average | 0.0 | 0.2 | 0.0 | 0.0 | 0.3 | 1.0 | 0.5 | 0.6 | 1.2 | 2.2 |
Fig. S1.
Pancreatic ductal cells and cancer cells express CXCL12. (A) Human PDAC tissues were immunostained against CXCL12, which was detected primarily in ductal cells. The black arrows point to CXCL12-immunoreactive intrapancreatic ducts. (B) In double immunolabeling with the ductal marker CK19, CXCL12 was detected in normal human pancreatic ductal cells. The white arrows point to double-labeled (yellow) intrapancreatic ducts. (C) Quantitative real-time PCR analysis was performed with human PDAC cell lines (Capan1, Su86.86, and T3M4), in human pancreatic stellate cells (hPSC), and in human Jurkat T cells to compare CXCL12 expression levels. (D) The CXCL12 mRNA levels of cancer cells isolated from murine PDAC models (i.e., KC and KPC mice) were compared. The p53-deficient KPC cancer cells tended to have higher CXCL12 expression. (E) At the tissue level, the expression of CXCL12 tended to be higher in 12- to 20-wk-old KC mice that have not yet progressed to overt invasive cancer than in KPC mice with overt cancer. (Scale bars: 100 μm.)
Table S3.
Semiquantitative analysis of CXCL12, CXCR4, and CXCR7 immunoreactivity in NP and in human PDAC
| Tissue | Localization of CXCL12, CXCR4, and CXCR7 | |||||||
| Acini | Ductal cells | Ganglia | Nerves | Vessels | Inflammatory cells | ECM | PDAC cells | |
| NP | ||||||||
| CXCL12 | ± | ++ | − | − | ± | − | − | |
| CXCR4 | ± | + | ++ | ++ | ± | ++ | − | |
| CXCR7 | ± | + | ++ | ++ | ± | ++ | − | |
| PDAC | ||||||||
| CXCL12 | − | − | − | + | +++ | |||
| CXCR4 | ++ | ++ | ± | +++ | − | ++ | ||
| CXCR7 | ++ | ++ | ± | ++ | ± | ++ | ||
−, absent; ±, absent to very weak; +, weak; ++, moderate; +++, strong immunoreactivity.
Table S4.
Histology scores for CXCL12 immunostaining
| Tissue | Acini | Acinar atrophy | Ducts | Tubular complex | Islets | Arteries | Nerves | Ganglion | PanIN 1 | PanIN 1B | PanIN 2 | PanIN 3 | Cancer | ECM | Immune cells | IHC score |
| NP | ||||||||||||||||
| BD 2376 | 0.0 | X | 0.5 | X | 0.5 | 0.0 | 0.0 | X | X | X | X | X | X | 0.0 | 0.0 | 1.0 |
| HD 1413-1 | 0.0 | X | 0.0 | X | 0.5 | 0.0 | 0.0 | X | X | X | X | X | X | 0.0 | 0.0 | 0.5 |
| BN 2016 | 0.0 | X | 0.0 | X | 0.0 | 0.0 | 0.0 | X | X | X | X | X | X | 0.0 | 0.0 | 0.0 |
| HD 1242-1 | 0.0 | X | 0.0 | X | 1.0 | 0.0 | 0.0 | X | X | X | X | X | X | 0.0 | 0.0 | 1.0 |
| HD 1207-1 | 0.0 | X | 0.0 | X | 0.5 | 0.0 | 0.0 | X | X | X | X | X | X | 0.0 | 0.0 | 0.5 |
| HD 804-1 | 0.0 | X | 0.0 | X | 0.0 | 0.0 | 0.0 | X | X | X | X | X | X | 0.0 | 0.0 | 0.0 |
| HD 1234-3 | 0.0 | X | 0.0 | X | 0.5 | 0.0 | 0.0 | X | X | X | X | X | X | 0.0 | 0.0 | 0.5 |
| HD 1434-1 | 0.0 | X | 0.0 | X | 0.0 | 0.0 | 0.0 | X | X | X | X | X | X | 0.0 | 0.0 | 0.0 |
| HD 1417-1 | 0.0 | X | 0.0 | X | 1.0 | 0.0 | 0.0 | X | X | X | X | X | X | 0.0 | 0.0 | 1.0 |
| HD 1611-2 | 0.0 | X | 0.0 | X | 0.0 | 0.0 | 0.0 | X | X | X | X | X | X | 0.0 | 0.0 | 0.0 |
| BN 1245 F | 0.0 | X | 0.0 | X | 0.0 | 0.0 | 0.0 | X | X | X | X | X | X | 0.0 | 0.0 | 0.0 |
| BN 1309F | 0.0 | X | 0.0 | X | 0.0 | 0.0 | 0.0 | X | X | X | X | X | X | 0.0 | 0.0 | 0.0 |
| BN 1608 | 0.0 | X | 0.5 | X | 0.5 | 0.0 | 0.0 | X | X | X | X | X | X | 0.0 | 0.0 | 1.0 |
| HD 2024-1 | 0.0 | X | 0.5 | X | 0.5 | 0.0 | 0.0 | X | X | X | X | X | X | 0.0 | 0.0 | 1.0 |
| Average | 0.0 | X | 0.1 | X | 0.4 | 0.0 | 0.0 | X | X | X | X | X | X | 0.0 | 0.0 | 0.5 |
| PDAC | ||||||||||||||||
| HD 282-1 | 0.0 | 0.5 | 0.5 | 0.5 | X | 0.0 | 0.0 | X | 0.0 | 0.0 | 0.0 | 0.5 | 0.5 | 0.0 | 0.0 | 2.5 |
| BP 2422 | 0.0 | 0.0 | 0.5 | 0.5 | 0.0 | 0.0 | 0.0 | X | 0.0 | 0.5 | 0.5 | 0.5 | 0.5 | 0.0 | 0.0 | 3.0 |
| HD 653-1 | 0.0 | 0.5 | 0.5 | 0.5 | 0.0 | 0.0 | 0.0 | X | X | X | X | X | X | 0.0 | 0.0 | 1.5 |
| HD 502-1 | 0.0 | 0.0 | 0.5 | 0.5 | 0.0 | 0.0 | 0.0 | X | 0.0 | 0.0 | 0.5 | 0.5 | 0.5 | 0.0 | 0.0 | 2.5 |
| 3824/09 | 0.0 | 0.5 | 0.5 | 0.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.5 | 0.5 | 1.0 | 1.0 | 0.0 | 0.0 | 4.5 |
| HD 562-1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 26806/04 | 0.0 | 0.0 | 0.5 | 0.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.5 | 0.5 | 0.0 | 1.0 | 0.0 | 0.0 | 3.0 |
| 22915/03 | 0.5 | 0.5 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | X | 0.5 | 0.5 | 0.5 | 0.5 | 1.0 | 0.0 | 0.0 | 5.5 |
| HD 555-1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | X | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 3173/09 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | X | 0.0 | 0.0 | 0.0 | 0.0 | 0.5 | 0.0 | 0.0 | 0.5 |
| 17259/09 | 0.0 | 0.5 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | X | 2.0 | 2.0 | 2.0 | 2.0 | 2.0 | 0.0 | 0.0 | 12.5 |
| 15754/09 | 0.0 | 1.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | X | 0.5 | 0.5 | 0.5 | 1.0 | 1.5 | 0.0 | 0.0 | 7.0 |
| 17329/09 | 0.0 | 0.5 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.0 | 0.0 | 5.0 |
| Average | 0.0 | 0.3 | 0.5 | 0.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.3 | 0.4 | 0.5 | 0.5 | 0.8 | 0.0 | 0.0 | 3.7 |
Table S5.
