Significance
The brain parenchyma is considered to be “immune privileged” based upon differences between the innate and adaptive immune responses of the brain and those of the periphery. This work provides a clear demonstration of an innate immune response to direct infection by a virus, a response that is capable of limiting the spread of the virus along neuronal circuitry. The question of the brain parenchyma’s response to a viral infection has implications for the use of viruses as tools by neuroscientists, for vaccine development, and for potential clinical applications. Additionally, the approach used here provides a framework for further examination of the immunologic state of the brain as well as the mechanisms by which encephalitic viruses circumvent this response.
Keywords: vesicular stomatitis virus, VSV, brain, innate immunity, interferon
Abstract
The brain has a tightly regulated environment that protects neurons and limits inflammation, designated “immune privilege.” However, there is not an absolute lack of an immune response. We tested the ability of the brain to initiate an innate immune response to a virus, which was directly injected into the brain parenchyma, and to determine whether this response could limit viral spread. We injected vesicular stomatitis virus (VSV), a transsynaptic tracer, or naturally occurring VSV-derived defective interfering particles (DIPs), into the caudate–putamen (CP) and scored for an innate immune response and inhibition of virus spread. We found that the brain parenchyma has a functional type I interferon (IFN) response that can limit VSV spread at both the inoculation site and among synaptically connected neurons. Furthermore, we characterized the response of microglia to VSV infection and found that infected microglia produced type I IFN and uninfected microglia induced an innate immune response following virus injection.
The brain has been described as a site of “immune privilege,” owing to the differences between the systemic immune response and that of the brain (1, 2). For example, intranasal influenza virus inoculation elicits T-cell priming and antibody production, whereas no response is detectable following intracerebral inoculation of the virus over the same time frame (3). Similar observations have been made for the innate immune response; lipopolysaccharide injection into the skin, but not the brain, elicits monocyte recruitment within 2 h (4). Recent studies have demonstrated that these differences are confined to the brain parenchyma, which has a specialized immune environment that is actively maintained, facilitated by compartmentalization of the brain, and promoted by the blood–brain barrier (BBB) (1, 5).
The innate immune response provides a rapid defense against infection by pathogens and enhances adaptive immune responses (6). A prominent branch of the systemic innate immune response that controls viral infection is the type I interferon (IFN) response. Virus infection induces expression of IFN-stimulated genes (ISGs) that interfere with viral replication and promote virus clearance. The IFN response is triggered by the recognition of viral pathogen-associated molecular patterns (PAMPs) by host-cell pattern recognition receptors (PRRs). In the canonical model for the type I IFN response, IFN-β production and secretion are first triggered by the recognition of viral PAMPs by PRRs. Then, IFN-β binds the IFN-α receptor (IFNαR) in an autocrine and paracrine manner to drive IRF7 gene expression and enable the full type I IFN response upon viral spread or secondary infection (7).
Recent in vivo studies have provided evidence supporting the induction of the type I IFN response in the brain by viruses. For instance, infection of the brain by Theiler’s virus and La Crosse virus (LACV) led to the production of type I IFN by ependymal cells, macrophages, and neurons; however, only 3% of infected neurons expressed type I IFN (8). A similar study with LACV found nearly no type I IFN-expressing neurons but rather that glia expressed type I IFN (9). In addition, other innate immune pathways, such as the type II and III IFN responses (10, 11), and inflammation (12), are activated by viral infection. These observations suggest a great deal of complexity in the brain’s response to viral infection and raise several issues in need of clarification. For example, which cell types mount a particular type of innate immune response and how different cell types respond to different innate immunity signals need to be clarified. Perhaps of most importance, however, is whether the immune-privileged parenchyma can limit virus spread through an innate immune response. We set out to answer this question as well as to explore the type of innate immune pathways that are activated in response to viral infection.
We tested whether the brain parenchyma can initiate an innate immune response that is capable of restricting the spread of a model neurotropic virus, vesicular stomatitis virus (VSV). VSV is the prototypic nonsegmented negative-strand RNA virus and has been used extensively to study the systemic innate immune response, which promotes its rapid clearance (13, 14). VSV was selected because it has a characteristic transsynaptic anterograde transmission pattern among neurons and is straightforward to track, as it can be engineered to express eGFP (15). As the spread of wild-type (WT) VSV is not well-controlled in the mouse brain and can rapidly lead to mortality (16), we sought to provide the host with a “head start” in the induction of an innate immune response following infection. This provided a sensitized system for detecting and measuring the effects of a host response on viral replication and transsynaptic transmission. For this purpose, we introduced naturally occurring VSV-derived defective interfering particles (DIPs). DIPs have been found in the sera of patients during multiple viral infections and in viral vaccine strains. At least in a subset of infections, it has been posited that DIPs promote antiviral host responses, and in cultured cells, DIPs can stimulate an innate immune response (17). The inhibition of WT VSV replication by DIPs is well established and thought to occur in coinfected cells due to the more rapid replication of the short DIPs’ genomes compared with the much longer full-length viral genome. Coinjection of DIPs with full-length virus into the nose (18) and brain (19) has been reported to extend the survival of mice, with protection corresponding to the subsequent detection of DIPs’ genomes (20). However, whether this effect was solely due to direct inhibition of VSV replication, or whether induction of an innate immune response by DIPs played a role, has not been fully explored. We used DIPs as a tool to probe for the promotion of an effective antiviral innate immune response in the brain. We found that DIPs could incite a paracrine response from the parenchyma that can limit the transsynaptic spread of VSV and determined that this response required the type I IFN pathway. In addition, we analyzed the response of microglia to VSV infection in vivo. Microglia are believed to be the major resident cell type to mediate innate immune responses within the brain (5, 10, 12, 21). We found that infected microglia produce type I IFN and that uninfected microglia are primed for a full type I IFN response in the context of brain infection.
A greater understanding of the brain parenchyma’s response to infection by a neurotropic virus should inform several areas of research and perhaps lead to improvements in therapies. Neurotropic viruses are increasingly used as transsynaptic tracers (22) but only label a fraction of the connections of infected neurons (23, 24). The data presented here raise the possibility that innate immunity is at least one factor that contributes to this inefficiency. In addition, the potential neurotropism of vaccines, including VSV-based vaccines (25), has raised concerns, even when there is a desperate need—for example, for Ebola vaccines (26). Finally, a greater understanding of virus–host interactions in the brain might enable the stimulation of a more effective response in the unfortunate cases of naturally occurring viral infections of the brain (27).
Results
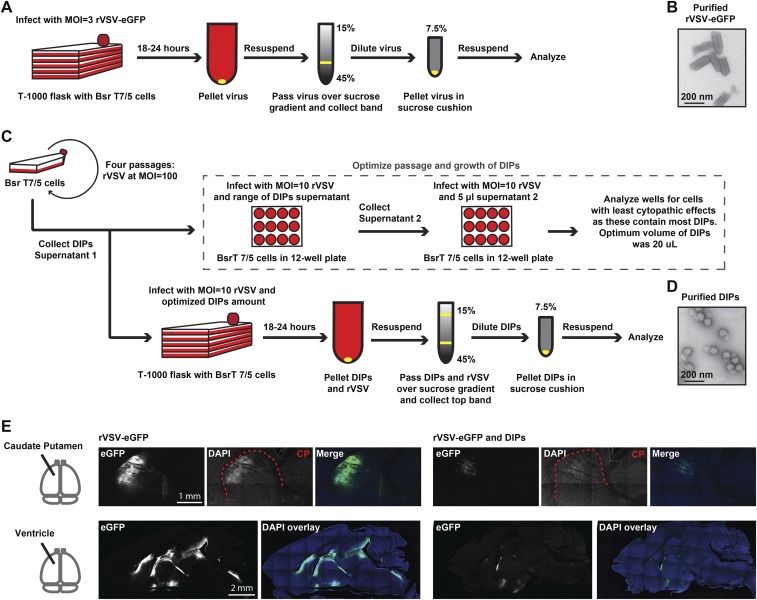
Generation of Recombinant VSV-eGFP and Recombinant VSV DIPs for in Vivo Use.
DIPs are infectious replication-defective particles that are generated by spontaneous genome deletions during the passaging of replication-competent parent viruses. DIPs have truncated genomes, smaller viral particle size, and can interfere with replication of the parent virus. We wished to use DIPs to test the response of the immune system to viral particles because DIPs have been shown to be potent stimulators of the innate immune response (17). This potency likely results from the inability of DIPs to shut off host transcription, including IFN induction, as is achieved by the parent virus (28). We first generated relatively pure stocks of recombinant (r) VSV-eGFP, with eGFP in the first genome position, and rVSV-derived DIPs (Fig. S1 A and C) (29–33). We used negative-stain transmission electron microscopy (TEM) to examine each stock (Fig. S1 B and D). We did not observe any DIPs in the purified rVSV-eGFP preparations and observed that the DIPs preparations had <0.1% full-length particles. Previous studies have shown that coinjection of DIPs with WT VSV into the brain delays mortality and reduces viral titers (19) but did not define the injection sites within the brain. To test whether DIPs could directly inhibit VSV propagation in immunologically distinct locations within the brain, we coinjected DIPs with rVSV-eGFP into the caudate–putamen (CP) or ventricles and evaluated the expression of eGFP in the brain at 3 d postinfection (dpi) (Fig. S1 E and F). As has been seen with work conducted in cell culture (34) and in vivo (18, 19), rVSV-eGFP propagation was indeed inhibited by DIPs in both locations.
Fig. S1.