Histology scores for CXCR4 immunostaining
| Tissue | Acini | Acinar atrophy | Ducts | Tubular complex | Islets | Arteries | Nerves | Ganglion | Cancer | ECM | Immune cells | IHC score |
| NP | ||||||||||||
| M198 | 1.00 | 0.00 | 1.00 | 0.00 | 1.00 | 0.00 | 1.00 | 1.00 | 0.00 | 0.00 | 2.00 | 7.00 |
| M644 | 2.00 | 0.00 | 2.00 | 0.00 | 1.00 | 0.00 | 1.00 | 1.00 | 0.00 | 0.00 | 2.00 | 9.00 |
| M150 | 1.00 | 0.00 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 2.00 | 5.00 |
| M2212 | 1.50 | 0.00 | 1.50 | 0.00 | 0.50 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 2.00 | 5.50 |
| M267 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 2.00 | 2.00 |
| M1229 | 1.50 | 0.00 | 1.50 | 0.00 | 0.50 | 0.00 | 0.50 | 0.00 | 0.00 | 0.00 | 2.00 | 6.00 |
| M282 | 0.50 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 2.00 | 3.50 |
| M296 | 1.50 | 0.00 | 1.50 | 0.00 | 0.50 | 0.00 | 0.50 | 0.00 | 0.00 | 0.00 | 2.00 | 6.00 |
| M283 | 1.00 | 0.00 | 1.00 | 0.00 | 0.50 | 0.00 | 0.50 | 0.00 | 0.00 | 0.00 | 2.00 | 5.00 |
| M690 | 1.00 | 0.00 | 1.00 | 0.00 | 0.50 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 2.00 | 4.50 |
| Average | 1.10 | 1.15 | 0.55 | 0.00 | 0.35 | 0.00 | 2.00 | 4.46 | ||||
| PDAC | ||||||||||||
| HD765 | 1.50 | 1.50 | 1.50 | 1.50 | 0.50 | 0.00 | 0.50 | 0.50 | 1.50 | 0.00 | 2.00 | 11.00 |
| HD176 | 0.50 | 0.50 | 0.50 | 0.50 | 0.50 | 0.00 | 0.00 | 0.50 | 1.50 | 0.00 | 2.00 | 6.50 |
| HD151 | 0.50 | 0.50 | 0.50 | 0.50 | 0.00 | 0.00 | 0.00 | 1.00 | 1.00 | 0.00 | 2.00 | 6.00 |
| HD280 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 2.00 | 0.00 | 2.00 | 0.00 | 0.00 | 4.00 |
| HD112 | 0.50 | 0.50 | 0.50 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 2.00 | 3.50 |
| HD242 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.50 | 2.00 | 0.00 | 1.50 | 5.00 |
| HD148 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 | 0.00 | 1.50 | 0.00 | 0.00 | 0.00 | 2.50 |
| HD646 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.50 | 0.00 | 2.00 | 0.00 | 1.50 | 4.00 |
| HD653 | 0.50 | 0.50 | 0.50 | 0.00 | 0.00 | 0.00 | 0.50 | 0.00 | 1.00 | 0.00 | 0.00 | 3.00 |
| HD464 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 | 0.50 | 0.00 | 1.00 | 0.00 | 0.00 | 2.50 |
| HD78 | 1.00 | 1.00 | 1.00 | 1.00 | 0.50 | 0.00 | 0.50 | 0.00 | 2.00 | 0.00 | 2.00 | 9.00 |
| HD502 | 0.50 | 1.00 | 0.50 | 1.00 | 0.50 | 0.00 | 0.50 | 0.00 | 1.50 | 0.00 | 2.00 | 7.50 |
| HD698 | 0.50 | 0.50 | 0.50 | 0.50 | 0.00 | 0.00 | 0.50 | 0.00 | 1.00 | 0.00 | 2.00 | 5.50 |
| M334 | 1.50 | 1.50 | 1.50 | 1.50 | 0.50 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 2.00 | 8.50 |
| Average | 0.50 | 0.50 | 0.46 | 0.32 | 0.00 | 0.39 | 1.18 | 0.00 | 1.36 | 5.38 | ||
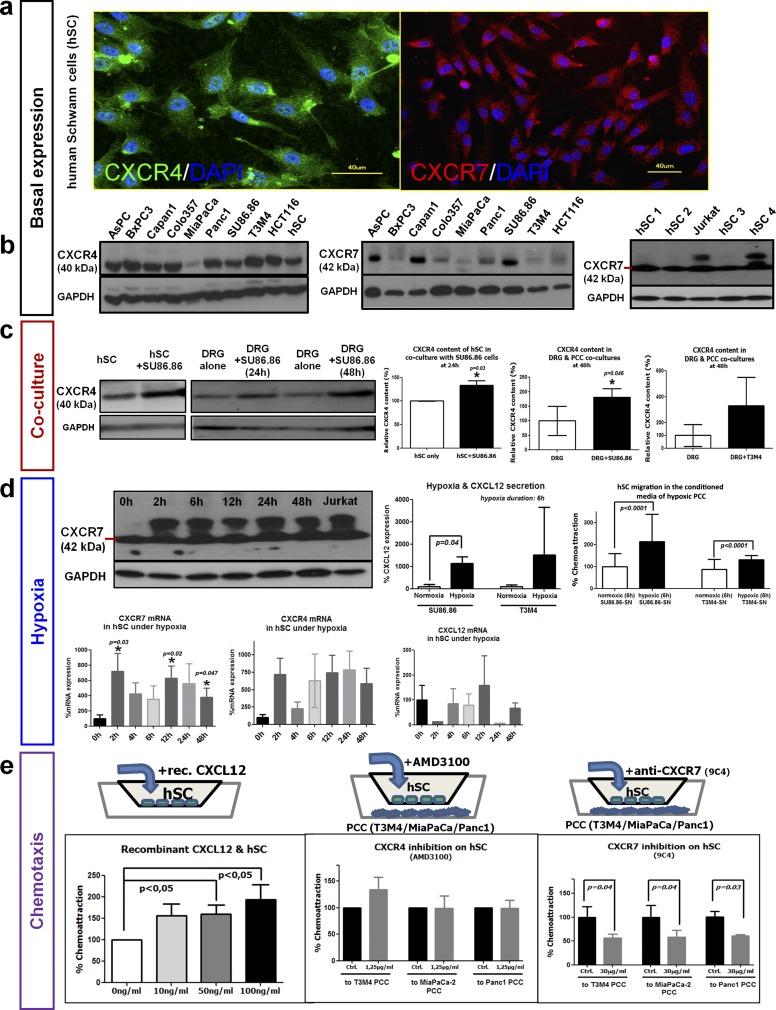
In accordance with the immunohistochemical detection in human PDAC nerves, both CXCR4 and CXCR7 were detected in cultivated primary human SC (hSC) via immunolabeling (Fig. S2A). At the protein level, CXCR4 and CXCR7 were consistently detectable in hSC and PDAC cell lines (Fig. S2B). To elucidate the potential regulation of CXCR4 or CXCR7 in hSC upon confrontation with PCC, we next cocultured hSC with PCC and performed immunoblotting to detect the dynamic changes in CXCR4/CXCR7 expression in hSC (Fig. S2C). CXCR4 (but not CXCR7) levels were elevated in hSC when they were cocultivated with SU86.86 PCC (CXCR4: 133.1 ± 17.4% of hSC monoculture) (Fig. S2C). In analogy, the CXCR4 and CXCR7 content of dorsal root ganglia (DRG) was prominently enhanced in coculture with the PCC line SU86.86 (CXCR4: 181.5 ± 71.9% of DRG monoculture) (Fig. S2C), and a similar tendency was seen in the T3M4 PCC line (329.0 ± 444.6% of DRG monoculture).
Fig. S2.
Basal and regulated expression of CXCR4 and CXCR7 in SC. (A and B) Primary hSC were found to express CXCR4 and CXCR7 via immunofluorescence (A) or via immunoblotting (B) in simultaneous comparison with human PCC lines and one human colon cancer cell line (HCT116). (C, Left) hSC were cocultured with the SU86.86 PCC line, and at 24 h their CXCR4 content was compared with that of monocultured hSC via immunoblotting. (Right) Newborn rat DRG that contain SC were also cocultured with SU86.86 or T3M4 PCC and at 48 h of coculture exhibited higher CXCR4 content than monocultured DRG (unpaired t test). (D, Upper Left) hSC that were exposed to increasing periods of hypoxia (0–48 h) exhibited both the 42-kDa native CXCR7 isoform and another larger isoform that also was present in Jurkat T cells. (Lower Left) At the mRNA level, 2- or 12-h hypoxia exposure led to significant up-regulation of CXCR7 in hSC, with a similar tendency for CXCR4. However, CXCL12 expression in hSC was not influenced by hypoxia. (Right) Hypoxia exposure enhanced CXCL12 expression in SU86.86 and T3M4 PCC lines. Accordingly, the addition of the supernatant of hypoxia-treated (6 h) PCC lines to the medium of hSC enhanced the hSC transmigration in Boyden chambers (Mann–Whitney u test). (E, Left) hSC were placed in the upper chamber of a Boyden Transwell chamber and were supplied with increasing doses of recombinant human CXCL12. The number of transmigrating hSC was determined and expressed as percent of control. (Center and Right) Similarly, T3M4 or MiaPaCa-2 human PCC were placed in the lower chamber, and hSC in the upper compartment were pretreated with the CXCR4 inhibitor AMD3100 or with a CXCR7-blocking antibody (9C4). The solvent for AMD3100 or a nonimmunized mouse IgG1 antibody was used as control (unpaired t test). Experiments were repeated three times.
Because of the intrinsic hypoxic trait of PDAC (19), we next investigated whether CXCR4 and CXCR7 levels in hSC are influenced by hypoxia. At the protein level, hypoxic hSC exhibited an additional, larger isoform of CXCR7 in addition to the native 42-kDa isoform (Fig. S2D). Accordingly, hypoxia led to the up-regulation of CXCR7 mRNA especially at 2 h (1,618 ± 1,489% of levels in normoxic hSC) and 12 h (629.3 ± 323%) of hypoxia exposure (Fig. 2D), and a similar tendency was seen CXCR4 expression (Fig. S2D). Hypoxia did not affect CXCL12 expression in hSC (Fig. S2D), but in both SU86.86 and T3M4 cells 6 h of hypoxia resulted in at least fivefold up-regulation of CXCL12 expression (SU86.86: 505.1 ± 310.3%; T3M4: 882.8 ± 1,180% of normoxic levels) (Fig. S2D). Accordingly, treatment of hSC in Boyden chambers containing the conditioned medium of hypoxia-treated PCC enhanced hSC transmigration (Fig. S2D, Right).
CXCL12/CXCR4/CXCR7 Signaling Mediates Chemoattraction of SC to Cancer Cells.
To dissect the potential chemoattracting role of PCC-derived CXCL12 on hSC, we first incubated hSC in Boyden chambers containing increasing amounts of CXCL12 in the lower compartment (Fig. S2E). We observed a dose-dependent increase in the amount of transmigrating hSC (10 ng/mL: 156.4 ± 86.4%; 50 ng/mL: 160.4 ± 66.4%; 100 ng/mL: 193.8 ± 110.5% of 0 ng/mL control hSC) (Fig. S2E). We next placed three different human PCC lines (T3M4, MiaPaCa-2, and Panc1) into the lower chamber of the Transwell migration assays and pretreated the hSC in the upper chamber with either the CXCR4 small-molecule inhibitor AMD3100 or with a CXCR7-blocking antibody. Blockade of CXCR4 did not affect hSC transmigration toward the PCC lines (Fig. S2E); however, when a CXCR7-neutralizing antibody (9C4) was added to the hSC, there was a significant dose-dependent attenuation of hSC transmigration toward PCC (toward T3M4 at 30 µg/mL: 56.1 ± 15.4% of control; toward MiaPaCa-2 at 30 µg/mL: 58.4 ± 29.1% of control; toward Panc1 at 30 µg/mL: 71.5 ± 21.8% of control) (Fig. S2E).
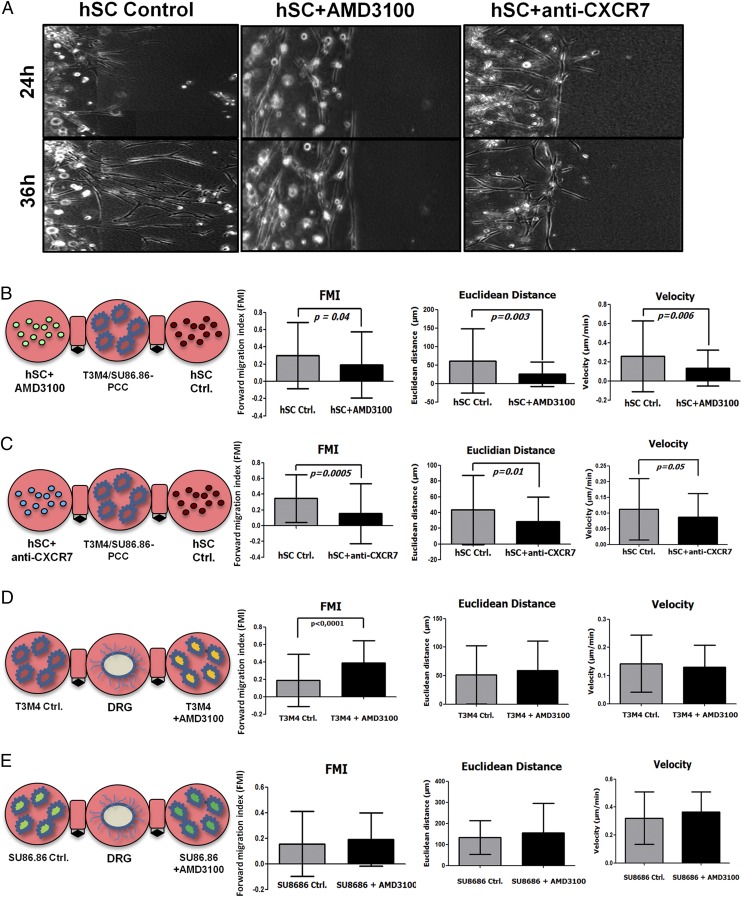
To test this impact of CXCL12 on hSC migration toward PCC further, we used a more complex 3D migration assay in which the respective cell types were resuspended in ECM to simulate the in vivo situation more closely. Here, PCC were simultaneously confronted with control hSC (pretreated with the solvent for AMD3100 or with a nonimmunized mouse IgG1 antibody) and with hSC pretreated with inhibitor (either AMD3100 for CXCR4 or with anti-CXCR7 antibody), and the cancer-directed migration of hSC was tracked by digital time-lapse microscopy (Fig. 3A). In this setting, hSC that were pretreated with AMD3100 or with anti-CXCR7 exhibited a much less targeted, shorter, and slower migration toward T3M4 or SU86.86 PDAC cells than control hSC [forward migration index (FMI) of hSC+AMD3100: 0.20 ± 0.39 vs. control hSC: 0.31 ± 0.38; FMI of hSC+anti-CXCR7: 0.15 ± 0.38 vs. control hSC: 0.35 ± 0.30) (Fig. 3 B and C). In additional 3D migration assays we also evaluated whether inhibition of CXCR4 or CXCR7 on PCCs influenced their targeted migration toward DRG. Inhibition of CXCR4 or CXCR7 on PCC did not inhibit the migration of these cells to DRG (Fig. 3D). In fact, inhibition of CXCR4 via AMD3100 even augmented the T3M4 cancer cell migration toward DRG (Fig. 3D). Overall, these observations suggest that CXCR4 and CXCR7 are mediators of hSC migration toward PCC, whereas CXCR4 seems to exert this effect in an ECM-containing environment.
Fig. 3.
Impact of CXCR4/CXCR7 on hSC migration to PCC in a 3D migration assay. (A–C) hSC pretreated with AMD3100 or CXCR7-blocking antibody were resuspended in an ECM drop, placed next to PCC, and recoded via live-cell imaging. Although solvent- or IgG1-treated (Control) hSC rapidly extend protrusions and migrate toward PCC (which do not possess any affinity to hSC) (A), hSC in which CXCR4 or CXCR7 was inhibited did not demonstrate any 3D migration, as evidenced by their prominently reduced FMI (B and C). (Magnification: A, 100×.) (D and E) In comparison, the pretreatment of the human PCC lines T3M4 or SU86.86 with AMD3100 or CXCR7-blocking antibody did not reduce their migration toward DRG neurons (Mann–Whitney u test for the FMI, unpaired t test for the remaining parameters). Experiments were repeated three times.
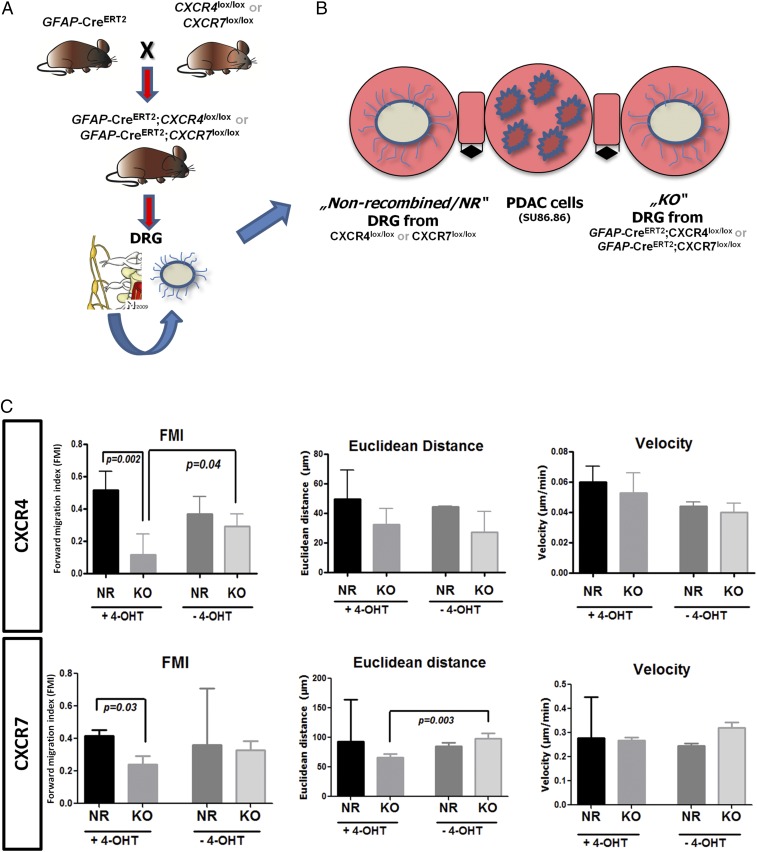
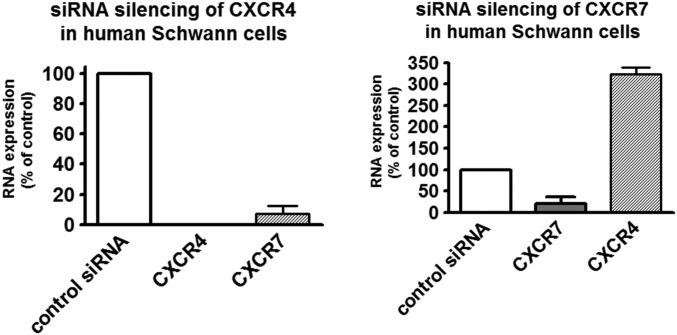
Theoretically, for SC migration to occur in PDAC, SC would need to grow out of the damaged intrapancreatic nerves toward PCC. Therefore in the current study, dissociated, SC-containing DRG of genetically engineered mice were confronted with PCC in the 3D migration assay (Fig. 4 A and B). After induction of the Cre recombinase with 4-hydroxytamoxifen (4-OHT) in the growth medium, SC from the DRG of GCE;CXCR4lox/lox and GCE;CXCR7lox/lox (KO DRG) mice (Fig. 4B) did not exhibit the same extent of targeted migration to PCC as those from CXCR4lox/lox and CXCR7lox/lox mice with no Cre recombinase (nonrecombined; NR DRG) (Fig. 4 B and C) or SC from KO DRG mice in the absence of 4-OHT (Fig. 4C and Table S7). Overall, these multiple models provided evidence for the CXCR4- and CXCR7-mediated specific migration of SC toward PCC. Mechanistically, upon siRNA-mediated silencing of CXCR4, the expression of CXCR7 was also prominently down-regulated to a mere 7.3% of control hSC. Conversely, when CXCR7 was silenced via siRNA, the expression of CXCR4 was strongly up-regulated to 322% of control levels (Fig. S3 and Table S8). Thus, it seems that CXCR7 expression is coupled to CXCR4 expression in a repressive-feedback mechanism. The absence of CXCR4 seems to obviate the expression of CXCR7, and the absence of CXCR7 seems to release the brake on the CXCR4 expression. This interaction is thus comparable to the previously described fine-tuning of CXCR4 activity (20), because in both cases CXCR7 seems to reduce excessive signaling from CXCL12 to CXCR4. Therefore, genetic CXCR4 depletion seems to decrease SC migration more potently than CXCR7 depletion.
Fig. 4.
Glia-specific knockout of CXCR4 or CXCR7 abrogates SC migration to cancer cells. (A) GFAP-CreERT2 mice were interbred with CXCR4lox/lox or CXCR7lox/lox mice to generate GFAP-CreERT2;CXCR4lox/lox (n = 3) or GFAP-CreERT2;CXCR7lox/lox (n = 3) mice. The DRG of these mice were explanted. (B) After 4-OHT supplementation of their medium, SC of dissociated DRG from GFAP-CreERT2;CXCR4ox/lox or GFAP-CreERT2;CXCR7lox/lox (collectively, KO) mice were compared with SC of dissociated DRG from CXCR4ox/lox or CXCR7lox/lox (NR) mice for their migration to PDAC cells. (C) The addition of 4-OHT significantly impaired the migration of SC from KO animals to human PCC as compared with SC from NR mice; this difference was not seen in the absence of 4-OHT (unpaired t test). Experiments were repeated three times.
Table S7.
FMI of SC that grow out of DRG from SC-specific CXCR4- or CXCR7-KO mice
| GFAP-CreERT2 | CXCR4lox/lox | CXCR7lox/lox | 4-OHT | FMI* |
| + | + | − | + | 0.11 ± 0.13 |
| − | + | − | + | 0.52 ± 0.11 |
| + | + | − | − | 0.29 ± 0.07 |
| − | + | − | − | 0.37 ± 0.11 |
| + | − | + | + | 0.24 ± 0.33 |
| − | − | + | + | 0.42 ± 0.27 |
| + | − | + | − | 0.33 ± 0.37 |
| − | − | + | − | 0.36 ± 0.35 |
Results are expressed as mean ± SD.
Fig. S3.
Reciprocal relationship between CXCR4 and CXCR7 expression in hSC. siRNA-mediated silencing of CXCR4 in hSC resulted in the suppression of both CXCR4 and CXCR7 expression, whereas siRNA-mediated silencing of CXCR7 in hSC up-regulated CXCR4 expression to ca. 300% of control.
Table S8.
siRNA sequences
| Type | Sequence |
| Scrambled no.1 | AUU GUA UGC GAU CGC AGA CUU |
| Scrambled no. 2 | GUC UGC GAU CGC AUA CAA UUU |
| CXCR4 no.1 | GGCAGUCCAUGUCAUCUACtt |
| CXCR4 no. 2 | GUAGAUGACAUGGACUGCCtt |
| CXCR7 no.1 | GGAUGACACUAAUUGUUAGtt |
| CXCR7 no. 2 | CUAACAAUUAGUGUCAUCCtt |
Ablation of Cancer Cell-Derived CXCL12 in PDAC Abrogates the SC Affinity Toward PanIN Lesions.