Effect of DIP coinjection on rVSV-eGFP infection and replication. (A) Schematic for gradient purification of rVSV-eGFP. (B) Negative-stain EM of gradient-purified rVSV-eGFP. (C) Schematic for production and purification of DIPs. (D) Negative-stain EM of gradient-purified DIPs. (E) Direct inhibition of rVSV-eGFP infection and spread in CP and ventricles; DIP stock or PBS was mixed 1:1 with 4 × 107 pfu/mL rVSV-eGFP, and 100 nL volume was injected. Infection was allowed to proceed for 3 d, at which point tissue was collected and sagittal sections were made and DAPI-stained.
rVSV-eGFP Spread Was Reduced by Contralateral Injection of DIPs in the Brain.
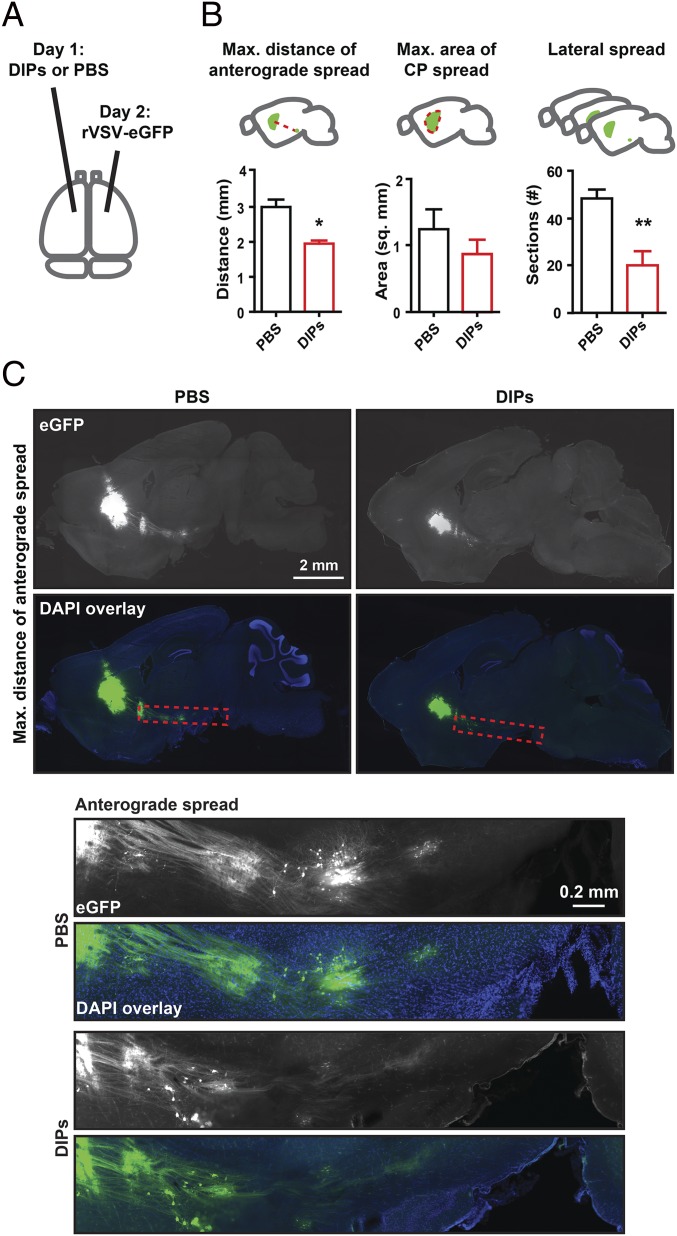
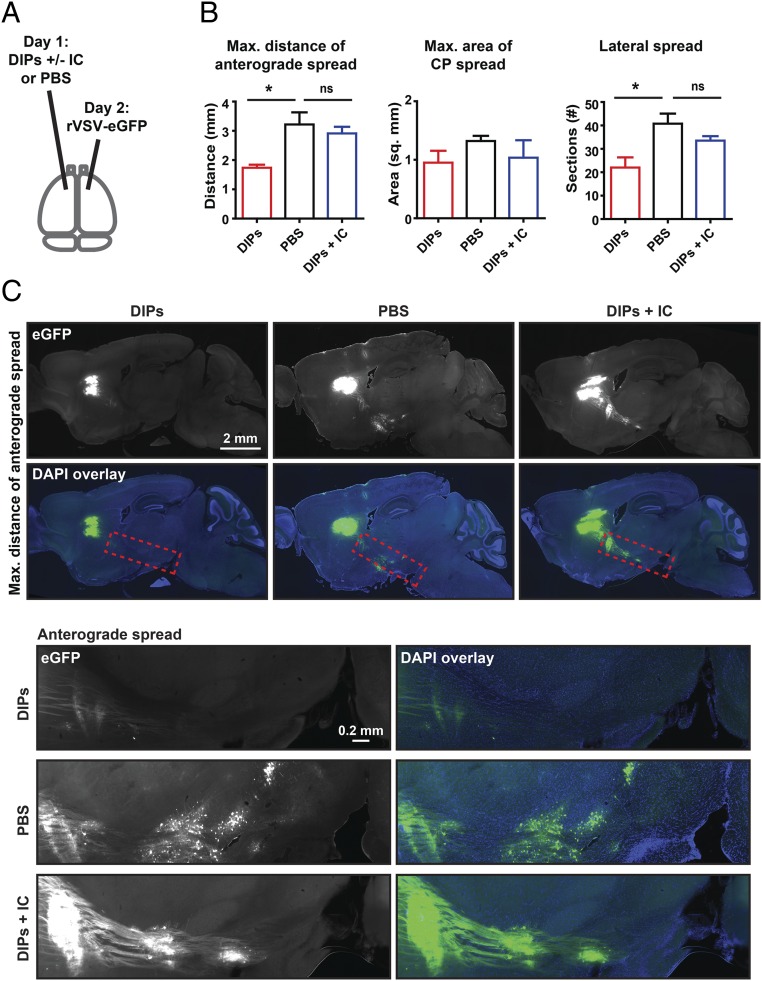
DIPs were used to test whether infectious particles can induce paracrine signaling capable of limiting virus infection and spread in the brain parenchyma. The infection and spread of replication-competent rVSV-eGFP was quantified. The CP was chosen as an inoculation site due to its unidirectional, anterograde connections to the globus pallidus (GP) and basal nuclei (NB) (15).
DIPs or PBS were injected into the right CP 1 d before rVSV-eGFP inoculation into the left CP. Sagittal sections were made from tissue collected at 3 dpi with rVSV-eGFP (Fig. 1A), and virus infection was scored using three metrics: maximum distance of anterograde spread (MDAS), maximum area of CP spread (MACS), and lateral spread (LS) (Fig. 1B). These metrics were chosen to allow relative quantification of infection and transmission throughout the initial inoculation site (MACS and LS) and transmission to more distant sites (MDAS) (Fig. 1B). MDAS was measured from the center of the infected CP area to the cell body of the furthest labeled anterograde cell. This metric likely assesses only transsynaptic transmission events. MACS allowed for quantification of spread within the initial inoculation site. It was determined by scoring the largest labeled area for each animal following review of all sagittal sections. LS was scored as the number of labeled sections spreading from the CP infection site. It encompasses spread within the parenchyma. MACS and LS likely encompass initial infection of neuronal and nonneuronal cells as well as transmission events within the inoculation site.
Fig. 1.
Effect of contralateral injection of DIPs on rVSV-eGFP spread. (A) Timing and infection sites for injection of DIPs and rVSV-eGFP into the CP are indicated. Effect of contralateral injection of DIPs on rVSV-eGFP spread was tested. DIPs and rVSV-eGFP stocks were diluted, and 100 nL (170 ng) of DIPs or PBS was injected on day 1 and 100 nL (2,000 pfu) of rVSV-eGFP was injected on day 2. Sagittal sections were made at 3 dpi. (B) Metrics for analysis are illustrated (Top). Graphs display means ± SEM for each metric and condition. n PBS = 5 and n DIPs = 4. Unpaired two-tailed Student’s t test, *P ≤ 0.05, **P ≤ 0.01. (C) Representative images of sagittal sections with the MDAS and Insets of anterograde spread from these sections are presented. Inset locations are indicated by dotted red line on sagittal sections.
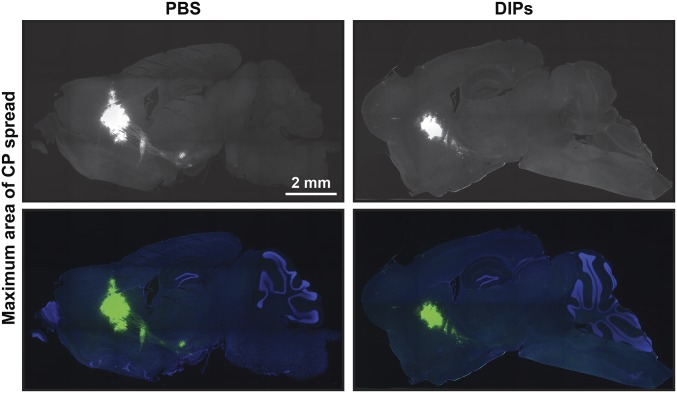
Preinfection by DIPs in one hemisphere limited the spread of rVSV-eGFP introduced into the contralateral hemisphere compared with PBS, with MDAS and LS reduced by ∼35% and ∼60%, respectively (Fig. 1 B and C); MACS was not significantly reduced (Fig. 1A and Fig. S2). Taken together, these data indicate that the brain can mount an effective and rapid antiviral response to limit the spread of a neurotropic virus, including limiting transsynaptic transmission.
Fig. S2.
MACS for rVSV-eGFP following contralateral injections of DIPs and PBS. Shown are corresponding MACS sections to animals presented in Fig. 1.
Type I IFN Can Limit rVSV-eGFP Spread.