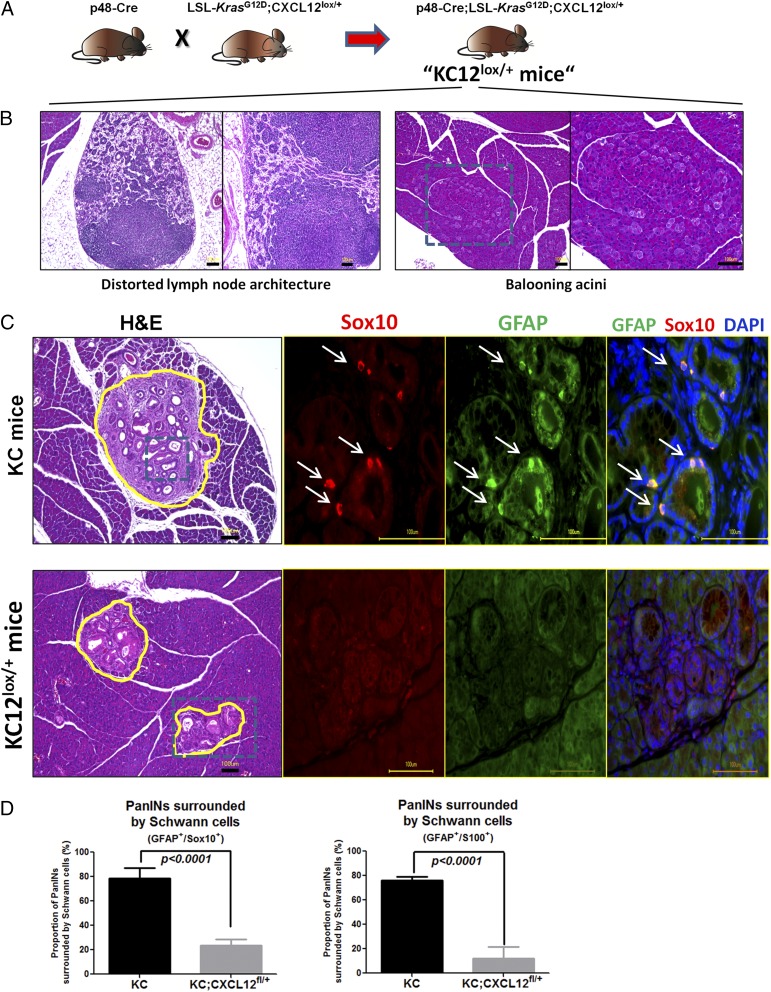
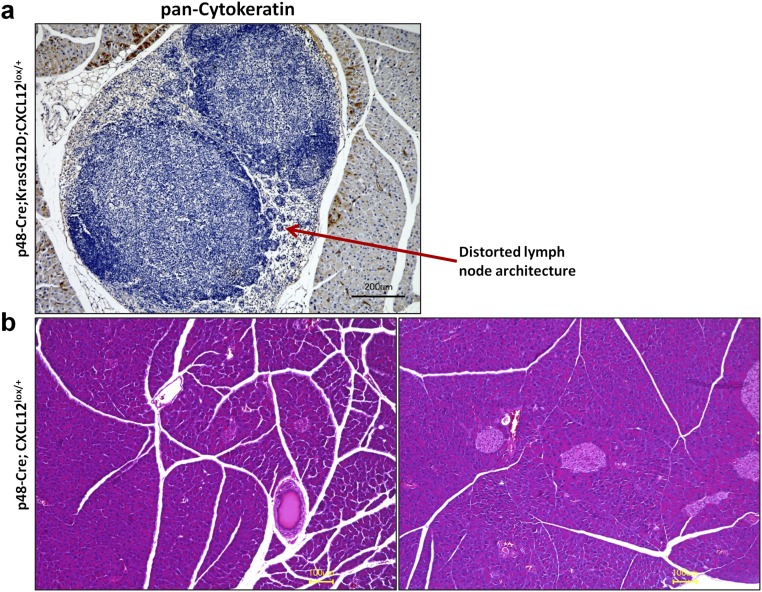
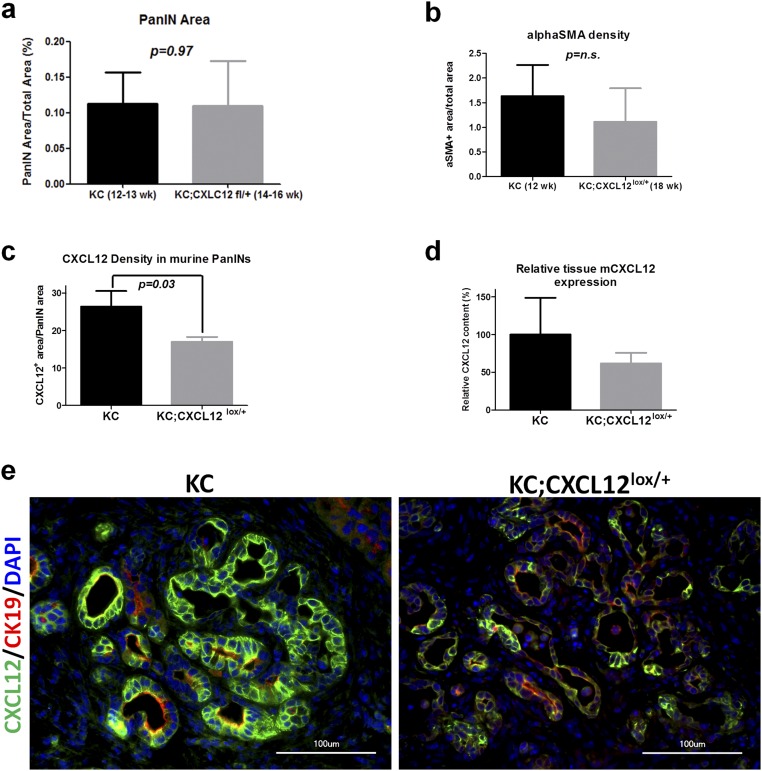
In the next step we investigated the impact of the disruption of CXCL12 signaling on glial activity in vivo in the murine KC model of PDAC (Fig. 5A). For this purpose, we generated mice in which CXCL12 secretion by pancreatic cells, including (pre)cancerous cells, was abrogated in a conditional manner (p48-Cre;LSL-KrasG12D;CXCL12lox/+, termed “KC12lox/+”) (Fig. 5A). KC12lox/+ mice exhibited distorted lymph node architecture and clusters of altered acinar cells that had a “ballooning” appearance (Fig. 5B and Fig. S4). Control p48-Cre;CXCL12lox/+ mice did not exhibit any obvious histological abnormality, and their pancreata did not differ from normal wild-type pancreas (Fig. S4). The decrease in CXCL12 content of PanIN lesions and the comparability of PanIN degree and stromal activity in KC and KC12lox/+ mice was confirmed by immunohistochemistry (Fig. S5). A typical feature of the KC model is the emergence of SC around the PanIN lesions, an observation that is assumed to be the precursor of NI (2). Here, SC can be detected by the expression of different glial markers (GFAP+Sox10+S100+-expressing cells) (Fig. 5C). In comparison with KC mice, we observed a prominent reduction in the number of SC that surrounded PanIN lesions in KC12lox/+ mice (KC: 75.9 ± 10.3% and KC12lox/+: 11.9 ± 9.9% for GFAP/S100 coexpression; KC: 78.5 ± 8.7% and KC12lox/+: 23.5 ± 11.0% for GFAP/Sox10 coexpression) (Fig. 5D). Hence, this model suggested that CXCL12 from the transformed PanIN lesions mediates the emergence of SC around these precursor lesions.
Fig. 5.
Pancreas-specific ablation of CXCL12 production disrupts SC accumulation around PanIN/precursor lesions. (A) p48-Cre mice were interbred with LSL-KrasG12D;CXCL12fl/+ mice to generate the KC12lox/+ mice with pancreas-specific reduction of the CXCL12 production. (B) The KC12lox/+ mice exhibited alterations in their lymph nodes, which appeared distorted, and ballooning of pancreatic acini. (C) These mice were killed between 12–22 wk of age when KC mice mice typically exhibit PanIN lesions (n = 9 mice). Immunolabeling against different glial markers (here Sox10/GFAP) revealed the presence of glial marker-expressing cells around PanIN lesions. (D) Quantification of glia-surrounded PanINs revealed that such cells were widely absent around the PanINs of KC12lox/+ mice (n = 5 mice). The number of analyzed PanINs was n = 289 in the KC12lox/+ mice and n = 423 in the KC mice (unpaired t test). (Scale bars: 100 μm.)
Fig. S4.
Histology of p48-Cre;KrasG12D;CXCL12lox/+ and p48-Cre;CXCL12lox/+ mice. (A) Because of the distorted appearance of the lymph nodes in the p48-Cre;KrasG12D;CXCL12lox/+ mice, we performed immunolabeling against the exocrine cell marker pan-cytokeratin to detect metastatic cells. There were no exocrine cells in these distorted lymph nodes, thus excluding metastasis-induced damage to the lymph node structure as a cause of distortion. (B) p48-Cre;CXCL12lox/+ mice were used as control animals to study the effects of heterozygous Cxcl12 ablation on the pancreatic morphology. There were no obvious differences in pancreatic morphology from normal wild-type pancreas.
Fig. S5.
Comparison of PanIN extent, stromal activity, and CXCL12 expression in KC vs. KC12lox/+ mice. (A) Immunolabeling against the tumor cell marker CK19 was performed to measure the PanIN area in KC and KC12lox/+ mice at different ages. Because of the slower growth dynamics of the newly generated KC12lox/+ mice, 12- to 13-wk-old KC mice and 14- to 16-wk-old KC12lox/+ mice with comparable PanIN progression were compared in the pain studies. (B) In the PanIN stage-matched mice, the extent of stromal activity as measured by the proportion of the alpha smooth muscle (alphaSMA)-immunoreactive tumor area was comparable, thus excluding differences in the stromal activity of the two cohorts. (C and D) Despite a heterozygous conditional knockout, the proportion of CXCL12-containing PanIN cells was reduced in KC12lox/+ mice compared with KC mice, and a similar tendency was seen for the overall CXCL12 expression. (E) Representative images of CXCL12-expressing PanIN lesions in KC and KC12lox/+ mice.
Disruption of the CXCL12-Mediated SC Recruitment Toward PDAC Increases Pain Sensation Through Reactivation of Spinal Glia and Up-Regulation of Multiple Nociceptive Mediators in SC.
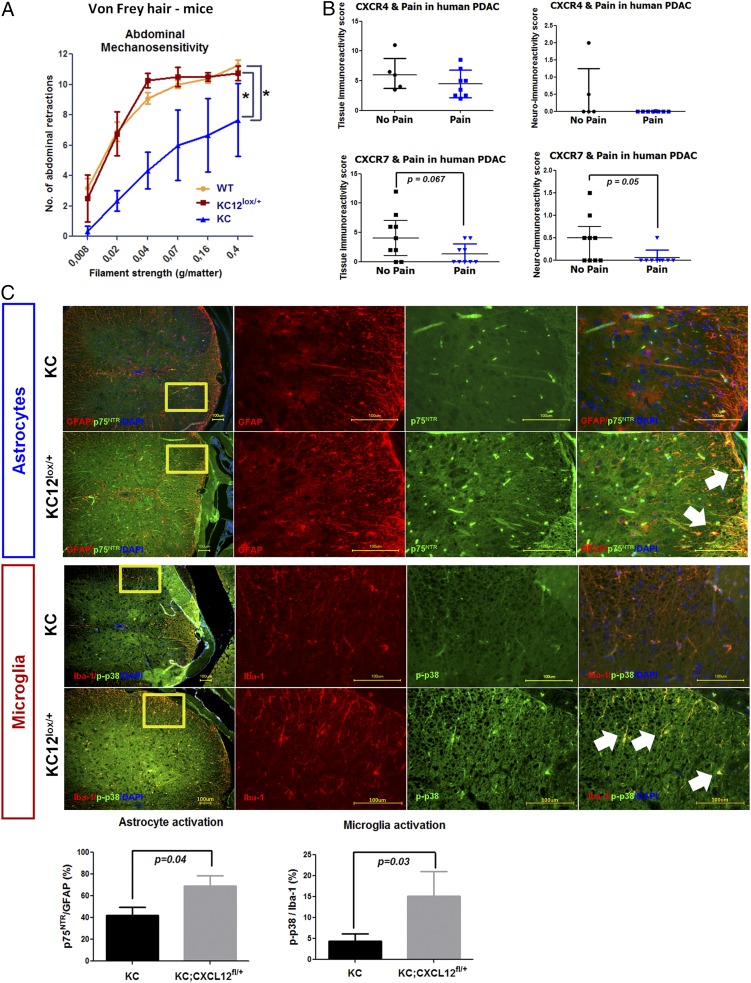
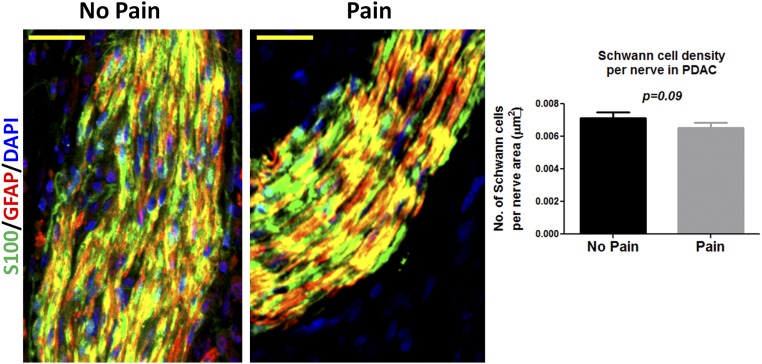
We next evaluated the possibility that disruption of CXCL12 signaling between PCC and SC may affect the clinical course of PDAC. For this purpose, we assessed the extent of mechanical sensitivity (8) as an indirect measure of potential pain sensation in the abdominal area of KC and KC12lox/+ mice (Fig. 6A). Intriguingly, KC12lox/+ mice had much greater abdominal hypersensitivity scores (8.6 ± 3.4) than KC mice (4.6 ± 2.8), as assessed by means of von Frey filaments. Interestingly, the mechanosensitivity of KC12lox/+ mice was comparable to wild-type C57BL6/J mice (von Frey score: 8.4 ± 2.9), implying the suppression of pain in KC mice rather than the promotion of pain in KC12lox/+ mice. This observation would suggest that PDAC patients with active CXCL12–CXCR4/CXCR7 signaling in SC would suffer less pain. We therefore analyzed the immunoreactivity of nerves among PDAC patients subgrouped into those with or without pain. In this analysis, we indeed detected a decreased mean neural immunoreactivity for CXCR7 among patients with pain as compared with patients without pain (neural CXCR7: no pain = 0.4 ± 0.5 vs. pain = 0.06 ± 02) (Fig. 6B), with a similar tendency for tissue CXCR7 immunoreactivity (tissue CXCR7: no pain = 4.4 ± 4.0 vs. pain = 1.3 ± 1.7) (Fig. 6B). The average neural CXCR4 immunoreactivity also appeared weaker among patients with pain than in patients without pain (neural CXCR4: no pain = 0.5 ± 0.9 vs. pain = 0.02 ± 0.004) (Fig. 6B). The tissue CXCR4 immunoreactivity did not differ between the no pain and pain groups (tissue CXCR4: no pain = 6.2 ± 3.0 vs. pain = 4.5 ± 2.3) (Fig. 6B). We also counted the nuclei that colocalized with S100/GFAP within the intrapancreatic nerves of PDAC patients and compared these counts in patients with or without pain. The average number of SC tended to be lower in intrapancreatic nerves of PDAC patients with pain than of patients without pain (Fig. S6). This observation is in harmony with our previous findings on the decreasing GFAP content of nerves of PDAC patients with pain (8).
Fig. 6.
Active CXCL12 signaling in the pancreas diminishes pain by suppressing spinal glial activity. (A) Abdominal mechanosensitivity to von Frey hairs was compared in KC (n = 3) and KC12lox/+ (n = 4) mice. The number of abdominal retractions when the abdomen was touched was quantified and compared with wild-type C57BL6/J mice. The enhanced SC activity in KC mice was associated with the suppression of abdominal retractions and thus less mechanosensitivity (unpaired t test). (B) The immunoreactivity of tumor tissue and tumor nerves for CXCR4 and CXCR7 was compared in PDAC patients with (n = 10) and without pain (n = 11). The neural immunoreactivity for CXCR7 was prominently lower in patients with painful disease (unpaired t test). (C) In the spinal cord of KC and KC12lox/+ mice, the proportion of activated dorsal horn astrocytes (p75NTR+ proportioned to GFAP+ cells) and microglia (p-p38+ proportioned to Iba-1+ cells) was compared via immunolabeling of the thoracic spinal segments 8–11 (three sections per spinal cord). Diminished CXCL12-signaling was associated with enhanced astrocyte and microglia activity in the dorsal horn (unpaired t test). The white arrows point toward double-stained astrocytes or microglia. (Scale bars: 100 μm.)
Fig. S6.
Analysis of SC density in the intrapancreatic nerves of PDAC patients with or without pain. PDAC patients were divided into two groups depending on their preoperative pain status (no pain vs. pain; n = 10 patients each). The paraffin-embedded histological specimens were double-immunostained against the SC markers S100 and GFAP, and the number of DAPI+ nuclei in each nerve was counted manually. (Scale bars: 40 µm.)
To decipher the implications of suppressed pain sensation in KC versus KC12lox/+ mice, we analyzed the activity status of the central spinal glia, i.e., astrocytes and microglia, which are well-established actors in chronic and neuropathic pain states (Fig. 6C) (8, 21, 22). A comparison of the proportion of activated astrocytes (coexpressing p75NTR+GFAP+) and microglia (coexpressing p-p38+Iba-1+) in the pancreas-innervating thoracic segments of the spinal cord of KC and KC12lox/+ mice revealed a considerably higher proportion of activated astrocytes and microglia in the dorsal, i.e., sensory, part of the spinal cord of KC12lox/+ mice (active astrocytes: 69.2 ± 20.5%; active microglia: 15.1 ± 13.4%) than in KC mice (active astrocytes: 42.3 ± 7.4%; active microglia: 4.3 ± 1.8%) (Fig. 6C). Collectively, these observations suggested that SC recruitment via cancer cell-derived CXCL12 attenuates pain by suppressing spinal glial activity.
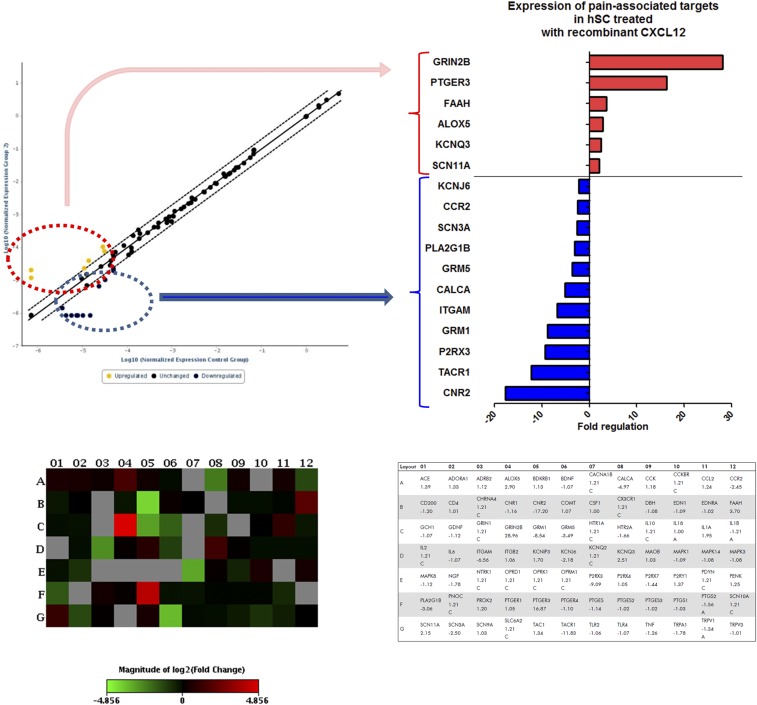
Diminished mechanosensitivity or pain resulting from CXCL12-mediated SC chemoattraction implied that CXCL12 also may suppress some pain-associated pathways in SC. To test this possibility, we analyzed the transcriptomic alterations in hSC treated with recombinant CXCL12 with a particular focus on the expression of 84 pain-associated targets. Here, 17 of 84 pain-associated genes were found to be differentially expressed (i.e., at least threefold up-regulated) in hSC after CXCL12 treatment. Among these, 6 of the 17 targets were up-regulated and thus were associated with potential pain-promoting consequences (Fig. S7). However, the larger portion, 11 of 17, were down-regulated and included major pain-associated targets such as the tachykinin receptor 1 (TACR1/NK1R, down-regulated 12.2-fold), the purinergic receptor P2RX3 (down-regulated 9.4-fold), the metabotropic glutamate receptor 1 (GRM1) (down-regulated eightfold), and the calcitonin gene-related peptide alpha (CGRPa/CALCA) (down-regulated 5.1-fold). Hence, these target alterations suggest that chemokine-mediated SC recruitment and the resulting masking of pain may be caused by the diminished transcription of pain-associated pathways in SC.