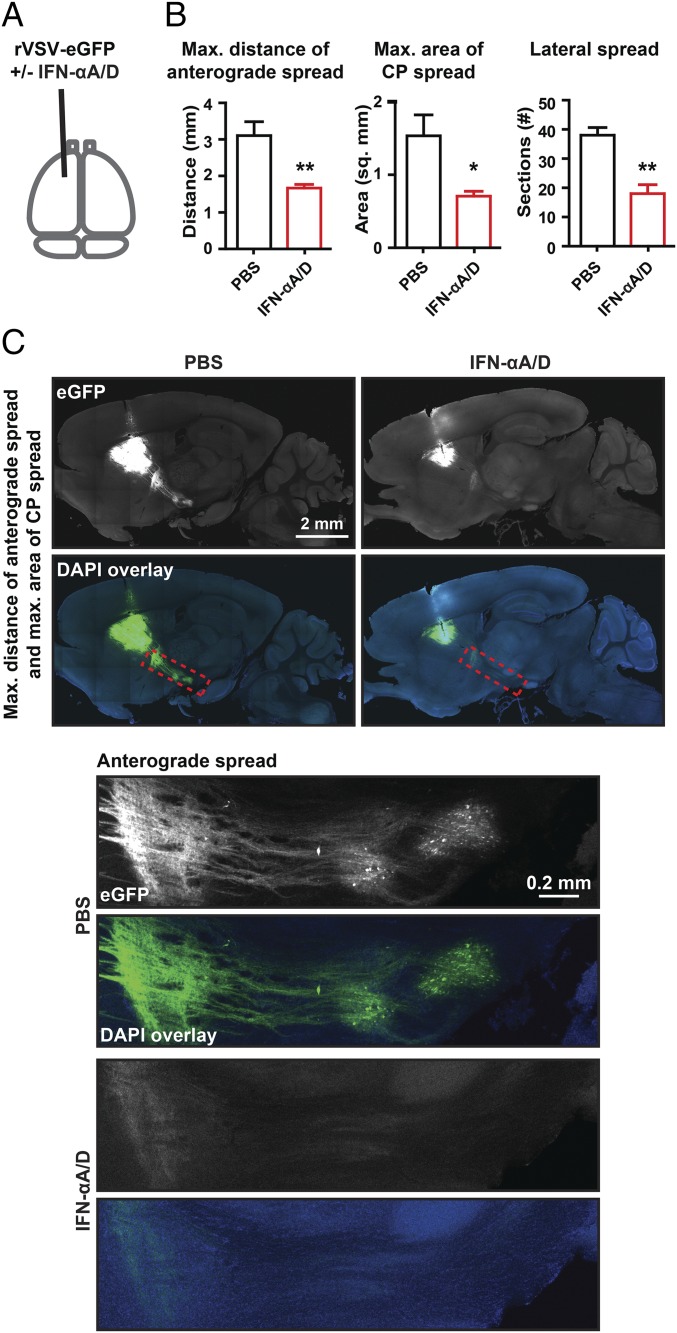
To define the mechanism by which the brain limited virus spread, we investigated the activity of type I IFN. The type I IFN response limits virus spread following i.v. VSV inoculation; homozygous IFNαR-null animals show significant VSV infection in multiple organs, including the brain. In contrast, animals with a defective type II IFN response do not have increased susceptibility to systemic VSV infection (14).
IFN-αA/D, known as universal type I IFN, is an IFN-α hybrid that elicits a type I response and is active in multiple species (35). We coinjected recombinant IFN-αA/D (80 units) and rVSV-eGFP (2,000 pfu) into the CP and scored viral spread at 3 dpi (Fig. 2A). Coinjection was used to directly assay the effect of type I IFN on rVSV-eGFP spread in the parenchyma. The MDAS, MACS, and LS were significantly reduced (Fig. 2 B and C), indicating that the type I IFN response elicited by IFN-αA/D coinjection limited both distant transsynaptic and local spread of rVSV-eGFP.
Fig. 2.
Effect of type I IFN coinjection on rVSV-eGFP spread. (A) Timing and infection site for injection of rVSV-eGFP with or without IFNα-A/D into the CP are indicated. rVSV-eGFP stocks were diluted in either PBS or PBS with a final concentration of 8 × 105 U/mL of IFNα-A/D. A volume of 100 nL (2,000 pfu) of each was injected into the CP, and infection was allowed to proceed for 3 d. (B) Graphs display mean ± SEM for each metric and condition. n PBS = 3 and n IFNα-A/D = 4. Unpaired two-tailed Student’s t test, *P ≤ 0.05, **P ≤ 0.01. (C) Representative images of sagittal sections with the MDAS and MACS shown and Insets of anterograde spread from these sections are presented. Inset locations are indicated by dotted red line on sagittal sections.
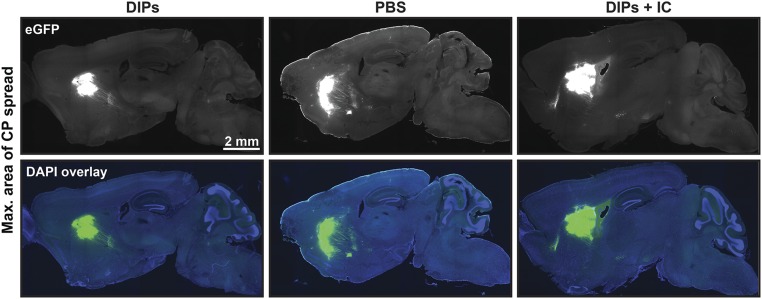
A Type I IFN Pathway Inhibitor Mixture Reduces Inhibition of rVSV-eGFP Spread by Contralateral DIPs.
To test whether DIPs initiated a type I IFN response and, if so, whether it was responsible for the observed inhibition of VSV spread (Fig. 1), we targeted both IFN gene expression and IFNαR signaling through the use of an inhibitor cocktail, comprising BX-795, CP-690550, and INCB018424. BX-795 inhibits TBK1/IKKε phosphorylation and subsequent IFN-β production (36). INCB018424 and CP-690550 inhibit JAK1–3 and prevent downstream signaling from the IFNαR (37, 38). DIPs were coinjected into the CP with the inhibitor mixture 1 d before injection of rVSV-eGFP into the contralateral CP (Fig. 3A). DIPs alone were injected in parallel as a positive control. PBS preinjection was used as a negative control (Fig. 3A). As observed previously, DIPs preinjection significantly reduced the MDAS and LS compared with PBS (Fig. 3 B and C and Fig. S3). In contrast, coinjection of the inhibitor mixture with DIPs ameliorated the inhibition by DIPs on rVSV-eGFP spread (Fig. 3 B and C). These data, taken together with the ability of IFN-αA/D to inhibit spread, suggest that DIPs induce the type I IFN pathway, which restricts virus spread.
Fig. 3.
Effect of inhibitor cocktail (IC) on DIP restriction of rVSV-eGFP spread. (A) Timing and infection sites for injection of DIPs, inhibitor cocktail (IC; BX795, CP-690550, and INCB018424), PBS, and rVSV-eGFP into the CP are indicated. Effect of IC on DIPs’ restriction of rVSV-eGFP spread was tested. A volume of 100 nL of either PBS, DIPs (170 ng), or DIPs with IC (0.13 mM for BX795, 0.27 mM for CP-690550, and 0.27 mM for INCB018424) was injected on day 1. Then, 100 nL of rVSV-eGFP (2,000 pfu) was injected on day 2. Tissue was examined at 3 dpi with rVSV-eGFP. (B) Graphs display means ± SEM for each metric and condition. n DIPs = 3, n PBS = 4, and n DIPs+IC = 4. ANOVA with Dunnett’s multiple comparison post test; *P ≤ 0.05; ns, not statistically significant. (C) Representative images of sagittal sections with the MDAS and Insets of anterograde spread from these sections are presented. Inset locations are indicated by the dotted red line on sagittal sections.
Fig. S3.
MACS for injections of DIPs with or without the inhibitor cocktail (IC), followed by rVSV-eGFP. Shown are corresponding MACS sections to animals presented in Fig. 3.
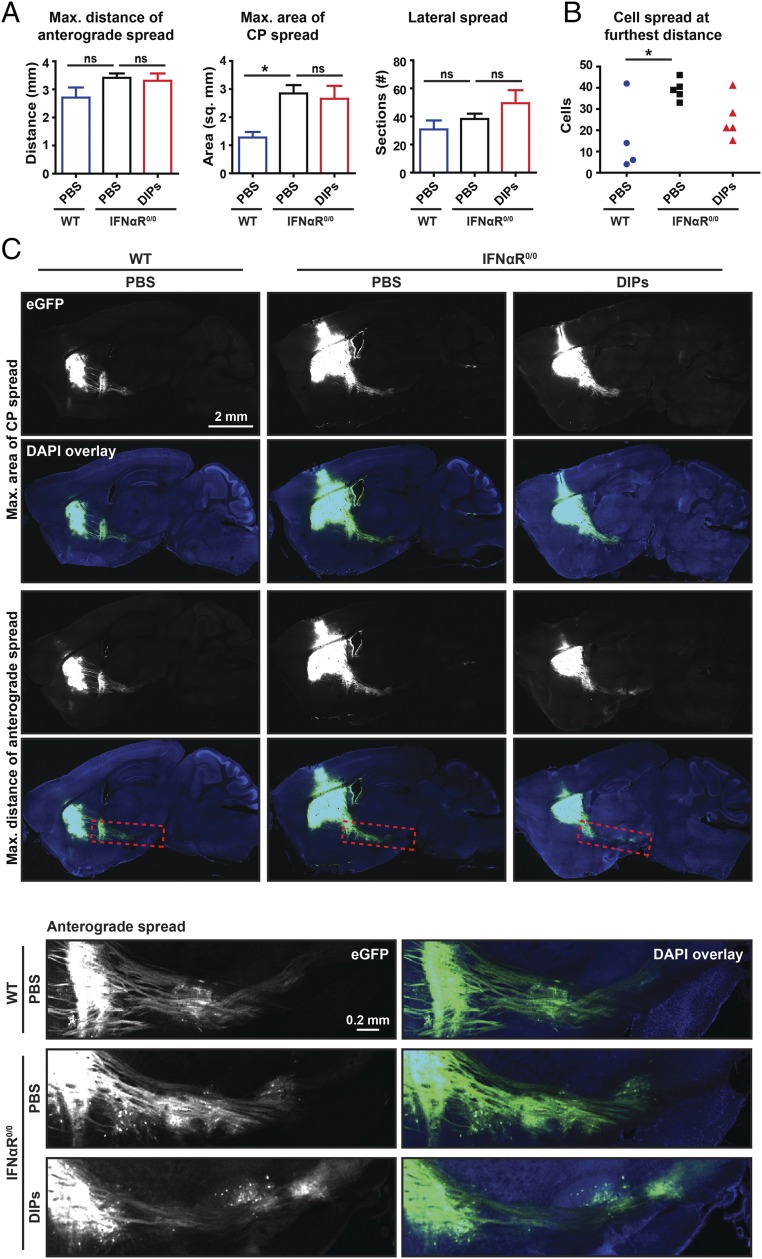
IFNαR Is Required for DIP-Mediated Inhibition of rVSV-eGFP Spread.