Fig. S7.
SC treated with recombinant CXCL12 exhibit alterations in pain-associated molecular mediators. hSC were treated for 24 h with human recombinant CXCL12 (10 ng/mL) and subsequently were analyzed for the mRNA expression of 84 different pain-related targets. In comparison with solvent-treated control SC, CXCL12 treatment resulted in at least threefold regulation in the expression of 17 targets listed on the right; up-regulated targets are circled in red; down-regulated targets are circled in blue. Alterations in six of these targets were associated with a potentially proalgesic outcome; the remaining 11 targets were associated with a potentially analgesic outcome. The scatter plot, heatmap, and table of targets were generated via the online Qiagen Data Analysis Center.
Discussion
Cancer cells readily exploit intrinsic developmental and defense mechanisms in their microenvironment to boost their growth and spread. In the present study, we show another example of the exploitation of the microenvironment by tumor cells, namely, the exploitation of glial cells, a cell type that had not previously been subject to investigation in this context. We show that PCC can chemoattract SC in vivo and in vitro in a CXCL12-dependent mechanism that seems to result in decreased pain sensation resulting from the suppression of SC-intrinsic molecular pain pathways and of spinal astrocytes and microglia in vivo. These observations add dimensions to the very recent investigations of cancer-associated neuropathy and provide a mechanistic perspective on the very early dissemination of PDAC.
Previous studies showed that CXCL12 and CXCR4 actively participate in nociception via direct stimulation of nociceptive neurons (23), via induction of mechanical hypersensitivity in rats (24), or via desensitization of opioid receptors, but all these effects are in the central nervous system (25). In the present study, we show this chemokine–receptor axis has a role in the activation of peripheral glia, i.e., SC, in the tumor microenvironment, which seems to entail an analgesic effect in the early disease course. Accordingly, we recently demonstrated decreased pain sensation among PDAC patients who bear increased pancreatic gliosis with cellular hypertrophy of pancreatic glia (8). In the light of the previously reported pro-nociceptive roles of CXCR4 and CXCL12, our observations suggest that peripheral and central CXCL12-mediated signaling exert contrasting effects for nociception, i.e., CXCL12 mediating analgesia via modulation of SC activity.
The tumorigenic effects of CXCL12–CXCR4 in PDAC include promotion of PCC invasiveness, migration, proliferation, epithelial–mesenchymal transition, and metastasis via Akt-, Erk-, and sonic hedgehog-dependent pathways (11, 14, 15, 26–31). High levels of tissue CXCL12 and CXCR4 were also shown to be associated with poor survival of PDAC patients (15, 26, 32). These studies thus uncovered the autocrine trophic effects of CXCL12 on cancer cells and disease progression and on the cancer-driven modeling of inflammation via selective chemoattraction of growth-promoting immune cell subtypes to the tumor (33). The present study expands this knowledge of chemokine-driven cancer progression by unraveling the chemoattraction of glia to cancer via CXCL12–CXCR4–CXCR7 as the initiator of nerve–cancer interactions and as a factor that delays the onset of symptoms such as pain.
This pathomechanistic concept for the generation of NI in PDAC turns the classical view of the active invasion of nerves by cancer cells on its head and provides an explanation for the very early dissemination of PDAC (2). As the peripheral glial cells, SC previously were shown to express both CXCR4 and CXCR7 in a cAMP- and TNFα-dependent manner (34, 35). In the current study, we show that this expression is subject to up-regulation by glia–cancer confrontation and especially by hypoxia as encountered in PDAC tissues. Both these receptors are essential for regulating SC survival, migration, and communication (e.g., with DRG neurons) (34, 35). However, beyond SC biology, CXCL12–CXCR4 interactions regulate multiple aspects of glial biology in general, including astrocytic and microglial migration and survival (36), neural recovery after ischemia or damage (23, 37), and neuron–glia communication (23). CXCR4 was further shown to be increasingly expressed on astrocytes after stimulation by interleukin-6 (38), which we recently demonstrated to be a key factor that generates and maintains reactive gliosis in PDAC (8). Therefore, one can assume that all the above attributes of CXCR4 and CXCR7 in glia biology may also influence the extent of nerve–cancer interactions in PDAC.
Specifically, we propose that CXCR4- and CXCR7-expressing SC are chemoattracted to PCC in the cancer precursor stage and, similar to their role in nerve regeneration, can serve as paths of axonal guidance toward cancer cells and physically initiate nerve–cancer cell contact. In a recent study, Pukrop et al. demonstrated a very similar cancer-promoting function for microglia in the central nervous system (39). In brain slice cocultures with breast cancer cells, microglia emerged at cancer infiltration zones and actively migrated toward breast cancer cells before the entrance of breast cancer cells into the brain. Upon establishment of physical contact, microglia “pulled” clusters of cancer cells into the brain and thereby initiated metastasis. Moreover, these transport interactions and brain metastases could be disrupted via the microglia inhibitor clodronate (39). Therefore, it is conceivable that CXCR4- and CXCR7-mediated SC chemoattraction of cancer cells may represent a corresponding example of the misuse of glia by cancer cells in PDAC and may serve as a future therapeutic target.
In conclusion, CXCL12 secretion from PCC can induce SC carcinotropism via both CXCR4 and CXCR7 and contribute to central hypoalgesia in PDAC. Modulating this chemokine–receptor axis thus may turn out to be a double-edged sword, limiting the initiation of NI in PDAC at the cost of potentially increased pain sensation. Although these findings need to be extended by studies in advanced stages of cancer, this mechanistic demonstration of the role of CXCL12–CXCR4–CXCR7 axis in reactive gliosis in PDAC may have implications for understanding NI, pain, and nerve–cancer interactions in several malignancies.
Materials and Methods
All animal experiments were carried out in accordance with the regulations of the governmental commission for animal protection of the Government of Upper Bavaria. All patients provided informed written consent for tissue collection. The study of the collected tissue was approved by the ethics committee of the Technische Universität München (approval no. 1926/07) and the University of Heidelberg (approval no. 301/2001). For details on the experimental methods, please refer to SI Materials and Methods.
SI Materials and Methods
Patients and Human Tissues.
Tissue samples from the pancreatic head were collected from Union for International Cancer Control (UICC) stage III PDAC patients (n = 25 patients) (Table S1) following tumor resection or from nonallocated NP from organ donors (n = 12) and were processed as described previously (5). All patients provided informed written consent for tissue collection. The study of the collected tissue was approved by the ethics committee of the Technische Universität München (approval no. 1926/07) and the University of Heidelberg (approval no. 301/2001). In all PDAC patients, the individual degree of pain (no pain, 0; mild pain, I; severe pain, II) was registered prospectively and calculated before the operation, as described previously (40).
Conditional Mouse Models.
To generate glia-specific CXCR4-KO (GCE;CXCR4lox/lox) mice that express the Cre-estrogen receptor (Cre-ERT2) fusion protein under the control of the GFAP promoter [B6.Cg-Tg(GFAP-cre/ERT2)505Fmv/J; GCE mice] were interbred with mice that homozygously carry loxP sites on either side of exon 2 of the Cxcr4 gene (B6.129P2-Cxcr4tm2Yzo; CXCR4lox/lox mice). GCE and CXCR4lox/lox mice were obtained from The Jackson Laboratory. To obtain glia-specific CXCR7-KO (GCE;CXCR7lox/lox) mice, CXCR7lox/+ mice that heterozygously carry loxP sites on either side of exon 2 of the Cxcr7 gene (41), kindly provided by ChemoCentryx, Inc., were crossed to obtain CXCR7lox/lox mice. CXCR7lox/lox mice then were interbred with GCE mice. The correct genotype was confirmed via PCR within the DNA isolated from the ear tissue (obtained during labeling of the mice). To activate the Cre recombinase, 4-OHT (Sigma-Aldrich) was added daily to the medium of the 3D migration assay at 1 µM from the time of seeding onwards for 5 consecutive days. Excision of CXCR4 or CXCR7 was confirmed in the protein lysate of the cells used for the in vitro assays.
Mutant LSL-KrasG12D;p48-cre (KC) mice develop the putative precursor of PDAC, i.e., PanIN lesions, and exhibit PDAC at around 1 y of age (42). To obtain pancreas-specific knockout of CXCL12, mice that heterozygously carry loxP sites around the exon 2 of the mouse Cxcl12 gene [B6(FVB)-Cxcl12tm1.1Link/J mice] were crossed to KC12lox/+ mice. Excision and reduced expression of acinar CXCL12 was confirmed via immunofluorescence labeling with CK19 and CXCL12. All animal experiments were carried out in accordance with the regulations of the governmental commission for animal protection of the Government of Upper Bavaria.
Histopathological Analysis, Immunohistochemistry, and Immunofluorescence.
Consecutive 3-µm sections from formalin-fixed, paraffin-embedded mouse and human tissues or monolayers of hSC were incubated overnight with the corresponding antibodies (Table S2) in a humid chamber at 4 °C, as shown previously (43). For immunofluorescence staining, Alexa Fluor 488 and 594 antibodies (Invitrogen) in combination with DAPI nuclear stain were used. Histopathological analysis was performed by two independent observers (K.K. and I.E.D.) blinded to patient diagnosis, followed by resolution of any differences by joint review and consultation with a third observer (G.O.C.), as performed previously (44). The degree of immunoreactivity on each human tissue section and each tissue substructure was scored and added using a numerical scale (0: no staining; 1: weak staining; 2: moderate staining; 3: strong staining) and was averaged among all patients to obtain the mean tissue immunoreactivity score, as described previously (40, 45).
Table S2.
Primary and secondary antibodies
| Antibody | Species | Type | Dilution | Source |
| Alexa Fluor goat anti-mouse IgG 488/594 | Goat | Polyclonal | 1:400 (IF) | Invitrogen |
| Alexa Fluor goat anti-rabbit IgG 488/594 | Goat | Polyclonal | 1:400 (IF) | Invitrogen |
| CXCL12 | Mouse | Monoclonal | 1:350 (IHC/IF) | R&D Systems |
| CXCR4 | Rabbit | Polyclonal | 1:400 (IHC/IF) 1:500 (WB) | Abcam |
| CXCR7 | Mouse | Monoclonal | 1:400 (IHC/IF),1:500 (WB) | MBL |
| CK19 | Mouse | Monoclonal | 1:200 (IF) | Santa Cruz |
| GAPDH | Mouse | Monoclonal | 1:5,000 (WB) | Santa Cruz |
| GFAP | Rabbit | Polyclonal | 1:1,000 (WB), 1:400 (IHC/IF) | DAKO |
| Iba-1 | Mouse | Monoclonal | 1:200 (IF) | Cell Signaling |
| p75NTR | Mouse | Monoclonal | 1:200 (IF) | Sigma-Aldrich |
| PGP 9.5 | Mouse | Monoclonal | 1:1,000 (IHC) | DAKO |
| phospho-ERK | Rabbit | Polyclonal | 1:200 (IF) | Cell Signaling |
| phospho-p38 | Rabbit | Polyclonal | 1:200 (IF) | Cell Signaling |
| S100 | Mouse | Monoclonal | 1:200 (IF) | Millipore |
| Sox10 | Rabbit | Polyclonal | 1:400 (IF) | Novus Biologicals |
IF, immunofluorescence; IHC, immunohistochemistry; WB, Western blot.