A mouse with deletion of the type I IFN response was available to directly test the requirement of this pathway for DIP-mediated inhibition. As the IFNαR is the only identified type I IFN receptor, a mouse strain homozygous for the IFNαR-null allele was used for this test (14). The right CP of IFNαR0/0 mice was injected with DIPs or PBS 1 d before rVSV-eGFP inoculation into the left CP. WT mice were injected in parallel with PBS and then rVSV-eGFP as a reference for virus spread. DIPs did not have a significant effect on rVSV-eGFP spread in IFNαR0/0 animals (Fig. 4), indicating that DIPs exert a paracrine restriction on rVSV-eGFP spread through the induction of the type I IFN response.
Fig. 4.
Effect of contralateral DIP injections on rVSV-eGFP spread in IFNαR0/0 mice. (A) The effect of contralateral DIPs on rVSV-eGFP spread in IFNαR0/0 mice was assessed. A volume of 100 nL of either DIPs (170 ng) or PBS was injected on day 1, and 100 nL of rVSV-eGFP (2,000 pfu) was injected on day 2. Infection was allowed to proceed for 3 d. Graphs display mean ± SEM for each metric and condition. n WT with PBS = 4, n IFNαR0/0 with PBS = 5, and n IFNαR0/0 with DIPs = 5. ANOVA with Dunnett’s multiple comparison post test, *P ≤ 0.05. (B) As the MACS was different between WT and IFNαR0/0 mice, the number of anterograde cells infected at the furthest distance on the section with the MDAS was assessed. Two-tailed Student’s t test, *P ≤ 0.05). (C) Representative images of sagittal sections with the MACS and MDAS and Insets of anterograde spread from these sections are presented. Inset locations are indicated by the dotted red line on sagittal sections.
The difference in the amount of eGFP labeling between WT and IFNαR0/0 animals was significant with more than double the MACS virus labeling in IFNαR0/0 animals compared with WT controls following PBS preinjection (Fig. 4 A and C). Although the MDAS and LS for the two genotypes were not significantly different, we asked whether there were more infected cells at the furthest site of anterograde spread, in correlation with the MACS values. Indeed, we observed 17 ± 9 cells in WT animals and 39 ± 2 cells in IFNαR0/0 animals (mean ± SEM, cells at MDAS site on a single section) (Fig. 4). The difference observed in rVSV-eGFP spread between these two genotypes indicates that WT animals restrict rVSV-eGFP spread through the type I IFN response, even without prestimulation by DIPs.
Evaluation of the Response Within Microglia Following Infection with DIPs and rVSV-eGFP.
Innate immune signaling pathways are activated in microglia by infections of the brain (10), during neurodegeneration (12), as part of neuropsychiatric disorders (39), and following systemic pathogenic stimuli (12). In addition, LACV has been found to elicit IFN production by microglia. However, surprisingly, many IFN-producing cells were observed near infected cells but were not themselves obviously infected (9). Additionally, lymphocytic choriomeningitis virus (LCMV) infection elicits changes in microglia morphology to an activated ramified structure (40). Here we evaluated the rapid response of both infected and uninfected microglia to infection.
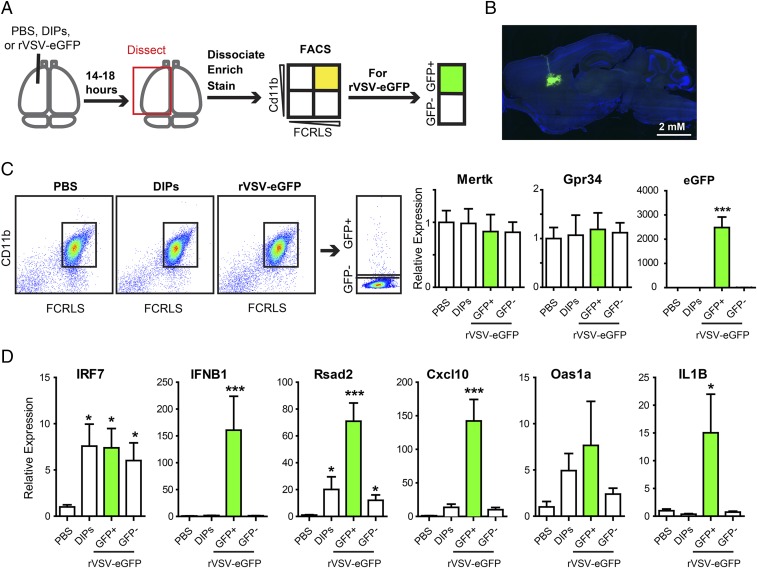
To test the response of microglia, we injected PBS, DIPs, or rVSV-eGFP into the CP and then isolated microglia and examined expression of candidate genes. rVSV-eGFP was used in addition to PBS and DIPs, because eGFP would label infected cells and allow for a comparison of infected versus uninfected cells. Infection proceeded for 14–18 h before tissue harvest (Fig. 5A). Due to this short time window, 105 pfu of rVSV-eGFP were used to obtain adequate numbers of labeled and infected cells. Histological examination of infected tissue following infection with 105 pfu rVSV-eGFP indicated modest infection with no obvious spread beyond the CP (Fig. 5B).
Fig. 5.
Microglia response to DIP and rVSV-eGFP infection. (A) Schematic of CP injection and microglia isolation, which was followed by qPCR. (B) Representative CP infection by 100 nL containing 105 pfu rVSV-eGFP at 18 hpi. (C, Left) Representative FACS plots for FCRLS+Cd11b+ microglia following PBS, DIP, or rVSV-eGFP injection and for eGFP+ microglia following rVSV-eGFP injection. (C, Right) Relative expression of microglia-enriched genes, Mertk and Gpr34, and virus-encoded eGFP. Kruskal–Wallis test with Dunn’s multiple comparison post test, ***P ≤ 0.001. (D) Relative expression of innate immune genes. Kruskal–Wallis test with Dunn’s multiple comparison post test, *P ≤ 0.05, ***P ≤ 0.001.
Microglia were isolated using a discontinuous gradient followed by fluorescent activated cell sorting (FACS) of CD11b+FCRLS+ cells (41); ramified microglia in the central nervous system (CNS) are CD11b+, and FCRLS is uniquely expressed by microglia (41). Microglia-enriched populations from all three injection conditions had similar CD11b and FCRLS FACS profiles (Fig. 5C). CD11b+FCRLS+ microglia from rVSV-eGFP–infected brains were further separated into eGFP+ and eGFP− populations (Fig. 5C). To test for the effectiveness of the isolation procedure, CD11b+FCRLS+ cells were evaluated using quantitative PCR (qPCR) for Mertk, Gpr34, and eGFP (Fig. 5C). Mertk and Gpr34 are highly enriched microglial genes with minimal, if any, expression in either organ-specific macrophages or other immune cells (41). Mertk and Gpr34 expression levels in microglia isolated from different injection conditions were similar, indicating that the isolated microglia were not differentially diluted by other cell types among the different experimental conditions. High eGFP expression was detected only in Cd11b+FCRLS+eGFP+ cells and not in Cd11b+FCRLS+eGFP− cells from rVSV-eGFP–infected brains. CD11b+FCRLS+ cells from PBS or DIP-injected brains did not show any eGFP expression, which indicated that eGFP+ cells were indeed infected (Fig. 5C).
Expression of IFN-β and IRF7 was selected for analysis in isolated microglia because IFN-β is an early marker of innate immune activation and IRF7 is a master regulator of the type I IFN response (42). Injection of DIPs and rVSV-eGFP led to increased expression of IRF7 in microglia compared with PBS but induction of IFN-β only in the infected eGFP+ microglia (Fig. 5D). GFP− and DIP-infected microglia did not have a significant difference in IFN-β expression compared with PBS-injected animals (Fig. 5D). It is likely that IFN-β induction was not detected following DIP injection because signal from infected cells was diluted by uninfected cells.
To further investigate the induction of the type I IFN response, the expression of several known ISGs—Rsad2, Cxcl10, and Oas1α—was assayed (43). Compared with PBS injection, DIPs led to an induction in Rsad2 (Fig. 5D). rVSV-eGFP led to an increase in expression of Rsad2 and Cxcl10 in eGFP+ cells. For eGFP− cells, Rsad2, but not Cxcl10, showed a significant increase in expression (Fig. 5D). For Oas1α, the average was higher following DIP and rVSV-eGFP injection than the PBS control, but significance was not reached due to variability between animals (Fig. 5D). It is likely that we observed greater induction of ISGs in infected cells than uninfected cells following rVSV-eGFP injection because Rsad2 and Cxcl10 expression can also be driven by IRF3 following PRR activation (44).
In addition to testing induction of the type I IFN response, we sought to investigate activation of an antiviral response through a parallel transcription pathway. In the canonical model for RNA virus detection by host cells, the RIG-I–like receptor (RLR) family activates the NF-κB family of transcription factors, in addition to IRFs (45). NF-κB is not necessary for IFN-β expression but does drive IL1β expression (45, 46). In tissue-resident macrophages, viral infection drives inflammation through IL1β (46), and in the CNS, IL1β up-regulation in microglia is a marker of inflammation (47). Observation of IL1β expression in infected microglia would provide additional evidence of virus activation of an innate immune response. IL1β expression was up-regulated in eGFP+ microglial cells compared with PBS injection (Fig. 5D).
The molecular characterization of gene expression following inoculation of the brain with DIPs and rVSV-eGFP demonstrated that the type I IFN response was activated in a CNS resident cell type. Microglia directly infected by virus-produced IFN-β. IRF7 was up-regulated in both infected and uninfected cells following inoculation of the brain with DIPs and rVSV-eGFP. A downstream response to type I IFN was also observed in both infected and uninfected microglia. Furthermore, up-regulation of IL1β in infected cells raises the possibility of other innate immune pathways acting in parallel to the type I IFN response.
Discussion
The data presented here show that the brain parenchyma can directly mount a functional innate immune response that can limit virus spread. Furthermore, this study demonstrates that a CNS resident cell type, microglia, can produce type I IFN in response to viral infection and that the type I IFN pathway can limit viral spread.
Use of a Transsynaptic Virus and Well-Defined Circuitry to Study the Brain Parenchyma’s Innate Immune Response.
We were able to evaluate the effectiveness of the antiviral response mounted within the brain parenchyma by exploiting the characteristics of a transsynaptic virus and the well-defined connections of the basal ganglia (15). This approach enabled the histological identification and quantification of the types of cells that were infected, which included neurons infected by transsynaptic spread. The ability to trace transsynaptic spread among neurons was crucial to the interpretation of these results, as the brain has complex structural intersections with both the lymphatic and vascular architecture. The meninges of the brain include lymphatic vessels that carry immune cells (48). The choroid plexuses are located within the ventricles and have a rich blood supply, located between the blood–brain and blood–ventricle barriers (1). The presence of these tissues within the brain prevents evaluation of virus spread among neurons and glia using methods such as titration of virus from homogenized tissue or qPCR for viral genes (14, 43, 49) because such methods cannot differentiate between virus load in CNS and non-CNS tissues. The presence of these structures also prevents conclusions regarding CNS innate immunity following peripheral infections, because peripheral infections can pass through both the brain’s vasculature and lymphatics, and these structures can mount innate immune responses (1).
Intranasal instillation of VSV has been used historically, and more recently, to probe the effects of host and viral factors on virus spread into the nervous system (16, 18, 50). Several recent studies used this approach to probe the brain’s innate immune response to virus (49, 51). Intranasal application of virus is technically simpler than intracerebral stereotactic injection and has the advantage that it interrogates a route of infection that may be a natural one. However, in terms of dissecting the specific responses of the CNS, it has several limitations arising from developmental and anatomic complexity. The nervous system has clear divisions into the central and peripheral nervous systems, based on developmental history. The olfactory nerve derives from cells that originate in the olfactory placode, which places it in the peripheral nervous system, in contrast to the derivatives of the neural tube, which comprise the CNS (52). This means that the initial innate immune responses and virus replication following intranasal infection are not attributable to the CNS. In addition, the olfactory nerve passes through the meninges at the cribriform plate, which provides a connection between the subarachnoid space and the nasal mucosa lymphatics (53). This structural complexity allows for signaling and virus propagation within non-CNS sources proximal to the brain. These features may account for the failure of intranasal infection to recapitulate intracerebral infection, as originally reported by Sabin and Olitsky (16), and then confirmed by Stevenson et al. (3).
The advantage of the approach taken here is that it is able to provide an unambiguous conclusion regarding the ability of the brain parenchyma to limit virus spread through an innate immune response. Importantly, this approach allows for the characterization and quantification of virus transmission and spread from a consistent and precise injection site within the parenchyma and identifies virus spread among neurons in well-defined circuitry. The approach and techniques used here are straight-forward and should allow further dissection of the mechanisms used by the brain in vivo to limit viral spread as well as the mechanisms used by neurotropic viruses to evade this response (54).
The Type I IFN Response Can Limit Virus Spread Within the Brain Parenchyma and Among Neurons.
Previous studies have demonstrated that viral infection can stimulate expression of ISGs and proinflammatory cytokines in cultured CNS cells and in the brain (8, 10, 55, 56) and that both neurons and glia are responsive to IFN treatment (57–59). Additionally, studies tracking virus spread into the brain from peripheral sites of inoculation have highlighted the importance of the innate immune response in preventing high virus load within the brain (14, 50, 60, 61). However, there have been no studies that resolved whether an innate immune response mounted within the brain parenchyma can limit viral spread, especially along neuronal circuitry. Studies using in vitro-cultured neurons have attempted to address whether an innate immune response can restrict virus spread among neurons but produced conflicting results, including for VSV (57, 58). These previous studies raised the issue of whether virus spread among neurons in the immunologically distinct parenchyma can be limited by the innate immune response.
The data presented here strongly support a model in which a paracrine type I IFN response initiated within the brain can limit virus spread among CNS neurons in vivo. The difference in viral spread between WT and IFNαR-null mice indicates that rVSV-eGFP spread is normally limited in WT mice by the type I IFN pathway. Type I IFN may also be sufficient to limit viral spread, at least in part, as injection of recombinant IFNαA/D showed a significant, but not complete, inhibition of viral spread. Other innate immune pathways, such as the type II IFN response and inflammation, may act in concert with the type I IFN response to restrict virus spread (62). This possibility is supported by our observation that infected microglia have increased levels of IL-1β expression, which can be involved in inflammation.
Use of DIPs to Attenuate Virus Spread.
The existence of DIPs has been known for quite some time, yet little is known about their in vivo activity in relation to replication-competent, full-length virus (28). The data presented here indicate that DIPs can inhibit rVSV-eGFP propagation by direct coinjection, as has been previously shown by other metrics (18, 19), and through paracrine signaling across the brain. These results inform studies regarding the development of viral vaccines and the potential role(s) of DIPs in inducing immunity. A variety of live-attenuated virus vaccines have been found to contain DIPs or RNA from DIPs, leading to the hypothesis that DIPs can contribute to the efficacy of some vaccines (28). Contributions could include attenuation of virus replication through direct interference, or through the stimulation of innate immune pathways, to facilitate stronger adaptive responses (28). Data presented here support both of these possibilities.
Microglia Response to Virus Infection.
Recent studies have started to reveal the important role of microglia in maintaining CNS health and in mediating disease pathology (10, 21, 47). We found that infected microglia produce type I IFN. Furthermore, microglia that are not infected express increased levels of IRF7. This observation is in agreement with previous work that indicated an increase in IRF7 expression in the brain following LCMV infection (56). The observation of paracrine IRF7 induction is interesting because the CNS has been reported to not contain cells with high constitutive IRF7 expression (8). In contrast, in the periphery, plasmacytoid dendritic cells express constitutively high IRF7 levels and, together with lymph node macrophages, produce the majority of type I IFN following VSV infection (13). Analogously, in the brain, it is possible that a rapid IRF7 response is necessary to prime cells for a full type I IFN response upon virus spread from infected cells. These observations suggest that the type I IFN response in the brain parenchyma may be similar to that of the periphery. Subsequent work will need to be carried out to further elucidate the parallels and the differences between the rapid innate immune responses of the brain and periphery to virus infection.
Recent work has also indicated that expression of innate immune genes such as IRF7 increases in microglia with age in a region-specific manner, including the striatum (63). Additionally, the spread of VSV along CNS circuitry is more robust in younger animals compared with older ones (64). Taken together, it is likely that the innate immune response of the brain parenchyma develops with age. It would be of interest to characterize these changes in future studies and determine their effects on virus infection and spread.
Future Directions
With constantly improving technology for the analysis of single cells (65, 66), it will soon be possible to evaluate the brain’s full antiviral innate immune response. Additionally, in combination with genetically modified animals, and perhaps in vivo imaging (40), it will be possible to dissect the cellular mechanisms by which viruses are detected in the brain as well as the immune responses that are elicited. It also will be interesting to interrogate the brain’s response to different types of viruses, which have evolved different mechanisms to combat the immune system in the arms race between pathogens and hosts (67).
Materials and Methods
Virus Production.
rVSV-eGFP.
Generation, concentration, and titration of rVSV-eGFP were carried out as previously described (68). Serial tissue culture passage of virus stocks was avoided to prevent an accumulation of novel mutations in the genome. Gradient-purified preparations had titers between ∼1010 and 1011 pfu/mL.
Defective-interfering particles from natural truncations.