In the analysis of mouse tissue, all visible PanIN lesions on one section per mouse were included in the analysis. KC and KC12lox/+ mice between the ages of 12 and 22 wk were used in the study. To rule out differences caused by potentially different PanIN progression rates in different genotypes (i.e., KC vs. KC12lox/+), data from mice with the same histologic degree of PanIN progression were included in the behavioral and spinal cord analyses. In addition to H&E staining, consecutive sections of the pancreatic tumor were immunostained with antibodies against GFAP, S100, or Sox10, and the nuclei were visualized by DAPI staining. Digital imaging was performed on the Keyence BioRevo BZ-9000 platform (Keyence).
After behavioral and hyperalgesia testing, mice were killed, and pancreas-innervating thoracic segments 8–11, together with the spinal cord, were removed en bloc, formalin-fixed, paraffin-embedded, and subjected to EDTA-based demineralization. Consecutive 3-µm sections of the spinal cord of each animal were immunostained for GFAP, p75NTR, Iba-1, phospho-p38/p-p38, p-ERK, and β-III-tubulin. Color thresholds in the ImageJ software were used to measure the area covered by the activation markers p75NTR for astrocytes and p-p38 for microglia, in proportion to the total area of GFAP-stained astroglia and the percent of Iba-1–stained microglia in both dorsal horns of each spinal cord section. Two sections were analyzed per mouse.
hSC and PDAC Cell Lines.
Human PCC lines AsPC-1, BxPC, Capan1, Colo357, MiaPaCa-2, Panc1, and SU86.86 were purchased from ATCC, and hSC isolated from human sciatic nerves (ScienCell) served as primary cells. T3M4 cells were a gift from R. Metzgar, Duke University, Durham, NC. The cell lines and the primary hSC were routinely grown in complete medium, as shown previously (2, 46). The cell lysates and supernatants were obtained at 100% cell confluence, and protein concentration was measured with the BCA protein assay (Pierce Chemical Co.).
Immunoblotting.
Thirty micrograms of protein from cell-culture monolayers or cells that were extracted from the ECM within the 3D migration assays were separated and electroblotted, and the membrane was exposed to CXCR4 or CXCR7 (1:500; MBL) antibodies at 4 °C overnight. Equal loading was assured by reprobing with anti-GAPDH antibodies, as shown previously (44). The densitometric analysis of the blot was performed via ImageJ version 1.44p (NIH) (40).
ELISA.
Protein levels of CXCL12 were measured in PCC and SC lysates via the human SDF-α Quantikine ELISA kit (R&D Systems) according to the manufacturer’s instructions.
Hypoxia Exposure of hSC and Tumor Cells.
The effect of hypoxia upon CXCR4 and CXCR7 levels in hSC and PCC was assessed by incubating these cells under hypoxic conditions (89.25% N2 + 10% CO2 + 0.75% O2) after reaching 80% confluence for up to 48 h at 37 °C in normal serum-free growth medium. Cells then were lysed with radioimmunoprecipitation (RIPA) buffer containing a protease inhibitor mixture (Roche), and protein content was measured with the BCA protein assay (Pierce Chemical Co.).
SC, PCC, and DRG Coculture Experiments.
hSC (150,000 cells per well) or DRG neurons (derived from five rat DRG per well) were seeded in poly-d-lysine (Sigma Aldrich)-coated six-well plates. After 24 h, coculture inserts (0.4-μm pores) (BD Falcon) were placed in the wells, and T3M4 or SU86.86 PDAC cells were seeded at 300,000 cells per insert. Cells in the upper and lower compartments were lysed after 48 h of cocultivation with RIPA buffer containing a Complete protease inhibitor tablet (Roche) for subsequent immunoblotting.
Transwell Chemotaxis Assay.
Five thousand PCC (T3M4 or MiaPaCa-2) were placed in each well of a 24-well plate and were incubated overnight. Five thousand hSC were added to 24-well 8-μm chemotaxis chamber inserts (BD Falcon), placed in the wells containing PCC, and kept for 22 h in a 1:1 mixture of PCC-SC medium supplied with the CXCR4 small-molecule inhibitor AMD3100 (0.5 µg/mL or 1.25 µg/mL) (Tocris) or mouse anti-CXCR7 neutralizing antibody (10 µg/mL or 30 µg/mL) (MBL). After 22 h, the inserts were removed, cleaned of nonmigrating cells, fixed in 4% (wt/vol) paraformaldehyde, stained with the Vybrant CFDA SE Cell Tracker Kit (Life Technologies), and scanned via an automated digital epifluorescence microscope (Keyence BioRevo BZ-9000). The number of stained (migrated) cells was counted via ImageJ version 1.44p (NIH).
3D Migration Assay.
The 3D migration assay was performed with human PCC, hSC, and dissociated DRG of newborn GCE;CXCR4lox/lox or GCE;CXCR7lox/lox mice. DRG from nonrecombined CXCR4lox/lox or CXCR7lox/lox mice (i.e., lacking Cre recombinase) were used as controls. Then 105 cells of each type were resuspended in an ECM gel drop (Sigma-Aldrich), placed at 1-mm distance from each other connected via an ECM “bridge” for the generation of a chemoattractive gradient, and analyzed via digital time-lapse microscopy, as described previously (2). In the assays with hSC, these cells were pretreated with the CXCR4 small-molecule inhibitor AMD3100 (0.5 µg/mL or 1.25 µg/mL) (Tocris) or mouse anti-CXCR7 neutralizing antibody (10 µg/mL or 30 µg/mL) (MBL) for 30 min at 37 °C in serum-free medium and subsequently resuspended in the ECM gel for the assay.
siRNA Transfection of hSC.
hSC were transfected with two different siRNA sequences against CXCR4 or CXCR7 or with scrambled control siRNA, as described previously (40). Forty-eight hours after transfection, hSC were detached from their culture flasks and subsequently lysed for RNA extraction via the RNeasy mini kit (Qiagen). All siRNA oligomers were obtained from Sigma-Aldrich. Their nucleotide sequences are indicated in Table S8.
Abdominal Mechanosensitivity Testing.
Mechanical hypersensitivity was tested via von Frey filaments (Stoelting Co.) of increasing tactile stimulus intensity by applying each filament type 10 consecutive times from the bottom of a grid to the abdomen of the mice and by adding the reaction scores of each mouse (no reaction: 0; slight reaction or licking:1; jumping: 2) (47).
RT2 Profiler PCR Array.
The transcriptional profile of the hSC after treatment with recombinant human CXCL12 (10 ng/mL) (R&D Systems) was compared within the total RNA via the RT2 Profiler PCR Array Human Pain: Neuropathic & Inflammatory (SABiosciences, Qiagen) according to the manufacturer’s instructions. The PCR plates were analyzed on a Roche LightCycler 480 system. Statistical comparisons were performed using the manufacturer’s online data analysis center. The changes in the expression profile were considered relevant for a gene if an absolute fold regulation of at least threefold was detected after t test-based statistical comparison of the reference group (RNA of untreated hSC) with the experimental groups (RNA of CXCL12-treated hSC).
Statistical Analysis.
Results are expressed as mean ± SD. Two-group analyses were performed using the unpaired t test for continuous values and with the Mann–Whitney u test for scores and indices. Analyses of more than two groups were conducted using one-way ANOVA followed by Bonferroni’s multiple comparison test. All tests were two-sided, and a P value < 0.05 was considered to indicate statistical significance.
Acknowledgments
We thank Prof. Andreas Schober for providing CXCR7fl/+ mice and Ms. Ulrike Bourquain for excellent technical assistance. CXCR7fl/fl mice were provided by ChemoCentryx, Inc., as part of a material transfer agreement. This work is part of K.K.’s M.D. thesis. I.E.D. was supported by an institutional KKF (Kommission für Klinische Forschung, B10-10) Grant of the Faculty of Medicine of the Technische Universität München.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1606909114/-/DCSupplemental.