This protocol was designed to optimize the ratio of naturally truncated DIPs to full-length replication-competent virus for production of a purified DIP preparation. rVSV WT was passaged on BSR T7/5 cells at a multiplicity of infection (MOI) of 100 for four passages to yield supernatant 1, which comprised 90% DIPs and 10% full-length VSV, as evaluated by TEM. Supernatant 1 was then optimized for growing DIPs; a range of 0–100 μL of supernatant 1 was combined with an MOI of 10 of rVSV WT to infect 70–90% confluent BSR T7/5 cells seeded in a flat-bottom 12-well plate (Corning). The supernatants from this 12-well dish were harvested and are referred to as supernatants 2. As DIPs interfere with the replication of WT VSV, an assay was devised to titer interference induced by each supernatant 2 to allow for the optimal ratio of DIPs to VSV for a preparation of DIPs to use for in vitro amplification. We combined 5 μL of each supernatant 2 with an MOI of 10 of rVSV WT to infect 70–90% confluent BSR T7/5 cells seeded in a flat-bottom 12-well plate. Then, at 8 hpi, the infected cells were evaluated by phase contrast microscopy for cytopathic effects. The well with the least cytopathic effect was the one deemed to have the most DIPs. As a negative control for DIPs, the well derived from no supernatant 1 combined with rVSV WT had minimal defective particles and thus the most cell death. From these titrations, the optimal amount of supernatant 1 was chosen, and a scaled-up preparation was made with 5.3 mL of supernatant 1 and rVSV (MOI of 10) in a T1000 flask (Corning) with 70–90% confluent BSR T7/5 cells. The resulting viral preparation was used to prepare the DIPs through a gradient purification. Ultimately, DIPs were resuspended in PBS with 50 mM Hepes, pH 7.4, and diluted to a titer of ∼105 pfu/mL and a concentration of 8.4 mg/mL, aliquoted, and frozen at –80 °C. It is likely that the DIPs had a virus equivalent of at least 5 × 109–5 × 1010 particles per mL as the ratio of DIPs to longer virus particles was >1,000:1 by negative-stain TEM and because longer particles have a particle/pfu ratio ranging between 1/50 and 1/500 (69).
Sucrose gradient purification of rVSV-eGFP.
This method is previously described (70). Briefly, a 70–90% confluent T-1000 flask (EMD Millipore) of BSR T7/5 cells was infected with an MOI of 3 of rVSV-eGFP in serum-free DMEM (Thermo Fisher Scientific). After 1 h, media was replaced with DMEM supplemented with 2% FBS and 50 mM Hepes, pH 7.4. Infection was allowed to proceed for 18–24 h. When cytopathic effects were apparent, supernatant was collected, centrifuged at a low speed to remove debris, and passed through a 0.45-μm, 100-mL filter (Corning). Virus was then pelleted by ultracentrifugation for 90 min at 4 °C at 17,200 rpm in a SW-28 rotor or 17,700 rpm in a SW-32 rotor (Beckman Coulter). Pellets were allowed to resuspend at 4 °C overnight in a total volume of 1 mL NTE (10 mM Tris, pH 7.4, 100 mM NaCl, 1 mM EDTA). Resuspended virus was transferred to a 1.5-mL tube and centrifuged for 1 min at 13,000 rpm (∼17,900 × g) in a tabletop microcentrifuge to clear large virus clumps. Next, supernatant was loaded onto a linear 15–45% (wt/vol) sucrose gradient prepared in NTE. The gradient was centrifuged for 5 h at 4 °C at 25,000 rpm in a SW-41 rotor (Beckman Coulter). A needle and syringe were used to puncture the tube and remove the virus band. The DIPs band appeared 1/4 down the gradient, and the rVSV-eGFP band appeared 2/3 down the gradient. Virus and sucrose were diluted 1:4 with NTE and ultracentrifuged for 1 h at 4 °C at 41,000 rpm in a SW-55 rotor (Beckman Coulter). The pellet was resuspended in 0.25–1 mL of PBS supplemented with 50 mM Hepes, pH 7.4 and then aliquoted and frozen at –80 °C until use. After freezing, virus was thawed and titered using a plaque-forming assay on VERO cells. Freeze-thaw cycles of virus were kept to a minimum.
Negative-stain TEM.
Virus was adhered to glow-discharged carbon and Formvar-coated copper grids and stained with 1% (wt/vol) phosphotungstic acid in H2O, pH 7.5. Imaging was performed using either a JEOL 1200EX – 80 kV (JEOL USA) or Tecnai G2 Spirit BioTWIN (FEI) transmission electron microscopes.
Mouse Strains.
Male 8–9-wk-old C57BL/6J mice (000664; The Jackson Laboratory) and IFNαR0/0 mice (032045-JAX; MMRRC) were used for experiments, which were approved by the Longwood Medical Area Institutional Animal Care and Use Committee. Breeding was conducted in biosafety containment level 1 conditions and infections in biosafety containment level 2 conditions.
Mouse Injections.
All experiments were performed using 8–9-wk-old male animals to control for age- and sex-dependent variations (71, 72). Animals were injected using a stereotaxic instrument (Narishige International USA) and pulled capillary microdispensers (Drummond Scientific) using the following coordinates for the CP: A/P, 1 from bregma, L/M, 1.8; D/V, −2.5. Injection volumes were 100 nL and contained either PBS, rVSV-eGFP with/without IFN-αA/D (I4401-100KU; Sigma-Aldrich), or DIPs with/without IC. rVSV-eGFP stocks were diluted in PBS to a concentration of 2 × 107 pfu/mL, which resulted in 2 × 103 pfu per injection of 100 nL. For rVSV-eGFP injections with IFN-αA/D, 1 × 108 pfu/mL rVSV-eGFP was diluted 1:5 in 1,000 U/uL IFN-αA/D, leading to a final concentration of 2 × 107 pfu/mL for virus and 800 U/uL for IFN-αA/D. For DIPs, stocks were diluted 1:5 in either PBS or PBS with the inhibitor mixture, leading to a final concentration of 1.7 mg/mL, which resulted in 170 ng injected in a volume of 100 nL. The inhibitor mixture was composed of (i) BX795 (tlrl-bx7; InVivoGen), which was diluted to a stock concentration of 10 mM in DMSO; (ii) CP-690550 (tlrl-cp69; InVivoGen), which was diluted to a stock concentration of 20 mM in DMSO; and (iii) INCB018424 (tlrl-rux; InVivoGen), which was diluted to 20 mM in DMSO. For injection, all three inhibitors were combined in a ratio of 1:1:1, and this mixture was then diluted 1:20 in PBS to obtain a working stock. Then DIPs were combined in a ratio of 1:5 with the working stock.
All initial DIPs and IFN-αA/D experiments were performed double-blinded. The scientist performing injections was blinded as to what was injected, and the scientist sectioning, imaging, and performing analysis was blinded to the condition until after all analyses were completed.
Tissue Processing and Preparation.
Perfusion was performed and brains were fixed in 4% paraformaldehyde (wt/vol) (in PBS) overnight at 4 °C. The tissue was sequentially passed through PBS solutions containing 7.5%, 15%, and 30% (wt/vol) sucrose (in PBS). The tissue was kept at 4 °C in each sucrose solution between 2 h and 24 h until it lost buoyancy and sank in the solution. It was subsequently flash-frozen in optimum cutting temperature compound (4583; VWR) using an ethanol and dry ice bath. Frozen tissue was stored at –80 °C until it was sectioned. Tissue sections were taken at 50 μM using a Leica CM 3050S cryostat (Leica). The entire brain was sectioned, and all sections were collected, mounted on slides, and DAPI-stained.
Microscopy.
All sections were screened using a Nikon Eclipse E1000 upright epifluorescent microscope and a 4× N.A. 0.13 dry objective. Sections containing the most significant spread were imaged for quantification. All sections for quantification within a given experimental group were imaged using the same settings and on the same day. All images for quantification were obtained using an inverted Zeiss LSM780 microscope (Carl Zeiss AG) with 405-nm and 488-nm lasers and a 10× N.A. 0.3 Zeiss Plan Neofluardry objective. Image montages were obtained using either the Zeiss LSM780 microscope and 10× objective or a Keyence BZ-9000 microscope and Nikon PlanApo 4× N.A. 0.20 dry objective lens (Nikon).
Quantification.
Quantification was performed on multiple imaged sections for each brain, as it was not possible to determine the correct sections to score for MDAS and MACS by eye. FIJI (73–76) was used for all analyses. The Bio-Formats plugin (77) was used to import all images. The background was measured in three consistent and separate areas near the CP for each image, and values were averaged. To determine MACS, the threshold for signal was set to three times the mean background, and the “Analyze particles” function was used to determine “Regions of Interest” (ROIs). The ROI encompassing the area of virus injection was selected, and the following measurements were taken: (i) Area to provide MACS and (ii) Centroid to provide the average x and y coordinate for all of the pixels measured. If the Area included extensive axons beyond the CP, these were excluded using the “Freehand selections tool” and followed by the “Analyze Particles” function within the defined area and subsequent ROI measurement for Area. Next a straight line was drawn from the centroid to the furthest infected anterograde cell body to determine the MDAS. Once the line was drawn, the “Measure” function was used to calculate the distance. The greatest MACS and MDAS for each animal are the data presented.
To quantify the LS, the number of eGFP-positive sections was counted for each animal. This quantification included spread within the CP and excluded any spread through the ventricles. Spread through the ventricles was determined by clear eGFP labeling reaching the ventricles from the CP and the presence of eGFP-positive ventricles lateral to the infection site.
For quantification of the number of infected anterograde cells, the section with the largest MDAS was selected for each animal. The number of green cell bodies at the furthest anterograde site was counted by eye within this section.
Mouse Microglia Isolation and Sorting.
Microglia isolation and sorting was performed as previously described (41). Briefly, mice were transcardially perfused with ice-cold Hanks’ Balanced Salt Solution (HBSS), and brains were separately dissected and dissociated. Single cell suspensions were prepared and centrifuged over a 37%/70% discontinuous Percoll gradient (GE Healthcare) for 25 min at 900 × g at 25 °C with the brake off. Mononuclear cells were then isolated from the interface. Isolated cells were stained with a combination of anti-FCRLS (3 μg·mL−1, clone 4G11, O.B. laboratory, Harvard Medical School, Boston, MA), followed by secondary detection with goat anti-rat IgG conjugated to APC (0.7 μg·mL−1, clone Poly4054, Biolegend) and then CD11b-PeCy7 (2 μg·mL−1, clone M1/70, BD Biosciences) antibodies to specifically sort resident microglia. Stained cells were sorted on a BD FACSAria Multicolor High Speed Sorter (Becton Dickinson) and data analyzed using FlowJo Software (TreeStar).
Gene Expression Analysis.
RNA isolation from FACS sorted cells was performed using the ARCTURUS PicoPure RNA Isolation Kit (KIT0204; Thermo Fisher Scientific). RNA was eluted in 11 μL for each sample.
The SuperScript III First-Strand Synthesis System for RT-PCR (18080051; Thermo Fisher Scientific) was used for reverse transcription of isolated RNA (30 ng). All real-time quantitative PCR was performed using Vii7 (Applied Biosystems) with TaqMan Gene expression Master Mix (Applied Biosystems) and the specific TaqMan Gene Expression Assay (Applied Biosystems). The following TaqMan Gene Expression assays (applied biosystems) were used: Cxcl10 (Mm00445235_m1), Gpr34 (Mm02620221_s1), Ifnb1 (Mm00439552_s1), Irf7 (Mm00516793_g1), Il1b (Mm00434228_m1), Mertk (Mm00434920_m1), mouse gapdh (Mm99999915_g1), Oas1a (Mm00836412_m1), and Rsad2 (Mm00491265_m1). For eGFP, the following primers were used (1): 5′-/56FAM/ACCTGAGCACCCAGTCCGCCCT/36-TAMSp/-3′ (250 nM Prime Time 5′6-FAM/3′TAMRA; IDT), 5′-CTGCTGCCCGACAACCAC-3′ (25 nM; IDT), and 5′-TGTGATCGCGCTTCTCGTT-3′ (25 nM; IDT) with final reaction concentration of 900 nM for the forward and reverse primers and 250 nM for the probe. Real-time eGFP primers (78) were verified for VSV-eGFP in 293T cells. All samples were run in duplicate. The mRNA expression levels between samples were normalized using GAPDH endogenous control (VIC dye labeled, Applied Biosystems). The quantification of mRNA expression in treatment groups relative to the saline control (fold change) was done using the 2-∆∆CT method.
Acknowledgments
We thank Maria Ericsson, Elizabeth Benecchi, and Louise Trakimas for their expertise and preparation of TEM samples at the Harvard Medical School Cell Biology Conventional Electron Microscopy Facility; Hunter Elliott for his expertise and guidance in image analysis at the Harvard Medical School Image and Data Analysis Core; Brian Rabe for technical assistance; and ChangHee Lee for help with statistics. We thank the staff of the Nikon Imaging Center at Harvard Medical School, Boston, for their expertise and use of their equipment. rVSV-eGFP and rVSV were kind gifts from Sean P.J. Whelan. This work was supported by National Institutes of Health Grants R01 NS083848 (to C.L.C.), F31 AG041582 (to E.D.), and R01NS088137 (to O.B.); National Multiple Sclerosis Society Grant 5092A1 (to O.B.); and the Nancy Davis Foundation Faculty Award (O.B.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1618157114/-/DCSupplemental.
References
- 1.Galea I, Bechmann I, Perry VH. What is immune privilege (not)? Trends Immunol. 2007;28(1):12–18. doi: 10.1016/j.it.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 2.Murphy JB, Sturm E. Conditions determining the transplantability of tissues in the brain. J Exp Med. 1923;38(2):183–197. doi: 10.1084/jem.38.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stevenson PG, Hawke S, Sloan DJ, Bangham CR. The immunogenicity of intracerebral virus infection depends on anatomical site. J Virol. 1997;71(1):145–151. doi: 10.1128/jvi.71.1.145-151.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersson PB, Perry VH, Gordon S. The acute inflammatory response to lipopolysaccharide in CNS parenchyma differs from that in other body tissues. Neuroscience. 1992;48(1):169–186. doi: 10.1016/0306-4522(92)90347-5. [DOI] [PubMed] [Google Scholar]
- 5.Bechmann I, Galea I, Perry VH. What is the blood-brain barrier (not)? Trends Immunol. 2007;28(1):5–11. doi: 10.1016/j.it.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 6.Le Bon A, Tough DF. Links between innate and adaptive immunity via type I interferon. Curr Opin Immunol. 2002;14(4):432–436. doi: 10.1016/s0952-7915(02)00354-0. [DOI] [PubMed] [Google Scholar]
- 7.Perry AK, Chen G, Zheng D, Tang H, Cheng G. The host type I interferon response to viral and bacterial infections. Cell Res. 2005;15(6):407–422. doi: 10.1038/sj.cr.7290309. [DOI] [PubMed] [Google Scholar]
- 8.Delhaye S, et al. Neurons produce type I interferon during viral encephalitis. Proc Natl Acad Sci USA. 2006;103(20):7835–7840. doi: 10.1073/pnas.0602460103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kallfass C, et al. Visualizing production of beta interferon by astrocytes and microglia in brain of La Crosse virus-infected mice. J Virol. 2012;86(20):11223–11230. doi: 10.1128/JVI.01093-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rock RB, et al. Role of microglia in central nervous system infections. Clin Microbiol Rev. 2004;17(4):942–964. doi: 10.1128/CMR.17.4.942-964.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sorgeloos F, Kreit M, Hermant P, Lardinois C, Michiels T. Antiviral type I and type III interferon responses in the central nervous system. Viruses. 2013;5(3):834–857. doi: 10.3390/v5030834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rivest S. Regulation of innate immune responses in the brain. Nat Rev Immunol. 2009;9(6):429–439. doi: 10.1038/nri2565. [DOI] [PubMed] [Google Scholar]
- 13.Iannacone M, et al. Subcapsular sinus macrophages prevent CNS invasion on peripheral infection with a neurotropic virus. Nature. 2010;465(7301):1079–1083. doi: 10.1038/nature09118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Müller U, et al. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264(5167):1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 15.Beier KT, et al. Anterograde or retrograde transsynaptic labeling of CNS neurons with vesicular stomatitis virus vectors. Proc Natl Acad Sci USA. 2011;108(37):15414–15419. doi: 10.1073/pnas.1110854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sabin AB, Olitsky PK. Influence of host factors on neuroinvasiveness of vesicular stomatitis virus: III. EFFEct of age and pathway of infection on the character and localization of lesions in the central nervous system. J Exp Med. 1938;67(2):201–228. doi: 10.1084/jem.67.2.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.López CB. Defective viral genomes: Critical danger signals of viral infections. J Virol. 2014;88(16):8720–8723. doi: 10.1128/JVI.00707-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plakhov IV, Aoki C, Reiss CS, Huang AS. Pathogenesis of murine encephalitis limited by defective interfering particles. An immunohistochemical study. J Neurovirol. 1995;1(2):207–218. doi: 10.3109/13550289509113967. [DOI] [PubMed] [Google Scholar]
- 19.Doyle M, Holland JJ. Prophylaxis and immunization in mice by use of virus-free defective T particles to protect against intracerebral infection by vesicular stomatitis virus. Proc Natl Acad Sci USA. 1973;70(7):2105–2108. doi: 10.1073/pnas.70.7.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cave DR, Hagen FS, Palma EL, Huang AS. Detection of vesicular stomatitis virus RNA and its defective-interfering particles in individual mouse brains. J Virol. 1984;50(1):86–91. doi: 10.1128/jvi.50.1.86-91.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ransohoff RM, El Khoury J. Microglia in health and disease. Cold Spring Harb Perspect Biol. 2015;8(1):a020560. doi: 10.1101/cshperspect.a020560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nassi JJ, Cepko CL, Born RT, Beier KT. Neuroanatomy goes viral! Front Neuroanat. 2015;9(July):80. doi: 10.3389/fnana.2015.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beier KT, et al. Transsynaptic tracing with vesicular stomatitis virus reveals novel retinal circuitry. J Neurosci. 2013;33(1):35–51. doi: 10.1523/JNEUROSCI.0245-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marshel JH, Mori T, Nielsen KJ, Callaway EM. Targeting single neuronal networks for gene expression and cell labeling in vivo. Neuron. 2010;67(4):562–574. doi: 10.1016/j.neuron.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lichty BD, Power AT, Stojdl DF, Bell JC. Vesicular stomatitis virus: Re-inventing the bullet. Trends Mol Med. 2004;10(5):210–216. doi: 10.1016/j.molmed.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 26.Geisbert TW, et al. Vesicular stomatitis virus-based ebola vaccine is well-tolerated and protects immunocompromised nonhuman primates. PLoS Pathog. 2008;4(11):e1000225. doi: 10.1371/journal.ppat.1000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parisi SG, et al. Viral infections of the central nervous system in elderly patients: A retrospective study. Int J Infect Dis. 2016;44:8–10. doi: 10.1016/j.ijid.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 28.Frensing T. Defective interfering viruses and their impact on vaccines and viral vectors. Biotechnol J. 2015;10(5):681–689. doi: 10.1002/biot.201400429. [DOI] [PubMed] [Google Scholar]
- 29.Cureton DK, Massol RH, Whelan SPJ, Kirchhausen T. The length of vesicular stomatitis virus particles dictates a need for actin assembly during clathrin-dependent endocytosis. PLoS Pathog. 2010;6(9):e1001127. doi: 10.1371/journal.ppat.1001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stauffer Thompson KA, Rempala GA, Yin J. Multiple-hit inhibition of infection by defective interfering particles. J Gen Virol. 2009;90(Pt 4):888–899. doi: 10.1099/vir.0.005249-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Drokhlyansky E, Soh TK, Cepko CL. Preferential budding of vesicular stomatitis virus from the basolateral surface of polarized epithelial cells is not solely directed by matrix protein or glycoprotein. J Virol. 2015;89(22):11718–11722. doi: 10.1128/JVI.01658-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whelan SP, Ball LA, Barr JN, Wertz GT. Efficient recovery of infectious vesicular stomatitis virus entirely from cDNA clones. Proc Natl Acad Sci USA. 1995;92(18):8388–8392. doi: 10.1073/pnas.92.18.8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schott DH, Cureton DK, Whelan SP, Hunter CP. An antiviral role for the RNA interference machinery in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2005;102(51):18420–18424. doi: 10.1073/pnas.0507123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adler R, Banerjee AK. Analysis of the RNA species isolated from defective particles of vesicular stomatitis virus. J Gen Virol. 1976;33(1):51–60. doi: 10.1099/0022-1317-33-1-51. [DOI] [PubMed] [Google Scholar]