References
- 1.Demir IE, Friess H, Ceyhan GO. Neural plasticity in pancreatitis and pancreatic cancer. Nat Rev Gastroenterol Hepatol. 2015;12(11):649–659. doi: 10.1038/nrgastro.2015.166. [DOI] [PubMed] [Google Scholar]
- 2.Demir IE, et al. Investigation of Schwann cells at neoplastic cell sites before the onset of cancer invasion. J Natl Cancer Inst. 2014;106(8):dju184. doi: 10.1093/jnci/dju184. [DOI] [PubMed] [Google Scholar]
- 3.Demir IE, Schäfer KH, Tieftrunk E, Friess H, Ceyhan GO. Neural plasticity in the gastrointestinal tract: Chronic inflammation, neurotrophic signals, and hypersensitivity. Acta Neuropathol. 2013;125(4):491–509. doi: 10.1007/s00401-013-1099-4. [DOI] [PubMed] [Google Scholar]
- 4.Liebl F, et al. The impact of neural invasion severity in gastrointestinal malignancies: A clinicopathological study. Ann Surg. 2014;260(5):900–907, discussion 907–908. doi: 10.1097/SLA.0000000000000968. [DOI] [PubMed] [Google Scholar]
- 5.Ceyhan GO, et al. Pancreatic neuropathy and neuropathic pain–a comprehensive pathomorphological study of 546 cases. Gastroenterology. 2009;136(1):177–186 e171. doi: 10.1053/j.gastro.2008.09.029. [DOI] [PubMed] [Google Scholar]
- 6.Demir IE, Friess H, Ceyhan GO. Nerve-cancer interactions in the stromal biology of pancreatic cancer. Front Physiol. 2012;3:97. doi: 10.3389/fphys.2012.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gil Z, et al. Paracrine regulation of pancreatic cancer cell invasion by peripheral nerves. J Natl Cancer Inst. 2010;102(2):107–118. doi: 10.1093/jnci/djp456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demir IE, et al. Activated Schwann cells in pancreatic cancer are linked to analgesia via suppression of spinal astroglia and microglia. Gut. 2016;65(6):1001–1014. doi: 10.1136/gutjnl-2015-309784. [DOI] [PubMed] [Google Scholar]
- 9.Sun X, et al. CXCL12 / CXCR4 / CXCR7 chemokine axis and cancer progression. Cancer Metastasis Rev. 2010;29(4):709–722. doi: 10.1007/s10555-010-9256-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zou YR, Kottmann AH, Kuroda M, Taniuchi I, Littman DR. Function of the chemokine receptor CXCR4 in haematopoiesis and in cerebellar development. Nature. 1998;393(6685):595–599. doi: 10.1038/31269. [DOI] [PubMed] [Google Scholar]
- 11.Roy I, et al. CXCL12 chemokine expression suppresses human pancreatic cancer growth and metastasis. PLoS One. 2014;9(3):e90400. doi: 10.1371/journal.pone.0090400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Domanska UM, et al. A review on CXCR4/CXCL12 axis in oncology: No place to hide. Eur J Cancer. 2013;49(1):219–230. doi: 10.1016/j.ejca.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 13.Burns JM, et al. A novel chemokine receptor for SDF-1 and I-TAC involved in cell survival, cell adhesion, and tumor development. J Exp Med. 2006;203(9):2201–2213. doi: 10.1084/jem.20052144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koshiba T, et al. Expression of stromal cell-derived factor 1 and CXCR4 ligand receptor system in pancreatic cancer: A possible role for tumor progression. Clin Cancer Res. 2000;6(9):3530–3535. [PubMed] [Google Scholar]
- 15.Maréchal R, et al. High expression of CXCR4 may predict poor survival in resected pancreatic adenocarcinoma. Br J Cancer. 2009;100(9):1444–1451. doi: 10.1038/sj.bjc.6605020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morimoto M, et al. Enhancement of the CXCL12/CXCR4 axis due to acquisition of gemcitabine resistance in pancreatic cancer: Effect of CXCR4 antagonists. BMC Cancer. 2016;16:305. doi: 10.1186/s12885-016-2340-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feig C, et al. Targeting CXCL12 from FAP-expressing carcinoma-associated fibroblasts synergizes with anti-PD-L1 immunotherapy in pancreatic cancer. Proc Natl Acad Sci USA. 2013;110(50):20212–20217. doi: 10.1073/pnas.1320318110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao Z, Wang X, Wu K, Zhao Y, Hu G. Pancreatic stellate cells increase the invasion of human pancreatic cancer cells through the stromal cell-derived factor-1/CXCR4 axis. Pancreatology. 2010;10(2-3):186–193. doi: 10.1159/000236012. [DOI] [PubMed] [Google Scholar]
- 19.Koong AC, et al. Pancreatic tumors show high levels of hypoxia. Int J Radiat Oncol Biol Phys. 2000;48(4):919–922. doi: 10.1016/s0360-3016(00)00803-8. [DOI] [PubMed] [Google Scholar]
- 20.Naumann U, et al. CXCR7 functions as a scavenger for CXCL12 and CXCL11. PLoS One. 2010;5(2):e9175. doi: 10.1371/journal.pone.0009175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scholz J, Woolf CJ. The neuropathic pain triad: Neurons, immune cells and glia. Nat Neurosci. 2007;10(11):1361–1368. doi: 10.1038/nn1992. [DOI] [PubMed] [Google Scholar]
- 22.Yamamoto Y, et al. Activated microglia contribute to convergent nociceptive inputs to spinal dorsal horn neurons and the development of neuropathic pain. Neurochem Res. 2015;40(5):1000–1012. doi: 10.1007/s11064-015-1555-8. [DOI] [PubMed] [Google Scholar]
- 23.Lazarini F, Tham TN, Casanova P, Arenzana-Seisdedos F, Dubois-Dalcq M. Role of the alpha-chemokine stromal cell-derived factor (SDF-1) in the developing and mature central nervous system. Glia. 2003;42(2):139–148. doi: 10.1002/glia.10139. [DOI] [PubMed] [Google Scholar]
- 24.Reaux-Le Goazigo A, Rivat C, Kitabgi P, Pohl M, Melik Parsadaniantz S. Cellular and subcellular localization of CXCL12 and CXCR4 in rat nociceptive structures: Physiological relevance. Eur J Neurosci. 2012;36(5):2619–2631. doi: 10.1111/j.1460-9568.2012.08179.x. [DOI] [PubMed] [Google Scholar]
- 25.Wilson NM, Jung H, Ripsch MS, Miller RJ, White FA. CXCR4 signaling mediates morphine-induced tactile hyperalgesia. Brain Behav Immun. 2011;25(3):565–573. doi: 10.1016/j.bbi.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang JJ, et al. High levels of expression of human stromal cell-derived factor-1 are associated with worse prognosis in patients with stage II pancreatic ductal adenocarcinoma. Cancer Epidemiol Biomarkers Prev. 2010;19(10):2598–2604. doi: 10.1158/1055-9965.EPI-10-0405. [DOI] [PubMed] [Google Scholar]
- 27.Marchesi F, et al. Increased survival, proliferation, and migration in metastatic human pancreatic tumor cells expressing functional CXCR4. Cancer Res. 2004;64(22):8420–8427. doi: 10.1158/0008-5472.CAN-04-1343. [DOI] [PubMed] [Google Scholar]
- 28.Saur D, et al. CXCR4 expression increases liver and lung metastasis in a mouse model of pancreatic cancer. Gastroenterology. 2005;129(4):1237–1250. doi: 10.1053/j.gastro.2005.06.056. [DOI] [PubMed] [Google Scholar]
- 29.Shen X, et al. Chemokine receptor CXCR4 enhances proliferation in pancreatic cancer cells through AKT and ERK dependent pathways. Pancreas. 2010;39(1):81–87. doi: 10.1097/MPA.0b013e3181bb2ab7. [DOI] [PubMed] [Google Scholar]
- 30.Singh AP, et al. CXCL12/CXCR4 protein signaling axis induces sonic hedgehog expression in pancreatic cancer cells via extracellular regulated kinase- and Akt kinase-mediated activation of nuclear factor κB: Implications for bidirectional tumor-stromal interactions. J Biol Chem. 2012;287(46):39115–39124. doi: 10.1074/jbc.M112.409581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhong W, et al. CXCL12/CXCR4 axis plays pivotal roles in the organ-specific metastasis of pancreatic adenocarcinoma: A clinical study. Exp Ther Med. 2012;4(3):363–369. doi: 10.3892/etm.2012.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gebauer F, et al. Prognostic impact of CXCR4 and CXCR7 expression in pancreatic adenocarcinoma. J Surg Oncol. 2011;104(2):140–145. doi: 10.1002/jso.21957. [DOI] [PubMed] [Google Scholar]
- 33.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454(7203):436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 34.Küry P, et al. Cyclic AMP and tumor necrosis factor-alpha regulate CXCR4 gene expression in Schwann cells. Mol Cell Neurosci. 2003;24(1):1–9. doi: 10.1016/s1044-7431(03)00132-5. [DOI] [PubMed] [Google Scholar]
- 35.Odemis V, Boosmann K, Heinen A, Küry P, Engele J. CXCR7 is an active component of SDF-1 signalling in astrocytes and Schwann cells. J Cell Sci. 2010;123(Pt 7):1081–1088. doi: 10.1242/jcs.062810. [DOI] [PubMed] [Google Scholar]
- 36.Dziembowska M, et al. A role for CXCR4 signaling in survival and migration of neural and oligodendrocyte precursors. Glia. 2005;50(3):258–269. doi: 10.1002/glia.20170. [DOI] [PubMed] [Google Scholar]
- 37.Schönemeier B, Schulz S, Hoellt V, Stumm R. Enhanced expression of the CXCl12/SDF-1 chemokine receptor CXCR7 after cerebral ischemia in the rat brain. J Neuroimmunol. 2008;198(1-2):39–45. doi: 10.1016/j.jneuroim.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 38.Odemis V, Moepps B, Gierschik P, Engele J. Interleukin-6 and cAMP induce stromal cell-derived factor-1 chemotaxis in astroglia by up-regulating CXCR4 cell surface expression. Implications for brain inflammation. J Biol Chem. 2002;277(42):39801–39808. doi: 10.1074/jbc.M200472200. [DOI] [PubMed] [Google Scholar]
- 39.Pukrop T, et al. Microglia promote colonization of brain tissue by breast cancer cells in a Wnt-dependent way. Glia. 2010;58(12):1477–1489. doi: 10.1002/glia.21022. [DOI] [PubMed] [Google Scholar]
- 40.Wang K, et al. The neurotrophic factor neurturin contributes toward an aggressive cancer cell phenotype, neuropathic pain and neuronal plasticity in pancreatic cancer. Carcinogenesis. 2014;35(1):103–113. doi: 10.1093/carcin/bgt312. [DOI] [PubMed] [Google Scholar]
- 41.Sierro F, et al. Disrupted cardiac development but normal hematopoiesis in mice deficient in the second CXCL12/SDF-1 receptor, CXCR7. Proc Natl Acad Sci USA. 2007;104(37):14759–14764. doi: 10.1073/pnas.0702229104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hingorani SR, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4(6):437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 43.Ceyhan GO, et al. Pancreatic neuropathy results in “neural remodeling” and altered pancreatic innervation in chronic pancreatitis and pancreatic cancer. Am J Gastroenterol. 2009;104(10):2555–2565. doi: 10.1038/ajg.2009.380. [DOI] [PubMed] [Google Scholar]
- 44.Ceyhan GO, et al. The neurotrophic factor artemin influences the extent of neural damage and growth in chronic pancreatitis. Gut. 2007;56(4):534–544. doi: 10.1136/gut.2006.105528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Demir IE, et al. Neuronal plasticity in chronic pancreatitis is mediated via the neurturin/GFRalpha2 axis. Am J Physiol Gastrointest Liver Physiol. 2012;303(9):G1017–1028. doi: 10.1152/ajpgi.00517.2011. [DOI] [PubMed] [Google Scholar]
- 46.Ceyhan GO, et al. The neurotrophic factor artemin promotes pancreatic cancer invasion. Ann Surg. 2006;244(2):274–281. doi: 10.1097/01.sla.0000217642.68697.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Michalski CW, et al. Cannabinoids ameliorate pain and reduce disease pathology in cerulein-induced acute pancreatitis. Gastroenterology. 2007;132(5):1968–1978. doi: 10.1053/j.gastro.2007.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]