- 35.Rehberg E, Kelder B, Hoal EG, Pestka S. Specific molecular activities of recombinant and hybrid leukocyte interferons. J Biol Chem. 1982;257(19):11497–11502. [PubMed] [Google Scholar]
- 36.Clark K, Plater L, Peggie M, Cohen P. Use of the pharmacological inhibitor BX795 to study the regulation and physiological roles of TBK1 and IkappaB kinase epsilon: A distinct upstream kinase mediates Ser-172 phosphorylation and activation. J Biol Chem. 2009;284(21):14136–14146. doi: 10.1074/jbc.M109.000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quintás-Cardama A, et al. Preclinical characterization of the selective JAK1/2 inhibitor INCB018424: Therapeutic implications for the treatment of myeloproliferative neoplasms. Blood. 2010;115(15):3109–3117. doi: 10.1182/blood-2009-04-214957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu W, Sun X-H. Janus kinase 3: The controller and the controlled. Acta Biochim Biophys Sin (Shanghai) 2012;44(3):187–196. doi: 10.1093/abbs/gmr105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blank T, Prinz M. Microglia as modulators of cognition and neuropsychiatric disorders. Glia. 2013;61(1):62–70. doi: 10.1002/glia.22372. [DOI] [PubMed] [Google Scholar]
- 40.Nayak D, et al. Type I interferon programs innate myeloid dynamics and gene expression in the virally infected nervous system. PLoS Pathog. 2013;9(5):e1003395. doi: 10.1371/journal.ppat.1003395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Butovsky O, et al. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat Neurosci. 2014;17(1):131–143. doi: 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Honda K, et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434(7034):772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 43.Lazear HM, et al. IRF-3, IRF-5, and IRF-7 coordinately regulate the type I IFN response in myeloid dendritic cells downstream of MAVS signaling. PLoS Pathog. 2013;9(1):e1003118. doi: 10.1371/journal.ppat.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grandvaux N, et al. Transcriptional profiling of interferon regulatory factor 3 target genes: Direct involvement in the regulation of interferon-stimulated genes. J Virol. 2002;76(11):5532–5539. doi: 10.1128/JVI.76.11.5532-5539.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Balachandran S, Beg AA. Defining emerging roles for NF-κB in antivirus responses: Revisiting the interferon-β enhanceosome paradigm. PLoS Pathog. 2011;7(10):e1002165. doi: 10.1371/journal.ppat.1002165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Negash AA, et al. IL-1β production through the NLRP3 inflammasome by hepatic macrophages links hepatitis C virus infection with liver inflammation and disease. PLoS Pathog. 2013;9(4):e1003330. doi: 10.1371/journal.ppat.1003330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Butovsky O, et al. Modulating inflammatory monocytes with a unique microRNA gene signature ameliorates murine ALS. J Clin Invest. 2012;122(9):3063–3087. doi: 10.1172/JCI62636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Louveau A, et al. Structural and functional features of central nervous system lymphatic vessels. Nature. 2015;523(7560):337–341. doi: 10.1038/nature14432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van den Pol AN, Ding S, Robek MD. Long-distance interferon signaling within the brain blocks virus spread. J Virol. 2014;88(7):3695–3704. doi: 10.1128/JVI.03509-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trottier MD, Lyles DS, Reiss CS. Peripheral, but not central nervous system, type I interferon expression in mice in response to intranasal vesicular stomatitis virus infection. J Neurovirol. 2007;13(5):433–445. doi: 10.1080/13550280701460565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Detje CN, et al. Upon intranasal vesicular stomatitis virus infection, astrocytes in the olfactory bulb are important interferon Beta producers that protect from lethal encephalitis. J Virol. 2015;89(5):2731–2738. doi: 10.1128/JVI.02044-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suzuki J, Osumi N. Neural crest and placode contributions to olfactory development. Curr Top Dev Biol. 2015;111:351–374. doi: 10.1016/bs.ctdb.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 53.Faber WM. The nasal mucosa and the subarachnoid space. Am J Anat. 1937;62(1):121–148. [Google Scholar]
- 54.Koyuncu OO, Hogue IB, Enquist LW. Virus infections in the nervous system. Cell Host Microbe. 2013;13(4):379–393. doi: 10.1016/j.chom.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lieberman AP, Pitha PM, Shin HS, Shin ML. Production of tumor necrosis factor and other cytokines by astrocytes stimulated with lipopolysaccharide or a neurotropic virus. Proc Natl Acad Sci USA. 1989;86(16):6348–6352. doi: 10.1073/pnas.86.16.6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ousman SS, Wang J, Campbell IL. Differential regulation of interferon regulatory factor (IRF)-7 and IRF-9 gene expression in the central nervous system during viral infection. J Virol. 2005;79(12):7514–7527. doi: 10.1128/JVI.79.12.7514-7527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kreit M, et al. Inefficient type I interferon-mediated antiviral protection of primary mouse neurons is associated with the lack of apolipoprotein l9 expression. J Virol. 2014;88(7):3874–3884. doi: 10.1128/JVI.03018-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trottier MD, Jr, Palian BM, Reiss CS. VSV replication in neurons is inhibited by type I IFN at multiple stages of infection. Virology. 2005;333(2):215–225. doi: 10.1016/j.virol.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 59.Wollmann G, Robek MD, van den Pol AN. Variable deficiencies in the interferon response enhance susceptibility to vesicular stomatitis virus oncolytic actions in glioblastoma cells but not in normal human glial cells. J Virol. 2007;81(3):1479–1491. doi: 10.1128/JVI.01861-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fensterl V, et al. Interferon-induced Ifit2/ISG54 protects mice from lethal VSV neuropathogenesis. PLoS Pathog. 2012;8(5):e1002712. doi: 10.1371/journal.ppat.1002712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lazear HM, et al. Interferon-λ restricts West Nile virus neuroinvasion by tightening the blood-brain barrier. Sci Transl Med. 2015;7(284):284ra59. doi: 10.1126/scitranslmed.aaa4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jensen S, Thomsen AR. Sensing of RNA viruses: A review of innate immune receptors involved in recognizing RNA virus invasion. J Virol. 2012;86(6):2900–2910. doi: 10.1128/JVI.05738-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grabert K, et al. Microglial brain region-dependent diversity and selective regional sensitivities to aging. Nat Neurosci. 2016;19(3):504–516. doi: 10.1038/nn.4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sabin AB, Olitsky PK. Influence of host factors on neuroinvasiveness of vesicular stomatitis virus: II. Effect of age on the invasion of the peripheral and central nervous systems by virus injected into the leg muscles or the eye. J Exp Med. 1937;66(1):35–57. doi: 10.1084/jem.66.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Macosko EZ, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161(5):1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Habib N, et al. Div-Seq: Single-nucleus RNA-Seq reveals dynamics of rare adult newborn neurons. Science. 2016;353(6302):925–928. doi: 10.1126/science.aad7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lafon M. Immune evasion, a critical strategy for rabies virus. Dev Biol (Basel) 2008;131:413–419. [PubMed] [Google Scholar]
- 68.Piccinotti S, Kirchhausen T, Whelan SPJ. Uptake of rabies virus into epithelial cells by clathrin-mediated endocytosis depends upon actin. J Virol. 2013;87(21):11637–11647. doi: 10.1128/JVI.01648-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dargan DJ, Galt CB, Subak-Sharpe JH. The effect of cicloxolone sodium on the replication of vesicular stomatitis virus in BSC-1 cells. J Gen Virol. 1992;73(Pt 2):397–406. doi: 10.1099/0022-1317-73-2-397. [DOI] [PubMed] [Google Scholar]
- 70.Cureton DK, Massol RH, Saffarian S, Kirchhausen TL, Whelan SPJ. Vesicular stomatitis virus enters cells through vesicles incompletely coated with clathrin that depend upon actin for internalization. PLoS Pathog. 2009;5(4):e1000394. doi: 10.1371/journal.ppat.1000394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Libert C, Dejager L, Pinheiro I. The X chromosome in immune functions: When a chromosome makes the difference. Nat Rev Immunol. 2010;10(8):594–604. doi: 10.1038/nri2815. [DOI] [PubMed] [Google Scholar]
- 72.Shaw AC, Goldstein DR, Montgomery RR. Age-dependent dysregulation of innate immunity. Nat Rev Immunol. 2013;13(12):875–887. doi: 10.1038/nri3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pietzsch T, Preibisch S, Tomancák P, Saalfeld S. ImgLib2--Generic image processing in Java. Bioinformatics. 2012;28(22):3009–3011. doi: 10.1093/bioinformatics/bts543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schindelin J, Rueden CT, Hiner MC, Eliceiri KW. The ImageJ ecosystem: An open platform for biomedical image analysis. Mol Reprod Dev. 2015;82(7-8):518–529. doi: 10.1002/mrd.22489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Linkert M, et al. Metadata matters: Access to image data in the real world. J Cell Biol. 2010;189(5):777–782. doi: 10.1083/jcb.201004104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bissonnette N, Lévesque-Sergerie J-P, Thibault C, Boissonneault G. Spermatozoal transcriptome profiling for bull sperm motility: A potential tool to evaluate semen quality. Reproduction. 2009;138(1):65–80. doi: 10.1530/REP-08-0503. [DOI] [PubMed] [Google Scholar]